Abstract
Chitin is a major component of fungal cell walls and serves as a molecular pattern for the recognition of potential pathogens in the innate immune systems of both plants and animals. In plants, chitin oligosaccharides have been known to induce various defense responses in a wide range of plant cells including both monocots and dicots. To clarify the molecular machinery involved in the perception and transduction of chitin oligosaccharide elicitor, a high-affinity binding protein for this elicitor was isolated from the plasma membrane of suspension-cultured rice cells. Characterization of the purified protein, CEBiP, as well as the cloning of the corresponding gene revealed that CEBiP is actually a glycoprotein consisting of 328 amino acid residues and glycan chains. CEBiP was predicted to have a short membrane spanning domain at the C terminus. Knockdown of CEBiP gene by RNA interference resulted in the suppression of the elicitor-induced oxidative burst as well as the gene responses, showing that CEBiP plays a key role in the perception and transduction of chitin oligosaccharide elicitor in the rice cells. Structural analysis of CEBiP also indicated the presence of two LysM motifs in the extracellular portion of CEBiP. As the LysM motif has been known to exist in the putative Nod-factor receptor kinases involved in the symbiotic signaling between leguminous plants and rhizobial bacteria, the result indicates the involvement of partially homologous plasma membrane proteins both in defense and symbiotic signaling in plant cells.
Keywords: elicitor, host–pathogen interaction, N-acetylchitooligosaccharides, pathogen-associated molecular patterns
Higher plants have the ability to initiate various defense reactions such as hypersensitive responses, production of phytoalexins and antimicrobial proteins, and reinforcement of cell walls when they are infected by various pathogens (1, 2). They can distinguish self and non-self, or detect specific pathogens, through the perception of signal molecules (elicitors) mostly generated/secreted from pathogens. Fragments of cell surface macromolecules typical of microorganisms such as cell wall polysaccharides, secreted proteins, as well as a flagella protein, often serve as a potent elicitor to induce defense reactions. They are classified as “general elicitors” that are commonly found in various microorganisms and induce defense responses in a wide range of plant species. Perception of general elicitors has been thought to play an important role in the basic resistance, or nonhost resistance, of plants to most potential pathogens. It has also been emerged in recent years that the defense systems mediated by the perception of these “general elicitors” have a considerable similarity with mammalian innate immunity, in the recognition of pathogen-associated molecular patterns as well as the molecules involved in the perception and transduction of these signal molecules (3).
Chitin oligosaccharides (N-acetylchitooligosaccharides) are a representative general elicitor inducing defense responses in a wide range of plant cells including both monocots and dicots (4). Chitin oligosaccharides were reported to induce defense responses also in mammalian and insect cells (4, 5). Interestingly, specific modifications of chitin oligosaccharides by fatty acids, sulfate, or some sugars, generate “Nod factors” that induce nodulation in legume roots in the symbiotic interaction with rhizobial bacteria (6). Thus, the recognition of chitin oligosaccharides and related compounds seems to play a fundamental role in the establishment of basal resistance to potential pathogens in plants and in some cases the symbiotic relationships between leguminous plants and rhizobial bacteria.
Concerning to the receptor for chitin oligosaccharide elicitor, we previously identified a high-affinity binding protein for this elicitor in the plasma membrane of rice cells by affinity labeling (7). Similar binding proteins were also detected in various plant cells that could respond to the elicitor (8, 9). Correlation between the presence of the binding proteins and the elicitor responsiveness of these cells, correlation between the binding specificity and the preference of the structure of chitin oligosaccharides in defense responses, strongly indicated that the binding proteins function as a receptor, or a part of receptor complex, for chitin oligosaccharide elicitor. Here we report the purification of this chitin oligosaccharide elicitor-binding protein (hereafter designated as CEBiP), cloning of the corresponding cDNA and its functional characterization. These results indicate that CEBiP plays a key role in the chitin elicitor signaling. The results also showed that CEBiP shares some structural similarity with the receptor kinases involved in the perception of Nod-factors in legume roots.
Results
Purification of CEBiP.
