Abstract
The origin of vertebrates was defined by evolution of a skeleton; however, little is known about the developmental mechanisms responsible for this landmark evolutionary innovation. In jawed vertebrates, cartilage matrix consists predominantly of type II collagen (Col2α1), whereas that of jawless fishes has long been thought to be noncollagenous. We recently showed that Col2α1 is present in lamprey cartilage, indicating that type II collagen-based cartilage evolved earlier than previously recognized. Here, we investigate the origin of vertebrate cartilage, and we report that hagfishes, the sister group to lampreys, also have Col2α1-based cartilage, suggesting its presence in the common ancestor of crown-group vertebrates. We go on to show that lancelets, a sister group to vertebrates, possess an ancestral clade A fibrillar collagen (ColA) gene that is expressed in the notochord. Together, these results suggest that duplication and diversification of ColA genes at the chordate–vertebrate transition may underlie the evolutionary origin of vertebrate skeletal tissues.
Keywords: development, gene duplication, skeleton evolution, chordate, notochord
The phylogenetic relationships of the vertebrates were established largely based on anatomical characters, particularly those of the skeleton. The skeletons of jawed vertebrates (gnathostomes) are composed of cartilage and bone, which contain high levels of type II and type I collagen (Col2α1 and Col1α1/2 protein, respectively). By contrast, the cartilaginous skeletons of lampreys and hagfishes, the only extant jawless fishes (agnathans), have been reported to be noncollagenous and to contain instead the elastin-like proteins lamprin and myxinin (1). This difference in the extracellular matrices of vertebrate skeletons led to the idea that type II collagen became the major structural component of gnathostome cartilage after the divergence of these two lineages (1). However, this view was challenged by our recent report that lampreys have two Col2α1 orthologs and that both genes are expressed during chondrogenesis. Adult lamprey cartilage was also shown to be rich in Col2α1 protein (2). Furthermore, we identified a lamprey ortholog of Sox9, a direct transcriptional regulator of Col2α1 in gnathostomes, and we showed that it is coexpressed with Col2α1 (2). This discovery revealed that the genetic pathway for vertebrate chondrogenesis predated the divergence of lampreys and gnathostomes, although precisely how early this character arose is unknown. Here, we investigate the evolutionary origin of Col2α1-based cartilage by expanding our analysis to the most inclusive clade of vertebrates, which includes the hagfishes, and to a sister group to the vertebrates, the lancelets (3, 4). We report that hagfishes also have Col2α1-based cartilage, suggesting that this type of cartilage was present in common ancestor of all crown vertebrates. Our analysis of lancelets revealed the presence of an ancestral clade A fibrillar collagen (ColA) gene that is expressed in the notochord. Thus, during the chordate–vertebrate transition, an ancestral ColA gene underwent duplication and diversification, and this process may underlie the evolutionary origin of vertebrate skeletal tissues.
Results
Identification of Col2α1 in Hagfishes.
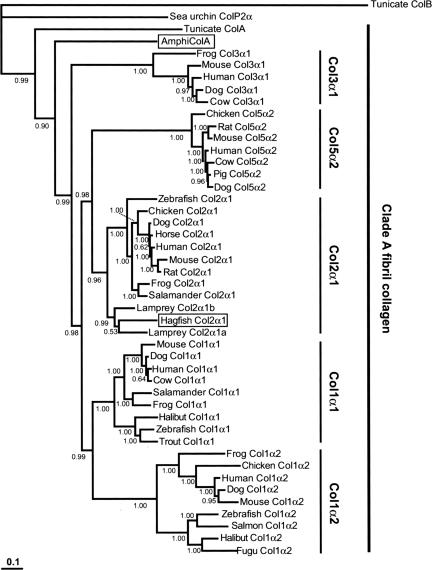
We searched for an expressed Col2α1 ortholog in hagfish using degenerate RT-PCR, and we recovered a 2,214-bp cDNA fragment with a sequence that corresponds to the region between the major triple helix and the C-propeptide domains of gnathostome Col2α1. The deduced amino acid sequence of the hagfish clone was 76% identical to mouse Col2α1, and comparison with the two lamprey Col2α1 orthologs showed 80% identity to lamprey Col2α1a and 77% identity to lamprey Col2α1b. We next conducted molecular phylogenetic analyses using Bayesian phylogenetics, minimum evolution, and maximum likelihood methods. All three analyses placed the hagfish sequence within the vertebrate Col2α1 clade, supporting the designation of this gene as hagfish Col2α1 (Fig. 1 and, which are published as supporting information on the PNAS web site). Interestingly, each tree further positioned hagfish Col2α1 as a sister to lamprey Col2α1a, with lamprey Col2α1b falling out as the sister branch (Fig. 1, 5, and 6). The topology of these trees suggests that Col2α1 was present in the common ancestor of agnathans and gnathostomes and that an additional duplication gave rise to Col2α1a and Col2α1b in the cyclostome (lampreys plus hagfishes) lineage.
