Abstract
Selection of initiation sites for DNA replication in eukaryotes is determined by the interaction between the origin recognition complex (ORC) and genomic DNA. In mammalian cells, this interaction appears to be regulated by Orc1, the only ORC subunit that contains a bromo-adjacent homology (BAH) domain. Since BAH domains mediate protein–protein interactions, the human Orc1 BAH domain was mutated, and the mutant proteins expressed in human cells to determine their affects on ORC function. The BAH domain was not required for nuclear localization of Orc1, association of Orc1 with other ORC subunits, or selective degradation of Orc1 during S-phase. It did, however, facilitate reassociation of Orc1 with chromosomes during the M to G1-phase transition, and it was required for binding Orc1 to the Epstein–Barr virus oriP and stimulating oriP-dependent plasmid DNA replication. Moreover, the BAH domain affected Orc1's ability to promote binding of Orc2 to chromatin as cells exit mitosis. Thus, the BAH domain in human Orc1 facilitates its ability to activate replication origins in vivo by promoting association of ORC with chromatin.
Keywords: cell cycle, DNA replication, Epstein–Barr virus, origin recognition complex, oriP
Introduction
Selection of initiation sites for DNA replication in metazoa involves two components: a DNA replication origin and an origin recognition complex (ORC). DNA replication origins are determined by DNA sequences referred to as replicators (Aladjem and Fanning, 2004) and by epigenetic parameters such as chromatin structure, nuclear organization, transcription, transcription factors, deoxyribonucleotide pools, DNA topology and DNA methylation (Gilbert, 2004; Cvetic and Walter, 2005). Consequently, the density of active replication origins can change from an average of one every 10–20 kb in the rapidly cleaving embryos of frogs, flies and fish (Mechali, 2001) to one every 50–300 kb in the differentiated cells of adult organisms (Berezney et al, 2000). Developmental changes in origin density also occur during specific stages in mammalian development (Norio et al, 2005). Thus, metazoan genomes contain many potential initiation sites for DNA replication, but during development, some of these sites are selectively activated while others are suppressed (‘Jesuit Model'; DePamphilis, 1999).
How do ORCs recognize DNA replication origins? This process appears to be different significantly between yeast and mammals. Yeast ORCs consist of a stable complex of six different subunits that remain bound to chromatin throughout cell division and target specific DNA sequences (Kong and DePamphilis, 2001 and references therein). In contrast, HsORC (species abbreviations; Figure 1B) consists of a stable core complex (ORC(2–5)) consisting of Orc2 through Orc5 that interacts weakly with Orc1 and Orc6 (Dhar et al, 2001a; Vashee et al, 2001; Giordano-Coltart et al, 2005; Ranjan and Gossen, 2006). Nevertheless, the association of Orc1 with ORC(2–5) is essential for prereplication complex assembly and DNA replication in vitro (Giordano-Coltart et al, 2005) and in vivo (Ohta et al, 2003). In vitro, however, metazoan ORCs exhibit little affinity for specific DNA sequences other than a preference for asymmetric A:T-rich regions of the type targeted by the SpORC (Kong et al, 2003; Vashee et al, 2003; Remus et al, 2004 and references therein). Nevertheless, in the differentiated cells of mammals and flies, ORCs appear to be localized at specific genomic sites coincident with DNA replication origins (Keller et al, 2002; Ladenburger et al, 2002; Abdurashidova et al, 2003; Todorovic et al, 2005 and references therein). Thus, the ability of ORC to activate a particular replication origin appears to depend on its ability to interact with DNA as it exists within the nucleus, an interaction that appears to be regulated by Orc1 (reviewed in Depamphilis et al, 2006).
Figure 1.

BAH domain structure and mutations. (A) Computer generated diagrams of the BAH domains in ScOrc1 (aa 53–176), ScRsc2 (aa 412–514) and ScSir3 (aa 53–176) were obtained using CPHmodels 2.0 Server and DeepView Swiss-Pdb Viewer. Green and blue ribbons indicate α-helix and β-sheet structures, respectively. Van der Waals surfaces of a highly conserved glutamic acid residue are indicated. (B) BAH domains were identified using ELM-‘functional sites in proteins' program. ScOrc1 and HsOrc1 BAH-domains consist of 10 β-sheets and three α-helices. Numbers refer to the amino-acid sequence. The conserved glutamic acid is in β-sheet no. 7 in ScOrc1 (Zhang et al, 2002) and no. 5 in chicken polybromodomain protein (Oliver et al, 2005). (C) HsOrc1 landmarks include BAH and AAA+ domains. A nuclear localization signal and a HP1 binding site have been reported (Lidonnici et al, 2004). FH-Orc1(wt)=aa 1–861. FH-Orc1(ΔBAH)=(Δ(1–169)). FH-Orc1(ΔH)=(Δ(98–107)), and FH-Orc1(E111K)=E → K at position 111), N315=aa 1–315. Abbreviations are Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; Xl, Xenopus laevis; Mm, Mus musculus; Cg, Cricetulus griseus; Rn, Rattus norvegicus; Cf, Canis familiaris; Pt, Pan troglodytes; and Hs, Homo sapiens.
All Orc1 proteins contain two highly conserved domains: a bromo-adjacent homology (BAH) near its N-terminus, and an AAA+ homology within its C-terminal half that binds and hydrolyzes ATP. The BAH domain is unique to the Orc1 subunit, but the AAA+ domain appears in Orc1, Orc4 and Orc5. Remarkably, the AAA+ domain is required for Orc1 to support DNA replication in several different organisms (Giordano-Coltart et al, 2005; Takeda et al, 2005 and references therein), whereas the BAH domain is not (Bell et al, 1995; Takeda et al, 2005). Instead, the BAH domain appears to be required only for regulating gene activity by interacting with chromatin binding proteins (Hou et al, 2005; Hsu et al, 2005). However, the same properties that facilitate the ability of Orc1 to regulate gene expression may also facilitate the ability of Orc1 to regulate initiation of DNA replication by regulating the ability of ORC to bind to chromatin.
To explore this hypothesis, mutations were constructed in HsOrc1 that either deleted or disrupted the BAH domain, and the resulting proteins were then characterized by expressing them either constitutively or transiently in human cells. The results revealed that the BAH domain in Orc1 facilitated its ability to bind to chromosomes, interact with DNA replication origins, and initiate DNA replication.
Results
Constitutively expressed recombinant Orc1 as a model for endogenous Orc1
Human 293EBNA1 epithelial cells were constructed that constitutively expressed either an N-FLAG-3(hemagglutinin (HA))-tagged HsOrc1 fusion protein (FH-Orc1) or a genetically altered FH-Orc1 protein. The results described below demonstrate that constitutively expressed FH-Orc1 modeled its endogenous counterpart: FH-Orc1 was assembled into ORCs in the nuclei of G1-phase cells where it preferentially bound a DNA replication origin (oriP) and stimulated oriP-dependent plasmid DNA replication. Then, like endogenous Orc1, FH-Orc1 was selectively degraded during S-phase. The fact that stimulation of oriP-dependent DNA replication required the BAH domain in FH-Orc1 confirmed that FH-Orc1 was functionally, as well as physically, active. The fact that all three FH-Orc1 BAH domain mutants exhibited the same phenotype in each experiment confirmed that the phenotype resulted from the mutation and not from differences in protein expression levels.
