Abstract
The founding member of the inhibitor of apoptosis protein (IAP) family was originally identified as a cell death inhibitor. However, recent evidence suggests that IAPs are multifunctional signaling devices that influence diverse biological processes. To investigate the in vivo function of Drosophila melanogaster IAP2, we have generated diap2 null alleles. diap2 mutant animals develop normally and are fully viable, suggesting that diap2 is dispensable for proper development. However, these animals were acutely sensitive to infection by gram-negative bacteria. In Drosophila, infection by gram-negative bacteria triggers the innate immune response by activating the immune deficiency (imd) signaling cascade, a NF-κB-dependent pathway that shares striking similarities with the pathway of mammalian tumor necrosis factor receptor 1 (TNFR1). diap2 mutant flies failed to activate NF-κB-mediated expression of antibacterial peptide genes and, consequently, rapidly succumbed to bacterial infection. Our genetic epistasis analysis places diap2 downstream of or in parallel to imd, Dredd, Tak1, and Relish. Therefore, DIAP2 functions in the host immune response to gram-negative bacteria. In contrast, we find that the Drosophila TNFR-associated factor (Traf) family member Traf2 is dispensable in resistance to gram-negative bacterial infection. Taken together, our genetic data identify DIAP2 as an essential component of the Imd signaling cascade, protecting the organism from infiltrating microbes.
Drosophila melanogaster lacks an adaptive immune system and relies exclusively on innate immune reactions for its defense against microbial infection. Activation of the innate immune response leads to the expression of hundreds of genes, some of which encode potent antimicrobial peptides that are synthesized in immunocompetent tissues, such as the tracheal epithelium, circulating “blood” cells, and fat body, the analogue of the mammalian liver (19).
Depending on the infecting microbe, Drosophila activates the Toll or immune deficiency (Imd) signaling pathway. Exposure to fungi or gram-positive bacteria activates a serine-protease cascade, through pattern recognition molecules (10), that triggers cleavage and activation of Spatzle. Spatzle, in turn, binds and activates the transmembrane Toll receptor, which engages an intracellular signaling cascade that results in nuclear translocation of the NF-κB-like transcription factors Dif and Dorsal. Dif and Dorsal then induce expression of drosomycin, a potent antifungal peptide. In contrast, the Imd pathway is activated in response to gram-negative bacteria. Diaminopimelic acid-type peptidoglycan (DAP-PG), a major component of the gram-negative bacterial cell wall, is recognized by the pattern recognition protein peptidoglycan recognition protein LE (PGRP-LE) and the transmembrane receptor PGRP-LC. Upon binding to DAP-PG, PGRP-LC triggers nuclear translocation of the NF-κB-like transcription factor Relish, which, among others, induces expression of the antibacterial peptide gene Diptericin via the Imd pathway (20, 38).
The Imd pathway shares striking similarities with the tumor necrosis factor receptor 1 (TNFR1) signaling cascade (48). Following exposure to pathogens, PGRP-LC activates Imd, which carries a C-terminal death domain that is similar to the domain of the mammalian adaptor protein receptor-interacting protein 1 (RIP1) (11). Through this death domain, Imd recruits dFADD and the Drosophila caspase 8 orthologue Dredd to the PGRP-LC receptor (4, 28, 29, 33). Microbe-driven complex formation triggers activation of the Drosophila mitogen-activated protein kinase kinase kinase Tak1 and the Relish kinase complex Ird5/Kenny (IκB kinase β [IKKβ]/IKKγ complex) (31, 39, 44, 51). The similarities between Imd and TNFR1 signaling also extend to ubiquitin-mediated activation of IKK (3). As in mammals, Drosophila Ubc13(Bendless)/UEV1A, an E2 ubiquitin-protein conjugase complex that promotes K63-linked polyubiquitylation, is required for the activation of Tak1 and the IKK complex (57). Relish activation requires at least two posttranslational modifications, phosphorylation and Dredd-dependent proteolytic cleavage (44-46). These changes enable translocation of Relish to the nucleus and expression of antibacterial peptide genes. However, recent in vivo evidence suggests that Tak1-mediated Jun N-terminal protein kinase (JNK) activation, in addition to Relish activation, is required for antimicrobial peptide gene expression (9). These data are consistent with a model whereby Imd signaling bifurcates at the level of Tak1, which activates both JNK and IKK signaling. Thus, a cooperative input from JNK and NF-κB signaling seems to be required for full induction of antibacterial peptide gene expression in response to bacterial infection (see Fig. 8). Loss-of-function mutations in any of the components of the Imd signaling cascade result in the same immune deficiency phenotype, in which animals become acutely susceptible to infection by gram-negative bacteria. Common to all these mutants is their failure to induce expression of antibacterial peptide genes and, therefore, to fend off bacterial infection (38).
FIG. 8.

Model of Imd signaling based on genetic epistasis data in vivo. Our genetic epistasis analysis places diap2 downstream of or in parallel to imd, Dredd, Tak1, and Relish. Intriguingly, loss of diap2 copies the phenotype of Tak1 mutant animals, since Diptericin induction, following enforced expression of imd and Dredd, is also blocked in Tak1 mutant animals, while this is not the case in kenny and ird5 mutant flies. The Imd signal transduction pathway bifurcates at the level of Tak1, which is required for the activation of the NF-κB signaling branch as well as the JNK signaling branch, both of which are necessary for expression of antibacterial peptide genes in the fat body. We currently favor the model whereby diap2 functions genetically at the level of Tak1. This view is supported by recent reports of Drosophila tissue culture cells which suggest that DIAP2 is required for Tak1-mediated JNK activation. Arrows indicate genetic interactions that rely on overexpression of individual components of the Imd pathway in vivo. Note that the ability to induce Diptericin expression varied substantially among heat shock-induced overexpression of imd, Dredd, Tak1, and Relish (see Fig. 6 for more details). AMPs, antimicrobial peptides.
Signaling through the mammalian TNFR1 also results in the recruitment of cellular inhibitor of apoptosis protein 1 (c-IAP1) and c-IAP2, two members of the evolutionarily conserved IAP family (37, 41). Although IAPs were originally identified as inhibitor of apoptosis proteins (7), recent evidence suggests that IAPs also fulfill functions that operate independently of their ability to control caspases and cell death (50). Thus, c-IAP1 and c-IAP2, through their ability to bind to TNFR1, TNFR-associated factor 2 (TRAF2), and RIP1, are implicated in modulating TNFR1 signaling. However, due to redundancy or compensatory mechanisms among these IAPs, mutant animals did not display aberrant TNFR1 signaling (5, 6, 42). Recent genome-wide RNA interference (RNAi) screens in cultured cells as well as in vivo RNAi analysis identified the Drosophila inhibitor of apoptosis 2 (DiAP2) as a potential component of the Drosophila Imd pathway (12, 23). However, due to lack of diap2 mutant animals, the physiological role of DIAP2 for Drosophila immune responses is not fully established.
To investigate the in vivo function of DIAP2, we have generated diap2 null alleles. diap2 mutant animals develop normally and are fully viable, suggesting that diap2 is dispensable for proper development. However, these animals were acutely sensitive to infection by gram-negative bacteria. Consistently, diap2 mutant flies failed to induce expression of the antibacterial peptide genes and, hence, to mount a proper innate immune response. Thus, our data unambiguously demonstrate that DIAP2 is an essential component of the Imd signaling cascade in vivo, protecting flies from microbial infection.
MATERIALS AND METHODS
Fly stocks.
