Abstract
Mammalian liver contains a Pregnane X Receptor (NR1I2), which binds drugs and other xenobiotics, and stimulates (or suppresses) expression of numerous genes involved in the metabolic elimination of foreign compounds and some toxic endogenous substances. In the present study we used microarray analysis to identify genes whose expression in rat liver was significantly altered by pregnenolone 16α-carbonitrile (PCN) treatment. PCN is a synthetic steroid that induces cytochrome P4503A expression and is hepatoprotective by increasing resistance to subsequent stressful insults. Significant induction was seen for 138 genes while expression of 82 genes was significantly repressed. We found induction of genes known to be induced by PCN, such as enzymes involved in drug metabolism and transport. In addition, many genes were differentially expressed whose functions concerned intracellular metabolism, transport of essential small molecules, cell cycle, and redox balance. Our results support the idea that the domain of PXR controlled gene networks may be even more extensive than currently thought and may extend to functions apart from xenobiotic metabolism.
Keywords: pregnenolone 16α-carbonitrile, pregnane X receptor, microarray, gene expression profiling
INTRODUCTION
It has been known for decades that animals treated with pregnenalone-16α-carbonitrile (PCN), a synthetic steroid, exhibit increased resistance to subsequent stressful insults. PCN appears to protect an animal by eliciting adaptive responses in the liver (and possibly in other tissues) manifested by such physiologic changes as increase in liver size (Garg et al., 1970), hypertrophy of hepatocytes (Japundzic et al., 1974), and increased bile production (Selye, 1971). Biochemical and molecular studies then verified that PCN induces expression of genes that encode mammalian liver enzymes involved in xenobiotic elimination. For example, PCN treatment of rats induces the levels of enzymes involved in oxidative metabolism (cytochromes P4503A) (Gonzalez et al., 1993), solubilization by conjugation (DIG- glucuronosyltransferase (Schuetz et al., 1986; Arlotto et al., 1987), glutathione-S-transferase (Maglich et al., 2002), sulfotransferase (Runge-Morris et al., 1999), and small molecule transport (product of the p-glycoprotein gene (P-gp or ABC1) (Maglich et al., 2002) and Na+-independent organic anion transporter 2 (Oatp2) (Staudinger et al., 2001a). These observations of multi-gene induction suggested the participation of a receptor-mediated pathway, an idea that has been verified by the discovery of the pregnane X receptor (PXR, NR1I2), a member of the superfamily of nuclear receptor transcription factors. PXR is expressed in liver and several other parenchymal tissues, specifically binds PCN, interacts with a specific DNA response element associated with PCN responsive genes (Goodwin et al., 2002), and accounts for the pharmacologic characteristics of induction of “drug-metabolizing” enzymes in the liver (Dussault and Forman, 2002). Indeed, many investigators accept that PXR functions as a “xenobiotic sensor” (Blumberg et al., 1998). Conversely, a variety of endogenous substances have been suggested as candidate physiological ligands for the PXR, including pregnanes, steroids, and bile acids (Dussault and Forman, 2002; Orans et al., 2005).
The wide variety of stresses which PCN counteracts suggests additional systems, apart from drug metabolism, that are activated in response to PCN treatment. To test this idea, we previously used the technique of differential display to search for mRNA species that were induced or repressed by PCN treatment. We found 76 mRNA species differential expressed in rat liver treated with PCN. One of these was identified as RT1.B(I)Beta, a component of the major histocompatability class II (MHC) gene family of the immune system (Jimenez et al., 2002). Another mRNA species induced by PCN was identified as IF1, an inhibitor peptide of the ATP synthase/ATPase complex (Jimenez et al., 2000). The finding that genes connected to immune regulation and mitochondrial energy production, respectively, are activated by PCN, suggested that functions unrelated to drug metabolism may be important in the protective adaptation program of the liver. This hypothesis prompted us to further investigate additional genes that fall under the inductive control of PCN/PXR.
In the present study, we used high-density oligonucleotide arrays to obtain a record of rat liver genes whose expression is affected by PCN. This technology can identify each gene, as well as the magnitude to which it is induced or repressed by PCN treatment when compared to a control. Statistical analysis of differential responses allows for identification of significant genes, and software analysis provides pathway information. Our results indicate that the domain of PCN-controlled genes in the liver is broader than previously recognized, and may provide novel insights into the physiological role of the PXR.
MATERIALS AND METHODS
Experimental animals and treatments
Adult female Sprague-Dawley rats (Harlan, Indianapolis, IN) weighing 150–200 g were maintained with free access to animal chow and water. After an overnight fast, animals (five treated, five controls) received a single intraperitoneal dose (100 mg/kg) of PCN suspended in corn oil or vehicle and were sacrificed 8 h later. Livers were isolated and total RNA was extracted as described below. Animals were maintained and research conducted in accordance with guidelines set forth by the Animal Care and Research Committee (University of Colorado). PCN was a gift from The Upjohn Company (Kalamazoo, MI).
Isolation and quality check of total RNA
Liver sections were excised, immediately homogenized in lysis buffer, and total RNA isolated by using the RNeasy Total RNA Isolation Kit (Qiagen Inc., Valencia, California). The integrity of the RNA samples was examined by the Agilent 2100 Bioanalyzer. Northern blot analysis of total RNA from each animal was performed to confirm PCN-induced expression of CYP3A. As expected, treatment of rats with 100 mg/kg of PCN for 8 h induced the expression of CYP3A mRNA in all 5 animals (Fig. 1).
Figure 1.
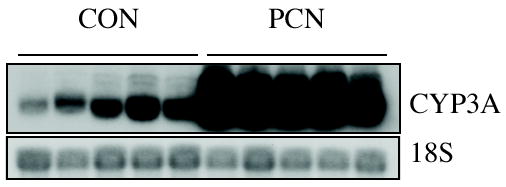
CYP3A expression from control and PCN-treated animals. Total RNA was extracted from the livers of rats treated with vehicle solvent or PCN as described in “Materials and Methods.” Total liver RNA was analyzed by northern blotting for expression of CYP3A using a cDNA probe that recognizes all CYP3A isoforms. The blot was subsequently probed with a cDNA encoding 18S to verify equal loading.
