Abstract
The development of highly pathogenic avian H5N1 influenza viruses in poultry in Eurasia accompanied with the increase in human infection in 2006 suggests that the virus has not been effectively contained and that the pandemic threat persists. Updated virological and epidemiological findings from our market surveillance in southern China demonstrate that H5N1 influenza viruses continued to be panzootic in different types of poultry. Genetic and antigenic analyses revealed the emergence and predominance of a previously uncharacterized H5N1 virus sublineage (Fujian-like) in poultry since late 2005. Viruses from this sublineage gradually replaced those multiple regional distinct sublineages and caused recent human infection in China. These viruses have already transmitted to Hong Kong, Laos, Malaysia, and Thailand, resulting in a new transmission and outbreak wave in Southeast Asia. Serological studies suggest that H5N1 seroconversion in market poultry is low and that vaccination may have facilitated the selection of the Fujian-like sublineage. The predominance of this virus over a large geographical region within a short period directly challenges current disease control measures.
Keywords: influenza A, molecular epidemiology, virus evolution
Extensive surveillance and genetic studies have revealed that highly pathogenic avian influenza H5N1 viruses have become first predominant and then endemic in poultry in southern China and Southeast Asia since 2003 (1). This endemicity resulted in the establishment of multiple distinct regional sublineages (2). The recognition of multiple different H5N1 sublineages makes it possible to identify the source and to understand the evolutionary and transmission pathways of H5N1 viruses that have become widespread in Southeast Asia, Europe, and Africa.
Since the H5N1 influenza virus caused the first outbreak in migratory waterfowls at Qinghai Lake in May 2005 (3), a new transmission and outbreak wave was initiated. The virus expanded its geographical distribution and caused outbreaks in poultry in over 30 countries from Central Asia, the Middle East, Europe, and Africa (4). This expansion led directly to a marked increase in human infection cases and escalated the pandemic threat. In the first 8 months of 2006, the World Health Organization confirmed 96 cases from 9 countries; whereas for the whole of 2005 there was a total of 95 cases from 5 countries (5). In addition, in Indonesia recently there were suspected cases of human-to-human transmission involving members of an extended family, and the infection sources of other human cases have not been identified (6).
In China, despite a compulsory program for the vaccination of all poultry commencing in September 2005 (7), H5N1 influenza virus has caused outbreaks in poultry in 12 provinces from October 2005 to August 2006 (4). At the same time, 22 human infection cases have been confirmed from 14 provinces since November 2005 (4, 5). Some of those cases were residents of metropolitan areas remote from poultry farms, such as Guangzhou, Wuhan, and Shanghai (4). Furthermore, there were no obvious poultry outbreaks reported in neighboring markets or farms before or after the onset of those human infections. Therefore, whether those people were infected locally and directly from affected poultry or other sources, including humans, is still unknown. This situation directly challenges current pandemic preparedness plans, raising concern that a pandemic could emerge not only from the countryside but also from an urban area, just as severe acute respiratory syndrome emerged from the live-animal markets of Guangzhou and the Pearl River delta (8, 9).
Here we report updated virological and epidemiological findings from our market surveillance in southern China. Epidemiological analysis showed that H5N1 influenza viruses were continued to be perpetuated in poultry in each of the provinces tested, mainly in domestic duck and geese. Genetic analysis revealed that an H5N1 influenza variant had emerged and become predominant in each of the provinces, replacing those previously established multiple sublineages in different regions of southern China. This virus had also transmitted to Hong Kong, Laos, Malaysia, and Thailand. Serological studies suggested that the seroconversion rate in poultry in China is low and that the emergence and predominance of this H5N1 strain may be associated with vaccination in poultry. Genetic findings also revealed that these viruses also were responsible for all recently reported human infection cases in China. The predominance of this virus over a large geographical region within a short period questions the efficacy of current disease control measures in poultry and revealed that a new transmission and outbreak wave has been initiated from China to Southeast Asia since early 2006.
