Abstract
CD36 and LIMPII analog 1, CLA-1, and its splicing variant, CLA-2 (SR-BI and SR-BII in rodents), are human high density lipoprotein receptors with an identical extracellular domain which binds a spectrum of ligands including bacterial cell wall components. In this study, CLA-1- and CLA-2-stably transfected HeLa and HEK293 cells demonstrated several-fold increases in the uptake of various bacteria over mock-transfected cells. All bacteria tested, including both Gram-negatives (Escherichia coli K12, K1 and Salmonella typhimurium) and Gram-positives (Staphylococcus aureus and Listeria monocytogenes), demonstrated various degrees of lower uptake in control cells. This result is consistent with the presence of high-density lipoprotein-receptor-independent bacterial uptake that is enhanced by CLA-1/CLA-2 overexpression. Bacterial lipopolysaccharides, lipoteichoic acid, and synthetic amphipathic helical peptides (L-37pA and D-37pA) competed with E. coli K12 for CLA-1 and CLA-2 binding. Transmission electron microscopy and confocal microscopy revealed cytosolic accumulation of bacteria in CLA-1/CLA-2-overexpressing HeLa cells. The antibiotic protection assay confirmed that E. coli K12 was able to survive and replicate intracellularly in CLA-1- and CLA-2-overexpressing HeLa, but both L-37pA and D-37pA prevented E. coli K12 invasion. Peritoneal macrophages isolated from SR-BI/BII-knockout mice demonstrated a 30% decrease in bacterial uptake when compared with macrophages from normal mice. Knockout macrophages were also characterized by decreased bacterial cytosolic invasion, ubiquitination, and proteasome mobilization while retaining bacterial lysosomal accumulation. These results indicate that, by facilitating bacterial adhesion and cytosolic invasion, CLA-1 and CLA-2 may play an important role in infection and sepsis.
Keywords: bacterial adhesion, internalization, phagocytes, cytosolic invasion, high-density lipoprotein receptor
CLA-1 and its splicing variant CLA-2, orthologues of rodent scavenger receptor class B type I (SR-BI) and its splicing variant SR-BII, are known as high-density lipoprotein (HDL) receptors. These receptors mediate HDL binding, followed by selective uptake of cholesteryl ester in the liver and steroidogenic tissues (for review, see refs. 1 and 2). Organs with the highest expression of CLA-1/SR-BI include the liver, adrenals, ovaries, and testes, whereas a relatively high expression has been demonstrated in intestinal cells, phagocytes, and endothelial cells (3, 4). In steroid-producing tissues, such as the adrenals, most of the cholesterol for steroid hormone production is delivered through CLA-1/SR-BI. In the liver, CLA-1/SR-BI is involved in bile formation and appears to be the major mechanism of cholesterol excretion. Recent findings point to a gender-dependent expression of SR-BI and SR-BII. SR-BI is predominantly expressed in males, whereas SR-BII is the predominant gene product in the female mouse liver (5).
Accumulating evidence suggests that the function of SR-BI gene products is not solely linked to cholesterol ester metabolism but involves a wide spectrum of activities. CLA-1/SR-BI has been recently shown to be involved in the uptake of apoptotic cells (6), triglyceride and phospholipid delivery to certain cell types (7), serum amyloid A (SAA) uptake (8, 9) and selective uptake of HDL associated with vitamin E (10). We recently demonstrated that CLA-1 binds bacterial cell-wall components such as endotoxin [lipopolysaccharides (LPS)] of Gram-negative bacteria and lipoteichoic acid (LTA) of Gram-positive bacteria (11, 12). These receptors also are able to bind and internalize other bacterial and animal proteins that contain amphipathic helices (8, 12), suggesting that, sharing the same extracellular loop receptor domain, CLA-1 and CLA-2 may also directly bind these bacterial cell-wall components mediating bacterial adhesion, invasion, intracellular survival, and proliferation. A recent report supports this possibility (13).
In this work, we expand our previous observations (11, 12, 14) by showing that the binding by the CLA-1 and CLA-2 receptors may represent important pathogenic mechanisms during bacterial infection/sepsis. These receptors mediate the interaction between bacterial cell wall components and host mammalian cells and allow for bacterial cytosolic invasion and survival in the host cells.
Results
Uptake of Live Bacteria in HeLa Cell Lines.
