Abstract
NF-E2-related factor 2 (Nrf2) regulates antioxidant-responsive element-mediated induction of cytoprotective genes in response to oxidative stress. The purpose of this study was to determine the role of BRG1, a catalytic subunit of SWI2/SNF2-like chromatin-remodeling complexes, in Nrf2-mediated gene expression. Small interfering RNA knockdown of BRG1 in SW480 cells selectively decreased inducible expression of the heme oxygenase 1 (HO-1) gene after diethylmaleate treatment but did not affect other Nrf2 target genes, such as the gene encoding NADPH:quinone oxidoreductase 1 (NQO1). Chromatin immunoprecipitation analysis revealed that Nrf2 recruits BRG1 to both HO-1 and NQO1 regulatory regions. However, BRG1 knockdown selectively decreased the recruitment of RNA polymerase II to the HO-1 promoter but not to the NQO1 promoter. HO-1, but not other Nrf2-regulated genes, harbors a sequence of TG repeats capable of forming Z-DNA with BRG1 assistance. Similarly, replacement of the TG repeats with an alternative Z-DNA-forming sequence led to BRG1-mediated activation of HO-1. These results thus demonstrate that BRG1, through the facilitation of Z-DNA formation and subsequent recruitment of RNA polymerase II, is critical in Nrf2-mediated inducible expression of HO-1.
When living organisms are exposed to chemical electrophiles, such as xenobiotics, drugs, toxins, or carcinogens, a battery of genes is induced via antioxidant-responsive elements (AREs) or electrophile-responsive elements to coordinate cellular defenses (45, 56). Nrf2, which belongs to the “cap'-n'-collar” (CNC) family of transcription factors, regulates cytoprotective gene expression via ARE binding (21, 22). The Nrf2-ARE system regulates expression of numerous cytoprotective enzymes (reviewed in reference 32), including NADPH:quinone oxidoreductase 1 (NQO1), heme oxygenase 1 (HO-1), and the subunits of γ-glutamylcysteine synthetase (γ-GCS).
Under unstressed conditions, Keap1 (Kelch-like ECH-associated protein 1) facilitates degradation of Nrf2 via proteasome and inhibits nuclear accumulation of Nrf2 (23, 24). In contrast, electrophiles and oxidants liberate Nrf2 from Keap1-dependent degradation, leading to Nrf2 accumulation in the nucleus. Importantly, when Nrf2 is overexpressed in cells by transfection, Nrf2 accumulates in the nucleus and activates transcription even in the absence of external stimuli (31, 40). Consistent with this observation, both in murine keap1 knockout and human KEAP1 knockdown cell lines, Nrf2 is stabilized and accumulates in the nucleus, which leads to ARE-mediated transactivation of cytoprotective genes in a stress-independent manner (10, 59). Thus, while Keap1 modification is important for its activity as a stress sensor and as a substrate recognition subunit of E3 ubiquitin ligase, modification of Nrf2 is not necessarily required for Nrf2 activation by oxidants or electrophiles.
We previously determined that Nrf2 contains two transactivation domains, Neh4 and Neh5, which cooperatively bind the coactivator CBP (CREB binding protein) in an oxidative stress-independent manner to activate Nrf2 (27). Another intriguing feature of the Nrf2-ARE transcription regulatory system is that, while cytoprotective genes show various patterns of gene expression, Nrf2 is required for expression of virtually all these genes. This suggests that additional cofactors likely generate the diversity observed in the cytoprotective gene expression profile.
HO-1 is a cytoprotective enzyme with potent anti-inflammatory, antioxidative, and antiproliferative effects. HO-1 is the rate-limiting enzyme in the catabolism of heme into biliverdin, and this reaction releases free iron and carbon monoxide (CO). The expression of the HO-1 gene is induced by oxidative or nitrosative stresses, cytokines, and other mediators produced during inflammation (3). Intracellular heme concentrations are tightly regulated, as free heme generates reactive oxygen species (34). Therefore, under homeostatic conditions, HO-1 is repressed; thus, subsequent derepression and transactivation occur upon Nrf2 stimulation (51, 53, 54).
It is interesting to note that several regulatory features distinguish the HO-1 gene from other Nrf2-ARE-regulated genes. For example, inducible expression of the HO-1 gene utilizes two distal enhancers, E1 and E2, that are located far upstream of the transcriptional initiation site (2). In contrast, most AREs in other Nrf2 target genes are located in close proximity to the transcriptional start site. Furthermore, the CNC transcription factor Bach1 specifically represses HO-1 gene expression by antagonizing Nrf2 binding (53). HO-1, in contrast to other ARE-regulated genes, is constitutively expressed in Bach1 knockout animals (53). Thus, to further delineate how the diversity in cytoprotective gene expression occurs, it is crucial to examine the contributions of transcriptional coactivators and corepressors, including their chromatin-remodeling activity, to inducible gene expression.
Chromatin remodeling influences nearly every step of gene transcription, including preinitiation complex formation, transcriptional initiation, and elongation (1, 5, 7, 13, 52). Currently, four distinct classes of remodeling complexes have been described: SWI/SNF, ISWI, Mi-2, and Ino80 (38). Each class is defined by a unique subunit composition and the presence of a distinct ATPase subunit. The yeast SWI/SNF complex was the first chromatin-remodeling complex to be described, and it contained Swi2/Snf2 as the ATPase subunit. Human SWI/SNF chromatin-remodeling complexes can be divided into two subclasses, BAF (BRG1-associated factors) and PBAF (polybromo- and BRG1-associated factors), which are defined by the specific subunits BAF250 and BAF180, respectively. Human cells contain two distinct Swi2/Snf2-like ATPase subunits, hBRM (human Brahma) and BRG1 (Brahma-related gene 1) (28). BAF complexes contain either BRG1 or hBRM as the ATPase subunit, whereas PBAF contains only BRG1. Since BRG1 or hBRM do not contain any canonical DNA binding domains, they must be recruited with the help of sequence-specific transcription factors, including c-Myc (4), EKLF (26), or C/EBPβ (33) and the nuclear receptors like the glucocorticoid receptor (14) or estrogen receptor (25).
In order to delineate the molecular basis for the diverse response in ARE-mediated transcriptional activation, we initiated characterization of coactivators and corepressors interacting with Nrf2. We determined that BRG1 interacts with Nrf2 and is specifically required for Nrf2-mediated activation of human HO-1 gene transcription. The human HO-1 gene promoter contains TG repeats that favor left-handed Z-DNA formation. Since Z-DNA formation in the promoter region has been shown to stimulate transcription (37, 42, 50) and BRG1 was reported to initiate the formation of Z-DNA (36, 37), in this study we examined the role of Z-DNA and BRG1 in influencing Nrf2-mediated induction of HO-1. The results demonstrate that in response to oxidative stress, Nrf2 recruits BRG1 to the HO-1 gene regulatory region, and BRG1, with the help of the Z-DNA structure, subsequently recruits RNA polymerase II (Pol II) for transcriptional initiation. Thus, we report for the first time that Nrf2 can influence transcription by interacting with cofactors involved in chromatin remodeling.
MATERIALS AND METHODS
Constructs.
A series of mammalian expression plasmids for GBD-NT, GBD-Neh2-4, and GBD-Neh5 were prepared as described previously (27). A series of Nrf2 deletion plasmids lacking the ETGE motif was derived from previously described deletion mutants (23) by replacing the Neh2 domain with that of plasmid Nrf2ΔETGE (29). Expression plasmids for BRG1 (pcDNA3.1-3×Flag-BRG1), BRG1 K785A (pcDNA3.1-3×Flag-BRG1 K785A), and hBRM (pCI-Neo-3×Flag-hBRM) were previously described (20). To generate a Gal4-luciferase reporter (pCEP4 Gal4-Luc), the BamHI and BsrB1 fragment of GB5-E1b-luciferase (23) was blunted by T4 DNA polymerase and subcloned into the SalI site of pCEP4 (Invitrogen). The human HO-1 promoter-reporter (pCEP4 hHO-1 Luc WT) was constructed by subcloning the KpnI-SalI/blunt fragment of phHOLUC45 (55) into the SalI site of pCEP4. PCR was performed to replace the 30-TG repeat sequence in the pCEP4 hHO-1 Luc WT promoter with 18 GC repeats or with a random sequence from the human NQO1 fifth exon. The primers (5′ to 3′) were as follows: 18 GC, TCA GAT TTC CTT AAA GGT TTG CGC GCG CGC GCG CGC GCG CGC GCG CGC GCG CGC GCT TTT CTC TAA AAG TCC TATG and CAT AGG ACT TTT AGA GAA AAG CGC GCG CGC GCG CGC GCG CGC GCG CGC GCG CGC GCA AAC CTT TAA GGA AAT CTGA; random, TCA GAT TTC CTT AAA GGT TTA TCC CAA CTG ACA ACC AGA TCA AAG CTA GAA AAT GAT TTT CTC TAA AAG TCC TATG and CAT AGG ACT TTT AGA GAA AAT CAT TTT CTA GCT TTG ATC TGG TTG TCA GTT GGG ATA AAC CTT TAA GGA AAT CTGA.