Mutational analysis of the Orc1 BAH domain
The BAH domain in mammalian Orc1 proteins consists of a 125 amino-acid sequence near the N-terminus that is 64–99% identical among mammals. However, the BAH domain in ScOrc1 shares only 7% sequence identity with the BAH domain in HsOrc1. Nevertheless, structure-guided sequence alignment has revealed the common elements between the BAH domains of ScOrc1 and HsOrc1 (Zhang et al, 2002), which along with similar data on the BAH domain in the chicken polybromo protein (Oliver et al, 2005) suggests a common 3-D structure (Figure 1A). Based on this information, three mutations were constructed in HsOrc1 that should inactivate BAH function (Figure 1C). ΔBAH deletes the N-terminal region of HsOrc1 that includes the BAH domain. ΔH deletes the H-domain that lies at the center of the BAH domain. In ScOrc1, the H-domain is required for binding of ScOrc1 to ScSir1 (Hsu et al, 2005). E111K contains K in place of E at amino acid 111. E111 lies at the center of an eight amino-acid identity found in six mammalian Orc1 proteins, and alignment of 343 BAH domain sequences (Pfam data base, http://www.sanger.ac.uk/Software/Pfam/) revealed that the glutamic acid residue corresponding to E111 in HsOrc1 is highly conserved (Figure 1B), suggesting that it plays a critical role in BAH domain function. In fact, changing E–K at this site in the BAH domain of ScSir3 (E131; Stone et al, 2000) and of ScRsc2 (E468; Wong et al, 2002) inactivates the function of these proteins (Figure 1A and B).
The BAH domain affects Orc1 stability
Changes in the BAH domain affected the level of Orc1 expression (Figure 2A). Serial dilution of nuclear extracts followed by immunoblotting revealed that deletion of the BAH domain increased Orc1 expression ∼5-fold, whereas the E111K and ΔH mutations diminished Orc1 expression ∼4-fold. To determine whether or not these differences reflected differences in protein stability, the half-life of each protein was measured by culturing cells first with 35S-Met for 2 h and then in the absence of radiolabel. ΔBAH (2.4 h) was more stable than wild-type protein (1.8 h), but E111K (1.1 h) and ΔH (1.1 h) were less stable, an effect that was previously observed with the corresponding mutation in ScSir3 (E131K; Figure 1; Stone et al, 2000). Furthermore, culturing cells briefly in the presence of MG132, a specific inhibitor of the 26S proteasome, increased the amounts of the BAH domain mutants to levels similar to wild-type (Figure 2B). Similarly, the ScSir3 E131K-mutation is degraded via a ubiquitin-dependent, quality control pathway (Gardner et al, 2005). Thus, misfolding of the BAH-domain would decrease protein stability, whereas elimination of the BAH-domain would increase protein stability. These results suggested that the BAH domain may affect the selective degradation of Orc1 when cells enter S-phase.
Figure 2.

Effects of BAH domain mutations in HsOrc1 on FH-Orc1 expression, degradation, and ORC assembly. (A) Relative levels of the indicated FH-Orc1 protein were determined by subjecting whole-cell lysates to immuno-blotting using anti-HA antibody to detect recombinant proteins and anti-HsOrc2 to detect endogenous Orc2. Cells transformed with pOP served as a control. (B) Cells cultured in 25 μM MG132 for 4 h before lysis were subjected to immuno-blotting. (C) Asynchronously proliferating cells were stained with anti-HA antibody to detect FH-Orc1(wt) (red), anti-PCNA antibody to detect PCNA (green), and DAPI to visualize nuclear DNA (Blue). (D) Cells expressing either FH-Orc1 wt or ΔBAH were fractionated by centrifugal elutriation, and individual fractions were analyzed by FACS. Nuclei from cells lysed in HB were subjected to immunoblotting. (E) FH-Orc1 wt and ΔBAH were affinity purified from NEB350 extracts of 293EBNA1 cells that constitutively expressed the indicated protein. FH-Orc1 was bound to anti-FLAG M2 agarose beads, eluted with FLAG peptide, fractionated by SDS-gel electrophoresis and stained with silver. The positions of FH-Orc1 (100 kDa), Orc3 (82 kDa), Orc2 (66 kDa), Orc4 (50.4 kDa), Orc5 (50.3 kDa) and Orc6 (28 kDa), and Mark 12 protein standards (Invitrogen) are indicated. Experimental details are provided in ‘Supplementary Materials and methods'.
The BAH domain is not required for nuclear localization and S-phase specific degradation of Orc1
To determine whether or not the BAH domain is required for S-phase specific degradation of HsOrc1, exponentially proliferating cells that constitutively expressed either FH-Orc1 wt, ΔBAH, E111K, or ΔH were fixed in situ and then stained with anti-HA antibody to visualize the recombinant protein, and with anti-PCNA antibody to visualize the S-phase specific protein proliferating cell nuclear antigen (PCNA). In each case, 50–60% of the cells exhibited high levels of nuclear FH-Orc1, but low levels of PCNA, whereas the remaining interphase cells exhibited high levels of nuclear PCNA with little, if any, FH-Orc1 (Figure 2C; data not shown). This result was consistent with previous observations on transiently overexpressed HsOrc1 (Lidonnici et al, 2004). Since all four cell lines were indistinguishable in this assay, the BAH domain was not required for either nuclear localization or S-phase dependent degradation of Orc1.
This conclusion was confirmed and extended by using centrifugal elutriation to fractionate cells according to their progress in cell division and then assaying individual fractions for Orc1 in the chromatin (nuclear) fraction. As previously reported for both ectopically expressed recombinant HsOrc1 and for endogenous HsOrc1 (Kreitz et al, 2001; Mendez et al, 2002; Ritzi et al, 2003; Tatsumi et al, 2003), FH-Orc1 was detected predominantly in G1-phase cells (Figure 2D). Furthermore, neither deletion (Figure 2D) nor mutation (data not shown) of the BAH domain altered this result. Similar results were obtained when whole-cell extracts from each fraction were analyzed. We conclude that the BAH domain strongly affects Orc1 stability in vivo, but not through the S-phase specific mechanism that targets Orc1 for selective modification and degradation. Presumably, the BAH domain mediates Orc1 stability during G1-phase through interaction with other proteins.
The BAH domain is not required for ORC assembly
To determine whether or not the HsOrc1 BAH domain is required for ORC assembly, FH-Orc1(wt) protein was affinity purified from cells that constitutively expressed it, and the purified proteins were fractionated by SDS–PAGE and stained with silver. The result was consistent with the presence of FH-ORC(1–5) complexes in vivo (Figure 2E). Only the Orc6 subunit was not detected, consistent with previous reports that HsOrc6 binds very weakly to the other ORC subunits (Ranjan and Gossen, 2006 and references therein). Results with the three BAH domain mutants were similar to those with FH-Orc1(wt), demonstrating that the HsOrc1 BAH domain was not required to form stable ORC(1–5) complexes (Figure 2E and data not shown).
