Abstract
Cardiac myosin binding protein C (cMyBP-C) has three phosphorylatable serines at its N terminus (Ser-273, Ser-282, and Ser-302), and the residues' phosphorylation states may alter thick filament structure and function. To examine the effects of cMyBP-C phosphorylation, we generated transgenic mice with cardiac-specific expression of a cMyBP-C in which the three phosphorylation sites were mutated to aspartic acid, mimicking constitutive phosphorylation (cMyBP-CAllP+). The allele was bred into a cMyBP-C null background (cMyBP-C(t/t)) to ensure the absence of endogenous dephosphorylated cMyBP-C. cMyBP-CAllP+ was incorporated normally into the cardiac sarcomere and restored normal cardiac function in the null background. However, subtle changes in sarcomere ultrastructure, characterized by increased distances between the thick filaments, indicated that phosphomimetic cMyBP-C affects thick–thin filament relationships, and yeast two-hybrid data and pull-down studies both showed that charged residues in these positions effectively prevented interaction with the myosin heavy chain. Confirming the physiological relevance of these data, the cMyBP-CAllP+:(t/t) hearts were resistant to ischemia–reperfusion injury. These data demonstrate that cMyBP-C phosphorylation functions in maintaining thick filament spacing and structure and can help protect the myocardium from ischemic injury.
Keywords: heart, ischemia
Cardiac myosin binding protein C (cMyBP-C) is localized to the sarcomere's thick filaments where it has structural and regulatory functions. MYBPC3 mutations account for 20–30% of all mutations linked to familial hypertrophic cardiomyopathy (1). cMyBP-C belongs to the intracellular Ig superfamily and is composed of Ig and fibronectin type-3 repeating domains (Fig. 1A). It is present not only in cardiac muscle, but also in skeletal muscle before the skeletal muscle-type isoforms are expressed, suggesting that the cardiac isoform is functional in early myofibrillogenesis and regenerating muscle (2, 3). cMyBP-C may modulate myosin assembly (4) and stabilize thick filaments (5). It binds titin via domains C8–C10 (6) and actin in the Pro-Ala-rich sequences between the C0 and C1 domains (7), which appear to be important for the precise arrangement of the actin–myosin filaments. Compared with the two skeletal muscle isoforms, the cardiac isoform contains an extra Ig domain at the N terminus (C0), an insertion of 28 residues within the C5 domain, and three potential phosphorylation sites (Ser-273, Ser-282, and Ser-302) that are substrates for cAMP-dependent PKA, Ca2+-calmodulin-activated kinase, and PKC (8). This region is located between the C1 and C2 domains of the N terminus, which binds to the subfragment 2 (S2) segment of myosin close to the lever arm domain (9–11), and this interaction may be dynamically regulated by the differential phosphorylation of cMyBP-C (12).
Fig. 1.
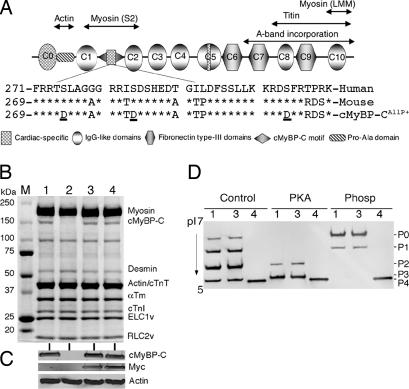
cMyBP-CAllP+ transgene expression. (A) Domain organization of cMyBP-C. The cardiac-specific phosphorylation motif (sequence shown) is located between domains C1 and C2. The three known phosphorylation sites (Ser-273, Ser-282, and Ser-302, underlined) were each substituted with aspartic acid. (B) Representative Coomassie blue-stained SDS/PAGE analysis shows the lack of cMyBP-C in the homozygous cMyBP-C(t/t) hearts (lane 2), replacement of cMyBP-C to normal levels in the cMyBP-CWT:(t/t) (lane 3) and cMyBP-CAllP+:(t/t) (lane 4) crosses, and conservation of the other sarcomeric protein levels in these mice compared with a NTG sample (lane 1). (C) Western blot analysis shows the absence of endogenous cMyBP-C in the homozygous cMyBP-C(t/t) hearts and the presence of myc-tagged TG cMyBP-C in cMyBP-CWT:(t/t) and cMyBP-CAllP+:(t/t) mice. Lanes are numbered as in B. (D) 1D isoelectric focusing showing complete replacement of cMyBP-CAllP+ in the cMyBP-C null background. Samples from NTG (lane 1), cMyBP-CWT:(t/t) (lane 3), and cMyBP-CAllP+:(t/t) (lane 4) hearts were either untreated (Control) or treated with PKA or phosphatase (Phosp). Protein derived from cMyBP-C(t/t) hearts was omitted because of a lack of detectable cMyBP-C signal.
cMyBP-C is the only thick filament protein that is differentially phosphorylated at multiple sites by the enzymes PKA, PKC, and Ca2+-calmodulin-activated kinase (13). Reconstitution studies showed that PKA-mediated phosphorylation of cMyBP-C extends the cross-bridges from the backbone of the thick filament, changes their orientation, increases the degree of order of the cross-bridges, and decreases cross-bridge flexibility (11). cMyBP-C phosphorylation can change both filament orientation and contractile mechanics (13), but the in vivo role of these posttranslational modifications on whole-organ function remains obscure.