Cell culture, transfection, and luciferase assay.
SW480, SW13, and 293T cells were cultured in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum (Gibco). For immunoprecipitation analysis, 10 μg of Nrf2 expression plasmid and 10 μg of BRG1 expression plasmid were transfected into 293T cells by calcium phosphate precipitation (48). For reporter assays, human SW480 and SW13 cells were transfected with LipofectAmine Plus (Invitrogen), according to the manufacturer's instructions. Luciferase assays were performed with the dual-luciferase reporter kit (Promega), according to the prescribed protocol. Luciferase activity was quantified with a Biolumat luminometer (Berthold), and transfection efficiency was normalized by cotransfection of the PRL Renilla construct. The mean of at least three independent experiments, each carried out in duplicate, is presented with the standard error of the mean (SE). For overexpression of BRG1 and hBRM, SW13 cells were cotransfected with 2 μg of pSUPER control vector possessing a puromycin resistance gene (see below) and 20 μg of either vector alone, 3×Flag-BRG1, 3×Flag-BRG1(K785A), or 3×Flag-hBRM expression plasmids and selected in 2 μg/ml of puromycin for 2 days.
Transient transfection of siRNA.
SW480 cells were transfected with Nrf2 small interfering RNA (siRNA) or control siRNA (QIAGEN) by using LipofectAmine 2000 (Invitrogen). At 24 h after transfection, the cells were treated with 100 μM diethylmaleate (DEM) for 3.5 h and examined by immunoblotting and chromatin immunoprecipitation (ChIP). The siRNA sequence for human Nrf2 was 5′-AAG AGT ATG AGC TGG AAA AAC-3′ (18).
Generation of stable cell lines.
The mammalian expression vector pSUPER.retro.puro (OligoEngine) was used for expression of siRNA in SW480 cells. A 19-nucleotide sequence corresponding to nucleotides 1406 to 1424 downstream of the transcription start site (GGC AGA AGC ACC AGG AATA) of human BRG1, followed by complementary 19-nucleotide sequence, which is separated by a 9-nucleotide sequence (TTC AAG AGA), was cloned into the BglII and HindIII site of pSUPER.retro.puro and referred to as pSUPER-BRG1. pSUPER-control (pSUPER-Con) vector was constructed by using a 19-nucleotide sequence (GCG CGC TTT GTA GGA TTCG) that has no significant homology to any mammalian gene sequence, thus serving as a nonsilencing control (kindly provided by Akira Kobayashi) (63). To generate stable transformants that express siRNA, SW480 cells were transfected with 10 μg of either pSUPER-Con or pSUPER-BRG1 by using LipofectAmine Plus reagent (Invitrogen), according to the manufacturer's instructions, and selected with puromycin (4 μg/ml).
Immunoprecipitation.
Whole-cell lysates of 293T cells were prepared in buffer A (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1.5 mM MgCl2, 0.25% Nonidet P-40, protease inhibitor cocktail [Roche Diagnostics], and 10 μM MG132). Cell lysates were incubated with anti-Flag M2-conjugated beads (Sigma) with gentle rocking at 4°C overnight. The immunoprecipitates were washed three times with buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1.5 mM MgCl2, 0.5% Nonidet P-40) and subjected to Western analysis with anti-Nrf2 (Santa Cruz) and anti-Flag (Sigma) antibodies.
Immunoblot analysis.
The nuclei of SW480 cells and SW13 cells were prepared as described previously (21). Briefly, cells were suspended in hypotonic buffer and vortexed for 15 s, and the nuclear fraction was precipitated at 10,000 rpm for 1 min. Nuclei were resuspended in sodium dodecyl sulfate (SDS) sample loading buffer (without dye or 2-mercaptoethanol) and boiled for 5 min, and protein concentrations were estimated by bicinchoninic acid protein assay (Pierce). Proteins were separated by SDS-polyacrylamide gel electrophoresis in the presence of 2-mercaptoethanol and transferred onto Immobilon membranes (Millipore). To detect immunoreactive proteins, the blots were probed with anti-BRG1 polyclonal rabbit serum (Santa Cruz) or lamin B (Santa Cruz), followed by the reaction with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (IgG). Signals were detected with ECL Plus (Amersham).
RNA blot analysis.
Total RNAs from SW480 and SW13 cells were isolated with Isogen (Nippon Gene). Total RNA (10 μg) was subjected to electrophoresis in 1.5% agarose-2.2 M formaldehyde gels and transferred onto Zeta-Probe GT membranes (Bio-Rad). Blots were probed with 32P-labeled cDNA for BRG1, HO-1, and NQO1. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as a positive control. Band intensities were measured by NIH Imaging software and normalized with GAPDH.
Quantitative RT-PCR analysis.
Total RNA (1 μg) was reverse transcribed into cDNA and used for real-time (RT)-PCR analysis (Invitrogen). For quantitative RT-PCR, the cDNA was analyzed in duplicate with qPCR Mastermix (Eurogentec) for 15 min at 95°C for initial denaturing, followed by 40 cycles of 95°C for 30 s and 60°C for 1 min in the ABI 7700 Sequence Detection System. The primers and TaqMan probe set (5′ to 3′) were as follows: human HO-1 primers, CCA GCA ACA AAG TGC AAG ATTC and TCA CAT GGC ATA AAG CCC TACAG; probe, TCT CCG ATG GGT CCT TAC ACT CAG CTT TCT; human NQO1 primers, GTC ATT CTC TGG CCA ATT CAG AGT and TTC CAG GAT TTG AAT TCGGG; probe, ACT GAC ATA TAG CAT TGG GCA CAC TCC AGC. 18S rRNA was used as a positive control for quantitative RT-PCR analysis. The primers and TaqMan probes for detection of AKR1C1, GCSL, and GCSH were previously described (10).
ChIP analysis.
ChIP analysis was performed as described previously (49). In brief, after 100 μM DEM treatment, the cells were fixed by 1% formaldehyde for 5 min at room temperature. Cells were then sonicated to prepare chromatin suspensions of 300 to 1,000 bp of DNA in length. Immunoprecipitation analysis was carried out with control rabbit IgG, anti-Nrf2 (sc-13032; Santa Cruz), rabbit polyclonal anti-BRG1 (43), and anti-RNA Pol II (sc-899; Santa Cruz) antibodies. PCRs were carried out with Blend Taq-Plus DNA polymerase (Toyobo). Primers (5′ to 3′) were as follows: human HO-1 E1, GCT GCC CAA ACC ACT TCTGT and GCC CTT TCA CCT CCC ACCTA; human HO-1 E2, TCC TTT CCC GAG CCA CGTG and TCC GGA CTT TGC CCC AGG; human HO-1 promoter, CCA GAA AGT GGG CAT CAGCT and GTC ACA TTT ATG CTC GGCGG; human HO-1 exon 3, CAC CCG CTA CCT GGG TGAC and GGA GCG GTA GAG CTG CTTGA; human NQO1 pr, AAG TGT GTT GTA TGG GCCCC and TCG TCC CAA GAG AGT CCAGG; human NQO1 exon 2, CCT GTA GCT GAA GGT TTG CTGG and CCT ACC TGT GAT GTC CTT TCTGG. Five percent of the chromatin DNA was also subjected to PCR analysis and indicated as input.
Statistical analysis.
Data were evaluated by Student's test. P values of less than 0.05 were considered to be statistically significant.
RESULTS
BRG1 enhances Nrf2-mediated transcription in SW13 cells.
To explore the role BRG1 plays in Nrf2-mediated gene regulation, we examined the effects of BRG1 on Nrf2 transactivation activity using a luciferase reporter plasmid that contains Nrf2 binding sites in triplicate in front of the minimal TATA box (23) in SW13 cells. This cell line possesses marginally detectable expression of BRG1 and hBRM (60, 62) (see Fig. 2A). We deleted the ETGE motif in each of the mutants (27) to avoid the possibility that Keap1-mediated repression contributes to reporter gene activity. The results demonstrate that BRG1 activates Nrf2 transcription in a dose-dependent manner, and this activation was partially attenuated in mutants without the Neh4 or Neh5 domain (Fig. 1A). The effect of BRG1 on reporter gene expression was decreased to the control level in the Nrf2 mutant that lacks both the Neh4 and Neh5 domains (Fig. 1A).
