Abstract
Outbreaks of Salmonella Dublin infections were recorded in 25 Danish mink and fox farms. All farms suffered extensive disease problems; clinical and pathological observations included abortion, stillbirths, necrotizing endometritis, and increased mortality. By genotyping with pulsed-field gel electrophoresis and amplified fragment length polymorphism, all isolates of S. Dublin had indistinguishable patterns. The outbreaks took place during April and May, around the time of whelping. During this period, mink are particularly susceptible to Salmonella infections. All affected farms were served by the same feed factory and it was concluded that a batch of contaminated feed was responsible for the outbreaks, although repeated culture of feed samples collected during the same period were negative. No other likely source could be identified. The results emphasize the importance of strict hygiene measures at feed factories and the proper use of ingredients of known Salmonella status, in particular during the whelping season. Infected mink farms did not have a higher risk of outbreak of salmonellosis in the year following the outbreak.
Résumé
Flambée d’avortements associée à Salmonella Dublin dans des élevages d’animaux à fourrure du Danemark. Des flambées d’infection à Salmonella Dublin ont été rapportées dans 25 élevages de visons et de renards du Danemark. Tous les élevages montraient de nombreux problèmes de santé; les observations cliniques et pathologiques mentionnaient des cas d’avortements, de morti-natalités, d’endométrites nécrosantes et de mortalité plus élevés. Le génotypage par électrophorèse en champ pulsé et le «amplified fragment length polymorphism» ont montré que tous les isolats du S. Dublin présentaient des profils indiscernables. Les flambées se sont produites en avril et en mai, en période de mise bas. Pendant cette période, les visons sont particulièrement susceptibles aux infections à Salmonella. Tous les élevages affectés recevaient leur nourriture de la même usine et il a été conclu qu’un lot d’aliments contaminés était responsable de la situation, même si des cultures répétées d’échantillons d’aliments recueillis pendant cette période se sont révélées négatives. Aucune autre source possible n’a pu être identifiée. Les résultats montrent toute l’importance des mesures strictes d’hygiène dans les usines d’aliments et de l’utilisation adéquate d’ingrédients dont l’état sanitaire vis-à-vis des salmonelles est connu, particulièrement en période de mise bas. Les élevages de vison affectés ne présentaient pas de plus hauts risques de flambées de salmonellose l’année suivante.
(Traduit par Docteur André Blouin)
Introduction
Salmonella infection as a cause of abortion in mink (Mustela vison) was first described by Gorham et al (1). Records from the Danish Institute for Food and Veterinary Research files indicate that isolation of Salmonella from admissions during 1941–1948 were very rare (17 out of 3079 case submissions); abortion in Danish mink caused by Salmonella Dublin was first recorded in 1957 (2). The disease was also reported in mink several years ago from Norway (3) and more recently from Sweden (4).
Salmonella Dublin is generally regarded as a host-adapted serotype, mostly restricted to bovine species. In cattle, S. Dublin is one of the most important serotypes and infects both young and adult animals, where it causes enteritis and systemic disease. Clinical signs include diarrhea, abortion, fever and drop in milk yield, and pneumonia. The mortality can be very high, and the infection has a marked tendency to persist with constant or intermittent excretion of Salmonella in the feces. Salmonella Dublin seldom infects humans, but when it occurs, it has a greater tendency to become invasive than most other serotypes (5,6).
Danish mink fur production consists of approximately 2250 mink farms with an average of around 1000 breeding females per farm. The average litter size has, for a number of years, been stable around 5.3 mink kits per dam and it varies only slightly between genotypes. A few (around 100) Danish farmers produce foxes and most foxes are raised in farms that also produce mink. Feed is produced freshly by 16 feed factories and delivered fresh daily. The number of breeding mink females fed by each feed factory varies from 40 000 up to several hundred thousands. The composition of the feed varies significantly during the growth period, being based on industrial fish products, fishmeal, fish offal, chicken pulp, and cereals—mainly barley. Blood products and slaughter offal are used in varying but usually limited amounts. The following would compose a typical mink feed in April: Fish offal 46%, industrial fish 14%, salmon offal 5%, chicken pulp 12%, swine pulp 5%, cereals 6%, corn gluten 2%, blood products 2%, soy oil 0.3%, and water added to 100%.
