Abstract
The NAD-dependent histone deacetylase Sir2 plays a key role in connecting cellular metabolism with gene silencing and aging. The androgen receptor (AR) is a ligand-regulated modular nuclear receptor governing prostate cancer cellular proliferation, differentiation, and apoptosis in response to androgens, including dihydrotestosterone (DHT). Here, SIRT1 antagonists induce endogenous AR expression and enhance DHT-mediated AR expression. SIRT1 binds and deacetylates the AR at a conserved lysine motif. Human SIRT1 (hSIRT1) repression of DHT-induced AR signaling requires the NAD-dependent catalytic function of hSIRT1 and the AR lysine residues deacetylated by SIRT1. SIRT1 inhibited coactivator-induced interactions between the AR amino and carboxyl termini. DHT-induced prostate cancer cellular contact-independent growth is also blocked by SIRT1, providing a direct functional link between the AR, which is a critical determinant of progression of human prostate cancer, and the sirtuins.
The incidence of prostate cancer, the most common noncutaneous malignancy affecting males in the United States, increases strikingly with age (13, 30). Prostate cancer has a heterogeneous clinical behavior with preoperative prostate-specific antigen (PSA), Gleason score, and global histone modifications being predictors of clinical outcome (24, 51). Abnormal function of the androgen receptor (AR) has been linked to both the pathogenesis and the progression of human prostate cancer. The AR is a DNA-binding transcription factor that governs male sexual development and differentiation. The induction of AR activity is regulated by hormones, including dihydrotestosterone (DHT), which enhances coactivator (p300 and SRC) and reduces corepressor protein (NCoR, histone deacetylase [HDAC], and Smad) association with the AR (13, 56). Cointegrator binding regulates modular intramolecular associations of the amino (N) and carboxyl (C) termini of the AR required for ligand-induced transactivation (7, 28). Posttranslational modification of the AR by phosphorylation, acetylation, and sumoylation govern its subcellular localization, cointegrator association, and DNA binding (20).
Acetylation of the AR by histone acetyltransferases (HATs), including p300, P/CAF, and TIP60, occurs through a conserved motif within the AR hinge region (17, 18). Point mutation of the lysine residues within this conserved motif abrogates acetylation of the full-length AR in cultured cells (17-19) and alters the association with corepressor (HDAC1, NCoR, and Smad complexes) and coactivator (p300) proteins (18). Acetylation mimic mutants of the AR display enhanced binding of the coactivator p300, both in vitro and in cultured cells, correlating with enhanced transactivation by multiple AR coactivators (SRC1, Ubc9, and ARA70) and increased access to androgen-responsive elements of endogenous target genes in chromatin immune precipitation assays (16, 18). In cultured cells, DHT induces AR acetylation. Stimuli such as bombesin that enhance DHT-induced AR activity further enhance AR acetylation (25). Together, these studies suggest that AR acetylation is a physiological consequence of hormone-activated AR signaling.
Acetylation of histones is a reversible process involved in the regulation of transcriptional activation and silencing. Nonhistone proteins are also acetylated on lysine residues to regulate their activities (reviewed in reference 20). The relative levels of acetylation are controlled by the actions of HATs and HDACs. Based on their homology to yeast transcriptional repressors, HDACs have been divided into three distinct classes, with class I and II deacetylases being homologous to Rpd3P and Hda1P proteins, respectively. Class III HDACs are homologous to the yeast transcriptional repressor Sir2p and are broadly defined as sirtuins. The proteins in class I and class II are characterized by their sensitivity to the inhibitor trichostatin A (TSA). In contrast, class III HDAC activity is NAD dependent and is not inhibited by TSA (8, 29, 32, 53).
The silent information regulator 2 (Sir2) proteins in yeast are necessary for gene silencing at distinct loci, including telomeres, the rRNA gene locus, and the mating-type locus (22). The sirtuin gene family is conserved from archaebacteria to eukaryotes (15, 57). Mammalian genomes contain seven Sir2 homologs (SIRT1 to -7). The NAD+-dependent deacetylation of proteins by sirtuins couples the removal of the acetyl group from the protein substrate with the cleavage of a high-energy bond in NAD to synthesis of a novel product, 2′-O-acetyl-ADP-ribose (48).
Recent studies have demonstrated that acetylated lysine residues of p53 (37, 55), MyoD, FOXO (10, 21, 41), and TAF168 (42) are substrates for NAD-dependent histone deacetylation by human SIRT1 (hSIRT1) and murine Sir2α (mSir2α), which are the closest homologs to yeast Sir2. Sir2-dependent deacetylation of p53 repressed p53 function and promoted cell survival in response to stress signals (4, 37, 55). Here, we investigated the role of hSIRT1 in AR function in vitro and in vivo. hSIRT1 functions as an NAD-dependent transcriptional repressor of DHT-induced prostate cancer cellular growth. AR activity is repressed by SIRT1, requiring the AR-acetylated lysine motif. An 18-amino-acid peptide containing the AR acetylation site is an efficient substrate for NAD-dependent deacetylation. In view of the dependence of class III HDAC function upon the metabolic precursor NAD, the current study suggests that metabolism may in turn directly contribute to ligand-induced hormone signaling.
MATERIALS AND METHODS
Reporter genes and expression vectors.
The human SIRT1 expression vectors in pcDNA3-SIRT1 and pcDNA3-Myc-SIRT1 were previously described (37). The SIRT2α antibody, which recognizes both human and mouse Sir2α, was prepared as a polyclonal antibody directed against the highly conserved C terminus of Sir2α and was previously described (9, 37). The androgen-responsive synthetic reporter constructions mouse mammary tumor virus (MMTV)-LUC, PSA-LUC, androgen-responsive element (ARE)4-LUC, p21CIP1-LUC, pSV40-LUC, and pG5-LUC (17, 18, 23); the expression vectors pCMVHA-p300, pcDNA3-AR-GFP, and hARwt; and the AR point mutants ARK630T and ARK630Q were described previously (17, 19, 38, 44, 47).
Cell culture, DNA transfection, and luciferase assays.
Reporter assays, cell culture, DNA transfection, and luciferase assays were performed as previously described (16). The prostate cancer cell lines LNCaP, DU145, and PC3; the HEK293 cell line; and mouse embryonic fibroblasts were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 1% penicillin, and 1% streptomycin. The cells were incubated in media containing 10% charcoal-stripped fetal bovine serum prior to experimentation using DHT (16). Statistical analyses were performed using the Mann-Whitney U test.
Western blotting and IP-Western blotting.
Western blotting and immunoprecipitation (IP)-Western blotting were performed as previously described (16). The antibodies used in Western blot analysis were anti-SIRT1 antibody (Upstate Biotechnology) and mouse anti-AR antibody (441) and anti-Myc (9E10) antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.) The guanine nucleotide dissociation inhibitor antibody (a generous gift from Perry Bickel, Washington University, St. Louis, Mo.) was used as an internal control for protein abundance. LNCaP cells were treated with vehicle (ethanol) or DHT (10 nM) for 24 h or treated with TSA (30 nM), nicotinamide (NAM) (50 mM), or sirtinol (50 μM) and splitomycin (50 μM) for 6 h. The cell lysates were harvested in cell lysis buffer (50 mM HEPES [pH 7.2], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 0.1% Tween 20, 0.1 mM phenylmethylsulfonyl fluoride, 2.5 μg of leupeptin/ml, 0.1 mM sodium orthovanadate), dissolved in 9% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer, and subjected to Western blotting. For the detection of protein, the membrane was incubated with anti-AR (1:1,000) and anti-human SIRT1 (1:1,000) at 4°C overnight. The blots were then washed three times with 0.5% Tween 20-phosphate-buffered saline and incubated with the appropriate horseradish peroxidase-conjugated secondary antibody. The proteins were visualized by the enhanced chemiluminescence system (Amersham Pharmacia Biotech). IP-Western blotting was performed as previously described (16) with the lysates from HEK293 cells transfected with pcDNA3-AR, pcDNA3-Myc-SIRT1, or an empty expression vector cassette as a control. The cells were treated for 24 h with 100 nM DHT or vehicle or treated with TSA (30 nM) or NAM (50 mM) for 6 h. The cells were lysed in cell lysis buffer. The extracts were cleared by centrifugation and further precleared by rocking them at 4°C with washed protein G-agarose beads (Roche Molecular Biochemicals, Indianapolis, Ind.). The precleared extracts were immunoprecipitated with 5 μg of AR antibody or 5 μg of anti-Myc antibody and 30 μl of protein G-agarose for 8 to 12 h at 4°C. Equivalent amounts of immunoglobulin G (IgG) were used as the control. The beads were washed five times with lysis buffer and boiled in SDS sample buffer, and the released proteins were resolved by SDS-PAGE. The gels were transferred to nitrocellulose, and Western blotting was performed.
Immunofluorescence staining.
HeLa cells were grown in four-well chamber slides and transfected with expression vector for green fluorescent protein (GFP)-AR, Myc-SIRT1, or control vector with Lipofectamine reagents. The cells were treated with vehicle (ethanol) or 100 nM DHT for 3 h, fixed with 4% paraformaldehyde for 20 min at room temperature, and washed once with phosphate-buffered saline. The primary antibody used was rabbit anti-Myc (Santa Cruz; 1:50). The secondary antibody used was Rhodamine Red-X-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories) (1:100). The samples were visualized on an Olympus IX70 laser scanning confocal microscope with a 60× PlanApo oil immersion objective using Olympus Fluoview FV-300 software. The images were processed with MetaMorph (Molecular Devices).