We reported that cMyBP-C phosphorylation decreases during the development of heart failure, ischemia–reperfusion (I-R) injury, or pathologic hypertrophy (14). This decrease may initiate changes in myofibril thick filament structure that decreases strong binding of myosin heads with the actin thin filaments. To test the hypothesis that cMyBP-C phosphorylation is essential for normal cardiac function, we generated transgenic (TG) mice in which three phosphorylation sites were mutated to nonphosphorylatable alanines (cMyBP-CAllP−). These animals showed altered sarcomeric structure and depressed cardiac contractility (14). The complete replacement of endogenous cMyBP-C with cMyBP-CAllP− failed to rescue the null cMyBP-C(t/t) mice and led to significant functional deficits, whereas overexpression of the normal cMyBP-C (cMyBP-CWT) effectively rescued the null phenotype.
In the present study, we examined the effects of total chronic phosphorylation of cMyBP-C by generating TG mice in which the known cMyBP-C phosphorylation sites are converted to aspartic acid (cMyBP-CAllP+) to mimic a constant state of constitutive phosphorylation. The mice were then bred into a homozygous cMyBP-C(t/t) background (15) to obtain complete replacement of endogenous cMyBP-C with cMyBP-CAllP+. The phosphomimetic effectively rescued the null phenotype and strikingly conferred cardioprotection when the hearts were subjected to I-R injury.
Results
Characterization of the cMyBP-CAllP+:(t/t) Mice.
Previously, we found that cMyBP-C was extensively phosphorylated under basal conditions, but became dephosphorylated during the development of cardiac pathology with the triphosphorylated form largely or completely absent in advanced heart failure (14). These results were consistent with our data showing the necessity of phosphorylation for normal cMyBP-C function (14). To investigate the effects of chronic cMyBP-C phosphorylation, we generated cardiac-specific TG mice in which the three known phosphorylation sites Ser-273, Ser-282, and Ser-302 were changed to aspartic acid (Fig. 1A), mimicking constitutive phosphorylation (cMyBP-CAllP+). A line was selected whose expression levels matched the previously generated cMyBP-CWT mice. Approximately 35% replacement with cMyBP-CAllP+ occurred in line 34, which is equivalent to the level of replacement previously measured in the cMyBP-CWT-expressing hearts [line 21 (16)] that expressed myc-tagged WT cMyBP-C (Fig. 6, which is published as supporting information on the PNAS web site). Expression of cMyBP-CAllP+ was benign with no signs of increased morbidity, mortality, or cardiac hypertrophy (data not shown). To ensure the absence of endogenous dephosphorylated cMyBP-C and to assess the functional significance of complete and chronic cMyBP-C phosphorylation in vivo, these mice were bred into a cMyBP-C null background in which <10% of normal levels of a truncated cMyBP-C are present [cMyBP-C(t/t)] (15). The cMyBP-CWT was also bred to the nulls to serve as a control (16). SDS/PAGE (Fig. 1B) and Western blot analysis using anti-cMyBP-C and anti-myc antibodies (Fig. 1C) showed the absence of cMyBP-C in null hearts and complete replacement of cMyBP-CAllP+ and cMyBP-CWT compared with the nontransgenic (NTG) controls. No changes in myofilament protein stoichiometry could be detected in the cMyBP-CAllP+:(t/t) hearts. To confirm that cMyBP-CAllP+ replacement in the cMyBP-C(t/t) background was complete, isoelectric focusing followed by Western blot analysis with anti-cMyBP-C antibodies was performed on untreated, PKA-treated, and phosphatase-treated samples (14). Results showed that the mimetic cMyBP-CAllP+ (P4) migrated closely to the triphosphorylated cMyBP-C species (P3) and that replacement with cMyBP-CAllP+ was complete (Fig. 1D).
cMyBP-CAllP+ Rescues the cMyBP-C Null Phenotype.
