Abstract
C-terminal binding proteins (CtBPs) are multifunctional proteins that can mediate gene repression. CtBPs contain a cleft that binds Pro-X-Asp-Leu-Ser (PXDLS) motifs. PXDLS motifs occur in numerous transcription factors and in effectors of gene repression, such as certain histone deacetylases. CtBPs have been depicted as bridging proteins that self-associate and link PXDLS-containing transcription factors to PXDLS-containing chromatin-modifying enzymes. CtBPs also recruit effectors that do not contain recognizable PXDLS motifs. We have investigated the importance of the PXDLS binding cleft to CtBP's interactions with various partner proteins and to its ability to repress transcription. We used CtBP cleft mutant and cleft-filled fusion derivatives to distinguish between partner proteins that bind in the cleft and elsewhere on the CtBP surface. Functional assays demonstrate that CtBP mutants that carry defective clefts retain repression activity when fused to heterologous DNA-binding domains. This result suggests that the cleft is not essential for recruiting effectors. In contrast, when tested in the absence of a fused DNA-binding domain, disruption of the cleft abrogates repression activity. These results demonstrate that the PXDLS binding cleft is functionally important but suggest that it is primarily required for localization of the CtBP complex to promoter-bound transcription factors.
The C-terminal binding proteins (CtBPs) function as transcriptional corepressors but also play roles in Golgi maintenance and in synaptic ribbon formation (6, 8, 49, 55, 58). They mediate transcriptional repression by associating with DNA-bound transcription factors and recruiting a complex of chromatin-modifying enzymes (46-48, 64). CtBPs have been shown to regulate gene expression in organisms from Drosophila to mammals. In Drosophila, CtBPs associate with developmental transcription factors such as Krüppel, Hairy, and Knirps (34-36). In mammals, CtBPs associate with a number (about 30) of different sequence-specific transcription factors (9, 25, 31, 57, 61). Knockout studies with mice, in combination with microarray analyses, have implicated the mammalian CtBPs in the repression of genes involved in apoptosis and in the epithelial-to-mesenchymal transition (13, 15, 16).
Structural analyses have revealed that CtBP is composed of three domains, the substrate-binding domain that primarily comprises the PXDLS binding cleft, the central nucleotide-binding domain that is responsible for NAD(H) binding and dimerization, and an intrinsically unstructured C-terminal region (32, 33). Details of the PXDLS cleft (hereafter referred to as the PIDLS cleft, as X is often Ile) have come from studies of one member of the CtBP family, rat CtBP3/BARS (recently renamed short-CtBP1 or CtBP1-S) (5, 32). Mutagenesis has confirmed that two residues in the CtBP cleft, A41 and V55, are critical to PIDLS recognition (32).
Although PIDLS motifs in transcription factors have been implicated in CtBP binding, the mechanisms through which CtBP then recruits chromatin-modifying enzymes are not well characterized (3, 47, 65). It has been established that CtBPs can self-associate and that this ability is critical to activity (27, 47). The crystal structures revealed the presence of dimers, but higher-order complexes, including tetramers (dimers of dimers) have been observed in gel filtration experiments and small-angle X-ray scattering in solution (2, 27, 32, 33, 47). The PIDLS binding cleft exists in each monomer subunit, so the self-association of CtBPs immediately suggests that CtBP dimers might bridge two distinct PIDLS motif-containing partners (32). Indeed, several papers on CtBPs, including our own, represent CtBP as a bridging molecule that links two PIDLS motif-containing proteins involved in gene repression (6, 24, 30, 56, 58). The crystal structure shows that CtBP forms an elongated dimer with a PIDLS binding cleft at each end, further suggesting that CtBP is well suited to functioning as a linking molecule (32). The observation that certain histone deacetylase proteins, HDAC4 and HDAC7, contain PIDLS motifs provided the first specific mechanistic explanation of how the dimerization and bridging activities of CtBP might effectively recruit histone deacetylases to DNA-bound transcription factors and silence promoter activity (64) (Fig. 1A).
FIG. 1.
Models of CtBP dimers interacting with PXDLS partners. (A) Model of CtBP as a bridging molecule linking a transcription factor (TF) and an effector. An elongated dimer of CtBP as a link between a promoter-bound transcription factor containing a PIDLS motif and the effector molecule HDAC4, which also contains a PIDLS motif. (B) Model where a CtBP dimer is localized to a promoter by binding two PIDLS motif-containing transcription factors and then recruits effectors through a different surface.
The following question arises: how does a CtBP dimer associated with a PIDLS motif-containing DNA-bound transcription factor direct its second PIDLS cleft to an effector (such as HDAC4) rather than to another of the 30 or so known PIDLS-containing DNA-binding proteins (Fig. 1B)? Furthermore, some transcription factors contain two or more PIDLS motifs (18, 37, 40), so it is possible that both clefts of a CtBP dimer would be filled, leaving no clefts available for recruiting effectors. In this case, the critical effectors would be likely to be non-PIDLS-containing molecules (Fig. 1B).
Importantly, a number of CtBP partner proteins do not have canonical PIDLS motifs. Some of these probably have related motifs that can presumably bind the PIDLS cleft (10, 40, 42, 52, 61, 62), but others may bind different surfaces of CtBP. Distinguishing between the two types of partners may help differentiate between partners involved in recruiting CtBP to promoters and those involved in mediating or regulating CtBP activity. Interestingly, the effector proteins in the CtBP repression complex, HDAC1, HDAC2, histone methyltransferase G9, and histone demethylase LSD1, have not been shown to contain PIDLS motifs. Thus, it seems that these proteins either bind a separate surface of CtBP or associate with it indirectly, via additional bridging molecules (47, 53, 60).
In this study, we have investigated the importance of the PIDLS cleft for CtBP's physical interaction with partner proteins and also for CtBP's repression activity. We used a number of different CtBP cleft mutants. We disrupted the cleft by mutation, and we also produced a CtBP fusion protein that contains a C-terminal PIDLS peptide that appears to fill CtBP's own cleft, termed the cleft-filled mutant protein.
With these mutant proteins, we confirmed that several recognized PIDLS-dependent partners rely on the cleft for binding to CtBP. We also confirmed that some proteins without recognizable PIDLS motifs do not require an intact CtBP cleft for binding and therefore presumably contact other faces of CtBP. Finally, we observed that certain protein domains that interact with CtBP but do not have recognizable PIDLS motifs do, nevertheless, rely on the PIDLS cleft and therefore propose that PIDLS-type motifs that cannot be recognized by visual sequence inspection may exist.
In terms of functional gene repression, our assays with Gal4 DNA-binding domain (Gal4DBD)-CtBP or zinc finger-CtBP fusions revealed that significant repression activity remains even when the PIDLS cleft is mutated. In contrast, the cleft is required for repression when CtBP is not fused to a heterologous DNA-binding domain. These results suggest that the cleft plays an essential role in recruiting CtBP to PIDLS-containing transcription factors.
MATERIALS AND METHODS
Plasmid constructs.
Full-length murine CtBP2 was amplified by PCR (mCtBP2Xma.F, TCCCCCCGGGAAAAAGAATGGCCCTTGTGGATAAGCA; mCtBP2NotStopSal.R, ACGCGTCGACCTATGCGGCCGCTTGCTCGTTGGGGTGCTCTC), and the product was cloned into the XmaI/SalI pGBT9 (Clontech) vector. This resulting construct, pGBT9-mCtBP2, was used as wild-type CtBP2 in yeast experiments. Secondly, murine BKLF residues 1 to 84, 1 to 75, and 30 to 75, each containing the 61PVDLT65 motif, were amplified by PCR (for example, BKLF 30-75 with mBKLF.30.Not.F [ATAAGAATGCGGCCGCAAAGCCCAACAAATATGGGGT] and mBKLF.75.Sal.R [ACGCGTCGACCTATGCAGCAGGCGGGGAGCCCC]) and cloned into the NotI/SalI sites of pGBT9-mCtBP2. The resulting yeast two-hybrid constructs expressed the BKLF segments fused to the C terminus of CtBP2. The pGBT9-mCtBP2-BKLF 30-75 construct is referred to as cleft filled. The control ΔDL fusion, which contains a DL-to-AS mutation in the PVDLT motif in the BKLF 30-75 portion of the fusion, was generated from the pGBT9-mCtBP2-BKLF 30-75 construct by overlap PCR mutagenesis (BKLFdeltaDL.F, CAGGTGGAGCCGGTGGCCAGCAGGTGAACAAGCGG; BKLFdeltaDL.R, CCGCTTGTTCACCGTGCTGGCCACCGGTCCACCTG). The mCtBP2, mCtBP2-BKLF 30-75, and mCtBP2-BKLF 30-75 ΔDL inserts were subcloned into the XmaI/SalI sites of the pGAD10(new) (derived from pGAD10 from Clontech) vector to allow them to be expressed in the yeast two-hybrid system as both Gal4 activation domain (Gal4AD) and Gal4DBD fusions.