Clustering of widespread, lethal infections in mink caused by feed-borne bacteria are rarely documented. However, during the spring of 2000, a number of Danish mink farms, all served by the same feed factory, experienced serious infections caused by S. Dublin. In the present report, clinical, pathological, bacteriological, and epidemiological aspects of the outbreak are presented and discussed.
Materials and methods
Animals and pathology procedures
Mink females, stillbirths, and aborted fetuses from Danish mink farms suffering a severe outbreak of salmonellosis in 2000 were sent to the Danish Institute for Food and Veterinary Research (DFVF) for diagnostic examination. Autopsy and thorough laboratory examination was carried out on a representative number of carcasses (n = 52), representing all affected farms. More carcasses were submitted to the laboratory, but they were not subjected to thorough laboratory examination due to pronounced cadaverosis or obvious identical disease conditions. Carcasses were subjected to a routine necropsy procedure with recording of weight, sex, and gross pathological changes. Bacteriological, serological, histopathological, and virological examinations were performed when considered relevant, as judged from the gross pathological changes. Intestinal tissue routinely taken at the level of the pancreas was placed in 10% buffered formaldehyde for examination for changes consistent with mink virus enteritis. Otherwise, histopathologic examination was performed when indicated (when gross pathological changes were noted in the parenchymal organs). Formalin-fixed tissue was processed, embedded in paraffin, cut 4 μm thick, and stained with hematoxylin and eosin before microscopic examination.
Bacteriological examination and culture methods
Examination of material from mink organs —
Material from the intestine, spleen, liver, uterus, and lung was sampled for bacteriological investigation, although material from all of these organs was not sampled from all animals. The material was subjected to standard culture methods by inoculation onto blood agar (Blood agar base No. 2; Oxoid, Basingstoke, UK, supplemented with 5% calf blood) and Drigalski agar (7). In addition, material from the intestines and liver was sampled for detection of Salmonella in selective media as follows: an approximately 1-cm long transverse section of the rectum and approximately 1 g of liver tissue were sampled and transferred to buffered peptone water (BPW; Merck, VWR International, Albertslund, Denmark) in a 1:10 ratio (w/vol) and subjected to preenrichment by incubation at 37ºC for 16–20 h, where upon 100 μL were transferred to 10 mL Rappaport-Vassiliadissoya peptone broth (RVS, Oxoid) and further incubated at 41.5ºC for 20–24 h. A loopful, corresponding to 10 μL, of both broth cultures was then each streaked onto Rambach agar (Merck) and incubated at 37ºC for 18–24 h.
Examination of feed samples —
Mink feed factories are obliged to collect and store a daily sample from the feed production and all the feed factories participate in a voluntary quality control program including an annual inspection. Samples of the mink feed from the suspected feed factory, spanning a period from April 10 through May 8, were examined for the presence of Salmonella. Feed was produced on 22 d during this period; consequently, 22 stored feed samples were tested individually. Samples, approximately 25 g, were transferred to BPW in a 1:10 (w/vol) ratio for preenrichment by incubation at 37ºC for 16–20 h. Three different media were inoculated with the BPW: 1) 100 μL was transferred to 10 mL RVS broth; 2) 100 μL was transferred to 10 mL of selenite broth (per liter medium:sodium hydrogen selenite [Oxoid L 121] 4g); bactopeptone (Difco 0118-17-0; Detroit, Michigan, USA) 5g; lactose (Merck 7652) 4g; disodium hydrogen phosphate × 12H2O (Merck 6579) 10g; pH 7.0, and 3) 3 drops of 50 mL were deposited onto modified semi-solid Rappaport-Vassiliadis medium (MSRV medium base [Oxoid CM910] with MSRV selective supplement [Oxoid SR161E]). The RVS was incubated at 41.5ºC for 20–24 h and the selenite broth at 37ºC for 18–24 h. A loopful, corresponding to 10 μL, of each broth culture was then streaked onto both Rambach agar and modified brilliant green agar (Oxoid CM329) and incubated at 37ºC for 18–24 h. The MSRV medium was incubated at 41.5ºC for 20–24 h. Some of the basic feed ingredients (raw herring, hearts from swine, vitamin mixture, and hemoglobin flour) still in stock at May 3–16 were examined for the presence of Salmonella by using the same method. Hemoglobin flour was stored in a large tower silo and holes were drilled in 10 different places to enable the sampling procedure. Furthermore, the results from a voluntary inspection carried out March 9, 2000 were included in the evaluation.