Because the presence of LTA or LPS in the outer bacterial cell wall is well documented, we hypothesized that CLA-1 and CLA-2 might be the sites of initial bacterial adhesion associated with subsequent downstream events such as internalization and cellular signaling. Increased HDL (Supporting Text, Appendix A, and Fig. 8, which are published as supporting information on the PNAS web site) and anti-CLA-1 loop antibody (Fig. 1) uptake by CLA-1 and CLA-2 HeLa cells confirmed the functional activity of the overexpressed HDL receptors. By using FACS analysis, the uptake (binding plus intracellular accumulation) of live Escherichia coli K12, K1, Salmonella typhimurium, Staphylococcus aureus and Listeria monocytogenes were 5- to 20-fold higher in CLA-1-HeLa and CLA-2-HeLa when compared with mock-HeLa (Fig. 1). CLA-1 and CLA-2 also mediated bacterial uptake in HEK293 cells (data not shown). These HDL receptors also mediated uptake of heat-killed bacteria (Supporting Text, Appendix A).
Fig. 1.
Uptake of Alexa Fluor 488-labeled live bacteria in HeLa cells by FACS analysis. Bacteria or 10 μg/ml Alexa Fluor 488-labeled anti-CLA-1/CLA-2 loop IgG were incubated with monolayers of HeLa cells for 1.5 h at 37° C and then harvested for FACS analysis. The uptake for each bacterial species is indicated at the bottom of the graphs in mock-transfected (blue) and CLA-1- (green) and CLA-2- (red) overexpressing HeLa cells.
CLA-1 and CLA-2 Ligands Compete Against Bacterial Uptake.
HDL and LPS inhibited the uptake of Alexa Fluor 488-labeled E. coli K12 by 80% and 60%, respectively (Fig. 2) in CLA-1 and CLA-2 HeLa cells. Two other ligands, L-37pA and D-37pA, also blocked bacterial adhesion, whereas no inhibition occurred when L3D-37pA, an L-37pA analogue with disrupted helical structure, was used (Supporting Text, Appendix C, and which are published as supporting information on the PNAS web site).
Fig. 2.
Competition of CLA-1/CLA-2 ligands against E. coli K12. Bacteria were incubated with confluent monolayer HeLa cells in the presence or absence of 100 μg/ml LPS (yellow), HDL (blue), or without competitor (green) for 1.5 h at 37 C° and then harvested for FACS analysis. Autofluorescence is seen in red. CLA-1-overexpressing (A), CLA-2-overexpressing (B), and mock HeLa (C) cells, are shown.
E. coli K12 Internalization in CLA-1 and CLA-2 HeLa Cells.
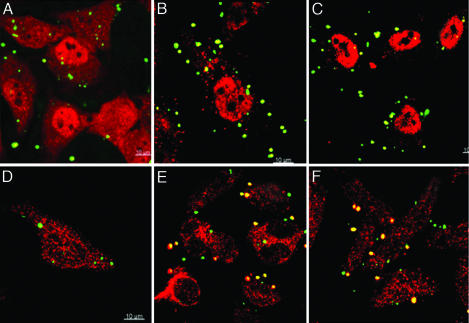
E. coli K12 uptake was studied by using transmission electron microscopy (TEM) in HeLa cells. Fig. 3 shows that, after initial adhesion to the cell surface, paraformaldehyde-fixed E. coli K12 was internalized in HeLa cells overexpressing either CLA-1 or CLA-2. The bacteria are seen as small cytoplasmic inclusions in the cells expressing either of the two receptors, but they are not present in mock-transfected HeLa cells (data not shown). The bacteria retained their inner and outer membranes while residing in the cytoplasm rather than presenting as visible subcellular structures within lysosomes, endocytic vesicles, or endoplasmic reticulum.
Fig. 3.
Transmission electron microscopic pictures showing E. coli K12 adhesion, engulfment, and internalization by CLA-1 and CLA-2 receptors in HeLa cells. CLA-1 (A–C) and CLA-2 (D and E) -overexpressing HeLa cells were incubated with paraformaldehyde-fixed E. coli K12 and processed as described in Materials and Methods. (B) A magnified image of bacterial adhesion sites in HeLa cells. Arrows show cytoplasmic and adherent bacteria.
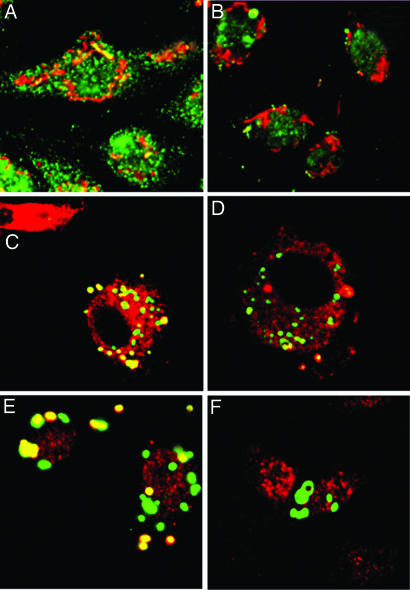
Bacterial Uptake Analyzed by Confocal Microscopy.
