Abstract
The Rad51 paralog Rad51C has been implicated in the control of homologous recombination. To study the role of Rad51C in vivo in mammalian cells, we analyzed short-tract and long-tract gene conversion between sister chromatids in hamster Rad51C−/− CL-V4B cells in response to a site-specific chromosomal double-strand break. Gene conversion was inefficient in these cells and was specifically restored by expression of wild-type Rad51C. Surprisingly, gene conversions in CL-V4B cells were biased in favor of long-tract gene conversion, in comparison to controls expressing wild-type Rad51C. These long-tract events were not associated with crossing over between sister chromatids. Analysis of gene conversion tract lengths in CL-V4B cells lacking Rad51C revealed a bimodal frequency distribution, with almost all gene conversions being either less than 1 kb or greater than 3.2 kb in length. These results indicate that Rad51C plays a pivotal role in determining the “choice” between short- and long-tract gene conversion and in suppressing gene amplifications associated with sister chromatid recombination.
Double-strand breaks (DSB) are a threat to genome stability, since their misrepair can cause chromosome translocations, gene amplifications, and genetic deletions, chromosomal rearrangements that are often seen in cancer cells. DSB formed during the S or G2 phase of the cell cycle may be repaired preferentially by homologous recombination (HR) between sister chromatids (sister chromatid recombination [SCR]) (37). The mechanism of DSB repair used depends upon how the DNA break is processed, whether Rad51 is efficiently loaded onto the processed DNA end, whether a homologous donor is available, and how recombination intermediates generated subsequent to strand exchange are resolved. Although the mechanisms of end processing, Rad51 loading, and strand invasion in mammalian cells are increasingly well understood, the regulation of steps subsequent to strand invasion are relatively obscure.
In Saccharomyces cerevisiae, somatic gene conversion typically copies a short tract (50 to 300 bp) of DNA sequence from the donor template, resolving by synthesis-dependent strand annealing (SDSA) without crossing over (CO) (26, 27, 34). An alternative copying mechanism termed break-induced replication (BIR) is engaged under some circumstances, including when gene conversion is defective (20). S. cerevisiae rad51, rad54, rad55, and rad57 mutants reveal a defect in gene conversion but increased use of BIR (20). In yeast, therefore, conventional gene conversion and BIR are separable pathways. BIR in S. cerevisiae can copy hundreds of kilobases from the donor chromosome, sometimes involving an entire chromosome arm and resulting in a nonreciprocal translocation with loss of heterozygosity (LOH) (16, 25). BIR has also been implicated in palindromic gene amplification, increased rates of LOH in aging yeast, telomere elongation in the absence of telomerase, and gene targeting (4, 23, 25, 29).
In mammalian cells, somatic gene conversion tracts are typically ∼50 to 200 bp in length (5, 7, 35), but a proportion of gene conversions extend beyond 1 kb (13, 30). These have been termed short-tract gene conversions (STGC) and long-tract gene conversions (LTGC), respectively. It is unknown whether STGC and LTGC arise by distinct mechanisms in mammalian cells. These classifications are therefore only descriptive. In cells lacking the Rad51 paralog XRCC3, mean gene conversion tract length is increased from ∼200 to ∼500 bp, suggesting a role for XRCC3 in late stages of recombination (5).
There are five mammalian paralogs of Rad51, sharing extensive homology only within the conserved Walker A and B motifs. They exist in at least two distinct complexes: the Rad51B/Rad51C/Rad51D/XRCC2 (BCDX2) complex and the Rad51C/XRCC3 (CX3) complex (22, 38). Rad51 paralogs are required for efficient HR in vertebrate cells (reviewed in reference 38); the BC heterodimer of the BCDX2 complex is known to assist Rad51-mediated strand exchange in vitro (32). A role for the paralogs in SCR is suggested by abundant chromatid-type errors in paralog-deficient cells, by reduced induction of sister chromatid exchange following DNA damage, and by reduced sister chromatid cohesion in cells lacking Rad51C (8, 10). Rad51C and XRCC3 have also been implicated in Holliday junction resolution and branch migration, suggesting a role in later stages of recombination (19, 40). The Rad51C gene itself is amplified in some human breast cancers, raising the possibility that alteration in Rad51C copy number promotes tumorigenesis (3, 41).
To test whether Rad51C regulates steps subsequent to strand exchange in vivo, we studied SCR regulation by Rad51C, using a system that allows independent but parallel measurement of short- and long-tract gene conversion between sister chromatids, induced by a site-specific chromosomal DSB (28, 42). Surprisingly, we found that Rad51C−/− cells generate a high proportion of long-tract gene conversions, an outcome suppressed by reexpression of wild-type Rad51C. The results indicate that Rad51C controls the extent of repair synthesis during gene conversion and normally suppresses certain types of gene amplification associated with sister chromatid recombination.
MATERIALS AND METHODS
Plasmids.
SCR reporter construction and the design of the I-SceI expression vector were described previously (28). Expression vectors for human Rad51 (hRad51) paralogs, including hRad51C and its mutants, were described previously (9-11, 36).
Antibodies and Western blotting.
Cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 8.0, 250 mM NaCl, 0.1% sodium dodecyl sulfate, 1% NP-40 with added protease and phosphatase inhibitors), and then 20 μg of extracted protein was separated on 4 to 12% bis-Tris sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (Invitrogen) and analyzed by Western blotting using mouse monoclonal antibodies against hRad51C, hRad51B, hRad51D, hXRCC2, and hXRCC3 (Novus) or rabbit polyclonal antibodies to Rad51. Patrick Sung kindly provided rabbit polyclonal XRCC3 antibodies. Beta-actin expression was analyzed using a specific monoclonal antibody (AbCam).
Cell lines and cell culture.
CL-V4B, V79B, CHO-AA8, and irs1SF cells were cultured as described previously (10, 11). To obtain single-copy integration of the SCR reporter, 3 × 106 cells were electroporated (Nucleofector kit T, program U23; Amaxa Biosystems) with 1 μg of AflIII-linearized SCR reporter and then cultivated in puromycin (2 μg/ml). Genomic DNA from puromycin-resistant colonies was analyzed by Southern blotting using multiple parallel restriction digests, allowing the identification of clones carrying a single, randomly integrated copy of the reporter. For stable expression of hRad51C and mutants, the relevant expression plasmids were transfected using Lipofectamine 2000 (Invitrogen). Pools of G418-resistant colonies were expanded, without recloning, for functional analysis.
Recombination assays.
Cells (5 × 106) were electroporated (Amaxa Biosystems) with 14 μg of pcDNA3β-myc-NLS-I-SceI or control pcDNA3β (28). Transfection efficiency (typically ∼99%) was measured in parallel as described previously (42). Cells were analyzed by fluorescence-activated cell sorting 72 h after transfection as described previously (28). In all experiments, the percentage of green fluorescent protein-positive (GFP+) cells (whether I-SceI-induced or background frequency) was measured in quadruplicate-transfected samples and I-SceI-transfected values were corrected for I-SceI transfection efficiency. The background GFP+ frequency (<0.002% in all clones) was subtracted from this number to give the corrected I-SceI-induced GFP+ frequency. To analyze SCR events, transiently transfected quadruplicate cultures, receiving either I-SceI or control empty vector, were trypsinized, counted, and replated at 2.5 × 106 to 3 × 106 cells per 10-cm plate. At 24 h after plating, 5 μg/ml blasticidin (BSD) was added and colonies were selected for 2 to 3 weeks, stained, and counted for analysis. Plating efficiency was determined in parallel for each experiment by plating defined numbers of cells sparsely, allowing colonies to form without antibiotic selection. The raw I-SceI-induced BsdR+ frequency was corrected for transfection efficiency and plating efficiency, with subtraction of the background BsdR+ frequency. The background BsdR+ frequency was always <0.001%, with the exception of clone HR103, for which this value was ≤0.004%. Statistical analysis of these experiments was performed using an unpaired t test with unknown variance. Southern blotting was performed as described previously (28).