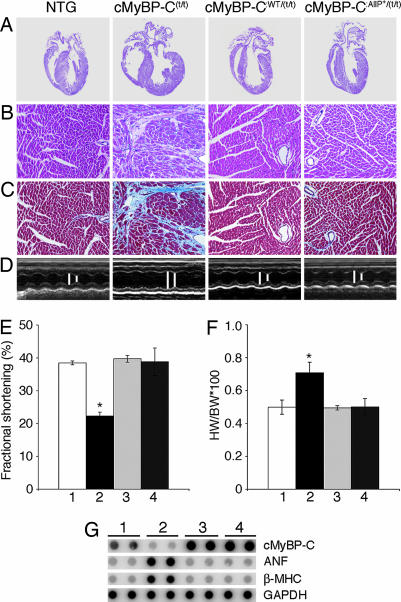
The cMyBP-C(t/t) mice are viable but soon after birth display a progressive heart failure with dilated cardiomyopathy, myocyte hypertrophy, disarray, fibrosis, and calcification (15). We reported previously that cMyBP-CWT TG expression effectively rescued the overt hypertrophy displayed by the cMyBP-C(t/t) hearts, but that expression of the nonphosphorylatable cMyBP-CAllP− was ineffective, with cMyBP-CAllP−:(t/t) mice displaying cardiac disease characteristic of the null homozygotes (14). The cMyBP-CAllP+:(t/t) hearts showed no hypertrophy and/or dilation (Fig. 2A). Histological analyses confirmed the lack of any pathology in these hearts, whereas the cMyBP-C(t/t) hearts showed markedly enlarged chambers, with significant cardiac hypertrophy and myocyte disarray (Fig. 2 B and C). Noninvasive M-mode echocardiographic measurements showed the cMyBP-CAllP+:(t/t) mice had normal left ventricle (LV) end-diastolic and end-systolic dimensions (Fig. 2D) and normal fractional shortening (Fig. 2E), and that expression of cMyBP-CAllP+ was as effective as cMyBP-CWT expression in rescuing the null phenotype (Table 1, which is published as supporting information on the PNAS web site). The heart/body weight ratios did not significantly differ between cMyBP-CAlP+:(t/t) and cMyBP-CWT:(t/t) animals compared with NTG littermates at 3 months, indicating that heart/body weights in the cMyBP-CAllP+:(t/t) mice were essentially normal (Fig. 2F). Furthermore, we confirmed restoration of normal RNA levels of β-myosin heavy chain (MHC) and atrial natriuretic factor, both of which serve as sensitive molecular markers for cardiac stress (Fig. 2G). In confirmation of these data, heart rate, LV pressure, dP/dtmax (maximum rate of LV pressure development), and dP/dtmin (maximum rate of LV pressure fall) were normal for both the cMyBP-CAllP+:(t/t) and cMyBP-CWT:(t/t) animals (Table 2, which is published as supporting information on the PNAS web site). To confirm that functional restoration was not caused by compensatory effects by the other contractile proteins subject to posttranslational modification, the phosphorylation status of these proteins was examined and found to be unchanged in the cMyBP-CAllP+:(t/t) hearts (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 2.
cMyBP-CAllP+ rescues the cMyBP-C(t/t) cardiac phenotype. All data and functional measurements were derived from mixed-gender, 12-week hearts. (A) Representative hearts stained with hematoxylin-eosin. (B) Hematoxylin-eosin staining. (C) Masson trichrome-stained myocardial sections to assess fibrosis. (D) M-mode echocardiographic tracings show the left ventricular chamber. B, C, and D correspond to the samples shown above in A. (E) Fractional shortening (%) measured by M-mode echocardiography (n = 6). (F) Heart (wet weight)-to-body weight ratios (n = 10). All data are presented as mean ± SE. ∗, Significant difference vs. NTG (P < 0.05). (G) RNA dot-blot analyses. In E-G, lanes 1, 2, 3, and 4 refer to NTG, cMyBP-C(t/t), cMyBP-CWT:(t/t), and cMyBP-CAllP+:(t/t) respectively. (Magnifications: A, ×4; B and C, ×20.)
cMyBP-CAllP+ Affects Sarcomere Spacing.
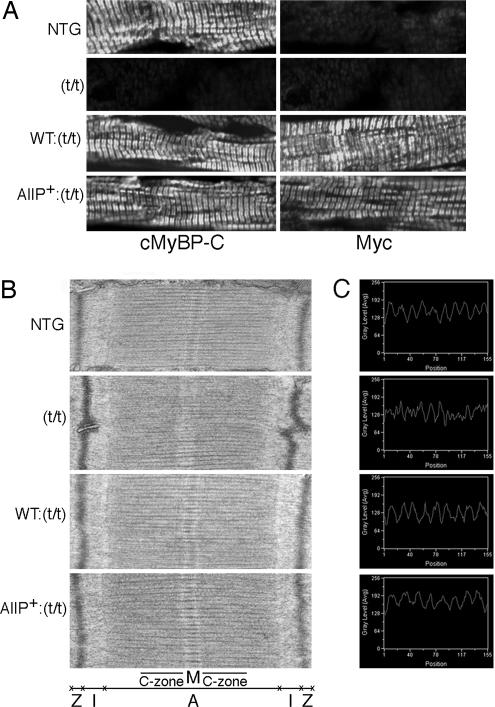
Immunohistochemical analysis using either cMyBP-C or myc antibody confirmed the absence of cMyBP-C in the null animals and normal incorporation of both the cMyBP-CAllP+ and cMyBP-CWT species in the cardiac sarcomeres (Fig. 3A). EM analysis showed well organized sarcomeres in the cMyBP-CAllP+:(t/t) and cMyBP-CWT:(t/t) hearts, although, as observed previously, the cMyBP-C(t/t) sarcomeres showed a loss of M-band definition (Fig. 3B) (15). In vitro experiments have shown that phosphorylation of cMyBP-C extends the cross-bridge to the surface of the thin filament and loosens the packing of the rod portion of the myosin molecule. Consistent with these data, incorporation of cMyBP-CAllP+:(t/t) affected the steric arrangement of the filament lattice as detected by measuring the distance between two thick myosin filaments. Results showed that interthick filament distances (Fig. 3C) in the NTG, cMyBP-CWT:(t/t), and cMyBP-CAllP+:(t/t) sarcomeres were 56.79 ± 0.37 nm (n = 464 peaks per 50 scans), 57.23 ± 0.62 nm (n = 462 peaks per 50 scans), and 65.29 ± 0.25 nm (n = 403 peaks per 50 scans; *, P < 0.0001), respectively. The cMyBP-CAllP+(t/t) sarcomeres have significantly increased distances between the thick filaments, and the absence of cMyBP-C in the cMyBP-C(t/t) sarcomeres resulted in irregular spacing of the thick filament such that peak–peak distances could not be determined. These data indicate that cMyBP-C phosphorylation plays an essential role in regulating thick filament structure.