Preparation of RNA for hybridization to microarrays
Total RNA (1–5 μg) was converted to double-stranded cDNA (ds-cDNA) by using the Superscript Choice System (Life Technologies Inc., Gaithersburg, MD). An oligo-dT primer containing a T7 RNA polymerase promoter (Proligo Primers & Probes, Boulder, Colorado) was utilized. The ds-cDNA was purified and recovered by using GeneChip sample cleanup module (Affymetrix Inc, Santa Clara, California). In vitro transcription was performed to generate biotin-labeled cRNA. In vitro transcription was performed using a RNA Transcript Labeling Kit (Enzo, Farmingdale, New York) and ds-cDNA template was transcribed in the presence of a mixture of biotin-labeled ribonucleotides. Biotin-labeled cRNA was purified using GeneChip sample cleanup module (Affymetrix) or an RNeasy affinity column (Qiagen). To ensure optimal hybridization to the oligonucleotide array, the cRNA was fragmented. Fragmentation is performed such that the cRNA fragments are between 50–200 bases in length by incubating the cRNA at 94 C for 35 min in a fragmentation buffer. The sample was then added to a hybridization solution containing 100 mM MES, 1 M NaCl, and 20 mM EDTA in the presence of 0.01% Tween 20. The final concentration of the fragmented cRNA was 0.05 μg/μL.
GeneChip processing
The Affymetrix GeneChip® Rat Expression Set 230 was used to determine the changes in mRNA expression in response to PCN. The microarray set incorporates data from over 30,200 transcripts and variants, including 28,000 well substantiated rat genes. Set 230A chip contains ESTs (Expressed Sequence Tags) plus full-length sequences while Set 230B contains only ESTs. Hybridization was performed by incubating 200 μL of the sample with the Affymetrix GeneChip® arrays. Hybridizations were performed at 45 C for 16 h using a GeneChip® Hybridization Oven 640 (Affymetrix). After hybridization, the hybridization solutions were removed and the arrays were washed and stained with Streptavidin-phycoerythrin using a GeneChip® Fluidics Station 450 (Affymetrix). Arrays were read at a resolution of 2.5 to 3 microns using the GeneChip Scanner 3000 (Affymetrix).
Data analysis and statistical and informatic analysis
Detailed protocols for data analysis of Affymetrix microarrays and extensive documentation of the sensitivity and quantitative aspects of the method are described in the documentation of the Affymetrix Microarray Analysis Suite version 5.0 (MAS 5.0, Affymetrix). Briefly, each gene is represented by the use of ~11 perfectly matched (PM) and mismatched (MM) control probes. The MM probes act as specificity controls that allow the direct subtraction of both background and cross-hybridization signals. Raw fluorescence data are processed into normalized expression values. Raw data from array scans were averaged across all gene probes on each array and a scaling factor was applied to bring the average intensity for all probes on the array to 500. This allows any sample to be normalized for comparison to any other comparable sample, i.e. control vs. PCN treatment. Rat 230 type A and B chips were combined, resulting in 31,142 gene fragments. These expression values output by MAS 5.0 were further processed by the method described below to produce a list of 220 genes that are differentially expressed with high confidence.
We applied two preprocessing filters to the output of MAS 5.0 to remove meaningless genes; a variance filter and an absolute call filter. The median filter removes genes without at least moderate variance; genes whose expression does not vary at all seem unlikely to be involved in these phenotypes. In most circumstances, including this one, it is reasonable to assume that only a modest proportion of genes changed expression levels, and therefore the gene with the median variance is a reasonable model of null variation; i.e., the variation due to other factors. We calculate the variance s2 for each gene; the null hypothesis is that these variances represent random and Normally distributed noise. We can then compute the statistic W = (N-1)s2/median(s2), where N is the number of observations of the gene, which is approximately chi-square distributed with N-1 degree of freedom (Hogg and Craig, 1978). We calculate a p value for rejecting the null hypothesis that the gene did not vary, and perform the False Discovery Rate (FDR) multiple testing correction (Benjamini and Hochberg, 1995), setting the FDR to be 0.10. The genes that pass through this filter have significantly greater variation than the median variation gene, with at most 10% of that list including genes having true variation less than or equal to median variation. The second filter uses Affymetrix’s mRNA detection call to exclude all genes with an Absolute Call of “Absent” in all experiments. These pre-processing steps screen out genes with low total variance, and low mRNA levels; the remainder, 9117 genes, is then analyzed for differences related to experimental condition.
The D’Agostino normality test was applied to each chip individually, and all had p values of less than 0.001, indicating normal distributions. We then performed a two-tailed t-test. The false discovery rate procedure is again used to correct for multiple testing, producing a final list of 220 differentially expressed genes at FDR of 0.07. Selection of a lower FDR results in no differentially expressed genes, and in the context of 220 genes, an FDR setting of 0.07 is conservative. The result can be interpreted using the formula: FDR = expected number of false positive/total number of positives, where total number of positives is 220 and FDR is 0.07, for an expected number of false positives equal to 15 of the 220 genes. Of these, 138 genes were induced and 82 repressed. The entire dataset is available at NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE4959.
The Affymetrix probe set identifiers for these genes were then mapped to GenBank accession numbers. The Gene Ontology database (http://www.ncbi.nlm.nih.gov/entrez) was used to identify known functional information regarding the differentially expressed genes, as presented in Tables 1 and 2 under the heading “Group.” The genes for which the annotation was unclear such as ESTs and those of weak identity were excluded from the table. The majority of annotated differentially expressed genes identified were ESTs with unknown functions, a finding most likely the result of an inadequate knowledge of the rat genome and transcriptome.
Table 1.