A58E and V72R mutations were introduced into murine CtBP2 by overlap PCR mutagenesis (mCtBP2.A58E.F, GACCTGGCCACTGTGGAATTCTGTGATGCACAG; mCtBP2.A58E.R, CTGTGCATCACAGAATTCCACAGTGGCCAGGTC; mCtBP2.V72R.F, GAAATCCATGAGAAGCGGTTGAATGAAGCTGTG; mCtBP2.V72R.R, CACAGCTTCATTCAACCGCTTCTCATGGATTTC). BglII/SalI-digested mutant inserts were ligated into the BamHI/SalI sites of the pGBT9 and pGAD10(new) vectors to generate pGBT9-mCtBP2-A58E, pGBT9-mCtBP2-V72R, pGAD10(new)-mCtBP2-A58E, and pGAD10(new)-mCtBP2-V72R.
Amino acids 1 to 362, 32 to 445, 32 to 362, 10 to 445, 20 to 445, 26 to 445, 1 to 55, 50 to 362, and 70 to 362 of murine CtBP2 were amplified by PCR with appropriate oligonucleotides, and BglII/SalI-digested products were ligated into the BamHI/SalI pGBT9 and pGAD10(new) vectors to generate pGBT9-mCtBP2 32-362, pGBT9-mCtBP2 50-362, pGBT9-mCtBP2 70-362, pGAD10(new)-mCtBP2 1-362, pGAD10(new)-mCtBP2 32-445, pGAD10(new)-mCtBP2 32-362, pGAD10(new)-mCtBP2 10-445, pGAD10(new)-mCtBP2 20-445, pGAD10(new)-mCtBP2 26-445, and pGAD10(new)-mCtBP2 1-55. pcDNA3-rCtBP1-S, encoding full-length rCtBP1-S, was kindly provided by Daniela Corda (Department of Cell Biology and Oncology, Consorzio Mario Negri Sud, Santa Maria Imbaro, Chieti, Italy) and has been described previously (50). From this template and with appropriate oligonucleotides, rCtBP1-S 1-430 (full length) and 15-345 were amplified by PCR and ligated into the EcoRI/SalI sites of the pGBT9 and pGAD10(new) vectors to generate pGBT9-rCtBP1-S 1-430, pGBT9-rCtBP1-S 15-345, pGAD10(new)-rCtBP1-S 1-430, and pGAD10(new)-rCtBP1-S 15-345. pCMV5 FLAG-hCtBP1-L, encoding full-length hCtBP1-L, was kindly provided by David Wotton (University of Virginia, Charlottesville) and has been described previously (20). From this template and with appropriate primers, hCtBP1-L 1-440 (full length) and 26-440 were amplified by PCR and ligated into the EcoRI/SalI sites of the pGBT9 vector to generate pGBT9-hCtBP1-L 1-440 and pGBT9-hCtBP1-L 26-440.
The yeast two-hybrid assay vectors pGAD10-BKLF 1-268 and pGBT9-BKLF 1-268, encoding murine BKLF, have been described previously (57). pcDNA3-FHL3 was kindly provided by Ju Chen (University of California at San Diego, La Jolla) and has been previously described (7). Full-length murine Four-and-a-Half LIM Domain Protein 3 (FHL3) was amplified from this vector by PCR (mFHL3.Bam.F, CGGGATCCATGAGCGAGGCATTTGAC; mFHL3.Eco.R, CGGAATTCTCAGGGGCCTGCTTGGC), and the product was cloned into the BamHI/EcoRI sites of the pGBT9(new) vector, allowing expression of Gal4DBD-FHL3 in yeast two-hybrid assays. The yeast two-hybrid assay vector for murine Ubc9, pGAD10-Ubc9, has been described previously (39). Murine homeodomain-interacting protein kinase 1 (HIPK1) amino acids 619 to 1040 and 619 to 1083 were amplified by PCR (5x HIPK Frag, GGAATTCTCGTCGGCAGCGCCAGTTCCT; 3x HIPK1Frag-PLNLS, CGGGATCCCTACACCGCGCTGGCTCCCCCT; 3x HIPK1 Frag, CGGGATCCCTAGAAGGCGTAGGGGGCTTGGGA) and cloned into the EcoRI/BamHI sites of the pGAD10 vector to generate pGAD10-HIPK1 619-1040 and pGAD10-HIPK1-619-1083. The NL-to-AS mutation was introduced into the 1042PLNLS1047 motif of the pGAD10-HIPK1 619-1083 construct by overlap PCR mutagenesis (5× HIPK1-PLASS,GTGCAGCCACTCGCCAGTAGCCAGAACCAG; 3× HIPK1-PLASS, CTGGTTCTGGCTACTGGCGAGTGGCTGCAC) to generate pGAD10-HIPK1 619-1083 ΔDL. Murine HIPK2 amino acids 615 to 1067 and 881 to 1067 were amplified by PCR (5× HIPK2 Frag, GGAATTCCCCTCAGCGGCATCCATGGC; HIPK2_I881_5′, CGGAATTCATCACCATCAGCAGTGAC; 3× HIPK2 Frag, CGGGATCCTAGAAGGTGTACGGAGCTTGCGC) and cloned into the EcoRI/BamHI sites of the pGAD10(new) vector to generate pGAD10(new)-HIPK2 615-1067 and pGAD10(new)-HIPK2 881-1067. The NL-to-AS mutation was introduced into the 1031PLNLS1035 motif of the pGAD10(new)-HIPK2 881-1067 and pGAD10(new)-HIPK2 615-1067 constructs by overlap PCR mutagenesis (5× HIPK2 PLASS, CAGCAGCAGCCCCTCGCTTCCAGCCAGGCCCAGCAG; 3× HIPK2 PLASS, CTGCTGGGCCTGGCTGGAAGCGAGGGGCTGCTGCTG) to generate pGAD10(new)-HIPK2 881-1067 ΔDL and pGAD10(new)-HIPK2 615-1067 ΔDL.
Wild-type CtBP2 and the A58E mutant form were amplified by PCR with appropriate oligonucleotides, and BglII/SalI-digested products were ligated into the BamHI/SalI pGEX 2TY (new) vector to generate pGEX 2TY (new)-CtBP2 and pGEX 2TY (new)-CtBP2 A58E.
The yeast two-hybrid assay vectors for murine Ikaros [pGBT9(new)-Ikaros 1-81 and pGBT9(new)-Ikaros 1-81 ΔDL], human Eos [pGAD10-Eos 101-532, pGAD10(new)-Eos 101-532 ΔDL, and pGAD10-Eos 364-400 ΔDL], and human TRPS1 (pGAD10-TRPS 1068-1186 and pGAD10 TRPS 1068-1186 ΔDL) have been described previously (38).
The Gal4DBD (147 amino acids) was amplified by PCR with appropriate primers and ligated into the PstI/NotI sites of the pMT3 (derived from pMT2) vector to generate pMT3-Gal4 without a stop codon. A separate pMT3-Gal4 plasmid with a stop codon was generated to act as a control in mammalian repression assays. Secondly, wild-type mCtBP2 and mCtBP2-A58E and mCtBP2-V72R mutant inserts were reamplified by PCR with appropriate primers and cloned into the NotI/SalI sites of pMT3-Gal4 without a stop codon 3′ of the Gal4 gene to generate pMT3-Gal4-mCtBP2, pMT3-Gal4-mCtBP2 A58E, and pMT3-Gal4-mCtBP2 V72R.
The DNA-binding Krüppel-like zinc fingers of BKLF (amino acids 254 to 344, BKLFf) were amplified by PCR with appropriate primers. The product was cloned into the XhoI/XbaI sites of the pcDNA3 vector (Invitrogen) to generate pcDNA3-BKLFf. Secondly, mCtBP2 wild-type and A58E and V72R mutant inserts were amplified by PCR with appropriate primers to remove the natural stop codon at the end of the CtBP coding sequence. These products were digested with BglII/SalI and ligated into the BamHI/XhoI sites of pcDNA3-BKLFf to generate pcDNA3-BKLFf-mCtBP2, pcDNA3-BKLFf-mCtBP2 A58E, and pcDNA3-BKLFf-mCtBP2 V72R.