Fig. 3.
Sarcomeric ultrastructural and immunohistochemical analyses. (A) Localization of cMyBP-CAllP+ into the cMyBP-C(t/t) background. Ventricular myocardial sections were immunostained for cMyBP-C by using either anti-cMyBP-C (Left) or anti-myc antibodies (Right). There is no staining of cMyBP-C in the cMyBP-C(t/t) background. (B) Transmission electron micrographs showing sarcomere ultrastructure. (C) Peaks show the distance between two thick myosin filaments, which was measured with a line scanner using Metamorph software with line length held constant at 525.83 nm (1.84501 nm per pixel). (Magnifications: A, ×60; B, ×30,000.)
cMyBP-C Phosphorylation and Myosin Interaction.
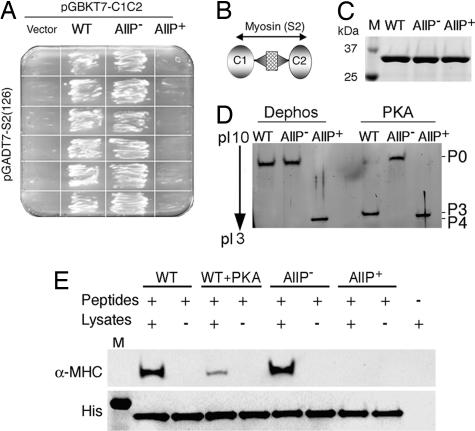
The N-terminal phosphorylation motif interacts with the S2 region of MHC, which may be dynamically regulated by phosphorylation/dephosphorylation, contributing to sarcomeric integrity (10, 12). We previously used alanine substitutions at the serine residues to create a cMyBP-C that could not be phosphorylated (cMyBP-CAllP−) (14). It has been shown (11, 13) that phosphorylation of cMyBP-C abolishes its interaction with myosin S2. To confirm that the alanine and aspartic acid substitutions in the phosphorylation motif have the expected effects on myosin S2 interaction, yeast two-hybrid experiments were carried out. Results show that cotransformation of C1-C2WT or C1-C2AllP− with myosin S2 resulted in robust growth, but that cotransformants with C1-C2AllP+ failed to grow (Fig. 4A). The C1-C2 domains with the phosphorylation motif and S2 binding sites (Fig. 4B) were used to produce soluble His-tagged C1-C2WT, C1-C2AllP−, and C1-C2AllP+ recombinant peptides (Fig. 4C), which show the expected isoelectric point values of 8.54, 8.54, and 7.42, respectively and expected shifts or lack thereof after PKA treatment (Fig. 4D). To confirm the yeast two-hybrid data, pull-down experiments were performed with total mouse heart lysates. C1-C2WT and C1-C2AllP− proteins were able to pull-down α-MHC as evidenced by Western blot analysis with α-MHC-specific antibodies, whereas PKA-treated C1-C2WT and untreated C1-C2AllP+ were not (Fig. 4E).
Fig. 4.
cMyBP-C phosphorylation and myosin interaction. (A) Yeast two-hybrid experiments confirm that C1-C2WT and C1-C2AllP− [phosphorylation negative mimetic (14)] interact with the myosin S2, but the phosphorylation mimetic cMyBP-CAllP+ does not. (B) Schematic diagram shows the region of cMyBP-C being tested for interaction. (C) SDS/PAGE analysis of purified C1-C2WT, C1-C2AllP, and C1-C2AllP+ recombinant peptides. (D) In vitro phosphorylation of C1-C2 by PKA and stained with SYPRO Ruby (Bio-Rad). (E) C1-C2 and myosin S2 associate in vitro. Twenty micrograms of His-tagged C1-C2 (Peptides) was mixed with 200 μg of total mouse heart lysates (Lysates), the C1-C2-complex, and associated protein purified with Ni-NTA resin (see Materials and Methods), and the proteins were separated by 4–15% SDS/PAGE and blotted onto a PVDF membrane. Western blots were treated with anti-α-MHC (BA-G5, ATCC, Rockville, MD) or anti-His antibodies (Roche, Indianapolis, IN).
cMyBP-CAllP+:(t/t) Mice Are Protected from I-R Injury.