Fig. 1.
Extended majority-rule consensus tree for the Bayesian phylogenetic analysis of clade A fibrillar collagen proteins. Numbers at each node indicate posterior probability (pp) values based on one million replicates. Branch lengths are proportional to means of the pp densities for their expected replacements per site. The tree is rooted by tunicate (C. intestinalis) clade B fibrillar collagen (ColB) and sea urchin fibrillar collagen (ColP2α). Hagfish Col2α1 (boxed) is grouped with lamprey Col2α1a and Col2α1b with a pp of 0.99. This cyclostome Col2α1 clade joins to the base of the gnathostome Col2α1 clade with a pp of 0.96. Lancelet clade A fibrillar collagen (AmphiColA; boxed) is joined to the vertebrate clade A collagen family with a pp of 0.99. Minimum evolution and maximum likelihood methods confirm these positions for hagfish Col2α1 and AmphiColA (see also).
Col2α1 Localizes to Hagfish “Soft” Cartilage.
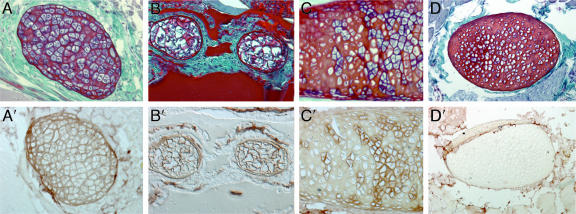
We next asked whether Col2α1 is expressed in hagfish cartilage. Little is known about hagfish embryonic development because of difficulties in obtaining fertilized embryos (5–7). Eptatretus embryos were last collected in 1930 (7), and only three embryos of Myxine have ever been found (5, 6). The unavailability of embryos therefore precludes analysis of hagfish Col2α1 expression during development; however, Col2α1 protein is known to be detectable in adult cartilage of gnathostomes and lampreys (2, 8). We therefore investigated whether Col2α1 is present in hagfish cartilage by using a human antibody against the highly specific N-terminal region of Col2α1 (2, 9–11). Immunohistochemical analysis revealed the presence of Col2α1 in the extracellular matrix of several cartilage elements in the hagfish head and tail (Fig. 2). Col2α1 protein was also detected in the notochord (data not shown), which is known to contain collagen fibers (12). Although a number of cartilage elements in hagfish were rich in Col2α1 protein (Fig. 2 A′ and B′), others showed a mosaic distribution (Fig. 2C′) or lacked Col2α1 altogether (Fig. 2D′). Hagfishes have been described as having two types of cartilage: “soft” cartilage, which contains large hypertrophic chondrocytes that stain with hematoxylin (blue) and are surrounded by a thin extracellular matrix; and “hard” cartilage, which contains smaller chondrocytes that are surrounded by an abundance of extracellular matrix (13, 14). Cartilage elements that were positive for Col2α1 had the cellular characteristics of soft cartilage (Fig. 2 A, A′, B, and B′), whereas cartilages that lacked Col2α1 exhibited the features of hard cartilage (Fig. 2 D and D′). Elements that showed a mosaic distribution of Col2α1 also showed a mosaicism of hard and soft features, and Col2α1 was restricted to the soft regions of these structures (Fig. 2 C and C′). Hagfish soft cartilage has been proposed to be structurally similar to lamprey cartilage (14), and our finding that each is composed of Col2α1 protein supports this idea. The presence of Col2α1 in the cartilages of hagfishes, lampreys (2), and gnathostomes (15) strongly suggests that their last common ancestor had a Col2α1-based endoskeleton.