Retroviral production and infection.
Retroviral production was described previously (34). Mouse stem cell virus (MSCV)-internal ribosome entry site (IRES)-GFP or MSCV-SIRT1-IRES-GFP retroviruses (34) were prepared by transient cotransfection with helper virus into 293T cells, using calcium phosphate precipitation. The retroviral supernatants were harvested 48 h after transfection and filtered through a 0.45-μm membrane. LNCaP, DU145, and PC3 cells were incubated with fresh retroviral supernatants in the presence of 8 μg/ml Polybrene for 24 h, cultured for 72 h, and subjected to fluorescence-activated cell sorting (FACS) (FACS Vantage SE; BD Biosciences) for GFP-positive cells.
Colony formation and MTT assay.
LNCaP cells were incubated with fresh retroviral supernatants of MSCV-IRES-GFP or MSCV-SIRT1-IRES-GFP. The cells were seeded at low density onto 35-mm plates (4,000 cells/plate; three plates per sample) in 0.3% agar in 2 ml complete medium, which was then covered with 0.5 to 1.0 ml medium containing 10% serum. The plates were incubated for up to 2 weeks. The sizes and numbers of GFP-positive colonies growing on soft-agar plates were scored on day 15. The thiazolyl blue (MTT) assay was performed as described previously (49).
In vivo acetylation.
LNCaP cells were treated with 50 μM sirtinol or 100 nM DHT for 24 h. The cell lysates were harvested and immunoprecipitated with anti-acetylated lysine antibody (Ac-K103; catalog no. 9681; Cell Signaling Technology). The immunoprecipitate was separated by SDS-PAGE and blotted with anti-AR antibody.
In vitro translation and GST pull down.
To detect interaction between AR and SIRT1 in vitro, glutathione S-transferase (GST) pull down was conducted. In vitro-translated AR proteins were prepared by coupled transcription-translation with a Promega TNT-coupled reticulocyte lysate kit (Promega, Madison, WI) using 0.5 μg of plasmid DNA (pcDNA3 vector or pcDNA3-AR) in a total volume of 50 μl. The in vitro-translated product (45 μl) was incubated with GST-SIRT1 (catalog no. 17-370; Upstate Biotechnology) or GST alone at 4°C for 2 h, and GST pull down was performed. The immunoprecipitate was resolved in a 9% SDS-PAGE and blotted with anti-AR, -SIRT1, or -GST antibody.
SIR2 reaction and LC/MS analysis.
Three hundred micromoles of the AR peptide (synthesized by the Albert Einstein College of Medicine peptide facility) Ac-GMTLGAR(KAc)L(KAc)(KAc)LGNLKLQ, identical to the androgen receptor sequence from 623 to 640, in acetylated form, was incubated with 600 μM NAD+ and 1 μM mSIR2α or 1 μM hSIRT1 in potassium phosphate (50 mM, pH 7.6). After 30 min of incubation at 35°C, the reaction solution was loaded onto a Vydac 1.0- by 50-mm C8 column (The Separations Group, Hesperia, CA). An HP 1100 high-performance liquid chromatograph (HPLC) equipped with a degasser and a binary pump was used to generate acetonitrile gradients at a flow rate of 50 μl/min. Solvent A contained 0.1% trifluoroacetic acid (TFA), and solvent B contained 90% (vol/vol) acetonitrile-0.1% trifluoroacetic acid. The total solvent composition for all chromatography is given by the equation A + B = 100% (where A and B refer to the respective solvents). The sample was desalted at a 5% concentration of solvent B for 20 min, and the peptides were separated by a 3-min gradient in which the solvent B concentration increased from 5% to 15%, followed by a 35-min gradient in which the solvent B concentration increased from 15% to 50% and a 5-min gradient in which the solvent B concentration increased from 50% to 95%. The column effluent was delivered directly to an LCQ quadruple ion trap mass spectrometer (ThermoFinnigan, Riviera Beach, FL) equipped with an electrospray ionization source. For liquid chromatography/mass spectrometry (LC/MS) analysis, the mass spectrometer was operated in normal MS scan mode to detect ions in the m/z range of 400 to 2,000. For LC/MS/MS analysis, the mass spectrometry was performed in the data-dependent mode. The mass spectrometer detected the intensity of the ions in the m/z range of 700 to 1,500 and was switched to the collision-induced dissociation mode to acquire an MS/MS spectrum when certain criteria were met. The mass isolation window for the collision-induced dissociation mode was set at 3 mass units, and the relative collision energy was set arbitrarily at 30%. Reactions and analyses for yeast Sir2p and Archaeoglobus fulgidus Sir2af2 were performed similarly (49).
To determine the deacetylation selectivity of human SIRT1 on triacetylated AR peptide, the AR peptide (5 μl; 100 μM) and SIRT1 enzyme (2 μl; 3.85 μM) were added to 45 μl of NAD+ (200 μM) dissolved in 50 mM phosphate buffer (pH 7.5). The reaction was incubated at 25°C for 15 min and quenched by the addition of 8 μl of TFA (760 μM); 10 μl each of the reaction mixture and control were injected onto a C18 column and analyzed as previously described.
To determine the Michaelis-Menten curves of SIRT1-catalyzed deacetylation for p53 (KKQSTSRHKK[AC]LMFKTEG) (48), p300 (ERSTELK[Ac]TEIK(Ac)EEEDQPSTS) (9, 37), and AR peptide, p53, p300, and AR peptide (from 5 μM to 525 μM) were added to NAD+ (500 μM) in 50 mM phosphate buffer (pH 7.5). Deacetylation was initiated by the addition of 3 μl of SIRT1 enzyme (3.85 μM). The reaction mixture was incubated at 25°C for 30 min and then quenched with 8 μl of 760 μM TFA. Ten microliters of each reaction mixture was injected onto the HPLC for analysis. The chromatograms were obtained by separation with 0.1% TFA in water using an analytical C18 column (Macherey-Nagel Nucleosil 100-3 720192.46) and 260-nm detection. The areas of resolved peaks for 2′- and 3′-O-acetyl-ADP-ribose were used to quantitate total deacetylation.
Homology model of SIR2-AR peptide complex.
Several homology models for the catalytic domain of human SIRT1 (residues 240 to 500) were built based on the following templates: SIR2-p53 peptide complex, Archaeoglobus fulgidus (Protein Data Bank [PDB] [http://www.rcsb.org, 6] code, 1MA3 [3]); Sir2 homolog-NAD complex, Archaeoglobus fulgidus (PDB code, 1ICI); 1SZC Sir2-NAD-H4 peptide complex, Saccharomyces cerevisiae (PDB code, 1SZC); and Sir2 adenosinephosphoribose-peptide complex, Saccharomyces cerevisiae (PDB code, 1SZD [58]). The peptide sequence of AR used in the modeling studies was 627-GARKLKKLG-635. Multiple sequence alignments were used as previously shown (40), and homology models were built using the comparative modeling program Modeler version 6.0 (14). The models were subjected to visual screening, and the models that were built using the high-resolution X-ray crystal structures 1SZD (with a resolution of 1.5 Å) and 1SZC (1.75-Å resolution) were selected for further in silico structure-function analyses. An acetate group was added to the Nɛ atom of K630, and the coordinates of the whole hSirT1-peptide-NAD complex were energy minimized using the DISCOVER module (Accelrys Inc., San Diego, CA) with a CFF91 force field and a distance-dependent dielectric constant to compute the electrostatic interactions.
RESULTS
SIRT1 inhibits ligand-induced androgen receptor transcriptional activity.
Nicotinamide is the first product from hydrolysis of the pyridinium-N-glycosidic bond of NAD, and it is an inhibitor of Sir2 enzymes from different organisms, including human, yeast, and Archaea enzymes (32, 37, 49). Evidence suggests that NAD+-dependent protein deacetylases are also inhibited by nicotinamide in vivo (49). Addition of NAM (5 mM) to the prostate cancer cell line LNCaP induced accumulation of AR in the cells (Fig. 1A). AR gene expression is responsive to androgens, and treatment with DHT induced AR abundance 10-fold over the control (Fig. 1A). Nicotinamide also enhanced DHT-induced AR expression, indicating that nicotinamide can modulate androgen hormone-dependent signaling.
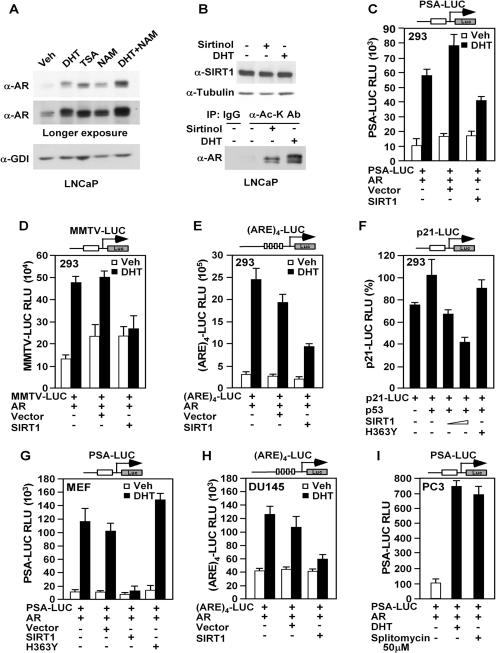
FIG. 1.