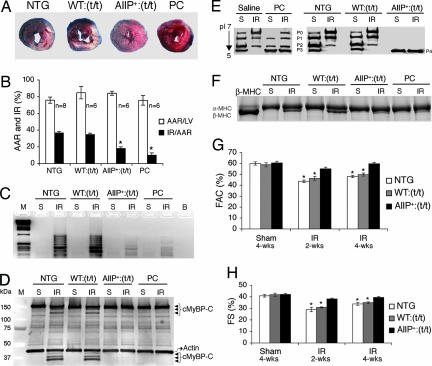
We recently reported that total cMyBP-C phosphorylation, particularly the triphosphorylated species, was decreased in mice with I-R injury (14). This finding is consistent with data from Decker et al. (17), who found that cMyBP-C is dephosphorylated and its degradation accelerated during low-flow ischemia. Thus, reduced phosphorylation of cMyBP-C is associated with contractile dysfunction. We wanted to determine whether cMyBP-CAllP+ might be cardioprotective during stress. cMyBP-CAllP+:(t/t) hearts were subjected to left ventricular cardiac ischemia for 1 h followed by 24 h of reperfusion to induce myocardial infarction and cell death. The β-adrenergic agonist isoproterenol was used as a positive control to protect the heart against ischemic injury (18). Remarkably, cMyBP-CAllP+:(t/t) mice showed a greater reduction (18 ± 2%) in infarcted area normalized to area at risk (AAR) (AAR was not different between the groups), compared with the cMyBP-CWT:(t/t) (35 ± 2%) and NTG (37 ± 2%) controls (Fig. 5A and B). Whereas significant cardiac injury is apparent in the cMyBP-CWT:(t/t) and NTG hearts, the cMyBP-CAllP+:(t/t) hearts were relatively unaffected. cMyBP-CAllP+:(t/t) and isoproterenol-treated hearts also showed significantly less DNA fragmentation, compared with either cMyBP-CWT:(t/t) or NTG hearts (Fig. 5C) and reduced TUNEL-positive cardiomyocytes (Fig. 8, which is published as supporting information on the PNAS web site). Degradation of the N terminus of cMyBP-C (17) in I-R hearts from the NTG and cMyBP-CWT:(t/t) groups, but not in the cMyBP-CAllP+:(t/t) and isoproterenol-treated hearts, is apparent (Fig. 5D). Protein extracts from these hearts were subjected to isoelectric focusing and Western blotting to determine the extent of phosphorylation. As expected, I-R treatment reduced cMyBP-C phosphorylation levels in cMyBP-CWT:(t/t) and NTG hearts but not in the cMyBP-CAllP+:(t/t) and preconditioned (PC) hearts (Fig. 5E). A shift in the myosin isoforms often accompanies cardiomyocyte injury resulting from ischemia and/or infarction (14, 19). Consistent with these observations, 24 h after I-R we detected significant amounts of β-MHC in the NTG and cMyBP-CWT:(t/t) hearts, whereas, in contrast, the cMyBP-CAllP+:(t/t) and isoproterenol-treated hearts were relatively unaffected (Fig. 5F).
Fig. 5.
Phosphorylation of cMyBP-C protects the hearts from I-R injury. (A) The left anterior descending artery was ligated for 1 h to induce ischemia and it subsequently was reperfused for 24 h. Representative cross-sections were stained with triphenyl tetrazolium chloride and Evans blue to determine the extent of injury. (B) Quantification of infarct area vs. AAR after I-R injury in the indicated groups. (C and D) Assessment of DNA fragmentation by DNA ladder assay (C) and Western blot analysis (D) showing cMyBP-C and its degradation products (arrowheads). Actin was included as a loading control. M, molecular weight marker. (E and F) cMyBP-C phosphorylation levels (E) and myosin isoform (F) shift after a sham (S) procedure or I-R (IR) injury as above. (G and H) M-mode echocardiography at the indicated time points was done to determine differences in fractional area change (FAC) (G) and fractional shortening (FS) (H) after an initial 1-h ischemic injury (∗, P < 0.05 vs. sham; n = 7 from each group).
Cardiac function was analyzed 2 and 4 weeks after ischemic injury by M-mode echocardiography for alterations in fractional area changes and fractional shortening, which reflect the degree of myocardial injury, secondary remodeling, and failure. Compared with sham-operated mice of each genotype, function was conserved in the cMyBP-CAllP+:(t/t) animals, whereas cMyBP-CWT:(t/t) and NTG mice were ≈15% significantly compromised (Fig. 5 G and H) at 4 wk. The data indicate that cMyBP-C phosphorylation protects the myocardium from cell injury and death and decreases subsequent infarction expansion, which secondarily preserves myocardial function and reduces secondary remodeling.
