Abstract
Genetic variation for seed dormancy in nature is a typical quantitative trait controlled by multiple loci on which environmental factors have a strong effect. Finding the genes underlying dormancy quantitative trait loci is a major scientific challenge, which also has relevance for agriculture and ecology. In this study we describe the identification of the DELAY OF GERMINATION 1 (DOG1) gene previously identified as a quantitative trait locus involved in the control of seed dormancy. This gene was isolated by a combination of positional cloning and mutant analysis and is absolutely required for the induction of seed dormancy. DOG1 is a member of a small gene family of unknown molecular function, with five members in Arabidopsis. The functional natural allelic variation present in Arabidopsis is caused by polymorphisms in the cis-regulatory region of the DOG1 gene and results in considerable expression differences between the DOG1 alleles of the accessions analyzed.
Keywords: natural variation, seed germination, abscisic acid, gibberellin, cis variation
Seed dormancy, defined as the inability of a viable seed to germinate under conditions that allow germination, is assumed to be an important adaptive trait in nature (1). It can prevent germination when environmental conditions are suitable for germination but where the probability of completing the life cycle is low (2, 3). In Arabidopsis a large number of mutants affecting seed dormancy have been generated artificially, and the genetic, physiological, and molecular characterization of these mutations is starting to shed light on the complexity of the regulation of this process. For instance, mutants in genes such as ABA-insensitive 3 (ABI3) (4, 5), FUSCA3 (FUS3) (6, 7), and LEAFY COTYLEDONS (LEC1 and LEC2) (8–10) with defective seed maturation are nondormant, indicating that dormancy is part of the developmental program established during the later phases of seed development. The analyses of testa mutants originally identified by their altered seed color or shape provided strong evidence for the importance of the testa in the control of seed germination (11). Nongerminating mutants affected in the biosynthesis of the plant hormone gibberellin (GA) (12) and the nondormant mutants deficient in abscisic acid (ABA) (13) have shown the important role of ABA in the induction and maintenance of dormancy and the opposing roles of GA and ABA in the control of dormancy and germination (reviewed in ref. 14). The downstream effects of the hormones are less well known. Light-induced stimulation of seed germination is affected in phytochrome photoreceptor-deficient mutants (15). Furthermore, genes that most likely do not specifically affect hormone or light signaling pathways have been described as dormancy regulators. These include genes encoding transcription regulators such as DOF affecting germination (DAG) (16, 17) and several genes with unknown functions such as those disrupted in the reduced dormancy 1–4 mutants (rdo 1–4) (18, 19). An additional set of genes that play roles in seed dormancy are those identified by the study of natural variation. The identification of the genes underlying this natural variation for seed dormancy may help to further increase our understanding of the mechanisms involved in this process. At the same time, it will provide insights into the way nature shaped genetic variability for this trait during adaptive evolution. Currently, quantitative trait locus (QTL) mapping is a standard procedure in quantitative genetics. The identity of individual QTL, i.e., the DNA sequences (coding or noncoding) responsible for the QTL, can be established (20).
Natural variation for seed dormancy has been described in many species (21, 22), and several dormancy QTL regions have been validated in introgression or near isogenic lines (NILs) for Arabidopsis, barley, and rice (refs. 23–26 and reviewed in ref. 21). However, in none of these cases has the molecular function of the genes underlying dormancy QTL been as yet identified.
Previous research demonstrated that there is considerable genetic variation for seed dormancy between the laboratory strain Landsberg erecta (Ler) with low seed dormancy and the dormant Cvi accession from the Cape Verde Islands (24). QTL analysis for seed dormancy on a recombinant inbred line population derived from a cross between these two accessions identified Delay of Germination 1 (DOG1) as an important determinant of seed dormancy within this population. DOG1, for which the Cvi allele increased the level of seed dormancy, explained 12% of the variance observed in seed dormancy in this recombinant inbred line population. Here we report the positional cloning and analysis of the seed dormancy QTL DOG1, which, based on the phenotype of the loss-of-function mutants, seems to specifically control seed dormancy whereas previously described seed dormancy and germination genes often affect multiple plant processes.
Results
Identification of a Mutant at the DOG1-Cvi Locus.
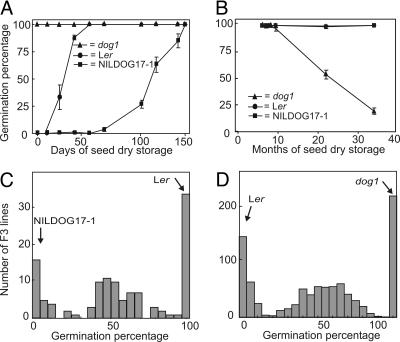
To study the role of DOG1 in seed dormancy we used a NIL containing a Cvi introgression at the position of this QTL in a Ler genetic background (called NILDOG17-1) (24). This line, which is more dormant than the Ler accession (Fig. 1A), confirming the presence of a dormant Cvi allele in that region, was submitted to a mutagenesis treatment. Mutants with no or strongly reduced seed dormancy were selected in this experiment. Among these were mutants at the ABA1 and ABA2 loci, indicating that ABA is needed for the functioning of DOG1. One line had a mutation in the DOG1 gene itself (dog1), as was concluded from the fact that no recombinants between this mutation and the dormant Cvi allele could be detected among 2,061 gametes analyzed in the F2 progeny of a Ler × dog1 cross. The dog1 mutant does not show any obvious pleiotropic effects except that it cannot be stored as long as Arabidopsis wild-type seeds at room temperature (Fig. 1B). To study the genetics of the different DOG1 alleles, the germination behavior of F3 seeds, obtained from F2 plants derived from crosses of Ler with either DOG1-Cvi(NILDOG17-1) or dog1, were tested. Segregation ratios were in agreement with monogenic inheritance and suggest semidominance of the DOG1 alleles (Fig. 1 C and D).