The BAH domain facilitates binding of Orc1 to chromatin
Since BAH domains appear to facilitate the action of chromatin binding proteins (Hou et al, 2005; Hsu et al, 2005; Katsuyama et al, 2005; Oliver et al, 2005; Connelly et al, 2006), the HsOrc1 BAH domain may facilitate interaction of Orc1 with chromatin. Previous studies have reported that HsOrc1 levels are restored in cells during the M to G1-phase transition of their cell division cycle, and that the new HsOrc1 is associated with the chromatin fraction (Mendez et al, 2002; Tatsumi et al, 2003). The data described above (Figure 2C and D) are consistent with that conclusion. Therefore, to determine whether or not the BAH domain affects the affinity of HsOrc1 for chromatin during the M to G1-phase transition, asynchronously proliferating cells that constitutively expressed a recombinant Orc1 were fixed in situ and then stained with anti-HA antibody to detect FH-Orc1. Three different fixation protocols were employed with the same results. When cells entered mitosis (prophase and metaphase; Figure 3), FH-Orc1 wt, ΔBAH, E111K and ΔH proteins were not associated with chromosomes, as noted by the scarcity of FH-tagged protein (green) at the sites occupied by DNA (red). However, as cells progressed from metaphase to anaphase, FH-Orc1(wt) associated predominantly, if not exclusively, with chromosomes, but the three Orc1 BAH mutants did not. Moreover, as cells entered telophase, all of the detectable FH-Orc(wt) was bound to chromosomes, whereas the three Orc1 BAH mutants remained distributed throughout the cell. Thus, the HsOrc1 BAH domain is required in vivo for efficient binding of Orc1 to chromatin. This conclusion was reinforced by noting that BAH mutants appeared to be completely absent from either prophase or metaphase chromosomes, as indicated by the black furrow corresponding to condensed chromosomes within the ‘green' Orc1 mutant protein. Conversely, the uniform distribution of BAH mutant proteins throughout anaphase and telophase cells indicated that some of the protein was bound to the chromosomes during the M to G1-phase transition. These data are consistent with the reappearance of significant amounts of Orc1 in M-phase cells (Figure 2D).
Figure 3.

The HsOrc1 BAH domain facilitated association of Orc1 with chromosomes in vivo. Exponentially proliferating cells expressing either FH-Orc1 wt, ΔBAH, E111K or ΔH were fixed with formalin, permeabilized with SDS buffer, and stained with propidium iodide to detect nuclear DNA (red) and with anti-HA to detect recombinant protein (green). Images were captured in a confocal microscope and then merged to reveal sites where DNA and FH-Orc1 protein were coincident (yellow). Interphase nuclei containing FH-Orc1 were in G1-phase (G1), because they lacked PCNA (see Figure 2C). Cells with a puddle of condensed chromatin were considered prophase (P). Cells with condensed chromosomes aligned in a single plate were considered metaphase (M). Cells with two metaphase plates were considered anaphase (A). Two adjacent cells, each with condensed chromosomes in a single plate of condensed chromosomes, were considered telophase (T). Similar results were obtained with cells fixed in formalin and permeabilized with Triton X-100, and with cells fixed in methanol only.
Although both wild-type and BAH mutants of Orc1 accumulated in the nuclei of G1-phase cells (Figure 2C and 3), the association of these proteins with G1-phase chromosomes could not be discerned by cytological methods. Therefore, exponentially proliferating cells were lysed in a hypotonic buffer (HB) containing Triton X-100 to solubilize the outer nuclear membrane, and the insoluble nuclear pellet was extracted with increasing concentrations of salt in a nuclear extraction buffer (NEB) to release FH-Orc1. Since FACS analysis revealed that 92% of these cells were either in G1 or S-phase, and ∼90% of the FH-Orc1 was localized in the nuclei of G1-phase cells (Figure 2C and D), these extraction profiles reflected the properties of Orc1 in G1-phase cells.
Consistent with previously reported elution profiles for HsOrc1 (Kreitz et al, 2001; Ohta et al, 2003), none of the FH-Orc1 was soluble in HB, but at least 80% of the FH-Orc1 was recovered with 350 mM NaCl, and the remainder appeared in the 170 and 500 mM NaCl fractions (Figure 4B, FH-Orc1). Similar results were obtained with endogenous HsOrc1 (Figure 4A, Orc1). The amount of FH-Orc1 extracted with NEB350 was about six-fold greater than the amount of endogenous Orc1 extracted from the same number of cells using the same buffer (Figure 4A).
Figure 4.

The HsOrc1 BAH domain stabilized the association between Orc1 and nuclear components in cell lysates. (A) The level of FH-Orc1 in 293EBNA1 cells that constitutively expressed this protein was compared directly with the level of endogenous Orc1 in 293EBNA1 cells that had been transformed with the expression vector pOP. Identical extracts from the same number of cells were fractionated on the same gel before subjecting it to immunoblotting with anti-HsOrc1 antibody. Extract from FH-Orc1 cells was serially diluted before fractionation. (B) Cells expressing the indicated protein were fractionated into cytosol and nuclei as previously described (Mendez and Stillman, 2000), with modifications (see Supplementary Materials and methods). Cells were lysed in HB and then sequentially extracted with NEB containing either 170, 350 or 500 mM NaCl. The soluble fractions from each extraction as well as the final nuclear pellet were then subjected to Western immunoblotting either with anti-HA antibody to detect FH-Orc1, or with anti-HsMcm3 antibody. HsMcm3 was assayed in the FH-Orc1(wt) cell line. Proteins were identified on the basis of their size and antibody crossreactivity.
Remarkably, when the BAH domain was either deleted (ΔBAH) or inactivated (ΔH, E111K), about half of the FH-Orc1 was soluble in HB alone; the remainder exhibited a salt elution profile similar to that of FH-Orc1(wt). These results were not a consequence of differences in expression levels among the four FH-Orc1 proteins, because ΔBAH was expressed in greater amounts than Orc1(wt), but ΔH and E111K were expressed in lesser amounts than Orc1(wt) (Figure 2A and B). Nevertheless, results were essentially the same with all three mutants. Furthermore, the same extraction conditions confirmed the existence of two populations of Mcm(2–7) complexes (Figure 4), one that is stably bound to chromatin and one that is not (Mendez and Stillman, 2000). Thus, the BAH domain appeared to be required for Orc1 binding to some portions of chromatin, but not to others.