Genes significantly up-regulated by PCN
| Fold Change | Gene/Description | Gene Symbol | GenBank Accession No. | Group |
|---|---|---|---|---|
| 10.36 | Growth arrest and DNA-damage-inducible 45 beta | Gadd45β | NM_001008321 | Binding-protein |
| 5.92 | Moderate similarity to p53-associated protein (H. sapiens) | MDM2 | CAA78055 | Binding-protein |
| 4.28 | Cytochrome P45026, retinoic acid | Cyp26 | NM_130408 | Metabolism
Transport-electron |
| 4.01 | Moderate similarity to suppression of tumorigenicity 5 | St5 | NM_005418 | Signal transduction
Tumor suppression |
| 3.67 | Aminolevulinic acid synthase 1 | Alas1 | NM_024484 | Metabolism-heme biosynthesis |
| 3.07 | P450 (cytochrome) oxidoreductase | Por | NM_031576 | Transport-electron |
| 2.74 | Moderate similarity to Ras association, domain family1 (H. sapiens) | RALGDS/AF6 | NM_007182 | Signal transduction
Tumor suppression |
| 2.74 | Cytochrome P450, 3a18 | Cyp3a18 | NM_145782 | Metabolism
Transport- electron |
| 2.55 | ATP-binding cassette, sub-family C (CFTR/MRP), member 3 | Abcc3 | NM_080581 | Binding-ATP
Transport |
| 2.45 | Moderate similarity to HUMAN sorting nexin 8 | SNX8 | NM_013321 | Intracellular signaling
Transport- protein |
| 2.40 | Similar to B-cell leukemia/lymphoma 3 (predicted) | Bcl3 (predicted) | XM_214856 | Cell cycle |
| 2.35 | Moderate similarity to yeast Sec31p homolog (H. sapiens) | SEC31p | NM_199530 | Unknown |
| 2.28 | Strong similarity to HUMAN calcium-binding mitochondrial carrier protein | SLC25A12 | NM_003705 | Binding-calcium ion
Transport |
| 2.24 | Similar to retinol dehydrogenase 10 (all-trans) [Mus musculus] | Rdh10 | NM_133832 | Oxidoreductase activity |
| 2.08 | ATP-binding cassette, sub-family B (MDR/TAP), member 1A | Abcb1a | NM_133401 | Binding-ATP
Transport |
| 2.03 | GTP cyclohydrolase 1 | Gch | NM_024356 | Hydrolase activity |
| 1.90 | Lactamase, beta 2 | Lactb2 | NM_00102427 | Unknown |
| 1.85 | Cyclin D1 | Ccnd1 | NM_171992 | Binding-protein
Cell cycle |
| 1.84 | Strong similarity to HUMAN ARL-6 interacting protein-2 | ARL6 | NM_032146 | Hydrolase activity
Binding-protein |
| 1.83 | BCL2/adenovirus E1B 19 kDa-interacting protein 3-like | Bnip3 | NM_053420 | Apoptosis |
| 1.79 | Liver UDP-glucuronosyltransferase, phenobarbital-inducible form | Ugt2b3 | NM_153314 | Metabolism-glucuronosy transferase |
| 1.78 | Strong similarity to mitogen inducible gene mig-2 (H. sapiens) | PLEKHC1 | Z24725 | Binding-protein |
| 1.78 | Protein phosphatase 2a, catalytic subunit, beta isoform | Ppp2cb | NM_017040 | Protein phosphatase |
| 1.74 | DnaJ homolog, subfamily b, member 9 | Dnajb9 | NM_012699 | Protein binding-heat shock |
| 1.71 | Mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase | Mgat2 | NM_053604 | Transferase activity |
| 1.69 | PDZ domain containing 1 | Pdzk2 | NM_133226 | Binding-Protein
Intracellular signalling |
| 1.69 | Tumor necrosis factor receptor superfamily, member 1a | Tnfrsf1a | NM_013091 | Receptor activity
Intracellular signalling |
| 1.67 | Glutathione S-transferase, mu 1 | Gstm1 | NM_017014 | Metabolism-glutathione |
| 1.67 | Purinergic receptor P2Y, G-protein coupled 2 | P2ry2 | NM_017255 | Binding-nucleotide |
| 1.66 | BCL2/adenovirus E1B 19 kDa-interacting protein 3-like | Bnip3l | NM_053420 | Apoptosis |
| 1.63 | Phospholipase A2, activating protein | Plaa | NM_053866 | Metabolism-phospholipid |
| 1.62 | TERF1 (TRF1)-interacting nuclear factor 2 | Tinf2 | NM_001006962 | Binding-protein |
| 1.60 | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 | Abcc2 | NM_012833 | Binding-ATP
Transport |
| 1.58 | RAB2, member RAS oncogene family | Rab2 | NM_031718 | Binding-nucleotide
Protein transport |
| 1.55 | Cytochrome P450, subfamily 3A, polypeptide 3 | Cyp3a3 | NM_013105 | Metabolism
Transport-electron |
| 1.55 | Heat shock 22kDa protein 8 | Hspb8 | NM_053612 | Protein binding-heat shock |
| 1.53 | Valosin-containing protein (p97)/p47 complex-interacting protein p135 | Ycip135 | NM_176857 | Binding-protein |
| 1.52 | Proteasome (prosome, macropain) 26S subunit, non-ATPase 4 | Psmd4 | NM_031331 | Transport-fluid |
| 1.49 | Moderate similarity to mitochondrial ribosomal protein L11 (H.sapiens) | MRPL11 | NM_016050 | Structural constituent of ribosome |
| 1.48 | Moderate similarity to elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 (H.sapiens) | ELOVL1 | NM_022821 | Fatty acid biosynthesis |
| 1.47 | Sjogren syndrome antigen B | Ssb | NM_031119 | Binding-RNA |
| 1.46 | Eukaryotic translation initiation factor 4 gamma, 1 (predicted) | Eif4g1 (predicted) | XM_213569 | Binding-RNA
Protein biosynthesis |
| 1.44 | t-Complex protein 1 | Tcp1 | NM_012670 | Binding-nucleotide
Protein folding |
| 1.44 | Lectin, galactose binding, soluble 3 | Lgals3 | NM_031832 | Binding-protein
Cell adhesion |
| 1.41 | Similar to SAR1a gene homolog mRNA | Sara1 | NM_001007739 | Binding-nucleotide |
| 1.40 | RAB6, member RAS oncogene family | Rab6 | XM_574503 | ATPase/GTPase activity |
| 1.38 | Moderate similarity to Poly(A) polymerase alpha (PAP) (H. sapiens) | PAPOLA | NM_032632 | Binding-RNA
Transferase activity |
| 1.37 | Fibronigen-like protein 1 | Fgl1 | NM_172010 | Cell growth |
| 1.33 | Strong similarity to small GTP-binding protein; RAB1B, member RAS oncogene family (H.sapiens) | RAB1B | NM_030981 | Binding-nucleotide
Signal transduction |
| 1.32 | Heterogeneous nuclear ribonucleoprotein U | Hnrpu | NM_057139 | Binding-nucleotide
RNA processing |
| 1.29 | Eukaryotic translation initiation factor 2, subunit 1 (alpha ) | Eif2s1 | NM_019356 | Binding-nucleic acid
Binding-protein Protein biosynthesis |
| 1.27 | DnaJ (Hsp40) homolog, subfamily A, member 2 | Dnaja2 | NM_032079 | Protein binding-heat shock |
| 1.26 | Leukocyte cell-derived chemotaxin 2 (predicted) | Lect2 (predicted) | XM_341486 | Metalloendopeptidase activity
Chemotaxis |
Table 2.