Crossover/noncrossover recombination analysis.
I-SceI-transfected cells were plated on 15-cm dishes to yield single-cell colonies consisting of ∼35 × 103 viable BsdR− colonies and ∼1 to 2 BsdR+ colonies per plate. BSD was added to cells 48 h after transfection and removed exactly 24 h later by extensive washing in phosphate-buffered saline, followed by cultivation in medium lacking BSD. After ∼14 days, sectored GFP+ and GFP− colonies were identified by microscopic screening, isolated, and expanded for Southern analysis.
RESULTS
Gene conversion defect in Rad51C−/− cells.
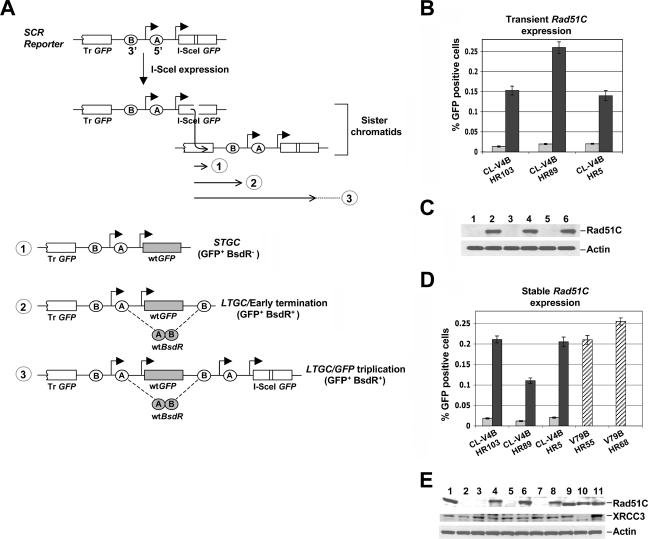
We introduced an SCR reporter into Rad51C−/− CL-V4B cells, which are derived from Rad51C+/+ V79B hamster fibroblasts but carry a homozygous deletion of Rad51C exon 5 (10). The SCR reporter (28) contains two mutant copies of the gene encoding enhanced GFP (here termed GFP): the first is truncated at the 5′ end, and the second is full length but is interrupted by an 18-bp restriction site recognized by the rare-cutting restriction endonuclease I-SceI (31). I-SceI-induced gene conversion between the two GFP copies generates wild-type (wt) GFP, and the resulting GFP+ cell is detectable by flow cytometry (fluorescence-activated cell sorting). STGC could arise by either interchromatid (SCR) or intrachromatid recombination. As originally proposed by L. Symington, LTGC or crossing over could each result in triplication of the GFP copies (Fig. 1A) (6, 13).
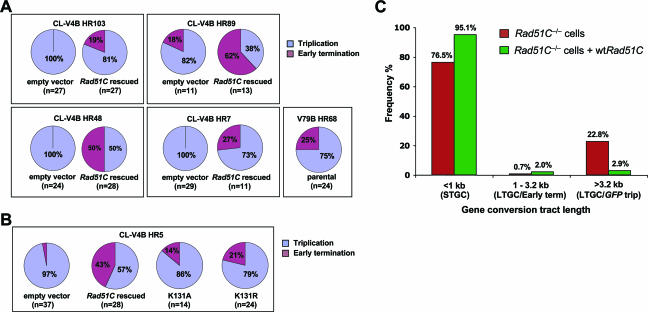
FIG. 1.
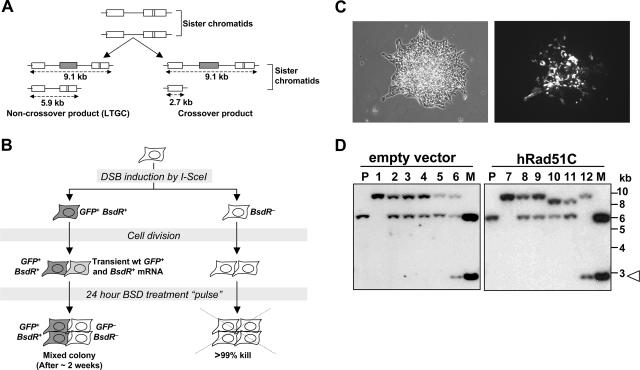
Impaired HR in Rad51C−/− CL-V4B cells. (A) Structure of the SCR reporter. The filled arrowheads indicate promoters. Ovals A and B depict the 5′ exon and 3′ exon of bsdR, respectively. The diagram shows two sister chromatids offset to represent unequal SCR, generating wtGFP (shaded). Three outcomes of I-SceI-induced recombination are shown: gene conversion that terminates prior to duplication of bsdR exon B (1, STGC); gene conversion that results in duplication of bsdR exon B but terminates prior to generating a third GFP copy (2, LTGC/early termination); and gene conversion that results in duplication of the bsdR cassette, generating a third GFP copy (3, LTGC/GFP triplication). Wild-type BsdR mRNA is shaded. Tr GFP, truncated GFP. (B) I-SceI-induced GFP+ frequencies in CL-V4B HR103, HR89, and HR5, transiently cotransfected with I-SceI expression vector and with either control empty vector (gray bars) or hRad51C expression plasmid (black bars). Error bars represent standard errors of the means. P < 0.002, according to a t test between all sample pairs expressing hRad51C and empty vector. (C) Rad51C protein abundance and actin loading control for the experiment whose results are depicted in panel B. Lanes 1, 3, and 5: empty vector in HR103, HR89, and HR5, respectively; lanes 2, 4, and 6: wtRad51C in the same three clones. (D) I-SceI-induced GFP+ frequency in the same three CL-V4B HR clones stably expressing control vector (gray bars) or wtRad51C (black bars). In all clones, P was <0.001 according to a t test between wtRad51C- and empty vector-transfected samples. Striped bars, I-SceI-induced GFP+ frequency in V79B HR55 and HR68. (E) Abundance of Rad51C, XRCC3, and actin proteins in V79B (lane 1), CL-V4B (lane 2), CL-V4B HR103, HR89, and HR5 expressing empty vector (lanes 3, 5, and 7) or wtRad51C (lanes 4, 6, and 8), CHO-AA8 cells (lane 9), irs1SF XRCC3−/− cells (derived from CHO-AA8) (lane 10), and irs1SF complemented with wthXRCC3 (lane 11).
To discriminate between STGC and LTGC, we developed a way to positively select the “GFP triplication” outcome (28). We introduced a cassette containing two artificial exons of the BSD resistance gene (bsdR) placed head to toe, between the two GFP copies of the reporter (Fig. 1A). As a single copy, bsdR exons A (5′ exon) and B (3′ exon) are incorrectly oriented and the cell is sensitive to BSD (BsdR−). In the context of STGC, no duplication of the bsdR cassette occurs and the cell remains BsdR− (Fig. 1A, outcome 1). Conversely, a gene conversion tract of ≥1,031 bp will result in the duplication of bsdR exon B during gene conversion, resulting in its placement 3′ of exon A (Fig. 1A). In this case, splicing between exon A and exon B of the neighboring (duplicated) cassette generates wtbsdR mRNA and the cell becomes resistant to BSD (BsdR+). Thus, our functional definition of STGC entails a gene conversion tract of <1,031 bp, while LTGC entails a tract of ≥1,031 bp. If the gene conversion tract extends beyond ≥3,250 bp, a third GFP copy is introduced during gene conversion and the entire bsdR cassette is duplicated (Fig. 1A). We previously validated this system for human osteosarcoma U2OS cells and for mouse embryonic stem (ES) cells (28, 42).