Results
Surveillance.
From July 2005 to June 2006 our influenza surveillance in live-poultry markets in six provinces of southern China showed that 1,294 of 53,220 (overall isolation rate 2.4%) poultry were H5N1-positive (Fig. 1; see also Fig. 5 and Table 3, which are published as supporting information on the PNAS web site). The main body of H5N1 isolates was from duck and goose, with only a small number isolated from chicken (chicken 0.5%, duck 3.3%, goose 3.5%). The prevalence of H5N1 viruses in southern China has increased when compared with the period July 2004 to June 2005 (overall, 0.9%; chicken, 0.2%; duck, 1.3%; and goose, 2.0%) (Fig. 1 and Table 3). A winter-seasonal peak was observed from October 2005 to March 2006 as in previous years (1, 2), during which H5N1 influenza viruses were isolated in each province tested (Fig. 1). However, an extension of the peak season was observed in April to June 2006 because isolation rates remained high in these warmer months (Fig. 1) (1, 2).
Fig. 1.
Comparison of H5N1 influenza virus isolation rate (%) in chicken (A), duck (B) and goose (C) from southern China, July 2004 to June 2006. Surveillance was conducted in live-poultry markets in Fujian, Guangdong, Guangxi, Guiyang, Hunan, and Yunnan.
Comparison between different types of poultry shows that H5N1 viruses were mainly isolated from domestic duck and goose wherein the viruses were prevalent year-round, whereas chicken tested positive mostly during the winter (Fig. 1 and Table 3). It is notable that H5N1-positive chicken were detected in 11 of the last 12 months, a marked increase from only 4 positive months in 2004/2005 (Fig. 1A). These findings indicate an escalation of H5N1 activity in poultry in 2005/2006 compared with previous years and suggest that H5N1 influenza viruses have not been effectively contained in this region and have maintained endemicity broadly in poultry, especially domestic duck and goose.
Regarding virus names and sublineages, the following nomenclature applies: BH goose, bar-headed goose; Ck, chicken; FJ, Fujian; Dk, duck; GD, Guangdong; Gf, Guinea fowl; Gs, goose; GX, Guangxi; GY, Guiyang; HK, Hong Kong; HN, Hunan; IDN, Indonesia; Mixed, southern China isolates; QH, Qinghai; ST, Shantou; YN, Yunnan; VNM, Vietnam; VNM2, second Vietnam introduction in March 2005;. The year the virus was identified is represented by the last two digits of the year: e.g., 02, 2002.
Antigenic Analysis.
Antigenic analysis with World Health Organization H5N1 reference antisera with representative viruses from different sublineages (see below) demonstrated a diversity of reaction patterns that generally corresponded to their phylogenetic relationships (Fig. 2; see also Table 4, which is published as supporting information on the PNAS web site). All tested FJ-like H5N1 viruses showed high HA inhibition (HI) titers to antiserum of Anhui/1/06, a virus from the same sublineage. Those viruses had moderate to low reactivity to antisera of IDN/357/06, Iraq/1/06, Dk/HN/101/04, and BH goose/QH/1A/05 but no reactivity with antisera of IDN/5/05, VNM/1203/04, Turkey/65596/06, and Whooping swan/Mongolia/244/05 (Table 4).
Fig. 2.
Numerical analysis of HI titers (see Table 4) by using hierarchical agglomerative clustering (A) and nonmetric multidimensional ordination in two dimensions (B). Colors indicate viruses from the FJ-like (red), GY1 (blue), GY2 (green), YN2 (purple), and GD/06 (orange) sublineages. MYS, Malaysia; Ph, pheasant; VTM, Vietnam/Thailand/Malaysia.