Serotyping
Salmonella-suspect colonies from Rambach or Drigalski agar were first tested for agglutination with polyvalent Salmonella O-serum (Statens Serum Institut; Copenhagen, Denmark). If they reacted positively, they were serotyped according to the Kauffmann-White scheme (8).
Pulsed-field gel electrophoresis
One or more isolates from each infected farm was subjected to genotyping by using pulsed-field gel electrophoresis (PFGE), as described by Nauerby et al (9). XbaI, NotI, and SpeI (New England BioLabs; Beverly, Massachusetts, USA) were used as restriction enzymes. The electrophoresis conditions were 6 V/cm at 12°C for 20 h. The ramping times for SpeI and XbaI were 7–12 s for 10 h, followed by 20–40 s for another 10 h; for NotI, the ramping time was 0.5–15 s for 18 h. Following electrophoresis, the gel was stained in aqueous ethidium bromide, destained in water, and photographed under 254 nm UV transillumination.
Amplified fragment length polymorphism
Amplified fragment length polymorphism (AFLP) was carried out on 1 isolate from each of 20 different farms and, for comparison, on 23 Danish isolates of S. Dublin from cattle, together with 3 S. Dublin reference strains, SARB12, SARB13, and SARB14 from the Salmonella Genetic Stock Centre (SGSC; Calgary, Alberta) (10).
By using a commercial kit (EASY-DNA kit; Invitrogen, Carlsbad, California, USA), DNA was extracted and AFLP analysis was then performed with 3 μL of the DNA preparations digested with 10 units of Bgl II and BspDI (New England Biolabs). Ligation to Bgl II and BspDI adaptor oligonucleotides (11), PCR with BGL2F-0 and BSPD1-0 primers (11), and detection of the amplification products were all done under conditions previously described by Kokotovic et al (12). Data collection and preprocessing was done by using a fragment analysis software (GENESCAN; Applied Biosystems, Foster City, California, USA). The analysis was performed by using analysis software (GENESCAN; Applied Biosystems) and a gel documentation software (BioNumerics; Applied Maths, Sint-Martens-Latem, Belgium).
Serological analysis for Aleutian disease virus
All farms were tested for Aleutian disease (AD) through a mandatory national surveillance program. For this program, ten percent of the breeding stock on all Danish farms is tested during May and June, including all barren females and dams whose entire litter dies during the first couple of weeks. Infected stocks are stamped out; after disinfection, new breeding stocks from AD-free farms can be introduced.
Blood samples or fluid from the thoracic cavity from all the carcasses were examined by a counter current electrophoresis method (13) for the presence of antibodies against AD virus.
Epidemiology
All farms (n = 25) with a laboratory-based diagnosis of S. Dublin infection during breeding season 2000 were included in the epidemiological study. Twenty-two farms were breeding only mink, 2 farms raised both mink and foxes, and 1 farm bred only foxes. The foxes constituted a minor fraction of the animals. Additionally, the breeding season of foxes is prolonged compared with that of mink, and there are differences between the different genotypes of foxes. Therefore, likely not all foxes were exposed at the time they were most susceptible to infection, so the foxes were left out of the epidemiological calculations. Six of the mink farms suffered from an intercurrent reinfection of AD. Farm data from the preceding breeding season could not be used for calculation of epidemiological data, as many of the farms had restocked between 1999 and 2000 as a result of an AD diagnosis in 1999. Therefore, average figures for 1999 from that particular region of Denmark were used as the reference.