Wild-type mCtBP2 and mCtBP2-A58E mutant inserts were reamplified by PCR with appropriate primers and cloned into the NotI/SalI sites of pMT3 to generate pMT3-mCtBP2 and pMT3-mCtBP2 A58E, respectively.
The firefly luciferase reporter vector pGL2-(Gal4)5-(LexA)2-E1B-Luc and LexA-VP16 mammalian expression plasmid pCMV-LexA (1-202)-VP16 (410-490) were generous gifts from Luke Gaudreau and Mark Ptashne (The Sloan-Kettering Institute, New York, NY). A second firefly luciferase reporter vector containing five Gal4 binding sites and the thymidine kinase (TK) promoter, pGL2-(Gal4)5-TK-Luc, has been described previously (38). The pØAγGH vector contains the human Aγ promoter upstream of the human growth hormone (GH) reporter gene in the pØ-GH vector (Nichols Institute Diagnostics). The Aγ promoter has been shown to be activated by the GATA-1 mammalian expression vector pXM.GF1 and contains binding sites for BKLFf (59). The pGL3 human E-cadherin (−427/+53)-luciferase reporter vector was a gift from Stephen Sugrue (Department of Anatomy and Cell Biology, Harvard Medical School) and has been previously described (1).
Yeast two-hybrid assays.
The Clontech two-hybrid system was used according to the manufacturer's instructions. Test proteins were expressed in yeast strain HF7c as either Gal4DBD or Gal4AD fusions. Transformant colonies were selected on Leu/Trp-deficient plates and patched onto His/Leu/Trp-deficient plates. Growth was scored following 72 h of incubation.
Mammalian cell culture.
NIH 3T3 cells were cultured and transfected by the calcium phosphate method as described previously (57). COS-1 cells were cultured as described previously (39) and transfected with the transfection reagent FuGEN6 (Roche Diagnostics) by following the manufacturer's instructions. CtBP1+/− CtBP2+/− and CtBP1−/− CtBP2−/− cells were cultured as previously described (16) and transfected with the Lipofectamine transfection reagent (Invitrogen) by following the manufacturer's instructions.
Mammalian repression assays.
To examine CtBP repression of reporter gene expression, six-well plates of COS-1 cells were transfected. To examine repression of basal expression, the following plasmids were used: 3 μg of the pGL2-(Gal4)5-TK-Luc reporter and 50 ng of either pMT3-Gal4 with stop, pMT3-Gal4-mCtBP2, pMT3-Gal4-mCtBP2-A58E, or pMT3-Gal4-mCtBP2-V72R. To examine repression of activated expression, the following plasmids were used: 3 μg of the pGL2-(Gal4)5-(LexA)2-E1B-Luc reporter, 1 μg of the pCMV-LexA (1-202)-VP16 (410-490) expression vector, and 25 ng of either pMT3-Gal4 with stop, pMT3-Gal4-mCtBP2, pMT3-Gal4-mCtBP2-A58E, or pMT3-Gal4-mCtBP2-V72R. In both experiments, 10 ng of the Renilla luciferase vector pRL-Luc (Promega) was cotransfected to allow the firefly luciferase measurements to be corrected to control for transfection efficiency. Luciferase activity was measured 48 h posttransfection in a Turner Designs model TD 20/20 luminometer with the dual-luciferase reporter assay system (Promega). Results shown are averaged firefly/Renilla luciferase ratios from four replicates of a representative experiment with 1-standard-deviation error bars.
To further examine CtBP repression, six-well plates of CtBP1+/− CtBP2+/− and CtBP1−/− CtBP2−/− cells were transfected with 4 μg of the pGL2-(Gal4)5-(LexA)2-E1B-Luc reporter plasmid, 2 μg of the pCMV-LexA (1-202)-VP16 (410-490) expression plasmid, and 2 μg of either pMT3-Gal4 with stop, pMT3-Gal4-mCtBP2, pMT3-Gal4-mCtBP2 A58E, or pMT3-Gal4-mCtBP2 V72R. Twenty nanograms of the Renilla luciferase vector pRL-Luc was cotransfected to allow the firefly luciferase measurements to be corrected to control for transfection efficiency. To examine CtBP2 repression of the E-cadherin promoter, six-well plates of CtBP−/− cells were transfected with 2 μg of pGL3-E-cadherin-Luc, 1 μg of the pMT3-CtBP2 or pMT3-CtBP2 A58E vector, and 10 ng of pRL-Luc. Various amounts of pMT3 plasmid DNA were used in each transfection to make the total amount of DNA transfected the same. Luciferase assays were performed as described above 48 h following transfection. Results shown are averaged firefly/Renilla luciferase ratios from two replicates of a representative experiment with range-of-values error bars.
NIH 3T3 cells in six-well plates were transfected with 200 ng of the pØAγGH reporter plasmid, 2 μg of the pXM-GF1 GATA-1 expression plasmid, and either 250 ng of pcDNA3-BKLFf or 50 ng or 250 ng of pcDNA3-BKLFf-mCtBP2, pcDNA3-BKLFf-mCtBP2 A58E, or pcDNA3-BKLFf-mCtBP2 V72R. The pcDNA3 empty vector was added to make the amount of DNA in each transfection the same. One hundred microliters of medium was removed 48 h after transfection, and human GH expression was assayed in each sample with a radioisotopic assay kit according to the manufacturer's (Nichols Institute Diagnostics) instructions. Gamma radiation was measured with a Wallace gamma counter. The average value for the GATA-1-activated expression samples was set to 100, and all other values were expressed as fractions of this activated level. The results shown are for four replicates from a representative experiment with 1-standard-deviation error bars.
Western blot assays.
Western blot assays were performed to assess the relative expression levels of the different CtBP2 mammalian expression constructs, as has been described previously for other proteins (39). Briefly, 1 μg of the empty pMT3, pMT3-Gal4 with stop, pMT3-Gal4-mCtBP2, pMT3-Gal4-mCtBP2 A58E, or pMT3-Gal4-mCtBP2 V72R expression construct was transfected into a 10-cm petri dish of COS-1 cells and nuclear extracts were prepared 48 h following transfection. The expression levels of proteins subjected to Western blot assays were detected with mouse CtBP2 monoclonal antibody (BD Transduction Laboratories).
GST pull-down assay.
Bacterially produced glutathione S-transferase (GST), GST-CtBP2, and GST-CtBP2 A58E fusion proteins were recovered on glutathione-Sepharose 4B beads (Amersham) and incubated for 3 h at 4°C with the lysates prepared from COS-1 cells transfected with 3 μg of pcDNA3-HA-Ubc9, pMT3-HA-FHL3, and pMT3-Flag-BKLF (1-268) by using NP-40 cell lysis buffer (50 mM Tri-HCl, pH 8.0, 150 mM NaCl, 1% NP-40). These three vectors have been previously described (39, 59). The beads were then washed three or four times with cell lysis buffer containing 0.5 M KCl and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by detection by immunoblotting with an anti-hemagglutinin (HA) (12CA5; Roche Corporation) or an anti-Flag (M2; Sigma) antibody.
RESULTS
Generation of CtBPs carrying defective PIDLS binding clefts.
We have worked primarily with one member of the CtBP family, murine CtBP2. We used two distinct strategies to generate derivatives of CtBP2 that contain a defective PIDLS binding cleft. Firstly, noting that the crystal structure of rat CtBP1-S implicated amino acids A41 and V55 in binding to a PIDLS peptide (32), we identified the corresponding residues in CtBP2 and similarly mutated them to E and R, respectively. The second strategy was to generate a fusion protein comprising CtBP2 and an extension containing a PIDLS motif. We chose to use regions encompassing the naturally occurring PIDLS motif in the N terminus of BKLF (specifically, PVDLT, residues 61 to 65 of murine BKLF) (57). We initially tested three different lengths of the N-terminal region and fused each to the C terminus of CtBP2. The crystal structure indicates that the C terminus should be in proximity to the N-terminal domain hosting the PIDLS binding cleft (32). As a control, we made similar fusions but incorporated a mutation directly into the PIDLS motif. We used a well-characterized mutation termed ΔDL, where the PIDLS motif is altered to PIASS (42, 43). This control mutant was designed to reveal whether the fusion extension was binding directly and occupying the cleft or sterically impeding access to the cleft in a nonspecific fashion.