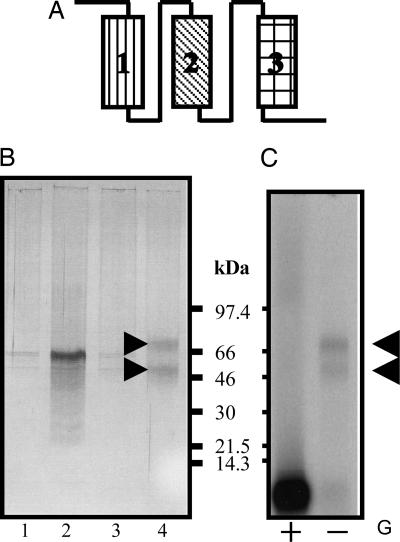
CEBiP, a chitin oligosaccharide elicitor- binding protein, was first identified as a 75-kDa plasma membrane (PM) protein from suspension-cultured rice cells by affinity labeling (7). We found that CEBiP, while retaining the elicitor binding activity, was effectively solubilized with Triton X-100 from the rice PM preparation. Thirty percent of the binding activity in the original PM preparation was solubilized and recovered by the treatment with 0.5% Triton X-100. To isolate the CEBiP from the solubilized fraction, an affinity matrix immobilized with a 2-(4-aminophenyl)ethylamine (APEA) derivative of N-acetylchitooctaose, (GlcNAc)8-APEA, was prepared. Two types of precolumns were also used to remove nonspecifically bound materials (Fig. 1A). After the application of the detergent solubilized PM protein and the removal of flow-through fractions, the bound proteins were recovered by the elution with an acidic buffer solution. SDS/PAGE followed by silver staining showed the presence of two protein bands, corresponding to 75 and 55 kDa (Fig. 1B). Affinity labeling with [125I]APEA-(GlcNAc)8 derivative showed that both of them carried the elicitor binding activity (Fig. 1C). These two proteins had identical N-terminal amino acid sequences, suggesting that the smaller-sized band might be generated from the 75-kDa protein by limited proteolysis during the purification. Indeed, when the molecular size of CEBiP was monitored by affinity labeling, the band of the [125I](GlcNAc)8-CEBiP was shifted gradually from 75 to 55 kDa by standing the solubilized PM protein at 4°C for 3 days, supporting this assumption. Purification yield was estimated to be 1.6% of the CEBiP originally present in the PM preparation based on the Scatchard analysis of the binding experiment and the molecular mass, 75 kDa, obtained from the affinity labeling. It was possible to obtain the sequence of N-terminal 32 amino acid residues as well as those of three peptide fragments obtained by lysyl-endopeptidase digestion.
Fig. 1.
Purification of CEBiP from the plasma membrane of suspension-cultured rice cell. (A) Affinity columns used for purification. Columns 1 and 2, precolumns packed with Sephadex G-75 and glycine-CH-Sepharose 4B, respectively. Column 3, major affinity column of (GlcNAc)8-APEA-CH-Sepharose 4B. (B) Silver staining of the SDS/PAGE gel. Lanes 1 and 2, proteins bound to precolumn 1 and 2, respectively. Lane 3, proteins eluted from the (GlcNAc)8-APEA-CH-Sepharose 4B column with elicitor-inactive sugar (cellohexaose and chitosan octasaccharide) solution. Lane 4, proteins finally eluted from the (GlcNAc)8-APEA-CH-Sepharose 4B column with glycine·HCl buffer. Arrowheads indicate CEBiP and its limited proteolysis product (see Results). (C) Affinity labeling of the proteins eluted from the (GlcNAc)8-APEA-CH-Sepharose 4B column with glycine·HCl buffer by a [125I]APEA-(GlcNAc)8 derivative, indicating that both 75- and 55-kDa bands retained the binding ability to the elicitor. Unlabeled (GlcNAc)8 (G, 25 μM) was added to ensure specific binding (+).
Cloning of CEBiP cDNA.
For the cloning of CEBiP cDNA, we designed 72 different kinds of primers corresponding to the N-terminal seven amino acid residues (K14 to T20) according to the information on the codon usage obtained from the rice EST database. These primers and a reverse primer for pBluescript SK− vector were used for the amplification of CEBiP-specific fragments by PCR using a rice cDNA library as a template. A PCR fragment (147 bp) was found to encode the N-terminal peptide sequence of CEBiP and further used to screen the full-length clone from the cDNA library. Three positive clones were finally obtained and all of them showed the identical nucleotide sequence for the ORFs. Analysis of the clone with the longest insertion showed a sequence of 1,436 nucleotide base pairs with an ORF encoding a polypeptide with 356 amino acid residues. Presence of a 28-aa signal peptide sequence as well as a 22-aa transmembrane domain at the C terminus was predicted for the polypeptide (Fig. 2B). Alignment of the N-terminal as well as the three internal peptide sequences of CEBiP showed a complete match with the deduced amino acid sequence, A1-A32, N142-Y148, G153-A158 and I176-K181, respectively (Fig. 2B). CEBiP has abundant serine, threonine, cysteine residues (34, 33, and 17 residues, respectively) and 11 possible glycosylation sites (S/T-X-N motif) in the molecule.
Fig. 2.