CLA-1-HeLa and CLA-2-HeLa were incubated with live Staphylococcus aureus, followed by colocalization with FK-2 antibody against polyubiquitins or anti-20S proteasome antibody. Visible colocalizations (yellow) were only found in CLA-1 and CLA-2 HeLa, further confirming bacterial sequestration to the cytosol in epithelial cells (Fig. 4). No significant bacterial colocalizations were found with endocytic compartment markers (Appendix C).
Fig. 4.
Confocal imaging of live Alexa Fluor 488 Staphylococcus aureus in HeLa cells. Mock-transfected (A and D), CLA-1 (B and E), and CLA-2 (C and F) HeLa cells were incubated with live Alexa Fluor 488 Staphylococcus aureus for 50 min. After washing with ice-cold PBS, fixing with 4% paraformaldehyde, immunostaining with monoclonal FK-2 antibody (A–C) or anti-20S proteasome antibody (D–F), slides were analyzed by confocal microscopy. The color images represent bacteria (green), polyubiquitins or 20S proteasome (both red), and bacteria associated with ubiquitins or proteasome (yellow).
Bacterial Uptake by SR-BI/SR-BII Knockout Mouse Peritoneal Macrophages.
Peritoneal macrophages (Mφ) isolated from control and SR-BI gene KO mice which lacks both SR-BI and its splicing variant SR-BII, were also used for bacterial uptake studies. The uptake of inflammation-related CLA-1/2 ligands, LPS and SAA, was decreased by 50%, E. coli K12 uptake was lower by 30%, whereas no difference was found for the uptake of denaturated HDL in the murine macrophages lacking the SR-BI gene (Supporting Text, Appendix D, and Fig. 11, which is published as supporting information on the PNAS web site).
Further analysis of bacterial phagocytosis by using live Alexa Fluor 488-labeled E. coli K12 and the lysosomal tracker, LysoTracker red, revealed a tendency for increased lysosomal accumulation in KO Mφ upon confocal microscopy of live Mφ cultures (61% colocalized bacteria in KO Mφ versus 52% in control Mφ), but the difference did not reach statistical significance. When analyzed after paraformaldehyde cell fixation, the colocalizations were slightly higher in the control than KO Mφ for both live Alexa Fluor 488-labeled E. coli K12 and Staphylococcus aureus (Fig. 5). Colocalization studies using fixed cells and anti-LIMP-2 as well as LysoTracker red revealed similar trends indicating retention of robust lysosomal phagocytosis in KO Mφ but, again, without reaching statistical significance (Supporting Text, Appendix E, and Fig. 12, which is published as supporting information on the PNAS web site).
Fig. 5.
Bacterial lysosomal phagocytosis in mouse peritoneal macrophages. Live Alexa Fluor 488-labeled E. coli K12 (A and B) and Staphylococcus aureus (C and D) were incubated with peritoneal Mφ isolated from normal (A and C) and SR-BI/SR-BII KO (B and D) mice for 45 min. After washing with PBS, the cells were further incubated with LysoTracker red for 15 min, fixed, and analyzed by confocal microscopy. The color images represent bacteria (green), lysosomal compartment (red), and bacteria in lysosomes (yellow).
Because both TEM and confocal microscopy showed cytoplasmic bacteria in CLA-1/CLA-2-overexpressing HeLa epithelial cells, the cytoplasmic accumulation was further analyzed in both normal and KO mouse Mφ. After a 50-min incubation, E. coli K12 and Staphylococcus aureus were colocalized with anti-polyubiquitin monoclonal FK-2 antibody in normal Mφ, whereas little or no colocalization was found in Mφ isolated from SR-BI/SR-BII KO mice (Fig. 6B and D). In normal Mφ that express the SR-BI gene but not in those from SR-BI/BII KO mice, the bacteria were also colocalized with an FK-1 antibody that recognizes both mono- and polyubiquitinated proteins (data not shown). In addition, bacteria selectively colocalized with 20S proteasome in normal Mφ, but not in KO Mφ, further confirming their cytosolic accumulation (Fig. 6 E and F).
Fig. 6.
Cytosolic bacterial accumulation, ubiquitination, and association with 20S proteasome in mouse peritoneal macrophages. Live Alexa Fluor 594-labeled E. coli K12 (A and B) and Alexa Fluor 488 Staphylococcus aureus (C–F) were incubated with peritoneal Mφ isolated from normal (A, C, and E) and SR-BI/SR-BII KO (B, D, and F) mice for 30 (E. coli K12) and 50 (Staphylococcus aureus) min. After washing with ice-cold PBS, fixing with 4% paraformaldehyde, immunostaining with monoclonal FK-2 antibody (A–D) or with monoclonal anti-20S proteasome antibody (E and F), slides were analyzed by confocal microscopy. The color images represent bacteria (Alexa Fluor 594-labeled E. coli K12, red; Alexa Fluor 488 Staphylococcus aureus, green), FK-2 antibody (green in A and B and red in C and D), anti-20S proteasome antibody (red), and bacteria in cytosol (yellow).