SIRT1 inhibits ligand-induced androgen receptor transcriptional activity and AR acetylation. (A) Western blot of endogenous AR abundance in LNCaP cells treated with vehicle (Veh), DHT, TSA or NAM. (B) LNCaP cells were treated with DHT (100 nM) or sirtinol (50 μM) as indicated for 24 h. The cell lysates were immunoprecipitated with anti-acetylated lysine antibody (α-Ac-K Ab; mouse monoclonal) and blotted with the AR antibody (AR-441). (C to E) HEK293 cells were transfected with expression vectors for pcDNA3-AR, hSIRT1, and the androgen-responsive reporter gene PSA-LUC (C), MMTV-LUC (D), or (ARE)4-LUC (E). The cells were treated with vehicle or 100 nM DHT, as indicated. The data are mean plus standard error of the mean (SEM) relative luciferase units for ≥9 transfections. (F) HEK293 cells were transfected with expression vectors for p53, hSIRT1, or hSIRT1 H363Y, together with p21CIP1-LUC. A luciferase assay was performed 30 h after transfection. (G) The role of SIRT1 in regulating the activities of the luciferase reporters PSA-LUC in mouse embryonic fibroblasts or (H) (ARE)4-LUC in DU145 cells was determined by coexpression of a SIRT1 expression plasmid or (I) addition of the SIRT1 inhibitor splitomycin. The data are means plus SEM for ≥9 separate experiments.
In order to determine the role of endogenous SIRT1 in regulating AR acetylation, LNCaP cells were treated with the SIRT1 inhibitor sirtinol. The anti-acetylated-lysine immune precipitated was subjected sequentially to Western blotting with the AR antibody, as previously described (25). The total amount of acetylated AR was increased by either sirtinol (Fig. 1B, bottom, lane 3 from the left) or DHT (Fig. 1B, bottom, fourth lane from the left) treatment, suggesting that SIRT1 regulates acetylation of the AR in LNCaP cells.
To examine the possible role of hSIRT1 in regulating AR function, androgen-responsive reporter genes were assessed in the AR-deficient HEK293 cell line. In the presence of transfected wild-type AR, DHT induced gene expression from the PSA promoter linked to a luciferase reporter gene (Fig. 1C). AR with DHT induced activities of the PSA promoter and the androgen-responsive reporter genes, MMTV-LUC and (ARE)4-LUC, five- to sixfold (Fig. 1C to E). Coexpression of hSIRT1 reduced DHT-induced AR transcriptional activity by 50% (Fig. 1C to E). Similar results were obtained in the AR-deficient cell line CV1 (see supplement 1A posted at http://www.pestelllab.org/publications/2006/fu&liu_ar_sirt1/supplement_1.pdf) (55). hSIRT1 did not affect the gene transcription activity at the simian virus 40 promoter (see supplement 1B posted at http://www.pestelllab.org/publications/2006/fu&liu_ar_sirt1/supplement_1.pdf) and repressed the p21CIP1 promoter (Fig. 1F), as previously shown (9, 37). In mouse embryo fibroblasts, SIRT1 repressed DHT-induced AR-dependent activation of the PSA promoter. Transcriptional repression by SIRT1 required the catalytic function and was abolished by a mutation of the catalytic domain of SIRT1 (SIRT1 H363Y) (Fig. 1G). To determine whether the AR binding site was sufficient to convey transcriptional repression in human prostate cancer cells, we examined a simple multimeric ARE enhancer linked to a luciferase reporter gene [(ARE)4-LUC]. SIRT1 expression reduced DHT-induced AR-dependent reporter gene activity in DU145 human prostate cancer cells (Fig. 1H). We next sought to determine whether endogenous SIRT1 repressed AR activity. For this purpose, we used a SIRT1 inhibitor, splitomycin (50 μM). DHT induced the activity of the androgen-responsive PSA promoter sevenfold. Addition of splitomycin (50 μΜ) also induced PSA promoter activity sevenfold. No induction was observed in the absence of the AR. These studies suggest that inhibition of endogenous SIRT1 activity induces the PSA promoter in the presence of the AR (Fig. 1I).
SIRT1 associates with the AR in vitro and in vivo.
To determine whether SirT1 physically coassociates with the AR, immunoprecipitation-Western blot analysis was conducted. The hAR and either SIRT1 or catalytically dead SIRT1 (SIRT1 H363Y) linked to the Myc epitope were coexpressed in 293T cells. Western blot analysis showed that SIRT1 reduced basal AR abundance by 10% and virtually abolished DHT-induced AR abundance (Fig. 2A). Western blotting, normalized for the loading control tubulin, showed that the SIRT1 H363Y mutant reduced DHT-induced AR abundance ∼20%, suggesting that the catalytic function of SIRT1 is required for full inhibition of DHT-induced AR abundance (Fig. 2A). Immunoprecipitation-Western blotting was conducted to examine the association of the AR and SIRT1 in cultured cells. The Myc antibody immunoprecipitated Myc-hSIRT1 and coprecipitated the AR, identified by staining for the 110-kDa protein with the antibody directed to either the AR or the Myc epitope of SIRT1 (Fig. 2D). Western blot analysis of the input used in IP demonstrated that DHT induced the AR without affecting SIRT1 expression.
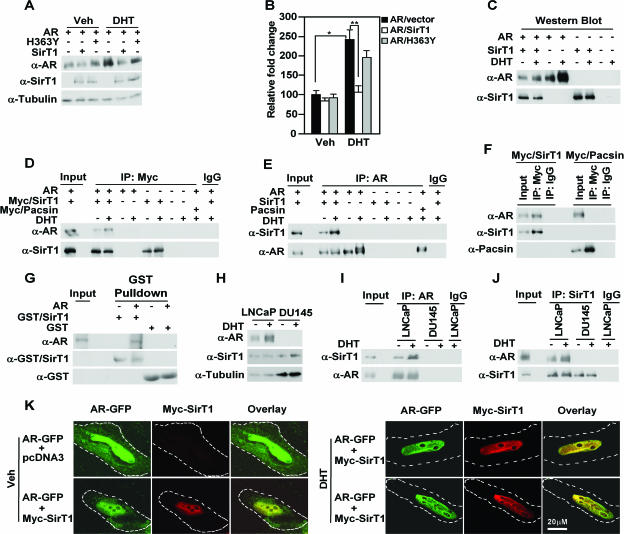
FIG. 2.
SIRT1 associates with the AR in vitro and in vivo. (A and B) HEK293T cells were transfected with expression vectors for SIRT1, the catalytic dead mutant SIRT1 (H363Y), and human AR and were then treated with vehicle (Veh) or DHT (100 nM), as indicated. (A) Western blot analysis and (B) mean (plus standard error of the mean) data for three separate transfections are shown. (C to F) Immunoprecipitation-Western blot analysis was conducted using antibodies directed to the Myc epitope of SIRT1 (D) or to the AR (E). HEK293T cells were transfected with Myc epitope-tagged pacsin as a form of negative control for the c-Myc immunoprecipitation (F), and AR and SIRT1 expression in cells was checked by Western blotting (C). (G) Western blot analysis of GST pull-down in which in vitro-translated full-length AR was incubated with purified full-length GST fusion protein for SIRT1. (H) Western blot and (I and J) immunoprecipitation with antibody directed to endogenous AR (I) or SIRT (J) in LNCaP cells, with sequential Western blotting for coassociated SIRT1 or AR. β-Tubulin is a loading control for the Western blot. (K) Confocal microscopy of AR and SIRT1 in HeLa cells transfected with AR-GFP and Myc-tagged SIRT1. The cells were treated with vehicle (left) or DHT (10−7 M) (right) for 3 h. Nuclear colocalization of AR (green) and SIRT1 (red) is shown in yellow. The Z-series reconstruction is shown as a supplemental movie at http://www.pestelllab.org/publications/2006/fu&liu_ar_sirt1/supplement_2.doc. Ten percent of the total amount of cell lysates used for IP was included as input in the IP-Western blot assay.
Reciprocal immunoprecipitation was next conducted. In reciprocal immunoprecipitation, IP-Western blotting with the AR antibody coprecipitated the AR and SIRT1 (Fig. 2E). DHT is known to induce a higher-molecular-weight hyperphosphorylated form of the AR (16). In the presence of DHT, immunoprecipitation with an AR antibody detected primarily a lower-molecular-weight form of the AR associated with SIRT1 in HEK293 cells (Fig. 2E). The cells were transfected with an expression vector encoding a Myc epitope-tagged protein (pacsin) as a form of negative control. IP with an antibody to the Myc epitope of SIRT1 precipitated the AR but did not coprecipitate pacsin (Fig. 2F). This study indicated that the coprecipitation of the AR with SIRT1 is not secondary to a nonspecific interaction of the antibody directed to the c-Myc epitope (Fig. 2F).
