Abstract
We used expression quantitative trait locus mapping in the laboratory rat (Rattus norvegicus) to gain a broad perspective of gene regulation in the mammalian eye and to identify genetic variation relevant to human eye disease. Of >31,000 gene probes represented on an Affymetrix expression microarray, 18,976 exhibited sufficient signal for reliable analysis and at least 2-fold variation in expression among 120 F2 rats generated from an SR/JrHsd × SHRSP intercross. Genome-wide linkage analysis with 399 genetic markers revealed significant linkage with at least one marker for 1,300 probes (α = 0.001; estimated empirical false discovery rate = 2%). Both contiguous and noncontiguous loci were found to be important in regulating mammalian eye gene expression. We investigated one locus of each type in greater detail and identified putative transcription-altering variations in both cases. We found an inserted cREL binding sequence in the 5′ flanking sequence of the Abca4 gene associated with an increased expression level of that gene, and we found a mutation of the gene encoding thyroid hormone receptor β2 associated with a decreased expression level of the gene encoding short-wavelength sensitive opsin (Opn1sw). In addition to these positional studies, we performed a pairwise analysis of gene expression to identify genes that are regulated in a coordinated manner and used this approach to validate two previously undescribed genes involved in the human disease Bardet–Biedl syndrome. These data and analytical approaches can be used to facilitate the discovery of additional genes and regulatory elements involved in human eye disease.
Keywords: bioinformatics, expression quantitative trait locus analysis, gene regulation, ophthalmology
Data generated by the Human Genome Project coupled with recent advances in microarray technology and bioinformatics have made it possible to perform experiments that examine the expression of thousands of genes in a large number of related individuals and to use these data to identify the chromosomal locations of the genetic elements that are responsible for the variation in gene expression among individuals (1–9). The first few of these expression quantitative trait locus (eQTL) mapping experiments (1–3) revealed three important features of eukaryotic gene regulation: (i) much of the variation in gene expression can be traced to loci that are distant from the genes they control; (ii) these remotely acting loci do not contain transcription factors more often than would be predicted by chance; and (iii) single loci can regulate multiple diverse genes.
A major challenge facing biomedical research is the discovery of specific disease mechanisms that underlie heritable disorders that display complex inheritance, including Mendelian disorders that exhibit variable expressivity and incomplete penetrance (10–12). The identification of genes involved in complex disorders and genes that modify Mendelian disorders has been difficult despite the use of a number of rational approaches. These approaches include the identification of genes causing rare Mendelian forms of common complex disorders such as hypertension (13), obesity (14–17), glaucoma (18, 19), and macular disease (20–23). In most cases, coding sequence variations in genes that cause high-penetrance Mendelian disorders do not account for a major portion of the more common complex diseases.
An appealing possibility is that mutations that alter gene expression may play an important role in complex disease. It has been shown in transgenic animal studies that gene dosage of mutant genes can have a profound effect on phenotype. In fact, improper regulation of structurally normal genes and alterations in gene dosage can each cause disease (24). An interesting example of gene expression variation leading to disease is the finding that both overexpression and haploinsufficiency of the FOXC1 gene can lead to developmental defects of the anterior chamber of the eye (18). Any genetic element that can be shown to alter the expression of a specific gene or gene family known to be involved in a specific disease is itself an excellent candidate for involvement in the disease, either primarily or as a genetic modifier. The main goals of this study were (i) to apply the eQTL mapping approach to the mammalian eye to gain insight into regulatory mechanisms that are involved in human eye disease and (ii) to evaluate a strategy for detecting functionally related genes by using gene expression data acquired from animals in a large genetic cross to identify genes with highly coordinated expression.
Results
Selection of Inbred Rat Strains.
The following criteria were used to select the most appropriate strains of rats from among the >600 strains with data in the Rat Genome Database (http://rgd.mcw.edu/strains). For this experiment, it was necessary for the two parental rat strains (i) to be highly inbred; (ii) to be commercially available or otherwise readily available from academic institutions; (iii) to have widely diverse genetic origins; (iv) to have genotyping data available so that genetic diversity between strains and the degree of inbreeding within strains could be assessed and so that informative markers could be selected for the mapping cross; (v) to be robust breeders; and (vi) to be free of early-onset systemic phenotypes or degenerative eye phenotypes. Eight strains that met all six screening criteria (ACI, BN, BUF, F344, LEW, SHRSP, SR/JrHsd, and WKY) were evaluated in detail in the preliminary phases of this experiment. Microarray analysis and extensive genotyping of these strains revealed that SR/JrHsd and SHRSP were ideally suited for this study. These two strains are highly inbred and genetically distant. In addition, microarray analysis revealed that, among the eight strains examined, SR/JrHsd and SHRSP exhibited the greatest interstrain differences in gene expression (data not shown). Electroretinography and histopathologic examination of the eyes of these two strains were performed at 12 weeks of age and revealed no abnormalities (data not shown).
eQTL Mapping.