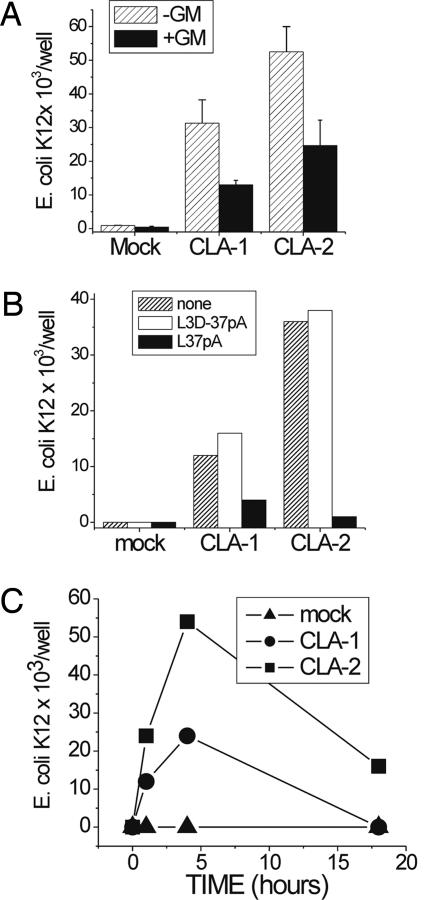
E. coli K12 Invasion and Intracellular Growth Assessed by the Antibiotic Protection Assay (ABPA).
As seen in Fig. 7A, HeLa cells overexpressing CLA-1 and CLA-2 demonstrated a 20- to 40-fold increase in adherent bacteria [no further incubation with gentamicin (GM)] when compared with mock-transfected cells. In the presence of GM, the number of growing E. coli K12 bacteria was reduced by ≈55%, indicating that ≈45% of the bacteria were inside the cells and thereby protected from the antibiotic (GM). When bacteria were incubated with CLA-1- and CLA-2-overexpressing HeLa cells in the presence of 50 μg/ml L-37pA, the amount of adherent and internalized bacteria was dramatically reduced (Fig. 7B). This result is in accord with our earlier findings (Supporting Text, Appendix B, and which are published as supporting information on the PNAS web site) that these ligands efficiently block bacterial adhesion to CLA-1 and CLA-2. No inhibitory effect was seen with the L3D-37pA (an analogue with disrupted helical structure), and none of the ligands had a direct effect on bacterial growth or viability (data not shown).
Fig. 7.
Entry and intracellular survival of E. coli K12 in CLA-1- and CLA-2-overexpressing HeLa cells. Monolayers of CLA-1- and CLA-2-overexpressing and mock-transfected HeLa cells were incubated with E. coli K12 in the presence or absence of L-37pA or L3D37-pA for 1.5 h in DMEM containing 10% FCS. Total cell-associated bacteria were determined by plating dilutions of hypotonic cell extracts on blood agar. For counting intracellular bacteria, the monolayers were washed first with PBS and then incubated in a medium containing GM (20 μg/ml) for another hour to kill extracellular bacteria. (A) Live bacteria before and after GM incubation. (B) Live bacteria incubated in the presence of peptides after GM incubation. (C) Live bacteria incubated for various time periods in the presence of GM.
Because various bacteria are characterized by their ability to survive and proliferate intracellularly in mammalian cells, we analyzed the survival and proliferation of E. coli K12 in HeLa cells overexpressing CLA-1 or CLA-2. After invasion of CLA-1 or CLA-2 HeLa cells, the bacteria were less sensitive to GM. The number of intracellular bacteria more than doubled within the first 4 h in the presence of the GM when compared with a 1-h incubation (Fig. 7C). Extension of the incubation to 18 h was associated with the decreasing numbers of bacteria, predominantly because of bacterium-induced HeLa cell killing. Other bacterial species including Salmonella typhimurium, E. coli K25, K1, and Staphylococcus aureus also demonstrated intracellular accumulation with the ABPA (Supporting Text, Appendix F, and Fig. 15, which is published as supporting information on the PNAS web site).