In order to calculate the loss of dams in the farms in 2000, the number of dams left for whelping was enumerated by April 15, as well as by July 1. The normal average mortality rate in mink farms in the year 1999 for that region of Denmark and for each genotype was subtracted from the total mortality of the infected farms in 2000 and the difference was denoted as the excessive dam mortality (EDM). Due to problems in distinguishing between late abortions, stillbirths, and early postnatal mortalities, the excessive kit mortality (EKM) had to be based on estimates. The average litter size for each genotype in the geographical region from breeding season 1999 was used as the baseline and the perinatal mortality rate was calculated as the difference between the actual number of mink kits in the farm by July 1st and the expected number of kits based on the number of breeding females. Due to the difference between genotypes in sensitivity to infections, as well as the average litter size, the losses were adjusted for the genotype. The impact of intercurrent AD relapse (n = 6) was compared between farms breeding wildtype (brown) mink (n = 23) by use of a t-test. The mean excess dam and kit loss with Wald-95% confidence limits was estimated with a general statistical procedure (GENMOD; Statistical Analysis System [14]).
In 3 selected mink farms, all carcasses, from the date of diagnosing the outbreak till pelting time in November, were necropsied at the DFVF in order to determine the persistence of S. Dublin infection at farm level. Likewise, 3 of the farms that suffered from the S. Dublin outbreak in 2000 were selected and closely supervised in 2001 for the occurrence of stillbirths and abortions, or any other type of unusual mortality.
Results
The 1st case of S. Dublin-infection in mink was detected in mink dead at a mink farm on April 27, 2000. Within 2 wk, 25 fur farms (22 mink farms, 2 fox farms, and 1 farm with both foxes and mink) were registered as suffering from severe disease problems due to infection with S. Dublin. The same feed kitchen served all of the affected fur farms. This feed kitchen served a total of 60 farms with 58 000 dams during the outbreak. No other cases of infection with S. Dublin were seen in fur animals in Denmark during that period. Information from the farms — as collected by the farmer and the practicing veterinarian during the outbreak — consistently included evidence of loss of appetite, increased numbers of barren females, abortions (dead fetuses covered with fetal membranes), stillbirths, perinatal mortality of pregnant mink, and early kit mortality. However, the clinical signs were unevenly distributed among the affected farms.
Postmortem findings
The mink females examined were, in general, in a good body condition. The most common macroscopic lesion at necropsy was a characteristic dark red, very fragile uterus with severe, necrotizing endometritis, often complicated with endometrial rupture and concurrent, diffuse, purulent peritonitis. The uterine contents consisted of crimson, thick, odorless fluid containing partly macerated fetuses and no sign of fetal membranes. The cervix was not dilated, but most of the females exhibited slight vaginal discharge with tangling of the vulval hairs. The spleen was very enlarged in some of the females and scattered pinpoint bleedings in the lungs were observed, but these pathological findings were not consistent. Histopathologically, the endometrium was dominated by a deep, necrotizing endometritis and myometritis.
Bacteriologic findings
From the index cases, S. Dublin was isolated from the uterus, liver, spleen, intestines, and lung. There was massive growth on blood agar plates and Drigalski agar when they were streaked directly from affected organs. Salmonella Dublin was recovered also from the intestines and liver on Rambach agar after enrichment. The majority of the samples submitted from the affected farms to DFVF during the outbreak period were positive for S. Dublin, either by streaking directly onto solid media or after medium enrichment. Isolates from all farms were stored by freeze-drying for subsequent typing. The examination of feed samples from the period April 10 – May 8 revealed no Salmonellae even though different test methods were applied. Also examination of the remaining feed ingredients (raw herring, vitamin mixture, haemoglobin flour and raw swine hearts) revealed no Salmonellae.