Fig. 1.
Phenotypes of the different DOG1 alleles. (A) Germination behavior of Ler, NILDOG17-1, and dog1 at different time points after seed harvest. The means and SE of four replicates are shown. (B) Seed storability phenotypes of the different DOG1 alleles. Germination behavior of the different DOG1 alleles, Ler, NILDOG17-1, and dog1 at different time points after seed harvest. The means and SE of triplicates are shown. (C and D) Segregation of DOG1 alleles. Shown are frequency distributions of germination percentages of F3 seed batches harvested from individual F2 plants derived from the crosses Ler × NILDOG17-1 (C) and Ler × dog1 (D).
Germination Characteristics of the Three DOG1 Alleles.
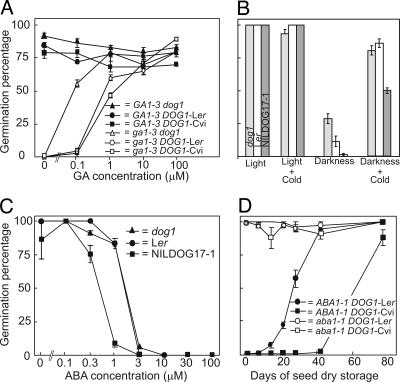
The germination behavior of the dog1 mutant resembles ABA-insensitive and ABA biosynthesis mutants (13). The aba1-1 mutant is characterized by the absence of dormancy but also by germination in darkness and germination without the need of de novo GA biosynthesis. This was shown by germination on GA biosynthesis inhibitors and the ability of double mutants with the GA biosynthesis mutant ga1-3 to germinate (13, 27). To determine whether dog1 has the same germination characteristics we performed similar experiments with dog1. This revealed that dog1 mutant seeds still require light and GA for germination, although less than Ler and NILDOG17-1 (Fig. 2A). Reduced germination in darkness can be partly overcome by cold stratification (Fig. 2B), probably because cold can increase GA sensitivity (27). DOG1 is not specifically involved in ABA signal transduction, because the dog1 mutant has a normal sensitivity to applied ABA (Fig. 2C). However, to achieve dormancy conferred by the DOG1-Cvi allele, ABA is required, because combining NILDOG17-1 with the aba1-1 mutant led to nondormant seeds (Fig. 2D). In conclusion, dog1 seeds still require light-induced GA biosynthesis to overcome inhibition by ABA present in imbibed nondormant seeds (28–30).
Fig. 2.
Germination behavior of DOG1 under different environmental circumstances and in different genetic backgrounds. (A) GA requirement for germination. Germination on various concentrations of GA4+7 of after-ripened seeds of GA1-3 dog1, GA1-3 DOG1-Ler, and GA1-3 DOG1-Cvi and on the GA-deficient seeds ga1-3 dog1, ga1-3 DOG1-Ler, and ga1-3 DOG1-Cvi. Seeds of GA1-3 dog1 and ga1-3 dog1 were 1 month after-ripened, and seeds of the other combinations were after-ripened for 25 months. Percentages are averages of four measurements ± SE. (B) Germination in darkness. Shown are germination percentages of after-ripened seeds of dog1, Ler, and NILDOG17-1 under different environmental conditions, in light, in light after a cold stratification, in darkness, and in darkness after a cold stratification. The means and SE of triplicates are shown. (C) ABA sensitivity of the three DOG1 alleles. Shown is germination in different ABA concentrations of after-ripened seeds of dog1, DOG1-Ler, and DOG1-Cvi. Seeds of dog1 and DOG1-Ler were after-ripened for 4 months, and seeds of DOG1-Cvi were after-ripened for 25 months. The percentages are means of triplicates ± SE. (D) ABA dependency of DOG1. Shown is germination behavior of aba1-1 in two different DOG1 backgrounds (DOG1-Ler and DOG1-Cvi) at different time points after seed harvest.
Isolation of DOG1.