Discussion
CLA-1 and CLA-2 are human HDL receptors that also bind and internalize bacterial cell wall components such as LPS and LTA (11, 12) as well as bacterial, viral, and animal proteins containing amphipathic helices (8, 12, 13). Negatively charged phospholipids (6, 15) and anionic class A amphipathic α-helixes are two known primary motifs for the CLA-1/SR-BI recognition (12, 16). It has been established that amphipathic helical peptides (1, 2) LPS (11) and LTA (12, 17) bind to CLA-1 with high affinity. The presence of one or both of these motifs has been associated with bacterial and viral recognition by class B scavenger receptors (13). In this work, two models were used including receptor overexpression in nonphagocytic human epithelial cells and receptor-deficient phagocytic mouse peritoneal macrophages. With this approach, it was possible to establish a direct role for HDL receptors in the cells of the initial pathogen contact (epithelial cells) as well as determine HDL-receptor impact on bacterial trafficking, sequestration, and processing in phagocytic cells that are characterized by multiple phagocytosis-related pathways of bacterial killing and processing. We confirmed bacterial uptake by HDL receptors for a variety of pathogens including Alexa Fluor 488-labeled live E. coli K12, K1, Salmonella typhimurium, Staphylococcus aureus and Listeria monocytogenes (Fig. 1). In addition to live bacteria, we also showed that CLA-1 and CLA-2 bind dead bacteria, pointing to a potential role of these receptors in the clearance of damaged bacteria. Supporting this possibility is the earlier observation of extensive colocalization of LTA with scavenger cells in the liver of patients with primary biliary cirrhosis (18).
Competition experiments where both LPS and LTA nearly completely blocked Alexa Fluor 488 E. coli K12 uptake indicate that LPS and LTA may play important roles as HDL-receptor recognition motifs. This observation is also consistent with the blocking effects of HDL, L-37pA and D-37pA; prototypes of potential anti-bacterial drugs that diminish LPS uptake by CLA-1 (11). Importantly, both D-37pA and L-37pA peptides reduced bacterial accumulation and, consequently, survival in the presence of GM (Fig. 7).
We selected E. coli K12 and Staphylococcus aureus for the study of receptor–mediated intracellular bacterial sequestration, trafficking, ubiquitination, and association with proteasome. In epithelial cells overexpressing CLA-1 and CLA-2, the bacteria did not colocalize with cellular vesicular compartments such as endosomes, endoplasmic reticulum, lysosomes, or Golgi complex. This finding is consistent with our TEM findings for both a lack of bacteria in vesicular structures and the presence of bacteria in the cytosol of CLA-1- and CLA-2-overexpressing HeLa cells. Furthermore, Staphylococcus aureus colocalized with cytosolic polyubiquitins and proteasome in HeLa cells (Fig. 3), pointing to CLA-1/CLA-2-dependent cytosolic sequestration of bacteria in nonphagocytic cells. Because it is difficult to achieve receptor knockout in human phagocytes, mouse Mφ from SR-BI KO mice were compared with cells from normal mice. There was a decreased cytosolic bacterial accumulation in SR-BI/SR-BII deficient Mφ, however both normal and KO Mφ retained robust lysosomal phagocytosis. We propose that bacteria can invade cells expressing CLA-1/CLA-2 through a vesicular-independent mechanism that leads to bacterial ubiquitination in the cytosol (Fig. 6). This mechanism can also lead to bacterial recognition and bacterial antigen processing through a proteasome-dependent mechanism that has been suggested in various studies demonstrating bacterial ubiquitination in phagocytes (19, 20).
A pattern-recognition receptor function does not follow the classically accepted mechanisms of interaction between ligand and receptor that require a receptor domain that sterically fits particular ligands and possesses a high level of ligand stereospecificity. In addition, ligands typically are significantly smaller than the receptor itself. In contrast, the CLA-1 and CLA-2 (SR-class BI/II) ligands typically are not simple molecules but rather multimolecular complexes in which the hydrophilic portions of the ligands are exposed, whereas the hydrophobic components are shielded within the complex. This is particularly true for phospholipid micelles where the hydrophilic heads are exposed and the hydrophobic fatty acids are inside the micelles. A micelle formation is characteristic for the majority of CLA-1 ligands such as HDL, low-density lipoproteins, liposomes, SAA, and LPS. With increasing charge of the hydrophilic heads, the interhead distance increases (21), exposing the hydrophobic moiety of the micelle. It is possible that CLA-1 and CLA-2 insert some portions of the extracellular loop into the micelle-ligand complex, functioning as a key, whereas the ligand works as a lock. Such a suggestion is reasonable, because the interhead distance increases in the order of micelles made of phosphatidylcholine (PC), phosphoinositides (PI), and phosphatidylserine (PS). Accordingly, the Kd for such micelles decreases in the order of PC, PI, and PS (15). Such a model can potentially explain the ability of CLA-1 and CLA-2 to interact with so many structurally diverse ligands, including apoA-I, lipoproteins, LPS, Gram-negative (and some Gram-positive) bacteria, viruses (e.g., HCV), and SAA.