Survival in BSD and GFP triplication does not differentiate between LTGC and crossing over between sisters. The experiments described below specifically address this point and indicate that LTGC is indeed the major pathway causing GFP triplication in the BsdR+ cell types examined here.
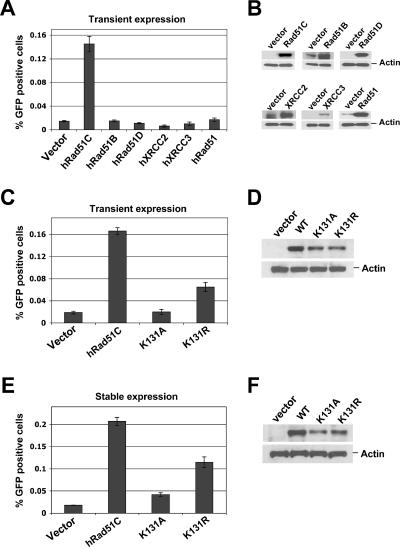
We electroporated CL-V4B and V79B cells, in parallel, with the linearized SCR reporter and identified individual clones that contain only one intact, randomly integrated copy of the reporter (see Materials and Methods). Following transient transfection of individual SCR reporter clones with a plasmid encoding I-SceI, we noted induction of both GFP+ cells and BsdR+ colonies in all clones. To assess the role of Rad51C in these HR outcomes, we transiently transfected individual reporter clones with either wt human Rad51C expression vector or control empty vector, cotransfecting, in parallel, either an I-SceI expression vector or a control vector (see Materials and Methods). In three independent clones (CL-V4B HR103, HR89, and HR5), cotransfection of control vector produced low levels of I-SceI-induced GFP+ cells (∼0.01 to 0.02% of transfected cells), but coexpression of wtRad51C increased I-SceI-induced HR efficiency ∼10-fold (0.1 to 0.2% of transfected cells) (Fig. 1B and C) to levels similar to those of parental V79B cells (Fig. 1D). This result confirms observations made previously by French et al. (9). The HR function of wtRad51C was specific, since transient expression of Rad51, Rad51B, Rad51D, XRCC2, or XRCC3 failed to restore efficient HR in CL-V4B cells (Fig. 2A and B). To determine whether stable expression of wtRad51C restores HR function in CL-V4B cells, we generated stable pools of transfectants of CL-V4B HR103, HR89, HR5, and HR48, expressing either wtRad51C or control vector, as described in Materials and Methods. The growth properties of CL-V4B HR clones were unaltered by wtRad51C (data not shown), and plating efficiency increased slightly from ∼55 to ∼60%. Consistent with the transient rescue experiments described above, stable expression of wtRad51C produced an ∼10-fold increase in HR efficiency compared to that of control cultures (Fig. 1D and E and 3A).
FIG. 2.
Lysine 131 is essential for Rad51C HR function. (A) Frequency of I-SceI-induced GFP+ cells in CL-V4B HR103 following transient cotransfection of I-SceI with empty vector, hRad51C, or the indicated hRad51 paralog expression plasmids. P < 0.0025, according to a t test between hRad51C-transfected and all other samples. (B) Abundance of Rad51 and paralog proteins or actin loading control for the experiment whose results are shown in panel A. (C) I-SceI-induced GFP+ frequencies in CL-V4B HR103 cells transiently transfected with control vector, wtRad51C, Rad51C K131A, or K131R expression plasmids. P < 0.01, according to a t test between all pairs of groups (except for vector versus K131A, which was not significant). (D) Abundance of Rad51C proteins or actin loading control for the experiment whose results are shown in panel C. (E) I-SceI-induced GFP+ frequencies in CL-V4B HR103 cells stably expressing empty vector, wtRad51C, or mutant Rad51C. P < 0.01, according to a t test between all pairs. (F) Abundance of Rad51C proteins or actin loading control for the experiment whose results are shown in panel E.
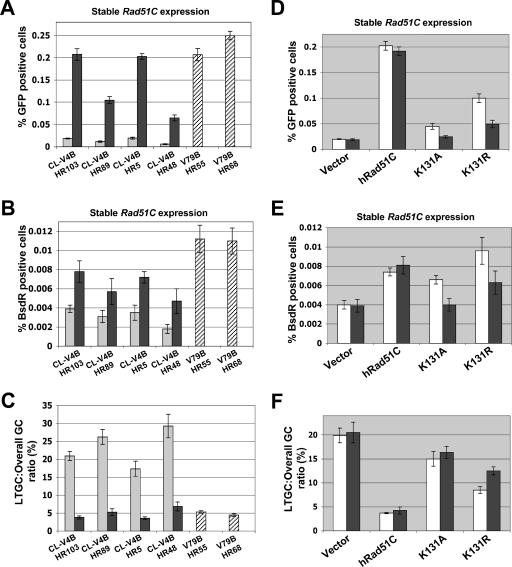
FIG. 3.
Bias in favor of LTGC in CL-V4B cells. (A) I-SceI-induced GFP+ frequencies for CL-V4B HR103, HR89, HR5, and HR48 and V79B HR55 and HR68 cells. Gray and black bars indicate stable expression of control vector and wtRad51C, respectively. Striped bars represent V79B HR cells. This figure partially repeats data from Fig. 1D. In all CL-V4B HR clones, P was <0.0025 according to a t test between hRad51C- and empty vector-transfected samples. (B) Frequencies of I-SceI-induced BsdR+ colonies (same experiment as that for panel A). (C) Ratio of I-SceI-induced BsdR+/GFP+ frequencies (LTGC/overall GC, expressed as a percentage) from the experiment whose results are shown in panels A and B. In all CL-V4B HR clones, P was <0.01 according to a t test between wtRad51C- and empty vector-transfected samples. (D) I-SceI-induced GFP+ frequencies in CL-V4B HR103 (white bars) and CL-V4B HR5 (black bars), stably expressing control vector, wtRad51C, or K131 mutants. (E) I-SceI-induced BsdR+ frequencies for the experiment whose results are shown in panel D. (F) Ratio of I-SceI-induced BsdR+/GFP+ frequency from the experiment whose results are shown in panels D and E. P < 0.01, according to a t test between all pairs of groups (except for vector versus K131A [P = 0.035] and K131A versus K131R [P = 0.027]).
Rad51C residue K131 is located on the phosphate-binding loop of the Walker A ATP-binding motif (9). To determine the role of ATP binding and hydrolysis in Rad51C HR function, we compared the function of wtRad51C with those of Rad51C ATP-binding (K131A) and ATP hydrolysis (K131R) mutants. Transient expression of Rad51C K131A in CL-V4B HR103 failed to restore HR function, whereas Rad51C K131R was partially functional (Fig. 2C and D). Stable expression of these mutants in CL-V4B HR103 and HR5 produced similar results (Fig. 2E and F and 3D). This suggests that ATP binding and hydrolysis are required for fully efficient Rad51C-mediated HR function.
Bias in favor of long-tract gene conversion in CL-V4B cells.