Genes significantly down-regulated by PCN
| Fold Change | Gene/Description | Gene Symbol | GenBank Accession No. | Group |
|---|---|---|---|---|
| −1.19 | Non-metastatic cell expressed protein 3 (nucleoside diphosphate kinase) | Mne3 | NM_053507 | Kinase activity
Nucleotide biosynthesis |
| −1.25 | Death-associated protein | Dap | NM_022526 | Apoptosis |
| −1.27 | RNA terminal phosphate cyclase domain 1 | Rtcd1 | NM_001004227 | Metabolism-RNA |
| −1.28 | Cytochrome P450 monooxygenase | Cyp2t1 | NM_134369 | Cytochrome-electron transport |
| −1.32 | Apoptotic peptidase activating factor 1 | Apaf1 | NM_023979 | Apoptosis |
| −1.35 | Low density lipoprotein receptor-related protein 11 (predicted) | Lrp11 (predicted) | XM_238072 | Receptor activity
Endocytosis |
| −1.35 | Ras homolog gene family, member E | Arhe | NM_001007641 | Binding-GTP |
| −1.39 | Dipeptidylpeptidase 4 | Dpp4 | NM_012789 | Catalytic activity aminopeptidase |
| −1.39 | Cytochrome P450, subfamily 2F, polypeptide 1 | Cyp2f1 | NM_019303 | Cytochrome-electron transport |
| −1.41 | Kinesin family member C2 | Kifc2 | NM_198752 | Binding -ATP
Microtubule motor activity |
| −1.41 | Acyl-CoA synthetase long-chain family member 4 | Acsl4 | NM_053623 | Catalytic activity
Metabolism-fatty acid |
| −1.43 | Glutamate dehydrogenase 1 | Glud1 | NM_012570 | Binding-nucleotide
Metabolism-amino acid |
| −1.45 | Moderate similarity to B-cell CLL/lymphoma 7A (H.sapiens) | BCL7A | NM_020993 | Actin binding |
| −1.49 | Methylmalonyl CoA epimerase (predicted) | Mcee (predicted) | XM_215213 | Metabolism- L-methylmalonyl-CoA |
| −1.59 | Carbonic anhydrase 2 | Ca2 | NM_019291 | Carbonic anhydrase activity |
| −1.61 | Liver glycogen phosphorylase | Pygl | NM_022268 | Catalytic activity
Metabolism-carbohydrate |
| −1.75 | Matrilin 1, cartilage matrix protein | Matn1 | NM_001006979 | Structure-extracellular |
| −1.79 | Aquaporin 8 | Aqp8 | NM_019158 | Transport-water |
| −1.79 | Aquaporin 9 | Aqp9 | NM_022960 | Transport-amine
Canalicular bile acid |
| −1.92 | Rab38, member of RAS oncogene family | Rab38 | NM_145774 | Binding-nucleotide
Transport-protein |
| −1.92 | Arginine vasopressin receptor 1A | Avpr1a | NM_053019 | G-protein coupled receptor activity |
| −2.04 | CTL target antigen | Cth | NM_017074 | Metabolism-amino acid |
| −2.04 | Strong similarity to secreted modular calcium-binding protein1 (H. sapiens) | SMOC1 | NM_001034852 | Binding-calcium ion |
| −2.08 | Similar to NOGO-interacting mitochondrial protein (H. sapiens) | RTN4IP1 | NM_032730 | Oxidoreductase activity |
| −2.27 | Glrx1 glutaredoxin 1 (thioltransferase) | Glrx 1 | NM_022278 | Transport-electron |
| −2.38 | Moderate similarity to protein vitrin (H. sapiens) | VIT | NP_444506 | Unknown |
| −2.50 | Myotubularin related protein 7 | Mtmr7 (predicted) | XM_240417 | Actin binding
Phospholipid dephosphorylation |
| −3.85 | Claudin 1 | Cldn1 | NM_031699 | Binding-protein |
| −7.14 | Low density lipoprotein receptor-related protein 5 (predicted) | Lrp5 (predicted) | XM_215187 | Receptor activity |
Quantitative real-time RT-PCR
Total RNA was treated with DNase I prior to analysis. Real time RT-PCR was performed using the ABI Prism 7700 sequence detector (Perkin Elmer Corp./Applied Biosystems) with the following primers: CYP3A1 (also referred to as CYP3A23) (accession no. X96721) forward primer, 5′-CTCACACTGGAAACCTGGGTC-3′, reverse primer, 5′-CGGGTCCCAAATCCGTAGA-3′, and TaqMan probe, 5′-TCCTGGCAGTCGTCCTGGTGCTC-3′, giving a product of 65 bp; Myd118/GADD45β (accession no. NM_001008321) forward primer, 5′-GTGACTGAATCATGACCCTGGA-3′, reverse primer, 5′-ACCGCCTGCATCTTCTGAAC-3′, and TaqMan probe, 5′- CTGGTGGCGAGCGACAACGC -3′, giving a product of 67 bp; claudin 1 (accession no. NM_031699) forward primer 5′-TTGCAGCTTCTGGGTTTCATC-3′, reverse primer, 5′-AGGGCAGTGCTGACGATAGAG-3′, and TaqMan probe, 5′- TGGCTTCGCTGGGATGGATCG-3′, giving a product of 65 bp. Each real time RT-PCR reaction was performed in duplicate and normalized to the 18S ribosomal RNA levels in the same sample. Since 18S ribosomal RNA is abundant when compared to the target, the RNA is diluted approximately 100 fold for 18S quantitation and the dilution factor taken in to account when calculating the quantities of 18S. The statistical analyses were performed using GraphPad Prism (version 4.0, GraphPad Software, San Diego, CA).