The yeast two-hybrid assay system has proved to be a reliable method for assessing the interactions between CtBP and its partners (17, 45, 57, 59, 62). We used this system to test the binding activity of the CtBP derivatives. We first tested the two cleft mutants CtBP2 A58E and CtBP2 V72R for the ability to interact with a known PIDLS motif-containing partner, BKLF. Yeast assays were carried out with the CtBPs present first as bait (Gal4DBD fusion, Fig. 2A) and then as prey (Gal4AD fusion, Fig. 2B) proteins. As expected, each of the mutations severely reduced binding to BKLF (Fig. 2). We also tested the mutants for the ability to dimerize with wild-type CtBP2 and found that this function was unaffected. The structural work has suggested that CtBP self-association is mediated by an extensive low-polarity dimerization interface (about 3,400 Å2) contributed by the central (nucleotide-binding) domain of the protein (27, 32). These results suggest that the general structure of the mutant CtBPs is not significantly affected by the cleft mutation. This is in agreement with the structural data, which locate the PIDLS cleft in the N-terminal domain (32) such that mutation would not be expected to interfere with the dimerization of CtBP.
FIG. 2.
Generation of CtBP2 derivatives with defective PIDLS binding clefts. CtBP2 derivatives containing either the A58E or the V72R mutation or with a PIDLS extension or a ΔDL mutant PIDLS motif extension were tested for the abilities to dimerize with wild-type CtBP2 and to interact with a recognized PIDLS motif-containing partner protein, BKLF. Yeast growth was monitored, and the results reflecting association of the proteins are shown. (A) Gal4DBD-CtBP2 cleft mutants were tested against Gal4AD-wild-type CtBP2 or BKLF 1-268. (B) Gal4AD-CtBP2 cleft mutant proteins were tested against Gal4DBD-wild-type CtBP2 or BKLF 1-268.
We also tested the fusion cleft-filled protein and the control ΔDL protein for binding to PIDLS partner proteins (Fig. 2). As mentioned above, we tried different-length fusion peptides but found that fusions containing BKLF residues 1 to 84 and 1 to 75 generated some autoactivation in yeast and so were not suitable for this analysis (data not shown). The shortest fusion protein, comprising residues 30 to 75 of BKLF fused to the C terminus of CtBP, did not produce autoactivation. This fusion was tested for the ability to bind a PIDLS partner; in this case, we again used BKLF. It was found that the fusion cleft-filled mutant could no longer associate with BKLF. Importantly, the control fusion containing a mutant ΔDL PVAST motif retained its ability to bind BKLF. This suggests that the peptide fusion extension is not nonspecifically impeding access to the cleft but that the PIDLS motif occupies the cleft and blocks the binding of external PIDLS motifs (Fig. 2).
Distinguishing between PIDLS-containing and non-PIDLS-containing CtBP partners.
We next sought to investigate a number of previously identified CtBP partners to determine whether they bound via PIDLS motifs slotting into CtBP's cleft or against another surface of CtBP. We first tested FHL3. FHL3 is a transcriptional adaptor protein composed almost entirely of highly structured LIM domains (59). No PIDLS motif has been identified by visual sequence inspection. Furthermore, it has been shown that FHL3 and the PIDLS motif-containing protein BKLF can coexist in a complex with CtBP2, suggesting that BKLF may fill the PIDLS cleft and FHL3 may interact with a different surface of CtBP2 (59). We tested whether FHL3 could bind the four CtBP2 cleft derivatives. The interaction was tested with CtBP-Gal4AD fusions and FHL3-Gal4DBD fusions. Wild-type CtBP2, the three cleft mutant proteins, and the control fusion protein all bound FHL3 (Fig. 3A). These results suggest that FHL3 does not bind in the PIDLS cleft but contacts a distinct surface of CtBP2.
FIG. 3.
Use of CtBP2 cleft derivatives to assess whether FHL3 and Ubc9 bind to CtBP2 in the cleft or elsewhere on the surface of CtBP2. (A) Murine FHL3 fused to the Gal4DBD was tested for the ability to interact with Gal4AD CtBP2 cleft derivatives in the yeast two-hybrid assay. (B) Murine FHL3 and BKLF 1-268 fused to the Gal4DBD were assessed for the ability to bind to the indicated fragments of CtBP2, rat CtBP1-S, and human CtBP1-L in the yeast two-hybrid assay. The two key cleft residues involved in binding to PIDLS motifs (A58 and V72 in CtBP2, A41 and V55 in rCtBP1-S, and A52 and V66 in hCtBP1-L) are indicated. (C) Murine Ubc9 fused to the Gal4AD was tested for the ability to interact with the CtBP2 cleft derivatives fused to the Gal4DBD in the yeast two-hybrid assay. (D) Murine Ubc9 fused to the Gal4AD was assessed for the ability to bind to the indicated fragments of CtBP2 and rat CtBP1-S in the yeast two-hybrid system. The two key cleft residues involved in binding to PIDLS motifs (A58 and V72 in CtBP2 and A41 and V55 in rCtBP1-S) are indicated. (E) Alignment of the N termini of murine CtBP2, rat CtBP1-S, and human CtBP1-L, indicating the regions of CtBP2 implicated in binding to FHL3 and Ubc9. One of the key cleft residues involved in binding to PIDLS motifs (A58 in CtBP2, A41 in rCtBP/BARS, or A52 in hCtBP1-L) is indicated. A summary of the protein-protein interaction results from panels A to D is also displayed on the right of the alignment.
To further investigate this possibility, we created CtBP2 deletions to map the regions of CtBP required for binding to FHL3 (Fig. 3B). The region comprising the N-terminal 55 amino acids of CtBP2 was identified as the minimal region required for interaction with FHL3. This region is distinct from the cleft, which includes key contact residues A58 and V72 of CtBP2. Furthermore, FHL3 shows interaction only with CtBP2 and not with the related family member CtBP1-L. CtBP2 and CtBP1-L are highly conserved in the cleft region, and both bind to PIDLS partners. However, these family members are divergent at the N terminus, where the mapping indicates that FHL3 binds (Fig. 3E). We conclude that FHL3 is a non-PIDLS-dependent CtBP2 partner.
We also tested other CtBP partners. The SUMO E2 conjugating enzyme Ubc9 is a recognized partner of CtBPs (20, 29). Ubc9 has been shown to bind CtBP1-L, and CtBP1-L is known to be subject to SUMOylation in vitro and in vivo (20, 29). We have also observed the binding of Ubc9 to CtBP2 in yeast two-hybrid assays (39). Sequence inspection does not reveal an obvious PIDLS motif in Ubc9, and the mechanism by which Ubc9 contacts CtBPs remains to be defined. We tested Ubc9 against wild-type CtBP and the four CtBP cleft derivatives. Ubc9 bound all five proteins (Fig. 3C). This result suggests that Ubc9 is another protein that does not require an intact CtBP PIDLS binding cleft but presumably binds to a distinct surface of CtBP2. Additionally, deletion mapping shows that amino acids 33 to 49 of CtBP2 are necessary for binding to Ubc9, suggesting that the regions of CtBP involved in contacting FHL3 and Ubc9 are distinct (Fig. 3D). This result demonstrates that there are at least two other regions of CtBP involved in partner protein binding in addition to the PIDLS binding cleft.
In order to confirm these results, we produced wild-type CtBP2 and an A58E mutant form fused to GST in bacteria. A pull-down assay was then performed with GST-CtBP2 and GST-CtBP2 A58E by using extracts from COS-1 cells transfected with tagged HA-Ubc9, HA-FHL3, and Flag-BKLF (1-268). As shown in Fig. 4 and in accordance with the yeast results, Ubc9 and FHL3 interact with both CtBP2 and CtBP2 A58E, while BKLF bound to GST-CtBP2 but not to GST-CtBP2 A58E. These results further indicate that Ubc9 and FHL3 are genuine non-PIDLS-containing CtBP partners, both in yeast and in mammalian cell extracts.
FIG. 4.
Interactions between CtBP2 and its partners. A pull-down assay was performed to detect binding of GST-CtBP2 and GST-CtBP2 A58E to Ubc9, FHL3, and BKLF (1-268). Inputs of whole-cell extracts from transfected COS-1 cells are shown (Input). Interactions with HA-Ubc9, HA-FHL3, and Flag-BKLF (1-268) were monitored by Western blot (WB) analysis with anti-HA (αHA) and anti-Flag (αFlag) antibodies as indicated. For Ubc9, the asterisk indicates an unknown band in the GST lane that does not comigrate with Ubc9.