We asked whether Rad51C mutation affects the balance between STGC and LTGC outcomes. Using the SCR reporter described above (Fig. 1A) (28), overall I-SceI-mediated HR is measured by induction of GFP+ cells, whereas I-SceI-mediated LTGC is measured by induction of BsdR+ GFP+ colonies. Since I-SceI-induced BsdR+ events are a subset of I-SceI-induced GFP+ events, it follows that the ratio of I-Sce-I-induced BsdR+ frequency to I-Sce-I-induced GFP+ frequency equals the estimated probability of an I-Sce-I-induced gene conversion resolving as LTGC. At the ROSA26 locus of mouse ES cells, this ratio was found to be ∼7% (42).
We quantified I-SceI-induced GFP+ and BsdR+ frequencies in CL-V4B HR clones stably expressing either wtRad51C or control vector and in parental V79B HR clones, as described in Materials and Methods. In four CL-V4B clones, HR103, HR89, HR5, and HR48, despite the ∼10-fold increase in I-SceI-induced GFP+ events attributable to expression of wtRad51C, expression of wtRad51C caused only a modest increase in I-SceI-induced BsdR+ events (Fig. 3A and B). The corresponding ratio of I-SceI-induced BsdR+ GFP+ events/total GFP+ events (LTGC/overall GC ratio), a direct measure of the probability of a gene conversion event resolving as LTGC, was 4 to 6% in cultures expressing wtRad51C or in parental V79B cells (Fig. 3C), similar to what was observed at the ROSA26 locus in mouse ES cells (42). In contrast, in “unrescued” CL-V4B cells, the ratio of I-SceI-induced BsdR+ GFP+ events to total GFP+ events was increased ∼5-fold to 20 to 30%. Thus, there is a bias in favor of LTGC in CL-V4B cells and this is suppressed by wtRad51C.
Stable expression of the ATP hydrolysis mutant, K131R, in CL-V4B HR103 and HR5 reduced this ratio from ∼20% to ∼10%, while the ATP-binding mutant, K131A, produced an even more modest suppression (Fig. 3D to F). Therefore, ATP binding and hydrolysis by Rad51C are required for efficient suppression of LTGC in CL-V4B cells.
Bimodal distribution of gene conversion tract lengths in CL-V4B cells.
Given that CL-V4B cells show a quantitative dysregulation in LTGC, we asked whether LTGC was qualitatively altered in these cells. For example, Rad51C dysfunction might skew the gene conversion tract length frequency distribution in favor of longer tracts, as was noted previously for cells lacking XRCC3 (5). If so, a higher proportion of BsdR+ colonies in Rad51C−/− cells might entail gene conversions of ≥1,031 bp but ≤3.2 kb. We tested this by assessing the frequency distribution of gene conversion tract length in I-SceI-induced BsdR+ GFP+ colonies derived from CL-V4B cells expressing either wtRad51C or control vector. In parallel, we assessed I-SceI-induced BsdR+ GFP+ colonies in parental V79B cells. These data were obtained using Southern blotting to analyze the pattern of rearrangements within the SCR reporter in BsdR+ colonies.
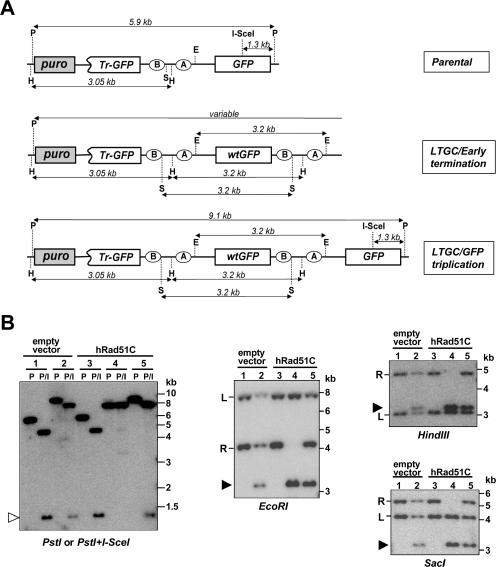
Figure 4A shows the expected restriction fragment sizes in genomic DNA (gDNA) Southern blots hybridized with a GFP cDNA probe for the parental reporter (Fig. 4A, upper panel), for a typical LTGC event that terminates prior to the third GFP copy (Fig. 4A, middle panel), and for the GFP triplication outcome (Fig. 4A, lower panel). Digestion of the parental configuration with PstI generates a single band of 5.9 kb, while combined digestion with PstI and I-SceI generates fragments of 4.6 kb and 1.3 kb, respectively (Fig. 4A and B, left panel, lanes 1 and 3). Either SacI or EcoRI digests generate two GFP-hybridizing fragments, a left and a right “arm” of the SCR reporter (Fig. 4B), the sizes of which vary between different HR clones, reflecting random integration of the SCR reporter. HindIII digestion also generates two GFP-hybridizing fragments, a left arm of 3.05 kb and a right arm, the size of which varies between different HR clones (Fig. 4A).
FIG. 4.
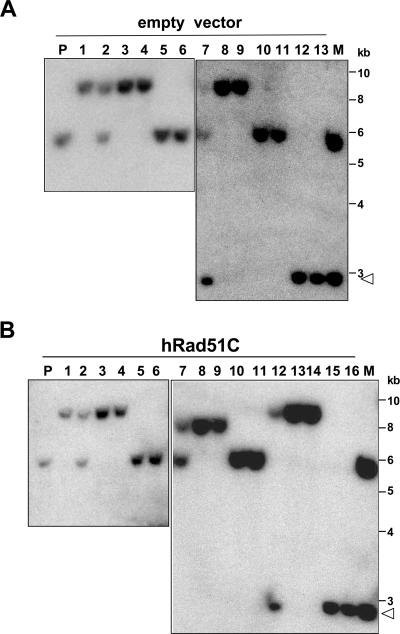
Southern analysis of I-SceI-induced LTGC in CL-V4B HR cells. (A) Structure of the SCR reporter with predicted DNA fragment sizes in Southern blots using GFP cDNA probe from unrearranged parental (upper panel), LTGC/early-termination (middle panel), and LTGC/GFP triplication (lower panel) outcomes (not drawn to scale). P, PstI; H, HindIII; S, SacI; E, EcoRI; puro, puromycin; Tr-GFP, truncated GFP. (B) Southern blot analysis of five individual CL-V4B HR7 clones stably expressing control vector or wtRad51C, as indicated. The lane number corresponds to the same clone in each panel. The restriction enzymes used are shown under each panel. (Left panel) P, PstI digest; P/I, PstI-plus-I-SceI digest; open arrowhead, 1.3-kb fragment released by PstI-plus-I-SceI double digestion. Lanes 1 and 3, parental configuration; lanes 2 and 5, GFP triplication; lane 4, early-terminating LTGC event (note the absence of an I-SceI site). In digests with EcoRI, HindIII, or SacI, the filled arrowhead indicates a 3.2-kb amplification product corresponding to duplication of part or all of the BsdR cassette. “L” and “R” indicate left and right arms of the reporter, respectively.
In the context of GFP triplication, the PstI fragment increases to 9.1 kb while PstI/I-SceI double digestion generates fragments of 7.8 kb and 1.3 kb, respectively (Fig. 4A, lower panel, and B, left panel, lanes 2 and 5). The presence of an intact I-SceI site in the third GFP copy is proof that the triplication event occurred by SCR, since the broken sister chromatid loses its I-SceI site during DSB end processing. Digestion with EcoRI, SacI, or HindIII generates unperturbed left and right arms but introduces a third GFP-hybridizing band of 3.2 kb, corresponding to duplication of the BsdR cassette (Fig. 4B).