Fig. 2.
Col2α1 in cranial and tail fin cartilages of Atlantic hagfish. Transverse sections through adult hagfish stained with hematoxylin/eosin and fast green (A–D) or with Col2α1 antibody (A′–D′) are shown. (A and A′) Soft cranial cartilage stained with hematoxylin contains large hypertrophic chondrocytes surrounded by thin layer extracellular matrix (A), and it is positive for Col2α1 (A′). (B and B′) Section through caudal fin shows cartilaginous fin rays with soft-cartilage characteristics (B) that are rich in Col2α1 (B′). (C and C′) Cranial cartilage element exhibiting a mosaic distribution of soft (blue-stained chondrocytes) and hard (red-stained chondrocytes) cartilage (C). Note that Col2α1 protein is restricted to the soft cartilage region (C′). (D and D′) Hard cranial cartilage stained with eosin but not hematoxylin contains small chondrocytes surrounded by a thick layer of extracellular matrix (D), and it is negative for Col2α1 (D′).
Duplication and Divergence of Clade A Fibrillar Collagen Genes Occurred in Stem Vertebrates.
Col2α1 belongs to the clade A fibrillar collagen family, which includes collagen types I, II, III, and Vα2 (16). To investigate the relationship between evolution of the clade A collagens and the origin of the vertebrate skeleton, we extended our analysis to lancelets, a sister group to the vertebrates (4). Comparative studies of a multitude of genes in lancelets and vertebrates show that the lancelet lineage diverged before the duplication events that occurred in the vertebrate genome (17). We screened for lancelet fibrillar collagen cDNAs using degenerate RT-PCR, and we isolated a 2,196-bp clone with a deducted amino acid sequence that is 54% identical to mouse Col2α1 and 53% identical to mouse Col1α1 and Col3α1. To determine the relationship of the lancelet clone to the vertebrate fibrillar collagen proteins, we carried out molecular phylogenetic analyses using C-propeptide of clade A, B, and C fibrillar collagens. Bayesian phylogenetics, minimum evolution, and maximum likelihood methods each supported its designation as a clade A fibrillar collagen (AmphiColA; Fig. 7, which is published as supporting information on the PNAS web site). We then used the C-propeptide and triple-helix domains to refine further the position of AmphiColA within the chordate clade A family, and all three methods placed it as the sister clade to the vertebrate (including cyclostome) clade A collagens (Fig. 1). These results suggest that duplication of the ancestral ColA, the precursor of the clade A fibrillar collagen multigene family, occurred in the vertebrate lineage after the divergence of lancelets.
AmphiColA Is Expressed in the Lancelet Notochord and Neural Tube.
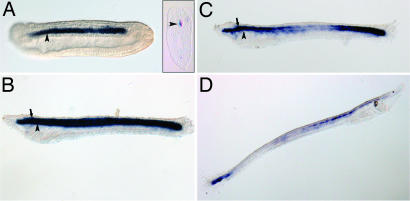
To gain insight into the expression pattern of the ancestral ColA gene, we investigated the developmental expression of AmphiColA by RNA in situ hybridization on lancelet embryos. During neurulation, AmphiColA was expressed in the notochord (Fig. 3A). At 30 h postfertilization, AmphiColA remained in the notochord, and a new domain of expression was detected in the neural tube (Fig. 3B). By 36 h, AmphiColA expression was being down-regulated in the middle third of the neural tube and notochord, but it remained strong anteriorly and posteriorly (Fig. 3C). In 5-day-old larvae, AmphiColA was detected in the tail bud and in the anterior region of the neural tube, up to the base of the cerebral vesicle (Fig. 3D). Expression of AmphiColA in the lancelet notochord and neural tube is strikingly similar to that of clade A fibrillar collagens in gnathostomes and lampreys. We did not detect AmphiColA in the embryonic and larval pharynx by in situ hybridization, although Rychel et al. (18) showed that the gill bars of adult cephalochordates can be stained with a chicken Col2α1 antibody. Our phylogenetic analyses indicate that AmphiColA is not a strict ortholog of Col2α1 but rather is a sister to the entire vertebrate ColA family, and whether the Col2α1 antibody can recognize AmphiColA protein is unclear. Nonetheless, our results cannot exclude the possibility that AmphiColA is activated in the pharynx during metamorphosis, when the gill region undergoes extensive remodeling.