Discussion
The location of the flexible phosphorylation motif at the cMyBP-C N terminus appears to be critical in permitting shuttling between the thick and thin filaments (20). Phosphorylation also can modulate the myosin–actin interactions that are necessary to maintain thick filament organization (15). cMyBP-CAllP+ does not exactly mimic either the endogenous cMyBP-C and cMyBP-CWT protein in that the percents of dephosphorylation, monophosphorylation, diphosphorylation, and triphosphorylation of cMyBP-C are ≈7%, ≈28%, ≈43%, and ≈24%, respectively (14). However, cMyBP-CAllP+ was equally effective as endogenous cMyBP-C in maintaining overall myofilament organization and cardiac function. In the absence of cMyBP-C phosphorylation, the C1-C2 domain of cMyBP-C is bound to myosin in the S2 region (21, 22), but when phosphorylated, it releases its interaction with myosin, binding to actin (23). Therefore, cMyBP-C probably helps regulate force generation by modulating thick–thin filament interactions. Together with our previous data, in which a phosphorylation-incompetent cMyBP-C species replaced the endogenous protein, the present data show that cMyBP-C phosphorylation is essential for normal cardiac function and its modulation impacts on thick filament packing by affecting cMyBP-C-myosin S2 binding.
cMyBP-C phosphorylation also appears to play a critical role in cardioprotection. Myocardial ischemia leading to the progressive death of heart muscle cells represents the most common cause of heart failure. The primary mechanisms responsible for decreased function after I-R is caused by acidosis and reduced Ca2+ sensitivity of the contractile myofilaments. There is also a transient elevation of intracellular Ca2+ that activates Ca2+-calmodulin-activated kinase and protease-like calpain I, leading to cleavage of cardiac troponin I, α-actinin, the myosin light chains (24), and cMyBP-C (17). cMyBP-C is dephosphorylated during low-flow ischemia (17) and in I-R-based heart failure models (14). Decreased phosphorylation would have the net effect of increasing thick filament packing density, possibly leading to reduced calcium-activated force generation (25).
The β-agonist isoproterenol can precondition the heart, reducing ischemic injury (26, 27). We hypothesized that PKA-mediated phosphorylation of cMyBP-C, via isoproterenol infusion or replacement of endogenous cMyBP-C with cMyBP-CAllP+ might be equally effective in protecting the heart from I-R injury if cMyBP-C phosphorylation plays a critical role in this process. Expression of cMyBP-CAllP+ resulted in significant protection from I-R injury with relatively conserved cardiac function and less cellular damage. cMyBP-CAllP+ was also highly resistant to the peptide cleavage associated with ischemia (17). Similar data have been obtained for PKA-mediated cardiac troponin I phosphorylation, which significantly reduced troponin I proteolysis (28). These data provide further support for the hypothesis that sarcomere-based gain of function provides long-term benefits in heart failure (29), although additional studies are needed to understand fully the role that cMyBP-C phosphorylation might play in cardioprotection. We think that understanding the consequences of myofilament protein phosphorylation is critical for the development of new pharmacological approaches to protect the heart and improve cardiac function in I-R injury.
Materials and Methods
TG and Targeted Mice.
The cDNA for mouse cMyBP-C (cMyBP-CWT) was used to convert Ser-273, Ser-282, and Ser-302 to aspartate (cMyBP-CAllP+) and the myc epitope was incorporated (14, 16). The construct was used to generate multiple TG founders with cardiomyocyte-specific expression. These were bred into the cMyBP-C(t/t) background (15). An animal that expressed the normal cardiac isoform at equivalent levels to cMyBP-CAllP+, cMyBP-CWT (16) was also bred to the nulls to serve as a control.
RNA Transcript and Protein Analyses.
cMyBP-C gene expression and expression of hypertrophic marker genes was quantitated by Northern and RNA dot blotting using antisense oligonucleotides (14). Histopathology, immunohistochemistry, and transmission EM analyses were performed as described (14). Enriched myofibrillar proteins were prepared by using F60 buffer, solubilized in urea buffer and treated with phosphatase and/or PKA (30). cMyBP-C was detected by SDS/PAGE followed by Western blots using anti-myc mAbs and anti-cMyBP-C rabbit polyclonal antibodies raised against the C0–C1 domains (14, 16). 1D isoelectric focusing was performed to identify the cMyBP-C phosphorylated forms (14, 17).
In Vivo Measurements of Cardiac Function.
Echocardiographic parameters were measured noninvasively as reported (14, 16). Closed-chest invasive hemodynamic studies were performed under ketamine-thiobutabarbital anesthesia, and β-adrenergic stress responsiveness was assessed after infusion of dobutamine as described (14, 16, 30).
Thick-Filament Distance Determination.
The distance between two thick myosin filaments was measured with a line scanner by using Metamorph software (version 6.2r6, Universal Imaging, Downingtown, PA). Line length was held constant at 525.83 nm (1.84501 nm per pixel). The distance between two peaks was considered as the distance between two thick filaments and the average was calculated.
Yeast Two-Hybrid Assay.