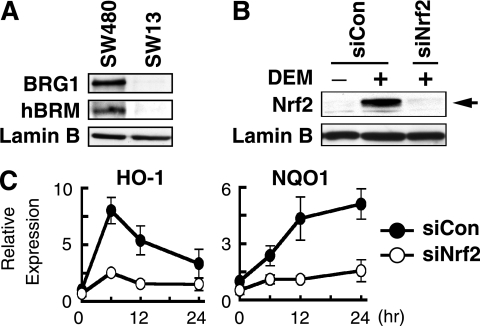
FIG. 2.
Knockdown of Nrf2 decreases DEM-inducible expression of HO-1 and NQO1. (A) Immunoblot analysis of BRG1 and hBRM in SW480 and SW13 cells. Nuclear extracts of SW480 and SW13 cells were analyzed by anti-BRG1, anti-hBRM, or anti-lamin B antibodies. (B) SW480 cells were transfected with control siRNA (siCon) or Nrf2 siRNA (siNrf2). At 24 h posttransfection, cells were treated with 100 μM DEM for 4 h and then subjected to immunoblot analysis using anti-Nrf2 (upper panel) or anti-lamin B (lower panel) antibodies. (C) HO-1 and NQO1 expression in siCon and siNrf2 cells after DEM treatment. SW480 cells were transfected with siCon or siNrf2. Following treatment with 100 μM DEM for the indicated time periods, total RNA was isolated and HO-1 and NQO1 mRNA expression was determined by quantitative RT-PCR, with 18S rRNA used as an internal standard. The means of three independent experiments performed in duplicate are shown, and error bars represent SE. HO-1 and NQO1 expression in siCon cells without DEM treatment was set at 1.
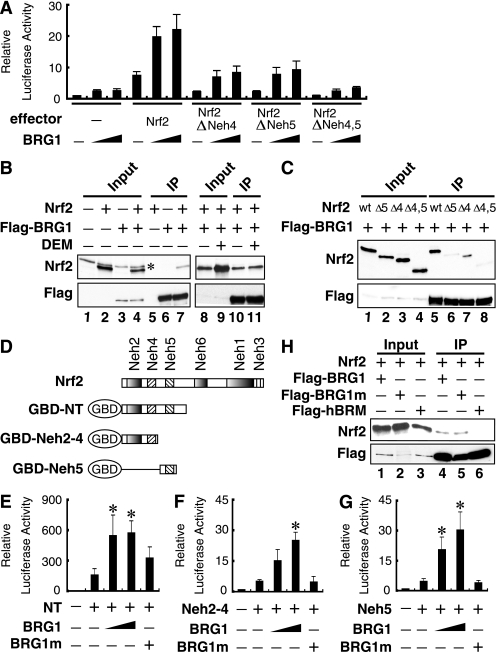
FIG. 1.
BRG1 interacts with Nrf2 and enhances Nrf2-mediated reporter gene expression through its ATPase activity. (A) BRG1 activates Nrf2 activity through the Neh4 and Neh5 domains. SW13 cells were transfected with a pRBGP2 luciferase reporter plasmid that has Nrf2 binding sites in triplicate in front of the minimal TATA box (23) and with increasing amounts of Nrf2 deletion plasmids that also lack the ETGE motif. Luciferase activity in the absence of an effector plasmid was arbitrarily set at 1, and the results shown represent the mean values from three independent experiments, with error bars representing SE. (B) 293T cells were transfected with the Nrf2 and/or Flag-BRG1 expression plasmids. Whole-cell lysates were immunoprecipitated with anti-Flag antibody-conjugated beads, followed by immunoblot analysis with anti-Nrf2 and anti-Flag antibodies (lanes 5 to 7 and lanes 10 and 11, respectively). Nrf2 and BRG1 expression levels were verified by immunoblot analysis with anti-Nrf2 and anti-Flag antibodies (lanes 1 to 4 and lanes 8 and 9). Cells of lanes 9 and 11 were treated with 100 μM DEM for 6 h. *, nonspecific antibody signal. (C) Nrf2 interacts with BRG1 through the Neh4 and Neh5 domains. 293T cells were transfected with the various Nrf2 deletion mutants (27) and Flag-BRG1 expression plasmids. Whole-cell lysates were immunoprecipitated with anti-Flag antibody-conjugated beads, followed by immunoblot analysis with anti-Nrf2 and anti-Flag antibodies (lanes 5 to 8). Nrf2 and BRG1 expression levels were verified by immunoblot analysis with anti-Nrf2 and anti-Flag antibodies (lanes 1 to 4). (D) Schematic presentation of the GBD-Nrf2 fusion proteins. (E to G) BRG1, but not the BRG1 K785A mutant (BRG1m), enhances Nrf2 transactivation activity in SW13 cells. SW13 cells were transfected with 20 ng of pCEP4 Gal4-Luc and 100 ng of GBD-NT (E), GBD-Neh2-4 (F), or GBD-Neh5 (G), along with 400 ng or 800 ng of the BRG1 or 800 ng of the BRG1 K785 mutant (BRG1m) expression plasmids. Luciferase activity of the reporter vector alone was set at 1, and relative values from three independent experiments each carried out in duplicate are shown with SE. *, significantly different from the activity of Gal4-Nrf2 alone (P < 0.05). (H) 293T cells were transfected with the Nrf2 expression plasmid together with Flag-BRG1 (lanes 1 and 4), Flag-BRG1m (lanes 2 and 5), or Flag-hBRM (lanes 3 and 6). Whole-cell lysates were immunoprecipitated with anti-Flag antibody-conjugated beads, followed by immunoblots with anti-Nrf2 and anti-Flag antibodies (lanes 4 to 6). Nrf2 and BRG1 expression levels were verified by immunoblotting with anti-Nrf2 and anti-Flag antibodies (lanes 1 to 3).
We next examined the protein-protein interactions between Nrf2 and BRG1. For this purpose, 293T cells were transfected with Nrf2 and/or Flag-tagged BRG1 expression vectors. Whole-cell lysates were immunoprecipitated with agarose-conjugated anti-Flag antibody and analyzed by immunoblotting with anti-Nrf2 antibody. Whereas Nrf2 coprecipitated with BRG1 in cells transfected with both Nrf2 and Flag-tagged BRG1 expression plasmids, no detectable Nrf2 was precipitated from cells transfected with Nrf2 or the Flag-tagged BRG1 expression plasmid alone (Fig. 1B, left panel). Importantly, DEM treatment did not alter the interactions between Nrf2 and BRG1 (Fig. 1B, right panel), indicating that Nrf2 interacts with BRG1 in an oxidative stress-independent manner. The importance of the Neh4 and Neh5 domains in this interaction was further examined. Immunoprecipitation using an anti-Flag antibody revealed that the Nrf2-BRG1 interaction was partially inhibited in ΔNeh4 and/or ΔNeh5 mutants (Fig. 1C). However, weak interactions between ΔNeh4 and/or ΔNeh5 mutants and BRG1 were observed, indicating the existence of a cryptic BRG1 binding site(s) other than Neh4 and Neh5.
We then tested the interaction between Nrf2 and BRG1 with pCEP4 Gal4-Luc, a replication-competent luciferase reporter, which possesses five GAL4 binding sites along with a series of Gal4 DNA binding domain (GBD)-Nrf2 fusion proteins (Fig. 1D). GBD-NT activated reporter gene expression 10-fold more than did the GBD-Neh2-4 or GBD-Neh5 constructs. The results demonstrated that BRG1 enhanced the reporter gene expression of GBD-NT, GBD-Neh2-4, and GBD-Neh5 in a dose-dependent manner, with a fourfold increase by NT, a sixfold increase by Neh2-4, and a sixfold increase by Neh5 when the maximal amount of BRG1 expression plasmid was used (Fig. 1E to G). GBD-Neh2 did not activate transcription of the reporter even when BRG1 was coexpressed (data not shown). In contrast, transfection of an ATPase-defective mutant of BRG1 (BRG1 K785A mutant or BRG1m) or hBRM (data not shown) into SW13 cells failed to significantly enhance the transactivation activity of any GBD-Nrf2 fusion proteins (Fig. 1E to G). Immunoprecipitation analysis further revealed that Nrf2 interacts with wild-type (WT) BRG1 and an ATPase-defective mutant of BRG1 but not hBRM (Fig. 1H). Taken together, these results indicate that BRG1 enhances Nrf2-mediated reporter gene transcription in an ATP-dependent manner by interacting with both Neh4 and Neh5.
Knockdown of BRG1 attenuates inducible expression of the HO-1 gene in SW480 cells.