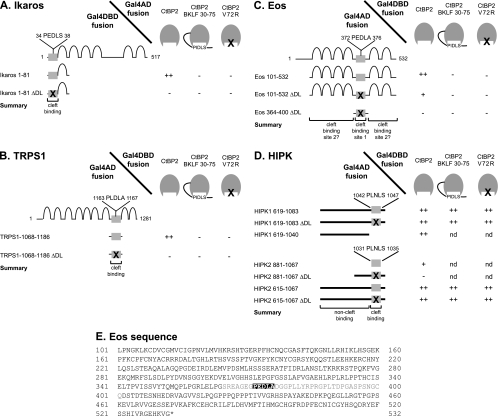
We next tested three members of the Ikaros family of zinc finger transcription factors, Ikaros, TRPS1, and Eos. All three proteins have PIDLS motifs that have been shown to be important for interaction with CtBPs (25, 26, 38). Ikaros contains a single PIDLS motif in its N terminus (34PEDLS38 in murine Ikaros). Mutation of this motif prevents binding to CtBP (25). We tested Ikaros against the CtBP cleft derivatives and observed that mutation of the PIDLS binding cleft (A58E, V72R, or cleft filled) prevented binding to Ikaros. This result confirms that Ikaros is a typical PIDLS-dependent CtBP partner that binds in the CtBP cleft (Fig. 5A). Similarly, TRPS1 contains a single PIDLS motif (1163PLDLA1167 in human TRPS1) and mutation of this motif prevents binding to CtBP (38). As with Ikaros, mutation of the CtBP PIDLS binding cleft prevented binding to TRPS1, indicating that TRPS1 is also a PIDLS-dependent partner of CtBP that binds in the cleft (Fig. 5B).
FIG. 5.
Use of CtBP cleft derivatives to distinguish between CtBP2 partners that bind in the cleft and elsewhere on the surface of CtBP. Yeast two-hybrid assays were performed to examine the interaction between the CtBP2 cleft derivatives and Ikaros (A), TRPS1 (B), Eos (C), and HIPK (D). The results for wild-type CtBP2 and for the CtBP2 fusion with the ΔDL mutant PIDLS extension were identical, and only the wild-type results are presented. A summary of the regions of the test proteins implicated in PIDLS cleft binding to CtBP and in binding to CtBP outside the cleft are indicated. nd, not determined. (E) Amino acid sequence of Eos residues 101 to 532. Residues 364 to 400 are in gray, and the known PIDLS motif, PEDLA, is shaded in black. Our yeast two-hybrid assay results indicate that there is a second PIDLS motif in residues 101 to 363 or 401 to 532.
In contrast, the situation with Eos is more complicated. Eos contains a PIDLS motif (372PEDLA376 in human Eos) but mutation of this motif does not abrogate binding to CtBP (38). This suggests that Eos contains a second CtBP contact surface. In order to investigate this second surface, we mutated the recognized PIDLS motif and then tested the resulting fragment of Eos against the CtBP cleft derivatives. The results indicate that the second surface is able to bind wild-type CtBP and the fusion control but not the cleft mutant forms (A58E, V72R, and cleft filled) (Fig. 5C). This suggests that the second contact motif also requires an intact and empty PIDLS cleft. Unexpectedly, sequence inspection does not reveal any obvious second PIDLS motif in Eos (Fig. 5E). Previous mapping has shown that both the N- and C-terminal regions of Eos may contain additional CtBP contact motifs (38). This result suggests that motifs that significantly diverge from recognizable PIDLS sequences but retain the ability to interact with the cleft occur in Eos.
We also tested HIPK1 and HIPK2. HIPK2 was shown to bind to CtBP1-L, and since it bound CtBP1-L in the presence of high concentrations of the E1A PIDLS motif, it was inferred that it did not bind in the PIDLS cleft (65). We also isolated HIPKs in yeast two-hybrid screens against CtBP (data not shown) and noted a potential PIDLS motif (PLNLS, residues 1042 to 1047 of murine HIPK1 and 1031 to 1035 of murine HIPK2). This result, together with the observation that HIPK2 can bind CtBP in the presence of E1A (65), suggests that HIPKs might be able to contact CtBP both through the PIDLS binding cleft and through an additional surface. As explained below, our results support this conclusion.
When HIPK1 (residues 619 to 1083) was tested against wild-type CtBP2 in the yeast two-hybrid assay, the protein interaction was evident (Fig. 5D). Deleting or mutating the PLNLS motif in HIPK1 did not abolish the interaction, supporting the view that a second CtBP2 contact surface exists. Furthermore, both wild-type HIPK1 619-1083 and HIPK1 619-1083 ΔDL bind to the CtBP cleft mutants (A58E, V72R, or cleft filled). This implies that the second interaction interface between HIPK1 and CtBP does not comprise a PIDLS motif fitting into the cleft of CtBP.
Fragments of HIPK2 were also tested for interaction with CtBP in the yeast two-hybrid system (Fig. 5D). HIPK2 (residues 881 to 1067) was able to bind to wild-type CtBP, but mutation of the PLNLS motif in this fragment of HIPK2 abolished the interaction. This demonstrated that the PLNLS motif in HIPK2 is functional and that it can mediate binding to CtBP. When larger fragments of HIPK2 were used, mutation of the PLNLS motif did not abolish binding to CtBP. Both the wild type and ΔDL HIPK2 615-1067 were able to interact with the CtBP cleft mutant forms (A58E, V72R, and cleft filled). This suggests that the second interaction site between HIPK2 and CtBP does not comprise a PIDLS motif fitting into the cleft of CtBP. Taking together these results obtained with HIPK1 and HIPK2, the data suggest that the HIPK family proteins contain both a PIDLS motif and a second, as yet unidentified, interaction site for CtBP.
The role of the CtBP PIDLS binding cleft in gene repression.
We first tested the repression function of CtBPs fused to heterologous DNA-binding domains. The fusions enable CtBP recruitment to the target promoter. Two different strategies were used. Firstly, wild-type CtBP2 and the CtBP2 mutant derivatives A58E and V72R were fused to the C terminus of the Gal4DBD. These proteins were tested against Gal4-dependent reporter genes. Secondly, fusion proteins consisting of the mutant forms or wild-type CtBP2 fused to the N terminus of the zinc fingers of BKLF were generated and tested against a BKLF-dependent reporter gene. We initially sought to test both the cleft mutants and the cleft-filled mutants, as we expected that they might have different effects. Unfortunately, the cleft-filled derivatives were expressed at lower levels in mammalian cells, so only limited information could be obtained from these assays (data not shown) but the cleft mutants were expressed at levels equivalent to that of wild-type CtBP2 (Fig. 6A).
FIG. 6.
Functional effects on gene repression activity of mutating the PIDLS cleft in CtBP2. (A) Western blot assay performed to examine the expression levels of the Gal4-CtBP2 wild-type and A58E and V72R mutant proteins in COS-1 cells. (B) Testing of Gal4-CtBP2 constructs for the ability to repress firefly luciferase reporter gene expression from the TK promoter in COS-1 cells following transient transfection. (C) Testing of Gal4-CtBP2 constructs for the ability to repress LexA-VP16-activated firefly luciferase reporter gene expression from the E1B promoter in COS-1 cells following transient transfection. (D) Testing of BKLFf-CtBP2 constructs for the ability to repress GATA-1-activated human GH reporter gene expression from the Aγ globin promoter in NIH 3T3 cells following transient transfection. (E) Testing of Gal4-CtBP2 constructs for the ability to repress LexA-VP16-activated firefly luciferase reporter gene expression from the E1B promoter in both CtBP1+/− CtBP2+/− and CtBP1−/− CtBP2−/− cells following transient transfection. (F) Testing of wild-type CtBP2 and the A58E cleft mutant form for the ability to repress the human E-cadherin (−427/+53)-luciferase promoter in CtBP1−/− CtBP2−/− cells. FF/R, firefly/Renilla.
The Gal4 fusions were tested first against a promoter containing five Gal4 binding sites upstream of the herpesvirus TK gene promoter. The (Gal4)5-TK promoter is a relatively weak promoter and is significantly repressed by Gal4-CtBP fusions. The assays were carried out with COS-1 cells. In this assay, Gal4-CtBP2 efficiently represses promoter activity (Fig. 6B). The A58E and V72R cleft mutant derivatives retain significant repression activity, although there is a modest reduction in repression.