Orc1 facilitates binding of ORC to chromatin
The results described above suggested that the Orc1 BAH domain may affect the ability of ORC to bind to chromatin during the M to G1-phase transition. This hypothesis was investigated by staining an asynchronous population of cells with anti-HA antibody to identify FH-Orc1 and with anti-HsOrc2 antibody to identify endogenous Orc2. Anti-HsOrc2 antibody detected a single protein in whole-cell extracts, and its size corresponded to HsOrc2 (Figure 5A). As previously reported for HeLa cells (Prasanth et al, 2004), endogenous Orc2 (red) was localized to the nucleus in G1-phase 293EBNA1 cells, but it was absent from chromosomal DNA (blue) in prophase, metaphase and anaphase cells, as evidence by the dark area where DNA resides (Figure 5B, control). In contrast, Orc2 was distributed uniformly throughout telophase cells, indicating that it had begun associating with chromatin. Remarkably, in cells constitutively expressing FH-Orc1(wt), the distribution of Orc2 was indistinguishable from that of FH-Orc1(wt), both were absent from prophase and metaphase chromosomes, and both began binding to chromatin during the transition from anaphase to telophase (Figure 5B), suggesting that increased levels of Orc1 increased the efficiency of Orc2 association with chromosomes. This hypothesis was confirmed by the discovery that the fraction of chromatin bound Orc2 in cells constitutively expressing either ΔBAH, E111K or ΔH was reduced relative to the fraction bound in cells expressing FH-Orc1(wt), but still greater than in cells expressing only endogenous Orc1 (Figure 5B; E111K and data not shown). This was consistent with the fact that only a fraction of each Orc1 BAH mutant bound to chromatin during the M to G1-phase transition (Figures 3 and 4). Thus, the ability of endogenous Orc2 to bind to chromatin during the M to G1-phase transition was dependent on the level of Orc1. Since Orc2 in low salt as well as high salt extracts exists as ORC (2–5) (Kreitz et al, 2001; Ohta et al, 2003; Radichev et al, 2006), the in vivo behavior of Orc2 reflects the behavior of the ORC(2–5) core complex.
Figure 5.
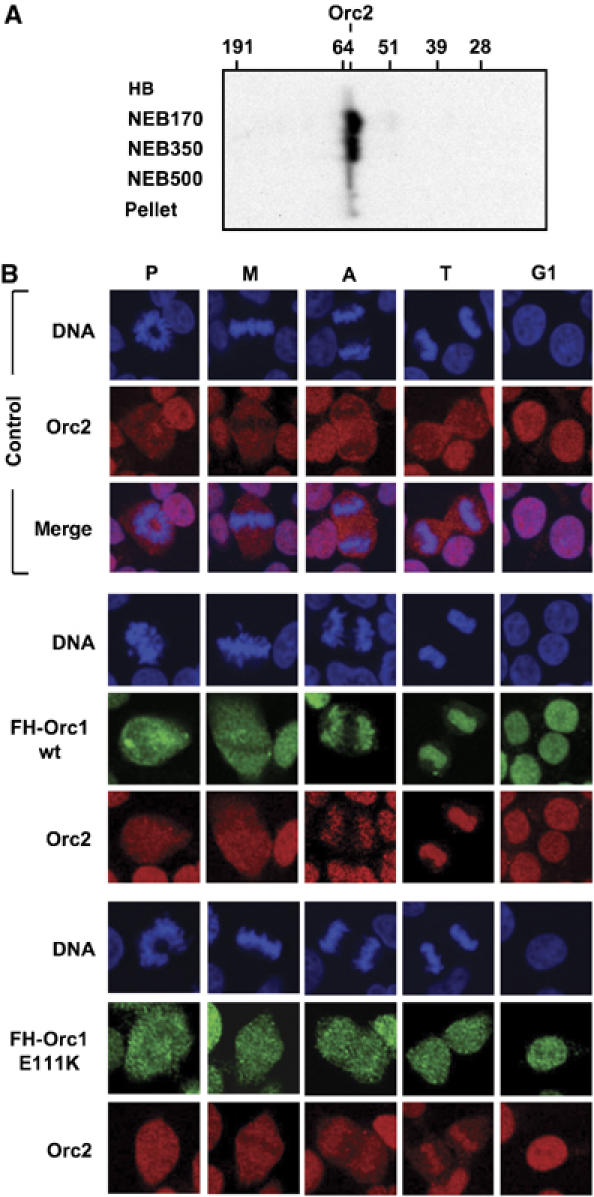
HsOrc1 facilitated association of Orc2 with chromosomes in vivo. (A) Cells expressing FH-Orc1(wt) were lysed, extracted with increasing amounts of salt, and then subjected to immuno-blotting using anti-Orc2 antibody, as in Figure 4. (B) Exponentially proliferating 293EBNA1 cells (control) or 293EBNA1 cells expressing either FH-Orc1 wt or E111K were stained with DAPI to detect nuclear DNA (blue), anti-HA to detect FH-Orc1 (green), and anti-HsOrc2 (red), as in Figure 3.
The BAH domain facilitates binding of Orc1 to DNA replication origins
FACS analysis revealed that none of the three Orc1 BAH mutants affected the fraction of cells in G1-phase (55±3%), S-phase (37±2%) and G2/M-phase (8±1%). Therefore, to determine whether or not the same mutations that affect the ability of Orc1 to bind chromosomes would also affect its ability to bind DNA replication origins and to support DNA replication, the effects of FH-Orc1 on Epstein–Barr virus oriP-dependent plasmid DNA replication was investigated.
OriP is an appropriate model for cellular DNA replication origins, and in addition, is more sensitive to ORC activity than are cellular origins (Dhar et al, 2001b). OriP-dependent plasmid DNA replication requires association of ORC with oriP (Chaudhuri et al, 2001; Schepers et al, 2001; Ritzi et al, 2003), and with the exception of virally encoded nuclear antigen-1 (EBNA1), relies entirely on cellular proteins to duplicate itself once and only once per cell division (reviewed in Hammerschmidt and Sugden, 2006). OriP consists of a tandem array of 20 EBNA1 binding sites (‘family of repeats' (FR)) and a dyad symmetry (DS) element. The FR element is required to partition the plasmid between daughter cells during mitosis. ORC binds preferentially to the DS element, which also contains four EBNA1-binding sites, and functions as an origin of bidirectional replication.
293EBNA1 cells, expressing both EBNA1 and FH-Orc1 and harboring an oriP-dependent episome (p818; Figure 6A), were subjected to chromatin immuno-precipitation analysis using anti-HA antibody. Under conditions where detectable levels of FH-Orc1 protein could be selectively immuno-precipitated from lysates of cells that had been pretreated briefly with formaldehyde (Figure 6B), DNA fragments that contained the DS locus (probe SC5) were preferentially cross-linked to FH-Orc1(wt) (Figure 6C), as previously reported (Chaudhuri et al, 2001; Schepers et al, 2001). However, deletion or mutation of the BAH domain in Orc1 eliminated the ability of formaldehyde to crosslink Orc1 to p818 DNA (Figure 5D), revealing that the BAH domain facilitates interaction between Orc1 and oriP in vivo.
Figure 6.