As was noted originally by Johnson and Jasin (13), a proportion of LTGC events generate rearrangements other than full triplication of the recombining reporter gene (13, 28, 42). We again noted such rearrangements in some CL-V4B I-SceI-induced BsdR+ GFP+ colonies. These entailed a gene conversion tract long enough to result in duplication of bsdR exon B (≥1,031 bp) but not long enough to generate a third GFP copy (<∼2,900 bp) (Fig. 1A, outcome 2, and 4A, middle panel). These “early-terminating” LTGC events are characterized by altered size of the PstI fragment, the absence of an I-SceI site, and alteration in the right arm of the reporter, for example, the colony shown in lane 4 of all panels of Fig. 4B. In this colony, the existence of an intact additional 3.2-kb GFP-hybridizing band following digestion with SacI, HindIII, or EcoRI, combined with complete loss of the right arm, indicates that the gene conversion tract was long enough to duplicate the entire bsdR cassette (≥2,216 bp) but was not long enough to generate a third copy of GFP (<∼2,900 bp). Another LTGC type also lacked an I-SceI site and showed alterations in the right arm of the reporter but differed from simple early-terminating LTGC events by the gain of additional restriction sites between the terminus of the long gene conversion tract and the right arm of the reporter (data not shown). These rearrangements reflect an early-terminating LTGC event involving the left-hand end of the DSB and, most likely, illegitimate recombination of the right-hand end of the break. In terms of gene conversion tract length, therefore, these events belong to the category of early-terminating LTGC events.
We assigned each BsdR+ colony analyzed to one of the two categories: GFP triplication and early termination. Strikingly, in CL-V4B HR clones 103, 48, and 7 that lacked wtRad51C, 100% of LTGC events (81/81 colonies examined) entailed GFP triplication, consistent with a gene conversion tract length of ≥3,250 bp (Fig. 5A). Stable expression of wtRad51C in the same HR clones altered LTGC in favor of early-terminating events, with proportions ranging between 19% and 50% of all LTGC events (Fig. 5A). In CL-V4B HR89 cells lacking wtRad51C, 18% (2/11) of LTGC events were early terminating and 82% (9/11) entailed GFP triplication (Fig. 5A). In CL-V4B HR5 cells lacking wtRad51C, 3% (1/37) of LTGC events were early terminating and 97% (36/37) entailed GFP triplication (Fig. 5B). As was noted for other CL-V4B HR clones, stable expression of wtRad51C shifted the distribution in CL-V4B HR89 and HR5 in favor of early-terminating events at the expense of GFP triplication (Fig. 5A and B). In all, only 2.5% (3/128) of LTGC tract lengths in CL-V4B HR clones lacking wtRad51C fell between 1,031 and 3,250 bp, while 97.5% (125/128) were ≥3,250 bp. For isogenic cultures expressing wtRad51C, 39% (42/107) of LTGC events were between 1,031 and 3,250 bp, while 61% (65/107) were ≥3,250 bp (a distribution similar to that seen in V79B HR68 cells). A χ2 analysis showed these differences to be highly significant (P < 0.001 for the pooled set of CL-V4B HR clones). Therefore, a lack of Rad51C skews LTGC in favor of tract lengths of ≥3,250 bp, and this phenotype is suppressed by wtRad51C.
FIG. 5.
Gene conversion tract length frequency distribution in CL-V4B cells. (A) Relative frequencies of I-SceI-induced GFP triplication (blue) and early-terminating (purple) outcomes of LTGC in CL-V4B HR clones expressing either empty vector or wtRad51C and in V79B HR68 cells. The number in parentheses shows the number of colonies analyzed. (B) Same data for CL-V4B HR5 expressing control vector, wtRad51C, or K131 mutants. P < 0.001, according to a χ2 analysis of a combined set of CL-V4B HR clones for wtRad51C versus control vector. (C) Mean frequencies of gene conversion tract lengths in CL-V4B HR103, HR89, HR5, and HR48 cells lacking Rad51C (red bars) or stably expressing wtRad51C (green bars). This figure combines data shown in panels A and B and Fig. 3C to indicate relative frequencies of STGC, LTGC/early termination (term), and LTGC/GFP triplication (trip), averaged across the four HR clones.
We pooled data from four clones (CLV-4B HR103, -89, -5, and -48), so as to represent the relative proportions of STGC (tract length, <1 kb), early-terminating LTGC (tract length, between 1 kb and 3.2 kb), and GFP triplication (tract length, >3.2 kb) events in isogenic cell lines expressing either wtRad51C or control empty vector (Fig. 5C). The data summarized in Fig. 5C take into account quantitative data from Fig. 3C, 5A, and 5B. As shown in Fig. 5C, gene conversion tract lengths in CL-V4B cells lacking wtRad51C fall into two quite distinct populations: those less than 1,031 bp (STGC) and those greater than 3,250 bp (LTGC/GFP triplication). The observation of a bimodal distribution of gene conversion tract lengths in Rad51C−/− cells (i.e., the near absence of gene conversion tracts between 1,013 and 3,250 bp) is an important finding that is further addressed in Discussion.
To determine how ATP binding and/or hydrolysis by Rad51C affects the distribution of gene conversion tract lengths in CL-V4B cells, we analyzed LTGC rearrangements in CL-V4B HR5 cells stably expressing wtRad51C, the ATP-binding mutant K131A, the ATP hydrolysis mutant K131R, or control vector (Fig. 5B). In cultures expressing the K131A mutant, 14% (2/14) were early terminating, while 86% (12/14) of the clones examined revealed GFP triplication (Fig. 5B). Cultures expressing Rad51C K131R revealed 21% (5/24) early-terminating and 79% (19/24) GFP triplication events. In the same clone, cultures expressing wtRad51C revealed 43% (12/28) early-terminating and 57% (16/28) GFP triplication events. These results are consistent with the quantitative data (Fig. 3D to F), indicating partial function of the Rad51C K131R mutant in suppressing LTGC and minimal residual function of the K131A mutant in suppressing LTGC.
Distinguishing between LTGC and crossover mechanisms of SCR.
As noted above, GFP triplication can arise by either LTGC or CO between sisters. Crossing over entails a complete exchange of material between sisters but need not necessarily involve extensive repair synthesis. Crossing over in this context would likely be the outcome of gap repair with double Holliday junction formation, a mechanism engaged frequently during meiotic gene conversion but seldom during somatic gene conversion (reviewed in reference 17). Clearly, if the majority of GFP triplication events noted in CL-V4B cells were due to crossing over, this would nullify the above interpretations regarding the extent of repair synthesis underlying LTGC. For these reasons, we set out to quantify the relative contribution of LTGC and CO to GFP triplication in CL-V4B cells.
LTGC and CO between sister chromatids can be distinguished by analyzing the structure of the donor sister subsequent to the recombination event. LTGC should leave the donor sister unaltered (Fig. 6A), whereas CO reduces the donor sister to a single 5′-truncated GFP copy, structurally identical to the outcome of a single-strand annealing (SSA) event (Fig. 6A). In each case, the cell that inherits the donor sister configuration will be genetically GFP negative and bsdR negative. During prolonged selection in BSD, products of this type should not survive. However, in the cell cycle during which the SCR event occurred, mRNA for both wtGFP and wtBsdR should be produced and transmitted to each daughter cell. Thus, the daughter cell that inherits the donor sister will be transiently functionally GFP+ and BsdR+, even though it is genetically GFP negative and bsdR negative. We reasoned that a brief pulse of selection in BSD might allow these donor sister-containing daughter cells to survive while killing background cells that never contained wtBsdR mRNA (Fig. 6B). Colonies formed later, after removal of BSD, should then be a mixture of BsdR+ GFP+ cells (containing LTGC or CO products) and BsdR− GFP− cells (containing the donor sister structure). Analysis of such sectored colonies would reveal the structure of both the donor and the recipient sister chromatids.