To examine the physical interaction of the AR and SIRT1 in vitro, the association of full-length in vitro-synthesized AR with full-length SIRT1 synthesized in bacteria was assessed. GST pull down and subsequent Western blotting showed that the AR, but not unprogrammed lysate, coassociated with purified SIRT1 (Fig. 2G). To examine further the physical association of the AR and SIRT1 in vivo, IP-Western analysis was conducted of the endogenous AR and SIRT1 in LNCaP cells. Immunoprecipitation using an anti-AR antibody and subsequent Western blotting with an antibody against SIRT1 demonstrated that SIRT1 is associated with the AR within LNCaP cells (Fig. 2I). The AR antibody did not coprecipitate with either AR or SIRT1 in the AR-deficient DU145 cell line. An equal dose of IgG did not precipitate AR or SIRT1. Western blotting demonstrated that DHT induced AR abundance in LNCaP cells without affecting SIRT1 expression. The reciprocal immunoprecipitation using an anti-SIRT1 antibody demonstrated the precipitation of SIRT1 and coprecipitation of the AR (Fig. 2J).
To examine if SIRT1 and the AR colocalize within cells, confocal microscopy was conducted of HeLa cells expressing an AR-GFP fusion protein and Myc-tagged SIRT1. In the absence of ligand, diffuse staining for the AR was observed in the nuclear and cytoplasmic compartments (Fig. 2K, left). Upon the addition of DHT, colocalization of the AR and SIRT1 was observed in the nuclear, extranucleolar compartment. The majority of colocalized immunofluorescence was observed in extranucleolar chordlike structures (Fig. 2K, right). This phenomenon is clearly observed in a three-dimensional reconstruction of the Z series confocal microscopy (see supplement 2 posted at http://www.pestelllab.org/publications/2006/fu&liu_ar_sirt1/supplement_2.doc). Collectively, these data indicate that the AR associates with SIRT1 both in vitro and in vivo.
SIRT1 represses p300-mediated AR intramolecular interaction.
The findings that AR transcriptional activity is repressed by SIRT1 and that AR associates with SIRT1 led us to examine further the basis for these phenomena. The AR coactivator p300 enhanced basal and DHT-induced activities of the PSA promoter, which was repressed 40% to 60% by SIRT1 compared with the control vector (Fig. 3A, bars 7 and 8 versus 9 and 10 from left). SIRT1 repression of AR-dependent PSA promoter activity was reversed by the H363Y mutation of the SIRT1 catalytic domain (Fig. 3A, bars 11 and 12 from left), showing the requirement for SIRT1 enzyme activity in repressing the AR transactivation function. The role of SIRT1 in regulating the AR intramolecular interaction was next assessed by using a mammalian two-hybrid approach in which the AR N terminus is linked as a chimeric fusion to Vp16 and the AR C terminus is linked to the heterologous Gal4 DNA-binding domain. Transient-expression studies were conducted using a luciferase reporter gene linked to multimeric DNA sequences that bind to the Gal4 DNA-binding domain (Fig. 3B). Consistent with previous studies (28), the AR N- and AR C-terminal interaction was augmented by p300 in the DHT ligand-induced state. Coexpression of hSIRT1 repressed the DHT-induced AR intramolecular interaction (Fig. 3B).
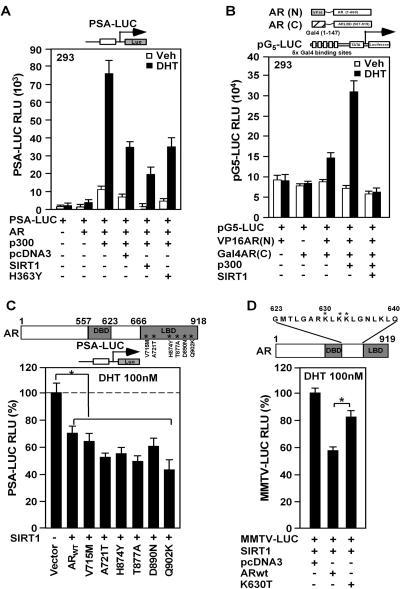
FIG. 3.
SIRT1 represses p300-mediated AR intramolecular interaction. (A) HEK293 cells were transfected with the androgen-responsive reporter gene (PSA) linked to the luciferase reporter and expression vectors for p300, SIRT1, or the catalytic dead SIRT1 mutant (SIRT1 H363Y). (B) The interaction between the AR N-terminal and AR C-terminal domains is shown as mean luciferase activity assessed by the heterologous reporter (pG5-LUC). (C and D) Activity of the PSA-LUC reporter was assessed with wild-type AR (ARWT) or point mutations found in patients with prostate cancer within the carboxyl terminus (C) or within the AR hinge region (K630T) (D). The effect of SIRT1 on DHT-induced AR activity is shown compared with the control vector. Significant differences (P < 0.05) are indicated by an asterisk. The data are mean plus standard error of the mean of 12 separate transfections.
The AR acetylation site is involved in SIRT1-mediated repression.
Point mutations arising within the AR in patients with prostate cancer may contribute to therapeutic resistance. The effect of SIRT1 to inhibit DHT-induced PSA-LUC activity was established at approximately 40% (Fig. 3C). Comparison was made between the AR wild type and the AR mutants found in a subset of patients with prostate cancer. The DHT-induced activity of each mutant was established as 100%, and the effect of SIRT1 on the DHT-induced activity of each mutant was determined. The DHT-induced activity of each of the carboxyl-terminal AR mutants was repressed 40% to 60% by SIRT1 expression (Fig. 3C). In contrast, the point mutant of the lysine residue in the AR acetylation motif found in prostate cancer, K630T, was resistant to SIRT1-mediated repression (Fig. 3D), suggesting that SIRT1 repression of liganded AR activity may involve the AR acetylation site.
SIRT1 inhibits AR-expressing prostate cancer cell growth.
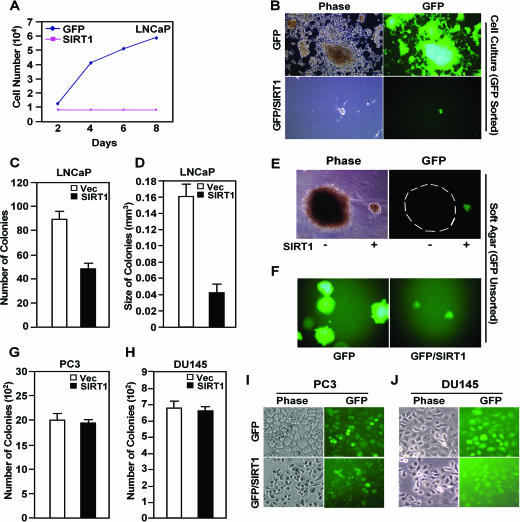
The basal and DHT-induced growth of the prostate cancer cell line LNCaP is dependent upon AR activity. The LNCaP cell line AR mutant (T877A) was sensitive to SIRT1 repression (Fig. 3C). LNCaP cell growth in tissue culture was inhibited >90% by SIRT1 expression delivered through retroviral transduction compared with the GFP vector control (Fig. 4A and B). LNCaP cells transduced with MSCV-SIRT1 (IRES-GFP) were next examined for anchorage-independent growth in semisolid agar medium. LNCaP/SIRT1 cells formed fewer and smaller colonies than the control vector cells (Fig. 4C to F). These results suggest that SIRT1 inhibits the tumorigenic phenotype of prostate cancer cells, blocking both cellular proliferation and contact-independent growth in soft agar. In order to determine whether the inhibition of prostate cancer cellular proliferation was selective to AR-expressing prostate cancer cell lines, we examined the growth properties of the PC3 and DU145 cell lines, which do not express the AR but grow in soft agar. Retroviral transduction of PC3 and DU145 cells with SIRT1-IRES-GFP demonstrated uniform transduction of the cell lines; however, SIRT1 expression did not affect colony formation (Fig. 4G to H).
FIG. 4.
SIRT1 inhibits prostate cancer cellular colony formation. (A and B) LNCaP cells were infected with MSCV-SIRT1-IRES-GFP (SIRT1) or control vector MSCV-IRES-GFP (Vec). GFP-positive cells were sorted by FACS and were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 100 nM DHT. (A) Growth curve of the LNCaP cells. (B) LNCaP cells transduced with either MSCV-SIRT1-IRES-GFP or with vector control. Phase contrast and green fluorescence from GFP are shown. (C to F) Colony formation assay of LNCaP cells transduced with MSCV-SIRT1-IRES-GFP or MSCV-IRES-GFP. The number (C) or size (D) of the colonies is shown. (E) Colonies of LNCaP cells (GFP negative) or LNCaP cells transduced with MSCV-SIRT1-IRES-GFP (green) are shown. (F) GFP-positive colonies of LNCaP cells transduced with MSCV-GFP vector or MSCV-SIRT1 are shown. (G to J) Colony formation assay of PC3 (G) and DU145 (H) cells transduced with MSCV-SIRT1-IRES-GFP or MSCV-IRES-GFP. The number of the colonies is shown. Phase contrast and green fluorescence from GFP in PC3 (I) and DU145 (J) cells are shown. The error bars indicate standard deviations.