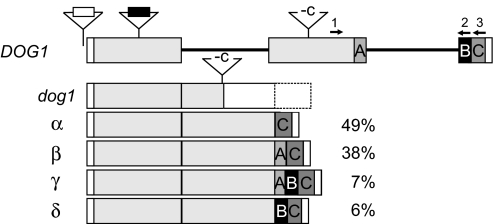
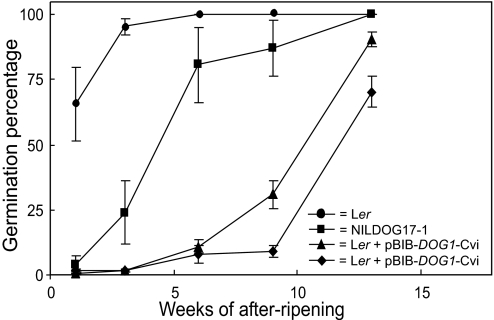
To understand the molecular function of DOG1, the gene was cloned by using positional information and the dog1 mutant described above. Recombination mapping, using the progeny of crosses between a Ler morphological marker line carrying the gl3 and tz mutation and NILDOG17-1 and Ler × dog1, placed the DOG1 locus between marker cMRA19z and marker K15I22 in the region where the QTL was mapped on chromosome five. This region contained 22 ORFs. Because the loss-of-function phenotype of DOG1 was known, we decided to look for the candidate gene in that region by the identification of a gene whose knockout phenotype resembled the phenotype of the dog1 mutant described above. Among the insertion mutants from the Salk collection (http://signal.salk.edu), Salk line 000867 was identified as completely nondormant. This line had a T-DNA insertion in the promoter region of At5g45830. Three additional lines (SM_3.20808, SM_3.20873, and SM_3.20886 from J. H. Clarke, John Innes Centre, Norwich, U.K.) with transposons inserted at position 213 of the same gene and therefore probably representing the same transposition event, also have nondormant phenotypes (positions of the insertions are indicated in Fig. 3). Sequence analyses of At5g45830 in NILDOG17-1 and dog1 identified a 1-bp deletion at position 914 (based on Col genomic sequence) in the dog1 mutant. This deletion changes the reading frame and terminates the ORF 6 aa after the deleted nucleotide (Fig. 3). To prove that At5g45830 caused the dormancy phenotype a 5.6-kb PCR fragment containing the Cvi allele of At5g45830 and both the 5′ and 3′ region was cloned into the binary vector pBIB-HYG (31) (pBIB-DOG1-Cvi) and transformed into Ler by using Agrobacterium tumefaciens. Two independent transformants homozygous for the construct had a strong dormant phenotype. This confirmed that the Cvi allele of At5g45830 is responsible for the strong seed dormancy in NILDOG17-1 (Fig. 4).
Fig. 3.
Schematic illustration and relative abundance of the DOG1 transcripts. At the top of the figure the DOG1 genomic DNA is shown. The insertions of SALK 000867 (white box), SM_3.20808, SM_3.20873, and SM_3.20886 (black box) and the position of the mutation in the dog1 mutant (-c) are indicted on top of this genomic structure. Boxes on the black solid line indicate the exons, white boxes indicate 3′ and 5′ untranslated regions, and the different grayscales are used to illustrate the different exon compositions of the transcripts α, β, γ, and δ, which are shown below. The percentages indicate the relative abundance of the different transcripts; this is an average of the abundance measured in dry dormant and nondormant seeds of several accessions analyzed. Primers used for PCR amplification are indicated by the black arrows (1, pyro F2; 2, pyro R2; 3, pyro R3).
Fig. 4.
Introduction of the DOG1-Cvi allele into the Ler genetic background. The germination of homozygous transformants obtained by Agrobacterium-mediated transformation of Ler with the genomic fragment of DOG1-Cvi is shown. Shown is germination behavior of Ler, NILDOG17-1, and two independent transformants at different time points after seed harvest. The means and SE of triplicates for Ler and NILDOG17-1 and 11 and 12 replicates for the two transformants, respectively, are shown.
DOG1 Is a Member of a Novel, Plant-Specific Gene Family.
DOG1 (At5g45830) belongs to a small gene family. Based on the gene structure of DOG1 we were able to identify four additional genes of this type in the Arabidopsis genome. We have called them DOG1-Like 1–4 (DOGL1–4), which encode At4g18660, At4g18680, At4g18690, and At4g18650, respectively. Four of the DOG1 family genes are highly conserved; the sequence similarity of DOGL1 to DOGL3 with DOG1 is, respectively, 54.3%, 43.1%, and 39.3% (Fig. 7, which is published as supporting information on the PNAS web site). DOGL4 is much more distinct, with only a 23.4% sequence similarity with DOG1. To test whether these DOGL genes affect seed dormancy we obtained presumed null mutants for DOGL1, DOGL2, and DOGL3 from the Salk T-DNA insertion project (http://signal.salk.edu). However, insertions in these genes did not result in a germination phenotype (data not shown). To assign a possible function for the Arabidopsis DOG1 gene family proteins we compared them with similar genes in other species. The highest percent similarity found is with a Brassica napus EST [embryo library; tBLASTx in NCBI EST database (CN827162); global identity 37.8%, local 53.4% in 206 aa]. This gene has not been annotated. The highest percent similarity with a gene that has a known function is that with the wheat transcription factor HBP-1b (CAA40102) (global 13.3%; local 42% in 33 aa). DOG1 contains three protein domains (PD870616, PD004114, and PD388003) as defined by ProDom (http://prodom.prabi.fr/prodom/current/html/home.php) (32). PD870616 is present only in the five related Arabidopsis genes mentioned above and has not been annotated. PD004114 is present in the D bZIP transcription factors described in ref. 33. This group of transcription factors contains the bZIP domain and an additional conserved motif (box 1). However, DOG1 does not show any homology in these two regions but only with the region between the bZIP domain and box 1. Protein domain PD388003 has been annotated as tumor-related protein-like (BAA05470.1) (23.7% global identity) because it was found in the hybrid Nicotiana glauca × Nicotiana langsdorffii, which shows tumorous cell growth. Another gene with the same DNA structure as DOG1 is BAB08196 from Oryza sativa (17.9% global identity). The functions of both these genes are unknown. This low percent similarity with genes of unknown function in other species means that a conclusion about the molecular function of DOG1 cannot be drawn as yet.