As well as testing repression of basal transcription from (Gal4)5-TK, the mutants were also assessed against VP16-activated transcription. A promoter containing two LexA sites and five Gal4 sites upstream of the adenovirus E1B promoter was used. This promoter can be strongly activated by LexA-VP16 fusion proteins and can then be efficiently repressed by Gal4-CtBP fusions. In this assay, Gal4-CtBP2 efficiently represses the promoter. The two cleft mutants A58E and V72R also repress transcription to a degree similar to that of wild-type CtBP2 (Fig. 6C).
We next tested the cleft mutant forms fused to the second DNA-binding domain (the zinc fingers of BKLF) for the ability to repress a BKLF-dependent reporter, the proximal Aγ fetal globin promoter. This promoter can be efficiently activated by the erythroid transcription factor GATA-1 in NIH 3T3 cells (59). It can then be repressed by zinc finger-CtBP2 fusion proteins. The cleft mutant zinc finger fusions A58E and V72R also repress this promoter but slightly less well than wild-type CtBP2 (Fig. 6D). This result is similar to the results obtained with Gal4 fusions and Gal4-dependent promoters in COS-1 cells.
It is known that CtBPs can dimerize (2, 45, 47, 54, 57). Thus, it was important to exclude the possibility that the mutant proteins were retaining repression activity by virtue of the ability to recruit wild-type endogenous CtBP. We therefore repeated the experiments with Gal4 fusions and reporters in murine embryonic fibroblasts derived from CtBP1/CtBP2 double-knockout murine embryos (16). The repression by wild-type CtBP2 and the A58E and V72R mutants seen in the CtBP−/− cells was indistinguishable from that seen in CtBP+/− cells (Fig. 6E). This suggests that dimerization with endogenous CtBPs does not make a major contribution in these assays. In conclusion, these results demonstrate that cleft mutants retain significant repression activity when CtBP is recruited to the promoter by a fused DNA-binding domain.
We finally tested CtBP that was not fused to a heterologous DNA-binding domain against a natural CtBP-dependent promoter. We chose the E-cadherin gene promoter, as the gene for E-cadherin has previously been shown to be a CtBP target (13, 14, 47). We transfected a CtBP2-encoding plasmid together with the E-cadherin reporter gene into CtBP−/− cells and observed, as expected, that CtBP2 significantly repressed expression (Fig. 6F). We also tested the mCtBP2 A58E cleft mutant and found in this case that the mutation abrogated repression.
DISCUSSION
The PIDLS motif has been found in a significant number of sequence-specific DNA-binding proteins (i.e., transcription factors). The importance of the PIDLS motif in individual transcription factors has generally been demonstrated by introducing the ΔDL (PIDLS-to-PIASS) mutation into the transcription factor and assessing the effects on binding to CtBP and on transcriptional repression (9, 40, 51, 57). In many cases, this type of experiment has confirmed that the PIDLS motif is necessary for CtBP recruitment. In some instances, mutation of a PIDLS motif does not abrogate CtBP binding or repression and a second PIDLS motif is identified by visual inspection of the sequence (4, 11, 19, 23, 40). In other cases, no obvious second motif is identifiable and it remains uncertain whether the transcription factor is contacting the CtBP cleft or another surface of CtBP (22, 44, 63, 64). The two CtBP cleft mutants and the cleft-filled derivatives described in this work are useful for determining whether CtBP partners interact with the CtBP cleft or not.
Our results confirm that Ikaros and TRPS1 are typical transcription factors that bind the CtBP cleft, while Eos also binds the CtBP cleft through one obvious PIDLS motif and another motif(s) that requires the cleft but which is not readily recognizable as a PIDLS motif by sequence inspection. We have also investigated the HIPKs. HIPKs were identified as partners of CtBP that do not require the PIDLS cleft (65). We have shown that HIPKs contain a PLNLS motif that interacts with the PIDLS cleft of CtBP and a second motif that does not require an intact cleft and presumably binds elsewhere on the surface of CtBPs.
We also show that both Ubc9 and FHL3 do not bind in the cleft and are likely to each recognize separate surfaces of CtBP. Furthermore, we found that FHL3 binds to CtBP2 but not to CtBP1. Our results indicate that FHL3 requires a portion of the N terminus of CtBP2 which is not well conserved with CtBP1 (Fig. 3). To our knowledge, this represents the first example of a CtBP2-specific partner that is able to distinguish between CtBP1 and CtBP2. Conversely, there is a published example of a CtBP1-specific partner, nNOS, which is a PDZ domain protein that binds to CtBP1 on a nonconserved carboxy-terminal sequence, DXL (41). The PIDLS binding cleft residues are conserved in CtBP family members, and proteins that contain PIDLS motifs are expected to bind both CtBP1 and CtBP2 (12). CtBP partner proteins that bind outside the CtBP cleft, like FHL3 or nNOS, may be important in imparting different functions to CtBP1 and CtBP2. CtBP1−/− mice are viable, whereas CtBP2−/− mice die in utero, and differences in partner proteins, in addition to differences in expression profiles, may contribute to the phenotypic difference (16).
In terms of functional repression, our results demonstrate that CtBPs retain significant repression activity when the PIDLS cleft is mutated, provided they are recruited to the promoter by some other means, such as the addition of a heterologous Gal4 or BKLF zinc finger DNA-binding domain. Thus, it is likely that the major effectors of CtBP function are proteins that do not contain PIDLS motifs and do not bind in the cleft but are recruited by binding to other surfaces of CtBP or by binding to bridging proteins that bind other surfaces of CtBP. This representation is shown in Fig. 1B and is consistent with recent work showing that CtBP associates with a large repression complex made up of proteins that do not contain recognized PIDLS motifs (28, 46, 47).
Our results with CtBP that is not fused to a heterologous DNA-binding domain demonstrate that the cleft is critical for CtBP-mediated gene repression. This result is in line with the view that the cleft is required for recruitment of CtBP to PIDLS-containing transcription factors (Fig. 1 and 6).
One can hypothesize that PIDLS-containing partners may include transcription factors, effectors, and regulators (particularly inhibitors) of CtBP activity. The role of PIDLS motifs in transcription factors is well characterized. PIDLS motifs are found in a large number of sequence-specific DNA-binding proteins (6, 58). They are also found in proteins, such as Friend of GATA-1 (17, 21, 57), which do not themselves bind DNA but are obligate cofactors of DNA-binding proteins like GATA-1. PIDLS motifs are also found in proteins such as RIP140 and Pc2 (45, 62), but again these proteins may be playing roles in the recruitment of CtBP to gene promoters rather than as effectors that mediate its activity. In this model, we propose that CtBP is not so much a linking protein but a capping protein that may dock to a promoter-bound transcription factor and bring in its large repression complex (Fig. 1B).
There is limited evidence that PIDLS motifs are found in effectors of CtBP function (64). Our finding that the cleft is not required for repression function when CtBP is directed to a promoter by a heterologous DNA-binding domain, together with the observation that enzymes present in the CtBP repression complex do not contain identifiable PIDLS motifs, suggests that PIDLS motif proteins like HDAC4 and -7 (64) may not be major effectors. It should be noted, however, that although the cleft mutants all retained repression activity, there were some slight differences in the level of residual repression observed in the different assays and promoter contexts (Fig. 6). This suggests that promoter context and the availability of cofactors have some influence and that the cleft may have a role in contacting effectors in some contexts (Fig. 1A).
Our work with the cleft mutant proteins is also relevant to the role of inhibitors, such as the cellular protein Pinin (1) and adenovirus protein E1A (42, 43). These might negatively regulate CtBP recruitment or activity by competing for the PIDLS motif binding cleft. The A58E and V72R fusions should be immune to such competitors and might be expected to be superrepressive if cellular inhibitors containing PIDLS motifs were abundant and active in the cell types we used for these experiments. The A58E and V72R fusions were found to display repression activity similar to but slightly less intense than that of wild-type CtBP2, arguing against endogenous competitive inhibitors playing a major role in our systems. Such inhibitors may, however, block recruitment of CtBP. The cleft-filled mutant derivatives were expressed at lower levels than wild-type CtBP2, so it is difficult to interpret the results obtained with these constructs (data not shown). Nevertheless, it is clear that they were also not superrepressive, again arguing against the presence of major inhibitory activities in the cellular systems we tested.
This absence of superrepression in the cleft-filled mutant form is also of interest because it suggests that a model whereby CtBP becomes functional in transcriptional corepresssion only after docking to a PIDLS partner and presenting a modified molecular surface (including the partner protein) is not likely to be valid. This result fits with the observation from the crystal structure that the PIDLS peptide does not significantly alter the overall structure of the protein (32).