To examine whether BRG1 is involved in Nrf2-mediated transcription activation in response to DEM, we examined the expression of Nrf2 target genes HO-1 and NQO1 in the SW480 human colon cancer cell line (57). In contrast to SW13 cells, both BRG1 and hBRM were highly expressed in SW480 cells (Fig. 2A), and the expression of HO-1 and NQO1 genes was induced by DEM in SW480 cells (data not shown). The expression of Nrf2-specific siRNA (18) caused a marked reduction of Nrf2 in SW480 cells (Fig. 2B), and the siRNA attenuated the induction of HO-1 and NQO1 mRNA by DEM (Fig. 2C).
To determine how BRG1 knockdown affects inducible expression of Nrf2-target genes, we stably transfected pSUPER-BRG1 and pSUPER-Con into SW480 cells, which express BRG1-specific and -irrelevant short-hairpin (SH)-type siRNAs, respectively. Of the eight cell lines transfected with pSUPER-BRG1, clones 4 (SH4) and 7 (SH7) showed the lowest expression of BRG1 after quantification by immunoblotting, but hBRM expression was not affected in these clones (Fig. 3A). Nrf2 expression was slightly increased both in SH4 and SH7 clones compared to SHCon cells, which are stably transfected with irrelevant pSUPER-Con. Thus, these SH4 and SH7 clones were utilized for all subsequent analyses.
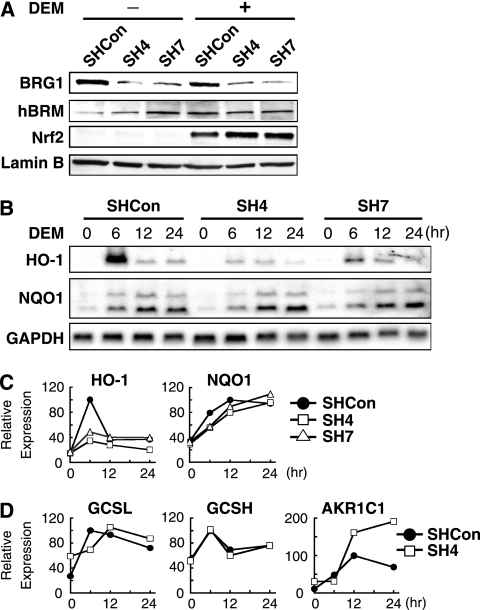
FIG. 3.
BRG1 knockdown selectively downregulates DEM-inducible expression of HO-1. (A) Immunoblot analysis of BRG1 knockdown cell lines. SHCon, SH4, and SH7 cells were treated with 100 μM DEM for 4 h, and the nuclear extracts were subjected to immunoblot analysis with antibodies against BRG1, hBRM, Nrf2, or lamin B. (B and C) RNA blot analysis of BRG1 knockdown cell lines. SHCon, SH4, and SH7 cells were treated with 100 μM DEM for the indicated time periods, and total RNA was isolated and analyzed by RNA blotting for HO-1, NQO1, and GAPDH (B). Band intensity was quantified by NIH Image and plotted after normalization with the GAPDH signal (C). The maximal induction level in SHCon cells was set at 100, and the mean relative expression levels from two independent experiments at each time point are presented. (D) Quantitative RT-PCR analysis of GCSL, GCSH, and AKR1C1. The induction levels of the GCSL, GCSH, and AKR1C1 genes at the 12-h time point in SHCon cells were set at 100, and the means of relative expression from two independent experiments each carried out in duplicate at each time point are presented; 18S rRNA was used as an internal standard.
We then treated SHCon, SH4, and SH7 cells with 100 μM DEM, and the expression of Nrf2 target genes was analyzed. We found that whereas the expression of HO-1, NQO1, the alodoketoreductase 1C1 gene (AKR1C1), and the genes encoding the heavy and light chains of glutathione synthetase (GCSH and GCSL, respectively) was induced by DEM in SHCon cells (Fig. 3B to D), the expression of HO-1 was markedly decreased in cells undergoing BRG1 knockdown (Fig. 3B and C). At 6 h after DEM treatment, HO-1 gene expression in SHCon cells showed peak induction, but the induction was reduced by 70% in SH4 cells and by 54% in SH7 cells. In contrast, the inducible NQO1, GCSL, and GCSH expression after DEM treatment was not affected substantially after BRG1 knockdown, but AKR1C1 expression was markedly increased at the 12- and 24-h time points (Fig. 3B to D). These results indicate that BRG1 is required for the maximal induction of HO-1 gene expression, but its contribution is not required for the expression of the other Nrf2 target genes examined in this study.
Chromatin-remodeling activity of BRG1 is required for the induction of HO-1.
To further delineate the role BRG1 plays in Nrf2 target gene induction by DEM, we examined the expression of the Nrf2 target genes in SW13 cells that substantially lack BRG1 and hBRM expression. It was previously demonstrated that BRG1 replenishment could reconstitute the functional BAF complex in SW13 cells (11, 64). In this study, therefore, we cotransfected into SW13 cells either the BRG1 expression or control plasmid concomitantly with a pSUPER-puro vector containing the puromycin resistance gene. Transfected cells were selected with puromycin for 2 days, followed by treatment with 100 μM DEM. RNA blot analysis demonstrated that the inducible expression of HO-1 by DEM was markedly enhanced in SW13 cells transfected with BRG1, with maximal induction occurring at 12 to 24 h (Fig. 4A and B). Consistent with the results of BRG1 knockdown analysis described in the previous section, BRG1 expression did not alter inducible NQO1 expression after DEM treatment at any of the time points observed (Fig. 4A and B). Importantly, hBRM expression did not alter HO-1 mRNA expression (Fig. 4C), indicating that the ability to enhance HO-1 gene expression is specific to BRG1.
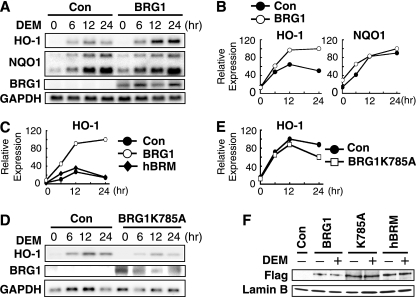
FIG. 4.
Expression of BRG1, but not the BRG1K785A mutant or hBRM, enhances inducible expression of the HO-1 gene by DEM in SW13 cells. (A and B) Reconstitution by BRG1 activity in SW13 cells. SW13 cells were cotransfected with either control vector or BRG1 expression plasmid along with pSUPER-puro control vector. Cells were selected with puromycin for 2 days, followed by treatment with 100 μM DEM for the indicated periods. Total RNA was subjected to RNA blot analysis for HO-1, NQO1, BRG1, and GAPDH (A). Band intensities (A) were quantified by NIH Image and plotted after normalization to the GAPDH signal (B). (C) hBRM does not activate HO-1 gene expression. SW13 cells were cotransfected with either control vector, Flag-BRG1, or Flag-hBRM expression plasmids, along with pSUPER-puro control vector. HO-1 mRNA levels were determined by quantitative RT-PCR using 18S rRNA as an internal standard. HO-1 expression in BRG1-overexpressing cells at 24 h after 100 μM DEM treatment was arbitrarily set at 100, and the means of two independent experiments performed in triplicate are presented. (D and E) ATPase activity of BRG1 is indispensable for transactivation. SW13 cells were cotransfected with either control vector or BRG1K785A mutant expression plasmid along with pSUPER-puro control vector. The cells were analyzed as described for panel A. Band intensities in panel D were quantified and plotted after normalization by GAPDH signals (E). (F) Expression of BRG1, BRG1K785A, and hBRM proteins in SW13 cells. SW13 cells were cotransfected with either control vector or BRG1, BRG1K785A mutant, or hBRM expression plasmid along with pSUPER-puro control vector. The nuclear extracts were subjected to immunoblot analysis with anti-Flag (upper panel) or anti-lamin B (bottom panel) antibodies.
To determine whether inducible HO-1 expression in SW13 cells requires BRG1 chromatin-remodeling activity, the ATPase-deficient mutant of BRG1 was also expressed transiently in SW13 cells, and the replenished cells were treated with DEM. Transfection of the ATPase-defective BRG1 into SW13 cells only slightly enhanced HO-1-inducible expression after DEM treatment (Fig. 4D and E). Furthermore, immunoblot analysis revealed that BRG1, hBRM, and BRG1m (i.e., BRG1 K785A) transgene products are all expressed at comparable levels in SW13 cells (Fig. 4F). These results further strengthen our contention that the chromatin-remodeling activity of BRG1 is required for HO-1 induction by DEM. Taken together, these results show that BRG1 regulates inducible expression of HO-1 and that the ATPase activity of BRG1 is critical in this regulatory process.