DOG1 Expression Is Seed-Specific.
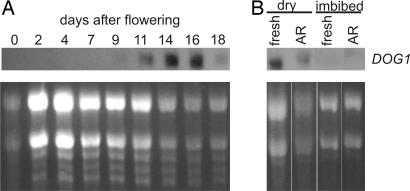
We analyzed the DOG1 expression in different plant tissues; seedlings, roots, flowers with closed buds and flowers with open buds, and developing siliques at different time points and in dormant and after-ripened dry and imbibed seeds. DOG1 transcription is seed-specific; transcription starts during the seed development 9 days after pollination and reaches its highest level during the last phases of seed development (as was detected by analyzing developing siliques) (Fig. 5). This is in agreement with data reported by the Genevestigator online search tool Gene Chronologer (www.genevestigator.ethz.ch) (34). DOG1 transcripts remain present in after-ripened (nondormant) dry seeds. Upon imbibition the transcripts rapidly disappear in both dormant and after-ripened seeds (Fig. 5). To determine whether the promoter region of DOG1 contains motifs that can explain its seed-specific expression we have run 2,349 bp of genomic sequence 5′ of the DOG1 ATG in PLACE (a database of plant cis-acting regulatory DNA elements) (35). We could identify a RY repeat (CATGCA; −500 from the ATG) required for seed-specific expression. In addition, we found ABRE motifs (TACGTGTC; −1671 and −1650 from the ATG) that are known to be required for ABA responsiveness and that have been found as well in 24 genes that are GA-down-regulated during Arabidopsis seed germination (36).
Fig. 5.
Northern blot analyses of the expression of DOG1 in seeds of NILDOG17-1. (A) The expression at different developmental stages, starting at the day of pollination until 18 days after pollination, when the seeds are mature and the silique opens (analyzed in developing siliques). (B) The expression in fresh (dormant) and after-ripened (AR; nondormant) dry and imbibed mature seeds is shown. (Upper) Autoradiograph. (Lower) RNA loading stained with ethidium bromide.
Alternative Splicing of the DOG1 Gene.
The At5g45830 gene is described as encoding a predicted ORF of 291 aa (GenBank accession no. NM123951). From sequence analyses performed on cDNA pools we could identify at least four different cDNAs (α–δ) present in Ler and in Cvi, suggesting that they are the result of alternative splicing. The four different cDNAs are different combinations of three exon fragments, A, B, and C (C only = cDNA α, A+C = β, A+B+C = γ, and B+C = δ) (Fig. 3). To investigate the possibility that the different phenotypes of the DOG1-Ler and NILDOG17-1 could be explained by different relative levels of the spliced cDNAs, we developed a pyrosequencing assay to quantify the relative abundance of the different transcripts in mature dry seeds (Fig. 3). This method did not detect a significant difference in amount of the different transcripts between the genotypes analyzed or between dormant (fresh) and nondormant (after-ripened) seeds. CDNA β and γ encode the same protein, which is the result of a stop codon in exon A. We do not know whether all of the cDNAs are translated into peptides or not.
Sequence Diversity of DOG1 in Different Accessions of Arabidopsis thaliana.
To understand the basis for the allelic differences at DOG1, we compared the nucleotide sequences of the DOG1-Ler and DOG1-Cvi alleles (in total ≈5.5 kb based on the Col sequence; starting 2,349 before the translational start). Three polymorphisms and one indel have been identified in the coding region, and two of the polymorphisms lead to an amino acid substitution, serine-927 to proline and proline-1694 to glutamine. The indel resulted in an extra glycine in DOG1-Ler compared with DOG1-Cvi. In addition, we sequenced DOG1 alleles of six additional accessions (Col, An-1, Kond, Sha, Kas-2, and Fei-0) of which recombinant inbred line populations derived from crosses with Ler have been constructed (37–39) (Carlos Alonso-Blanco, personal communication). In four of these populations (Sha, Kond, Kas-2, and Fei-0) a QTL at DOG1 or a closely linked locus has been identified (Table 1, which is published as supporting information on the PNAS web site) (ref. 38 and L.B., unpublished results). The relative dormancy levels of the different accessions are indicated in Table 1. The sequence comparison showed various polymorphisms in the coding regions of DOG1 alleles; however, we did not observe a correlation between the sequence and the dormancy level of the accessions. Therefore, we assume that these polymorphisms are not responsible for the functional allelic differences found for DOG1.
Expression Diversity of DOG1.