In conclusion, we have found that, in addition to bacterial binding, CLA-1 and CLA-2 receptors mediate bacterial cytosolic accumulation, proliferation, and evasion of the lysosomal complex. This mechanism may either facilitate infection with certain intracellular pathogens and/or represent a mechanism of bacterial cytosolic recognition, ubiquitination, and degradation. Pathogens involved with this mechanism(s) may include both bacterial and viral agents. For instance, hepatitis C virus has already bee shown to require CLA-1 for entry (22, 23). By using the principle of CLA-/CLA-2 targeting, our findings allow for creation of a number of receptor antagonists such as the model peptides L- and D-37pA that block bacterial invasion, making the bacteria accessible to antibiotics. The most effective compounds could be identified by evaluating CLA-1/CLA-2 bacterial binding in vitro using simple and robust bacterial-binding assays as described in this article.
Materials and Methods
Alexa Fluor 488-labeled heat-killed E. coli K1, K12, and Zymozan particles and all cell trackers and reactive fluorescent dyes were obtained from Invitrogen/Molecular Probes (Eugene, OR). Rabbit anti-loop CLA-1/CLA-2 antibody was produced against the region of the CLA-1 protein between amino acids 36–439 expressed as a fusion protein with N-terminal His Tag (Primm Biotech, Cambridge, MA). Rabbit anti-CLA-1 antibody, anti-CLA-2, and LIMP-2 antibodies were custom produced by using CTSAPKGSVLQEAKL for CLA-1 (Anaspec, San Jose, CA), CLPDSPSRQPPSPTA for CLA-2, and CDEGTADERAPLIRT for LIMP-2 (Sigma, St. Louis, MO), respectively. Peptides were synthesized and characterized as reported (12). Monoclonal FK-1 and FK-2 anti-ubiquitin antibodies were purchased from BioMol International (Plymouth Meeting, PA). Rabbit anti-20S proteasome antibody was from Novus Biologicals, Littleton, CO. LTA, Re-LPS, BSA, FCS, and LPS from E. coli B4:110 were purchased from Sigma. SAA and HDL were obtained from PeproTech (Rocky Hill, NJ) and Calbiochem (Darmstadt, Germany), respectively.
CLA-1- and CLA-2-Overexpressing HEK293 and HeLa Cells.
HeLa (Tet-off) cells (Clontech, Palo Alto, CA) overexpressing CLA-1 were generated, selected, and cultured as reported (11). Wild-type HeLa cells were also transfected with human SR-BI (CLA-1) and SR-BII (CLA-2) expressing pcDNA 3.1 plasmids by using lipofectamine reagent and further selected in the presence of 800 μg/ml G418 to generate CLA-1- and CLA-2-expressing HeLa clones (CLA-1-HeLa and CLA-2-HeLa). HeLa cells demonstrating an increased uptake of Alexa Fluor 488-HDL were selected and further analyzed by Western blotting using anti-CLA-1 and anti-CLA-2 antibodies. HEK293 cells overexpressing CLA-1 and CLA-2 were also generated as described for HeLa cells (CLA-1-HEK293 and CLA-2-HEK293).
Peritoneal Macrophage (Mφ) Preparation.
Peritoneal macrophages were isolated from KO Scarb1tm1Kri and normal C57BL/6 mice (The Jackson Laboratory, Bar Harbor, ME) and plated into glass microscopy chambers in DMEM containing 10% FCS, PepStrep, and 50 ng/ml mouse macrophage colony-stimulating factor (mCSF), Sigma. After a 5- to 7-day incubation period, differentiated macrophage cultures were used for confocal microscopy experiments.
Preparation of Alexa Fluor HDL and Alexa Fluor Bacteria.
HDL labeling was performed as reported (11). Live and 4% paraformaldehyde-fixed E. coli K1, K12, K25, Salmonella typhimurium, Listeria monocytogenes, and Staphylococcus aureus were labeled with Alexa Fluor 568/488, SE, a succinimide-based reactive fluorescent compound targeting primary amines (Molecular Probes), following the protein-labeling kit instructions.
Uptake of Labeled Bacteria.
All bacterial uptake and phagocytosis studies were performed by using DMEM containing 2 mg/ml BSA without antibiotics. HeLa and HEK293 cells overexpressing CLA-1 and CLA-2 were incubated with bacteria at ≈50 labeled bacteria per cultured cell. After a brief centrifugation of the plate to accelerate bacterial sedimentation, the cells were incubated at 37°C for 1.5 h and then washed extensively with PBS, detached with Cellstripper dissociation solution (Cellgro, Herndon, VA), fixed with 4% paraformaldehyde, and analyzed by a fluorescence-activated cell sorter (FACS, model A; Hitachi). In some experiments, bacterial uptake was studied without cell dissociation by using the multilabel counter Victor3 (PerkinElmer, Wellesley, MA) in 96-well-plate cultured cells.
Confocal Microscopy.