Based on these preliminary results, a cross of SR/JrHsd males and SHRSP females was performed to generate F1 and F2 animals. F1 animals were intercrossed, and 120 12-week-old male F2 offspring were selected for tissue harvesting, microarray analysis, and genotyping. The microarrays used to analyze the RNA from the eyes of these F2 animals contain >31,000 different probes. Transcripts complementary to more than one-half of these probes (18,976) were detected in the eyes of the F2 animals at a level sufficient to be considered “expressed” and with sufficient variation to allow mapping of eQTLs (see Materials and Methods). Among the set of 18,976 expressed probes, there were 3,057 that exhibited a 4-fold or greater change in expression among the F2 rats and 521 that exhibited at least an 8-fold change in expression. This degree of expression variation is very similar to that seen in the data of Chesler et al. (8) when the expression levels are analyzed as described in this study (data not shown).
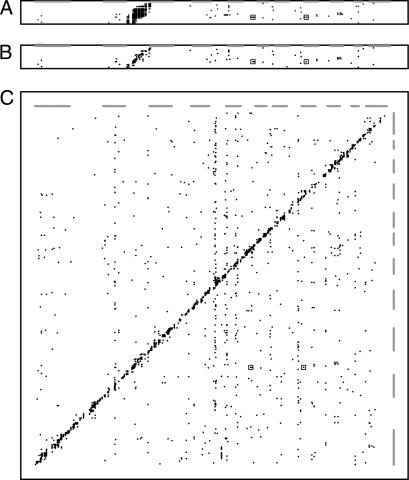
The 120 F2 rats were genotyped with 399 fully informative markers distributed across the genome (5 cM resolution) to evaluate the effect of genotype on the 18,976 expressed transcripts using ANOVA (25). Fig. 1 summarizes the results of this analysis by displaying significant linkages on a two-dimensional map (probes versus markers). Fig. 1A depicts all of the linkages we observed for transcripts on rat chromosome 4, with the level of significance indicated by the density of the points corresponding to the individual linkages. When the data are depicted in this way, one can see that many individual transcripts exhibit strong linkage with a number of contiguous markers (seen as short horizontal stripes in Fig. 1A). To avoid counting a single eQTL multiple times simply because it is physically linked to multiple markers, we assumed that any group of contiguous genetic markers that exhibits linkage to the expression of a single transcript is a single eQTL and that the location of this eQTL is best represented by the location of the marker with the highest ANOVA score (see ref. 5 for a similar approach). In Fig. 1B, this rule has been applied to the linkage data for transcripts on chromosome 4, and only linkages that are statistically significant at an α of 0.05 are shown. In addition, these linkages are shown with symbols of identical density regardless of the level of significance. With this approach, there are many fewer points to depict, and this allows each data point to be enlarged for greater clarity. In Fig. 1C, the linkage data from the entire rat genome are depicted in this manner.
Fig. 1.
Maps of eQTLs observed in the rat eye. Expressed transcripts are represented in genome order on the y axis, whereas genetic markers are represented in genome order on the x axis. (A) Linkage data from transcripts on chromosome 4. The relative statistical significance of the relationship between a given marker and a given transcript is shown with different densities of the plotted points (black = α of 0.00001; lightest gray = α of 0.05). The boxes indicate the locations of data corresponding to the significant noncontiguous linkages to the Opn1sw transcript. (B) Linkage data from chromosome 4 but with redundant linkages (multiple genetic markers detecting the same eQTL) removed as described. In addition, all linkages that are significant at an α level of 0.05 are shown with symbols of identical size and density. (C) Linkage data from the entire genome shown in the same fashion as in B. The genomic positions of the odd-numbered chromosomes are depicted as gray bars along both axes.
Two interesting features of the data can be seen in Fig. 1C. First, most of the statistically significant linkages fall along a diagonal band that corresponds to the set of genetic elements that map within 50 cM of the genes they control. It is often assumed that such physically contiguous elements would only affect the expression of the allele on which they reside, and thus they are often referred to as “cis-acting.” However, it has been noted that some regulatory elements that map very near the genes they control may actually act through a “trans” mechanism (26). Therefore, in this report, to avoid making any mechanistic assumption, these are referred to as contiguous regulatory elements. Fig. 1C also demonstrates that many significant linkages also occur between markers (x axis) and transcripts (y axis) that are on different chromosomes or that are separated by >50 cM. These noncontiguous regulatory elements are often assumed to act via some diffusible intermediate that in turn would be expected to act on both alleles of a given regulated gene. Thus, they are often referred to as trans-acting. However, in this report they are referred to as noncontiguous regulatory elements.
Table 1 summarizes the data from Fig. 1C numerically. The expression levels of 1,330 transcripts showed significant linkage to at least one locus at a genome-wide significance level of α = 0.001 (27). The false discovery rate was estimated based on these empirical P values to be only 1.4% of the detected linkages.