RESULTS
The profile of hepatic gene expression in rats exposed to PCN was analyzed using the Affymetrix GeneChip® Rat Expression Set 230. Recognizing that each of the large number of genes being tested is effectively a separate hypothesis about contribution to phenotype we applied multiple testing corrections to ensure meaningful results. The array technology we used allows for a comprehensive display of the primary genes affected by PCN, without the need for any prediction concerning which genes to look for. We also recognized that the use of a single animal could result in variability in the expression profile. To accommodate this, we used RNA from 5 vehicle-treated and 5 PCN-treated animals to screen 10 microarray chips as well as statistical analysis to maximize the accuracy of the array data. We found several hundred genes that demonstrated significant increases or decreases in the expression of mRNAs in PCN-treated liver. Standard statistical programs were applied using receiver/operator characteristics to maximize specificity of changes in gene expression and minimize false positives. A threshold for induction and repression was implemented to meet these requirements (see Tables 1 and 2).
Significant induction or repression was seen for 220 genes based on the analysis of hybridization to ten microarray chips. We found induction of 138 genes, including those known to be induced by PCN, such as cytochrome P4503A, aminolevulinic acid synthase (the rate limiting enzyme for heme biosythesis), UDP glucuronosyltransferase, glutathione S-transferase, and drug-transporting proteins such as Abcb1a. In addition, 82 genes were significantly repressed, including cytochrome P4502F1, for which ethoxycoumarin is a traditional substrate. Many of the genes identified involve functions unrelated to drug metabolism, including various aspects of intracellular metabolism and transport of ions, proteins, and other small molecules. These functions encompass various organelles including the mitochondria, golgi, and plasma membrane. Of the 220 annotated differentially expressed genes, 82 have known process functions (Tables 1 and 2). We found the majority of genes altered by PCN were involved in binding (e.g. protein, nucleotide, nucleic acid, calcium ion), metabolism (e.g. amino acid, phospholipid, fatty acid), and transport (e.g. bile acid, electron).
The expression of CYP3A1 and selected genes identified as being differentially expressed by microarray analysis was validated by quantitative RT-PCR. We chose CYP3A1 for validation because its expression, known to be highly induced by PCN in rat liver (Quattrochi and Guzelian, 2001), was not identified in the array analysis as being differentially expressed, although PCN-stimulated expression was found for rat CYP3A18 (2.74-fold) and transcripts similar to human CYP3A3 (1.50-fold). We found by RT-PCR analysis of RNA from 5 control and 5 PCN-treated animals (the same RNA preparations used for the microarray) and using primers specific to the CYP3A1 sequence that CYP3A1 expression was induced 8.2-fold over control animals (Fig. 2). This discrepancy is most likely due to cross-hybridization of multiple, homologous CYP3A transcripts, some of which are highly expressed constitutively, to the arrayed CYP3A1 target oligonucleotides.
Figure 2.
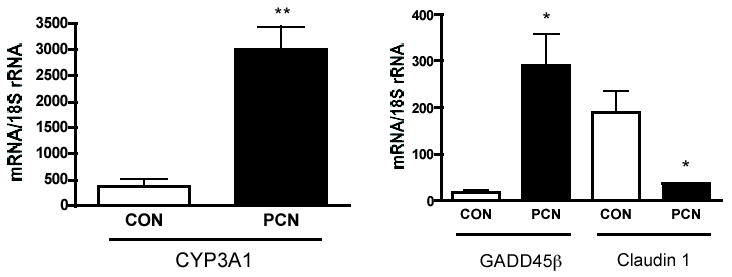
Validation by TaqMan quantitative RT-PCR of selected genes that were altered in the livers of PCN-treated animals. Expression of CYP3A1, GADD45β, and Claudin 1 was determined by quantitative RT-PCR. Data are presented as the mean ± standard deviation of the level of specific gene mRNA expression per 18S ribosomal mRNA as described in “Materials and Methods.” Quantitative RT-PCR was performed on RNA from 5 control and 5 PCN-treated animals (CYP3A1, left graph) and 3 control and 3 PCN-treated animals (GADD45β and Claudin 1, right graph) (the same RNA preparations used for the microarray) and using primers specific to the CYP3A1 sequence (also referred to as CYP3A23, accession no. X96721), GADD45β sequence (NM_001008321), and Claudin 1 sequence (NM_031699). P< 0.05* and P< 0.001** compared to control animals with unpaired t test.