Confocal microscopy experiments designed to demonstrate colocalization of bacteria with various cellular compartments were conducted as reported (1, 12, 13). Briefly, 8-well microscopy slides with cultured cells were added with Alexa Fluor-labeled bacteria, centrifuged (3000 × g for 5 min) to accelerate bacterial sedimentation, and further incubated for 30–60 min. After three washings with PBS to remove unattached bacteria, slides were incubated with fluorescent trackers (concentrations of fluorescent trackers were selected individually to achieve similar to bacterial intensity of the fluorescent signal). After a 5- to 30-min incubation with trackers, slides were washed with PBS and fixed with 4% paraformaldehyde and sealed or viewed immediately without fixation. When bacterial compartmentalization was analyzed by immunochemistry, cells were fixed after bacterial incubation, permeabilized with 0.1% Triton X-100 in PBS, blocked with 2% BSA, and further incubated with antibodies (anti-LIMP-2, 20S, FK-1, or FK-2). Visualization was achieved by using Alexa Fluor 488/594-labeled anti-mouse/rabbit IgG secondary antibodies (Molecular Probes). The Alexa Fluor 488 or Alexa Fluor 594 anti-IgG and tracker colors were selected based on whether Alexa Fluor 488 or 594 bacteria was used. To assess subcellular localization, images were obtained with a Zeiss 510 confocal system (Zeiss, Jena, Germany). Images were acquired sequentially by using a 488-nm laser line and emission between 505 and 580 nm for Alexa Fluor 488 and a 594-nm laser line and emission >610 nm for Alexa Fluor 594. Z-series of high-resolution (100 nm per pixel) images were obtained throughout the cells with a ×63, 1.4-numerical-aperture Plan-Apochromat oil-immersion objective. The 3D data sets were quantitatively analyzed for colocalization by using Imaris 5.0 software package (Bitplane, Zurich, Switzerland). Pixel codistribution was calculated for green and red staining patterns, and percentage of colocalized material (intensity and volume) was assessed.
Electron Microscopy.
Confluent monolayers of HeLa cells were incubated with ≈50 paraformaldehyde-fixed E. coli K12 per cell for 1.5 h in a CO2 incubator. After three washings with ice-cold PBS, cells were fixed in PBS-buffered 2.5% glutaraldehyde (PBSG), scrubbed into the PBSG followed by double fixing in PBSG and osmium tetroxide (0.5%), dehydrated, and embedded into Spurr's epoxy resin. Ultrathin (90-nm) sections were double stained with uranyl acetate and lead citrate and viewed in a Philips CM10 transmission electron microscope (TEM) (Philips Electronics, Mahway, NJ).
ABPA.
The ABPA was conducted as reported (24). Briefly, after adding of various numbers of live bacteria to cells in antibiotic-free DMEM and brief centrifugation of the plate to accelerate bacterial sedimentation, the cells were further incubated for 1 h in a CO2 incubator, followed by intensive PBS washing. Afterward, the cells were incubated in fresh culture media in the presence or absence of 20 μg/ml GM for an additional 1, 4, 8, and 24 h. The cells were then washed to remove GM and lysed in ice-cold water for 30 min. The lysates were serially diluted in PBS and plated to yeast–peptone–dextrose (YPD) agar plates (Teknova, Rockville, MD) for colony counting.
Supplementary Material
Acknowledgments
We thank Dr. Susan Harrington, Senior Fellow at the Department of Laboratory Medicine, Clinical Center, National Institutes of Health (NIH) for providing various bacteria; Dr. J. Philip McCoy, Jr., Director of the Flow Cytometry Core Facility, National Heart, Lung, and Blood Institute (NHLBI), NIH, for conducting cell sorting; and Dr. Edward B. Neufeld, Director, Lipid Trafficking Core Cell Biology and Physiology Center NHLBI, NIH, for helpful discussions.