Table 1.
eQTLs detectable at the α = 0.001 level for various sample sizes from 30–120 animals
| No. of animals | |||
|---|---|---|---|
| 120 | 60 | 30 | |
| Linked loci | |||
| 1 locus | 1,259 | 710 | 207 |
| 2 loci | 69 | 44 | 8 |
| 3 loci | 2 | 2 | 1 |
| Total | 1,330 | 756 | 216 |
| Linked transcripts | |||
| 0 | 52 | 115 | 266 |
| 1 | 69 | 92 | 82 |
| 2 | 64 | 65 | 36 |
| 3 | 51 | 56 | 8 |
| 4 | 51 | 30 | 4 |
| 5–10 | 99 | 36 | 1 |
| 11–20 | 11 | 5 | 2 |
| 21+ | 2 | 0 | 0 |
Linked loci data show the number of distinct genetic loci significantly associated with the expression level of individual transcripts. Linked transcripts data show the number of different transcripts whose expression levels are significantly associated with individual genetic loci.
eQTLs can be meaningfully related to one another in at least two ways: (i) a single genetic element can affect the expression of multiple genes; and (ii) multiple genetic loci can affect the expression of a single gene (often referred to as complex inheritance). These different relationships can be seen in Fig. 1C as vertical and horizontal alignments of the data points, respectively. Horizontal alignments of data points in Fig. 1C occur when two different genetic loci are significantly linked to the expression of a single transcript. These horizontal alignments have the interesting property that, for a given number of animals in the eQTL mapping experiment, there is a limit to the number of different loci that can exhibit significant linkage to a single transcript. This is because there is only a single expression value obtained for each transcript in each animal, and this value reflects the sum of all of the genetic influences on the expression of that gene in that animal. Thus, observations that support a linkage at a given locus will confound the ability to detect linkage at a second locus. Although the sample size of 120 is reasonably large for an eQTL mapping experiment and is sufficient to detect the major loci that are linked to a transcript, it has limited power to detect additional secondary loci that account for only a small percentage of the variation in the expression of the transcript. Table 1 shows the practical result of this limitation. When considering the data from all 120 F2 animals in this experiment, the maximum number of distinct loci exhibiting significant linkage (P < 0.001) to a single transcript is three, and only 2 of 18,976 transcripts exhibit this degree of regulatory complexity.
Another interesting relationship among noncontiguous eQTLs occurs when a single genetic locus significantly affects the expression of multiple genes (seen as a vertical column of points in Fig. 1). In contrast to the horizontal relationship of multiple loci affecting a single transcript, there is no limit to the number of transcripts that one could observe to be associated with the genotypes of a single locus. This is because the expression of each transcript is independently assayed in each animal, and as a result detecting a significant association of a given marker to one transcript does not limit the ability to detect another association for the same marker to a different transcript. In this experiment 278 markers showed significant linkage to two or more transcripts at α = 0.001 (Table 1). The genetic marker that was linked to the expression levels of the largest number of different transcripts was D20RAT2, which was significantly linked to the expression of 33 genes at α = 0.001. However, all 33 of these genes lie within 10 Mb of the marker and are thus actually “contiguous” in nature. This clustering of significant linkages is quite unlikely to occur by chance. If the 1,330 significant linkages that we observed were randomly distributed among all of the markers in this experiment, one would expect the marker with the greatest number of linkages to be linked to perhaps 10 or 11 transcripts. If one considers the fact that the region surrounding D20RAT2 has approximately twice the gene density of the genome as a whole, one might be able to explain as many as 20 of the contiguous linkages using a combination of chance and increased gene density. However, when the annotation of the genes in this region was examined in detail, we noted that 10 of the linked transcripts belonged to the same gene family (RT1) and had very large (>10-fold) variations in gene expression. We therefore entertained the possibility that all of these genes might be regulated by a common genetic element. However, the fact that five of these genes were more highly expressed in one parental strain and five were more highly expressed in the other makes this explanation unlikely. The RT1 genes encode major histocompatibility antigens, and their expression has been shown to vary widely among animal strains and tissues (28). Thus, it seems that the most plausible explanation for the statistically unlikely clustering of contiguous linkages at the rat major histocompatibility locus is the evolutionary advantage of variability among genes that encode components of the immune system, coupled with the large genetic distance that separates the two parental strains in this experiment.
The genetic marker that was significantly linked to the largest number of different “noncontiguous” transcripts (those whose genes are >50 cM distant from the marker) was D9RAT46, which was associated with the expression of 21 genes at α = 0.05. A modest association was noted with these genes to the Gene Ontology terms for regulation of nucleic acid metabolism (GO:0019219; P = 0.034) and induction of apoptosis by extracellular signals (GO:0008624; P = 0.039).
One of the primary motivations for this experiment was to investigate the role that variations in gene expression play in human eye disease. We therefore investigated 114 genes (RetNet, www.sph.uth.tmc.edu/RetNet) that have been identified to date to be associated with human hereditary diseases of the retina. Sixty-two of these were sufficiently expressed and variable in the adult rat eye to permit further analysis. Seven of these showed evidence of contiguous regulation alone, four had evidence of both contiguous and noncontiguous regulation, and 11 had only noncontiguous linkages, including two genes whose expressions were each linked to two noncontiguous loci (Table 2).