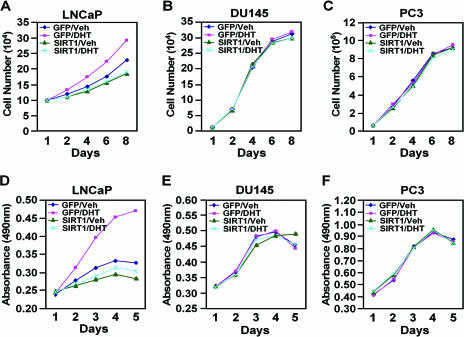
In order to examine further the role of SIRT1 in regulating prostate cancer cellular growth, either AR-expressing (LNCaP) or AR-deficient (PC3 and DU145) prostate cancer cell lines were transduced with retrovirus expressing SIRT1, and growth curves were conducted. DHT-induced cellular proliferation was abrogated by SIRT1 expression in LNCaP cells, as assessed by either cell numbers or MTT assay (Fig. 5A and D). In contrast, the proliferation rates of the AR-deficient cells, PC3 and DU145, were unaffected by the addition of DHT, and their cellular proliferation rates were unaffected by SIRT1 expression (Fig. 5B, E, C, and F). Collectively, these studies demonstrate that SIRT1 inhibits DHT-induced cellular proliferation in AR-expressing prostate cancer cells.
FIG. 5.
SIRT1 inhibits AR-dependent cellular proliferation. Cellular proliferation was assessed either by counting cells (A, B, and C) or through the MTT assay (D, E, and F). LNCaP, DU145, or PC3 cells were transduced with either MSCV-SIRT1-IRES-GFP or control vector (MSCV-IRES-GFP) and treated with DHT (100 nM) or vehicle (Veh), and monolayer proliferation was assessed daily for 8 days. DHT-induced growth of LNCaP cells that express the AR is inhibited by SIRT1 expression.
Deacetylation of AR by SIRT1 in vitro.
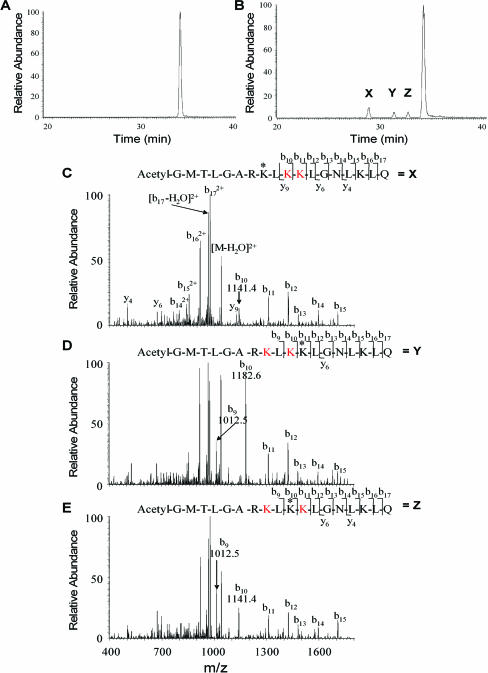
p300 augmentation of the AR intramolecular interaction between the amino and carboxyl termini was inhibited by SIRT1. Several lines of evidence suggested a role for the AR acetylation site in SIRT1 signaling. First, the physical association of the AR with p300 was dependent upon the AR acetylation site (17, 18). Second, acetylation of the AR was regulated by the SIRT1-specific inhibitor sirtinol. Third, point mutation at the AR acetylation site (K630T) resulted in resistance to SIRT1 repression. We therefore investigated the possibility that the AR could serve as a direct substrate for the SIRT1 NAD-dependent deacetylase. To determine whether hSIRT1 protein could deacetylate human AR in vitro, we used the mouse orthologue, mSir2a, hSIRT1, and two other enzymes, S. cerevisiae Sir2p and Archaeoglobus fulgidus Sir2af2 (Fig. 6 and 7) (see supplement 3 posted at http://www.pestelllab.org/publications/2006/fu&liu_ar_sirt1/supplement_3.pdf). These enzymes were examined for deacetylation activity against the synthetic peptide substrate comprised of residues 623 to 640 of the human AR protein, in which the lysine residues 630, 632, and 633 are acetylated. The corresponding lysine residues in the human AR are known to be acetylated by p300 and P/CAF or TIP60 (16, 20). LC/MS analysis of the deacetylation reaction mixtures showed that the archaeal and bacterial enzymes formed both monodeacetylated and dideacetylated products (see supplement 3A to C posted at http://www.pestelllab.org/publications/2006/fu&liu_ar_sirt1/supplement_3.pdf). The identities of peptide species were determined from correspondence to typical mass signals (see supplement 3D to F posted at http://www.pestelllab.org/publications/2006/fu&liu_ar_sirt1/supplement_3.pdf) for double-charged species corresponding to the triacetylated, diacetylated, and monoacetylated forms of the peptide.
FIG. 6.
Sir2a deacetylation of the androgen receptor. (A and B) LC/MS chromatograms of deacetylation reaction mixtures containing the trietracetyl lysine-containing peptide Ac-GMTLGAR(KAc)L(KAc)(KAc)LGNLKLQ homologous to the AR acetylation sequence. LC/MS peaks are labeled according to the dominant mass observed and are tri, di, and mono to indicate loss of zero, one, and two acetyl residues from the eluted peptide, as determined by mass measurement. (A) LC/MS of the AR peptide without added enzyme. (B) LC/MS of the peptide after incubation with mouse Sir2a, showing the production of three monodeacetylated species, X, Y, and Z. (C) MS/MS of species X identifying the site of deacetylation, as indicated by the asterisk, for the peak labeled X in the chromatogram. (D) MS/MS of species Y identifying the site of deacetylation, as indicated by the asterisk, for the peak labeled Y in the chromatogram. (E) MS/MS of species Z identifying the site of deacetylation, as indicated by the asterisk, for the peak labeled Z in the chromatogram. The “K” in red indicates acetylated lysine.
FIG. 7.
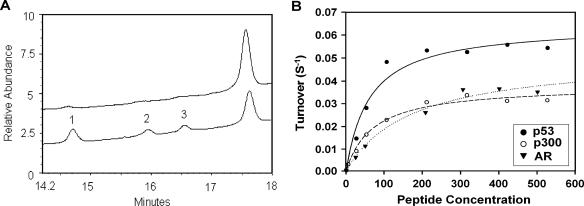
Deacetylation selectivity of human SIRT1. (A) HPLC chromatograms of AR peptide deacetylation with human SIRT1 enzyme. Unreacted triacetylated AR peptide (top chromatogram) and three monodeacetylated products (peaks 1 to 3) (bottom chromatogram) are shown. The areas of peak 1:peak 2:peak 3 are 2.31:1.13:1. (B) The deacetylation rate of SIRT1 enzyme as a function of peptide concentration (p53, p300, and AR peptide). The lines are best fits to the Michaelis-Menten equation and determined the following values for Vmax and Km: p53, Vmax = 0.064 s−1, Km = 61 μM; p300, Vmax = 0.038 s−1, Km = 71 μM; and AR, Vmax = 0.05 s−1, Km = 174 μM.
For the mouse enzyme, the reaction was terminated to produce only monodeacetylated peptides. Thus, three new LC/MS peaks were observed in injected reaction mixtures compared to the unreacted control (see supplement 3G and H posted at http://www.pestelllab.org/publications/2006/fu&liu_ar_sirt1/supplement_3.pdf). All three monodeacetylated peptides were identified by their parent mass, confirming that mouse Sir2a deacetylates all three acetyl lysines in the sequence. MS/MS fragmentation analysis identified the products (Fig. 6). As shown in Fig. 6C to E, the first peak (X) eluting in Fig. 6B is the deacetylation product corresponding to loss of the acetyl group from lysine 630 in AR. Peaks Y and Z were determined to be products arising from removal of acetate from lysines 632 and 633 in the full-length AR sequence, respectively. Integrations of the LC/MS chromatograms showed that peak X was twice peaks Y and Z (Fig. 6B). Based on the peak areas, the preferred deacetylation site in the peptide sequence corresponded to lysine 630 in the full-length sequence, with roughly equal sequence specificities observed for deacetylation at the other two acetyl lysines (K632 and K633).
To determine the deacetylation selectivity of human SIRT1 on triacetylated AR peptide, the AR peptide (5 μl; 100 μM) and hSIRT1 enzyme (2 μl; 3.85 μM) were incubated (Fig. 7A). The preferred deacetylation site was that of acetyl lysine corresponding to K630 in the full-length AR sequence. The preferred deacetylation was twofold over the other two acetyl lysine groups in the AR sequence and is consistent with the results obtained for deacetylation of the same sequence with mouse Sir2a (mouse SIRT1). Comparison of AR reactivities to p53 and p300 sequences established that all react at a maximum rate of metabolism (Vmax) that is within a factor of 2 and vary in Km value within a range of 3 (Fig. 7B). (The lines of best fit to the Michaelis-Menten equation determined the following values for Vmax and Km: p53, Vmax = 0.064 s−1, Km = 61 μM; p300, Vmax = 0.038 s−1, Km = 71 μM; and AR, Vmax = 0.05 s−1, Km = 174 μM.) This established that AR is similar in reactivity to previously published target sequences of human SIRT1 (31).