OregonR and CantonS flies were used as wild-type controls. spatzlerm7, Tak11, RelishE20, and Traf2Ex1 alleles, Hsp-GAL4, Act5c-GAL4, Da-GAL4 drivers, upstream activation sequence (UAS)-imd, hsp-GAL4, UAS-Tak1, UAS-Relish (full-length) and UAS-Dredd transgenic flies were described previously (2, 18, 29, 51). The EP(G2326) line was purchased from Genexel Inc. (Daejeon, South Korea). Df(2R)exel7138 spans the diap2 locus (35). diap27c and diap27a alleles were generated by transposase-mediated imprecise excision of EP(G2326) and mapped as indicated below. The UAS-diap2 construct was generated by cloning the diap2 open reading frame in EcoRI/XhoI-digested pUAST vector. w1118 flies were used to generate UAS-diap2 transgenic flies. An insertion of this construct on the third chromosome was used in this study. Drosophila stocks and crosses were maintained at 25°C. Following septic injury, flies were incubated at either 25°C for quantitative reverse transcription-PCR (RT-PCR) analysis or 29°C for survival assays. Heat shock-mediated induction of imd, Tak1, Relish, and Dredd overexpression was performed at 37°C for 1 h, followed by a recovery phase at 25°C.
Analysis of genomic lesions.
Genomic DNA from homozygous diap27c and diap27a flies was extracted from an adult individual as described previously (13). Five microliters of genomic DNA was used for PCR amplification using Easy-A High-Fidelity PCR cloning enzyme (Stratagene, United Kingdom). The following oligonucleotide primers were used to amplify the diap2 locus: 5′-CGGGGCACATCACTTGAAGACCG-3′ and 5′-GGCATTGCCCATGGGCTTAAGC-3′). The resulting PCR product was purified, cloned into pGEMt vector (Promega), and analyzed by DNA sequencing.
Immunoblot analysis.
Protein extracts were prepared from five adults or third-instar larvae by snap-freezing, homogenizing, and boiling in Laemmli buffer. Protein samples were analyzed by immunoblotting with anti-DIAP1 (55), anti-DIAP2, and antitubulin antibodies (Sigma, United Kingdom). Anti-DIAP2 was generated in rabbit using a purified, recombinant DIAP2 fragment spanning the baculovirus IAP repeat 3 (BIR3) region (amino acids 215 to 281). For immunoblot detection and quantification of signals, Odyssey technology was used according to the manufacturer's instructions (Licor Biosciences, United Kingdom).
Bacterial strains, infection experiments, and survival analysis.
Microbial septic injuries were performed by pricking third-instar larvae in the posterior region or adults in the lateral part of the thorax with a thin needle previously dipped into a concentrated (optical density of ≈200) culture of Erwinia carotovora subsp. carotovora 15, Micrococcus luteus, Enterococcus faecalis, or Candida albicans. For natural infection by Erwinia carotovora subsp. carotovora 15, Drosophila third-instar larvae were exposed to a mixture of crushed banana and bacteria as described previously (1). For the survival experiments, flies were examined at different time points to monitor survival after septic injury. The infected flies were transferred to fresh vials daily. The experiments were performed using at least 40 flies for each genotype.
Quantitative real-time PCR.
For quantitative analysis of Attacin-A, Cecropin-A1, Defensin, Diptericin, Drosocin, Drosomycin, Metchnikowin, and rp49 mRNA expression, RNA was extracted from whole animals using RNA TRIzol (Invitrogen). cDNAs were synthesized using SuperScript II (Invitrogen) and quantitative PCR was performed using double-stranded DNA dye SYBR green I (Roche Diagnostics). Primer pairs were as follows: for Attacin-A, sense, 5′-CCCGGAGTGAAGGATG-3′, antisense, 5′-GTTGCTGTGCGTCAAG-3′; for Cecropin-A1, sense, 5′-GAACTTCTACAACATCTTCGT-3′, antisense, 5′TCCCAGTCCCTGGATT-3′; for Defensin, sense, 5′-GTTCTTCGTTCTCGTGG-3′, antisense, 5′-CTTTGAACCCCTTGGC-3′; for Diptericin, sense, 5′-GCTGCGCAATCGCTTCTACT-3′, antisense, 5′-TGGTGGAGTGGGCTTCATG-3′; for Drosocin, sense, 5′-CCATCGTTTTCCTGCT-3′, antisense, 5′-CTTGAGTCAGGTGATCC-3′; for Drosomycin, sense, 5′-CGTGAGAACCTTTTCCAATATGATG-3′, antisense, 5′-TCCCAGGACCACCAGCAT-3′; for Metchnikowin, sense, 5′-AACTTAATCTTGGAGCGA-3′, antisense, 5′-CGGTCTTGGTTGGTTAG-3′; and for rp49, sense, 5′-GACGCTTCAAGGGACAGTATCTG-3′, antisense, 5′-AAACGCGGTTCTGCATGAG-3′. SYBR green analysis was performed on a Lightcycler (Roche Diagnostics). The amount of mRNA detected was normalized to control rp49 mRNA values. We used normalized data to quantify the relative levels of a given mRNA according to cycling threshold analysis.
RESULTS
Generation of diap2 null mutant flies.
To elucidate the in vivo function of DIAP2, we created a null allele of diap2 by screening for imprecise excisions of an existing P element, EP(G2326) (Genexel Inc.) (Fig. 1A). EP(G2326) is inserted at position 11449676 (Drosophila genome release 4.2.1), which is located 12 nucleotides (nt) 3′ of the transcriptional start site of the diap2-RB transcript, and 69 nt 5′ of the transcriptional start site of the diap2-RA transcript. We obtained two mutants, diap27a and diap27c, which carried deletions that removed the start codon and extended 371 nt (diap27a) and 735 nt (diap27c) into the coding region of DIAP2. Thus, diap27a lacks the entire first exon (778-bp genomic deletion, from 11449676 to 11448898), while diap27c lacks exon one and most of exon two (1,357-bp genomic deletion, from 11449676 to 11448319) (Fig. 1A). Immunoblot analysis with an anti-DIAP2 antibody, raised against the BIR3 domain of DIAP2, indicated that DIAP2 protein was not detectable in diap27a and diap27c homozygous, diap27a/diap27c transheterozygous, or diap27c/Def(2R)exel7138 (diap27c/def) hemizygous mutant animals. In contrast, DIAP2 protein was readily detectable in animals of the parental EP(G2326) line as well as in diap27c/+ or def/+ individuals (Fig. 1B and data not shown). Importantly, the deletions in diap27a and diap27c did not affect the transcriptional start site and open reading frame of the nearby gene CG8297 (Fig. 1A and data not shown). Taken together, these results suggest that diap27c and diap27a are null alleles of diap2.
FIG. 1.
Generation of DIAP2-deficient flies. (A) Imprecise excision of EP(G2326) created diap27c and diap27a alleles that carry deletions, which removed large portions of the diap2 locus. Schematic representation depicting the diap2 locus, the insertion site and orientation of EP(G2326), and the genomic DNA. The positions of the primers used to clone the respective genomic DNA fragments are indicated. (B) DIAP2 protein levels were undetectable in diap2 mutant flies. The presence of DIAP2 protein was examined by immunoblot analysis using anti-DIAP2 antibodies. Protein extracts from the following genotypes were used to monitor DIAP2 expression: diap27c/Cyo-actGFP (diap27c/+, third-instar larvae [L3]) (lane 1), diap27c (third-instar larvae) (lane 2), CantonS (wild type [WT], adult) (lane 3), EP(G2326) (adult) (lane 4), diap27c/Df(2R)exel7138 (diap27c/def, adult) (lane 5), diap27c (adult) (lane 6), diap27a (adult) (lane 7), or diap27a/diap27c (adult) (lane 8). Antitubulin immunoblot analysis was used to determine equal protein loading. (C) DIAP1 protein levels remain unchanged in diap2 mutant flies. The level of DIAP1 protein was examined by immunoblot analysis using anti-DIAP1 and antitubulin antibodies. Quantification of signals was performed using the LICOR system. Protein extracts from the following genotypes were analyzed: CantonS (WT, lane 1), diap27c (lane 2), diap27a (lane 3), or diap27a/diap27c (lane 4).
Knockout studies and biochemical characterization of mammalian IAPs have revealed homeostatic cross-regulation among certain IAPs, whereby loss of one IAP can cause compensatory upregulation of family members (6, 15, 42). To address whether loss of DIAP2 resulted in increased levels of DIAP1, we analyzed DIAP1 protein levels in diap2 mutant flies by quantitative Western blot analysis. As shown in Fig. 1C, DIAP1 levels remained unchanged in diap2 mutant flies compared to wild-type flies.