Table 2.
Significant linkages to known retinal disease genes
| Gene | Probe ID | Marker | P value |
|---|---|---|---|
| Abca4 | 1384603_at | D2Rat55 | 3.0E-04 |
| Aipl1 | 1387166_at | D10Mgh6 | 6.9E-11 |
| Bbs1 | 1383417_at | D1Rat112 | 1.9E-10 |
| Bbs4 | 1383007_at | D8Rat141 | 4.3E-05 |
| Bbs8 | 1378416_at | D8Rat46 | 4.6E-04 |
| Ca4 | 1368437_at | D10Mgh6 | 2.0E-04 |
| Cacna1f | 1387846_at | DXRat40 | 2.4E-06 |
| Cacna1f | 1387846_at | DXRat38 | 6.6E-09 |
| Grk1 | 1369257_at | D14Rat94 | 1.9E-04 |
| Jag1 | 1368725_at | D3Rat26 | 4.4E-04 |
| Lrp5 | 1380242_at | D15Rat40 | 8.1E-04 |
| Opn1 sw | 1388025_at | D10Rat27 | 8.0E-07 |
| Opn1 sw | 1388025_at | D15Mit2 | 1.1E-06 |
| Pcdh15 | 1378408_at | D6Mit3 | 2.0E-04 |
| Pde6a | 1393426_at | D18Rat51 | 0.0 |
| Pex1 | 1376595_at | D3Rat70 | 5.5E-04 |
| Pex7 | 1379784_at | D1Rat3 | 1.3E-04 |
| Pex7 | 1379784_at | D8Mit5 | 9.6E-05 |
| Rbp4 | 1371762_at | D1Rat119 | 2.6E-07 |
| Rdh12 | 1382949_at | D1Rat97 | 4.0E-04 |
| Rdh5 | 1379587_at | D7Rat154 | 0.0 |
| Rdh5 | 1379587_at | D2Mit5 | 4.2E-04 |
| Rs1 | 1396807_at | D20Rat21 | 2.8E-04 |
| Tead1 | 1389287_at | D4Rat61 | 4.1E-04 |
| Tead1 | 1389287_at | DXRat40 | 5.9E-04 |
| Timm8a | 1368400_at | DXRat73 | 1.8E-05 |
| Timm8a | 1368400_at | D11Mgh5 | 2.5E-04 |
| Timm8a | 1368400_at | D8Mit5 | 7.1E-04 |
| Tulp1 | 1393071_at | D1Rat371 | 6.9E-04 |
For each linked region, the genetic marker with the most significant P value is shown. A Bonferroni-adjusted P value of 0.05 was used based on correction for the number of genes tested.
We chose one of the 11 contiguously regulated genes (Abca4) and one of the noncontiguously regulated genes (Opn1sw) for more careful scrutiny for the following reasons. In humans, ABCA4 is associated with one of the more common forms of heritable blindness among children and young adults (Stargardt disease) (29, 30), but a substantial number of disease alleles (>35%) in humans do not harbor plausible high-penetrance disease-causing variations in the ABCA4 coding sequences, suggesting that variations that affect gene expression may be involved in some cases (23). Opn1sw encodes the opsin that is found in the subset of cone photoreceptors that are maximally sensitive to blue light. The developmental fate of cone photoreceptors is known to be affected by at least two different transcription factors, NR2E3 (31) and THRB (32).
For Abca4, the highest logarithm of odds score was obtained with the marker closest to the Abca4 coding sequence (i.e., a classic contiguous linkage). Reasoning that the most likely location for a contiguous regulatory element affecting gene expression would be in the 5′ flanking sequences, we sequenced 10 kb of genomic DNA 5′ to the Abca4 transcription start site. When we did this, we observed a 38-bp insertion in the SHRSP parental strain 6.6 kb upstream from the transcription start site. This insertion includes a perfect 10-bp cREL transcription factor binding site. cREL binding is typically associated with increased transcription (33, 34), and the parental strain that harbors this insertion has significantly higher Abca4 expression than the other parental strain.
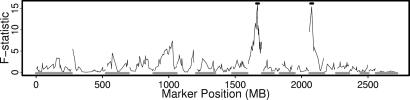
Expression of the gene encoding the blue sensitive opsin (Opn1sw) was significantly linked to two noncontiguous markers: D10RAT27 and D15MIT2. These linkages are marked with a box in Fig. 1C, and the exact linkage values of Opn1sw to every marker are shown in Fig. 2. The 11 Mb of the genome that is nearer D15MIT2 than any other genetic marker in this experiment harbors 65 genes, whereas the 4.5 Mb flanking D10RAT27 harbors 72 genes. Of interest, the chromosome 15 locus contains the gene that encodes thyroid hormone receptor β2, a transcription factor known to affect the determination of cone cell fate (32). We therefore sequenced the Thrb gene in both parental rat strains and discovered two amino acid-altering variations (Ser56Asn and His58Arg) in the transactivation domain of the SHRSP strain. The serine at codon 56 is conserved among human, chimp, rat, mouse, dog, cow, and chicken. Rats homozygous for these two Thrb variations exhibit a 30% lower expression of Opn1sw than animals that are homozygous for the wild-type sequence present in the SR/JrHsd rat strain. Heterozygous animals exhibit an expression of Opn1sw that is intermediate between the expression levels of animals with the homozygous genotypes.