The HsOrc1 BAH domain facilitated binding of Orc1 to oriP DNA. Cells constitutively expressing either FH-Orc1 wt, ΔBAH, E111K or ΔH and maintaining p818 as an episome were subjected to chromatin immunoprecipitation (ChIP) assays using anti-HA antibody, as previously described (Chaudhuri et al, 2001; Schepers et al, 2001), with modifications (see Supplementary Materials and methods). (A) The relative locations of PCR probes E15′ and SC5 in plasmid p818 are indicated along with the EBNA1 and hygromycin phosphotransferase (hpt) genes (transcription indicated by arrow), and the oriP FR and dyad symmetry (DS) elements. SC5 is about 4 kb from E15′. (B) FH-Orc1 and Orc2 recovered in ChIP assays were detected by immunoblotting with a mixture of anti-HsOrc1 and anti-HsOrc2 antibodies. (C) Relative amounts of p818 DNA containing probe E15′ (gray) and SC5 (cross-hatched) sequences recovered from cells expressing FH-Orc1(wt) were determined by real-time quantitative PCR and expressed as a fraction of p818 episomal DNA. (D) The experiment in (C) was carried out with cells expressing either FH-Orc1 wt, ΔBAH, E111K or ΔH. Error bars indicate s.e.m. for four independent experiments.
The BAH domain facilitates the ability of Orc1 to support DNA replication
To determine whether or not the Orc1 BAH domain facilitates the ability of Orc1 to support DNA replication in vivo, the effects of BAH domain mutations on oriP-dependent DNA replication were examined using three different assays. To measure the ability of cells to maintain the replication and partitioning of a plasmid over several weeks, 293EBNA1 cells were co-transfected with p818, an oriP-plasmid that expressed the hygromycin-resistance gene, and pEGFP, a non-replicating plasmid that expressed the green fluorescence protein (GFP). Transfection efficiency was determined as the fraction of cells expressing GFP. Colony formation was dependent on the presence of p818 (Figure 7A), and the number of colonies obtained was normalized to the transfection efficiency for each cell line. The resulting colonies contained an average of 50–100 plasmids, as previously reported (Yates and Guan, 1991). In contrast, p571, a plasmid similar to p818 but lacking the DS element (Yates et al, 2000), did not produce hygromycin-resistant colonies, confirming that plasmid maintenance required the oriP DNA replication function. Constitutive expression of FH-Orc1(wt) did not increase the number of hygromycin-resistant colonies, but constitutive expression of either FH-Orc1 ΔBAH, E111K or ΔH did reduce the number of colonies obtained (Figure 7A), suggesting that mutant Orc1 molecules interfered with the initial rounds of plasmid DNA replication driven by endogenous ORC.
Figure 7.

BAH domain in HsOrc1 facilitated oriP-dependent plasmid DNA replication in human cells. (A) Plasmid maintenance assays: 293EBNA1 cells constitutively expressing either FH-Orc1 wt, ΔBAH, E111K or ΔH were co-transfected with the oriP-dependent plasmid p818 (Figure 6A) and pEGFP, a plasmid that lacks oriP. Cells were selected for their resistance to hygromycin B. Cells transformed with pOP, the parental expression vector, were used as a control. Results were normalized to the fraction of eGFP expressing cells. (B) Transient plasmid DNA replication assays: 293EBNA1 cells constitutively expressing either FH-Orc1 wt, ΔBAH, E111K or ΔH were co-transfected with p818 to determine the effect of the ectopic protein on oriP-dependent DNA replication, and with pEGFP to measure transfection efficiency. Plasmid DNA was isolated 4 days post-transfection, and then digested with BamH1 to convert it into linear monomers and with DpnI to degrade unreplicated DNA. DNA was detected by blotting-hybridization using appropriate 32P-DNA probes and quantified using a phosphorimager. Each lane contains DNA from an equivalent number of transfected cells. ‘STD' is purified p818. Transfections were carried out in duplicate. Error bars indicate s.e.m. for four independent experiments.
To test this hypothesis, the effects of HsOrc1 BAH mutants on plasmid DNA replication were assayed directly. 293EBNA1 cells were again co-transfected with p818 and pEGFP, but this time the amount of p818 DNA replication was assayed soon after transfection by its resistance to the DpnI restriction endonuclease. DpnI cleaves DNA only when it is methylated on both strands. Since mammalian cells lack the ability to methyate DpnI restriction sites, a single round of DNA replication prevents cleavage by DpnI. The results revealed that FH-Orc1(wt) stimulated p818 DNA replication, but ΔBAH, E111K and ΔH did not (Figure 7B), confirming that wt Orc1, but not BAH mutated Orc1, could stimulate oriP activity.
To demonstrate that the Orc1 BAH domain was required for this Orc1 activity, the ability of the Orc1 BAH domain to inhibit p818 DNA replication was assayed. If interaction between the Orc1 BAH domain and another protein is required to initiate DNA replication, then overexpression of the BAH domain alone may interfere with interaction between Orc1 and the target of its BAH domain. Cells were co-transfected with p818, pEGFP, and pN315, a plasmid that expressed the N-terminal 315 amino acids of HsOrc1 fused to the FLAG epitope. N315 contained the complete BAH domain as well as a putative nuclear localization signal (Figure 1C). N315 was expressed efficiently (Figure 8A) and localized to the nucleus (Figure 8B). Based on the requirements for N315 release from isolated nuclei (NEB+350 mM NaCl), it appeared to be bound to chromatin (Figure 8C). Expression of N315 suppressed p818 DNA replication, but it did not reduce the amount of pEGFP DNA (Figure 8D), consistent with a role for the HsOrc1 BAH domain in stimulating oriP-dependent DNA replication.
Figure 8.

The N-terminal fragment of HsOrc1 containing the BAH domain inhibited oriP-dependent plasmid DNA replication. 293EBNA1 cells were transfected in triplicate with p818 (Figure 6A), either in the presence or absence of pF-N315, a plasmid expressing FLAG-tagged N315, the N-terminal 315 amino acid portion of FH-HsOrc1 (Figure 1C). Four days post-transfection, whole-cell extracts were subjected to immunoblotting using anti-FLAG and anti-CcnA2 antibodies (A), stained with anti-FLAG antibody (green) to determine whether or not N315 localized to the nucleus (B), and extracted first with HB and then with NEB containing the indicated mM amount of NaCl (C). (D) Transient plasmid DNA replication assays were carried out as in figure 7B. (E) Total p818 and pEGFP DNA (−DpnI) in either the presence or absence of pF-N315 was quantified using a phophorimager. (F) Replicated p818 (black bars) and pEGFP (gray bars) DNA was quantified as the amount of DNA resistant to DpnI digestion relative to the total amount detected in the absence of pF-N315.