BRG1 is recruited to regulatory regions of the HO-1 gene in an Nrf2-dependent manner.
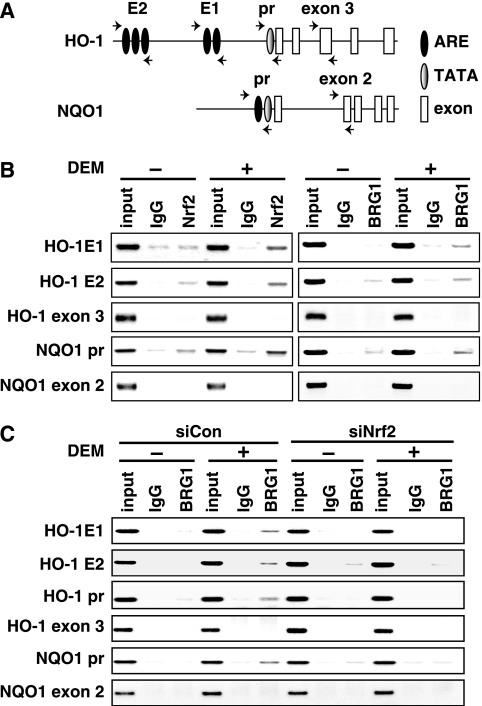
Differential regulation of Nrf2 target genes by BRG1 raises the possibility that BRG1 might be selectively recruited to the regulatory regions of Nrf2 target genes. To test this hypothesis, we treated SW480 cells with DEM, and DNA binding of BRG1 was examined by ChIP. As shown in Fig. 5A, only one functional ARE has been reported in the NQO1 gene proximal regulatory region (i.e., around bp −520; NQO1 promoter [41]). In contrast, human HO-1 contains two enhancers, E1 and E2, located at approximately 4 and 10 kb, respectively, upstream from the transcription start site. These two enhancers harbor multiple AREs (2, 3), but there is no ARE in its proximal regulatory region (HO-1 promoter). SW480 cells were treated with 100 μM DEM for 3.5 h, and ChIP analysis was performed with the anti-Nrf2 or anti-BRG1 antibodies and primer sets shown in Fig. 5A. Exon 3 of HO-1 and exon 2 of NQO1 were used as controls along with normal rabbit IgG in this experiment. ChIP analysis revealed that the recruitment of BRG1 and Nrf2 to the AREs found in the HO-1 enhancers and NQO1 promoter was markedly enhanced in response to DEM (Fig. 5B). On the contrary, recruitment of Nrf2 to the HO-1 gene promoter region was not observed in the ChIP analysis (data not shown), excluding the possibility that a cryptic ARE may reside in the promoter region.
FIG. 5.
Nrf2 recruits BRG1 to both the HO-1 and NQO1 genes in response to DEM. (A) Schematic presentation of the primer locations used to amplify genomic regions. Human HO-1 E1 and E2 enhancer regions contain AREs, and the promoter region (pr) encompasses a TATA box. HO-1 exon 3 was used as a control. The human NQO1 promoter region (pr) harbors both an ARE and a TATA box. The genomic region encompassing NQO1 exon 2 was used as a control. (B) ChIP analyses of the HO-1 and NQO1 genes using antibodies against Nrf2 and BRG1. SHCon cells were treated with 100 μM DEM for 3.5 h, and ChIP analysis was performed with antibodies against Nrf2 and BRG1. Normal rabbit IgG was used as a control. (C) ChIP analyses of HO-1 and NQO1 genes in the presence of Nrf2 siRNA. SHCon cells were transfected with control siRNA (siCon) or Nrf2 siRNA (siNrf2) and treated with 100 μM DEM for 3.5 h. ChIP analysis was performed with antibodies against BRG1 and primers for HO-1 and NQO1. In the input lane, 5% unprecipitated chromosomal DNA was amplified.
To determine whether Nrf2 actively recruits BRG1 to the regulatory regions of HO-1 and NQO1, we transfected Nrf2-specific siRNA or control siRNA to SW480 cells and treated the cells with DEM for 3.5 h. DNA binding of BRG1 was subsequently examined by ChIP analysis. We found that recruitment of BRG1 to AREs in the HO-1 enhancers and NQO1 promoter was enhanced in response to DEM in SW480 cells transfected with control siRNA (siCon) (Fig. 5C). In stark contrast, this enhancement of BRG1 binding was significantly reduced in SW480 cells transfected with Nrf2-specific siRNA (siNrf2), indicating that Nrf2 recruits BRG1 to these AREs.
Importantly, we also found that the recruitment of BRG1 to the proximal HO-1 promoter, which does not possess any AREs, also occurred after DEM treatment (Fig. 5C). This process required the presence of Nrf2, as the transfection of siRNA specific for Nrf2 abrogated BRG1 binding to the HO-1 promoter. These results suggest that the proximal HO-1 promoter may interact with distal E1 and/or E2 enhancer motifs that contain known AREs.
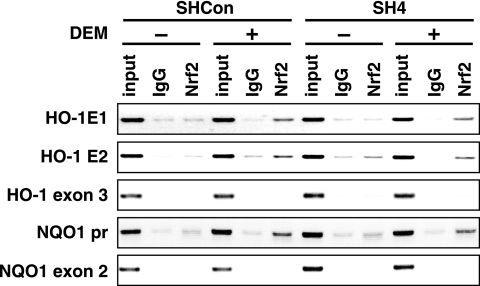
In vivo Nrf2 binding to AREs is independent of BRG1 activity.
It is generally accepted that chromatin-remodeling activity is required for transcription factors to bind to specific response elements in chromatin (35). We therefore examined whether Nrf2 binding to AREs requires BRG1 activity. To this end, additional ChIP analysis was performed using DNA samples from SW480-derived SHCon and SH4 stable cell lines treated with DEM. Although the expression of BRG1 was knocked down in SH4 cells, Nrf2 was found to associate with the AREs in the HO-1 enhancers and NQO1 promoter (Fig. 6). Similarly, we found that expression of BRG1 in SW13 cells did not affect Nrf2 binding to the HO-1 and NQO1 gene AREs (data not shown). These results thus indicate that the Nrf2 binding to ARE is independent of BRG1 and that BRG1-mediated induction of HO-1 gene expression occurs after the binding of Nrf2 to the AREs.
FIG. 6.
In vivo binding of Nrf2 to AREs is independent of BRG1. SHCon or SH4 cells were treated with 100 μM DEM for 3.5 h and analyzed by ChIP with antibodies against Nrf2 and primers that amplify the HO-1 E1, HO-1 E2, HO-1 exon 3, NQO1 promoter (pr), or NQO1 exon 2 regions. Normal rabbit IgG was used as a control. The input lane represents PCR using 5% chromosomal DNA without immunoprecipitation.
Contribution of Z-DNA-forming sequence in the HO-1 promoter to BRG1 activity.
The finding that BRG1 selectively activates HO-1 gene expression prompted us to hypothesize that the HO-1 gene possesses some intrinsic regulatory property that causes the differential response to BRG1. Through analysis of the HO-1 promoter, a set of 30 TG dinucleotide repeats was identified at 200 bases (to −260) upstream of the transcriptional start site (Fig. 7A). This long stretch of TG repeats harbors significant potential to form left-handed Z-DNA (16). BRG1 assistance in Z-DNA formation is often needed to form open-chromatin structures in gene regulatory regions (36, 37). Thus, we hypothesized that BRG1 may be required for Nrf2-mediated induction of the HO-1 gene because of the necessity of BRG1 in Z-DNA formation. To test this concept, a luciferase reporter vector containing 4.5 kb of the 5′ flanking sequence of HO-1 was constructed (pCEP4-hHO-1-Luc WT [Fig. 7B]). We transfected the reporter into SW480 cells together with the Nrf2 expression vector and found that Nrf2 activates reporter gene expression in a dose-dependent manner (Fig. 7C).
FIG. 7.