Fig. 3.
Expression of AmphiColA during lancelet development. Whole-mount in situ hybridizations of lancelet embryos with an AmphiColA antisense riboprobe are shown. Anterior is to the left in A–C and to the right in D. Stages shown are 18 h (A), 30 h (B), 36 h (C), and 5 days (D) postfertilization. (A Inset) Transverse section of 18-h embryo. The arrowhead indicates notochord, and the arrows point to the neural tube.
Discussion
Col2α1-Based Cartilage Is a Shared Character of Crown-Group Vertebrates.
Cartilage based on type II collagen has long been considered a unifying character of gnathostomes that separates them from lampreys and hagfishes (1, 19). Recently, we reported that lampreys possess two Col2α1 orthologs and that adult lamprey cartilage is composed of Col2α1 protein (2). Here, we have gone on to show that hagfishes, the sister group to lampreys, also possess a Col2α1 ortholog, and we demonstrate that Col2α1 protein is localized to their soft cartilage. Taken together, these results suggest that the common ancestor of all crown-group (the living jawed and jawless) vertebrates had Col2α1-based cartilage. The presence of an undifferentiated clade A fibrillar collagen in lancelets and tunicates (20) suggests that the expansion of the ColA gene family occurred in stem vertebrates after the divergence of lancelets and tunicates. Thus, Col2α1-based cartilage is a synapomorphy of all crown-group vertebrates.
Our finding that hagfish hard cartilage lacks Col2α1 indicates that hagfishes also possess some non-Col2α1-based cartilage. This finding highlights a relationship between the profile of collagen expression and the structure of skeletal tissues in hagfishes. In gnathostomes, cartilage matrix is composed predominantly of Col2α1, whereas bone matrix is mostly Col1α1. During endochondral ossification, which involves the transition from cartilage to bone, Col2α1 is replaced by Col1α1 in the skeletal matrix. Our observation that a single skeletal element in the hagfish can have a mosaic structure (both soft and hard cartilage) that corresponds to mosaic distribution of Col2α1 raises the question of whether one type of cartilage may develop from the other, perhaps by altering the proportion of Col2α1 relative to other types of collagen or noncollagen matrix proteins. Alternatively, soft and hard cartilage elements may arise from distinct chondrocyte lineages (Col2α1-positive and -negative), with mosaic cartilages having a mixed lineage. Further characterization of hagfish hard cartilage will clarify whether it is composed of other types of collagen or whether this subset of the skeleton is noncollagenous.
Did Vertebrate Chondrocytes Evolve from the Notochord?
It has been suggested that the notochord may represent a primitive form of cartilage, based on their many shared structural, cellular, and molecular properties, and that vertebrate chondrocytes may have evolved from notochordal cells (21, 22). In gnathostomes, the notochord and/or notochordal sheath expresses most of the vertebrate clade A fibrillar collagen genes (15, 23–25). Our finding that AmphiColA is expressed in the notochord and notochordal sheath of Branchiostoma floridae, taken together with the recently reported data on Ciona intestinalis CiFCol1 and Branchiostoma belcheri BbFCol1 (both clade A fibrillar collagens), supports the idea that an ancestral ColA gene was expressed in the notochord of stem-group chordates (14, 20, 26). We suggest that duplication and divergence of the clade A collagen genes in stem-group vertebrates may have facilitated the evolutionary diversification of chondrocytes and notochordal cells. This hypothesis deals specifically with the origin of vertebrate chondrocytes, and it is important to note that cartilage is also found in several invertebrates, including cephalopods, snails, and horseshoe crabs (1, 21). Future work on the molecular basis of invertebrate chondrogenesis should reveal whether fibrillar collagens also were used in these independent evolutionary events.