Loss of diap2 renders flies susceptible to septic injury with gram-negative bacteria.
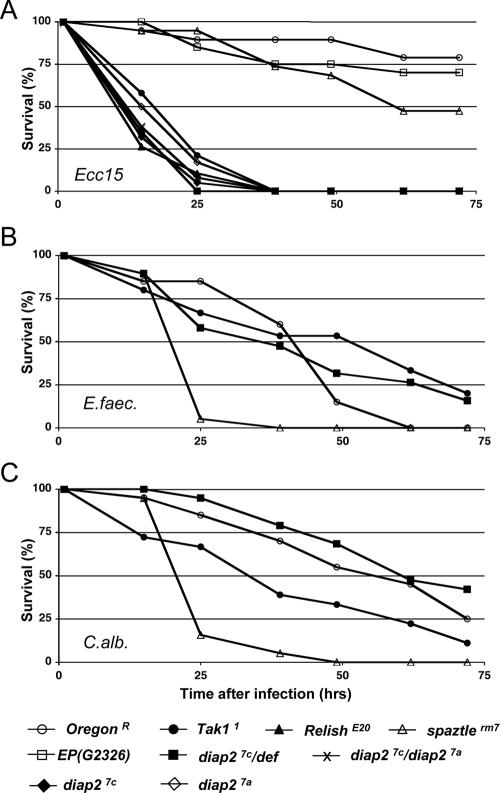
Most diap27c/def hemizygous mutant individuals survived embryogenesis and developed normally (86% survival, n = 506 for diap27c/def). Thus, in contrast to diap1 mutant animals that die early during embryogenesis with deregulated caspase activity (14, 30, 54), loss of zygotic expression of diap2 did not confer such a phenotype. Although diap2 mutants showed no obvious developmental defects, we noticed that diap2 mutant flies were acutely sensitive to infections. To investigate the potential implication of diap2 in the regulation of Drosophila immune response in vivo, we analyzed the survival profile of diap2 mutant flies in different models of microbial infection by septic injury, an established system to analyze Drosophila immune phenotypes (49). To this end, we infected wild-type and mutant flies with the gram-negative bacterium Erwinia carotovora subsp. carotovora 15 (Fig. 2A), the gram-positive bacterium Enterococcus faecalis (Fig. 2B), or the fungus Candida albicans (Fig. 2C). Infection by gram-negative bacteria activates the Imd signal transduction pathway, which results in the expression of antibacterial peptide genes. Flies with mutations in Tak1 (Tak11) and Relish (RelishE20), two components of the Imd pathway, failed to mount such an Imd response and consequently succumbed to infection by E. carotovora subsp. carotovora 15 (Fig. 2A) (51). By contrast, a mutation in the spatzle gene (spatzlerm7), which blocks Toll activation, sensitized animals only to infection by gram-positive bacteria and fungi (Fig. 2B and C) (25, 40). Interestingly, we found that, similar to Tak11 and RelishE20 mutant flies, diap2 mutant individuals (diap27c/def) were highly susceptible to injection of the gram-negative bacterium E. carotovora subsp. carotovora 15 (Fig. 2A) (51), but not gram-positive bacteria (Fig. 2B) and fungi (Fig. 2C) (36). The survival rate of diap27c/def hemizygous mutant animals was almost identical to those of diap27c and diap27a homozygous or diap27a/diap27c transheterozygous mutant animals (Fig. 2A), establishing that diap27c and diap27a are genetically null alleles. Moreover, flies from the parental EP(G2326) line, which was used to generate the diap27c and diap27a alleles, showed no susceptibility to E. carotovora subsp. carotovora 15 injection (Fig. 2A). This confirms that the observed immune deficiency phenotype relies on the deletion generated by imprecise excision of EP(G2326).
FIG. 2.
DIAP2 is required to resist gram-negative bacterial infection. The survival rates of adult males in response to different types of septic injuries are presented. Animals were pricked with a needle previously dipped into Erwinia carotovora subsp. carotovora 15 (Ecc15) (A), Enterococcus faecalis (E.faec.) (B), or Candida albicans (C.alb.) (C). The following genotypes were examined for susceptibility to microbes: wild-type (OregonR), Tak11, RelishE20, spatzlerm7, EP(G2326), diap27c/def, diap27c, diap27a, and diap27c/diap27a. Note that diap2 mutant flies behaved as Tak1 and Relish mutant flies, which are known to be highly susceptible to Erwinia carotovora subsp. carotovora 15 infection (A), but not to Enterococcus faecalis (B) or Candida albicans (C) infection. Both diap2 alleles, diap27c and diap27a, hemizygous (diap27c/def) or transheterozygous flies (diap27c/diap27a) showed similar susceptibility to E. carotovora subsp. carotovora 15 infection, while animals of the parental EP(G2326) line were fully resistant (A).
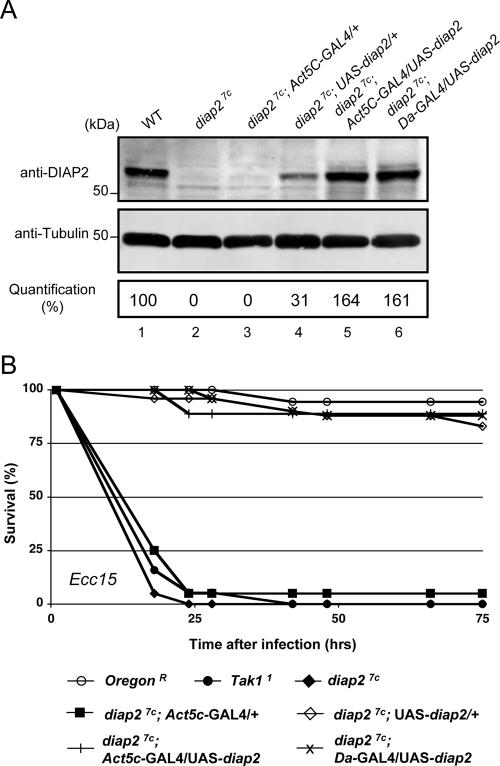
Next, we addressed whether the observed susceptibility to E. carotovora subsp. carotovora 15 infection in diap2 mutant flies was indeed due to mutation in diap2. We took advantage of the yeast UAS/GAL4 binary system to constitutively express a wild-type diap2 transgene in otherwise diap2 mutant flies. Expression of the UAS-diap2 transgene was driven by either the GAL4 driver Daughterless-GAL4 (Da-GAL4) or Actin5c-GAL4 (Act5c-GAL4), which express GAL4 constitutively in all cells. Quantitative Western blot analysis was used to determine diap2 transgene expression levels in otherwise diap27c homozygous mutant flies. While diap27c mutant animals were devoid of DIAP2 protein, “leaky” expression (in the absence of a GAL4 driver) of the UAS-diap2 transgene resulted in small, but significant, amounts of DIAP2 protein levels (31% of the DIAP2 protein levels observed in wild-type flies [Fig. 3A, lane 4]). In contrast, Act5c- or Da-GAL4-driven expression of UAS-diap2 resulted in DIAP2 protein levels that were 64% and 61% above wild-type levels (Fig. 3A, lanes 5 and 6). Intriguingly, the “leaky,” low level of DIAP2 transgene expression in diap27c;UAS-diap2/+ animals was sufficient to completely rescue the immunodeficient phenotype observed in diap27c mutant animals. Even in the absence of GAL4 drivers, diap27c;UAS-diap2/+ transgenic flies were fully resistant to infection by gram-negative bacteria and survived microbial exposure like their wild-type counterparts (Fig. 3B). Similarly, diap27c mutant flies with Act5c- or Da-GAL4-driven expression of UAS-diap2 were fully resistant to gram-negative bacterial infection, while diap27c flies died within 25 h of septic injury.
FIG. 3.