TG repeats can be replaced with an alternative Z-DNA-forming sequence to sustain induction of the hHO-1 gene. (A) The sequence of the human HO-1 promoter with TG repeats. The numbers above and below the sequence are relative positions from the transcription start site (+1). (B) Schematic representation of the mutant reporter construct. Thirty TG repeats are replaced with 18 GC repeats or the NQO1 fifth exon (random). (C) Activities of the pCEP4 hHO-1 luc (WT), 18GC, and random constructs. The pCEP4 hHO-1 luc, 18GC, and random constructs were cotransfected with the Nrf2 expression plasmid into SW480 cells. The means of three independent experiments each carried out in duplicate are shown, and error bars represent SE. Luciferase activity in the WT HO-1 promoter reporter plasmid alone was set at 1. *, significantly different from the activity of the WT construct in the presence of the same amount of Nrf2 expression plasmid (P < 0.05). (D) Activities of the pCEP4 hHO-1 luc (WT), 18GC, and random constructs in response to DEM. The pCEP4 hHO-1 luc, 18GC, and random constructs were transfected into SW480 cells. After transfection, cells were treated with 100 μM DEM for 24 h. Luciferase activity in the WT HO-1 promoter-reporter plasmid in the absence of DEM was set at 1. The results are presented as described for panel C. *, significantly different from the activity of the WT construct in the presence of DEM (P < 0.05). (E) TG repeats can be replaced by GC repeats for activation of the HO-1 gene by BRG1. The 30 TG repeats in the HO-1 promoter in pCEP4 hHO-1 luc (WT) were replaced by 18 GC repeats (18GC) or sequence from the NQO1 fifth exon (random). SW13 cells were transfected with 20 ng of each reporter construct along with 100 ng of Nrf2 expression plasmid (white bars) or both with Nrf2 and 800 ng of BRG1 expression plasmids (black bars). Luc activity of each reporter construct cotransfected with Nrf2 expression plasmid alone was arbitrarily set at 1. *, significantly different from the activity of the WT construct in the presence of BRG1 (P < 0.05).
To determine whether this TG repeat sequence facilitates Z-DNA formation in a manner that enables the HO-1 promoter to be activated by BRG1, we constructed additional Luc reporters by replacing the TG repeat sequence with an alternate 18-GC repeat Z-DNA-forming sequence, (18GC) or a random sequence (random) and tested their activities by cotransfection of Nrf2 in SW480 cells (Fig. 7B). The reporter gene expression by Nrf2 overexpression was significantly lower in the random construct than in the WT and 18GC constructs, indicating that Z-DNA formation positively regulates Nrf2-mediated HO-1 transcription (Fig. 7C). Furthermore, the DEM-inducible expression of luciferase genes from the random construct in SW480 cells is significantly lower than from the WT and 18GC constructs (Fig. 7D). Next, we tested the effect of BRG1 on reporter gene expression in SW13 cells. While BRG1 overexpression increased the WT, 18GC, and random constructs, the magnitude of induction of the random sequence after BRG1 overexpression was attenuated (Fig. 7E). These results suggest that Z-DNA formation is important in Nrf2-BRG1-mediated activation of HO-1 gene transcription.
BRG1 is important for the recruitment of RNA Pol II to the regulatory region.
Finally, the recruitment of RNA Pol II to the HO-1 and NQO1 gene regulatory regions was examined in the context of BRG1 knockdown and DEM induction. For this purpose, SHCon and SH4 cells were treated with DEM for 3.5 h for HO-1 or 12 h for NQO1, and ChIP analyses were performed with antibodies against Pol II and primers for the HO-1 and NQO1 promoter regions. Normal rabbit IgG and the 5′ upstream region of the CSF-1 gene (37) were used as a control for this experiment, and we amplified 5% of the chromosomal DNA before immunoprecipitation in the input lane.
Recruitment of Pol II to the NQO1 gene regulatory region was not affected by BRG1 knockdown (Fig. 8). In contrast, the recruitment of Pol II to the HO-1 gene regulatory region was decreased approximately 70% in SH4 cells compared to SHCon cells (Fig. 8). These results thus demonstrate that BRG1-mediated chromatin remodeling is essential for Pol II recruitment to HO-1 promoter but not to the NQO1 promoter.
FIG. 8.
Knockdown of BRG1 reduces the recruitment of RNA Pol II to the HO-1 promoter but not the NOQ1 promoter. SHCon and SH4 cells were treated with 100 μM DEM for 3.5 h for HO-1 or 12 h for NQO1, and ChIP analyses were performed with antibodies against RNA Pol II and primers for the HO-1 and NQO1 promoter regions (pr). Normal rabbit IgG was used as a control. The 5′ upstream region of the CSF-1 gene (37) was used as a control. In the input lane, 5% unprecipitated chromosomal DNA was amplified by PCR.
DISCUSSION
In this study we found that, in response to oxidative stress, Nrf2 recruits BRG1 to the HO-1 enhancers and the promoter, and subsequently RNA Pol II is recruited for transcription initiation. Inducible expression of the HO-1 gene is impaired by the endogenous BRG1 knockdown with SH-type siRNA. Furthermore, reconstitution of BRG1 activity in SW13 cells, which lack BRG1 activity, with WT BRG1 markedly enhanced HO-1 induction in response to oxidative stress, whereas neither the functionally defective BRG1 mutant nor hBRM could enhance gene induction. These results thus demonstrate that BRG1 is a critical component of Nrf2-mediated HO-1 induction. An important observation in this study is that BRG1 is selectively involved in inducible expression of Nrf2 target genes. Whereas BRG1 knockdown markedly decreased the inducible expression of the HO-1 gene in SW480 cells, it did not attenuate induction of the other Nrf2 target genes, including NQO1, GCSL, and GCSH. Thus, BRG1 was essential for HO-1 inducible expression but dispensable for induction of other Nrf2 target genes. In fact, BRG1 is often inactivated in tumors (46); thus, HO-1 expression may be selectively lost in such cells.
Consistent with the present observations, BRG1 has been reported to modulate the expression of a subset of genes through interactions with specific transcription factors (8, 9, 19). For example, BRG1 interacts with STAT2 to selectively potentiate the expression of a subset of alpha interferon-inducible genes (19). However, selective recruitment of BRG1 by Nrf2 to AREs located in the regulatory regions of the HO-1 and NQO1 genes does not seem to fully explain the difference in the induction of these genes, since BRG1 was recruited comparably to the AREs in the HO-1 and NQO1 genes after treatment with DEM.
In contrast to the other cytoprotective enzymes, the inducible expression of AKR1C1 by DEM is increased in BRG1 knockdown cells. In this regard, it should be noted that BRG1 has been shown to be involved in the transcriptional repression of genes such as those encoding c-Fos, metallothionein, and CAD (6, 39, 44). Indeed, the hSWI/SNF complex associates with mSIN3A and arginine methyltransferase PRMT5, both of which directly repress transcription (44). A transcription factor complex containing the DNA methylase Dmnt3a, a repressor of transcription, also harbors several BRG1 complex members, such as BRG1, Baf155, and Baf57 (6). Therefore, the BRG1 complex may cause transcriptional repression in a context-dependent manner, and our current results suggest that BRG1 is involved in the negative regulation of AKR1C1.
Nrf2-mediated HO-1 activation is schematically explained in Fig. 9. In this model, the first step of HO-1 gene induction is Nrf2 nuclear accumulation in response to oxidative stress (30). Secondly, Nrf2 interacts with BRG1 in a stress-independent manner and recruits BRG1 to the distal E1 and E2 enhancers of the HO-1 promoter, which aids in the recruitment of BRG1 to the proximal promoter region via a looping mechanism. BRG1 then assists in Z-DNA formation to open the chromatin structure around the HO-1 transcriptional start site.
FIG. 9.
Schematic presentation of the role that BRG1 and Nrf2 play in the DEM-inducible expression of HO-1. In response to oxidative stress, Nrf2 accumulates in the nucleus. Subsequently, Nrf2 recruits BRG1 to the HO-1 regulatory region and facilitates chromatin remodeling through interactions with BRG1. BRG1 remodels the nucleosome (shown in red) to generate Z-DNA formation. Formation of Z-DNA facilitates the opening of chromatin and the recruitment of RNA Pol II.
The Z-DNA-forming TG repeat sequence in the HO-1 promoter, which is not found in the regulatory regions of other Nrf2 target genes, is a critical factor in the differential regulation of the Nrf2 gene battery. This TG repeat sequence can be replaced with an alternative Z-DNA-forming sequence to sustain BRG1-mediated transactivation of human HO-1. Thus, we surmise that this Z-DNA-forming structure facilitates recruitment of RNA Pol II to the HO-1 promoter. Z-DNA is inherently unstable, and its formation needs negative supercoiling or strain for stability. Thus, the Z-DNA structure is typically generated as the RNA Pol II passes over a potential Z-DNA-forming site. Snf2 ATPases including BRG1 may generate the α-helical torsion to initiate and stabilize Z-DNA formation (15). However, the mechanism as to how BRG1-containing chromatin remodeling enhances Z-DNA formation still requires elucidation.