In our study, PCN stimulated the highest level of expression of a gene product not involved in drug metabolism, namely Myd118/GADD45β. There was good agreement between microarray and RT-PCR data for expression of GADD45β 10.36-fold vs. 16.77-fold, respectively (Table 1 and Fig. 2). GADD45β is a member of a family of genes that has been shown to be induced by DNA damage and other genotoxic stresses, as well as by terminal differentiation and apoptotic cytokines (Liebermann and Hoffman, 1998). Indeed, a large number of differentially expressed genes were found in the functional category characterized by pathways involved in cell cycle, growth suppression, apoptosis, cell signaling (Table 1 and 2), providing additional evidence that the activated PXR functions as a sensor of xenobiotic stress. Such findings also evoke important therapeutic considerations. If PCN can bypass immediate early transcription factors to directly stimulate the process of DNA replication and mitosis as has been proposed for the constitutive androstane nuclear receptor (CAR) (Columbano et al., 2005), then clinically relevant PXR agonists such as rifampicin may be useful for stimulating proliferation of transplanted donor livers, provided that drug-drug interactions can be avoided (Costa et al., 2005).
Claudin 1 was also selected for validation because it was identified as a gene that statistically displayed one of the highest levels of suppression, (see Table 2). There was also good agreement between microarray and RT-PCR data for changes in expression of claudin 1: -3.85 versus -5.26, respectively (Table 2 and Fig. 2). Claudins are a family of integral membrane proteins important for tight junction formation (Van Itallie and Anderson, 2004). Gap junctional intercellular communication is essential in the maintenance of tissue homeostasis and for the maintenance of differentiated liver functions; it also contributes to the protection against various cellular stresses (e.g. radiation, oxidative, and heat shock). Our findings of substantial suppression of claudin 1 expression in intact rat liver by PCN treatment may provide an explanation for the observation that PCN inhibits gap junctional intercellular communication in rat liver (Kolaja et al., 2000).
DISCUSSION
Design, analysis, and interpretation of gene expression microarray experiments requires careful attention to statistical and bioinformatic considerations. Uncorrected p values for each gene over a certain threshold can be deceptive; testing 10,000 genes at a critical value of p <0.05 means that 500 genes would be expected to appear positive by chance. We used the false discovery rate correction (Benyamini & Hochberg, 1995), a well accepted method in microarray analysis, to create statistically reliable gene lists with a reliable (and controllable) estimate of the number of false positives.
Molecular studies of the effects of PCN on the liver have traditionally focused on the structure, function and inducibility of enzymes involved in xenometabolism. However, recent work has identified specific genes involved in mitochondrial metabolism and immune response. In addition, reports that activation of the SXR (human PXR) appears to elicit expression of genes related to bone homeostasis (Tabb et al., 2003) and that PXR mRNAs (and its splice variants) are present in many human extrahepatic tissues such as brain, bone marrow and heart (Lamba et al., 2004) suggest a broad metabolic role for this receptor and the domain of genes it controls. The present results give further evidence for the multi-system effect of PCN. This steroid, acting specifically through the PXR receptor, simultaneously induces or represses literally hundreds of genes, most of which remain unidentified at this time. This suggests that the adaptive response of the liver requires the involvement of a surprisingly wide group of cellular systems, aside from metabolism and excretion of foreign toxic compounds.
Gene profiling studies confirm that PCN induces or represses expression of a large number of mRNA species. For example, transgenic mice expressing a constitutively active variant of the PXR indicate 168 ESTs are differentially expressed (about half induced) as compared to wild-type mice (Rosenfeld et al., 2003). The authors emphasize that the induced gene products appear to function in activation, solubilization, or elimination of xenobiotics. Still, their results do identify gene products whose functions are unrelated to drug metabolism such as those involved in cell cycle kinetics and growth factors. It is important to note that few of the identified sequences affected by PCN treatment of the rat in the present study appear as differentially expressed genes in these transgenic mice. Another study of wild-type mice focusing on 38 genes selected for their involvement in liver xenobiotic metabolism and elimination found that PCN induced expression 1.5 fold or greater of 16 genes analyzed by real time quantitative PCR analysis (suppression of mRNA was not examined) in the liver, whereas only 4 genes were differentially expressed in PCN-treated PXR-null mice (Maglich et al., 2002). Finally, examination of the effects of rifampin on mRNA expression in primary cultures of human hepatocytes (an inducer of the human PXR) found that 32 genes were induced as analyzed by either cDNA-based or oligonucleotide-based microarrays (Rae et al., 2001). Although left without comment by the authors, 1 of the induced genes is not involved in drug metabolism or transport (a DNA mismatch repair protein, PMS1). This finding could be important given that PCN is an anticarcinogen (Argus et al., 1978).
Because we sampled the liver at 8 h of exposure, we believe that most of the observed effects are likely to be primary changes effected by PCN, rather than secondary consequences of primary stimulated genes. However, further analysis is required to demonstrate the direct effect of PXR on the transcriptional activation of individual genes. Furthermore, we recognize that statistical significance of PCN altered gene expression (as demonstrated here) does not equate necessarily to the biological significance. Even modest changes can be important if the gene product limits the rate of a metabolic pathway, whereas in other functional contexts large changes may be biologically inconsequential. There is little evidence that in the rat, PCN interacts with CAR or other liver X-receptors, which may have overlapping functions in regulation of drug metabolism. Moreover, our previous studies of the MHCII gene by PCN in the liver indicated that induction could be distinguished from inductive effects of glucocorticoids (dexamethasone), which are also known to interact with the PXR, by comparing the 6 and 18 h treatments (Jimenez et al., 2002) (verification of this hypothesis will require isolation of each gene and detailed mechanistic studies of its response to the PCN/PXR activation). While transgenic mice have the advantage of avoiding pharmacologic non-specificities, the observed differentially expressed genes could reflect secondary responses to the longstanding activation of PXR, beginning with prenatal development. Such models also are unable to simulate the acute activation of the adaptive response of the liver and unable to recognize effects of induction and deinduction that accompany challenges by endogenous or exogenous inducers. Such differences in timing, approach, and in species may account, in part, for the lack of extensive correspondence between PCN-mediated differential expression of genes. Indeed, for reasons that are not clear at this time, our array results failed to return positive signals for MHCII as was expected from our studies of PCN induction of this gene in rat liver and in primary cultures of rat hepatocytes (Jimenez et al., 2002), even though treatment conditions (e.g. dose, time) were identical. IF1 inhibitor peptide, previously identified as being differentially expressed in rat liver (Jimenez et al., 2000) was also not identified by the microarray analysis, but this was most likely due to differences in its time-dependent expression. Accordingly, a combined approach of genetic models, pharmacologic challenges, and cell culture studies offers the best prospect of elucidating the complex mechanisms that undoubtedly underlie the highly conserved PXR pathways.