Structural features of CEBiP. (A) Schematic representation of the genomic DNA (AC099399) (a) and cDNA (b) encoding CEBiP. SP, signal peptide; ORF, open reading frame; NCR, noncoding region; LysM 1/LysM 2, LysM motif; TM, transmembrane region; RNAi, region inserted to the RNAi vector. (B) Amino acid sequence of CEBiP predicted from the cDNA. Solid and dashed underlines indicate the sequences corresponding to the N-terminal and internal peptide sequences obtained from the purified CEBiP, respectively. (C) Alignments of the two CEBiP LysM motifs to the consensus sequences of LysM motifs in the chitinases from Volvox (14), Lotus NFR1 (10), and Lotus NFR5 (11). The amino acid residues conserved in two or more sequences were underlined. Asterisks indicate the conserved residues in all four sequences.
The molecular weight calculated from the deduced peptide sequence of mature CEBiP was 34,640 for 328 aa residues, which is very different from the molecular weight, 75 k, expected from SDS/PAGE. However, MALDI TOF-MS analysis of the CEBiP preparation obtained by the affinity chromatography showed the presence of a broad peak approximately corresponding to 40 kDa, which corresponded well to the size of CEBiP obtained from the cDNA sequence, considering the presence of sugar chains in CEBiP. We already noticed the presence of sugar chains in CEBiP from the observation that CEBiP was adsorbed on Con A-agarose gel and eluted with methyl-α-mannoside. Glycoprotein nature of CEBiP was further confirmed by the treatment of the PM fraction by trifluoromethanesulfonic acid (TMSF), which chemically removes all sugar chains from glycoproteins. Western blotting analysis of the TMSF-treated PM proteins with an antibody against CEBiP detected a single band of 34-kDa polypeptide, which corresponded well to the size calculated from the deduced sequence of CEBiP. These observations clearly showed that the cloned cDNA encodes CEBiP and also this glycoprotein behaves very abnormally on SDS/PAGE because of the presence of sugar chains.
The sequence of CEBiP showed the highest homology with a rice cDNA clone (GenBank accession no. AK060664, 41% identical amino acid residues) that encodes a protein with unknown function and also Erwinia-induced protein 1 of Solanum tuberosum (GenBank accession no. AY187625, 38% identical amino acid residues). All of the cysteine residues in the Erwinia induced protein were conserved in CEBiP but not completely conserved in the homologous rice protein. On the other hand, CEBiP showed little homology with chitin-binding lectins, which have hevein motifs, such as rice agglutinin, wheat germ agglutinin, and chitinases from rice and other plants. Very interestingly, CEBiP had two extracellular LysM motifs in the region of Y85-P131 and Y149-P192 (Fig. 2B), which were also found in Nod-factor receptor kinases from Lotus japonicus, NFR1 (10), and NFR5 (11) (Fig. 2C). The LysM motif was first identified in bacterial lysins and muramidases, enzymes that degrade cell wall peptidoglycans (12), and also found in the chitinases from various origins, including Caenorhabditis elegans (13), Volvox carteri (14), and Kluyveromyces lactis (15), but not from higher plants. A BAC clone in the rice genome database (GenBank accession no. AC099399) was found to encode the CEBiP gene, which was consisted of three introns and four exons (Fig. 2A), located on chromosome 3.
Functional Analysis of CEBiP by Gene-Specific Knockdown.
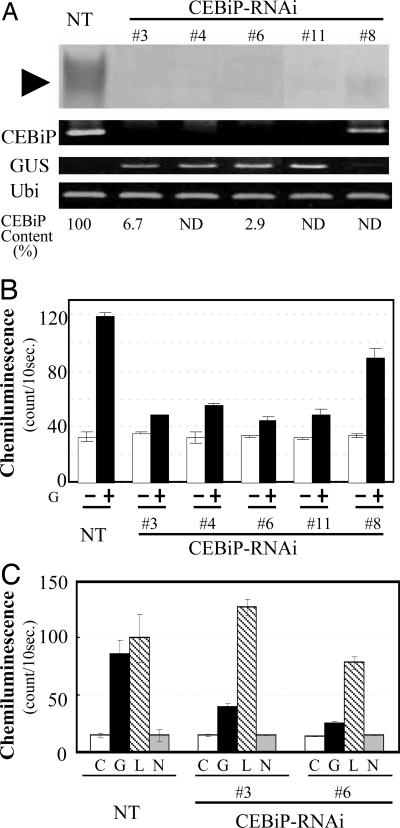
To see the function of CEBiP in the chitin elicitor signaling in rice cells, a plasmid for the gene-specific knockdown by RNA interference (RNAi) was constructed by using a sequence for the 3′-terminal region of CEBiP and transformed into the rice cells by using Rhizobium radiobacter (Agrobacterium tumefaciens). Four cell lines were selected as successful knockdown cell lines. RT-PCR and Western blotting analyses showed that the expression of CEBiP as well as the CEBiP protein were largely suppressed in these cell lines (Fig. 3A). Quantitative real-time PCR analysis indicated that the quantities of CEBiP mRNA of two CEBiP-RNAi cell lines were 2.9% and 6.7% of the amount found in the nontransformed cells.
Fig. 3.