FIG. 6.
GFP triplication arises by a noncrossover mechanism in CL-V4B cells. (A) Schematic diagram showing I-SceI-mediated LTGC and crossover products. Boxes represent GFP copies; the shaded box is wtGFP. The bsdR cassette is not shown. (B) Experimental design for capture of cells containing donor sister chromatids. Shaded cells represent the GFP+ BsdR+ outcome. (C) Phase-contrast image (left panel) and fluorescent image (right panel) of a sectored GFP+/GFP− colony. (D) Southern blot analysis of PstI-digested sectored GFP+/GFP− colonies of CL-V4B HR5 cells expressing control empty vector (lanes 2 to 6) or wtRad51C (lanes 8 to 12) cells. P, parental reporter structure; lanes 1 and 7, GFP triplication; M, 20 pg of PstI-digested plasmid, containing a mixture of parental reporter structure and a 2.7-kb crossover product (open arrowhead).
After plating of I-SceI-transfected cells sparsely onto large tissue culture dishes (see Materials and Methods) and a 7-day pulse of BSD, GFP+ colonies that formed after removal of BSD were 100% GFP+. Therefore, 7 days of treatment in BSD killed the GFP-negative, BsdR-negative daughter cells that inherited the donor sister. In contrast, following a 24-hour pulse of BSD, GFP+ colonies that formed after removal of BSD were a mixture of GFP+ and GFP− cells (Fig. 6C). The 24-h pulse of BSD nonetheless prevented colony formation in >99.9% of background cells (i.e., cells that never produced wtBsdR mRNA), indicating that the 24-h BSD pulse enabled retrieval of both recipient and donor products of the SCR event.
Using this method, we retrieved 13 sectored colonies from CL-V4B HR5 cells stably transfected with empty vector and 11 from CL-V4B HR5 cells stably expressing wtRad51C. We analyzed the sectored colonies by Southern blotting, using PstI-digested genomic DNA and a GFP probe (Fig. 6D). In CL-V4B cells lacking wtRad51C, 11/13 colonies revealed bands of 9 kb and 6 kb, consistent with GFP triplication arising by LTGC (Fig. 6D, left panel, lanes 2 to 5). Two sectored colonies in this group revealed three GFP-hybridizing bands of 9 kb, 6 kb, and 2.9 kb (Fig. 6D, left panel, lane 6). These latter colonies could have arisen from either LTGC or CO coupled with a second rearrangement and are therefore not informative. In CL-V4B cells stably expressing wtRad51C, 8/11 colonies revealed bands of 9 kb and 6 kb, consistent with GFP triplication arising by LTGC (Fig. 6D, right panel, lanes 8 and 9); 2/11 colonies revealed bands of 6 kb and 8 kb, reflecting early-terminating LTGC events (Fig. 6D, right panel, lanes 10 and 11). One remaining colony revealed bands of 9 kb and 2.9 kb (Fig. 6D, right panel, lane 12). The 2.9-kb band in this clone precisely comigrated with the 2,734-bp GFP-hybridizing band detected following PstI digestion of a plasmid mimicking the CO donor structure (Fig. 6D, lanes M). This colony therefore appears to represent a true crossover event. Recloning of a number of the typical sectored colonies generated the expected subclone types, which were either GFP+ BsdR+ (repaired sister) or GFP− BsdR− (donor sister) (Fig. 7).
FIG. 7.
Southern blot analysis of GFP-sectored colonies and their subclones. GFP-sectored colonies (from Fig. 6) were recloned without BSD selection to allow examination of repaired and donor sisters. Genomic DNA was digested with PstI and analyzed by Southern blotting. Lanes P, parental unrearranged reporter; lanes 1: GFP triplication outcome; lanes 2, sectored colony showing LTGC with GFP triplication; lanes 3 and 4, GFP+ BsdR+ subclones; lanes 5 and 6, GFP− BsdR− subclones; lanes M, PstI-digested plasmid showing the parental reporter (6 kb) and CO/SSA product (2.7 kb; open arrowhead) as markers. (A) CL-V4B HR5 cells expressing empty vector. Lane 7, sectored colony showing bands of 9 kb, 6 kb, and 2.7 kb; lanes 8 and 9, GFP+ BsdR+ subclones; lanes 10 to 13, GFP− BsdR− subclones. (B) CL-V4B HR5 cells expressing hRad51C. Lane 7, sectored colony showing LTGC with early termination; lanes 8 and 9, GFP+ BsdR+ subclones; lanes 10 and 11, GFP− BsdR− subclones; lane 12, sectored colony showing likely crossover associated with GFP triplication; lanes 13 and 14, GFP+ BsdR+ subclones; lanes 15 and 16, GFP− BsdR− subclones. Note the comigration of the 2.7-kb band in lanes 15 and 16 with the defined CO/SSA product of lane M (open arrowhead).
In summary, in CL-V4B cells lacking Rad51C, 11/11 (100%) interpretable sectored colonies were shown to arise by LTGC and 0/11 (0%) arose by crossing over. In CL-V4B cells stably expressing wtRad51C, 10/11 (91%) interpretable sectored colonies were shown to arise by LTGC and 1/11 (9%) arose by crossing over. The absence of crossover events in cells lacking Rad51C strongly argues against the idea that the GFP triplication events observed in these cells arose by gap repair with double Holliday junction formation, since some gap repair events would be expected to resolve by crossing over. Therefore, GFP triplication events observed at high frequency in CL-V4B cells lacking Rad51C reflect LTGC-associated repair synthesis extending >3.2 kb in length.
DISCUSSION
In addition to the known role of Rad51C in promoting overall gene conversion, we show here that Rad51C suppresses long-tract gene conversion between sister chromatids. The LTGC events identified here are not associated with crossing over. This suggests that gap repair, a process normally restricted to meiotic cells and associated with crossing over, is not employed at high frequency in Rad51C−/− cells. The LTGC events noted here must therefore entail extensive repair synthesis (>3.2 kb). This work identifies for the first time a gene in higher eukaryotes that differentially regulates long- and short-tract gene conversion during SCR (13, 28, 30).
Rad51C has been proposed to facilitate Rad51 function, perhaps analogous to the roles played by S. cerevisiae Rad55 and Rad57 in supporting ScRad51 function (32). Overexpression of S. cerevisiae RAD51 suppresses the recombination defects of rad55 and rad57 mutants (12, 14). In contrast, the gene conversion defect in CL-V4B cells was reversed by expression of Rad51C but not by Rad51 or other Rad51 paralogs. This points to a unique contribution of Rad51C to HR function. The impaired rescue of function noted in Rad51C lysine 131 mutants suggests that ATP binding and hydrolysis are essential for full Rad51C HR function. This is consistent with a previous report demonstrating that resistance to mitomycin C is restored in Rad51C−/− irs3 cells by wtRad51C but not by ATP-binding/hydrolysis mutants (9).