Clade A Fibrillar Collagen Duplication Facilitated Evolution of the Vertebrate Skeleton.
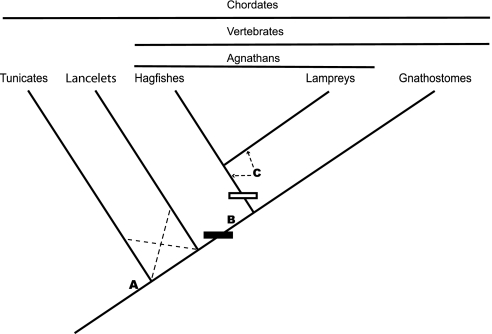
The identification of an undifferentiated, ancestral ColA gene in lancelets indicates that the duplication event that gave rise to Col2α1 occurred after the divergence of lancelets and vertebrates. We have focused our analysis on hagfishes and lancelets; however, it has recently been suggested that tunicates are the closest sister group to vertebrates (27). As noted above, tunicates also have an undifferentiated ColA gene (20), indicating that the duplication also postdates their divergence from the lineage leading to vertebrates. Taken together, the data show that the origin of Col2α1 from the ancestral ColA gene must have occurred in stem vertebrates. We propose that this duplication event may have been critical for the origin of the vertebrate skeleton (Fig. 4).
Fig. 4.
Origin of type II collagen-based cartilage. Phylogenetic relationships of tunicates, lancelets, hagfishes, lampreys, and gnathostomes (37, 38) are shown. Dashed lines indicate alternative branch positions (27). Letters point out major steps in Col2α1-based cartilage evolution. The solid rectangle indicates the origin of the Col2α1 gene by ColA gene duplication(s). The hollow rectangle indicates the origin of Col2α1a and Col2α1b by Col2α1 gene duplication. (A) Ancestral clade A collagen gene expression in notochord of stem chordates. (B) Origin of Col2α1-based cartilage in stem vertebrates. (C) Subfunctionalization of Col2α1, in which the expression domain of the ancestral gene is partitioned between the duplicate genes (Col2α1a and Col2α1b); arrows indicate possible positions of this event.
Our data fit with the hypothesis that at least one round of genome duplication occurred between the origin of chordates and the origin of vertebrates (28, 29). The expansion of the clade A fibrillar collagen gene family, particularly the origin of Col2α1 and Col1α1, may account for the unique skeletal matrices of vertebrate cartilage and bone. Although cartilage has evolved multiple times in metazoa, it is unclear from the fossil record whether cartilage or bone evolved first in the vertebrates (30). Our results raise the possibility that Col1α1 and Col2α1 arose from the same duplication event, and thus the major matrix components of bone and cartilage may have evolved at the same time in vertebrates. Finally, it is noteworthy that clade A fibrillar collagens are physically linked with Hox gene clusters in vertebrates and in echinoderms (31). Duplication of the clade A fibrillar collagen genes therefore may have coincided with the Hox cluster duplications (32, 33). Coordinated expansion of the ColA and Hox gene families may have facilitated the diversification of vertebrate connective tissue types and provided a mechanism for differentially patterning them.
Materials and Methods
Animals.
Adult lancelets (B. floridae) were collected in Tampa Bay, FL. Spawning was induced in the laboratory according to published methods (34). Atlantic hagfishes (Myxine glutinosa) were purchased from Ward's Natural Science (Rochester, NY).
Gene Cloning.