Characterization of CEBiP-RNAi cell lines. (A) Western blot analysis of total proteins from the CEBiP-RNAi and nontransformed (NT) cell lines using anti-CEBiP antiserum (first row). The amount of mRNA for CEBiP (second row), GUS (third row), and ubiquitin (fourth row) genes in the CEBiP-RNAi and nontransformed cell lines was analyzed by RT-PCR. The amount of CEBiP mRNA was also determined by quantitative RT-PCR and shown as relative amount to the nontransformant (fifth row). ND, not determined. (B) ROS generation induced by (GlcNAc)8 elicitor (G, 100 μg/ml) in the CEBiP-RNAi and nontransformed rice cells. One hundred milligrams of the suspension-cultured rice cells were incubated in 1 ml of the medium with (+) or without (−) the elicitor for 30 min at 25°C. (C) Specific suppression of ROS generation induced by chitin oligosaccharide elicitor in the CEBiP-RNAi cell line. Sixty milligrams of the CEBiP-RNAi and nontransformed rice cells were incubated in 1 ml of the medium containing (GlcNAc)8 (G, 100 ng/ml), lipopolysaccharides (L, 50 μg/ml), chitosan octasaccharide (N, 2 μg/ml), or sterile H2O (C) for 2 h at 25°C. All of the data were the average of three independent experiments. Error bars indicate standard deviation.
Chitin oligosaccharide elicitor induces a biphasic generation of reactive oxygen species (ROS) in suspension cultured rice cells, which shows maximum peaks at 30 min and 2 h (16, 17). When the CEBiP-RNAi cell lines 3 and 6 were treated with (GlcNAc)8 elicitor for 30 min, the amount of H2O2 induced in both cell lines decreased to 1/4 to 1/7 of the amount induced in the nontransformed cells (Fig. 3B). On the other hand, the H2O2 generation induced by a bacterial LPS, which also induces defense responses in rice cells (Y. Dasaki, V. Balakrishnan, S. Tsuyumu, H. Yamane, H.K., et al., unpublished data), was not affected in the CEBiP-RNAi cell lines (Fig. 3C). The results clearly showed that the knock down of the CEBiP gene resulted in the specific suppression of the reactive oxygen generation induced by the chitin oligosaccharide elicitor.
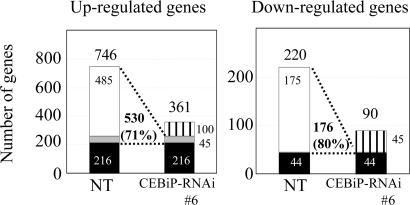
To see the effect of knockdown of CEBiP gene on elicitor-induced gene response, expression pattern of rice genes treated with (GlcNAc)8 elicitor was analyzed by using a microarray carrying 22 k rice genes (Fig. 4 and Table 1). The basal gene expression of the CEBiP-RNAi cell lines was very similar to that of nontransformed rice cells as evidenced by microarray analysis (data not shown). In the nontransformed rice cells, 746 genes were up-regulated and 220 genes were down-regulated by the elicitor treatment (Fig. 4). In the CEBiP-RNAi cell lines, 71% of the up-regulated genes and 80% of the down-regulated genes in the nontransformed cells became not responsive to the (GlcNAc)8 elicitor, or significantly decreased the degree of up/down-regulation, indicating a fundamental importance of CEBiP for the regulation of these chitin elicitor-responsive genes. The genes whose up-regulation was cancelled in CEBiP-knockdown cells included the genes encoding the enzymes for the biosynthesis of phytoalexins, lignin and ethylene, typical defense genes such as chitinase and thaumatin (Table 1). On the other hand, relatively small part of the up/down-regulated genes in the original nontransformed cells still responded to the elicitor similarly in the CEBiP-RNAi cell lines. A group of genes seem to be up/down-regulated by the elicitor only in the CEBiP-RNAi cell lines.
Fig. 4.
Microarray analyses of elicitor-responsive genes in the CEBiP-RNAi and nontransformed (NT) rice cells. Open box, genes responded to the elicitor only in NT; gray box, genes significantly decreased responsiveness to the elicitor (ratio of NT/CEBiP-RNAi >2) in CEBiP-RNAi; black box, genes equally responded to the elicitor in both NT and CEBiP-RNAi; hatched box, genes responded to the elicitor only in CEBiP-RNAi.
Table 1.