Numerical analysis of HI titers conducted to visualize similarity between the antigenic reactivity of different viruses showed that those FJ-like H5N1 viruses had a distinguishable antigenic reaction pattern. However, three viruses from this sublineage (Ck/GX/463/06, Dk/HN/856/06, and Gs/ST/18442/05) differed slightly (Fig. 2). This analysis also revealed two other major reactivity groups, one of Indonesia isolates and another that contained viruses from three different sublineages (GD/06, QH-like, and Mixed/VNM2) (Fig. 2). Four viruses (VNM/1203/06, Ck/GY/3570/05, Gs/YN/5539/05, and Gs/GY/337/06) representing four other distinct H5N1 sublineages had different reactivity patterns and did not group with other viruses (Fig. 2 and Table 4).
Phylogenetic Analysis.
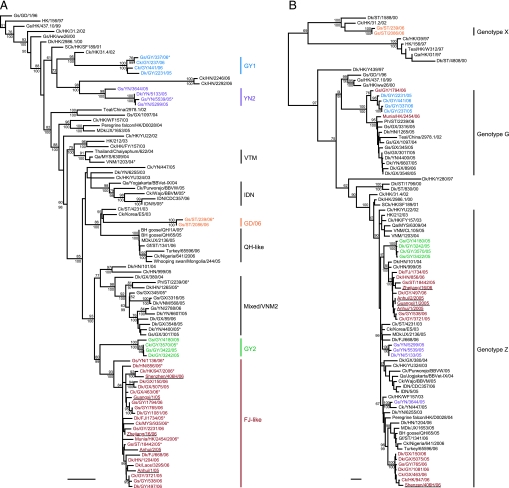
To better understand the increased prevalence of H5N1 in poultry and the emergence of human infection in China, 390 (30% of total new isolates) of those avian H5N1 influenza viruses isolated from July 2005 to June 2006 plus 16 viruses isolated from smuggled poultry and dead wild birds in Hong Kong in early 2006 were sequenced and analyzed together with sequences available from public databases. Phylogenetic analysis of the HA gene revealed that 266 of 390 (68%) of those recent H5N1 viruses from southern China formed a previously uncharacterized and distinct H5N1 sublineage (FJ-like) (Fig. 3 and 4A). Twenty-eight viruses isolated in Guiyang from November 2005 to January 2006 formed a sublineage (GY2) that is the sister group to the FJ-like sublineage (Figs. 3 and 4A). Another three sublineages from Guangdong (GD/06, n = 6), Guiyang (GY1, n = 14), and Yunnan (YN2, n = 13) also were identified. A further 59 viruses grouped in the Mixed/VNM2 sublineage, and only a single virus (Gf/ST/1341/06) belonged to the QH-like sublineage currently circulating in Africa and Europe (Fig. 3 and 4A). The remaining viruses analyzed in this study belonged to previously reported sublineages from China and Southeast Asia, except two isolates from Hunan (Ck/HN/2246/06 and Ck/HN/2292/06), isolated in May 2006 that do not fall with any of these sublineages (Fig. 4A).
Fig. 3.
Phylogenetic relationships of the HA genes of representative influenza A viruses isolated in Asia. Trees were generated by the neighbor-joining method in the PAUP* program (27). Numbers above or below branches indicate neighbor-joining bootstrap values. Not all supports are shown because of space constraints. Analysis was based on nucleotides 1–1011, and the tree was rooted to A/tern/South Africa/61. Colors indicate viruses from the FJ-like (red), GY1 (blue), GY2 (green), YN2 (purple), and GD/06 (orange) sublineages. VTM, Vietnam/Thailand/Malaysia. (Scale bar, 0.01 substitutions per site.)
Fig. 4.