The mouse cMyBP-C domains C1-C2 were cloned (C1-C2WT) and the known phosphorylation sites (Ser-273, Ser-282, and Ser-302) were mutated to alanine (nonphosphorylated form; C1-C2AllP−) or aspartate (phosphorylation mimetic; C1-C2AllP+) by standard PCR methods. The C1-C2WT, C1-C2AllP−, and C1-C2AllP+ regions were subcloned into pGBKT7 (Clontech, Mountain View, CA). The mouse cardiac α-MHC-S2 (126 aa) was cloned into the GAL4 activation domain of pGADT7. Interactions (C1-C2WT, C1-C2AllP−, or C1-C2AllP+ with S2) were confirmed by growth on His/Leu/Trp− plates per the manufacturer's instruction with growth on His/Leu− and His/Trp− plates used to test for false positives.
Pull-Down Assays.
The soluble His-tagged C1-C2WT, C1-C2AllP−, and C1-C2AllP+ recombinant peptides were purified by using the pET expression system (Novagen, San Diego, CA). A total of 200 μg of NTG mouse heart lysate with 10 μg of the C1-C2 peptides and 20 μl of Ni-NTA agarose beads was incubated for 2 h at 4°C and washed with Tris buffer (50 mM) containing 0.5% Triton X-100, 100 mM NaCl, 10 mM MgCl2, 0.1 mM PMSF, and 1× protease inhibitor. Proteins bound to the beads were eluted in Laemmli sample buffer (Bio-Rad, Hercules, CA) and subjected to Western blot analysis.
I-R Injury.
Cardiac I-R injury was performed at 10 weeks as described (31). Twenty four hours before I-R for the ischemic preconditioning controls, Alzet miniosmotic pumps (Durect, Cupertino, CA) containing either isoproterenol (60 mg/kg per day) in 0.02% ascorbic acid or vehicle only (saline) were surgically implanted (32). After the thoracotomy was closed, the mice were revived for 1 h of ischemia after which the knot was released and the heart was reperfused for 24 h or 4 weeks. Upon completion of the reperfusion, mice were killed and the hearts were analyzed for infarction injury with 2% triphenyl tetrazolium chloride. Images were quantified for AAR and infarcted area. The DNA ladder assay PCR kit procedure (MBI, San Francisco, CA) was used to analyze DNA fragmentation. Twenty micrograms of total heart lysate was analyzed by Western blotting to detect cMyBP-C degradation.
Statistical Analysis.
Results are presented as mean ± SE. For comparisons of data from two groups, Student's t test was used. For comparisons of multiple groups, one-way ANOVA or ANOVA for repeated measurements followed by the Tukey-Kramer multiple comparisons test was used (SigmaStat V3.0). A value of P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01 HL69799, HL60546, HL52318, HL60546, and HL56370 (to J.R.) and the American Heart Association, Ohio Valley Affiliate (S.S.).
Abbreviations
- cMyBP-C
cardiac myosin binding protein C
- TG
transgenic
- NTG
nontransgenic
- MHC
myosin heavy chain
- S2
subfragment 2
- I-R
ischemia–reperfusion
- AAR
area at risk
- PC
preconditioned.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Spirito P, Seidman CE, McKenna WJ, Maron BJ. N Engl J Med. 1997;336:775–785. doi: 10.1056/NEJM199703133361107. [DOI] [PubMed] [Google Scholar]
- 2.Bahler M, Moser H, Eppenberger HM, Wallimann T. Dev Biol. 1985;112:345–352. doi: 10.1016/0012-1606(85)90405-1. [DOI] [PubMed] [Google Scholar]
- 3.Kawashima M, Kitani S, Tanaka T, Obinata T. J Biochem (Tokyo) 1986;99:1037–1047. doi: 10.1093/oxfordjournals.jbchem.a135567. [DOI] [PubMed] [Google Scholar]
- 4.Offer G, Moos C, Starr R. J Mol Biol. 1973;74:653–676. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
- 5.Moos C, Mason CM, Besterman JM, Feng IN, Dubin JH. J Mol Biol. 1978;124:571–586. doi: 10.1016/0022-2836(78)90172-9. [DOI] [PubMed] [Google Scholar]
- 6.Freiburg A, Gautel M. Eur J Biochem. 1996;235:317–323. doi: 10.1111/j.1432-1033.1996.00317.x. [DOI] [PubMed] [Google Scholar]
- 7.Herron TJ, Rostkova E, Kunst G, Chaturvedi R, Gautel M, Kentish JC. Circ Res. 2006;98:1290–1298. doi: 10.1161/01.RES.0000222059.54917.ef. [DOI] [PubMed] [Google Scholar]
- 8.Mohamed AS, Dignam JD, Schlender KK. Arch Biochem Biophys. 1998;358:313–319. doi: 10.1006/abbi.1998.0857. [DOI] [PubMed] [Google Scholar]