Given the conclusive evidence that PXR mediates both induction and repression of genes involved in complex metabolic pathways of not only xenobiotics but also endogenous substrates such as lipids (Edwards et al., 2002) and bilirubin (Roy-Chowdhury et al., 2003), it might be expected that targeted disruption of the PXR gene would produce animals which, if viable at all, would exhibit significant metabolic disturbances. However, PXR-null mice appear normal and exhibit few, if any, manifestations of altered metabolism (Xie et al., 2000); Staudinger et al., 2001b). These findings suggest that PXR may function, in part, as a standby receptor to elicit a unique set of responses under conditions of stress. Many forms of stress raise levels of glucocorticoids, which in high concentration can directly activate the PXR. Such stress need not represent xenobiotic challenges. Considering the variety of systems induced by PCN and that PXR is activated by glucocorticoids (dexamethasone), it is likely that the role of PXR is to trigger the liver to adapt to stresses that affect high levels of endogenous glucocorticoids. Use of array technology for screening large numbers of genes to detect changes stimulated by PCN treatment represents, at best, an approach termed hypothesis generation rather than hypothesis testing. Genes of potential interest, unexpectedly discovered by this approach to be regulated by PCN treatment, as for example our findings of GADD45β and claudin 1, must be verified and evaluated for significance by detailed follow-up studies. For instance, while GADD45β up-regulation in livers of rats exposed to PCN conforms to Seyle’s thesis that PCN affords protection from toxic agents (i.e., GADD45β protects hepatocytes from apoptosis), suppression of claudin 1, a cell-cell adhesion molecule, is counterintuitive, since intercellular junctions contribute to the protection against various toxic stresses. Given our findings that PXR may stimulate expression of many genes outside the drug metabolizing system, we continue to pursue our hypothesis that PXR may serve as the stress receptor. Indeed, our initial results may simply be the molecular representations of the physiologic findings 50 years ago by Hans Selye who predicted that “catatoxic” steroids such as PCN would be found to activate a system of multiple cellular and hormonal changes (as opposed to effects restricted to a single specific tissue such as the liver) in maintaining body homeostasis in an individual exposed to “environmental stress”.
Acknowledgments
This work was supported by National Institutes of Health Grant ES05744 (PSG) and AA13524 (LH). Microarray analysis was performed by the University of Colorado at Denver and Health Sciences Center Microarray Core Laboratory. Quantitative RT-PCR was performed by the University of Colorado Cancer Center PCR Core Facility.
References
- Argus MF, Hoch-Ligeti C, Arcos JC, Conney AH. Differential effects of beta-naphthoflavone and pregnenolone-16alpha-carbonitrile on dimethylnitrosamine-induced hepatocarcinogenesis. J Natl Cancer Inst. 1978;61:441–449. [PubMed] [Google Scholar]
- Arlotto MP, Sonderfan AJ, Klaassen CD, Parkinson A. Studies on the pregnenolone-16 alpha-carbonitrile-inducible form of rat liver microsomal cytochrome P-450 and UDP-glucuronosyltransferase. Biochem Pharmacol. 1987;36:3859–3866. doi: 10.1016/0006-2952(87)90450-3. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Blumberg B, Sabbagh W, Jr, Juguilon H, Bolado J, Jr, van Meter CM, Ong ES, Evans RM. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 1998;12:3195–3205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Columbano A, Ledda-Columbano GM, Pibiri M, Cossu C, Menegazzi M, Moore DD, Huang W, Tian J, Locker J. Gadd45beta is induced through a CAR-dependent, TNF-independent pathway in murine liver hyperplasia. Hepatology. 2005;42:1118–1126. doi: 10.1002/hep.20883. [DOI] [PubMed] [Google Scholar]
- Costa RH, Kalinchenko VV, Tan Y, Wang I. The CAR nuclear receptor and hepatocyte proliferation. Hepatology. 2005;42:1004–1008. doi: 10.1002/hep.20953. [DOI] [PubMed] [Google Scholar]
- Dussault I, Forman BM. The Nuclear Receptor PXR: A master regulator of “homeland” defense. Crit Rev Eukaryot Gene Expr. 2002;12:53–64. doi: 10.1615/critreveukaryotgeneexpr.v12.i1.30. [DOI] [PubMed] [Google Scholar]
- Edwards PA, Kast HR, Anisfeld AM. BAREing it all: the adoption of LXR and FXR and their roles in lipid homeostasis. J Lipid Res. 2002;43:2–12. [PubMed] [Google Scholar]
- Garg BD, Kovacs K, Blascheck JA, Selye H. Ultrastructural changes induced by pregnenolone nitrile in the rat liver. J Pharm Pharmacol. 1970;22:872–873. doi: 10.1111/j.2042-7158.1970.tb08463.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Liu SY, Yano M. Regulation of cytochrome P450 genes: molecular mechanisms. Pharmacogenetics. 1993;3:51–57. doi: 10.1097/00008571-199302000-00006. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Redinbo MR, Kliewer SA. Regulation of CYP3A gene transcription by the pregnane X receptor. Annu Rev Pharmacol Toxicol. 2002;42:1–23. doi: 10.1146/annurev.pharmtox.42.111901.111051. [DOI] [PubMed] [Google Scholar]