Phylogenetic relationships of the HA (A) and PB2 (B) genes of representative influenza A viruses isolated in Asia. Trees were generated by the neighbor-joining method in the PAUP* program (27) (Bayesian analysis revealed the same relationships.) Numbers above and below branches indicate neighbor-joining bootstrap values and Bayesian posterior probabilities, respectively. Not all supports are shown because of space constraints. Analysis was based on nucleotides 1–1011 of the HA gene and 985-2233 of the PB2 gene. The HA and PB2 trees were rooted to A/tern/South Africa/61 and A/equine/Prague/1/56, respectively. Colors indicate viruses from the FJ-like (red), GY1 (blue), GY2 (green), YN2 (purple), and GD/06 (orange) sublineages. Recent human isolates from China are underlined. ∗, Viruses included in antigenic analysis (Fig. 2 and Table 4). JX, Jiangxi; MDK, migratory duck; MYS, Malaysia; Ph, pheasant; Qa, quail; SCK, silky chicken; VTM, Vietnam/Thailand/Malaysia. (Scale bar, 0.01 substitutions per site.)
The prototype virus of the FJ-like sublineage (Dk/FJ/1734/05) was detected in March 2005. From July to September 2005, only a single strain of 33 sequenced viruses was FJ-like (Table 1). Remarkably, from October 2005 onwards the percentage of FJ-like viruses detected increased dramatically, until from April to June 2006, 103 of the 108 H5N1 poultry isolates tested (95%) were FJ-like (Table 1). Viruses from other sublineages (YN2, GY2, GY1, GD/06, and Mixed/VNM2) were not detected in our surveillance since October 2005, November 2005, March 2006, April 2006, and May 2006, respectively. These findings reflect the process of FJ-like viruses gradually becoming predominant in this region.
Table 1.
Number of viruses from the Fujian-like sublineage in China
| Collection date | No. of FJ-like viruses |
|---|---|
| 2005 | |
| July–September | 1/33 (3) |
| October–December | 72/136 (53) |
| 2006 | |
| January–March | 90/113 (80) |
| April–June | 103/108 (95) |
| Total | 266/390 (68) |
Shown are the number of FJ-like sequences/total number (percentage) of H5N1-positive samples sequenced.
Phylogenetic analysis also revealed that the HA gene of five recent human H5N1 viruses from different provinces of China (4, 10, 11) belong to this FJ-like sublineage and were most closely related to poultry isolates (Fig. 4A). This finding suggests that H5N1 human infection from China since November 2005 was directly from affected poultry. Furthermore, H5N1 viruses isolated in early 2006 from neighboring regions of mainland China, including the 16 Hong Kong viruses and two poultry isolates from Laos and Malaysia, all joined the FJ-like sublineage (Fig. 4A).
Of those genotyped viruses, 99 of 137 (72%) were from this predominant FJ-like sublineage, all of which belonged to H5N1 genotype Z, except for a single genotype G virus (Gs/GY/1794/06) (Fig. 4B) (1, 2). Thirty-two viruses from other sublineages belonged to genotypes G and Z, whereas six GD/06 viruses belonged to genotype X, which has only previously detected in Hong Kong and Guangdong (Fig. 4B) (1). Of the 16 H5N1 virus isolates from Hong Kong in 2006, there were 7 genotype Z viruses, whereas 9 genotype G viruses were isolated from dead wild birds, suggesting a different ecology. Genotype G has seven gene segments in common with genotype Z but has a Gs/GD-like PB2 gene (Fig. 4B) (1, 2). These findings show that FJ-like H5N1 influenza viruses have become predominant and panzootic in southern China since mid-October 2005, have gradually replaced other H5N1 sublineages, and have been transmitted to Laos and Malaysia.
Molecular Characterization.
All viruses characterized in this study maintained the motif of multiple basic amino acids at the HA cleavage site characteristic of highly pathogenic avian influenza. Furthermore, these viruses also kill embryonated eggs within 24 h of inoculation, which also is a characteristic of highly pathogenic avian influenza. However, those viruses from the FJ-like sublineage have a Gln→Leu substitution at position −9 from the cleavage site (LRERRR-KR/G). The receptor-binding pocket of HA1 retains amino acid residues Gln-222 and Gly-224 (H5 numbering used throughout) that preferentially bind to α2,3-NeuAcGal linkages of avian cell-surface receptors (12, 13). Other amino acid residues relevant to receptor binding were identical to those of Gs/GD-like viruses (14), with the exception of Dk/HN/5128/05, which had Val-131-Met substitution.