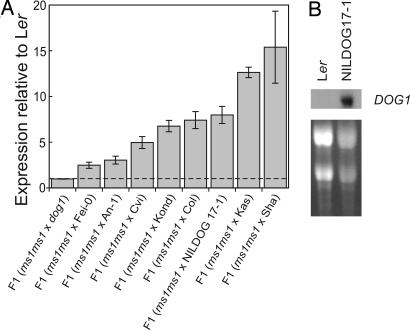
In addition to the sequence diversity in the coding region we observed 19 polymorphisms and 10 indels in the putative cis-regulatory region of the DOG1 gene. To determine whether these differences could cause the difference in phenotype we analyzed the functional variation of the DOG1 cis-regulatory region. To quantify this functional variation as a result of the cis-regulation of DOG1 mRNA levels specifically, we used F1 seeds in which distinct promoter alleles were compared within a common trans-regulatory background and quantified the relative levels of the two parental mRNA forms in the hybrid by pyrosequencing (as described in refs. 40 and 41). F1 seeds were analyzed by comparing Ler with each of nine other genotypes (the accessions Fei-0, An-1, Cvi, Kond, Col, Kas-2, and Sha and NILDOG17-1 and dog1). The Ler coding region harbored a single nucleotide polymorphism allowing us to distinguish this DOG1 allele (DOG1-Ler). The relative amount of each parental allele was quantified by pyrosequencing (Fig. 6A). This procedure did not distinguish between the differentially spliced transcripts (α–δ). Differences in DOG1 expression found between Ler and NILDOG17-1 were confirmed by RT-PCR (data not shown) and Northern blot analyses (Fig. 6B). Cis-regulatory alleles have a 2- to 15-fold difference in activity relative to the Ler cis-regulatory allele, which is much larger than what was previously observed in A. thaliana at another locus (41). DOG1-Ler and the dog1 mutant have the lowest expression levels. DOG1-Cvi in the background of the Cvi accession has clearly a lower expression level than DOG1-Cvi in the background of NILDOG17-1, which could be because of loci present outside the introgression in Cvi that reduce the mRNA level of DOG1 (24).
Fig. 6.
Expression analyses of DOG1 in different genetic backgrounds. (A) Relative expression levels were measured by pyrosequencing of cDNAs derived from RNA extracted from F1 seeds made by pollinating a male sterile 1 (ms1) mutant in a Ler background with pollen of each of nine other genotypes (the accessions Fei-0, An-1, Cvi, Kond, Col, Kas-2, and Sha and NILDOG17-1 and dog1). The relative values are means of triplicates ± SE. The horizontal dashed line indicates a relative expression level of 1. (B) Northern blot analyses of the expression of DOG1 in mature seeds of Ler and NILDOG17-1. (Upper) Autoradiograph. (Lower) RNA loading stained with ethidium bromide.
Discussion
The genetics of seed dormancy has been studied mainly by analyzing mutants that were selected based on a germination phenotype. This revealed several mutants defective in hormonal pathways.
Natural variation provides another genetic resource to identify genes that are involved in the control of seed dormancy. One of these genes which determines the genetic variation for seed dormancy in nature in Arabidopsis is the DOG1 locus, identified by QTL analysis of the progeny from a cross between the accessions Ler and Cvi (24). In the present work we have identified the underlying gene of the DOG1 QTL by fine (high-resolution) mapping of the QTL and by the identification of a loss-of-function allele, which resulted in loss of dormancy. The latter allowed us to screen for T-DNA insertion mutants in the available collections in the low-dormancy Col accession. DOG1 is encoded by At5g45830 and represents a novel gene with unknown function. DOG1 belongs to a small family of proteins in Arabidopsis that contains three conserved domains (PD870616, PD004114, and PD388003) of which the functions are unknown. DOG1 by it self has a strong effect on seed dormancy whereas the DOG1-like genes, which are highly similar to each other, have no proven influence on seed dormancy.
DOG1, of which the different alleles behave in a codominant manner, is a gene that specifically controls seed dormancy. Apart from a seed longevity phenotype in dog1 mutant seeds, no other pleiotropic phenotypes have been identified. DOG1 function is clearly related to ABA, based on the double mutant analysis. Preliminary data suggest that it may affect ABA levels in dry seeds (J. Zeevaart, personal communication) and slightly changes ABA sensitivity (Fig. 2C). The observation that the DOG1 transcript levels drop during seed imbibition (Fig. 5) and that ABA also has an effect in imbibed seeds where it antagonizes the effect of GA (induced by light) might indicate that DOG1 functions only in the establishment of dormancy during seed maturation, whereas ABA may function during imbibition also. This might be in agreement with the observation that the dog1 mutant still requires light and GA to induce germination in the nondormant seeds that need the latter to initiate germination. Another nondormant mutant that still requires light and GA is fusca 3 (42). This mutant belongs to a group of genes that control the seed maturation program.
The amino acid substitutions that were detected in the structural part of the DOG1 gene when comparing different accessions most likely are not functional. Therefore, the natural variation, present in DOG1, may be controlled by differences in cis-regulatory region(s). Expression analyses performed on F1 seeds of Ler crossed with the different accessions indeed show clearly that the natural allelic variation in the DOG1 gene between these accessions causes differences in gene expression level. The 5′ upstream cis-acting region controlling the expression has also been sequenced, and, although many polymorphisms were shown to be present in this region upstream of the putative translational start codon, it is difficult to define the responsible polymorphism(s) that affect the expression of the DOG1 gene. To judge whether certain haplotypes are responsible for the effect on the expression, more accessions need to be analyzed.
With the isolation of DOG1 the first seed dormancy gene accounting for variation occurring in natural populations has been identified at the molecular level. The identification of the first seed dormancy QTL is an important step toward the elucidation of a genetic pathway that controls this important agronomical trait.