Pulsed-field gel electrophoresis and amplified fragment length polymorphism typing
Pulsed-field gel electrophoresis revealed only a single pattern with all 3 restriction enzymes (data not shown). By AFLP, all isolates from mink were indistinguishable (data not shown).
Epidemiology
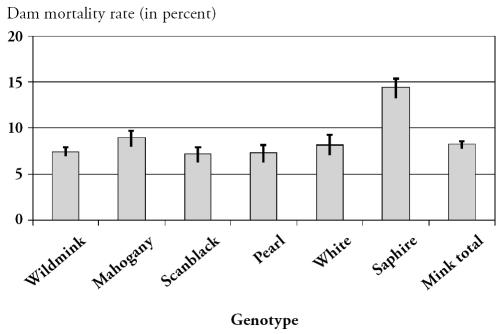
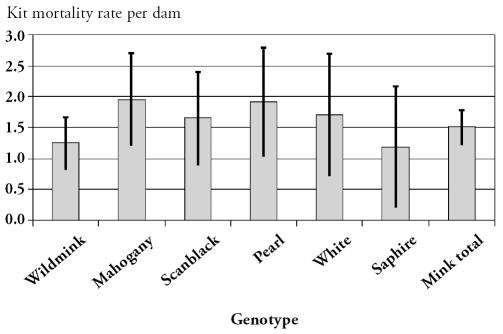
There were 42 840 mink dams on the affected farms. During the outbreak, 3677 died, together with 59 278 kits. The dam mortality is shown in Figure 1 and the kit mortality is shown in Figure 2. Out of the 23 farms raising wildmink genotypes, 6 were infected with AD virus. However, there was no statistically different EDM (P = 0.48) or EKM (P = 0.98) between AD-infected and uninfected farms. In the 3 follow-up farms, no clinical signs of salmonellosis were noted and morbidity and mortality during the following breeding season were at a normal level. No dead females were submitted to the laboratory, the mink kits examined showed no signs of salmonellosis, and Salmonella could not be cultured from organs. Furthermore, the 3 follow-up farms had normal production and the farmers experienced no significant disease problems during the growth period.
Figure 1.
The overall mean excess dam mortality rate (in percent) for each genotype with 95% confidence intervals.
Figure 2.
The overall mean excess kit mortality rate per litter for each genotype with 95% confidence intervals.
Discussion
This is the first recorded outbreak of massive abortions, stillbirths, and deaths among pregnant mink in Scandinavia. Infection with S. Dublin was recorded a few times in the late 1940s in Danish mink and fox farms. During that time, the finding was incidental, and only 1 outbreak, in 1957, with a limited number of abortions has been described (3). The origin of the infection in all cases was believed to be slaughter offal from cattle (3,15). Empirical data suggest that the mink is only susceptible to Salmonella infection during pregnancy (1); therefore, raw slaughter offal should not be used in the mink diet during the pregnancy period.
Although extensive testing was carried out, Salmonellae were not detected in the stored, frozen feed samples, or in any of the remaining dry matter constituents from the feed factory. As mink feed is a day-to-day production, this negative finding is not necessarily indicative of the bacterium not being transmitted via the feed, rather it reflects that the proper test material was not available. The Salmonella bacterium may have been unevenly distributed in the feed, with some parts being free from Salmonella and others being severely infected. However, Salmonella infection has characteristic and often dramatic clinical consequences and pathological signs in mink, we assume that clinically, the infection is not easily overlooked, and that all cases in the actual outbreak were detected. All farmers served by the feed factories in the geographical region of the outbreak were urged to submit material from suspected cases, but all mink submitted to DFVF from farms served by feed factories other than the one serving the affected farms were tested and found to be negative for Salmonella spp. during the whelping period.
According to an investigation carried out by the Danish Veterinary Health Council, slaughter offal from unspecified animal species and bovine rumens were found on the premises of the feed factory serving the affected farms. The outcome of a subsequent court case was that the feed factory was found guilty from circumstantial evidence and liable to pay the farmers full compensation for their losses. During previous, much smaller, outbreaks in Norway (3) and Sweden (4), the bacterium was detected in specific feed ingredients.