Acknowledgments
We thank J. Hildebrand for the CtBP knockout cells, L. Gaudreau and M. Ptashne for the pGL2-(Gal4)5-(LexA)2-E1B-Luc and LexA-VP16 mammalian expression plasmids, D. Corda for rCtBP1-S, D. Wotton for hCtBP1-L, J. Chen for mFHL3 expression vectors, and S. Sugrue for the E-cadherin-Luc promoter.
K.Q. and A.K. are supported by Australian Postgraduate Awards. S.L. is supported by an International Postgraduate Award. M.B. is grateful to the Italian MIUR FIRB project, AIRC, and Fondazione Cariplo for continuous support. This work was supported by NIH grant NHLBI HL073443-02 and a grant from the Australian NHMRC to M.C.
Footnotes
Published ahead of print on 28 August 2006.
REFERENCES
- 1.Alpatov, R., G. C. Munguba, P. Caton, J. H. Joo, Y. Shi, Y. Shi, M. E. Hunt, and S. P. Sugrue. 2004. Nuclear speckle-associated protein Pnn/DRS binds to the transcriptional corepressor CtBP and relieves CtBP-mediated repression of the E-cadherin gene. Mol. Cell. Biol. 24:10223-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balasubramanian, P., L. J. Zhao, and G. Chinnadurai. 2003. Nicotinamide adenine dinucleotide stimulates oligomerization, interaction with adenovirus E1A and an intrinsic dehydrogenase activity of CtBP. FEBS Lett. 537:157-160. [DOI] [PubMed] [Google Scholar]
- 3.Barnes, C. J., R. K. Vadlamudi, S. K. Mishra, R. H. Jacobson, F. Li, and R. Kumar. 2003. Functional inactivation of a transcriptional corepressor by a signaling kinase. Nat. Struct. Biol. 10:622-628. [DOI] [PubMed] [Google Scholar]
- 4.Brannon, M., J. D. Brown, R. Bates, D. Kimelman, and R. T. Moon. 1999. XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development 126:3159-3170. [DOI] [PubMed] [Google Scholar]
- 5.Chinnadurai, G. 2006. CtBP family proteins: unique transcriptional regulators in the nucleus with diverse cytosolic functions. In G. Chinnadurai (ed.), CtBP family proteins. Landes Biosciences, Georgetown, Tex. [Online.] http://eurekah.com/chapter.php?chapid=2824&bookid=198&catid=30.
- 6.Chinnadurai, G. 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell 9:213-224. [DOI] [PubMed] [Google Scholar]
- 7.Chu, P. H., P. Ruiz-Lozano, Q. Zhou, C. Cai, and J. Chen. 2000. Expression patterns of FHL/SLIM family members suggest important functional roles in skeletal muscle and cardiovascular system. Mech. Dev. 95:259-265. [DOI] [PubMed] [Google Scholar]
- 8.Corda, D., A. Colanzi, and A. Luini. 2006. The multiple activities of CtBP/BARS proteins: the Golgi view. Trends Cell Biol. 16:167-173. [DOI] [PubMed] [Google Scholar]
- 9.Criqui-Filipe, P., C. Ducret, S. M. Maira, and B. Wasylyk. 1999. Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and de-acetylation. EMBO J. 18:3392-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deltour, S., S. Pinte, C. Guerardel, B. Wasylyk, and D. Leprince. 2002. The human candidate tumor suppressor gene HIC1 recruits CtBP through a degenerate GLDLSKK motif. Mol. Cell. Biol. 22:4890-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandes, I., Y. Bastien, T. Wai, K. Nygard, R. Lin, O. Cormier, H. S. Lee, F. Eng, N. R. Bertos, N. Pelletier, S. Mader, V. K. Han, X. J. Yang, and J. H. White. 2003. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol. Cell 11:139-150. [DOI] [PubMed] [Google Scholar]
- 12.Furusawa, T., H. Moribe, H. Kondoh, and Y. Higashi. 1999. Identification of CtBP1 and CtBP2 as corepressors of zinc finger-homeodomain factor ΔEF1. Mol. Cell. Biol. 19:8581-8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grooteclaes, M., Q. Deveraux, J. Hildebrand, Q. Zhang, R. H. Goodman, and S. M. Frisch. 2003. C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc. Natl. Acad. Sci. USA 100:4568-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grooteclaes, M. L., and S. M. Frisch. 2000. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene 19:3823-3828. [DOI] [PubMed] [Google Scholar]
- 15.Hildebrand, J. D. 2006. CtBP proteins in vertebrate development. In G. Chinnadurai (ed.), CtBP family proteins. Landes Biosciences, Georgetown, Tex. [Online.] http://eurekah.com/chapter.php?chapid=2696&bookid=198&catid=30.
- 16.Hildebrand, J. D., and P. Soriano. 2002. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol. Cell. Biol. 22:5296-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes, M., J. Turner, A. Fox, O. Chisholm, M. Crossley, and B. Chong. 1999. hFOG-2, a novel zinc finger protein, binds the co-repressor mCtBP2 and modulates GATA-mediated activation. J. Biol. Chem. 274:23491-23498. [DOI] [PubMed] [Google Scholar]
- 18.Izutsu, K., M. Kurokawa, Y. Imai, K. Maki, K. Mitani, and H. Hirai. 2001. The corepressor CtBP interacts with Evi-1 to repress transforming growth factor beta signaling. Blood 97:2815-2822. [DOI] [PubMed] [Google Scholar]
- 19.Kaczynski, J., T. Cook, and R. Urrutia. 2003. Sp1- and Krüppel-like transcription factors. Genome Biol. 4:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kagey, M. H., T. A. Melhuish, and D. Wotton. 2003. The polycomb protein Pc2 is a SUMO E3. Cell 113:127-137. [DOI] [PubMed] [Google Scholar]
- 21.Katz, S. G., A. B. Cantor, and S. H. Orkin. 2002. Interaction between FOG-1 and the corepressor C-terminal binding protein is dispensable for normal erythropoiesis in vivo. Mol. Cell. Biol. 22:3121-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kegel, K. B., A. R. Meloni, Y. Yi, Y. J. Kim, E. Doyle, B. G. Cuiffo, E. Sapp, Y. Wang, Z. H. Qin, J. D. Chen, J. R. Nevins, N. Aronin, and M. DiFiglia. 2002. Huntingtin is present in the nucleus, interacts with the transcriptional corepressor C-terminal binding protein, and represses transcription. J. Biol. Chem. 277:7466-7476. [DOI] [PubMed] [Google Scholar]
- 23.Kim, S. C., Y. S. Kim, and A. M. Jetten. 2005. Krüppel-like zinc finger protein Gli-similar 2 (Glis2) represses transcription through interaction with C-terminal binding protein 1 (CtBP1). Nucleic Acids Res. 33:6805-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koipally, J., and K. Georgopoulos. 2002. Ikaros-CtIP interactions do not require CtBP and participate in a deacetylase-independent mode of repression. J. Biol. Chem. 277:23143-23149. [DOI] [PubMed] [Google Scholar]
- 25.Koipally, J., and K. Georgopoulos. 2000. Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J. Biol. Chem. 275:19594-19602. [DOI] [PubMed] [Google Scholar]
- 26.Koipally, J., and K. Georgopoulos. 2002. A molecular dissection of the repression circuitry of Ikaros. J. Biol. Chem. 277:27697-27705. [DOI] [PubMed] [Google Scholar]
- 27.Kumar, V., J. E. Carlson, K. A. Ohgi, T. A. Edwards, D. W. Rose, C. R. Escalante, M. G. Rosenfeld, and A. K. Aggarwal. 2002. Transcription corepressor CtBP is an NAD+-regulated dehydrogenase. Mol. Cell 10:857-869. [DOI] [PubMed] [Google Scholar]
- 28.Lee, M. G., C. Wynder, N. Cooch, and R. Shiekhattar. 2005. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437:432-435. [DOI] [PubMed] [Google Scholar]
- 29.Lin, X., B. Sun, M. Liang, Y. Y. Liang, A. Gast, J. Hildebrand, F. C. Brunicardi, F. Melchior, and X. H. Feng. 2003. Opposed regulation of corepressor CtBP by SUMOylation and PDZ binding. Mol. Cell 11:1389-1396. [DOI] [PubMed] [Google Scholar]