Ubiquitous expression of DIAP2 fully rescues the immune deficiency phenotype associated with diap27c. (A) Expression level of diap2 transgene in otherwise diap2 mutant flies. The presence of DIAP2 protein was examined by immunoblot analysis using anti-DIAP2 and antitubulin antibodies. Protein extracts from the following genotypes were used to monitor DIAP2 protein expression: CantonS (wild type [WT]) (lane 1), diap27c (lane 2), diap27c; Act5C-GAL4/+ (lane 3), diap27c; UAS-diap2/+ (lane 4), diap27c; UAS-diap2/Act5C-GAL4 (lane 5), and diap27c; UAS-diap2/Da-GAL4 (lane 6). Quantification of signals was performed using the LICOR system. (B) diap2 transgene expression rescued diap27c mutant flies from the lethal effects of Erwinia carotovora subsp. carotovora 15 (Ecc15)-mediated septic injury. The following genotypes were examined for susceptibility to microbes: wild type (OregonR), Tak11, diap27c, diap27c; Act5C-GAL4/+, diap27c; UAS-diap2/+, diap27c; Act5c-GAL4/UAS-diap2, and diap27c; Da-GAL4/UAS-diap2.
Taken together, these data demonstrate that mutations in diap2 are phenotypically similar to mutations in Tak1 and Relish and that these genes are essential for the resistance to infection by gram-negative bacteria but are dispensable to fend off gram-positive bacterial or fungal infections. Given that diap2 mutant animals mounted a normal response to infection by gram-positive bacteria and fungi, these data indicate that the immunoresponsive fat body is fully functional in diap2 mutants and that loss of diap2 does not generally affect fat body development or survival.
DIAP2 is essential for Imd-mediated expression of antibacterial peptide genes.
Infection by gram-negative bacteria triggers the Imd signal transduction pathway, which culminates in the transcriptional expression of immune genes, including those encoding antibacterial peptides (8). The expression of one such gene, Diptericin, has been established as a reliable and accurate readout to monitor Imd signaling in response to infection by gram-negative bacteria (24). On the other hand, gram-positive bacterial and fungal infections trigger Toll activation and induced expression of Drosomycin (25, 40). To test the activation of these pathways in adult diap2 mutant flies, we monitored Diptericin and Drosomycin expression by quantitative RT-PCR in response to septic injury with gram-negative (E. carotovora subsp. carotovora 15 [Fig. 4A ]) or gram-positive bacteria (Micrococcus luteus [Fig. 4B]). Consistent with the enhanced susceptibility to gram-negative bacteria, we found that diap2 mutant adult animals displayed a severely compromised immune response to gram-negative bacterial infection. Like Tak11 and RelishE20 mutant flies, diap2 mutant adult flies failed to induce expression of the Diptericin gene following E. carotovora subsp. carotovora 15 septic injury. A compromised Imd-mediated immune response was already apparent at the larval stage, since diap2 mutant larvae, like Tak11 and RelishE20 mutants, completely failed to induce Diptericin expression upon septic injury (data not shown). In contrast, diap2 mutant flies were normal in their ability to signal through the Toll pathway. M. luteus-mediated induction of Drosomycin remained unaffected by the diap2 mutation. Likewise, Drosomycin expression was normal in Tak1 mutant flies following M. luteus septic injury (Fig. 4B). Induced expression of Drosomycin in response to gram-positive bacterial infection was abrogated only in spatzle mutant flies. Constitutive transgene-mediated expression of diap2 fully rescued the diap27c mutant immune deficiency phenotype. diap27c;UAS-diap2/+ flies, like diap27c;Da-GAL4/UAS-diap2 or diap27c;Act5c-GAL4/UAS-diap2 flies, showed close to wild-type induction of Diptericin expression upon E. carotovora subsp. carotovora 15 infection, while diap27c flies failed to significantly induce Diptericin expression (Fig. 4C and data not shown).
FIG. 4.
diap2 mutant individuals fail to induce antibacterial peptide genes following Erwinia carotovora subsp. carotovora 15 infection. (A and C) Quantitative RT-PCR analysis of Diptericin (Dipt) induction after E. carotovora subsp. carotovora 15 septic injury (Ecc15 SI) in diap27c, diap27c/def, Tak11, and RelishE20 mutants, diap27c; UAS-diap2/+ and diap27c; UAS-diap2/Da-GAL4 flies, and wild-type (OregonR) adult males. Results are shown for control (unchallenged) (C) flies and flies 6 and 24 hours after infection. (B) Drosomycin (Drs) induction after Micrococcus luteus septic injury (M.lut. SI) of wild-type (OregonR), diap27c/def, Tak11, and spatzlerm7 adult males. Results are shown for control (unchallenged) (C) flies and flies 24 and 48 hours after infection. (D) Attacin-A (AttA), Cecropin-A1 (CecA1), Defensin (Def), Drosocin (Dro), and Metchnikowin (Mtk) induction 24 h after E. carotovora subsp. carotovora 15 septic injury of wild-type (OregonR), diap27c/def, Tak11, and RelishE20 adult males. Similar to Tak1 and Relish mutants, diap2 mutant flies were significantly impaired in their ability to induce antibacterial peptide genes in response to gram-negative bacterial septic injury. However, these mutants showed normal Drosomycin induction following exposure to gram-positive bacteria. Diptericin induction after E. carotovora subsp. carotovora 15 infection was restored in diap27c mutant flies expressing the UAS-diap2 transgene. rp49 was used as the experimental expression standard. Shown are the relative expression ratios of Dipt/rp49 (A and C), Drs/rp49 (B), AttA/rp49, CecA1/rp49, Def/rp49, Dro/rp49, and Mtk/rp49 (D).
To study the requirement of diap2 for the expression of other antibacterial peptide genes, we assessed the expression of Attacin-A, Cecropin-A1, Defensin, Drosocin, and Metchnikowin, all of which have been previously reported to be induced in an Imd-dependent manner following gram-negative bacterial infection (26). Following E. carotovora subsp. carotovora 15 septic injury, expression of Attacin-A, Cecropin-A1, Defensin, Drosocin, and Metchnikowin were severely impaired in diap27c/Def, Tak11, and RelishE20 mutant flies (Fig. 4D). These results, therefore, indicate that DIAP2 is generally required for activation of Imd-mediated immune responses, while it is dispensable for Toll signaling.
Next, we used an oral infection model to study the effects of the diap2 mutation. In this model, larvae are naturally infected via the digestive tract through exposure to food contaminated with E. carotovora subsp. carotovora 15 (Fig. 5) (1). Similar to the results obtained by septic injury, diap2 mutant larvae that were exposed to E. carotovora subsp. carotovora 15-contaminated food failed to induce expression of Diptericin (Fig. 5A), Drosocin (Fig. 5B), or Attacin-A (Fig. 5C). diap2 mutant larvae were similarly immunocompromised as known mutants (Tak11 and RelishE20) of the Imd pathway. Using a natural model of infection, these data corroborate the notion that DIAP2 is required for Imd-mediated immune response in vivo.
FIG. 5.

DIAP2 is required to mount a systemic antibacterial immune response to oral infection by Erwinia carotovora subsp. carotovora (Ecc15). Quantitative RT-PCR analysis of Diptericin (Dipt) (A), Drosocin (Dro) (B), and Attacin-A (AttA) (C) induction after E. carotovora subsp. carotovora 15 natural infection (Ecc15 NI) in wild-type (OregonR), diap27c/def, Tak11, and RelishE20 mutant third-instar larvae. Larvae were orally infected by exposing animals to food contaminated with E. carotovora subsp. carotovora 15. Similar to Tak1 and Relish mutants, diap2 mutant individuals failed to significantly induce Attacin-A, Diptericin, and Drosocin expression following natural infection. rp49 was used as an experimental expression standard. The relative Dipt/rp49 (A), Dro/rp49 (B), and AttA/rp49 (C) expression values for control (noninfected) (C) flies and flies 24 h after feeding are shown.
Epistatic positioning of DIAP2.