The number of up- or down-regulated genes of CEBiP-RNAi and nontransformed rice cells by the treatment with (GlcNAc)8
| Accession no. | Feature | Fold increase |
|
|---|---|---|---|
| NT | RNAi #6 | ||
| Up-regulated genes | |||
| AK072588 | Cinnamate 4-hydrocylase | 22.8 | ND |
| AK105971 | Pirin | 20.6 | ND |
| AK101431 | Patatin-like protein | 19.6 | ND |
| AK071482 | Caffeoy1-CoA 3-O-methyltransferase | 11.3 | ND |
| AK069456 | Peroxidase | 10.9 | ND |
| AK061042 | Endochitinase | 10.4 | ND |
| AK066687 | Shikimate kinase | 9.5 | ND |
| AK064250 | 1-Aminocyclopropane-1-carboxylate synthase | 4.8 | ND |
| AK073072 | CEBiP | 2.6 | ND |
| AK068993 | PAL | 48.4 | 2.9 |
| AK099946 | Thaumatin-like protein | 16.5 | 6.6 |
| Down-regulated genes | |||
| AK060423 | Alanine:glyoxylate aminotransferase-like protein | 11.2 | ND |
| AK071598 | Lipid transfer protein | 10.0 | ND |
| AK060625 | GDSL-motif lipase/hydrolase | 6.4 | ND |
| AK060582 | Replication protein A1(Os-RPA1) | 4.6 | 2.8 |
Typical genes affected for the elicitor responsiveness by the knockdown of CEBiP. ND, not detectable; NT, nontransformed.
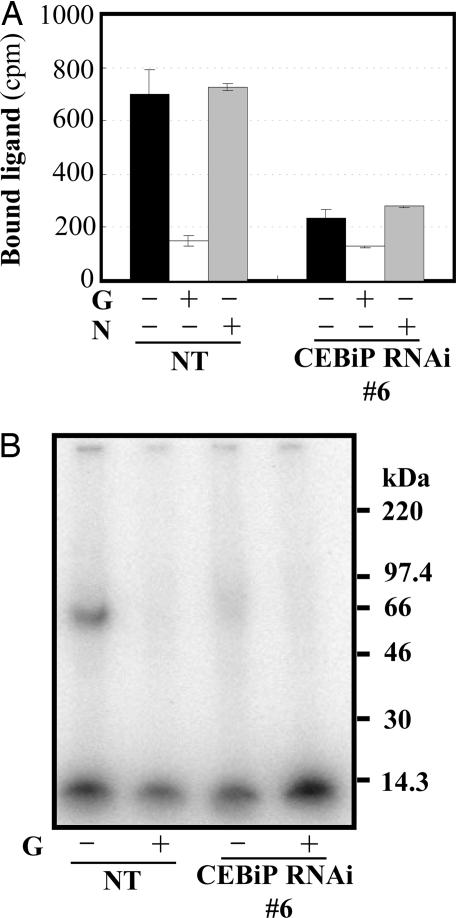
Binding assay as well as affinity labeling with [125I]APEA-(GlcNAc)8 derivative showed that the PM preparation from CEBiP-RNAi rice cells carried clearly reduced elicitor binding sites/protein (Fig. 5).
Fig. 5.
Decrease of the elicitor binding activity in the plasma membrane preparation from CEBiP-RNAi rice cells. (A) Binding activity of the PM preparation from CEBiP-RNAi and nontransformed (NT) rice cells to the [125I]APEA-(GlcNAc)8 derivative. Twenty micrograms of PM preparation was pretreated with elicitor-active [G, (GlcNAc)7] or inactive (N, chitosan heptamer) sugars (+) for 30 min before incubation with [125I]APEA-(GlcNAc)8 derivative. All of the data were the average of three independent experiments. Error bars indicate standard deviation. (B) Detection of the elicitor binding protein by affinity labeling with the [125I]APEA-(GlcNAc)8 derivative. Unlabeled (GlcNAc)8 (G, 25 μM) was added to ensure specific binding (+).
Discussion
Here we showed that a chitin elicitor binding protein, CEBiP, from the plasma membrane of rice cells plays a key role for the perception and transduction of chitin elicitor signal for defense responses. CEBiP-specific knockdown by RNA interference (CEBiP-RNAi) resulted in the suppression of the chitin elicitor-induced ROS generation in these cell lines. However, the ROS generation induced by another potent elicitor, LPS, was not affected, indicating a specific disruption of the chitin elicitor signaling in these cell lines. Microarray analysis of the expression pattern of rice genes in the CEBiP-RNAi cell lines also demonstrated that the knockdown of CEBiP greatly affected the gene responses induced by the chitin oligosaccharide elicitor. A large number of genes, which are normally induced/suppressed by the elicitor treatment, did not respond to the elicitor in the CEBiP-RNAi cell lines, or the responses were much weaker compared to the nontransformed cell lines. Elicitor binding activity of the PM preparation from the CEBiP-RNAi rice cells was drastically reduced compared to that of control cells, as shown by both binding assay as well as affinity labeling, further supporting that CEBiP is a molecule responsible for the elicitor binding. These results clearly indicate that CEBiP plays a critical role in the perception and transduction of chitin oligosaccharide elicitor in rice cells.