RNA was extracted by using TRIzol Reagent (Invitrogen, Carlsbad, CA). cDNA was made by reverse transcription reactions by using a SuperScript II first-strand cDNA synthesis kit (Invitrogen). Fibrillar collagen genes were amplified with degenerate primers designed by using the CODEHOP program (35). PCRs were carried out by using BD Advantage 2 PCR enzyme system (Clontech, Mountain View, CA) from cDNA. In 50-μl reactions containing 1-μl forward (5′-GGCCCTCCCGGCCTGCARGGNATGCC-3′) and 1-μl reverse (5′-GGGGCCGATGTCCACGCCRAAYTCYTG-3′) primers (20 pmol/μl), 5 μl of 10× buffer, 1 μl of cDNA template, 1 μl of dNTP at 10 mM (each), 1 μl of Taq polymerase mixture, and 40 μl of double-distilled water. Reactions were amplified as follows: 94°C for 1 min followed by 35 cycles of 94°C for 45 s, 65°C for 45 s, 68°C for 3 min, and a final 10-min elongation at 68°C. PCR products were purified by using QIAGEX II gel extraction kit (Qiagen, Valencia, CA) then cloned into pCRII-TOPO (Invitrogen) for sequencing.
Sequence Analysis and Molecular Phylogenetics.
Inferred protein sequences for hagfish and amphioxus cDNAs were initially assigned to the clade A fibrillar collagen families on the basis of BLAST searches and conserved domains. These preliminary assignments were followed by estimates of their amino acid identities and phylogenetic relationships. Multiple sequence alignments for available fibrillar A, B, and C collagen proteins, including the new hagfish and amphioxus sequences, were generated with ClustalX. Phylogenetic analyses of these multiple protein alignments were conducted with Bayesian phylogenetics, minimum evolution, and maximum likelihood methods, as described previously (2). The following amino acid sequences were retrieved from GenBank for inclusion in our phylogenetic analyses: mouse Col2α1, B41182; rat Col2α1, NP_037061; dog Col2α1, NP_001006952; chick Col2α1, NP_989757; horse Col2α1, T45467; salamander Col2α1, BAA82043; frog Col2α1, B40333; zebrafish Col2α1, XP_692723; lamprey Col2α1a, DQ136024; lamprey Col2α1b, DQ136025; human Col1α1, BAD92834; mouse Col1α1, CAI25880; dog Col1α1, NP_001003090; cow Col1α1, AAI05185; salamander Col1α1, BAA36973; frog Col1α1, BAA29028; halibut Col1α1, BAD77968; zebrafish Col1α1, AAH63249; rainbow trout Col1α1, BAB55661; human Col1α2, AAH42586; mouse Col1α2, NP_034060; dog Col1α2, NP_001003187; chick Col1α2, XP_418665; frog Col1α2, AAH49287; salmon Col1α2, BAB79229; halibut Col1α2, BAD77969; fugu Col1α2, CAG11117; zebrafish Col1α2, NP_892013; human Col3α1, AAL13167; mouse Col3α1, NP_031763; dog Col3α1, XP_851009; cow Col3α1, XP_588040; frog Col3α1, AAH60753; human Col5α2, NP_000384; mouse Col5α2, NP_031763; rat Col5α2, XP_343565; dog Col5α2, XP_535998; cow Col5α2, XP_581318; pig Col5α2, BAD91584; chick Col5α2, XP2421846; sea urchin ColP2α; NP_999675; acorn worm ColA, DQ233249; human Col5α1, NP_000084; mouse Col5α1, NP_056549; ColA; chick Col5α1, NP_990121; human Col5α3, NP_056534; mouse Col5α3, P25940; human Col11α1, NP_001845; mouse Col11α1, NP_031755; chick Col11α1, XP_422303; human Col11α2, CAA20240; mouse Col11α2, NP_034056; human Col24α1, NP_690850; mouse Col24α1, NP_082046; chick Col24α1, XP_422363; human Col27α1, NP_116277; mouse Col27α1, NP_079961; chick Col27α1, XP_415514. The tunicate fibrillar collagen genes (clade A fibrillar collagen, ci0100150759; clade B fibrillar collagen, ci0100154301) were retrieved from the JGI C. intestinalis genome website.
Histology, Immunohistochemistry, and in Situ Hybridization.
Formalin-fixed hagfish specimens were washed in 70% ethanol, embedded in paraffin, and sectioned (6 μm). Sections were either used for immunohistochemistry, or they were stained with hematoxylin/eosin and fast green (Mallory's trichrome) by using standard staining methods. Immunohistochemical staining for Col2α1 was carried out by using an antibody against human Col2α1 protein, as described previously (2). Whole-mount in situ hybridization of amphioxus embryos was performed following published methods (36).