Only six viruses from the FJ-like lineage (Ck/ST/3840/06, Ck/ST/3923/06, Ck/FJ/11933/05, Ck/FJ/12239/05, Ck/FJ/584/06, and Dk/GX/1550/06) had the Ser-31-Asn mutation in the matrix protein 2, which confers resistance to amantadine (15). All other viruses have residues indicating sensitivity to amantadine. All viruses characterized from the FJ-like lineage had His at position 274 of the neuraminidase protein and are predicted to be sensitive to oseltamivir (16).
Serological Analysis.
To estimate the seroconversion rate of market poultry after vaccination, 1,113 chicken sera from Guangdong and Guiyang Provinces were collected from November 2005 to April 2006. HI assay revealed that 180 sera (16%) were positive (HI titer ≥ 20) against Ck/HK/YU22/02 (H5N1).
Of 180 positive sera, 76 were randomly selected for a neutralization test against three representative viruses from the FJ-like, GY2, and Mixed/VNM2 sublineages. Most HI-positive sera tested showed neutralizing titers to Ck/GY/3570/05 and Dk/YN/4400/05, whereas 55 had low or no neutralization to Dk/FJ/1734/05 (Table 2; see also Table 5, which is published as supporting information on the PNAS web site). Almost all sera had 2- to 4-fold higher titers to Ck/GY/3570/05 and Dk/YN/4400/05, which are from the GY2 and Mixed/VNM2 sublineages, in comparison with Dk/FJ/1734/05 (Tables 4 and 5). Only four sera were neutralizing-negative against all three representative strains, although they were positive in the HI assay (Table 5). These findings suggest that chicken in southern China are poorly immunized against FJ-like viruses in comparison with other sublineages.
Table 2.
Summary of serological analysis of chicken serum collected in southern China from April to November 2006 against H5N1 influenza viruses from different sublineages
| Virus | Sublineage | NI titer range |
|||
|---|---|---|---|---|---|
| < | 20–40 | 80–160 | ≥320 | ||
| Dk/FJ/1734/05 | FJ-like | 30 | 25 | 18 | 3 |
| Ck/GY/3570/05 | GY2 | 9 | 16 | 35 | 16 |
| Dk/YN/4400/05 | YN2 | 4 | 14 | 41 | 17 |
Virus sublineages were defined by phylogenetic analysis of the HA gene (Fig. 3). <, lowest dilution tested (1:20); NI, neutralizing inhibition.
Discussion
The highly pathogenic H5N1 influenza virus currently panzootic in Eurasian and African poultry populations is considered the most likely candidate for a new pandemic influenza. The development of more and more avian-to-human interspecies transmission events in the last 12 months seems to favor such a hypothesis (4, 5). Our results demonstrate that the emergence and predominance of a H5N1 influenza virus sublineage in China has initiated a new transmission wave in Southeast Asia.
The emergence of this FJ-like sublineage has had similar consequences to the first wave of virus transmission throughout Southeast Asia in early 2004 (1) and the second wave to Europe and Africa that followed the Qinghai Lake H5N1 outbreak (3, 4). The findings of our study show that this virus has replaced most of those previously established regional sublineages across a large geographical area in China (2). The predominance of this FJ-like virus appears to be responsible for the increased prevalence of H5N1 in poultry since October 2005 and recent human infection cases in China (4, 5). Furthermore, it has already caused poultry outbreaks in Laos, Malaysia, and Thailand and human disease in Thailand (4, 5). As such, it is likely that this variant has already initiated a third wave of transmission throughout Southeast Asia and may spread further in Eurasia. It is also probable that this virus will continue to evolve to form other regionally distinct sublineages, as witnessed with the H5N1 genotype viruses in the first and second transmission waves (1, 2, 17).