However, the expression pattern of some elicitor-responsive genes was not affected in the CEBiP-RNAi cell lines. This seems to indicate the presence of additional machinery for chitin perception, other than CEBiP, although the presence of such additional receptor(s) has not been detected by affinity labeling used in this work. It might be also possible that the effectiveness of the suppression of CEBiP gene expression somehow affected the gene responses induced by chitin oligosaccharide elicitor. The answer to this question remains to be solved in future studies involving the experiments with a mutant lacking a functional CEBiP gene as well as the survey of additional system(s) for the perception of chitin oligosaccharide elicitor. Some group of genes seems to become responsive to chitin oligosaccharide elicitor only in the transformant, although a rational explanation is difficult at present.
CEBiP is predicted to have a transmembrane domain at the C terminus but seems not to have any intracellular domains that are normally present in various membrane receptors. This observation strongly suggests that CEBiP requires “partner protein(s)” to form a functional receptor complex for successful signal transduction. Presence of such a receptor complex has been documented in the CLAVATA system involved in the regulation of meristem development in plants. In this case, a serine/threonine receptor kinase, CLAVATA1, is thought to form a heterodimeric receptor complex with another membrane protein, CLAVATA2, which has the similar extracellular domain but lacks the intracellular kinase domain. The CLAVATA1/CLAVATA2 heterodimer is thought to bind the ligand, CLAVATA3, and activate downstream responses (18). Presence of a complicated receptor system has also been documented in pathogen-associated molecular pattern recognition in vertebrates. In the case of LPS recognition in vertebrates, for example, LPS is initially bound by LPS-binding protein in serum and then transferred to a GPI-anchored membrane protein, CD14. The CD14-bound LPS seems to be further transferred to a complicated receptor complex including TLR4 and MD-2 on the plasma membrane to activate downstream defense responses (19). CEBiP might be a component of such a receptor complex for the perception of chitin oligosaccharide elicitor, possibly as a binding polypeptide.
Finding of two LysM motifs in the CEBiP sequence is very interesting. Originally, LysM motif was identified as a characteristic structural feature in the enzymes for bacterial cell wall degradation and also in the chitinases from yeast (K. lactis) (15) and alga (V. carteri) (14). Presence of LysM motif in CEBiP indicates the involvement of this motif in the perception of chitin oligosaccharide elicitor as similar to the substrate recognition in these enzymes. Interestingly, LysM motif was also found in the extracellular domain of putative Nod factor receptor kinases, NFR1 and NFR5, suggesting that the motif is also involved in the perception of the lipochitin–oligosaccharides, nodulation signals in the symbiosis between specific leguminous plants and corresponding rhizobial bacteria (10, 11). Thus, these LysM motif-containing membrane proteins seem to function for the perception of chitin-related signal molecules derived from fungi or bacteria on plant cell surface, although the downstream responses are different in the case of defense reactions and symbiotic responses.
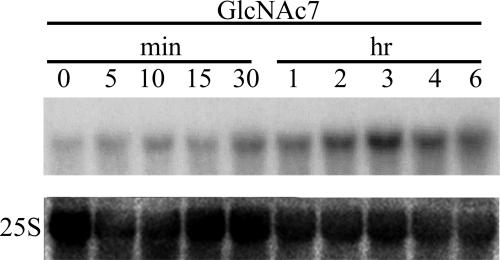
Interestingly, the expression of CEBiP gene itself was induced by the elicitor treatment (Fig. 6). Together with the observation that the expression of a number of genes seemingly involved in defense signaling were also induced by the elicitor treatment (20, 21), it seems to be a part of “sensitization” of defense machinery for further pathogen invasion.
Fig. 6.
Induction of CEBiP expression by chitin oligosaccharide elicitor. Expression of CEBiP induced by (GlcNAc)7 treatment (1 μg/ml) in the nontransformed rice cells was analyzed by Northern blotting using a cDNA probe.
Chitin elicitor-binding proteins similar to CEBiP are not only found in suspension-cultured rice cells but also found in the plasma membranes from various plant cells responsive to chitin oligosaccharide elicitor, such as barley, carrot, soybean, and wheat cells (8, 9), although the molecular sizes were slightly different depending on the origin. These results indicate that the defense systems based on chitin recognition is widely conserved among plant species. CEBiP is the first component identified for the perception of chitin oligosaccharide elicitor and should contribute for the understanding of the molecular machinery involved in the recognition of this molecular pattern for potential pathogens.
Materials and Methods
General Method.
PM was prepared by aqueous two-phase partitioning with the use of dextran T-500 and polyethyleneglycol 3350 from the suspension-cultured rice cells (22). The conjugate of (GlcNAc)8 with APEA derivative was synthesized as reported (7). The binding experiments and affinity labeling with the [125I]labeled ligand were conducted as described (7). The affinity matrix, (GlcNAc)8-APEA-CH-Sepharose, was prepared by immobilizing the (GlcNAc)8-APEA derivative on activated CH-Sepharose 4B (Amersham Pharmacia) according to the manufacturer’s protocol.