Homology model of hSIRT1 and AR “KLKK” peptide complex.
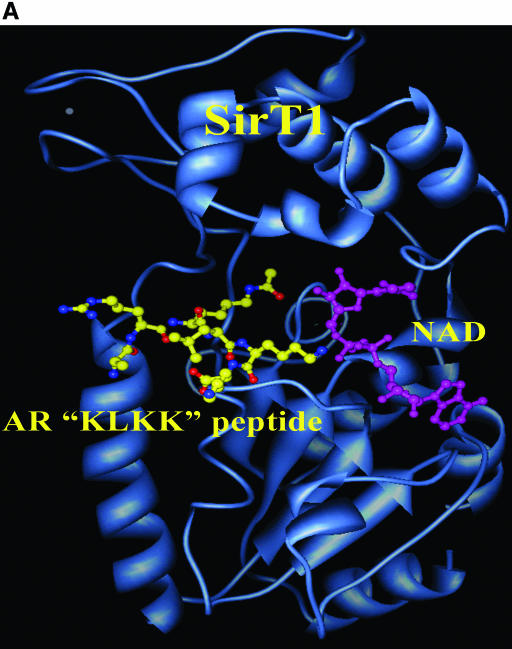
The final model of the minimized structures of hSirT1 and the AR “KLKK” peptide-NAD complex is shown in Fig. 8A as a ribbon model generated using the molecular display program CHIMERA from the University of San Francisco, San Francisco, CA (43). The “KLKK” peptide and the NAD molecule are shown as a “ball-and-stick” model. The acetyl lysine (K630) packs itself in the vicinity of critical residues H363 (catalytic histidine) and F309, and the acetate group is in close proximity to the ribosyl moiety of NAD. The hydrophobic part of the lysine K630 side chain packs favorably against the aromatic ring in the side chain of F309. The extended backbone conformation of the bound AR peptide to hSirT1 and additional favorable electrostatic interactions between the positively charged side chain of K632 and the negatively charged 5′-phosphate group of the adenine nucleotide part (in NAD) could possibly explain the preferential deacetylation of K630 in comparison with K632 and K633.
FIG. 8.
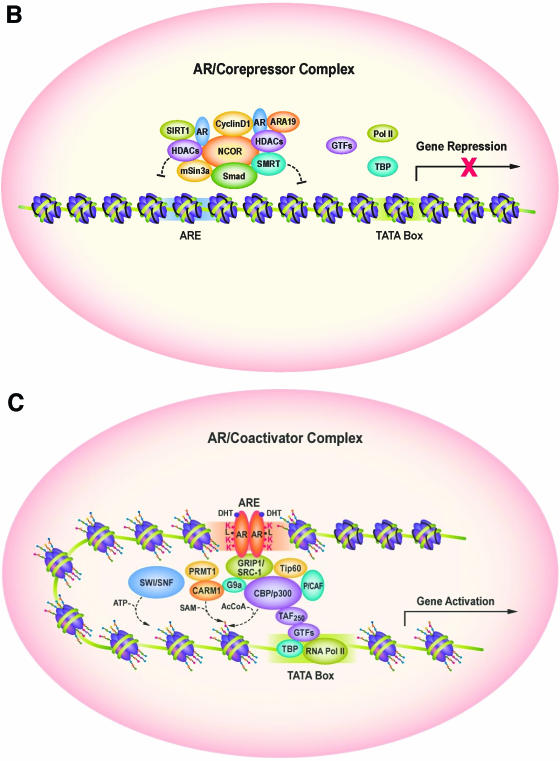
SIRT1 binds and deacetylates the AR. (A) Homology model of AR “KLKK” peptide with NAD bound SIRT1. The structures of the hSirT1 and AR “KLKK” peptide-NAD complex are shown as a ribbon model generated using the molecular display program CHIMERA. The “KLKK” peptide (yellow) and the NAD molecule (cyan) are shown as a “ball-and-stick” model. (B) Schematic representation of AR bound to corepressors in the absence of ligand in the context of local chromatin, dependent in part upon the AR lysine motif. SIRT1 inhibits AR activity. SIRT1 is shown to associate with the NCoR/HDAC repression complex. (C) Upon the addition of ligand (DHT), disengagement of the corepressor complex and recruitment of the coactivator complex engages gene expression, dependent in part upon the AR lysine motif.
DISCUSSION
The current study provides the first evidence for an AR regulatory pathway controlled by mammalian SIRT1. Endogenous androgen-regulated gene expression was induced by the SIRT1 inhibitor nicotinamide. hSIRT1 colocalized with the AR by confocal microscopy and physically associated with the AR in vitro and in vivo. AR function was repressed by hSIRT1, requiring the NAD-dependent enzymatic activity of hSIRT1. SIRT1 inhibited androgen-dependent gene expression in a ligand-dependent, AR and DNA sequence-specific manner. Acetylation of the AR at a conserved motif in the AR hinge region enhanced the physical association between the AR and coactivators in vitro and in cultured cells (17, 18). SIRT1 regulated the acetylation levels of the AR in LNCaP cells, and an 18-amino-acid peptide encoding the AR hinge region acetylation site was sufficient to interact with SIRT1. SIRT1 deacetylated the AR with Km and Kcat values similar to those of p53 and p300. SIRT1 expression inhibited contact-independent cellular growth of AR-expressing prostate cancer cells. Thus, the NAD-dependent histone deacetylase SIRT1 represses androgen receptor signaling and function.
hSIRT1 inhibited AR-induced reporter gene activity. Repression of AR activity was abrogated by point mutation of the acetylated AR lysine resides (ARK63OT). hSIRT1 inhibition of AR activity involved the deacetylation function of SIRT1. hSIRT1 inhibited DHT-induced AR activity in a DNA sequence-dependent manner, repressing expression of AR and activities of the PSA and MMTV promoters. An ARE site was sufficient for hSIRT1 repression. Point mutation of the hSIRT1 core domain histidine residue (H363Y), which abolishes the deacetylase activity of SIRT1, abrogated the repression of DHT-induced androgen signaling. The conserved lysine motifs of the AR, which are acetylated by p300 and other histone acetylases, were deacetylated by Sir2 in vitro. The murine orthologue of hSirT1, mSir2α, deacetylated an 18-mer peptide containing the three acetyl lysines (630, 632, and 633) corresponding to the native AR sequence. Enzymes derived from Archaea (Archaeoglobus fulgius AF2SIR2) and yeast (Saccharomyces cerevisiae SIRT) sources, closely related to hSir2, also deacetylated these residues. The deacetylation of the AR at 0.5 nM NAD+ and 202 nM SIRT1 enzyme demonstrated a Km value of 174 μM and a Kcat of 0.0395−1. These association and catalysis constants are comparable to those obtained for p300 and p53 peptide substrates.
The current findings extend previous observations that both histones and nonhistone proteins are targets for deacetylation by hSIRT1 proteins (57) and establish a role for SIRT1 in regulating cellular growth through the AR. SIRT1 inhibited DHT-induced prostate cancer cellular proliferation and contact-independent cellular growth. The endogenous AR was required for both DHT-induced proliferation and the effect of SIRT1. The ability of SIRT1 to deacetylate AR and repress AR activity mechanistically resembles the deacetylation of p53, RelA/p65, and FOXO but contrasts with the effects of SIRT1 on MyoD. SIRT1 directly bound and deacetylated the AR, whereas the effect of Sir2 on MyoD is indirect, as Sir2 deacetylates P/CAF (21) to reduce MyoD function (21). SIRT1 bound the AR both in vitro and in vivo in cultured prostate cancer cells and repressed and deacetylated the AR directly. The repression of AR coactivator proteins (P/CAF and p300) by SIRT1 (9, 21) may also contribute to repression of AR activity. When coexpressed in cultured cells, p300 and the p160 coactivator proteins are recruited to AREs in the context of local chromatin. In addition, arginine methyltransferases form part of the complex that coordinates cyclical recruitment of AR nuclear receptor complexes (33, 39). It is known that the AR-acetylated lysine residues serve as key docking sites for the association with p300 versus NCoR-HDAC-Smad3 complexes of the AR (18, 19) (Fig. 8B and C). The role that SIRT1-dependent deacetylation plays in coordinating recruitment of specific complexes to the AR remains to be determined.
The current study provides evidence for functional antagonism between SIRT1 and AR activities at the site acetylated by p300. The acetylated motif residues of the AR are predicted to reside in proximity to the superior aspect of the AR hydrophobic pocket. Previous studies have suggested functional antagonism between p300 and histone deacetylase. In Caenorhabditis elegans, inactivation of the cbp1 gene, an orthologue of p300/CBP, causes developmental arrest of C. elegans embryos except for neuronal differentiation. Coinactivation of genes encoding components of the HDAC complex, such as hda-1, rba-1, and rba-2, which antagonize HAT activity, rescues some of the cbp1 null phenotype (52), suggesting that CBP specifies differentiation by functioning along with the early cell fate-determining transcription factors, and the conserved histone deacetylase complex blocks CBP-mediated cellular differentiation by antagonizing the functions of CBP (52). SIRT1-dependent repression of AR activity was reduced by a point mutation (K630T) found in prostate cancer cells at the lysine residue acetylated by p300 but was not affected by mutations within the carboxyl terminus of AR. Here, SIRT1 repressed the ligand-augmented AR amino- and carboxyl-terminal intramolecular interaction. The N- and C-terminal interaction within the AR is enhanced by CBP/p300 (28). The ligand-binding domain of the AR recruits its AF-1 function via an N-terminal FXXXLF motif. The hydrophobic cleft of the AR ligand-binding domain created by ligand binding in the C-terminal domain interacts strongly with the AR N-terminal motif. As p300 acetylates the AR, the independent role of p300 repression by SIRT1 versus the direct repression of AR by SIRT1 cannot be dissociated in p300−/− cells or using small interfering RNA.