TG repeats in the human HO-1 promoter are replaced with an insertion of a polypyrimidine tract, composed mainly of pentamer TCTCT repeats, in the promoter region of the mouse HO-1 gene. However, this sequence tract, as well as the TG repeats, is absent in the rat HO-1 gene. Purine-pyrimidine repeats (e.g., GC or GT repeats) are known to form the Z-DNA structure (47). Thus, only the human HO-1 gene has acquired the Z-DNA-forming sequence specifically during molecular evolution. In humans, Z-DNA-forming microsatellite polymorphisms in the HO-1 promoter correlate with an increase in disease susceptibility (12, 17, 61). Notably, shorter TG repeat polymorphisms tend to increase inducible HO-1 expression and decrease the incidence of pulmonary emphysema and angioplastic restenosis (17, 61). Since our present data support the contention that Z-DNA formation increases HO-1 expression, the results are somewhat unexpected and contradictory. However, when we decreased the TG repeats in the reporter contracts from 30 (as in the WT gene) to 18 (GC repeats), the reduction did not affect the response to BRG1. This observation suggests that the repeat length may be saturated after certain repeats in terms of the Z-DNA formation (42).
In conclusion, this is the first report that the transcription factor Nrf2 regulates downstream target gene expression through interaction with cofactors involved in chromatin remodeling. This study also demonstrates that BRG1-mediated chromatin-remodeling activity is essential for maximal HO-1 induction during oxidative stress, most likely by enhancing Z-DNA formation. Selective recruitment of RNA Pol II to the HO-1 promoter through sequential formation of the initiation complex may explain in part why the HO-1 gene is differentially regulated from the rest of the Nrf2-mediated gene battery. As Z-DNA-forming microsatellite regions have been shown to influence the development of diseases, the interaction of Nrf2 with chromatin-remodeling complexes may serve as an important regulatory checkpoint in modulating disease states that occur in response to oxidative stress.
Acknowledgments
We thank Kiyohito Motojima, Yasutake Katoh, Katsuyuki Iida, Hozumi Motohashi, and Makoto Kobayashi for help and discussion and Akira Kobayashi for providing the pSUPER-control vector and the Nrf2ΔETGE expression vector.
This work was supported in part by grants from JST-ERATO; the Ministry of Education, Culture, Sports, Science and Technology; the Ministry of Health, Labor and Welfare; and the Naito foundation.
Footnotes
Published ahead of print on 21 August 2006.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Alam, J., and J. L. Cook. 2003. Transcriptional regulation of the heme oxygenase-1 gene via the stress response element pathway. Curr. Pharm. Des. 9:2499-2511. [DOI] [PubMed] [Google Scholar]
- 3.Alam, J., K. Igarashi, S. Immenschuh, S. Shibahara, and R. M. Tyrrell. 2004. Regulation of heme oxygenase-1 gene transcription: recent advances and highlights from the International Conference (Uppsala, 2003) on Heme Oxygenase. Antioxid. Redox Signal. 6:924-933. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, S. W., K. P. Davies, E. Yung, R. J. Beltran, J. Yu, and G. V. Kalpana. 1999. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat. Genet. 22:102-105. [DOI] [PubMed] [Google Scholar]
- 5.Corey, L. L., C. S. Weirich, I. J. Benjamin, and R. E. Kingston. 2003. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes Dev. 17:1392-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datta, J., S. Majumder, S. Bai, K. Ghoshal, H. Kutay, D. S. Smith, J. W. Crabb, and S. T. Jacob. 2005. Physical and functional interaction of DNA methyltransferase 3A with Mbd3 and Brg1 in mouse lymphosarcoma cells. Cancer Res. 65:10891-10900. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Debril, M. B., L. Gelman, E. Fayard, J. S. Annicotte, S. Rocchi, and J. Auwerx. 2004. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J. Biol. Chem. 279:16677-16686. [DOI] [PubMed] [Google Scholar]
- 8.De La Serna, I. L., K. A. Carison, D. A. Hill, C. J. Guidi, R. O. Stephenson, S. Sif, R. E. Kingston, and A. N. Imbalzano. 2000. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol. Cell. Biol. 20:2839-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De La Serna, I. L., K. Roy, K. A. Carlson, and A. N. Imbalzano. 2001. MyoD can induce cell cycle arrest but not muscle differentiation in the presence of dominant negative SWI/SNF chromatin remodeling enzymes. J. Biol. Chem. 276:41486-41491. [DOI] [PubMed] [Google Scholar]
- 10.Devling, T. W., C. D. Lindsay, L. I. McLellan, M. McMahon, and J. D. Hayes. 2005. Utility of siRNA against Keap1 as a strategy to stimulate a cancer chemopreventive phenotype. Proc. Natl. Acad. Sci. USA 102:7280-7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunaief, J. L., B. E. Strober, S. Guha, P. A. Khavari, K. Alin, J. Luban, M. Begemann, G. R. Crabtree, and S. P. Goff. 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79:119-130. [DOI] [PubMed] [Google Scholar]
- 12.Exner, M., E. Minar, O. Wagner, and M. Schillinger. 2004. The role of heme oxygenase-1 promoter polymorphisms in human disease. Free Radic. Biol. Med. 37:1097-1104. [DOI] [PubMed] [Google Scholar]
- 13.Fry, C. J., and C. L. Peterson. 2002. Unlocking the gates to gene expression. Science 295:1847-1848. [DOI] [PubMed] [Google Scholar]
- 14.Fryer, C. J., and T. K. Archer. 1998. Chromatin remodeling by the glucocorticoid receptor requires the BRG1 complex. Nature 393:88-91. [DOI] [PubMed] [Google Scholar]
- 15.Havas, K., A. Flaus, M. Phelan, R. Kingston, P. A. Wade, D. M. Lilley, and T. Owen-Hughes. 2000. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell 103:1133-1142. [DOI] [PubMed] [Google Scholar]
- 16.Herbert, A., and A. Rich. 1999. Left-handed Z-DNA: structure and function. Genetica 106:37-47. [DOI] [PubMed] [Google Scholar]
- 17.Hirai, H., H. Kubo, M. Yamaya, K. Nakayama, M. Numasaki, S. Kobayashi, S. Suzuki, S. Shibahara, and H. Sasaki. 2003. Microsatellite polymorphism in heme oxygenase-1 gene promoter is associated with susceptibility to oxidant-induced apoptosis in lymphoblastoid cell lines. Blood 102:1619-1621. [DOI] [PubMed] [Google Scholar]
- 18.Hosoya, T., A. Maruyama, M. I. Kang, Y. Kawatani, T. Shibata, K. Uchida, K. Itoh, and M. Yamamoto. 2005. Differential responses of the Nrf2-Keap1 system to laminar and oscillatory shear stresses in endothelial cells. J. Biol. Chem. 280:27244-27250. [DOI] [PubMed] [Google Scholar]
- 19.Huang, M., F. Qian, Y. Hu, C. Ang, Z. Li, and Z. Wen. 2002. Chromatin-remodeling factor BRG1 selectively activates a subset of interferon-alpha-inducible genes. Nat. Cell Biol. 4:774-781. [DOI] [PubMed] [Google Scholar]
- 20.Ishida, M., S. Tanaka, M. Ohki, and T. Ohta. 2004. Transcriptional co-activator activity of SYT is negatively regulated by BRM and Brg1. Genes Cells 9:419-428. [DOI] [PubMed] [Google Scholar]
- 21.Ishii, T., K. Itoh, S. Takahashi, H. Sato, T. Yanagawa, Y. Katoh, S. Bannai, and M. Yamamoto. 2000. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J. Biol. Chem. 275:16023-16029. [DOI] [PubMed] [Google Scholar]
- 22.Itoh, K., T. Chiba, S. Takahashi, T. Ishii, K. Igarashi, Y. Katoh, T. Oyake, N. Hayashi, K. Satoh, I. Hatayama, M. Yamamoto, and Y. Nabeshima. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant responsive elements. Biochem. Biophys. Res. Commun. 236:313-322. [DOI] [PubMed] [Google Scholar]
- 23.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, K. Igarashi, J. D. Engel, and M. Yamamoto. 1999. Keap1 represses nuclear activation of antioxidant responsive element by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13:76-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, T. O'Connor, and M. Yamamoto. 2003. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells 8:379-391. [DOI] [PubMed] [Google Scholar]
- 25.James, D., Y, Shang., M. Phelan, S. Sif, M. Myers, R. Kingston, and M. Brown. 2000. BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol. Cell. Biol. 20:7541-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadam, S., G. S. McAlpine, M. L. Phelan, R. E. Kingston, K. A. Jones, and B. M. Emerson. 2000. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 14:2441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katoh, Y., K. Itoh, E. Yoshida, M. Miyagishi, A. Fukamizu, and M. Yamamoto. 2001. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 6:857-868. [DOI] [PubMed] [Google Scholar]