The mechanism for the emergence and prevalence of FJ-like H5N1 variant is still unknown. The compulsory vaccination of all poultry was ordered in China beginning September 2005 (7), but our data indicate that seroconversion rates are still low and that poultry are poorly immunized against FJ-like viruses, which suggests that the poultry vaccine currently used in China may only generate very low neutralizing antibodies to FJ-like viruses in comparison to other previously cocirculating H5N1 sublineages. This situation could have helped to select for the FJ-like sublineage in poultry, because our results also show that these viruses had replaced the GY2 and YN2 virus sublineages, both of which had high titers in the serological tests. As such, this information suggests that the predominance of FJ-like viruses may be associated with immune escape from the current vaccine strain in poultry.
Previously, we described the establishment of multiple sublineages of H5N1 virus in southern China and Southeast Asia (2). The emergence and replacement of these sublineages by FJ-like viruses within a short period highlights the difficulties faced in controlling H5N1 virus in China. A complex ecology and highly diverse virus populations make it almost impossible to capture each circulating virus sublineage, even with the application of mass vaccination. This complexity has resulted in recurrent H5N1 outbreaks in poultry in different regions and has led to occasional human infection.
Since November 2005, 22 H5N1 human infection cases from 14 provinces of China have been reported (4, 5). It is noteworthy that four of the provinces (Fujian, Guangdong, Shanghai, and Zhejiang) with human cases have not recorded any outbreaks in poultry (4). However, phylogenetic and antigenic analyses in this study clearly show that those recent human H5N1 isolates from different provinces are FJ-like viruses, which suggests that this virus may be prevalent in an area much larger than we have identified. Given the lack of systematic influenza surveillance in poultry at a national level, the timely identification of the source of human infection is almost impossible. Therefore, to understand and identify possible infection sources and to avert a potential pandemic, comprehensive influenza surveillance in both human and animal populations is urgently required in H5N1-affected regions.
The repeated emergence of H5N1 variants from southern China and their subsequent spread to other parts of the world (1–3) makes it increasingly apparent that implementation of effective control measures in this region is of paramount importance. Such a system of control measures could be achieved by integrated real-time virological and genetic information with rapid diagnostic approaches and vaccine production accompanied by strict quality control. Our surveillance network in southern China is the longest running, and the data this network has generated have provided the most comprehensive insight into the ecology and evolution of H5N1 virus in its natural host (1, 2, 18–20). Despite these efforts, there remains a lack of information in the broader region, and it is critical that similar surveillance programs begin in other areas, including Indonesia, Vietnam, Thailand, and India. Perhaps most importantly, information from northern China is required, because it could answer key questions regarding the movement of H5N1 in and out of southern China, the hypothetical influenza epicenter (21).
Methods
Virological Surveillance, Isolation, and Characterization.
Cloacal, tracheal, and fecal samples were collected once every 7–10 days from apparently healthy poultry in live-poultry markets in Fujian, Guangdong, Guangxi, Guiyang, Hunan, and Yunnan (Fig. 5). Specimens were first screened by RT-PCR for H5 subtype influenza virus. All PCR-positive swabs were shipped to the State Key Laboratory of Emerging Infectious Diseases at The University of Hong Kong and grown in embryonated eggs. All isolates were identified and subtyped by using a panel of reference antisera as previously described (22).
Antigenic Analysis.
The antigenic characteristics of the H5N1 influenza viruses from different sublineages were compared by HI assay with ferret antisera to the World Health Organization reference H5 subtype viruses as previously described (20). The Department of Infectious Diseases at St. Jude Children's Research Hospital produced the ferret antisera against Dk/HN/101/04, IDN/5/05, BH goose/QH/1A/05, and VNM/1203/04. Antisera to Anhui/1/06, IDN/357/06, Iraq/1/06, Whooping swan/Mongolia/244/05, and Turkey/65596/06 were kindly provided by Nancy Cox (Centers for Disease Control and Prevention, Atlanta, GA). The HI assay started at 1:20 dilution.