Fig. 2.
ANOVA for Opn1sw expression levels for all markers evaluated. The x axis corresponds to the locations of the genetic markers, and the y axis corresponds to the ANOVA-based linkage of the Opn1sw expression level at each marker. The bars above the peaks correspond to the locations of markers D10RAT27 and D15MIT2.
Nonpositional Analysis of Correlated Gene Expression.
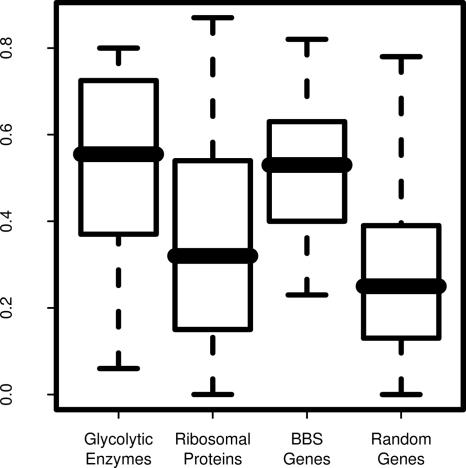
In previous eQTL mapping studies, the expression data have been used primarily to identify the genomic locations of specific genetic elements involved in gene regulation. However, in this study we also used these data in a nonpositional fashion to identify gene networks on the basis of their correlated response to genetic permutation. We hypothesized that pairwise correlation of gene expression might reveal biologically relevant functional relationships. We tested this hypothesis by comparing the pairwise correlations of gene expression for three groups of functionally related proteins and compared these to the correlations of randomly selected groups of expressed genes (Fig. 3). Specifically, genes encoding members of the glycolytic pathway, components of the small ribosomal subunit, and genes associated with a multisystem human disease known as Bardet–Biedl syndrome (BBS) were examined. The ribosomal subunit genes and the BBS genes showed much higher correlation of expression than randomly selected expressed genes (Fig. 3). Genes from the glycolytic pathway as a group were less well correlated. However, the correlation of glycolytic enzyme genes increased noticeably when pairs of genes that are adjacent in the glycolytic pathway were analyzed. These results suggest that correlation of expression may be useful in deducing the sequence of unknown metabolic pathways.
Fig. 3.
Distribution of pairwise correlation values in several sets of functionally related genes. Shown are the absolute values of the Pearson correlation coefficients for genes involved in the glycolytic pathway (adjacent pairs), the small ribosomal subunit, and BBS. A size-matched group of random genes is presented for comparison.
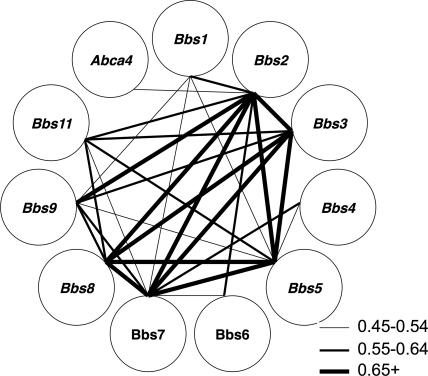
BBS is a genetically heterogeneous disease of multiple organ systems including the retina. When this study was begun, mutations in eight different genes were known to cause the disorder (35, 36). Collectively, these eight genes are responsible for approximately half of the cases of this disease, indicating that additional BBS genes remain to be discovered. Although recent evidence suggests that many of these genes are involved in cilia function, there is little structural or sequence similarity among the genes currently known to cause this disease. We hypothesized that expression of these genes would be highly correlated in the 120 F2 rats and that correlation of expression among BBS genes could aid in the identification of novel BBS genes. Evaluation of pairwise gene expression correlations among the 18,976 probe sets in the 120 F2 rats revealed that the expression levels of the rat orthologs of the eight known BBS genes were significantly and positively correlated with one another. Specifically, the expression of Bbs1, Bbs2, and Bbs7 were significantly correlated with each other and with all of the other known BBS genes. The expression of the least correlated gene, Bbs6, was significantly correlated with three BBS genes (Bbs1, Bbs2, and Bbs7) (Fig. 4). Based on a Monte Carlo simulation, the expression correlation among the eight BBS genes is highly significant (P < 0.001).
Fig. 4.
Pairwise correlations among the 10 BBS genes. The strength of each pairwise correlation is depicted by the thickness of the line connecting the symbols of the two genes. Abca4, a gene associated with an autosomal recessive macular disease, is shown for comparison.