- 9.Garvey JL, Kranias EG, Solaro RJ. Biochem J. 1988;249:709–714. doi: 10.1042/bj2490709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruen M, Gautel M. J Mol Biol. 1999;286:933–949. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- 11.Weisberg A, Winegrad S. Proc Natl Acad Sci USA. 1996;93:8999–9003. doi: 10.1073/pnas.93.17.8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruen M, Prinz H, Gautel M. FEBS Lett. 1999;453:254–259. doi: 10.1016/s0014-5793(99)00727-9. [DOI] [PubMed] [Google Scholar]
- 13.Gautel M, Zuffardi O, Freiburg A, Labeit S. EMBO J. 1995;14:1952–1960. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sadayappan S, Gulick J, Osinska H, Martin LA, Hahn HS, Dorn GW, 2nd, Klevitsky R, Seidman CE, Seidman JG, Robbins J. Circ Res. 2005;97:1156–1163. doi: 10.1161/01.RES.0000190605.79013.4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McConnell BK, Jones KA, Fatkin D, Arroyo LH, Lee RT, Aristizabal O, Turnbull DH, Georgakopoulos D, Kass D, Bond M, et al. J Clin Invest. 1999;104:1235–1244. doi: 10.1172/JCI7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Q, Sanbe A, Osinska H, Hewett TE, Klevitsky R, Robbins J. J Clin Invest. 1998;102:1292–1300. doi: 10.1172/JCI3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Decker RS, Decker ML, Kulikovskaya I, Nakamura S, Lee DC, Harris K, Klocke FJ, Winegrad S. Circulation. 2005;111:906–912. doi: 10.1161/01.CIR.0000155609.95618.75. [DOI] [PubMed] [Google Scholar]
- 18.Frances C, Nazeyrollas P, Prevost A, Moreau F, Pisani J, Davani S, Kantelip JP, Millart H. J Cardiovasc Pharmacol. 2003;41:396–405. doi: 10.1097/00005344-200303000-00008. [DOI] [PubMed] [Google Scholar]
- 19.Mercadier JJ, Lompre AM, Wisnewsky C, Samuel JL, Bercovici J, Swynghedauw B, Schwartz K. Circ Res. 1981;49:525–532. doi: 10.1161/01.res.49.2.525. [DOI] [PubMed] [Google Scholar]
- 20.Oakley CE, Hambly BD, Curmi PM, Brown LJ. Cell Res. 2004;14:95–110. doi: 10.1038/sj.cr.7290208. [DOI] [PubMed] [Google Scholar]
- 21.Kunst G, Kress KR, Gruen M, Uttenweiler D, Gautel M, Fink RH. Circ Res. 2000;86:51–58. doi: 10.1161/01.res.86.1.51. [DOI] [PubMed] [Google Scholar]
- 22.Levine R, Weisberg A, Kulikovskaya I, McClellan G, Winegrad S. Biophys J. 2001;81:1070–1082. doi: 10.1016/S0006-3495(01)75764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulikovskaya I, McClellan G, Flavigny J, Carrier L, Winegrad S. J Gen Physiol. 2003;122:761–774. doi: 10.1085/jgp.200308941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Eyk JE, Powers F, Law W, Larue C, Hodges RS, Solaro RJ. Circ Res. 1998;82:261–271. doi: 10.1161/01.res.82.2.261. [DOI] [PubMed] [Google Scholar]
- 25.McClellan G, Kulikovskaya I, Winegrad S. Biophys J. 2001;81:1083–1092. doi: 10.1016/S0006-3495(01)75765-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lochner A, Genade S, Tromp E, Podzuweit T, Moolman JA. Circulation. 1999;100:958–966. doi: 10.1161/01.cir.100.9.958. [DOI] [PubMed] [Google Scholar]
- 27.Tong H, Bernstein D, Murphy E, Steenbergen C. FASEB J. 2005;19:983–985. doi: 10.1096/fj.04-3067fje. [DOI] [PubMed] [Google Scholar]
- 28.Di Lisa F, De Tullio R, Salamino F, Barbato R, Melloni E, Siliprandi N, Schiaffino S, Pontremoli S. Biochem J. 1995;308:57–61. doi: 10.1042/bj3080057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Day SM, Westfall MV, Fomicheva EV, Hoyer K, Yasuda S, La Cross NC, D'Alecy LG, Ingwall JS, Metzger JM. Nat Med. 2006;12:181–189. doi: 10.1038/nm1346. [DOI] [PubMed] [Google Scholar]
- 30.Sakthivel S, Finley NL, Rosevear PR, Lorenz JN, Gulick J, Kim S, VanBuren P, Martin LA, Robbins J. J Biol Chem. 2005;280:703–714. doi: 10.1074/jbc.M409513200. [DOI] [PubMed] [Google Scholar]
- 31.Kaiser RA, Bueno OF, Lips DJ, Doevendans PA, Jones F, Kimball TF, Molkentin JD. J Biol Chem. 2004;279:15524–15530. doi: 10.1074/jbc.M313717200. [DOI] [PubMed] [Google Scholar]
- 32.Krenz M, Robbins J. J Am Coll Cardiol. 2004;44:2390–2397. doi: 10.1016/j.jacc.2004.09.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.