In a British study by Williams and Bellhouse (16), several serotypes (S. Senftenberg, S. Typhimurium, S. Dublin, S. Livingstone, S. Menston, S. Enteritidis, S. Bredeney, and S. Infantis) were found in 995 healthy mink from 4 different farms without disease problems. On 1 farm, Salmonella was isolated from male mink in March, but otherwise the animals were examined outside the critical period of gestation. The study also included 316 dead, adult mink from 12 farms suffering disease problems. Salmonellosis was diagnosed in 2 mink (S. Typhimurium and S. Dublin, respectively), of which 1 had died suddenly and the other had died from urolithiasis. It is noteworthy that the British mink were fed a ration of raw broiler offal, day-old chickens, bovine rumens, fish offal, and cereals, and that Salmonella was regularly isolated from the stomach contents of healthy mink, indicating a frequent supply of Salmonella in the feed. Raw slaughter offal is generally not used in Danish mink feed due to the risk of botulism, and the cooked slaughter offal constituents of the feed are continuously examined for Salmonella. Since did not succeed in isolating Salmonella from any of the samples of the produced feed from the actual feed factory, it is an open question how the feed became contaminated. However, it seems reasonable to hypothesize from the circumstantial evidence that the infection was caused by the same source in all farms (the 1st national recording for decades of a very rare disease outbreak in several farms at the same time of the year, severe and identical clinical problems in all affected farms, and all farms served by the same feed factory). Several routes of infection may be thought of, including cross-contamination from raw materials to the finished feed at the feed factory, insufficiently processed slaughter offal, and Salmonella-infected dry products included in the feed. Incidental spread from wild birds or from S. Dublin-infected cattle herds are more remote and quite unlikely possibilities, as these sources would hardly respect the pattern of different feed suppliers in the same geographical area. Water as a common source of S. Dublin is not likely, as the farms were not supplied from the same wells.
Olsen et al (17) found 8 different genomic lines among 35 isolates of S. Dublin of different origin from the United Kingdom, Denmark, Sweden, France, and Tanzania on the basis of a number of typing methods, including PFGE. However, when scoring only bands larger than 100 kb, PFGE with NotI as the restriction enzyme revealed only 4 patterns. In the present study, all isolates from mink displayed the same PFGE pattern with NotI. This pattern was similar to pattern I and IV, maybe identical to pattern IV of Olsen et al (17). Amplified fragment length polymorphism has been shown to be a more discriminatory technique than PFGE for S. Dublin (18). However, the AFLP results also revealed only a single clonal line. The AFLP and PFGE patterns obtained in the present study indicated that only a single clonal line of S. Dublin was involved; therefore, possibly only a single source was involved, but it is not possible to reach a conclusion about the origin of the contamination.
To prevent even rare outbreaks of salmonellosis in mink during the susceptible period of gestation and lactation, all the ingredients, raw as well as processed, for mink wet feed should be subjected to thorough bacteriological control, including culture for Salmonella on selective media. Furthermore, controlled hygiene at the feed factories in the form of Hazard Analysis Critical Control Points (HACCP) principles should be considered to eliminate/reduce the risk of contamination of the final product. CVJ
References
- 1.Gorham JR, Cordy DR, Quortrup ER. Salmonella infections in mink and ferrets. Am J Vet Res. 1949;10:183–192. [PubMed] [Google Scholar]
- 2.Knox Seith B. 1966. Infectious diseases, parasitic diseases, feed intoxication. In: Nordisk Håndbog for Minkopdrættere (Mink production. Nordic handbook for mink ranchers; in Danish), 2nd ed. Copenhagen: Det Kgl. Danske Landhusholdningsselskab, 1966:188–213.