Elicitors and Plant Material.
Chitoheptaose and octaose were kindly supplied by Yaizu Suisankagaku Industrial (Shizuoka, Japan) and reacetylated before use. LPS from Pseudomonas aeruginosa was purchased from Sigma. Suspension culture of Oryza sativa L. cv. nipponbare was maintained by using modified N6 medium and subcultured as reported (23).
Purification of CEBiP from Rice Plasma Membrane.
Detailed procedures of the solubilization of the PM protein and purification of CEBiP were described in Supporting Text, which is published as supporting information on the PNAS web site. Briefly, CEBiP in the solubilized membrane fraction was purified by using an affinity column of (GlcNAc)8-APEA-CH-Sepharose, which was connected with two precolumns of Sephadex G-75 and glycine-CH-Sepharose 4B to remove nonspecifically adsorbed proteins.
Cloning of CEBiP cDNA.
Poly(A) RNA was obtained from the cultured rice cells by phenol/SDS method followed by the separation on oligo(dT) cellulose (Amersham Pharmacia) as reported (24). A cDNA library was constructed from the poly(A) RNA with λZAP II (Stratagene) as a vector. PCR was carried out with a reverse primer (5′-GGAAACAGCTATGACCATG-3′) for pBluescript SK− vector and 72 different primers corresponding to the seven amino acid residues of CEBiP (from K14 to T20), using the rice cDNA library as a template. The amplified fragments were subcloned into TA-vector of TA cloning kit (Invitrogen) and sequenced by a DNA sequencer. The full-length CEBiP cDNA was screened for the rice cDNA library (4 × 105 plaque-forming units) by using a 147-bp PCR product for the N-terminal sequence.
Antiserum Against CEBiP.
The coding region of CEBiP cDNA, except the sequence for the C-terminal transmembrane region, was ligated to the NdeI and BamHI sites of the His-tagged pET-16b vector (Novagen) and expressed in Escherichia coli according to the manufacturer’s protocol. The expressed protein was separated by SDS/PAGE, and the pieces of the gel containing CEBiP were used directly to raise an antiserum in rabbits. The antiserum was used without further purification.
CEBiP-Specific Knockdown by RNAi.
The CEBiP-specific RNAi vector was constructed by using Gateway pENTR/D-TOPO cloning kit (Invitrogen) and pANDA destination vector supplied by K. Shimamoto (Nara Institute of Science and Technology, Ikoma, Japan) (25). Detailed procedures of transformation and quantitative RT-PCR are described in Supporting Text.
ROS Assay.
Sixty or 100 mg of rice cells were transferred to 1 ml of the fresh medium in a 2-ml Eppendorf tube and incubated at 25°C with Thermomixer Comfort (Eppendorf) at 700 rpm. After a preincubation for 15 h, 4 μl of various elicitor solutions, (GlcNAc)8 (100 ng/ml), LPS (50 μg/ml), (GlcNH2)7 (2 μg/ml), or sterile H2O as a control, was added separately in each tube and incubated for 30 min or 2 h at 25°C. The experiment was carried out in triplicate. The ROS generated in the reaction mixture were determined by the luminol-dependent chemiluminescence assay (26). In some cases, the cells used for the ROS assay were immediately frozen in liquid nitrogen, smashed to powder, and used for RNA extraction for microarray analysis.
Microarray Analysis.
Microarray analyses were carried out by using a 60-mer rice oligo microarray containing 22,575 features (Agilent Technologies). The RNAs from rice cells were fluorescence labeled according to the manufacturer’s protocol. Each set of RNAs was labeled by swapping the dyes (Cy3 and Cy5) to normalize for dye bias. The slide glasses were scanned by an Agilent scanner. The data below 1,000 counts and under 2-fold fluorescent counts between Cy5 and Cy3 were not used for further analysis. The data set is available in the Gene Expression Omnibus (accession no. GSE4645).
Supplementary Material
Acknowledgments
We thank Drs. Y. Ito, K. Sakano, and S. Tanabe of the National Institute of Agrobiological Sciences (NIAS) for valuable advice; Dr. Y. Nagamura and Ms. R. Motoyama (Rice Genome Resource Center, NIAS) for technical assistance for microarray analyses; and Dr. I. J. Goldstein of the University of Michigan for the critical reading of the manuscript. This research was supported in part by the Program for Promotion of Basic Research Activities for Innovative Bioscience (PROBRAIN); Ministry of Agriculture, Fisheries, and Food Program for Rice Genome Research; and a grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Abbreviations
- PM
plasma membrane
- APEA
2-(4-aminophenyl)ethylamine
- RNAi
RNA interference
- ROS
reactive oxygen species.