In a recent study we used genetic mapping and comparative genomic analysis to identify a ninth BBS gene (BBS9) (37). The identification of this gene allowed us to test in a retrospective manner whether expression correlation with the eight known BBS genes could aid in the identification of a previously undescribed gene. We tested whether the Bbs9 gene was highly correlated with the eight previously identified BBS genes. This analysis revealed that expression of the Bbs9 gene was significantly correlated with the expression of the eight previously known BBS genes. Fewer than 4% of all genes expressed in the eye (723) are as well correlated to the original eight BBS genes.
In another study we used expression correlation with the nine known BBS genes (including Bbs9) in a prospective manner to aid in the identification of BBS11 (38). In this experiment, a BBS11 locus was identified by genetic mapping to a 2.4-Mb interval on human chromosome 9. Only one gene (Trim32) within the BBS11 locus was shown to have significant expression correlation in the F2 rat animals with eight of nine BBS genes, including the recently identified Bbs9 gene (Fig. 4). No other positional candidate gene within the BBS11 interval had more than one significant expression correlation with the previously identified known BBS genes. Mutation analysis and functional studies were performed and supported TRIM32 as a BBS gene (BBS11) (38). A 10th BBS gene (BBS10) has also been reported but is not represented on the Affymetrix chip (39).
Discussion
Large-scale gene expression analysis using microarray technology has made it evident that abundant levels of variation in gene expression exist, suggesting that a substantial fraction of phenotypic variation results from variation in gene expression. Moreover, when large-scale gene expression analyses in multiple members of a genetic cross are combined with linkage analysis (“eQTL mapping”) (1–3) it becomes apparent that the regulation of gene expression is somewhat more complicated than previously imagined. For example, Yvert et al. (2) used eQTL mapping to study two inbred strains of yeast and found that the majority of the expression differences between these strains could be traced to noncontiguous (and presumably transacting) factors rather than to the genes themselves.
The experiments reported in this paper are the first to apply the eQTL mapping approach to microarray expression data collected from the eye. More than half of the genes in the mammalian genome are detectably expressed in the eye, a molecular reflection of the diversity of cell types found in this important organ. As in previous eQTL mapping experiments, we observed a large fraction of variation in gene expression to be traceable to heritable genetic factors. Many of the genetic variations that affect gene expression in the eye appear to reside near the genes themselves, but a substantial number also exist in remote genomic locations. Many genes appear to be controlled by several genetic loci, and some genetic loci control many genes. Using the eQTL mapping data as a guide, we were able to identify plausible transcription-altering sequence variations in two retinal disease genes, suggesting that the eQTL mapping approach will be an effective way to investigate the role of gene regulation in a number of human diseases.
Another interesting feature of this study was the way in which we used correlated gene expression to reveal biological relationships and specifically to identify and validate previously undescribed disease-causing genes. The principle behind this approach is that, as organisms evolve, there is an evolutionary advantage in linking the expression of functionally related genes to the biological situation for which their function is needed. There is also an advantage in providing a mechanism for expressing functionally related genes in the correct stoichiometric ratios. Given the evolutionary value of this regulatory connectivity, if one permutes the environment of an organism and observes which genes respond to that permutation, one can infer to some degree that these genes are functionally linked to each other and to the processes required to respond to the permutation. This strategy has been used by many investigators over the years to identify previously unknown members of specific biological pathways (40, 41).
In the experiment described in this article, a specific type of permutation, a genetic one, was performed. The genomes of two distantly related strains of a species have been shuffled by genetic recombination, resulting in 120 combinations of “normal” gene expression control signals. When such shuffling results in more of a given biological variable in one animal and less in another under the uniform laboratory conditions in which they were maintained, it should cause the conserved control mechanisms for the pathway related to that variable to respond in kind. When one evaluates the correlation of expression between every possible pair of transcripts in all 120 animals, one has the opportunity to observe genes whose expression levels are tightly correlated to one another independent of the mechanism of this correlation. Given the results of this study, it seems plausible that many statistically significant correlations between pairs of transcripts are the result of an evolutionarily advantageous control mechanism just as conservation of a given codon is evidence of the evolutionary advantage of that codon. This phenomenon was useful in the identification and validation of two previously undescribed genes involved in BBS, and it seems likely that these expression data and analytical approaches can be used in a similar fashion to identify a number of additional genes that cause human eye disease.
Materials and Methods
Statistical Analysis.
For eQTL mapping of the 31,042 noncontrol probes on the array we first excluded probes that were not expressed in the eye or that lacked sufficient variation. For a probe to be considered expressed, the maximum expression value observed for that probe among the 120 F2 rats was required to be greater than the 25th percentile of the entire set of RMA expression values. For a probe to be considered “sufficiently variable” it had to exhibit at least 2-fold variation in expression level among the 120 F2 animals.
eQTL mapping was performed by using a package developed in R (available from the authors upon request). For the 18,976 probes, F-statistics using ANOVA were used to evaluate the significance of linkage at each marker locus. For each probe, genome-wide empirical P values were estimated by using Churchill's permutation test to correct for multiply evaluating the presence of eQTLs at 399 markers across the genome. Once all of the empirical P values were calculated, we estimated the q value, a false discovery rate analog of the P value (42). For the probes corresponding to RetNet genes, uncorrected P values were corrected for multiple comparisons by using the Bonferroni method. A significance level of 0.05 was used both for the RetNet candidate assessment and to determine the marker with the largest transregulatory effect.