- Hogg R, Craig A. Introduction to Mathematical Statistics. Macmillan Publishing Company; New York: 1978. [Google Scholar]
- Japundzic M, Garg BD, Kovac K, Japundcic I. Effect of pregnenolone-16alpha-carbonitrile on mitotic activity in the intact and regenerating rat liver. Experientia. 1974;30:562–563. doi: 10.1007/BF01926353. [DOI] [PubMed] [Google Scholar]
- Jimenez BD, Quattrochi LC, Yockey CB, Guzelian PS. Identification by differential display of the IF1 inhibitor peptide of ATP synthase/ATPase as a gene inducible in rat liver by pregnenolone 16alpha-carbonitrile. Life Sci. 2000;67:1825–1832. doi: 10.1016/s0024-3205(00)00769-4. [DOI] [PubMed] [Google Scholar]
- Jimenez BD, Maldonado L, Dahl RH, Quattrochi LC, Guzelian PS. Ectopic expression of MHC class II genes (RT1. B(I) beta/alpha) in rat hepatocytes in vivo and in culture can be elicited by treatment with the pregnane X receptor agonists pregnenolone 16 alpha-carbonitrile and dexamethasone. Life Sci. 2002;71:311–323. doi: 10.1016/s0024-3205(02)01643-0. [DOI] [PubMed] [Google Scholar]
- Kolaja KL, Engelken DT, Klaassen CD. Inhibition of gap-junctional-intercellular communication in intact rat liver by nongenotoxic hepatocarcinogens. Toxicology. 2000;146:15–22. doi: 10.1016/s0300-483x(00)00162-1. [DOI] [PubMed] [Google Scholar]
- Lamba V, Yasuda K, Lamba JK, Assem M, Davila J, Strom S, Schuetz EG. PXR (NR1I2): splice variants in human tissues, including brain, and identification of neurosteroids and nicotine as PXR activators. Toxicol Appl Pharmacol. 2004;199:251–265. doi: 10.1016/j.taap.2003.12.027. [DOI] [PubMed] [Google Scholar]
- Liebermann DA, Hoffman B. MyD genes in negative growth control. Oncogene. 1998;17:3319–3329. doi: 10.1038/sj.onc.1202574. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown BD, Moore JT, Kliewer SA. Nuclear pregnane X receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol. 2002;62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- Orans J, Teotico DG, Redinbo MR. The nuclear xenobiotic receptor pregnane X receptor: recent insights and new challenges. Mol Endocrinol. 2005;19:2891–2900. doi: 10.1210/me.2005-0156. [DOI] [PubMed] [Google Scholar]
- Quattrochi LC, Guzelian PS. Cyp3A regulation: From pharmacology to nuclear receptors. Drug Metab Dispos. 2001;29:615–622. [PubMed] [Google Scholar]
- Rae JM, Johnson MD, Lippman ME, Flockhart DA. Rifampin is a selective, pleiotropic inducer of drug metabolism genes in human hepatocytes: Studies with cDNA and oligonucleotide expression arrays. J Pharmacol Exp Ther. 2001;299:849–857. [PubMed] [Google Scholar]
- Rosenfeld JM, Vargas R, Jr, Xie W, Evans RM. Genetic profiling defines the xenobiotic gene network controlled by the nuclear receptor pregnane X receptor. Mol Endocrinol. 2003;17:1268–1282. doi: 10.1210/me.2002-0421. [DOI] [PubMed] [Google Scholar]
- Roy-Chowdhury J, Locker J, Roy-Chowdhury N. Nuclear receptors orchestrate detoxification pathways. Dev Cell. 2003;4:607–608. doi: 10.1016/s1534-5807(03)00131-x. [DOI] [PubMed] [Google Scholar]
- Runge-Morris M, Wu W, Kocarek TA. Regulation of rat hepatic hydroxysteroid sulfotransferase (SULT2-40/41) gene expression by glucocorticoids: Evidence for a dual mechanism of transcriptional control. Mol Pharmacol. 1999;56:1198–1206. doi: 10.1124/mol.56.6.1198. [DOI] [PubMed] [Google Scholar]
- Schuetz EG, Hazelton GA, Hall J, Watkins PB, Klaassen CD, Guzelian PS. Induction of digitoxigenin monodigitoxoside UDP-glucuronosyltransferase activity by glucocorticoids and other inducers of cytochrome P-450p in primary monolayer cultures of adult rat hepatocytes and in human liver. J Biol Chem. 1986;261:8270–8275. [PubMed] [Google Scholar]
- Selye HJ. Hormones and resistance. J Pharm Sci. 1971;60:1–28. doi: 10.1002/jps.2600600102. [DOI] [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, Willson TM, Koller BH, Kliewer SA. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci USA. 2001a;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger J, Liu Y, Madan A, Habeebu S, Klaassen CD. Coordinate regulation of xenobiotic and bile acid homeostasis by pregnane X receptor. Drug Metab Dispos. 2001b;29:1467–1472. [PubMed] [Google Scholar]
- Tabb MM, Sun A, Ahou C, Grun F, Errandi JL, Romero KM, Pham H, Inoue S, Mallick S, Lin M, Forman BM, Blumberg B. Vitamin K2 regulation of bone homeostasis is mediated by the steroid and xenobiotic receptor. SXR J Biol Chem. 2003;278:43919– 43927. doi: 10.1074/jbc.M303136200. [DOI] [PubMed] [Google Scholar]
- Van Itallie CM, Anderson JM. The molecular physiology of tight junction pores. Physiology (Bethesda) 2004;19:331–338. doi: 10.1152/physiol.00027.2004. [DOI] [PubMed] [Google Scholar]
- Xie W, Barwick JL, Downes M, Blumberg B, Simon CM, Nelson MC, Neuschwander-Tetri BA, Brunt EM, Guzelian PS, Evans RM. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature. 2000;406:435–439. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]