To visualize similarity between the antigenic reaction patterns of different viruses, numerical analysis of HI titers was conducted by using PRIMER version 5.2.9 (PRIMER-E, Plymouth, United Kingdom). The data were standardized and square-root-transformed, and the Bray–Curtis coefficient (23) was used to construct a similarity matrix. Hierarchical agglomerative clustering with group-average linking (24) was conducted, and a dendrogram was produced. Nonmetric multidimensional scaling (25) also was used to produce two- and three-dimensional ordinations over 100 iterations. The two-dimensional configuration with lowest overall stress was presented.
Phylogenetic Analysis and Molecular Characterization.
We sequenced the HA gene of 390 of the 1,294 (30%) H5N1 influenza viruses isolated from poultry market surveillance in southern China from July 2005 to June 2006 along with 16 viruses isolated from smuggled poultry and dead wild birds in Hong Kong in January and February 2006. In addition, 137 of those 1,294 (11%) viruses, plus the 16 Hong Kong isolates, were partially sequenced for each of the 8 gene segments and genotyped as previously described (1, 2, 18). Sequence assembly, editing, alignment, and residue analysis were performed as previously described (2). Phylogenetic analysis using MrModelTest 2.2 (26), PAUP* 4.0 (27), and MrBayes 3.1 (28) also was carried out as previously described (2).
Serological Analysis.
Chicken sera (n = 1,113) were collected from live-poultry markets in different provinces from November 2005 to April 2006. The antibodies for H5 subtype influenza virus were detected by HI assay, with Ck/HK/YU22/02 as the antigen. A serum was considered positive for H5N1 virus if its HI titer was ≥20. Some of those HI-positive sera were randomly selected and further tested by neutralization assay for antibodies against the representative isolates: Ck/FJ/1734/05, Ck/GY/3570/05, and Dk/YN/4400/05, which belong to the FJ-like, GY2, and Mixed/VNM2 sublineages, respectively. Sera were screened at a 1:10 series dilution against 100 TCID50 (50% tissue culture infective dose) of those three representative viruses to exclude negative samples, as previously described (2). Titers of ≥20 were regarded as positive.
Supplementary Material
Acknowledgments
We thank L. J. Zhang, J. Wong, L. Duan, and W. S. Hong for excellent technical support. Sequence data from human cases in Indonesia were kindly provided by the Indonesian Department of Health. This work was supported by the Li Ka Shing Foundation and by National Institute of Allergy and Infectious Disease Contract AI95357.
Abbreviation
- HI
HA inhibition.
Footnotes
References
- 1.Li KS, Guan Y, Wang J, Smith GJD, Xu KM, Duan L, Rahardjo AP, Puthavathana P, Buranathai C, Nguyen TD, et al. Nature. 2004;430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- 2.Chen H, Smith GJD, Li KS, Wang J, Fan XH, Rayner JM, Vijaykrishna D, Zhang JX, Zhang LJ, Guo CT, et al. Proc Natl Acad Sci USA. 2006;103:2845–2850. doi: 10.1073/pnas.0511120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen H, Smith GJD, Zhang SY, Qin K, Wang J, Li KS, Webster RG, Peiris JSM, Guan Y. Nature. 2005;436:191–192. doi: 10.1038/nature03974. [DOI] [PubMed] [Google Scholar]
- 4.Centre for Health Protection. Global Statistics of Avian Influenza (As of 30 August 2006) China: Centre for Health Protection, Department of Health, Hong Kong SAR; 2006. available at www.info.gov.hk/info/flu/eng/global.htm. [Google Scholar]