The Pearson pairwise correlation (r2) was used to assess correlated expression among the genes. Correlation values were computed for all 18,976 expressed probes in the experiment using all 120 F2 animals. These values were used in a Monte Carlo simulation with 100,000 iterations to measure the significance for the BBS interactions.
Additional Details.
For additional details see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Paula Moore, Susan Jones, Charles Searby, and Garry Hauser for technical assistance; Drs. Darryl Nishimura and Robert Mullins for their help in preparing the manuscript; and Linda Koser for administrative assistance. This work was supported by the Foundation Fighting Blindness, the Carver Endowment for Molecular Ophthalmology, the Grousbeck Family Foundation, the Elmer and Sylvia Sramek Charitable Foundation, the Macula Vision Research Foundation, and Research to Prevent Blindness. V.C.S. and E.M.S. are investigators of the Howard Hughes Medical Institute. T.E.S. was supported by a Career Development Award from Research to Prevent Blindness.
Abbreviations
- eQTL
expression quantitative trait locus
- BBS
Bardet–Biedl syndrome.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus repository, www.ncbi.nlm.nih.gov/geo (accession no. GSE5680).
References
- 1.Brem RB, Yvert G, Clinton R, Kruglyak L. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 2.Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN, Mackelprang R, Kruglyak L. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 3.Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 4.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hubner N, Wallace CA, Zimdahl H, Petretto E, Schulz H, Maciver F, Mueller M, Hummel O, Monti J, Zidek V, et al. Nat Genet. 2005;37:243–253. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- 6.Mehrabian M, Allayee H, Stockton J, Lum PY, Drake TA, Castellani LW, Suh M, Armour C, Edwards S, Lamb J, et al. Nat Genet. 2005;37:1224–1233. doi: 10.1038/ng1619. [DOI] [PubMed] [Google Scholar]
- 7.Bystrykh L, Weersing E, Dontje B, Sutton S, Pletcher MT, Wiltshire T, Su AI, Vellenga E, Wang J, Manly KF, et al. Nat Genet. 2005;37:225–232. doi: 10.1038/ng1497. [DOI] [PubMed] [Google Scholar]
- 8.Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, Hsu HC, Mountz JD, Baldwin NE, Langston MA, et al. Nat Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- 9.Vazquez-Chona FR, Khan AN, Chan CK, Moore AN, Dash PK, Hernandez MR, Lu L, Chesler EJ, Manly KF, Williams RW, et al. Mol Vis. 2005;11:958–970. [PMC free article] [PubMed] [Google Scholar]
- 10.Lander ES, Schork NJ. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 11.Collins FS, Guyer MS, Charkravarti A. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- 12.Zwick ME, Cutler DJ, Chakravarti A. Annu Rev Genomics Hum Genet. 2000;1:387–407. doi: 10.1146/annurev.genom.1.1.387. [DOI] [PubMed] [Google Scholar]
- 13.Lifton RP, Gharavi AG, Geller DS. Cell. 2001;104:545–556. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 14.Slavotinek AM, Stone EM, Mykytyn K, Heckenlively JR, Green JS, Heon E, Musarella MA, Parfrey PS, Sheffield VC, Biesecker LG. Nat Genet. 2000;26:15–16. doi: 10.1038/79116. [DOI] [PubMed] [Google Scholar]
- 15.Nishimura DY, Searby CC, Carmi R, Elbedour K, Van Maldergem L, Fulton AB, Lam BL, Powell BR, Swiderski RE, Bugge KE, et al. Hum Mol Genet. 2001;10:865–874. doi: 10.1093/hmg/10.8.865. [DOI] [PubMed] [Google Scholar]
- 16.Mykytyn K, Braun T, Carmi R, Haider NB, Searby CC, Shastri M, Beck G, Wright AF, Iannaccone A, Elbedour K, et al. Nat Genet. 2001;28:188–191. doi: 10.1038/88925. [DOI] [PubMed] [Google Scholar]
- 17.Mykytyn K, Nishimura DY, Searby CC, Shastri M, Yen HJ, Beck JS, Braun T, Streb LM, Cornier AS, Cox GF, et al. Nat Genet. 2002;31:435–438. doi: 10.1038/ng935. [DOI] [PubMed] [Google Scholar]
- 18.Nishimura DY, Swiderski RE, Alward WL, Searby CC, Patil SR, Bennet SR, Kanis AB, Gastier JM, Stone EM, Sheffield VC. Nat Genet. 1998;19:140–147. doi: 10.1038/493. [DOI] [PubMed] [Google Scholar]