Discussion
Orc1 is the only one of the six ORC subunits that has a BAH domain, but the function of this domain in DNA replication, if any, has been a mystery. It is not required for DNA replication in the budding yeast S. cerevisiae, although it does have a <2-fold effect on plasmid maintenance (Bell et al, 1995). It is not required for DNA replication in human cells when HsOrc1 is tethered to DNA via a sequence-specific DNA binding protein (Takeda et al, 2005). Therefore, it is not required for subsequent binding of Cdc6, Cdt1 and the MCM helicase. The only known function for the BAH domain is binding to proteins that affect chromatin structure. In S. cerevisiae, the Orc1 BAH domain binds specifically to Sir1 and thereby recruits the Sir(2–4) gene silencing complex to specific genomic loci. The Sir3 BAH domain does not bind Sir1, but it contributes to an interaction between Sir3 and nucleosomes (Connelly et al, 2006). A similar role has been suggested for the BAH domains in two subunits of the RSC chromatin remodeling complex (Wong et al, 2002) and for the winged eye protein in Drosophila melanogaster (Katsuyama et al, 2005). The fact that mammals lack a homolog of Sir1, and the fact that there is little sequence homology between the BAH domains of ScOrc1 and HsOrc1, suggest that the BAH domain in HsOrc1 serves a function different from that in ScOrc1. Conversely, the fact that the Orc1 BAH domain sequence is highly conserved among mammals (Figure 1B) suggests that its function is required throughout the mammalian kingdom.
Taken together with previously published work, the results presented here support a role for mammalian Orc1 in regulating ORC activity and in origin recognition, with a unique role for the Orc1 BAH domain in origin recognition (Figure 9).
Figure 9.

‘ORC cycle' in mammalian cells (reviewed in Depamphilis et al, 2006). ORC is bound to chromatin during G1-phase of the cell cycle where it is part of a prereplication complex. When S-phase begins, human Orc1 is selectively degraded by the 26S proteasome via a ubiquitin-dependent mechanism, and then reappears during the M to G1-phase transition. During metaphase, Orc1 is hyperphosphorylated, an event that prevents ORC assembly. As cells exit metaphase, Orc1 is dephosphorylated and, together with other ORC subunits, binds to chromatin, an event that is facilitated by the Orc1 BAH domain.
Orc1 as a regulator of ORC activity
Previous studies have shown that HsOrc1 binds to HsORC(2–5), that ORC is bound to chromatin in G1-phase cells, and that HsOrc1 is selectively degraded by a ubiquitin-dependent mechanism when cells enter S-phase, a step that presumably contributes to preventing reinitiation of DNA replication during a single cell division cycle (see Introduction). The present studies confirmed these conclusions and extended them to show that the BAH domain was not required for either event (Figures 2C–E, 3 and 4). The cellular level of HsOrc1 is then reestablished during the M to G1-phase transition. It is doing this period of cell division that the BAH domain facilitates the ability of Orc1 to bind ORC(2–5) to chromosomes.
In contrast to Orc1, ORC(2–5) remains bound to chromatin until G2/M-phase, at which time HsOrc2 can no longer be crosslinked to DNA in metaphase arrested cells (Abdurashidova et al, 2003), and it is not associated with chromosomes during prophase, metaphase and anaphase (Figure 5; Prasanth et al, 2004). Solubility data from hamster cells, however, suggest that ORC(2–5) remain bound to chromatin throughout the cell cycle, including mitosis (Natale et al, 2000; Okuno et al, 2001). One explanation is that Orc subunits can also associate with nuclear components such as centrosomes (Craig et al, 2003; Prasanth et al, 2004), perinuclear structures (Saha et al, 2006), and nuclear matrix (Kreitz et al, 2001; Ohta et al, 2003), and these associations could account for their retention in the ‘nuclear fraction'. Although we could not confirm that Orc2 is localized at centrosomes in human 293EBNA1 cells (Figure 5B), we were able to confirm this event in mitotic hamster CHO cells (data not shown). These differences may be due to differences in the cellular levels of ORC proteins or in the antibodies used.
Solubility data from hamster cells (Li et al, 2004 and references therein) and crosslinking studies in HeLa cells (Abdurashidova et al, 2003) have suggested that Orc1 is not stably bound to chromosomes in metaphase arrested cells. Data presented here confirm and extend these studies by demonstrating that FH-Orc1 reappears in unsynchronized mitotic 293 cells (Figures 2D and 3), and that it is not bound to either prophase or metaphase chromosomes (Figure 3). Thus, it appears that mammalian ORC, like Xenopus ORC (Depamphilis et al, 2006), is not stably associated with chromatin during mitosis.
Since the Orc1 BAH domain was not required for ORC assembly (Figure 2E), we conclude that it was required for association of ORC with chromatin in vivo. This is consistent with reports that neither HsOrc1 nor HsORC(2–5) alone can bind to DNA in vitro (Vashee et al, 2003; Giordano-Coltart et al, 2005), and results suggesting that Orc1 is required to bind ORC(2–5) to chromatin in vivo (Kreitz et al, 2001; Ohta et al, 2003; Radichev et al, 2006). In fact, HsOrc1 appears to recruit ORC(2–5) to DNase I-insoluble nuclear structures (Kreitz et al, 2001; Ohta et al, 2003). Once bound, however, ORC(2–5) remains with the chromatin fraction even when Orc1 is selectively degraded during S-phase (Figure 2D; Mendez et al, 2002; Ohta et al, 2003).
Expression of FH-Orc1 stimulated the binding of endogenous Orc2 to chromatin as mitosis progressed from anaphase to telophase, and this phenomenon was facilitated by the Orc1 BAH domain. The proportion of Orc2 bound to telophase chromosomes was proportional to the amount of Orc1 (endogenous plus recombinant) that could bind to chromatin. Since only a portion of each of the three BAH domain mutants bound to chromatin (Figure 4), the extent of Orc2 binding in cells expressing FH-Orc1 BAH mutants was less than in FHOrc1(wt) cells, but greater than in control cells. Thus, the BAH mutants confirm the hypothesis that elevated levels of Orc1 drives the association of Orc2 (presumably ORC(2–5)) with chromatin during the M to G1-phase transition. Therefore, differences could exist in the time during cell division when prereplication complex assembly occurs simply because of differences in levels of ORC subunits.
The fact that ΔH and E111K Orc1 mutants could be constitutively expressed in cells with no discernible effects on cell proliferation, but with demonstrable effects on plasmid replication may seem surprising. However, the levels of ΔH and E111K were equivalent to those of endogenous Orc1, and previous studies (Dhar et al, 2001b) have shown that levels of ORC subunits (e.g. Orc2) can be reduced 90% with minimal effect on cell proliferation but with dramatic effect on oriP-dependent plasmid DNA replication. G1-phase of the cell cycle was prolonged under these conditions, but there was no effect on the utilization of either the c-Myc or β-globin gene DNA replication origins. However, the same cells failed to support oriP-dependent replication of p818, a defect that could be rescued by reintroduction of Orc2. Therefore, for reasons not yet understood, oriP-dependent plasmid DNA replication is much more sensitive to the level of ORC activity than is chromosomal DNA replication.