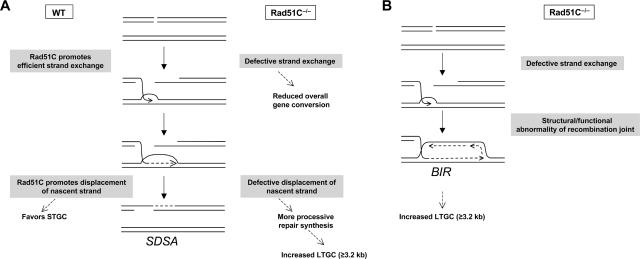
Loss of Rad51C skews gene conversion in favor of LTGC. This might suggest a role for Rad51C in terminating repair synthesis during conventional gene conversion. During conventional SDSA-mediated somatic gene conversion, termination is thought to occur by homology-directed annealing of the displaced nascent strand to the free second end of the DSB (17, 27). In the context of SDSA, this annealing process is termed pairing, although it might have some biochemical similarity to second-end capture invoked in the gap repair model of DSB repair (17). If LTGC events in Rad51C−/− cells do indeed arise by SDSA, this would suggest that Rad51C normally plays a role in facilitating pairing of the displaced nascent strand with the second end or in coupling pairing with the termination of repair synthesis. This model therefore supposes that STGC and LTGC are products of the same SDSA-type repair mechanism but that Rad51C dysfunction allows a greater proportion of gene conversions to “escape” beyond the initial tract of homology. Indeed, in flies, conventional SDSA can entail several kilobases of repair synthesis (1).
How might a defect in termination of repair synthesis explain the bimodal distribution of gene conversion tract lengths in Rad51C−/− cells? For gene conversions in which repair synthesis has extended beyond 1 kb but less than 3.2 kb, the displaced nascent strand lacks homology to the second end and therefore termination is achieved without pairing, presumably mediated by nonhomologous end joining (13). For gene conversions that have extended beyond 1 kb, the probability of termination prior to 3.2 kb is 39% in Rad51C+/+ cells but only 2.5% in Rad51C−/− cells (Fig. 5). One hypothesis that reconciles these observations is that gene conversion normally proceeds by a series of invasion-displacement cycles, as proposed recently (24), and that Rad51C normally plays a role in displacement of the nascent strand during such cycles (Fig. 8A). Such a role might increase the probability of effective pairing with the free homologous end of the DSB (if a homologous end is present), thereby favoring STGC over LTGC. (Note that the length of homology available for pairing is the same for each of the two GFP copies.) However, more-efficient displacement of the nascent strand during repair synthesis might also increase the chance of termination in regions where no homologous end is available (in this system, between 1 and 3.2 kb). In this way, a specific defect in displacement of the nascent strand during SDSA might lead to extensive repair synthesis and could account for both the increased frequency of LTGC events and the bimodal distribution of gene conversion tract lengths in Rad51C−/− cells.
FIG. 8.
Two alternative models of Rad51C-mediated LTGC suppression. (A) Nascent strand displacement hypothesis. Rad51C assists Rad51-mediated strand exchange with the neighboring sister chromatid. Repair synthesis is primed from the 3′ end of the invading strand, and Rad51C promotes displacement of the nascent strand, facilitating pairing with the second end of the DSB (SDSA). The failure to displace the nascent strand in Rad51C−/− cells leads to more-processive repair synthesis and increased LTGC in Rad51C−/− cells. (B) BIR hypothesis. A defect in the structure or function of the recombination joint leads to the establishment of a replication fork at the point of strand invasion in Rad51C−/− cells. Repair synthesis during BIR should be highly processive and could account for increased LTGC in Rad51C−/− cells.
At first glimpse, the 10-fold decrease in gene conversion efficiency in Rad51C−/− cells does not appear to be explained by the nascent strand displacement hypothesis of Rad51C function. This overall defect in HR most likely reflects Rad51C's known role in supporting efficient Rad51 function (32). It is notable that Rad51C exists in two distinct complexes, BCDX2 and CX3 (22, 38). Conceivably, one complex could function specifically in assisting Rad51-mediated strand exchange and the other specifically in displacement of the nascent strand during repair synthesis. The LTGC suppression function of Rad51C showed a pattern of dependence on ATP binding and hydrolysis similar to that observed for Rad51C-mediated promotion of overall gene conversion. Thus, we do not at present have evidence that these two putative Rad51C functions are genetically separable, and it therefore remains possible that they are reflections of a single biochemical function of Rad51C. Conceivably, Rad51C could catalyze each half of the putative invasion-displacement cycle, with the invasion half of the cycle being identical to the Rad51 assist function of Rad51C. One might speculate that Rad51C “melts” the donor template to assist both strand invasion and nascent strand displacement.
A second major hypothesis suggested by our findings is that STGC in Rad51C−/− cells is an outcome of conventional gene conversion, but LTGC in these cells is the product of a distinct mechanism of repair synthesis, perhaps a mechanism resembling BIR in S. cerevisiae or recombination-dependent replication in Escherichia coli (15, 20). BIR/recombination-dependent replication would entail the formation of a replication fork at the site of strand invasion; it should be more processive than repair synthesis associated with conventional gene conversion and therefore less prone to early termination (Fig. 8B) (21). The BIR hypothesis readily explains the bimodal distribution of gene conversion tract lengths observed in Rad51C−/− cells. BIR might be engaged by default if the recombination joint in Rad51C−/− cells were structurally or functionally abnormal (2); for example, maturation from paranemic to plectonemic joints might conceivably be impaired in Rad51C−/− cells. However, to date there has been no rigorous demonstration of BIR in mammalian cells. Although our data does not distinguish between the nascent strand displacement and the BIR hypotheses considered here, each model predicts that events subsequent to strand exchange are abnormal in Rad51C−/− cells (Fig. 8).
It is notable that Rad51C interacts with XRCC3 and that gene conversion tract length in XRCC3−/− cells is increased from ∼200 bp to ∼500 bp in an assay system that can detect gene conversions up to 1,000 bp in length (5). It will be important to determine whether Rad51C−/− cells phenocopy XRCC3−/− cells in this regard and, conversely, how XRCC3 mutant cells perform in LTGC by using the system described here. Contrary to a previous study in which RNA interference-treated HeLa cells acutely depleted of Rad51C showed reduced XRCC3 protein abundance (18), we did not find that either protein's abundance is dependent upon the other in the steady state (Fig. 1E). Therefore, it cannot be assumed that XRCC3 is dysfunctional in CL-V4B cells. It would be desirable to reexamine LTGC in primary cells carrying specific mutations in Rad51C. Our attempts at RNA interference-mediated depletion of Rad51C in mouse ES cells have generated only mild defects in HR, likely reflecting incomplete depletion of Rad51C (our unpublished data). It will also be important to identify other mammalian genes that normally suppress LTGC; not all genes that regulate SCR have such a role (42).
Our observations suggest a specific type of genomic instability in cells lacking Rad51C, one in which duplications or amplifications may arise through dysregulated LTGC between sister chromatids. Interestingly, under certain conditions, we have observed concatemer formation between sister chromatids in U2OS cells (28) and in CL-V4B cells (G. Nagaraju, S. Odate, and R. Scully, unpublished observations). End sequence profiling of a breast cancer genome has revealed complex patterns of chromosomal instability involving variegated amplifications of heterologous sequences (39). Such rearrangements could have arisen from LTGC during recombination between heterologous chromosomes. It will therefore be important to determine whether Rad51C suppresses LTGC during recombination between distant genetic loci. It will also be interesting to examine the role of Rad51C in suppressing LTGC between homologs, in view of the role of LTGC in promoting LOH during interhomolog recombination (33).
Acknowledgments
We are very grateful to Margaret Zdzienicka, John Thacker, Patrick Sung, and Roland Kanaar for generous gifts of cell lines, plasmids, and antibodies. We thank Jim Haber, Lorraine Symington, David Livingston, Jac Nickoloff, Steve Elledge, Steve West, and members of the Scully lab for critical comments and discussion.
This work was supported by NIH grants CA95175 and GM073894, an ACS Scholars award, a Leukemia and Lymphoma Society Scholar award, and a Pew Scholars award (to R.S.).
Footnotes
Published ahead of print on 5 September 2006.