What might be the significance of the finding that AR expression and DHT signaling are regulated by nicotinamide and NAD-dependent deacetylation? The requirement for NAD in Sir2 enzymatic activity has led to suggestions that Sir2 activity may be regulated by the intracellular concentration of NAD, by the NAD/NADH ratio, or by the intracellular concentration of nicotinamide (2, 35, 36, 45). Endogenous levels of nicotinamide may limit Sir2 activity (49), suggesting that the concentration of nicotinamide in response to physiological changes could affect Sir2 function. Metabolic changes in muscle induced by pyruvate, for example, increase the NAD+/NADH ratio and inhibit muscle gene expression, whereas lactate reduces the NAD/NADH ratio and stimulates muscle gene expression (21). Androgens maintain male muscle mass (10a, 26, 50) and induce muscle cellular gene expression. During prostate cancer progression, metabolism shifts toward cytosolic glycolysis (1, 12). The increased production of lactate that occurs during prostate cancer progression (5, 46) is predicted to inhibit SIRT1 and thereby enhance AR function. A recent study also suggested that global histone modification in prostate tumor tissues, including acetylation of H3K18 and H4 K12, dimethylation of H3K4 and H4R3, and acetylation of H3K9, a target of SirT1 (54), predict a risk of prostate cancer recurrence (51).
Here, SIRT1 inhibited androgen-dependent prostate cancer cellular growth and repressed the endogenous androgen-responsive target gene, AR. AR is an androgen-responsive gene, and sirtinol, a Sir2-specific inhibitor, increased the abundance of acetylated-AR acetylation. Nicotinamide, a noncompetitive inhibitor of Sir2, induced expression of the endogenous androgen-responsive AR gene. The selective chemical inhibitor of SIRT activity, splitomycin, enhanced AR-dependent gene expression, suggesting that endogenous SirT1 contributes to maintenance of the AR in a repressed state. The androgen receptor's function is a critical determinant of human prostate cancer pathogenesis and progression (11). Androgen ablation therapy remains a major therapeutic intervention in metastatic disease. Reexpression of androgen-responsive genes is a characteristic of progression of human prostate cancer, as monitored clinically by the androgen-regulated PSA gene. Although AR gene amplification or mutation contributes to reexpression of androgen-responsive genes in a subset of patients, in the majority of patients, the mechanism remains poorly defined. Multiple mechanisms, including AR overexpression and mutations, activation of growth factor signaling, and altered cointegrator expression and function, have been implicated in therapy resistance (13). Increased AR activity through loss of AR corepressors has been implicated in prostate cancer growth, and acetylation mimic mutants of the AR promote prostate cancer cellular growth (17, 18). The current study provides evidence for an important role for SIRT1 as a regulator of AR expression and function.
Acknowledgments
We thank Rajendram Rajnarayanan for help with homology modeling and S. Balk, J. J. Palvimo, and S. Chang for plasmids. We thank D. Scardino for assistance with manuscript preparation and Nanita Barchi for the preparation of schematic illustrations of AR-coregulator complexes.
Support for these studies was received from NIH R01CA70896, R01CA75503, R01CA86072, R01CA93596, R01CA107382, and the Kimmel Cancer Center Support Grant P30 CA56036 (R.G.P.); from the Breast Cancer Alliance through an Exceptional Project Grant (A.Q.); and from 1 R21DK065220-01 (NIDDK) (to M.F.). This project is supported in part by the Dr. Ralph and Marian C. Falk Medical Research Trust and a grant from the Pennsylvania Department of Health.
The Department of Health specifically disclaims responsibility for any analysis, interpretations, or conclusions.
Footnotes
Published ahead of print on 21 August 2006.
REFERENCES
- 1.Altenberg, B., and K. O. Greulich. 2004. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics 84:1014-1020. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R. M., K. J. Bitterman, J. G. Wood, O. Medvedik, and D. A. Sinclair. 2003. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423:181-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avalos, J. L., I. Celic, S. Muhammad, M. S. Cosgrove, J. D. Boeke, and C. Wolberger. 2002. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol. Cell 10:523-535. [DOI] [PubMed] [Google Scholar]
- 4.Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243-1254. [DOI] [PubMed] [Google Scholar]
- 5.Baron, A., T. Migita, D. Tang, and M. Loda. 2004. Fatty acid synthase: a metabolic oncogene in prostate cancer? J. Cell Biochem. 91:47-53. [DOI] [PubMed] [Google Scholar]
- 6.Berman, H. M., T. Battistuz, T. N. Bhat, W. F. Bluhm, P. E. Bourne, K. Burkhardt, Z. Feng, G. L. Gilliland, L. Iype, S. Jain, P. Fagan, J. Marvin, D. Padilla, V. Ravichandran, B. Schneider, N. Thanki, H. Weissig, J. D. Westbrook, and C. Zardecki. 2002. The Protein Data Bank. Acta Crystallogr. D 58:899-907. [DOI] [PubMed] [Google Scholar]
- 7.Bevan, C. L., S. Hoare, F. Claessens, D. M. Heery, and M. G. Parker. 1999. The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol. Cell. Biol. 19:8383-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blander, G., and L. Guarente. 2004. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73:417-435. [DOI] [PubMed] [Google Scholar]
- 9.Bouras, T., M. Fu, A. A. Sauve, F. Wang, A. A. Quong, N. D. Perkins, R. T. Hay, W. Gu, and R. G. Pestell. 2005. SIRT1 deacetylation and repression of P300 involves lysine residues 1020/1024 within the cell-cycle regulatory domain 1. J. Biol. Chem. 280:10264-10276. [DOI] [PubMed] [Google Scholar]
- 10.Brunet, A., L. B. Sweeney, J. F. Sturgill, K. F. Chua, P. L. Greer, Y. Lin, H. Tran, S. E. Ross, R. Mostoslavsky, H. Y. Cohen, L. S. Hu, H. L. Cheng, M. P. Jedrychowski, S. P. Gygi, D. A. Sinclair, F. W. Alt, and M. E. Greenberg. 2004. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303:2011-2015. [DOI] [PubMed] [Google Scholar]
- 10a.Catlin, D. 1995. Anabolic steroids, p. 2362-2369. In L. J. De Groot (ed.), Endocrinology, 3rd ed. W. B. Saunders and Co., Philadelphia, Pa.