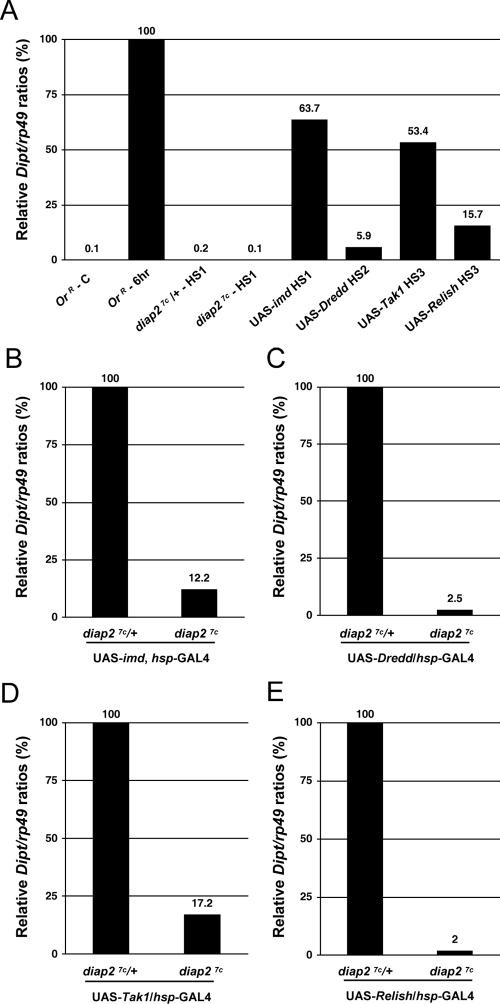
UAS/GAL4-mediated expression of imd, Dredd, Tak1, and Relish results in the activation of the Imd signal transduction pathway causing constitutive Diptericin expression, even in the absence of infecting microbes (11, 18, 51). We used this system to establish the epistatic position of DIAP2 in the Imd pathway. Heat shock-mediated overexpression of imd, Dredd, Tak1, and Relish resulted in induction of Diptericin expression (Fig. 6A). The ability to induce Diptericin expression varied substantially among imd, Dredd, Tak1, and Relish, which is consistent with previous reports (9, 11, 18, 29, 51). Overexpression of imd and Tak1 was most efficient in inducing Diptericin, with Imd inducing 63.7% and Tak1 53.4% of Diptericin levels observed after septic injury of control animals. In contrast, overexpression of Dredd and Relish merely achieved 5.9% and 15.7% of normal Diptericin expression levels after infection (Fig. 6A). To visualize the contribution of DIAP2 in this system, we set these Diptericin expression levels as 100% for all subsequent panels. Intriguingly, in the absence of DIAP2, imd-, Dredd-, Tak1-, and Relish-mediated induction of Diptericin was blocked (Fig. 6B to E). This indicates that diap2 is required for Diptericin induction by ectopic expression of all these components, corroborating its vital role in Imd signaling. Thus, in this system, diap2 appears to function downstream of or parallel to imd, Dredd, Tak1, and Relish.
FIG. 6.
DIAP2 functions genetically downstream of or in parallel to imd, Dredd, Tak1, and Relish. Quantitative RT-PCR analysis of Diptericin expression following overexpression of imd, Dredd, Tak1, and Relish was performed. (A) Diptericin (Dipt) expression levels of the following animals are shown: control OregonR animals that were not challenged (OrR - C) and 6 h after Erwinia carotovora subsp. carotovora 15 septic injury (OrR - 6hr); diap27c/+ and diap27c mutant flies 3 h after 1 h of heat shock at 37°C (diap27c/+-HS1 and diap27c-HS1) or heat shock-mediated overexpression of imd (UAS-imd HS1), Dredd (UAS-Dredd HS2), Tak1 (UAS-Tak1 HS3), and Relish (UAS-Relish HS3). (B) In the absence of infection, heat shock-mediated imd overexpression caused high levels of Diptericin (Dipt) expression (63.7% of the level of Diptericin observed 6 h after E. carotovora subsp. carotovora 15 septic injury in panel A) that was significantly thwarted in diap27c mutant flies (87.8% reduction). (C) Heat shock-mediated Dredd overexpression triggered weak but reproducible Diptericin expression (5.9% of the levels of Diptericin observed after 6 h of E. carotovora subsp. carotovora 15 septic injury in panel A), which was blocked in diap27c mutant flies. (D) Heat shock-mediated Tak1 overexpression triggered strong Diptericin expression (53.4% of Diptericin levels observed after 6 h of E. carotovora subsp. carotovora 15 septic injury in panel A) which was blocked in diap27c mutants. (E) Heat shock-mediated Relish overexpression triggered weak but reproducible Diptericin expression (15.7% of the levels of Diptericin observed after 6 h of E. carotovora subsp. carotovora 15 septic injury in panel A) which was not observed in diap27c mutant flies. Flies were heat shocked for 1 hour at 37°C and left to recover at 25°C for 3 h (A and B) (HS1), 1 h (A and C) (HS2), or 24 h (A, D, and E) (HS3) prior to analysis. The analyzed genotypes were as follows: (i) diap27c/Cyo, act-GFP (diap27c/+), diap27c; (ii) diap27c/Cyo; UAS-imd, hsp-GAL4/TM6Tb (UAS-imd), diap27c; UAS-imd, hsp-GAL4/TM6Tb; (iii) diap27c/Cyo; UAS-Dredd/hsp-GAL4 (UAS-Dredd), diap27c; UAS-Dredd/hsp-GAL4; (iv) diap27c, UAS-Tak1/+; hsp-GAL4/+ (UAS-Tak1), diap27c, UAS-Tak1/diap27c; hsp-GAL4/+; (v) diap27c, UAS-Relish/+; hsp-GAL4/+ (UAS-Relish); (vi) diap27c, UAS-Relish/diap27c; hsp-GAL4/+.
Traf2 is not required for an effective immune response triggered by gram-negative bacterial infection.
In mammals, TRAF2 plays a key role in TNFR1-mediated NF-κB activation (3). Following its recruitment to TNFR1, TRAF2 promotes conjugation of Lys63-linked polyubiquitin chains, which allows the recruitment and activation of Tak1 and IKK (3). Recently, it was suggested that the Drosophila Traf2 orthologue contributes to NF-κB-dependent signaling pathways in Drosophila (2). To test whether Traf2 is required for Imd-mediated immune responses, we challenged Traf2 null mutant flies with gram-negative bacteria. Surprisingly, Traf2 mutant flies were fully resistant to E. carotovora subsp. carotovora 15 septic injury (Fig. 7). Under the same conditions, diap2 or Tak1 mutant animals rapidly succumbed to the same microbial load. Thus, our results demonstrate that Drosophila Traf2 is dispensable to mount an efficient immune response to infection by gram-negative bacteria.
FIG. 7.
Traf2 is dispensable in resisting gram-negative bacterial infection. Shown are the survival rates of Traf2 null mutant adult males exposed to septic injury with E. carotovora subsp. carotovora 15 (Ecc15). The following genotypes were examined: wild type (OregonR), Tak11, diap27c, and Traf2ex1. Note that Traf2-deficient flies behaved like wild-type flies, while diap2 and Tak1 mutant flies were highly susceptible to E. carotovora subsp. carotovora 15-mediated septic injury.
DISCUSSION
The Drosophila innate immune response relies mainly on the differential expression of a variety of small peptides with antimicrobial activities (19). Depending on the infiltrating microbe, Drosophila selectively activates two distinct signaling pathways. While infections by fungi or gram-positive bacteria stimulate the Toll pathway, infection by gram-negative bacteria triggers the activation of the immune deficiency (Imd) signaling cascade. Activation of both Toll- and Imd signaling results in the activation of NF-κB-like transcription factors leading to the expression of specific sets of antimicrobial peptides (38). Here, we demonstrate through mutation analysis that DIAP2 plays a pivotal role in the Drosophila innate immune response. We find that, in vivo, the Drosophila inhibitor of apoptosis protein DIAP2 is indispensable for Imd-mediated expression of antibacterial peptide genes. Like known mutants of the Imd pathway, flies with a mutation in the diap2 gene failed to induce expression of Attacin-A, Cecropin-A1, Defensin, Diptericin, Drosocin, and Metchnikowin and mount an efficient immune reaction in response to infection by gram-negative bacteria. Consequently, diap2 mutant flies succumbed to gram-negative bacterial infection. In contrast, such flies mounted a normal Toll-dependent immune response and were resistant to infection by fungi and gram-positive bacteria. Our diap2 null mutant phenotype, therefore, demonstrates that DIAP2 is an essential component of the Imd pathway. Thus, our data are consistent with recent RNAi studies that have implicated diap2 in the Imd pathway (12, 23).