Supplementary Material
Acknowledgments
We thank Nick and Linda Holland (Scripps Institute of Oceanography, La Jolla, CA) and Simone Shuster and Ben Olaivar (University of Florida) for assistance in collecting lancelets; Mike Miyamoto (University of Florida) for advice on phylogenetic methods; M. M. and Billie Swalla (University of Washington, Seattle, WA) for comments on the manuscript; J. Havird, K. Choe, and D. Evans (University of Florida) for hagfish cDNA; and Hiroshi Wada (University of Tsukuba, Tsukuba, Ibaraki, Japan) for exchanging information before publication.
Abbreviations
- Col1α1
type I collagen
- Col2α1
type II collagen
Footnotes
References
- 1.Wright GM, Keeley FW, Robson P. Cell Tissue Res. 2001;304:165–174. doi: 10.1007/s004410100374. [DOI] [PubMed] [Google Scholar]
- 2.Zhang G, Miyamoto MM, Cohn MJ. Proc Natl Acad Sci USA. 2006;103:3180–3185. doi: 10.1073/pnas.0508313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donoghue PC, Forey PL, Aldridge RJ. Biol Rev Camb Philos Soc. 2000;75:191–251. doi: 10.1017/s0006323199005472. [DOI] [PubMed] [Google Scholar]
- 4.Zeng LY, Swalla BJ. Can J Zool. 2005;83:24–33. [Google Scholar]
- 5.Ota KG, Kuratani S. Zoolog Sci. 2006;23:403–418. doi: 10.2108/zsj.23.403. [DOI] [PubMed] [Google Scholar]
- 6.Powell ML, Kavanaugh SI, Sower SA. Integr Comp Biol. 2005;45:158–165. doi: 10.1093/icb/45.1.158. [DOI] [PubMed] [Google Scholar]
- 7.Wicht H, Northcutt RG. Philos Trans R Soc Lond B. 1995;349:119–134. doi: 10.1098/rstb.1995.0098. [DOI] [PubMed] [Google Scholar]
- 8.Hamerman D. N Engl J Med. 1989;320:1322–1330. doi: 10.1056/NEJM198905183202006. [DOI] [PubMed] [Google Scholar]
- 9.Cotrufo M, Della Corte A, De Santo LS, Quarto C, De Feo M, Romano G, Amarelli C, Scardone M, Di Meglio F, Guerra G, et al. J Thorac Cardiovasc Surg. 2005;130:504–511. doi: 10.1016/j.jtcvs.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 10.Guo X, Day TF, Jiang X, Garrett-Beal L, Topol L, Yang Y. Genes Dev. 2004;18:2404–2417. doi: 10.1101/gad.1230704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang AH, Chen JY, Lin JK. Kidney Int. 2003;63:1530–1539. doi: 10.1046/j.1523-1755.2003.00861.x. [DOI] [PubMed] [Google Scholar]
- 12.Welch U, Chiba A, Honma Y. In: The Biology of Hagfishes. Jorgensen JM, Lomholt JP, Weber RE, Malte H, editors. London: Chapman & Hall; 1998. pp. 145–159. [Google Scholar]
- 13.Cole FJ. Trans R Soc Edinburgh. 1905;41:749–791. [Google Scholar]
- 14.Robson P, Wright GM, Keeley FW. Anat Embryol. 2000;202:281–290. doi: 10.1007/s004290000113. [DOI] [PubMed] [Google Scholar]
- 15.Yan YL, Hatta K, Riggleman B, Postlethwait JH. Dev Dyn. 1995;203:363–376. doi: 10.1002/aja.1002030308. [DOI] [PubMed] [Google Scholar]
- 16.Aouacheria A, Cluzel C, Lethias C, Gouy M, Garrone R, Exposito JY. J Biol Chem. 2004;279:47711–47719. doi: 10.1074/jbc.M408950200. [DOI] [PubMed] [Google Scholar]