- 11.Chen, C. D., D. S. Welsbie, C. Tran, S. H. Baek, R. Chen, R. Vessella, M. G. Rosenfeld, and C. L. Sawyers. 2004. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 10:33-39. [DOI] [PubMed] [Google Scholar]
- 12.Chowdhury, S. K., A. Gemin, and G. Singh. 2005. High activity of mitochondrial glycerophosphate dehydrogenase and glycerophosphate-dependent ROS production in prostate cancer cell lines. Biochem. Biophys. Res. Commun. 333:1139-1145. [DOI] [PubMed] [Google Scholar]
- 13.Edwards, J., and J. M. Bartlett. 2005. The androgen receptor and signal-transduction pathways in hormone-refractory prostate cancer. Part 2: Androgen-receptor cofactors and bypass pathways. BJU Int. 95:1327-1335. [DOI] [PubMed] [Google Scholar]
- 14.Fiser, A., and A. Sali. 2003. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 374:461-491. [DOI] [PubMed] [Google Scholar]
- 15.Frye, R. A. 2000. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273:793-798. [DOI] [PubMed] [Google Scholar]
- 16.Fu, M., M. Rao, K. Wu, C. Wang, X. Zhang, M. Hessien, Y. G. Yeung, D. Gioeli, M. J. Weber, and R. G. Pestell. 2004. The androgen receptor acetylation site regulates cAMP and AKT but not ERK-induced activity. J. Biol. Chem. 279:29436-29449. [DOI] [PubMed] [Google Scholar]
- 17.Fu, M., C. Wang, A. T. Reutens, R. Angeletti, L. Siconolfi-Baez, V. Ogryzko, M. L. Avantaggiati, and R. G. Pestell. 2000. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J. Biol. Chem. 275:20853-20860. [DOI] [PubMed] [Google Scholar]
- 18.Fu, M., C. Wang, J. Wang, T. Sakamaki, D. Di Vizio, X. Zhang, C. Albanese, S. Balk, C. Chang, S. Fan, E. Rosen, J. J. Palvimo, O. A. Janne, S. Muratoglu, M. Avantaggiati, and R. G. Pestell. 2003. Acetylation of the androgen receptor enhances coactivator binding and promotes prostate cancer cell growth. Mol. Cell. Biol. 23:8563-8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu, M., C. Wang, J. Wang, T. Sakamaki, X. Zhang, Y.-G. Yeung, C. Chang, T. Hopp, S. A. W. Fuqua, E. Jaffray, R. T. Hay, J. J. Palvimo, O. A. Jänne, and R. G. Pestell. 2002. The androgen receptor acetylation governs transactivation and MEKK1-induced apoptosis without affecting in vitro sumoylation and transrepression function. Mol. Cell. Biol. 22:3373-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu, M., C. Wang, J. Wang, B. Zafonte, M. P. Lisanti, and R. G. Pestell. 2002. Acetylation in hormone signaling and the cell-cycle. Cytokine Growth Factor Rev. 13:259-276. [DOI] [PubMed] [Google Scholar]
- 21.Fulco, M., R. L. Schiltz, S. Iezzi, M. T. King, P. Zhao, Y. Kashiwaya, E. Hoffman, R. L. Veech, and V. Sartorelli. 2003. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell 12:51-62. [DOI] [PubMed] [Google Scholar]
- 22.Gasser, S. M., and M. M. Cockell. 2001. The molecular biology of the SIR proteins. Gene 279:1-16. [DOI] [PubMed] [Google Scholar]
- 23.Gingras, S., R. Moriggl, B. Groner, and J. Simard. 1999. Induction of 3β-hydroxysteroid dehydrogenase/δ5-δ4 isomerase type 1 gene transcription in human breast cancer cell lines and in normal mammary epithelial cells by interleukin-4 and interleukin-13. Mol. Endocrinol. 13:66-81. [DOI] [PubMed] [Google Scholar]
- 24.Gleason, D. F. 1966. Classification of prostatic carcinomas. Cancer Chemother. Rep. 50:125-128. [PubMed] [Google Scholar]
- 25.Gong, J., J. Zhu, O. B. Goodman, R. G. Pestell, P. N. Schlegel, D. M. Nanus, and R. Shen. 2006. Activation of p300 histone acetyltransferase activity and acetylation of the androgen receptor by bombesin in prostate cancer cells. Oncogene 25:2011-2021. [DOI] [PubMed] [Google Scholar]
- 26.Grinspoon, S., C. Corcoran, K. Lee, B. Burrows, J. Hubbard, L. Katznelson, M. Walsh, A. Guccione, J. Cannan, H. Heller, N. Basgoz, and A. Klibanski. 1996. Loss of lean body and muscle mass correlates with androgen levels in hypogonadal men with acquired immunodeficiency syndrome and wasting. J. Clin. Endocrinol. Metab. 81:4051-4058. [DOI] [PubMed] [Google Scholar]
- 27.Reference deleted.
- 28.Ikonen, T., J. J. Palvimo, and O. A. Janne. 1997. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J. Biol. Chem. 272:29821-29828. [DOI] [PubMed] [Google Scholar]
- 29.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 30.Jemal, A., T. Murray, A. Samuels, A. Ghafoor, E. Ward, and M. J. Thun. 2003. Cancer statistics, 2003. CA Cancer J. Clin. 53:5-26. [DOI] [PubMed] [Google Scholar]
- 31.Kaeberlein, M., T. McDonagh, B. Heltweg, J. Hixon, E. A. Westman, S. D. Caldwell, A. Napper, R. Curtis, P. S. DiStefano, S. Fields, A. Bedalov, and B. K. Kennedy. 2005. Substrate-specific activation of sirtuins by resveratrol. J. Biol. Chem. 280:17038-17045. [DOI] [PubMed] [Google Scholar]
- 32.Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins, L. Pillus, and R. Stemglanz. 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97:5807-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, D. Y., C. Teyssier, B. D. Strahl, and M. R. Stallcup. 2005. Role of protein methylation in regulation of transcription. Endocr. Rev. 26:147-170. [DOI] [PubMed] [Google Scholar]
- 34.Li, Z., C. Wang, X. Jiao, Y. Lu, M. Fu, A. A. Quong, C. Dye, J. Yang, M. Dai, X. Ju, X. Zhang, A. Li, P. Burbelo, E. R. Stanley, and R. G. Pestell. 2006. Cyclin D1 regulates cellular migration through the inhibition of thrombospondin 1 and ROCK signaling. Mol. Cell. Biol. 26:4240-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, S. J., P. A. Defossez, and L. Guarente. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289:2126-2128. [DOI] [PubMed] [Google Scholar]
- 36.Lin, S. J., E. Ford, M. Haigis, G. Liszt, and L. Guarente. 2004. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 18:12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo, J., A. Y. Nikolaev, S. Imai, D. Chen, F. Su, A. Shiloh, L. Guarente, and W. Gu. 2001. Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107:137-148. [DOI] [PubMed] [Google Scholar]
- 38.Matsumura, I., T. Kitamura, H. Wakao, H. Tanaka, K. Hashimoto, C. Albanese, J. Downward, R. G. Pestell, and Y. Kanakura. 1999. Transcriptional regulation of cyclin D1 promoter by STAT5: its involvement in cytokine-dependent growth of hematopoietic cells. EMBO J. 18:1367-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metzger, E., M. Wissmann, N. Yin, J. M. Muller, R. Schneider, A. H. Peters, T. Gunther, R. Buettner, and R. Schule. 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437:436-439. [DOI] [PubMed] [Google Scholar]
- 40.Min, J., J. Landry, R. Sternglanz, and R. M. Xu. 2001. Crystal structure of a SIR2 homolog-NAD complex. Cell 105:269-279. [DOI] [PubMed] [Google Scholar]
- 41.Motta, M. C., N. Divecha, M. Lemieux, C. Kamel, D. Chen, W. Gu, Y. Bultsma, M. McBurney, and L. Guarente. 2004. Mammalian SIRT1 represses forkhead transcription factors. Cell 116:551-563. [DOI] [PubMed] [Google Scholar]
- 42.Muth, V., S. Nadaud, I. Grummt, and R. Voit. 2001. Acetylation of TAF168, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J. 20:1353-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 44.Reutens, A. T., M. Fu, G. Watanabe, C. Albanese, M. J. McPhaul, S. P. Balk, O. A. Janne, J. J. Palvimo, and R. G. Pestell. 2001. Cyclin D1 binds the androgen receptor and regulates hormone-dependent signaling in a p300/CBP-associated factor (P/CAF)-dependent manner. Mol. Endocrinol. 15:797-811. [DOI] [PubMed] [Google Scholar]
- 45.Revollo, J. R., A. A. Grimm, and S. Imai. 2004. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 279:50754-50763. [DOI] [PubMed] [Google Scholar]
- 46.Rossi, S., E. Graner, P. Febbo, L. Weinstein, N. Bhattacharya, T. Onody, G. Bubley, S. Balk, and M. Loda. 2003. Fatty acid synthase expression defines distinct molecular signatures in prostate cancer. Mol. Cancer Res. 1:707-715. [PubMed] [Google Scholar]
- 47.Sathya, G., C. Y. Chang, D. Kazmin, C. E. Cook, and D. P. McDonnell. 2003. Pharmacological uncoupling of androgen receptor-mediated prostate cancer cell proliferation and prostate-specific antigen secretion. Cancer Res. 63:8029-8036. [PubMed] [Google Scholar]
- 48.Sauve, A. A., I. Celic, J. Avalos, H. Deng, J. D. Boeke, and V. L. Schramm. 2001. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochem. 40:15456-15463. [DOI] [PubMed] [Google Scholar]
- 49.Sauve, A. A., R. D. Moir, V. L. Schramm, and I. M. Willis. 2005. Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition. Mol. Cell 17:595-601. [DOI] [PubMed] [Google Scholar]
- 50.Schroeder, E. T., M. Terk, and F. R. Sattler. 2003. Androgen therapy improves muscle mass and strength but not muscle quality: results from two studies. Am. J. Physiol. Endocrinol. Metab. 285:E16-E24. [DOI] [PubMed] [Google Scholar]
- 51.Seligson, D. B., S. Horvath, T. Shi, H. Yu, S. Tse, M. Grunstein, and S. K. Kurdistani. 2005. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 435:1262-1266. [DOI] [PubMed] [Google Scholar]
- 52.Shi, Y., and C. Mello. 1998. A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev. 12:943-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, et al. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaquero, A., M. Scher, D. Lee, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2004. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell 16:93-105. [DOI] [PubMed] [Google Scholar]
- 55.Vaziri, H., S. K. Dessain, E. Ng-Eaton, S. I. Imai, R. A. Frye, T. K. Pandita, L. Guarente, and R. A. Weinberg. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149-159. [DOI] [PubMed] [Google Scholar]
- 56.Wang, L., C. L. Hsu, and C. Chang. 2005. Androgen receptor corepressors: an overview. Prostate 63:117-130. [DOI] [PubMed] [Google Scholar]
- 57.Yang, T., M. Fu, R. Pestell, and A. A. Sauve. 2006. SIRT1 and endocrine signaling. Trends Endocrinol Metab. 17:186-191. [DOI] [PubMed] [Google Scholar]
- 58.Zhao, K., X. Chai, and R. Marmorstein. 2004. Structure and substrate binding properties of CobB, a Sir2 homolog protein deacetylase from Escherichia coli. J. Mol. Biol. 337:731-741. [DOI] [PubMed] [Google Scholar]