DIAP2 is a member of the evolutionarily conserved IAP family (17). IAPs are classified by the presence of the BIR domain through which they interact with various “client” proteins (50). Genetic analysis of the Drosophila IAP DIAP1 has provided some of the most compelling insights into the in vivo function of this protein family. DIAP1, the first and most extensively studied Drosophila IAP, is essential for cell survival and acts as a potent caspase inhibitor (16, 17, 32, 54). Mutations that abrogate physical association of DIAP1 with caspases cause widespread and unrestrained caspase activation, leading to cell and organismal death (14, 30, 54, 55). In contrast to diap1, diap2 null mutants do not show an apparent cell death phenotype and develop normally. This is unexpected, because both these IAPs interact with caspases and IAP antagonists with similar affinities (27, 52, 53). Moreover, when overexpressed, DIAP2 can rescue diap1 RNAi-mediated apoptosis, suggesting that DIAP2 can functionally substitute for DIAP1 in its ability to regulate caspases (27). Nevertheless, diap2 mutant animals do not show any apparent apoptosis-related phenotypes during development. However, these animals appear to be sensitized to Reaper-mediated killing in the eye (F. Leulier and P. Meier, unpublished data). The lack of any apparent gross developmental phenotype may be due to sufficiently high levels of DIAP1 that may thwart unscheduled caspase activation in response to loss of DIAP2. In this respect, it is noteworthy that during embryonic development, the levels of diap1 mRNA dramatically exceed those of diap2 (17-fold difference, Berkeley Drosophila Genome Project [BDGP] expression profiles). Moreover, similarly to c-IAP2 knockout mice, where a cell death phenotype is revealed only after lipopolysaccharide challenge (5), phenotypic manifestation may become apparent only under certain conditions or in selective tissues. In agreement with this notion, RNAi-mediated depletion of DIAP2 has no effect on cell viability in unchallenged tissue culture cells (12, 23) but significantly sensitizes S2 cells to stress-induced apoptosis (59).
Although IAPs have originally been identified as apoptosis inhibitors (7), recent evidence suggests that IAPs are multifunctional signaling devices that, depending on the protein they interact with, influence diverse biological processes. In this respect, it is noteworthy that IAPs also carry C-terminal RING finger domains providing them with E3 ubiquitin-protein ligase, and hence, signaling activity (50). Thus, in addition to inhibiting apoptosis, IAPs also fulfill functions that operate independently of their ability to control caspases and cell death (50). Therefore, BIR-containing proteins are more precisely referred to as BIRCs rather than IAPs (43). Consistent with the notion that BIRCs are multifunctional proteins, the mammalian c-IAP1 and c-IAP2 bind to caspases as well as RIP1 and TRAF2, two components of the TNF receptor signaling complex (34, 37, 41). c-IAP1 or c-IAP2, or both, can promote ubiquitylation and degradation of TRAF2, RIP1, and NF-κB kinase (IKKγ)/NF-κB essential modulator (NEMO) (34, 50). Hence, these BIRC proteins are thought to modulate the response to TNF. More recently, another BIRC protein was identified as an important regulator of innate immune surveillance in mammals. BIRC1e (NAIP5) was found to control the intracellular pathogen Legionella pneumophila, a gram-negative microbe that causes severe bacterial pneumonia known as Legionnaires' disease (56). BIRC1e protects infected host macrophages by restricting intracellular replication of this pathogen.
We now find that the Drosophila BIRC protein DIAP2 is similarly required for innate immune responses and the resistance to gram-negative bacterial infection. diap2 null mutants become highly susceptible to gram-negative bacteria and fail to induce antibacterial peptide gene expression. Intriguingly, the Imd pathway, which is required for antibacterial peptide gene expression in response to gram-negative microbes, shares significant similarities with the TNFR1 signaling cascade. The notion that the BIRC proteins c-IAP1, c-IAP2, and DIAP2 are core components of the TNFR1 and Imd pathway, respectively, further reinforces the parallels between the mammalian TNFR1 pathway and the Imd pathway of Drosophila, pointing to an evolutionary conservation of these pathways in NF-κB activation (21, 22, 48). Moreover, both pathways seem to rely on ubiquitin-mediated protein modifications. As in human cells, where activation of TAK1 and IKK requires the E2 ubiquitin-conjugating enzyme complex Ubc13/UEV1A (3), Drosophila Ubc13(Bendless)/UEV1A are similarly required for activating Tak1 and the Drosophila IKK complex (57). Moreover, recent RNAi data from cultured cells suggest that Drosophila Tab contributes to Imd signaling, although this still awaits in vivo validation (12, 23, 58). Therefore, similar to the TNFR1 pathway, ubiquitin-mediated protein modification is likely to activate the Tak1/Tab complex via Tab's ability to bind to Lys63-linked polyubiquitin chains, thereby recruiting Tak1 to activator platforms. In contrast to the E2 ubiquitin-conjugating enzyme complex, little is known about the nature of the E3 ubiquitin-ligase of the Imd pathway. While TRAF2 is crucial for Ubc13/UEV1A-mediated ubiquitylation in the mammalian TNFR1 pathway, it seems that for Imd signaling Traf2, the TRAF2/6 orthologue in flies, is not a critical component. Traf2 null mutation did not completely block NF-κB activation in Drosophila (2). Moreover, our data clearly indicate that Traf2 mutant flies are fully competent to mount an immune response and resist gram-negative bacterial infection. Hence, Traf2 appears not to be essential for an effective Imd-mediated immune response. Since the Drosophila genome encodes at least three TRAF family members, it is possible that the loss of Traf2 function is complemented by other TRAF family members. Alternatively, other signaling pathways that bypass Traf2 to transmit the infection signal to NF-κB may exit in Drosophila. In agreement with this notion, RNAi-mediated knockdown of all three Drosophila TRAFs also did not abrogate Imd-signaling in S2 cells (23, 47, 57). Thus, an E3 ubiquitin ligase different from or in addition to Traf2 may be responsible for Imd signaling in Drosophila. Since DIAP2 carries a RING finger domain, it represents a likely candidate.
Our genetic epistatic analysis places diap2 downstream of or parallel to imd, Dredd, Tak1, and Relish. Overexpression of imd, Dredd, Tak1, and Relish failed to induce Diptericin expression in diap2 mutant animals, while in wild-type animals, enforced expression of these genes, in the absence of any infection, resulted in reproducible Diptericin induction. Intriguingly, Diptericin induction following enforced expression of imd and Dredd is also blocked in Tak1 mutant animals (9, 29), indicating that both DIAP2 and Tak1 are required downstream of Dredd. In contrast, kenny and ird5 seem not to be required for Diptericin induction when Dredd is overexpressed (51). A recent report indicates that Relish cleavage and nuclear translocation on its own are not sufficient for Diptericin expression and that, at least in vivo, a further cooperative input from the JNK signaling pathway is required (9). According to this scenario, the Imd signaling pathway bifurcates at the level of Tak1, with Tak1 activating the NF-κB signaling branch as well as JNK signaling branch, both of which are required for expression of antibacterial peptide genes in the fat body. In light of this model, the observation that diap2 acts genetically downstream of imd, Dredd, Tak1, and Relish may indicate that DIAP2 functions at the level of Tak1 (Fig. 8). This view is in agreement with recent reports from Drosophila tissue culture cells, which suggest that DIAP2 is required for Tak1-mediated JNK activation (12, 23). In this respect, DIAP2 functions at the same epistatic position as the putative E3 ubiquitin ligase of the Imd pathway. Future biochemical experiments will be required to test whether DIAP2 is indeed the E3 ubiquitin ligase that functions together with Ubc13/UEV1A to stimulate Tak1.