Footnotes
References
- 1.Boller T. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995;46:189–214. [Google Scholar]
- 2.Dangl J. L., Jones J. D. G. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 3.Nürnberger T., Brunner F., Kemmerling B., Piater L. Immunol. Rev. 2004;198:249–266. doi: 10.1111/j.0105-2896.2004.0119.x. [DOI] [PubMed] [Google Scholar]
- 4.Shibuya N., Minami E. Physiol. Mol. Plant Pathol. 2001;59:223–233. [Google Scholar]
- 5.Furukawa S., Taniai K., Yang J., Shono T., Yamakawa M. Insect Mol. Biol. 1999;8:145–148. doi: 10.1046/j.1365-2583.1999.810145.x. [DOI] [PubMed] [Google Scholar]
- 6.Truchet G., Roche P., Lerouge P., Vasse J., Camut S., de Billy F., Promè J.-C., Dènariè J. Nature. 1991;351:670–673. [Google Scholar]
- 7.Ito Y., Kaku H., Shibuya N. Plant J. 1997;12:347–356. doi: 10.1046/j.1365-313x.1997.12020347.x. [DOI] [PubMed] [Google Scholar]
- 8.Day R. B., Okada M., Ito Y., Tsukada K., Zaghouani H., Shibuya N., Stacey G. Plant Physiol. 2001;126:1162–1173. doi: 10.1104/pp.126.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okada M., Matsumura M., Ito Y., Shibuya N. Plant Cell Physiol. 2002;43:505–512. doi: 10.1093/pcp/pcf060. [DOI] [PubMed] [Google Scholar]
- 10.Radutoiu S., Madsen L. H., Madsen E. B., Felle H. H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., et al. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- 11.Madsen E. B., Madsen L. H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., et al. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- 12.Joris B. FEMS Microbiol. Lett. 1992;70:257–264. doi: 10.1016/0378-1097(92)90707-u. [DOI] [PubMed] [Google Scholar]
- 13.Kraulis P. J. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 14.Amon P., Haas E., Sumper M. Plant Cell. 1998;10:781–789. doi: 10.1105/tpc.10.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butler A. R., O’Donnell R. W., Martin V. J., Gooday G. W., Stark M. J. R. Eur. J. Biochem. 1991;199:483–488. doi: 10.1111/j.1432-1033.1991.tb16147.x. [DOI] [PubMed] [Google Scholar]
- 16.Tsukada K., Ishizaka M., Fujisawa Y., Iwasaki Y., Yamaguchi T., Minami E., Shibuya N. Physiol. Plant. 2002;116:373–382. [Google Scholar]
- 17.Yamaguchi T., Minami E., Ueki J., Shibuya N. Plant Cell Physiol. 2005;46:579–587. doi: 10.1093/pcp/pci065. [DOI] [PubMed] [Google Scholar]
- 18.Haffani Y. Z., Silva N. F., Goring D. R. Can. J. Bot. 2004;82:1–15. [Google Scholar]
- 19.Triantafilou M., Triantafilou K. Trends Immunol. 2002;23:301–304. doi: 10.1016/s1471-4906(02)02233-0. [DOI] [PubMed] [Google Scholar]
- 20.Akimoto T. C., Sakata K., Yazaki J., Nakamura K., Fujii F., Shimbo K., Yamamoto K., Sasaki T., Kishimoto N., Kikuchi S., et al. Plant Mol. Biol. 2003;52:537–551. doi: 10.1023/a:1024890601888. [DOI] [PubMed] [Google Scholar]
- 21.Day R. B., Tanabe S., Koshioka M., Mitsui T., Itoh H., Ueguchi-Tanaka M., Matsuoka M., Kaku H., Shibuya N., Minami E. Plant Mol. Biol. 2004;54:261–272. doi: 10.1023/B:PLAN.0000028792.72343.ee. [DOI] [PubMed] [Google Scholar]
- 22.Shibuya N., Ebisu N., Kamada Y., Kaku H., Cohn J., Ito Y. Plant Cell Physiol. 1996;37:894–898. doi: 10.1093/oxfordjournals.pcp.a029002. [DOI] [PubMed] [Google Scholar]
- 23.Yamada A., Shibuya N., Kodama O., Akatsuka T. Biosci. Biotechnol. Biochem. 1993;57:405–409. [Google Scholar]
- 24.Watanabe A., Price C. A. Proc. Natl. Acad. Sci. USA. 1982;79:6304–6308. doi: 10.1073/pnas.79.20.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miki D., Shimamoto K. Plant Cell Physiol. 2004;45:490–495. doi: 10.1093/pcp/pch048. [DOI] [PubMed] [Google Scholar]
- 26.Schwacke R., Hager R. Planta. 1992;187:136–141. doi: 10.1007/BF00201635. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.