Abbreviations
- ABPA
antibiotic protection assay
- GM
gentamicin
- HDL
high-density lipoprotein
- LPS
lipopolysaccharide
- LTA
lipoteichoic acid
- Mφ
peritoneal macrophage
- SAA
serum amyloid A
- TEM
transmission electron microscope.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Krieger M. J Clin Invest. 2001;108:793–797. doi: 10.1172/JCI14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Connelly MA, Williams DL. Curr Opin Lipidol. 2004;15:287–295. doi: 10.1097/00041433-200406000-00008. [DOI] [PubMed] [Google Scholar]
- 3.Reboul E, Klein A, Bietrix F, Gleize B, Malezet-Desmoulins C, Schneider M, Margotat A, Lagrost L, Collet X, Borel P. J Biol Chem. 2006;281:4739–4745. doi: 10.1074/jbc.M509042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balazs Z, Panzenboeck U, Hammer A, Sovic A, Quehenberger O, Malle E, Sattler W. J Neurochem. 2004;89:939–950. doi: 10.1111/j.1471-4159.2004.02373.x. [DOI] [PubMed] [Google Scholar]
- 5.Graf GA, Roswell KL, Smart EJ. J Lipid Res. 2001;42:1444–1449. [PubMed] [Google Scholar]
- 6.Murao K, Terpstra V, Green SR, Kondratenko N, Steinberg D, Quehenberger O. J Biol Chem. 1997;272:17551–17557. doi: 10.1074/jbc.272.28.17551. [DOI] [PubMed] [Google Scholar]
- 7.Schulthess G, Compassi S, Werder M, Han CH, Phillips MC, Hauser H. Biochemistry. 2000;39:12623–12631. doi: 10.1021/bi0011633. [DOI] [PubMed] [Google Scholar]
- 8.Cai L, de Beer MC, de Beer FC, van der Westhuyzen DR. J Biol Chem. 2005;280:2954–2961. doi: 10.1074/jbc.M411555200. [DOI] [PubMed] [Google Scholar]
- 9.Baranova IN, Vishnyakova TG, Bocharov AV, Kurlander R, Chen Z, Kimelman ML, Remaley AT, Csako G, Thomas F, Eggerman TL, et al. J Biol Chem. 2005;280:8031–8040. doi: 10.1074/jbc.M405009200. [DOI] [PubMed] [Google Scholar]
- 10.Goti D, Hrzenjak A, Levak-Frank S, Frank S, van der Westhuyzen DR, Malle E, Sattler W. J Neurochem. 2001;76:498–508. doi: 10.1046/j.1471-4159.2001.00100.x. [DOI] [PubMed] [Google Scholar]
- 11.Vishnyakova TG, Bocharov AV, Baranova IN, Chen Z, Remaley AT, Csako G, Eggerman TL, Patterson AP. J Biol Chem. 2003;278:22771–22780. doi: 10.1074/jbc.M211032200. [DOI] [PubMed] [Google Scholar]
- 12.Bocharov AV, Baranova IN, Vishnyakova TG, Remaley AT, Csako G, Thomas F, Patterson AP, Eggerman TL. J Biol Chem. 2004;279:36072–36082. doi: 10.1074/jbc.M314264200. [DOI] [PubMed] [Google Scholar]
- 13.Philips JA, Rubin EJ, Perrimon N. Science. 2005;309:1251–1253. doi: 10.1126/science.1116006. [DOI] [PubMed] [Google Scholar]
- 14.Bocharov AV, Baranova IN, Csako G, Eggerman TL, Patterson AP, Remaley AT, Vishnyakova TG. International patent WO. 2004 2004/041179 A2. [Google Scholar]
- 15.Nakagawa A, Shiratsuchi A, Tsuda K, Nakanishi Y. Mol Reprod Dev. 2005;71:166–177. doi: 10.1002/mrd.20278. [DOI] [PubMed] [Google Scholar]
- 16.Williams DL, de La Llera-Moya M, Thuahnai ST, Lund-Katz S, Connelly MA, Azhar S, Anantharamaiah GM, Phillips MC. J Biol Chem. 2000;275:18897–18904. doi: 10.1074/jbc.M002411200. [DOI] [PubMed] [Google Scholar]
- 17.J Morath JS, von Aulock S, Hartung T. Endotoxin Res. 2005;11:348–356. doi: 10.1179/096805105X67328. [DOI] [PubMed] [Google Scholar]
- 18.Tsuneyama K, Harada K, Kono N Hiramatsu K, Zen Y, Sudo Y, Gershwin ME, Ikemoto M, Arai H, Nakanuma Y. J Hepatol. 2001;35:156–163. doi: 10.1016/s0168-8278(01)00084-8. [DOI] [PubMed] [Google Scholar]
- 19.Perrin AJ, Jiang X, Birmingham CL, So NS, Brumell JH. Curr Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 20.Canadien V, Tan T, Zilber R, Szeto J, Perrin AJ, Brumell JH. J Immunol. 2005;174:2471–2475. doi: 10.4049/jimmunol.174.5.2471. [DOI] [PubMed] [Google Scholar]
- 21.Hauser H, Pascher I, Pearson RH, Sundell S. Biochim Biophys Acta. 1981;650:21–51. doi: 10.1016/0304-4157(81)90007-1. [DOI] [PubMed] [Google Scholar]
- 22.Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, Scarselli E, Cortese R, Nicosia A, Cosset FL. J Biol Chem. 2003;278:41624–41630. doi: 10.1074/jbc.M305289200. [DOI] [PubMed] [Google Scholar]
- 23.Voisset C, Callens N, Blanchard E, Op De Beeck A, Dubuisson J, Vu-Dac N. J Biol Chem. 2005;280:7793–7799. doi: 10.1074/jbc.M411600200. [DOI] [PubMed] [Google Scholar]
- 24.Cowan C, Jones HA, Kaya YH, Perry RD, Straley SC. Infect Immun. 2000;68:4523–4530. doi: 10.1128/iai.68.8.4523-4530.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.