Although the underlying mechanism for the impaired induction of antibacterial peptide gene expression by loss of DIAP2 remains to be defined, the genetic observations made here are likely to have relevance not only for innate immune responses in Drosophila but also for TNFR1 signaling in mammals. While in flies DIAP2 is indispensable for Imd signaling, genetic studies in mice have, so far, failed to uncover a physiological role for c-IAP1 and c-IAP2 in TNFR1 signaling (5, 6). Since c-iap1 knockout mice carry significantly elevated levels of c-IAP2 protein, it is feasible that the increased c-IAP2 levels functionally compensate for the loss of c-IAP1 (6). Consistently, mammalian IAPs have been reported to be under strict homeostatic control by regulating each other's protein levels, which provides a mechanistic explanation for the cross talk among IAPs (42). Thus, in mammals, double-knockout mice lacking both c-iap1 and c-iap2 genes will be required to study the roles of c-IAP1 and c-IAP2 in TNFR1 signaling. Therefore, Drosophila, where redundancies and compensatory mechanisms are less problematic, provides an ideal model system to study caspase-independent functions of IAPs in an in vivo setting.
Acknowledgments
We thank Bruce Hay and Kristin White for stimulating discussions and sharing unpublished information; Paulo S. Ribeiro, Brigitte Maroni, and Anna Zachariou for assistance; members of the Meier lab for critical reading of the manuscript; and Dan Hultmark, Jongkyeong Chung, and the Bloomington and Harvard Medical School Drosophila stock centers for fly lines.
B.L. is supported by CNRS, the “Agence Nationale pour la Recherche,” and the “Agence pour la Recherche sur le Cancer.” F.L. is funded by a long-term HFSP fellowship.
Footnotes
Published ahead of print on 7 August 2006.
REFERENCES
- 1.Basset, A., R. Khush, A. Braun, L. Gardan, F. Boccard, J. Hoffmann, and B. Lemaitre. 2000. The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc. Natl. Acad. Sci. USA 97:3376-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cha, G. H., K. S. Cho, J. H. Lee, M. Kim, E. Kim, J. Park, S. B. Lee, and J. Chung. 2003. Discrete functions of TRAF1 and TRAF2 in Drosophila melanogaster mediated by c-Jun N-terminal kinase and NF-κB-dependent signaling pathways. Mol. Cell. Biol. 23:7982-7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, Z. J., V. Bhoj, and R. B. Seth. 2006. Ubiquitin, TAK1 and IKK: is there a connection? Cell Death Differ. 13:687-692. [DOI] [PubMed] [Google Scholar]
- 4.Choe, K. M., H. Lee, and K. V. Anderson. 2005. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc. Natl. Acad. Sci. USA 102:1122-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conte, D., M. Holcik, C. A. Lefebvre, E. Lacasse, D. J. Picketts, K. E. Wright, and R. G. Korneluk. 2006. Inhibitor of apoptosis protein cIAP2 is essential for lipopolysaccharide-induced macrophage survival. Mol. Cell. Biol. 26:699-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conze, D. B., L. Albert, D. A. Ferrick, D. V. Goeddel, W. C. Yeh, T. Mak, and J. D. Ashwell. 2005. Posttranscriptional downregulation of c-IAP2 by the ubiquitin protein ligase c-IAP1 in vivo. Mol. Cell. Biol. 25:3348-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crook, N. E., R. J. Clem, and L. K. Miller. 1993. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 67:2168-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Gregorio, E., P. T. Spellman, P. Tzou, G. M. Rubin, and B. Lemaitre. 2002. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 21:2568-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delaney, J. R., S. Stoven, H. Uvell, K. V. Anderson, Y. Engstrom, and M. Mlodzik. 2006. Cooperative control of Drosophila immune responses by the JNK and NF-kappaB signaling pathways. EMBO J. 25:3068-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrandon, D., J. L. Imler, and J. A. Hoffmann. 2004. Sensing infection in Drosophila: Toll and beyond. Semin. Immunol. 16:43-53. [DOI] [PubMed] [Google Scholar]
- 11.Georgel, P., S. Naitza, C. Kappler, D. Ferrandon, D. Zachary, C. Swimmer, C. Kopczynski, G. Duyk, J. M. Reichhart, and J. A. Hoffmann. 2001. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev. Cell 1:503-514. [DOI] [PubMed] [Google Scholar]
- 12.Gesellchen, V., D. Kuttenkeuler, M. Steckel, N. Pelte, and M. Boutros. 2005. An RNA interference screen identifies Inhibitor of Apoptosis Protein 2 as a regulator of innate immune signalling in Drosophila. EMBO Rep. 6:979-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gloor, G. B., C. R. Preston, D. M. Johnson-Schlitz, N. A. Nassif, R. W. Phillis, W. K. Benz, H. M. Robertson, and W. R. Engels. 1993. Type I repressors of P element mobility. Genetics 135:81-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goyal, L., K. McCall, J. Agapite, E. Hartwieg, and H. Steller. 2000. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 19:589-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harlin, H., S. B. Reffey, C. S. Duckett, T. Lindsten, and C. B. Thompson. 2001. Characterization of XIAP-deficient mice. Mol. Cell. Biol. 21:3604-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawkins, C. J., S. J. Yoo, E. P. Peterson, S. L. Wang, S. Y. Vernooy, and B. A. Hay. 2000. The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. J. Biol. Chem. 275:27084-27093. [DOI] [PubMed] [Google Scholar]
- 17.Hay, B. A., D. A. Wassarman, and G. M. Rubin. 1995. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell 83:1253-1262. [DOI] [PubMed] [Google Scholar]
- 18.Hedengren, M., B. Asling, M. S. Dushay, I. Ando, S. Ekengren, M. Wihlborg, and D. Hultmark. 1999. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol. Cell 4:827-837. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann, J. A. 2003. The immune response of Drosophila. Nature 426:33-38. [DOI] [PubMed] [Google Scholar]
- 20.Kaneko, T., and N. Silverman. 2005. Bacterial recognition and signalling by the Drosophila IMD pathway. Cell. Microbiol. 7:461-469. [DOI] [PubMed] [Google Scholar]
- 21.Khush, R. S., F. Leulier, and B. Lemaitre. 2001. Drosophila immunity: two paths to NF-kappaB. Trends Immunol. 22:260-264. [DOI] [PubMed] [Google Scholar]
- 22.Khush, R. S., F. Leulier, and B. Lemaitre. 2002. Immunology. Pathogen surveillance—the flies have it. Science 296:273-275. [DOI] [PubMed] [Google Scholar]
- 23.Kleino, A., S. Valanne, J. Ulvila, J. Kallio, H. Myllymaki, H. Enwald, S. Stoven, M. Poidevin, R. Ueda, D. Hultmark, B. Lemaitre, and M. Ramet. 2005. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. EMBO J. 24:3423-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemaitre, B., E. Kromer-Metzger, L. Michaut, E. Nicolas, M. Meister, P. Georgel, J. Reichhart, and J. Hoffmann. 1995. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc. Natl. Acad. Sci. USA 92:9465-9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemaitre, B., E. Nicolas, L. Michaut, J. Reichhart, and J. Hoffmann. 1996. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86:973-983. [DOI] [PubMed] [Google Scholar]
- 26.Lemaitre, B., J. Reichhart, and J. Hoffmann. 1997. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. USA 94:14614-14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leulier, F., P. S. Ribeiro, E. Palmer, T. Tenev, K. Takahashi, D. Robertson, A. Zachariou, F. Pichaud, R. Ueda, and P. Meier. 17 February 2006, posting date. Systematic in vivo RNAi analysis of putative components of the Drosophila cell death machinery. Cell Death Differ. [Online.] doi: 10.1038/sj.cdd.4401868. [DOI] [PubMed]