- 19.Fingert JH, Heon E, Liebmann JM, Yamamoto T, Craig JE, Rait J, Kawase K, Hoh ST, Buys YM, Dickinson J, et al. Hum Mol Genet. 1999;8:899–905. doi: 10.1093/hmg/8.5.899. [DOI] [PubMed] [Google Scholar]
- 20.Stone EM, Lotery AJ, Munier FL, Heon E, Piguet B, Guymer RH, Vandenburgh K, Cousin P, Nishimura D, Swiderski RE, et al. Nat Genet. 1999;22:199–202. doi: 10.1038/9722. [DOI] [PubMed] [Google Scholar]
- 21.Heon E, Piguet B, Munier F, Sneed SR, Morgan CM, Forni S, Pescia G, Schorderet D, Taylor CM, Streb LM, et al. Arch Ophthalmol. 1996;114:193–198. doi: 10.1001/archopht.1996.01100130187014. [DOI] [PubMed] [Google Scholar]
- 22.Stone EM, Nichols BE, Kimura AE, Weingeist TA, Drack A, Sheffield VC. Arch Ophthalmol. 1994;112:765–772. doi: 10.1001/archopht.1994.01090180063036. [DOI] [PubMed] [Google Scholar]
- 23.Webster AR, Heon E, Lotery AJ, Vandenburgh K, Casavant TL, Oh KT, Beck G, Fishman GA, Lam BL, Levin A, et al. Invest Ophthalmol Vis Sci. 2001;42:1179–1189. [PubMed] [Google Scholar]
- 24.Chiba-Falek O, Touchman JW, Nussbaum RL. Hum Genet. 2003;113:426–431. doi: 10.1007/s00439-003-1002-9. [DOI] [PubMed] [Google Scholar]
- 25.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 26.Ronald J, Brem RB, Whittle J, Kruglyak L. PLoS Genet. 2005;1:e25. doi: 10.1371/journal.pgen.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doerge RW, Churchill GA. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dressel R, Walter L, Gunther E. Immunol Rev. 2001;184:82–95. doi: 10.1034/j.1600-065x.2001.1840108.x. [DOI] [PubMed] [Google Scholar]
- 29.Blacharski P. In: Retinal Dystrophies and Degenerations. Newsome D, editor. New York: Raven; 1988. pp. 135–159. [Google Scholar]
- 30.Allikmets R, Singh N, Sun H, Shroyer NF, Hutchinson A, Chidambaram A, Gerrard B, Baird L, Stauffer D, Peiffer A, et al. Nat Genet. 1997;15:236–246. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- 31.Haider NB, Jacobson SG, Cideciyan AV, Swiderski R, Streb LM, Searby C, Beck G, Hockey R, Hanna DB, Gorman S, et al. Nat Genet. 2000;24:127–131. doi: 10.1038/72777. [DOI] [PubMed] [Google Scholar]
- 32.Ng L, Hurley JB, Dierks B, Srinivas M, Salto C, Vennstrom B, Reh TA, Forrest D. Nat Genet. 2001;27:94–98. doi: 10.1038/83829. [DOI] [PubMed] [Google Scholar]
- 33.Delhalle S, Blasius R, Dicato M, Diederich M. Ann NY Acad Sci. 2004;1030:1–13. doi: 10.1196/annals.1329.002. [DOI] [PubMed] [Google Scholar]
- 34.Bernard D, Monte D, Vandenbunder B, Abbadie C. Oncogene. 2002;21:4392–4402. doi: 10.1038/sj.onc.1205536. [DOI] [PubMed] [Google Scholar]
- 35.Chiang AP, Nishimura D, Searby C, Elbedour K, Carmi R, Ferguson AL, Secrist J, Braun T, Casavant T, Stone EM, et al. Am J Hum Genet. 2004;75:475–484. doi: 10.1086/423903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan Y, Esmail MA, Ansley SJ, Blacque OE, Boroevich K, Ross AJ, Moore SJ, Badano JL, May-Simera H, Compton DS, et al. Nat Genet. 2004;36:989–993. doi: 10.1038/ng1414. [DOI] [PubMed] [Google Scholar]
- 37.Nishimura DY, Swiderski RE, Searby CC, Berg EM, Ferguson AL, Hennekam R, Merin S, Weleber RG, Biesecker LG, Stone EM, et al. Am J Hum Genet. 2005;77:1021–1033. doi: 10.1086/498323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiang AP, Beck JS, Yen H-J, Tayeh MK, Scheetz TE, Swiderski R, Nishimura D, Braun TA, Kim K-Y, Huang J, et al. Proc Natl Acad Sci USA. 2006;103:6287–6292. doi: 10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoetzel C, Laurier V, Davis EE, Muller J, Rix S, Badano JL, Leitch CC, Salem N, Chouery E, Corbani S, et al. Nat Genet. 2006;38:521–524. doi: 10.1038/ng1771. [DOI] [PubMed] [Google Scholar]
- 40.Eisen MB, Spellman PT, Brown PO, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang W, Morris QD, Chang R, Shai O, Bakowski MA, Mitsakakis N, Mohammad N, Robinson MD, Zirngibl R, Somogyi E, et al. J Biol. 2004;3:21. doi: 10.1186/jbiol16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Storey JD. J R Stat Soc Ser B. 2002;64:479–498. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.