A role for Orc1 in origin recognition
We propose that the BAH domain in Orc1 allows it to facilitate binding of ORC(1–5) to sites within chromatin that may otherwise be inaccessible. In this sense, Orc1 plays a key role in origin recognition. Thus, the cell cycle dependent loss of Orc1 activity, most dramatically embodied in the selective degradation of HsOrc1 during S-phase and Drosophila Orc1 during M-phase (Depamphilis et al, 2006), allows initiation site selection to be reprogrammed in these organisms after each cell division, and whenever quiescent cells (which lack Orc1) are stimulated to proliferate.
Orc1-dependent association of ORC with chromatin as cells exited mitosis appears to be critical to the activation of DNA replication origins. Stable association of Orc1 with the chromatin fraction precedes the appearance of site-specific prereplication complexes (‘origin decision point') that occurs during G1-phase (DePamphilis, 2003), and the same Orc1 BAH domain mutants that interfered with binding of Orc1 to chromatin also interfered with binding to oriP (Figure 6) and with the ability of Orc1 to stimulate oriP-dependent DNA replication (Figures 7 and 8). During mitosis, chromatin is organized into loops that appear to be associated at their base by attachment to nuclear matrix, and shorter loops are most efficient at binding ORC (Lemaitre et al, 2005). The role of EBNA1 in activating oriP may be analogous to that of nuclear matrix in that EBNA1 binding to the FR and DS components of oriP can form a short DNA loop that may facilitate recruitment of ORC (Mackey et al, 1995). Taken together, these observations suggest that establishment of ORC-chromatin sites may be allowed only after mitotic chromatin remodeling, and that the BAH domain of Orc1 may target ORC to specific chromosomal loops.
The fact that loss of BAH domain activity resulted in about half of the Orc1 binding stably to chromatin during late mitosis (Figure 3) and G1-phase (Figure 4) and half remaining in the nucleoplasm, suggests that replication origins differ in their protein composition. Therefore, the BAH domain in Orc1 may interact with a chromatin associated protein that allows ORC access to specific loci such as heterochromatin (Leatherwood and Vas, 2003). One candidate is HP1, a protein tightly associated with heterochromatic regions, and one that has been shown to bind GST-tagged HsOrc1 and Drosophila Orc1 in vitro (Pak et al, 1997; Lidonnici et al, 2004). However, while these HP1 binding sites partially overlap the BAH domain, they are not coincident, and neither the E111K nor the ΔH mutations lie within the HP1 binding site (Figure 1C). Moreover, affinity purification of FH-Orc1 from cells treated with protein crosslinking agents did not detect an Orc1–HP1 complex (data not shown). Other possibilities are proteins that interact with methylated DNA, since DNA methylation is confined largely, although not exclusively, to mammals and plants where it is associated with transcriptionally repressive heterochromatic regions of the genome. Finally, the fact that the BAH domain facilitated the ability of Orc1 to bind oriP and stimulate oriP activity suggests that it may interact with one of the chromosomal proteins that bind EBNA1 (Ito and Yanagi, 2003). Further investigation is required to discover the protein targets of the Orc1 BAH domain and thereby elucidate the nature of metazoan replication origins.
Materials and methods
Supplementary Material
Supplementary Materials and methods
Acknowledgments
We thank K Kaneko, M Kohn and R Yagi for technical advice, and A Dey and A Nishiyama for help with confocal microscopy. K Noguchi was supported by a National Research Council Research Associateship Award.
References
- Abdurashidova G, Danailov MB, Ochem A, Triolo G, Djeliova V, Radulescu S, Vindigni A, Riva S, Falaschi A (2003) Localization of proteins bound to a replication origin of human DNA along the cell cycle. EMBO J 22: 4294–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aladjem MI, Fanning E (2004) The replicon revisited: an old model learns new tricks in metazoan chromosomes. EMBO Rep 5: 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Mitchell J, Leber J, Kobayashi R, Stillman B (1995) The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell 83: 563–568 [DOI] [PubMed] [Google Scholar]
- Berezney R, Dubey DD, Huberman JA (2000) Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108: 471–484 [DOI] [PubMed] [Google Scholar]
- Chaudhuri B, Xu H, Todorov I, Dutta A, Yates JL (2001) Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein–Barr virus. Proc Natl Acad Sci USA 98: 10085–10089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JJ, Yuan P, Hsu HC, Li Z, Xu RM, Sternglanz R (2006) Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain. Mol Cell Biol 26: 3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig JM, Earle E, Canham P, Wong LH, Anderson M, Choo KH (2003) Analysis of mammalian proteins involved in chromatin modification reveals new metaphase centromeric proteins and distinct chromosomal distribution patterns. Hum Mol Genet 12: 3109–3121 [DOI] [PubMed] [Google Scholar]
- Cvetic C, Walter JC (2005) Eukaryotic origins of DNA replication: could you please be more specific? Semin Cell Dev Biol 16: 343–353 [DOI] [PubMed] [Google Scholar]
- DePamphilis ML (1999) Replication origins in metazoan chromosomes: fact or fiction? Bioessays 21: 5–16 [DOI] [PubMed] [Google Scholar]
- DePamphilis ML (2003) The ‘ORC cycle': a novel pathway for regulating eukaryotic DNA replication. Gene 310: 1–15 [DOI] [PubMed] [Google Scholar]
- Depamphilis ML, Blow JJ, Ghosh S, Saha T, Noguchi K, Vassilev A (2006) Regulating the licensing of DNA replication origins in metazoa. Curr Opin Cell Biol 18: 231–239 [DOI] [PubMed] [Google Scholar]
- Dhar SK, Delmolino L, Dutta A (2001a) Architecture of the human origin recognition complex. J Biol Chem 276: 29067–29071 [DOI] [PubMed] [Google Scholar]
- Dhar SK, Yoshida K, Machida Y, Khaira P, Chaudhuri B, Wohlschlegel JA, Leffak M, Yates J, Dutta A (2001b) Replication from oriP of Epstein–Barr virus requires human ORC and is inhibited by geminin. Cell 106: 287–296 [DOI] [PubMed] [Google Scholar]
- Gardner RG, Nelson ZW, Gottschling DE (2005) Degradation-mediated protein quality control in the nucleus. Cell 120: 803–815 [DOI] [PubMed] [Google Scholar]
- Gilbert DM (2004) In search of the holy replicator. Nat Rev Mol Cell Biol 5: 848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano-Coltart J, Ying CY, Gautier J, Hurwitz J (2005) Studies of the properties of human origin recognition complex and its Walker A motif mutants. Proc Natl Acad Sci USA 102: 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt W, Sugden B (2006) Epstein–Barr virus. In DNA Replication and Human Disease, DePamphilis, ML (ed), pp 687–706. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Hou Z, Bernstein DA, Fox CA, Keck JL (2005) Structural basis of the Sir1-origin recognition complex interaction in transcriptional silencing. Proc Natl Acad Sci USA 102: 8489–8494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HC, Stillman B, Xu RM (2005) Structural basis for origin recognition complex 1 protein-silence information regulator 1 protein interaction in epigenetic silencing. Proc Natl Acad Sci USA 102: 8519–8524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Yanagi K (2003) Epstein–Barr virus (EBV) nuclear antigen 1 colocalizes with cellular replication foci in the absence of EBV plasmids. J Virol 77: 3824–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuyama T, Sugawara T, Tatsumi M, Oshima Y, Gehring WJ, Aigaki T, Kurata S (2005) Involvement of winged eye encoding a chromatin-associated bromo-adjacent homology domain protein in disc specification. Proc Natl Acad Sci USA 102: 15918–15923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Ladenburger EM, Kremer M, Knippers R (2002) The origin recognition complex marks a replication origin in the human TOP1 gene promoter. J Biol Chem 277: 31430–31440 [DOI] [PubMed] [Google Scholar]