- 30.Meloni, A. R., E. J. Smith, and J. R. Nevins. 1999. A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc. Natl. Acad. Sci. USA 96:9574-9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murakami, A., S. Ishida, J. Thurlow, J. M. Revest, and C. Dickson. 2001. SOX6 binds CtBP2 to repress transcription from the Fgf-3 promoter. Nucleic Acids Res. 29:3347-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nardini, M., S. Spano, C. Cericola, A. Pesce, A. Massaro, E. Millo, A. Luini, D. Corda, and M. Bolognesi. 2003. CtBP/BARS: a dual-function protein involved in transcription co-repression and Golgi membrane fission. EMBO J. 22:3122-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nardini, M., D. Svergun, P. V. Konarev, S. Spano, M. Fasano, C. Bracco, A. Pesce, A. Donadini, C. Cericola, F. Secundo, A. Luini, D. Corda, and M. Bolognesi. 2006. The C-terminal domain of the transcriptional co-repressor CtBP is intrinsically unstructured. Protein Sci. 15:1042-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nibu, Y., H. Zhang, E. Bajor, S. Barolo, S. Small, and M. Levine. 1998. dCtBP mediates transcriptional repression by Knirps, Krüppel and Snail in the Drosophila embryo. EMBO J. 17:7009-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nibu, Y., H. Zhang, and M. Levine. 1998. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science 280:101-104. [DOI] [PubMed] [Google Scholar]
- 36.Nibu, Y., H. Zhang, and M. Levine. 2001. Local action of long-range repressors in the Drosophila embryo. EMBO J. 20:2246-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmer, S., J. P. Brouillet, A. Kilbey, R. Fulton, M. Walker, M. Crossley, and C. Bartholomew. 2001. Evi-1 transforming and repressor activities are mediated by CtBP co-repressor proteins. J. Biol. Chem. 276:25834-25840. [DOI] [PubMed] [Google Scholar]
- 38.Perdomo, J., and M. Crossley. 2002. The Ikaros family protein Eos associates with C-terminal-binding protein corepressors. Eur. J. Biochem. 269:5885-5892. [DOI] [PubMed] [Google Scholar]
- 39.Perdomo, J., A. Verger, J. Turner, and M. Crossley. 2005. Role for SUMO modification in facilitating transcriptional repression by BKLF. Mol. Cell. Biol. 25:1549-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Postigo, A. A., and D. C. Dean. 1999. ZEB represses transcription through interaction with the corepressor CtBP. Proc. Natl. Acad. Sci. USA 96:6683-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riefler, G. M., and B. L. Firestein. 2001. Binding of neuronal nitric-oxide synthase (nNOS) to carboxyl-terminal-binding protein (CtBP) changes the localization of CtBP from the nucleus to the cytosol: a novel function for targeting by the PDZ domain of nNOS. J. Biol. Chem. 276:48262-48268. [DOI] [PubMed] [Google Scholar]
- 42.Schaeper, U., J. M. Boyd, S. Verma, E. Uhlmann, T. Subramanian, and G. Chinnadurai. 1995. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. USA 92:10467-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaeper, U., T. Subramanian, L. Lim, J. M. Boyd, and G. Chinnadurai. 1998. Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J. Biol. Chem. 273:8549-8552. [DOI] [PubMed] [Google Scholar]
- 44.Senyuk, V., K. K. Sinha, and G. Nucifora. 2005. Corepressor CtBP1 interacts with and specifically inhibits CBP activity. Arch. Biochem. Biophys. 441:168-173. [DOI] [PubMed] [Google Scholar]
- 45.Sewalt, R. G., M. J. Gunster, J. van der Vlag, D. P. Satijn, and A. P. Otte. 1999. C-terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol. Cell. Biol. 19:777-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi, Y., F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, R. A. Casero, and Y. Shi. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941-953. [DOI] [PubMed] [Google Scholar]
- 47.Shi, Y., J. Sawada, G. Sui, B. Affar el, J. R. Whetstine, F. Lan, H. Ogawa, M. P. Luke, Y. Nakatani, and Y. Shi. 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422:735-738. [DOI] [PubMed] [Google Scholar]
- 48.Shi, Y., and Y. Shi. 2006. CtBP corepressor complex—a multienzyme machinery that coordinates chromatin modifications. In G. Chinnadurai (ed.), CtBP family proteins. Landes Biosciences, Georgetown, Tex. [Online.] http://eurekah.com/chapter.php?chapid=2751&bookid=198&catid=30.
- 49.Spano, S., C. H. Carcedo, and D. Corda. 2006. CtBP3/BARS and membrane fission. In G. Chinnadurai (ed.), CtBP family proteins. Landes Biosciences, Georgetown, Tex. [Online.] http://eurekah.com/chapter.php?chapid=2638&bookid=198&catid=30.
- 50.Spano, S., M. G. Silletta, A. Colanzi, S. Alberti, G. Fiucci, C. Valente, A. Fusella, M. Salmona, A. Mironov, A. Luini, D. Corda, and S. Spanfo. 1999. Molecular cloning and functional characterization of brefeldin A-ADP-ribosylated substrate. A novel protein involved in the maintenance of the Golgi structure. J. Biol. Chem. 274:17705-17710. [DOI] [PubMed] [Google Scholar]
- 51.Srinivasan, L., and M. L. Atchison. 2004. YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev. 18:2596-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramanian, T., and G. Chinnadurai. 2003. Association of class I histone deacetylases with transcriptional corepressor CtBP. FEBS Lett. 540:255-258. [DOI] [PubMed] [Google Scholar]
- 53.Sundqvist, A., K. Sollerbrant, and C. Svensson. 1998. The carboxy-terminal region of adenovirus E1A activates transcription through targeting of a C-terminal binding protein-histone deacetylase complex. FEBS Lett. 429:183-188. [DOI] [PubMed] [Google Scholar]
- 54.Thio, S. S., J. V. Bonventre, and S. I. Hsu. 2004. The CtBP2 co-repressor is regulated by NADH-dependent dimerization and possesses a novel N-terminal repression domain. Nucleic Acids Res. 32:1836-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tom Dieck, S., F. Schmitz, and J. H. Brandstatter. 2006. CtBPs as synaptic proteins. In G. Chinnadurai (ed.), CtBP family proteins. Landes Biosciences, Georgetown, Tex. [Online.] http://eurekah.com/chapter.php?chapid=2646&bookid=198&catid=30.
- 56.Tripathi, M. K., S. Misra, S. V. Khedkar, N. Hamilton, C. Irvin-Wilson, C. Sharan, L. Sealy, and G. Chaudhuri. 2005. Regulation of BRCA2 gene expression by the SLUG repressor protein in human breast cells. J. Biol. Chem. 280:17163-17171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turner, J., and M. Crossley. 1998. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Krüppel-like factor and other mammalian transcriptional regulators. EMBO J. 17:5129-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turner, J., and M. Crossley. 2001. The CtBP family: enigmatic and enzymatic transcriptional co-repressors. Bioessays 23:683-690. [DOI] [PubMed] [Google Scholar]
- 59.Turner, J., H. Nicholas, D. Bishop, J. M. Matthews, and M. Crossley. 2003. The LIM protein FHL3 binds basic Krüppel-like factor/Krüppel-like factor 3 and its co-repressor C-terminal-binding protein 2. J. Biol. Chem. 278:12786-12795. [DOI] [PubMed] [Google Scholar]
- 60.Ueda, J., M. Tachibana, T. Ikura, and Y. Shinkai. 2006. Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. J. Biol. Chem. 281:20120-20128. [DOI] [PubMed] [Google Scholar]
- 61.Valenta, T., J. Lukas, and V. Korinek. 2003. HMG box transcription factor TCF-4's interaction with CtBP1 controls the expression of the Wnt target Axin2/Conductin in human embryonic kidney cells. Nucleic Acids Res. 31:2369-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vo, N., C. Fjeld, and R. H. Goodman. 2001. Acetylation of nuclear hormone receptor-interacting protein rip140 regulates binding of the transcriptional corepressor CtBP. Mol. Cell. Biol. 21:6181-6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wen, Y., D. Nguyen, Y. Li, and Z. C. Lai. 2000. The N-terminal BTB/POZ domain and C-terminal sequences are essential for Tramtrack69 to specify cell fate in the developing Drosophila eye. Genetics 156:195-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, C. L., T. A. McKinsey, J. R. Lu, and E. N. Olson. 2001. Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J. Biol. Chem. 276:35-39. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, Q., Y. Yoshimatsu, J. Hildebrand, S. M. Frisch, and R. H. Goodman. 2003. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell 115:177-186. [DOI] [PubMed] [Google Scholar]