- 28.Leulier, F., A. Rodriguez, R. S. Khush, J. M. Abrams, and B. Lemaitre. 2000. The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Rep. 1:353-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leulier, F., S. Vidal, K. Saigo, R. Ueda, and B. Lemaitre. 2002. Inducible expression of double-stranded RNA reveals a role for dFADD in the regulation of the antibacterial response in Drosophila adults. Curr. Biol. 12:996-1000. [DOI] [PubMed] [Google Scholar]
- 30.Lisi, S., I. Mazzon, and K. White. 2000. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics 154:669-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, Y., L. P. Wu, and K. V. Anderson. 2001. The antibacterial arm of the Drosophila innate immune response requires an IkappaB kinase. Genes Dev. 15:104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meier, P., J. Silke, S. J. Leevers, and G. I. Evan. 2000. The Drosophila caspase DRONC is regulated by DIAP1. EMBO J. 19:598-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naitza, S., C. Rosse, C. Kappler, P. Georgel, M. Belvin, D. Gubb, J. Camonis, J. A. Hoffmann, and J. M. Reichhart. 2002. The Drosophila immune defense against gram-negative infection requires the death protein dFADD. Immunity 17:575-581. [DOI] [PubMed] [Google Scholar]
- 34.Park, S. M., J. B. Yoon, and T. H. Lee. 2004. Receptor interacting protein is ubiquitinated by cellular inhibitor of apoptosis proteins (c-IAP1 and c-IAP2) in vitro. FEBS Lett. 566:151-156. [DOI] [PubMed] [Google Scholar]
- 35.Parks, A. L., K. R. Cook, M. Belvin, N. A. Dompe, R. Fawcett, K. Huppert, L. R. Tan, C. G. Winter, K. P. Bogart, J. E. Deal, M. E. Deal-Herr, D. Grant, M. Marcinko, W. Y. Miyazaki, S. Robertson, K. J. Shaw, M. Tabios, V. Vysotskaia, L. Zhao, R. S. Andrade, K. A. Edgar, E. Howie, K. Killpack, B. Milash, A. Norton, D. Thao, K. Whittaker, M. A. Winner, L. Friedman, J. Margolis, M. A. Singer, C. Kopczynski, D. Curtis, T. C. Kaufman, G. D. Plowman, G. Duyk, and H. L. Francis-Lang. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36:288-292. [DOI] [PubMed] [Google Scholar]
- 36.Pili-Floury, S., F. Leulier, K. Takahashi, K. Saigo, E. Samain, R. Ueda, and B. Lemaitre. 2004. In vivo RNA interference analysis reveals an unexpected role for GNBP1 in the defense against Gram-positive bacterial infection in Drosophila adults. J. Biol. Chem. 279:12848-12853. [DOI] [PubMed] [Google Scholar]
- 37.Rothe, M., M. G. Pan, W. J. Henzel, T. M. Ayres, and D. V. Goeddel. 1995. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell 83:1243-1252. [DOI] [PubMed] [Google Scholar]
- 38.Royet, J., J. M. Reichhart, and J. A. Hoffmann. 2005. Sensing and signaling during infection in Drosophila. Curr. Opin. Immunol. 17:11-17. [DOI] [PubMed] [Google Scholar]
- 39.Rutschmann, S., A. C. Jung, R. Zhou, N. Silverman, J. A. Hoffmann, and D. Ferrandon. 2000. Role of Drosophila IKKγ in a Toll-independent antibacterial immune response. Nat. Immunol. 1:342-347. [DOI] [PubMed] [Google Scholar]
- 40.Rutschmann, S., A. Kilinc, and D. Ferrandon. 2002. The Toll pathway is required for resistance to gram-positive bacterial infections in Drosophila. J. Immunol. 168:1542-1546. [DOI] [PubMed] [Google Scholar]
- 41.Shu, H. B., M. Takeuchi, and D. V. Goeddel. 1996. The tumor necrosis factor receptor 2 signal transducers TRAF2 and c-IAP1 are components of the tumor necrosis factor receptor 1 signaling complex. Proc. Natl. Acad. Sci. USA 93:13973-13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silke, J., T. Kratina, D. Chu, P. G. Ekert, C. L. Day, M. Pakusch, D. C. Huang, and D. L. Vaux. 2005. Determination of cell survival by RING-mediated regulation of inhibitor of apoptosis (IAP) protein abundance. Proc. Natl. Acad. Sci. USA 102:16182-16187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silke, J., and D. L. Vaux. 2001. Two kinds of BIR-containing protein-inhibitors of apoptosis, or required for mitosis. J. Cell Sci. 114:1821-1827. [DOI] [PubMed] [Google Scholar]
- 44.Silverman, N., J. Zhou, S. Stöven, N. Pandey, D. Hultmark, and T. Maniatis. 2000. A Drosophila IkB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 14:2461-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stöven, S., I. Ando, L. Kadalayil, Y. Engström, and D. Hultmark. 2000. Activation of the Drosophila NF-κB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 1:347-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stoven, S., N. Silverman, A. Junell, M. Hedengren-Olcott, D. Erturk, Y. Engstrom, T. Maniatis, and D. Hultmark. 2003. Caspase-mediated processing of the Drosophila NF-kappa B factor Relish. Proc. Natl. Acad. Sci. USA 100:5991-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun, H., B. N. Bristow, G. Qu, and S. A. Wasserman. 2002. A heterotrimeric death domain complex in Toll signaling. Proc. Natl. Acad. Sci. USA 99:12871-12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanji, T., and Y. T. Ip. 2005. Regulators of the Toll and Imd pathways in the Drosophila innate immune response. Trends Immunol. 26:193-198. [DOI] [PubMed] [Google Scholar]
- 49.Tzou, P., M. Meister, and B. Lemaitre. 2002. Methods for studying infection and immunity in Drosophila. Methods Microbiol. 31:507-529. [Google Scholar]
- 50.Vaux, D. L., and J. Silke. 2005. IAPs, RINGs and ubiquitylation. Nat. Rev. Mol. Cell Biol. 6:287-297. [DOI] [PubMed] [Google Scholar]
- 51.Vidal, S., R. S. Khush, F. Leulier, P. Tzou, M. Nakamura, and B. Lemaitre. 2001. Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NF-kappaB-dependent innate immune responses. Genes Dev. 15:1900-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vucic, D., W. J. Kaiser, A. J. Harvey, and L. K. Miller. 1997. Inhibition of reaper-induced apoptosis by interaction with inhibitor of apoptosis proteins (IAPs). Proc. Natl. Acad. Sci. USA 94:10183-10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vucic, D., W. J. Kaiser, and L. K. Miller. 1998. Inhibitor of apoptosis proteins physically interact with and block apoptosis induced by Drosophila proteins HID and GRIM. Mol. Cell. Biol. 18:3300-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, S. L., C. J. Hawkins, S. J. Yoo, H. A. Muller, and B. A. Hay. 1999. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell 98:453-463. [DOI] [PubMed] [Google Scholar]
- 55.Zachariou, A., T. Tenev, L. Goyal, J. Agapite, H. Steller, and P. Meier. 2003. IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding. EMBO J. 22:6642-6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zamboni, D. S., K. S. Kobayashi, T. Kohlsdorf, Y. Ogura, E. M. Long, R. E. Vance, K. Kuida, S. Mariathasan, V. M. Dixit, R. A. Flavell, W. F. Dietrich, and C. R. Roy. 2006. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat. Immunol. 7:318-325. [DOI] [PubMed] [Google Scholar]
- 57.Zhou, R., N. Silverman, M. Hong, D. S. Liao, Y. Chung, Z. J. Chen, and T. Maniatis. 2005. The role of ubiquitination in Drosophila innate immunity. J. Biol. Chem. 280:34048-34055. [DOI] [PubMed] [Google Scholar]
- 58.Zhuang, Z. H., L. Sun, L. Kong, J. H. Hu, M. C. Yu, P. Reinach, J. W. Zang, and B. X. Ge. 2006. Drosophila TAB2 is required for the immune activation of JNK and NF-kappaB. Cell Signal. 18:964-970. [DOI] [PubMed] [Google Scholar]
- 59.Zimmermann, K. C., J. E. Ricci, N. M. Droin, and D. R. Green. 2002. The role of ARK in stress-induced apoptosis in Drosophila cells. J. Cell Biol. 156:1077-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]