- 5.World Health Organization. Cumulative Number of Confirmed Human Cases of Avian Influenza A (H5N1) Geneva: WHO; 2006. available at www.who.int/csr/disease/avian_influenza/country/en. [Google Scholar]
- 6.World Health Organization. Avian Influenza: Situation in Indonesia. Geneva: WHO; 2006. available at www.who.int/csr/don/archive/country/idn/en. [Google Scholar]
- 7.Cyranowski D. Nature. 2005;438:406. [Google Scholar]
- 8.Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, et al. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 9.Guan Y, Zhong NS, Chen H, Smith GJD, Zheng BJ. In: Challenges of Severe Acute Respiratory Syndrome. Chan JCK, Taam Wong VCW, editors. Singapore: Elsevier; 2006. pp. 93–100. [Google Scholar]
- 10.Yu H, Shu Y, Hu S, Zhang H, Gao Z, Chen H, Dong J, Xu C, Zhang, Yxiang N, et al. Lancet. 2006;367:84. doi: 10.1016/S0140-6736(05)67894-4. [DOI] [PubMed] [Google Scholar]
- 11.Shu Y, Yu H, Li D. N Engl J Med. 2006;354:1421–1422. doi: 10.1056/NEJMc053524. [DOI] [PubMed] [Google Scholar]
- 12.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. Proc Natl Acad Sci USA. 2001;98:11181–11186. doi: 10.1073/pnas.201401198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 14.Claas EC, Osterhaus AD, van Beek R, De Jong JC, Rimmelzwaan GF, Senne DA, Krauss S, Shortridge KF, Webster RG. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 15.Scholtissek C, Quack G, Klenk HD, Webster RG. Antiviral Res. 1998;37:83–95. doi: 10.1016/s0166-3542(97)00061-2. [DOI] [PubMed] [Google Scholar]
- 16.Treanor JJ, Hayden FG, Vrooman PS, Barbarash R, Bettis R, Riff D, Singh S, Kinnersley N, Ward P, Mills RG. J Am Med Assoc. 2000;283:1016–1024. doi: 10.1001/jama.283.8.1016. [DOI] [PubMed] [Google Scholar]
- 17.The World Health Organization Global Influenza Program Surveillance Network. Emerg Inf Dis. 2005;11:1515–1521. doi: 10.3201/eid1110.050644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guan Y, Peiris JS, Lipatov AS, Ellis TM, Dyrting KC, Krauss S, Zhang LJ, Webster RG, Shortridge KF. Proc Natl Acad Sci USA. 2002;99:8950–8955. doi: 10.1073/pnas.132268999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan Y, Peiris M, Kong KF, Dyrting KC, Ellis TM, Sit T, Zhang LJ, Shortridge KF. Virology. 2002;292:16–23. doi: 10.1006/viro.2001.1207. [DOI] [PubMed] [Google Scholar]
- 20.Guan Y, Poon LLM, Cheung CY, Ellis TM, Lim W, Lipatov AS, Chan KH, Sturm-Ramirez KM, Cheung CL, Leung YHC, et al. Proc Natl Acad Sci USA. 2004;101:8156–8161. doi: 10.1073/pnas.0402443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shortridge KF, Stuart-Harris CH. Lancet. 1982;2:812–813. doi: 10.1016/s0140-6736(82)92693-9. [DOI] [PubMed] [Google Scholar]
- 22.Guan Y, Shortridge KF, Krauss S, Chin PS, Dyrting KC, Ellis TM, Webster RG, Peiris M. J Virol. 2000;74:9372–9380. doi: 10.1128/jvi.74.20.9372-9380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bray RJ, Curtis JT. Ecol Monogr. 1957;27:325–349. [Google Scholar]
- 24.Sokal RR, Michener CD. Univ Kans Sci Bull. 1958;38:1409–1438. [Google Scholar]
- 25.Kruskal JB. Psychometrika. 1964;29:115–129. [Google Scholar]
- 26.Nylander JAA. Uppsala, Sweden: Evolutionary Biology Centre, Uppsala University; 2004. MrModeltest. Version 2. [Google Scholar]
- 27.Swofford DL. Sunderland, MA: Sinauer; 2001. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods) Version 4.0 beta. [Google Scholar]
- 28.Ronquist F, Huelsenbeck JP. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.