REFERENCES
- 1.Adams, M. D., M. McVey, and J. J. Sekelsky. 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299:265-267. [DOI] [PubMed] [Google Scholar]
- 2.Aguilera, A. 2001. Double-strand break repair: are Rad51/RecA-DNA joints barriers to DNA replication? Trends Genet. 17:318-321. [DOI] [PubMed] [Google Scholar]
- 3.Barlund, M., O. Monni, J. Kononen, R. Cornelison, J. Torhorst, G. Sauter, O.-P. Kallioniemi, and A. Kallioniemi. 2000. Multiple genes at 17q23 undergo amplification and overexpression in breast cancer. Cancer Res. 60:5340-5344. [PubMed] [Google Scholar]
- 4.Bosco, G., and J. E. Haber. 1998. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 150:1037-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenneman, M. A., B. M. Wagener, C. A. Miller, C. Allen, and J. A. Nickoloff. 2002. XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination. Mol. Cell 10:387-395. [DOI] [PubMed] [Google Scholar]
- 6.Chen, W., and S. Jinks-Robertson. 1998. Mismatch repair proteins regulate heteroduplex formation during mitotic recombination in yeast. Mol. Cell. Biol. 18:6525-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott, B., C. Richardson, J. Winderbaum, J. A. Nickoloff, and M. Jasin. 1998. Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cell. Biol. 18:93-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.French, C. A., J. Y. Masson, C. S. Griffin, P. O'Regan, S. C. West, and J. Thacker. 2002. Role of mammalian RAD51L2 (RAD51C) in recombination and genetic stability. J. Biol. Chem. 277:19322-19330. [DOI] [PubMed] [Google Scholar]
- 9.French, C. A., C. E. Tambini, and J. Thacker. 2003. Identification of functional domains in the RAD51L2 (RAD51C) protein and its requirement for gene conversion. J. Biol. Chem. 278:45445-45450. [DOI] [PubMed] [Google Scholar]
- 10.Godthelp, B. C., W. W. Wiegant, A. van Duijn-Goedhart, O. D. Scharer, P. P. van Buul, R. Kanaar, and M. Z. Zdzienicka. 2002. Mammalian Rad51C contributes to DNA cross-link resistance, sister chromatid cohesion and genomic stability. Nucleic Acids Res. 30:2172-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin, C. S., P. J. Simpson, C. R. Wilson, and J. Thacker. 2000. Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nat. Cell Biol. 2:757-761. [DOI] [PubMed] [Google Scholar]
- 12.Hays, S. L., A. A. Firmenich, and P. Berg. 1995. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl. Acad. Sci. USA 92:6925-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, R. D., and M. Jasin. 2000. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19:3398-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, R. D., and L. S. Symington. 1995. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol. Cell. Biol. 15:4843-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kogoma, T. 1996. Recombination by replication. Cell 85:625-627. [DOI] [PubMed] [Google Scholar]
- 16.Kraus, E., W. Y. Leung, and J. E. Haber. 2001. Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krogh, B. O., and L. S. Symington. 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38:233-271. [DOI] [PubMed] [Google Scholar]
- 18.Lio, Y. C., D. Schild, M. A. Brenneman, J. L. Redpath, and D. J. Chen. 2004. Human Rad51C deficiency destabilizes XRCC3, impairs recombination, and radiosensitizes S/G2-phase cells. J. Biol. Chem. 279:42313-42320. [DOI] [PubMed] [Google Scholar]
- 19.Liu, Y., J. Y. Masson, R. Shah, P. O'Regan, and S. C. West. 2004. RAD51C is required for Holliday junction processing in mammalian cells. Science 303:243-246. [DOI] [PubMed] [Google Scholar]
- 20.Malkova, A., E. L. Ivanov, and J. E. Haber. 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93:7131-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malkova, A., M. L. Naylor, M. Yamaguchi, G. Ira, and J. E. Haber. 2005. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol. Cell. Biol. 25:933-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masson, J. Y., M. C. Tarsounas, A. Z. Stasiak, A. Stasiak, R. Shah, M. J. McIlwraith, F. E. Benson, and S. C. West. 2001. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 15:3296-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMurray, M. A., and D. E. Gottschling. 2003. An age-induced switch to a hyper-recombinational state. Science 301:1908-1911. [DOI] [PubMed] [Google Scholar]
- 24.McVey, M., M. Adams, E. Staeva-Vieira, and J. J. Sekelsky. 2004. Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics 167:699-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrow, D. M., C. Connelly, and P. Hieter. 1997. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 147:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson, H. H., D. B. Sweetser, and J. A. Nickoloff. 1996. Effects of terminal nonhomology and homeology on double-strand-break-induced gene conversion tract directionality. Mol. Cell. Biol. 16:2951-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puget, N., M. Knowlton, and R. Scully. 2005. Molecular analysis of sister chromatid recombination in mammalian cells. DNA Repair (Amsterdam) 4:149-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rattray, A. J., B. K. Shafer, B. Neelam, and J. N. Strathern. 2005. A mechanism of palindromic gene amplification in Saccharomyces cerevisiae. Genes Dev. 19:1390-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richardson, C., M. E. Moynahan, and M. Jasin. 1998. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 12:3831-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rouet, P., F. Smih, and M. Jasin. 1994. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 14:8096-8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sigurdsson, S., S. Van Komen, W. Bussen, D. Schild, J. S. Albala, and P. Sung. 2001. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 15:3308-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stark, J. M., and M. Jasin. 2003. Extensive loss of heterozygosity is suppressed during homologous repair of chromosomal breaks. Mol. Cell. Biol. 23:733-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sweetser, D. B., H. Hough, J. F. Whelden, M. Arbuckle, and J. A. Nickoloff. 1994. Fine-resolution mapping of spontaneous and double-strand break-induced gene conversion tracts in Saccharomyces cerevisiae reveals reversible mitotic conversion polarity. Mol. Cell. Biol. 14:3863-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taghian, D. G., and J. A. Nickoloff. 1997. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol. Cell. Biol. 17:6386-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takata, M., M. S. Sasaki, E. Sonoda, T. Fukushima, C. Morrison, J. S. Albala, S. M. Swagemakers, R. Kanaar, L. H. Thompson, and S. Takeda. 2000. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol. 20:6476-6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takata, M., M. S. Sasaki, E. Sonoda, C. Morrison, M. Hashimoto, H. Utsumi, Y. Yamaguchi-Iwai, A. Shinohara, and S. Takeda. 1998. Homologous recombination and nonhomologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17:5497-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thacker, J. 2005. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 219:125-135. [DOI] [PubMed] [Google Scholar]
- 39.Volik, S., S. Zhao, K. Chin, J. H. Brebner, D. R. Herndon, Q. Tao, D. Kowbel, G. Huang, A. Lapuk, W. L. Kuo, G. Magrane, P. De Jong, J. W. Gray, and C. Collins. 2003. End-sequence profiling: sequence-based analysis of aberrant genomes. Proc. Natl. Acad. Sci. USA 100:7696-7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, R. C., A. Smogorzewska, and T. de Lange. 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119:355-368. [DOI] [PubMed] [Google Scholar]
- 41.Wu, G. J., C. S. Sinclair, J. Paape, J. N. Ingle, P. C. Roche, C. D. James, and F. J. Couch. 2000. 17q23 amplifications in breast cancer involve the PAT1, RAD51C, PS6K, and SIGma1B genes. Cancer Res. 60:5371-5375. [PubMed] [Google Scholar]
- 42.Xie, A., N. Puget, I. Shim, S. Odate, I. Jarzyna, C. H. Bassing, F. W. Alt, and R. Scully. 2004. Control of sister chromatid recombination by histone H2AX. Mol. Cell 16:1017-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]