Methods
Plant Material.
NILDOG17-1 has a 14-cM Cvi introgression around DOG1 in a Ler background as previously described by Alonso-Blanco et al. (24). Ler (NW20), Cvi (N8580), An-1 (N944), Kond (CS6175), Col (CS907), Kas-2 (N1264), Sha (CS929), and the transgenic lines SALK 000867 (N500867), SM_3.20808 (N105944), SM_3.20873 (N105997), and SM_3.20886 (N106006) were provided by the Nottingham Arabidopsis Stock Centre, and Fei-0 (CS22645) was a gift from Carlos Alonso-Blanco (Centro Nacional de Biotecnología, Madrid, Spain). The morphological marker line carrying the gl3 and tz mutation was constructed in our laboratory (43).
Mutant Isolation.
Approximately 10,000 M0 seeds of NILDOG17-1 were soaked in tubes containing 10 mM KNO3 and 0.2% agar for 20 h at 4°C, mutagenized with 300 Gy of γ-irradiation, and dispersed on soil in an air-conditioned greenhouse. M2 seeds were harvested from groups of ≈100 M1 plants. In total there were 90 bulks, and a few hundred seeds of every bulk were sown on water-saturated filter paper (no. 595; Schleicher & Schuell, Dassel, Germany) in Petri dishes on the day of harvest. Seeds that germinated within 3 days after seed harvest were selected as putative mutants. Freshly harvested NILDOG17-1 seeds did not germinate under these conditions. The mutant phenotype was confirmed by retesting the germination of the seeds harvested from the plants grown from these selected seedlings. To discard possible nondormant contaminants, the selected mutants were checked for the presence of the Cvi introgression by using Cleaved Amplified Polymorphic Sequence marker DFR (TT3 gene; the Arabidopsis Information Resource, www.arabidopsis.org).
Growth Conditions.
The growth conditions used were described in ref. 39. The ga1 mutants were germinated on 10 μM GA4+7, and the resulting seedlings were planted in the greenhouse, where they were sprayed once a week with 100 μM GA4+7 to stimulate growth, anther development, and seed production. Seeds were harvested in cellophane bags and stored in a cardboard box at room temperature.
Seed Dormancy Measurements and Germination Assays.
Germination tests in water under white light were performed at each time point by incubating seeds for 1 week as follows. Between 50 and 100 seeds of a genotype were evenly sown on a filter paper soaked with 0.7 ml of demineralized water, 0–100 μM GA4+7, or 0–100 μM ABA (mixed isomers; Sigma-Aldrich, St. Louis, MO) in a 6-cm Petri dish. GA4+7 was dissolved in a few drops of 1 M KOH and diluted with a phosphate citrate buffer containing 3.3 mM K2HPO4·3H2O and 1.7 mM citric acid (pH 5.0).
To allow germination the Petri dishes were placed in moisture chambers consisting of plastic trays containing a filter paper saturated with tap water and closed with transparent lids. These moisture chambers were transferred to a climate room (25°C, 16-h light/day, model TL57; Philips, Eindhoven, The Netherlands), and germination was scored after 7 days. The total number and the number of germinating seeds were scored, and the percentage of germinating seeds was calculated.
For germination in the dark, seeds were sown under green-light conditions, and the Petri dishes were wrapped in two layers of aluminum foil and stored in a closed box under the same conditions as the light-germinated seed.
Cold stratification was performed by placing the imbibed seeds on water-soaked filter paper for 7 days at 4°C in darkness.
Germination analyses to study the genetic behavior of the different DOG1 alleles were performed on F3 seeds from individual F2 plants of the crosses Ler × NILDOG17-1 and Ler × dog1. Seeds of Ler × NILDOG17-1 were sown 28 days after seed harvest to allow the Ler seeds to germinate 100% and seeds of Ler × dog1 were sown directly after seed harvest.
DNA Extractions.
DNA was extracted by using the Wizard magnetic 96 (FF3760; Promega, Madison, WI) DNA isolation kit.
Fine-Mapping of DOG1.
Details of the molecular markers used for the fine-mapping of DOG1 can be found in Table 2, which is published as supporting information on the PNAS web site.
Complementation of DOG1.
A genomic DNA fragment of 5.6 kb of DOG1-Cvi was amplified by PCR using primers EcoRI-5791 (CCGAATTCTCGTCTTGGAATGTGTTTCCCATGG) and KpnI-10944 (CCGGTACCAAATTGTTTGTGCATGCTTCAGC) and AccuPrime Pfx DNA polymerase (Invitrogen Life Technologies, Carlsbad, CA). This PCR product was cloned into the pCR-Blunt II-TOPO vector (Invitrogen Life Technologies). From this vector it was moved to pBIBkan-hyg (31) and inserted at the EcoRI/KpnI restriction sites to create pBIB DOG1-Cvi. pBIB DOG1-Cvi was introduced by electroporation to A. tumefaciens (AGLO strain) (44). The transformed AGLO strain was used to transform Ler plants by floral dip. T1 primary transformants selected on the basis of their resistance to hygromycin B were transferred to the greenhouse to set T2 seeds that were collected to perform germination experiments.
Sequencing.
PCR products were sequenced by using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). Sequence products were separated by Greenomics (Wageningen, The Netherlands).