- 3.Nordstoga K, Loftsgård G. Et utbrudd av smittsom abort — salmonel-lose — hos mink (An outbreak of contagious abortions — salmonellosis — in mink; in Norwegian) Norsk Pelsdyrblad. 1970;44:425–426. [Google Scholar]
- 4.Berndtsson L, Mejerland T. Salmonella Dublin påvisad hos mink (Salmonella Dublin found in mink; in Swedish) Våra Pälsdjur. 1995;66:86. [Google Scholar]
- 5.Kingsley RA, Bäumler AJ. Host adaptation and the emergence of infectious disease: The Salmonella paradigm. Mol Microbiol. 2000;36:1006–1014. doi: 10.1046/j.1365-2958.2000.01907.x. [DOI] [PubMed] [Google Scholar]
- 6.Uzzau S, Brown JD, Wallis T, et al. Host adapted serotypes of Salmonella enterica. Epidemiol Infect. 2000;125:229–255. doi: 10.1017/s0950268899004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vulfson L, Pedersen K, Chriél M, et al. Serogroups and antimicrobial susceptibility among Escherichia coli isolated from farmed mink (Mustela vison Schreiber) in Denmark. Vet Microbiol. 2001;79:143–153. doi: 10.1016/s0378-1135(00)00343-6. [DOI] [PubMed] [Google Scholar]
- 8.Popoff MY, Le Minor L. Antigenic formulas of the Salmonella serovars, 7th revision. Paris: WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur, Paris, 1999.
- 9.Nauerby B, Pedersen K, Dietz HH, Madsen M. Comparison of Danish isolates of Salmonella enteritidis PT9a and PT11 from hedgehogs (Erinaceus europaeus) and humans by plasmid profiling and pulsed-field gel electrophoresis. J Clin Microbiol. 2000;38:3631–3635. doi: 10.1128/jcm.38.10.3631-3635.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyd EF, Wang F-S, Beltran P, Plock SA, Nelson K, Selander RK. Salmonella reference collection B (SARB): Strains of 37 serovars of subspecies I. J Gen Microbiol. 1993;139:1125–1132. doi: 10.1099/00221287-139-6-1125. [DOI] [PubMed] [Google Scholar]
- 11.Torpdahl M, Skov MN, Sandvang D, Baggesen DL. Genotypic characterization of Salmonella by multilocus sequence typing, pulsed-field gel electrophoresis and amplified fragment length polymorphism. J Microbiol Methods. 2005;63:173–184. doi: 10.1016/j.mimet.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Kokotovic B, Friis NF, Jensen JS, Ahrens P. Amplified-fragment length polymorphism fingerprinting of Mycoplasma species. J Clin Microbiol. 1999;37:3300–3307. doi: 10.1128/jcm.37.10.3300-3307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho HJ, Ingram DG. Antigen and antibody in Aleutian disease in mink. I. Precipitation reaction by agar-gel electrophoresis. J Immunol. 1972;108:555–557. [PubMed] [Google Scholar]
- 14.SAS/STAT User’s Guide, version 8, Cary, North Carolina: SAS Institute Inc. 1999.
- 15.Momberg-Jørgensen HC. Pelsdyrsygdomme (Fur Animal Diseases; in Danish). Copenhagen: Carl Fr. Mortensen, 1952:82–85.
- 16.Williams DR, Bellhouse R. The prevalence of salmonellas in mink. J Hyg (Camb) 1974;72:71–78. doi: 10.1017/s0022172400023238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olsen JE, Skov MN. Genomic lineage of Salmonella enterica serovar Dublin. Vet Microbiol. 1994;40:271–282. doi: 10.1016/0378-1135(94)90116-3. [DOI] [PubMed] [Google Scholar]
- 18.Lindstedt BA, Heir E, Vardund T, Kapperud G. Fluorescent amplified-fragment length polymorphism genotyping of Salmonella enterica subsp. enterica serovars and comparison with pulsed-field gel electrophoresis typing. J Clin Microbiol. 2000;38:1623–1627. doi: 10.1128/jcm.38.4.1623-1627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]