- 28.Khavari, P. A., C. L. Peterson, J. W. Tamkun, D. B. Mendel, and G. R. Crabtree. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170-174. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi, A., M. I. Kang, H. Okawa, M. Ohtsuji, Y. Zenke, T. Chiba, K. Igarashi, and M. Yamamoto. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24:7130-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi, A., M. I. Kang, Y. Watai, K. I. Tong, T. Shibata, K. Uchida, and M. Yamamoto. 2006. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol. Cell. Biol. 26:221-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobayashi, M., K. Itoh, T. Suzuki, H. Osanai, K. Nishikawa, Y. Katoh, Y. Takagi, and M. Yamamoto. 2002. Identification of the interactive interface and phylogenetic conservation of the Nrf2-Keap1 system. Genes Cells 7:807-820. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi, M., and M. Yamamoto. 2005. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid. Redox Signal. 7:385-394. [DOI] [PubMed] [Google Scholar]
- 33.Kowenz-Leutz, E., and A. Leutz. 1999. A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell 4:735-743. [DOI] [PubMed] [Google Scholar]
- 34.Kumar, S., and U. Bandyopadhyay. 2005. Free heme toxicity and its detoxification systems in human. Toxicol. Lett. 157:175-188. [DOI] [PubMed] [Google Scholar]
- 35.Kwon, H., A. N. Imbalzano, P. A Khavari, R. E. Kingston, and M. R. Green. 1994. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 370:477-481. [DOI] [PubMed] [Google Scholar]
- 36.Liu, H., N. Mulholland, H. Fu, and K. Zhao. 2006. Cooperative activity of BRG1 and Z-DNA formation in chromatin remodeling. Mol. Cell. Biol. 26:2550-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, R., H. Liu, X. Chen, M. Kirby, P. O. Brown, and K. Zhao. 2001. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 106:309-318. [DOI] [PubMed] [Google Scholar]
- 38.Mohrmann, L., and C. P. Verrijzer. 2005. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta 1681:59-73. [DOI] [PubMed] [Google Scholar]
- 39.Murphy, D. J., S. Hardy, and D. A. Engel. 1999. Human SWI-SNF component BRG1 represses transcription of the c-fos gene. Mol. Cell. Biol. 19:2724-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nioi, P., T. Nguyen, P. J. Sherratt, and C. B. Pickett. 2005. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol. Cell. Biol. 25:10895-10906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nioi, P., and J. D. Hayes. 2004. Contribution of NAD(P)H:quinone oxidoreductase 1 to protection against carcinogenesis, and regulation of its gene by the Nrf2 basic-region leucine zipper and the arylhydrocarbon receptor basic helix-loop-helix transcription factors. Mutat. Res. 555:149-171. [DOI] [PubMed] [Google Scholar]
- 42.Oh, D. B., Y. G. Kim, and A. Rich. 2002. Z-DNA-binding proteins can act as potent effectors of gene expression in vivo. Proc. Natl. Acad. Sci. USA 99:16666-16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otsuki, T., Y. Furukawa, K. Ikeda, H. Endo, T. Yamashita, A. Shinohara, A. Iwamatsu, K. Ozawa, and J. M. Liu. 2001. Fanconi anemia protein, FANCA, associates with BRG1, a component of the human SWI/SNF complex. Hum. Mol. Genet. 10:2651-2660. [DOI] [PubMed] [Google Scholar]
- 44.Pal, S., R. Yun, A. Datta, L. Lacomis, H. Erdjument-Bromage, J. Kumar, P. Tempst, and S. Sif. 2003. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol. 23:7475-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Primiano, T., T. R. Sutter, and T. W. Kensler. 1997. Antioxidant-inducible genes. Adv. Pharmacol. 38:293-328. [DOI] [PubMed] [Google Scholar]
- 46.Reisman, D. N., J. Sciarrotta, W. Wang, W. K. Funkhouser, and B. E. Weissman. 2003. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 63:560-566. [PubMed] [Google Scholar]
- 47.Rich, A., and S. Zhang. 2003. Timeline: Z-DNA: the long road to biological function. Nat. Rev. Genet. 4:566-572. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Sawado, T., K. Igarashi, and M. Groudine. 2001. Activation of beta-major globin gene transcription is associated with recruitment of NF-E2 to the beta-globin LCR and gene promoter. Proc. Natl. Acad. Sci. USA 98:10226-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scroth, G. P., P. J. Chou, and P. S. Ho. 1992. Mapping Z-DNA in the human genome. Computer-aided mapping reveals a nonrandom distribution of potential Z-DNA-forming sequences in human genes. J. Biol. Chem. 267:11846-11855. [PubMed] [Google Scholar]
- 51.Shan, Y., R. W. Lambrecht, T. Ghaziani, S. E. Donohue, and H. L. Bonkovsky. 2004. Role of Bach1 in regulation of heme oxygenase-1 in human liver cells: insights from studies with small interfering RNAs. J. Biol. Chem. 279:51769-51774. [DOI] [PubMed] [Google Scholar]
- 52.Soutoglou, E., and I. Talianidis. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901-1904. [DOI] [PubMed] [Google Scholar]
- 53.Sun, J., H. Hoshino, K. Takaku, O. Nakajima, A. Muto, H. Suzuki, S. Tashiro, S. Takahashi, S. Shibahara, J. Alam, M. M. Taketo, M. Yamamoto, and K. Igarashi. 2002. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 21:5216-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun, J., M. Brand, Y. Zenke, S. Tashiro, M. Groudine, and K. Igarashi. 2004. Heme regulates the dynamic exchange of Bach1 and NF-E2-related factors in the Maf transcription factor network. Proc. Natl. Acad. Sci. USA 101:1461-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takeda, K., S. Ishizawa, M. Sato, T. Yoshida, and S. Shibahara. 1994. Identification of a cis-acting element that is responsible for cadmium-mediated induction of the human heme oxygenase gene. J. Biol. Chem. 269:22858-22867. [PubMed] [Google Scholar]
- 56.Talalay, P., A. T. Dinkova-Kostova, and W. D. Holtzclaw. 2003. Importance of phase 2 gene regulation in protection against electrophile and reactive oxygen toxicity and carcinogenesis. Adv. Enzyme Regul. 43:121-134. [DOI] [PubMed] [Google Scholar]
- 57.Trainer, D. L., T. Kline, F. L. McCabe, L. F. Faucette, J. Feild, M. Chaikin, M. Anzano, D. Rieman, S. Hoffstein, D. J. Li, D. Gennaro, C. Busccarino, M. Lynch, G. Poste, and R. Greig. 1988. Biological characterization and oncogene expression in human colorectal carcinoma cell lines. Int. J. Cancer 41:287-296. [DOI] [PubMed] [Google Scholar]
- 58.Reference deleted.
- 59.Wakabayashi, N., K. Itoh, J. Wakabayashi, H. Motohashi, S. Noda, S. Takahashi, S. Imakado, T. Kotsuji, F. Otsuka, D. R. Roop, T. Harada, J. D. Engel, and M. Yamamoto. 2003. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 35:238-245. [DOI] [PubMed] [Google Scholar]
- 60.Wong, A. K., F. Shanahan, Y. Chen, L. Lian, P. Ha, K. Hendricks, S. Ghaffari, D. Iliev, B. Penn, A. M. Woodland, R. Smith, G. Salada, A. Carillo, K. Laity, J. Gupte, B. Swedlund, S. V. Tavtigian, D. H. Teng, and E. Lees. 2000. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 60:6171-6177. [PubMed] [Google Scholar]
- 61.Yamada, N., M. Yamaya, S. Okinaga, K. Nakayama, K. Sekizawa, S. Shibahara, and H. Sasaki. 2000. Microsatellite polymorphism in the heme oxygenase-1 gene promoter is associated with susceptibility to emphysema. Am. J. Hum. Genet. 66:187-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamamichi-Nishina, M., T. Ito, T. Mizutani, N. Yamamichi, H. Watanabe, and H. Iba. 2003. SW13 cells can transition between two distinct subtypes by switching expression of BRG1 and Brm genes at the posttranscriptional level. J. Biol. Chem. 278:7422-7430. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, L., D. K. Fogg, and D. M. Waisman. 2004. RNA interference-mediated silencing of the S100A10 gene attenuates plasmin generation and invasiveness of Colo 222 colorectal cancer cells. J. Biol. Chem. 279:2053-2062. [DOI] [PubMed] [Google Scholar]
- 64.Zhao, K., W. Wang, O. J. Rando, Y. Xue, K. Swiderek, A. Kuo, and G. R. Crabtree. 1998. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95:625-636. [DOI] [PubMed] [Google Scholar]