RNA Extractions.
RNA extractions were performed according to ref. 45, with small modifications. The phenol, phenol/chloroform, and chloroform extractions were performed only once. The RNA pellet was dissolved in 50 μl of DEPC-treated MQ and subsequently applied to a RNeasy column to clean the RNA and to perform DNA treatment (RNA purification; Qiagen, Valencia, CA).
Northern Blot Analyses.
Northern blot analysis were performed following the protocol supplied with the Hybond-N nylon membranes (Amersham Pharmacia, Uppsala, Sweden) by using 10 μg of RNA isolated as described above. DOG1 expression was detected by using an 866-bp probe corresponding to the coding region of the DOG1 gene.
Pyrosequencing.
Pyrosequencing was used to quantify allele frequencies and transcript forms (alternative splicing) in RT-PCR products. Sequence primers were developed by using SNP Primer Design software (version 1.01) from Biotage (Uppsala, Sweden).
cDNA synthesis.
For the first-strand cDNA synthesis 800 ng total RNA was used in a volume of 20 μl by using M-MLV reverse transcriptase (Invitrogen Life Technologies) according to the manufacturer's protocol. The standard dT12–18 adapted primer was used.
Alternative splicing.
For RT-PCR 10 μl of 8× diluted cDNA was used with 10 pmol of each primer, 100 μM of each deoxynucleotide, 1 unit of Super TaqDNA polymerase, and the Super TAQ reaction buffer provide by Sphaero Q (Gorinchem, The Netherlands). For PCR the following primers were used: pyro F2, biotinylated GAGTGGGGAACTATGAGAGATCGT; pyro R2, CCCACTATTCACAGTTGTACATGC. Primer positions are indicated in Fig. 3.
The SNP was analyzed by using sequence primer Seq2 (TGTACATGCATCGAATATTA). To discriminate between cDNA transcripts γ and δ a second PCR was performed by using the following: pyro F2, biotinylated GAGTGGGGAACTATGAGAGATCGT; pyro R3, GCAAAATGCCACGACGTGA. The SNP was analyzed by using sequence primer Seq3 (TGCCACGACGTGAATAAA).
Quantification of allele frequencies.
For RT-PCR 2 μl of 5× diluted cDNA was used under the same PCR conditions as mentioned above for the alternative splicing. For PCR the following primers were used: pyro F1, TACAAGAAGACGCAGCGGATAT; pyro R1, biotinylated CGGAGATAGAATCCCGAGGA. The SNP was analyzed by using sequence primer Seq1 (GGAGAATGTCGGAGAG).
As controls, DNA of F1 plants was used. The relative expression of DOG1-Ler to the other DOG1 of the other accessions should be 1 in the F1 genomic DNA.
To control for possible position effects in the thermocycler, cDNA samples together with DNA extracted from heterozygous plants were randomly distributed across 96-well plates before PCR.
Pyrosequencing was carried out according to the manufacturer's standard protocols (Biotage).
Supplementary Material
Acknowledgments
We thank Dr. J. Schaart for help in pyrosequencing and Dr. W. Soppe and Dr J. de Meaux for critically reading the manuscript. This work was supported by grants from The Earth and Life Sciences Foundation subsidized by the Netherlands Organization for Scientific Research and European Union program NATURAL (Contract QLG2-CT-2001-01097).
Abbreviations
- QTL
quantitative trait locus
- ABA
abscisic acid
- GA
gibberellin
- NIL
near isogenic line
- Ler
Landsberg erecta.
Footnotes
References
- 1.Bewley JD. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donohue K. Seed Sci Res. 2005;15:175–187. [Google Scholar]
- 3.Fenner M, Thompson K. The Ecology of Seeds. Cambridge, UK: Cambridge Univ Press; 2005. [Google Scholar]
- 4.Ooms JJJ, Léon-Kloosterziel KM, Bartels D, Koornneef M, Karssen CM. Plant Physiol. 1993;102:1185–1191. doi: 10.1104/pp.102.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nambara E, Keith K, McCourt P, Naito S. Development (Cambridge, UK) 1995;121:629–636. [Google Scholar]
- 6.Bäumlein H, Miséra S, Luersen H, Kölle K, Horstmann C, Wobus U, Müller AJ. Plant J. 1994;6:379–387. [Google Scholar]
- 7.Luerssen H, Kirik V, Herrmann P, Misera S. Plant J. 1998;15:755–764. doi: 10.1046/j.1365-313x.1998.00259.x. [DOI] [PubMed] [Google Scholar]
- 8.Meinke DW, Franzmann LH, Nickle TC, Yeung EC. Plant Cell. 1994;6:1049–1064. doi: 10.1105/tpc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Cell. 1998;93:1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- 10.Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. Proc Natl Acad Sci USA. 2001;98:11806–11811. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debeaujon I, Léon-Kloosterziel KM, Koornneef M. Plant Physiol. 2000;122:403–414. doi: 10.1104/pp.122.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koornneef M, Van der Veen JH. Theor Appl Genet. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- 13.Koornneef M, Jorna ML, Brinkhorst-van der Swan DLC, Karssen CM. Theor Appl Genet. 1982;61:385–393. doi: 10.1007/BF00272861. [DOI] [PubMed] [Google Scholar]