- 17.Panopoulou G, Hennig S, Groth D, Krause A, Poustka AJ, Herwig R, Vingron M, Lehrach H. Genome Res. 2003;13:1056–1066. doi: 10.1101/gr.874803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rychel AL, Smith SE, Shimamoto HT, Swalla BJ. Mol Biol Evol. 2006;23:541–549. doi: 10.1093/molbev/msj055. [DOI] [PubMed] [Google Scholar]
- 19.Wright GM, Youson JH. Am J Anat. 1983;167:59–70. doi: 10.1002/aja.1001670106. [DOI] [PubMed] [Google Scholar]
- 20.Wada H, Okuyama M, Satoh N, Zhang S. Evol Dev. 2006;8:370–377. doi: 10.1111/j.1525-142X.2006.00109.x. [DOI] [PubMed] [Google Scholar]
- 21.Cole AG, Hall BK. Acta Zool. 2004;85:69–80. [Google Scholar]
- 22.Stemple DL. Development (Cambridge, UK) 2005;132:2503–2512. doi: 10.1242/dev.01812. [DOI] [PubMed] [Google Scholar]
- 23.Dubois GM, Haftek Z, Crozet C, Garrone R, Le Guellec D. Gene. 2002;294:55–65. doi: 10.1016/s0378-1119(02)00770-9. [DOI] [PubMed] [Google Scholar]
- 24.Ghanem E. Cell Biol Int. 1996;20:681–685. doi: 10.1006/cbir.1996.0090. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Q, Eberspaecher H, Lefebvre V, De Crombrugghe B. Dev Dyn. 1997;209:377–386. doi: 10.1002/(SICI)1097-0177(199708)209:4<377::AID-AJA5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 26.Satou Y, Takatori N, Yamada L, Mochizuki Y, Hamaguchi M, Ishikawa H, Chiba S, Imai K, Kano S, Murakami SD, et al. Development (Cambridge, UK) 2001;128:2893–2904. doi: 10.1242/dev.128.15.2893. [DOI] [PubMed] [Google Scholar]
- 27.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 28.Panopoulou G, Poustka AJ. Trends Genet. 2005;21:559–567. doi: 10.1016/j.tig.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Boot-Handford RP, Tuckwell DS. Bioessays. 2003;25:142–151. doi: 10.1002/bies.10230. [DOI] [PubMed] [Google Scholar]
- 30.Donoghue PC, Kouchinsky A, Waloszek D, Bengtson S, Dong XP, Val'kov AK, Cunningham JA, Repetski JE. Evol Dev. 2006;8:232–238. doi: 10.1111/j.1525-142X.2006.00093.x. [DOI] [PubMed] [Google Scholar]
- 31.Cameron RA, Rowen L, Nesbitt R, Bloom S, Rast JP, Berney K, Arenas-Mena C, Martinez P, Lucas S, Richardson PM, et al. J Exp Zool. 2006;306:45–58. doi: 10.1002/jez.b.21070. [DOI] [PubMed] [Google Scholar]
- 32.Bailey WJ, Kim J, Wagner GP, Ruddle FH. Mol Biol Evol. 1997;14:843–853. doi: 10.1093/oxfordjournals.molbev.a025825. [DOI] [PubMed] [Google Scholar]
- 33.Morvan-Dubois G, Le Guellec D, Garrone R, Zylberberg L, Bonnaud L. J Mol Evol. 2003;57:501–514. doi: 10.1007/s00239-003-2502-x. [DOI] [PubMed] [Google Scholar]
- 34.Stokes MD, Holland ND. Acta Zool. 1995;76:105–120. [Google Scholar]
- 35.Rose TM, Henikoff JG, Henikoff S. Nucleic Acids Res. 2003;31:3763–3766. doi: 10.1093/nar/gkg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimeld SM. Dev Biol. 1997;183:74–85. doi: 10.1006/dbio.1996.8481. [DOI] [PubMed] [Google Scholar]
- 37.Blair JE, Hedges SB. Mol Biol Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- 38.Donoghue PC, Sansom IJ, Downs JP. J Exp Zoolog. 2006;306:278–294. doi: 10.1002/jez.b.21090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.