- Kong D, Coleman TR, DePamphilis ML (2003) Xenopus origin recognition complex (ORC) initiates DNA replication preferentially at sequences targeted by Schizosaccharomyces pombe ORC. EMBO J 22: 3441–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D, DePamphilis ML (2001) Site-specific DNA binding of the Schizosaccharomyces pombe origin recognition complex is determined by the Orc4 subunit. Mol Cell Biol 21: 8095–8103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitz S, Ritzi M, Baack M, Knippers R (2001) The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J Biol Chem 276: 6337–6342 [DOI] [PubMed] [Google Scholar]
- Ladenburger EM, Keller C, Knippers R (2002) Identification of a binding region for human origin recognition complex proteins 1 and 2 that coincides with an origin of DNA replication. Mol Cell Biol 22: 1036–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherwood J, Vas A (2003) Connecting ORC and heterochromatin: why? Cell Cycle 2: 573–575 [PubMed] [Google Scholar]
- Lemaitre JM, Danis E, Pasero P, Vassetzky Y, Mechali M (2005) Mitotic remodeling of the replicon and chromosome structure. Cell 123: 787–801 [DOI] [PubMed] [Google Scholar]
- Li CJ, Vassilev A, DePamphilis ML (2004) Role for Cdk1 (Cdc2)/cyclin A in preventing the mammalian origin recognition complex's largest subunit (Orc1) from binding to chromatin during mitosis. Mol Cell Biol 24: 5875–5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidonnici MR, Rossi R, Paixao S, Mendoza-Maldonado R, Paolinelli R, Arcangeli C, Giacca M, Biamonti G, Montecucco A (2004) Subnuclear distribution of the largest subunit of the human origin recognition complex during the cell cycle. J Cell Sci 117: 5221–5231 [DOI] [PubMed] [Google Scholar]
- Mackey D, Middleton T, Sugden B (1995) Multiple regions within EBNA1 can link DNAs. J Virol 69: 6199–6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechali M (2001) DNA replication origins: from sequence specificity to epigenetics. Nat Rev Genet 2: 640–645 [DOI] [PubMed] [Google Scholar]
- Mendez J, Stillman B (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol 20: 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Zou-Yang XH, Kim SY, Hidaka M, Tansey WP, Stillman B (2002) Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol Cell 9: 481–491 [DOI] [PubMed] [Google Scholar]
- Natale DA, Li CJ, Sun WH, DePamphilis ML (2000) Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M-G(1) transition in mammals. EMBO J 19: 2728–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P, Kosiyatrakul S, Yang Q, Guan Z, Brown NM, Thomas S, Riblet R, Schildkraut CL (2005) Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell 20: 575–587 [DOI] [PubMed] [Google Scholar]
- Ohta S, Tatsumi Y, Fujita M, Tsurimoto T, Obuse C (2003) The ORC1 cycle in human cells: II. dynamic changes in the human ORC complex during the cell cycle. J Biol Chem 278: 41535–41540 [DOI] [PubMed] [Google Scholar]
- Okuno Y, McNairn AJ, den Elzen N, Pines J, Gilbert DM (2001) Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J 20: 4263–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver AW, Jones SA, Roe SM, Matthews S, Goodwin GH, Pearl LH (2005) Crystal structure of the proximal BAH domain of the polybromo protein. Biochem J 389: 657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR (1997) Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91: 311–323 [DOI] [PubMed] [Google Scholar]
- Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B (2004) Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J 23: 2651–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radichev I, Kwon SW, Zhao Y, DePamphilis ML, Vassilev A (2006) Genetic analysis of human Orc2 reveals specific domains that are required in vivo for assembly and nuclear localization of the origin recognition complex. J Biol Chem 281: 23264–23273 [DOI] [PubMed] [Google Scholar]
- Ranjan A, Gossen M (2006) A structural role for ATP in the formation and stability of the human origin recognition complex. Proc Natl Acad Sci USA 103: 4864–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beall EL, Botchan MR (2004) DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J 23: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzi M, Tillack K, Gerhardt J, Ott E, Humme S, Kremmer E, Hammerschmidt W, Schepers A (2003) Complex protein–DNA dynamics at the latent origin of DNA replication of Epstein–Barr virus. J Cell Sci 116: 3971–3984 [DOI] [PubMed] [Google Scholar]
- Saha T, Ghosh S, Vassilev A, Depamphilis ML (2006) Ubiquitylation, phosphorylation and Orc2 modulate the subcellular location of Orc1 and prevent it from inducing apoptosis. J Cell Sci 119: 1371–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers A, Ritzi M, Bousset K, Kremmer E, Yates JL, Harwood J, Diffley JF, Hammerschmidt W (2001) Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein–Barr virus. EMBO J 20: 4588–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EM, Reifsnyder C, McVey M, Gazo B, Pillus L (2000) Two classes of sir3 mutants enhance the sir1 mutant mating defect and abolish telomeric silencing in Saccharomyces cerevisiae. Genetics 155: 509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda DY, Shibata Y, Parvin JD, Dutta A (2005) Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev 19: 2827–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi Y, Ohta S, Kimura H, Tsurimoto T, Obuse C (2003) The ORC1 cycle in human cells: I. Cell cycle-regulated oscillation of Human ORC1. J Biol Chem 278: 41528–41534 [DOI] [PubMed] [Google Scholar]
- Todorovic V, Giadrossi S, Pelizon C, Mendoza-Maldonado R, Masai H, Giacca M (2005) Human origins of DNA replication selected from a library of nascent DNA. Mol Cell 19: 567–575 [DOI] [PubMed] [Google Scholar]
- Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC (2003) Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev 17: 1894–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashee S, Simancek P, Challberg MD, Kelly TJ (2001) Assembly of the human origin recognition complex. J Biol Chem 276: 26666–26673 [DOI] [PubMed] [Google Scholar]
- Wong MC, Scott-Drew SR, Hayes MJ, Howard PJ, Murray JA (2002) RSC2, encoding a component of the RSC nucleosome remodeling complex, is essential for 2 microm plasmid maintenance in Saccharomyces cerevisiae. Mol Cell Biol 22: 4218–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JL, Camiolo SM, Bashaw JM (2000) The minimal replicator of Epstein–Barr virus oriP. J Virol 74: 4512–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JL, Guan N (1991) Epstein–Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol 65: 483–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hayashi MK, Merkel O, Stillman B, Xu RM (2002) Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J 21: 4600–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and methods