- 14.Kucera B, Cohn MA, Leubner-Metzger G. Seed Sci Res. 2006;15:281–307. [Google Scholar]
- 15.Casal JJ, Sanchez RA. Seed Sci Res. 1998;8:317–329. [Google Scholar]
- 16.Papi M, Sabatini S, Bouchez D, Camilleri C, Costantino P, Vittorioso P. Genes Dev. 2000;14:28–33. [PMC free article] [PubMed] [Google Scholar]
- 17.Gualberti G, Papi M, Bellucci L, Ricci I, Bouchez D, Camilleri C, Costantino P, Vittorioso P. Plant Cell. 2002;14:1253–1263. doi: 10.1105/tpc.010491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Léon-Kloosterziel KM, van de Bunt GA, Zeevaart JAD, Koornneef M. Plant Physiol. 1996;110:233–240. doi: 10.1104/pp.110.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peeters AJ, Blankestijn-De Vries H, Hanhart CJ, Léon-Kloosterziel KM, Zeevaart JA, Koornneef M. Physiol Plant. 2002;115:604–612. doi: 10.1034/j.1399-3054.2002.1150415.x. [DOI] [PubMed] [Google Scholar]
- 20.Salvi S, Tuberosa R. Trends Plant Sci. 2005;10:297–304. doi: 10.1016/j.tplants.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Bentsink L, Soppe W, Koornneef M. In: Seed Development, Dormancy, and Germination. Bradford KJ, Nonogaki H, editors. Oxford: Blackwell; 2007. pp. 113–132. [Google Scholar]
- 22.Koornneef M, Bentsink L, Hilhorst HWM. Curr Opin Plant Biol. 2002;5:33–36. doi: 10.1016/s1369-5266(01)00219-9. [DOI] [PubMed] [Google Scholar]
- 23.Han F, Ullrich SE, Clancy JA, Romagosa I. Plant Sci. 1999;143:113–118. [Google Scholar]
- 24.Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-De Vries H, Koornneef M. Genetics. 2003;164:711–729. doi: 10.1093/genetics/164.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao W, Clancy JA, Han F, Prada D, Kleinhofs A, Ullrich SE. Theor Appl Genet. 2003;107:552–559. doi: 10.1007/s00122-003-1281-5. [DOI] [PubMed] [Google Scholar]
- 26.Takeuchi Y, Lin SY, Sasaki T, Yano M. Theor Appl Genet. 2003;107:1174–1180. doi: 10.1007/s00122-003-1364-3. [DOI] [PubMed] [Google Scholar]
- 27.Debeaujon I, Koornneef M. Plant Physiol. 2000;122:415–424. doi: 10.1104/pp.122.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamaguchi S, Sun T, Kawaide H, Kamiya Y. Plant Physiol. 1998;116:1271–1278. doi: 10.1104/pp.116.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grappin P, Bouinot D, Sotta B, Miginiac E, Jullien M. Planta. 2000;210:279–285. doi: 10.1007/PL00008135. [DOI] [PubMed] [Google Scholar]
- 30.Ali-Rachedi S, Bouinot D, Wagner MH, Bonnet M, Sotta B, Grappin P, Jullien M. Planta. 2004;219:479–488. doi: 10.1007/s00425-004-1251-4. [DOI] [PubMed] [Google Scholar]
- 31.Becker D. Nucleic Acids Res. 1990;18:203. doi: 10.1093/nar/18.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Servant F, Bru C, Carrere S, Courcelle E, Gouzy JP, Peyruc D, Kahn D. Brief Bioinform. 2002;3:246–251. doi: 10.1093/bib/3.3.246. [DOI] [PubMed] [Google Scholar]
- 33.Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- 34.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S. Plant Cell. 2003;15:1591–1604. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lister C, Dean C. Plant J. 1993;4:745–750. [Google Scholar]
- 38.Clerkx EJM, El Lithy ME, Vierling E, Ruys GJ, Blankestijin-De Vries H, Groot SPC, Vreugdenhil D, Koornneef M. Plant Physiol. 2004;135:432–443. doi: 10.1104/pp.103.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El-Lithy ME, Bentsink L, Hanhart CJ, Ruys GJ, Rovito D, Broekhof JL, van der Poel HJ, van Eijk MJ, Vreugdenhil D, Koornneef M. Genetics. 2006;172:1867–1876. doi: 10.1534/genetics.105.050617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wittkopp PJ, Haerum BK, Clark AG. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- 41.de Meaux J, Goebel U, Pop A, Mitchell-Olds T. Plant Cell. 2005;17:676–690. doi: 10.1105/tpc.104.027839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raz V, Bergervoet JH, Koornneef M. Development (Cambridge, UK) 2001;128:243–252. doi: 10.1242/dev.128.2.243. [DOI] [PubMed] [Google Scholar]
- 43.Koornneef M, Van Eden J, Hanhart CJ, Stam P, Braaksma FJ, Feenstra WJ. J Hered. 1983;74:265–272. [Google Scholar]
- 44.Lazo GR, Stein PA, Ludwig RA. Biotechnology. 1991;9:963–967. doi: 10.1038/nbt1091-963. [DOI] [PubMed] [Google Scholar]
- 45.Vicient CM, Delseny M. Anal Biochem. 1999;268:412–413. doi: 10.1006/abio.1998.3045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.