Abstract
Recent work with mouse models and human leukemic samples has shown that gain-of-function mutation(s) in Notch1 is a common genetic event in T-cell acute lymphoblastic leukemia (T-ALL). The Notch1 receptor signals through a γ-secretase-dependent process that releases intracellular Notch1 from the membrane to the nucleus, where it forms part of a transcriptional activator complex. To identify Notch1 target genes in leukemia, we developed mouse T-cell leukemic lines that express intracellular Notch1 in a doxycycline-dependent manner. Using gene expression profiling and chromatin immunoprecipitation, we identified c-myc as a novel, direct, and critical Notch1 target gene in T-cell leukemia. c-myc mRNA levels are increased in primary mouse T-cell tumors that harbor Notch1 mutations, and Notch1 inhibition decreases c-myc mRNA levels and inhibits leukemic cell growth. Retroviral expression of c-myc, like intracellular Notch1, rescues the growth arrest and apoptosis associated with γ-secretase inhibitor treatment or Notch1 inhibition. Consistent with these findings, retroviral insertional mutagenesis screening of our T-cell leukemia mouse model revealed common insertions in either notch1 or c-myc genes. These studies define the Notch1 molecular signature in mouse T-ALL and importantly provide mechanistic insight as to how Notch1 contributes to human T-ALL.
Mutations in the Notch1 receptor have been detected in primary human T-cell acute lymphoblastic leukemia (T-ALL) samples and cell lines and in several mouse models of T-ALL (12, 26, 32, 36, 43). In human T-ALL, mutations are observed in the heterodimerization (HD) and/or PEST regulatory domains (43). In contrast to human T-ALL, HD mutations are rare and insertions/frameshift mutations in the PEST region of Notch1 predominate in mouse T-ALL models (12, 26, 32, 36). Consistently, 74% of our spontaneous mouse tal1 tumors exhibit high intracellular levels of Notch1 and evidence of sustained Notch1 signaling (32). Upon inhibition of Notch1 signaling, mouse tal1 leukemic cell lines undergo cell cycle arrest and/or apoptosis, demonstrating that leukemic growth requires a sustained, as-yet-undefined Notch1 signal. Taken together, these studies reveal that Notch1 activation is a common and critical event in T-ALL and raise the possibility that Notch1 pathway inhibitors may have efficacy in the treatment of T-ALL (12, 26, 32, 36, 43).
Upon ligand binding, the highly conserved Notch1 transmembrane receptor undergoes two successive proteolytic cleavages that result in the translocation of the intracellular domain of Notch1 (Notch1IC) to the nucleus (10). Within the nucleus, Notch1IC binds to and displaces the corepressors from CSL/RBP-Jκ [also known as CBF1, Su(H), or Lag-1], thereby relieving transcriptional repression (24, 34). Notch1IC then recruits a member of the Mastermind (MAM) family and other transcriptional activators, such as CBP/p300, GCN5, and PCAF, to activate transcription of target genes such as hes1, deltex, and pre-Tα (8, 24, 34). CycC-cyclin-dependent kinase 8 (CDK8) and CycT1-CDK9/p-TEFb are also found recruited to the Notch1 target gene promoters, and recent work suggests that MAM not only functions in transcriptional activation but also regulates Notch1IC turnover by facilitating CycC:CDK8 phosphorylation of the C-terminal PEST domain of Notch1 (15, 16, 42). Notch1IC phosphorylation by CycC:CDK8 (16) and potentially other kinases promotes PEST recognition and subsequent degradation by the E3 ubiquitin ligases Numb, Fbw7/Sel10, and Itch (28, 35, 41).
Although clearly implicated in the etiology and pathogenesis of mouse and human T-ALL, the precise molecular nature of the Notch1 signal(s) in T-cell leukemogenesis remains unknown. Previous studies of other cell types have implicated Notch1 in the regulation of cyclin D1 and the cell cycle inhibitor p21 (30, 37). Inducible expression of Notch1 has been shown to enhance the expression of c-myc during the culture of mouse hematopoietic progenitors. Reporter assays and electrophoretic mobility shift assay analysis narrowed the Notch1 responsive region within the mouse c-myc promoter; however, the region defined did not contain a typical RBP-J (or CSL) binding sequence, and antibodies to the transfected FLAG-RBJ-VP16 fusion protein failed to supershift the complex, leading the authors to conclude that the activation of c-myc by Notch1 may be indirect (38).
To specifically identify Notch1IC target genes in mouse T-cell leukemia, we developed doxycycline (Dox)-regulated Notch1IC T-ALL cell lines. To generate these leukemic cell lines, we isolated thymomas from a doxycycline-regulatable intracellular Notch1 transgenic mouse (4). Administration of Dox to these leukemic animals inhibited Notch1IC expression and caused rapid tumor regression by induction of apoptosis (4). Similar to what was observed in vivo, addition of doxycycline to the culture medium suppressed Notch1IC expression and caused the leukemic cells to undergo G1 arrest and apoptosis, directly demonstrating the requirement for Notch1, opposed to other γ-secretase-dependent proteins, in leukemic growth/survival. We then used gene expression profiling to reveal the Notch1 signature in mouse T-cell leukemia. Consistent with published work, several known Notch1 target genes and pathways were induced. Microarray analyses and chromatin immunoprecipitation (ChIP) studies identified c-myc as a direct Notch1 target gene in mouse leukemic cells. Importantly, we demonstrate the functional consequences of Notch1 inhibition and identify c-myc as a critical Notch1 target gene in Notch1-mediated leukemogenesis. Consistent with these findings, retroviral insertional mutagenesis (RIM) screening of our tal1 leukemic mouse model reveals common insertions in notch1 or c-myc genes. These studies define the Notch1 molecular signature in mouse T-ALL and importantly provide mechanistic insight into how Notch1 contributes to T-cell leukemia.
MATERIALS AND METHODS
Mice and T-ALL cell lines.
For the RIM screen, wild-type or mut tal1 (R188G;R189G) neonates were injected with 50 μl of Moloney murine leukemia virus (MoMLV) and monitored daily for signs of disease. Kaplan-Meier analysis was performed on MoMLV-infected mut tal1 mice compared to uninfected mut tal1 mice. To develop a Dox-regulated NotchIC T-ALL line, a cohort of Eμ/tTA/notchIC mice (4) were aged and monitored for disease. To convert primary tumors to culture, Eμ/tTA/notchIC tumors were minced into a single-cell suspension using frosted slides and cultured in RPMI with 10% fetal bovine serum, l% glutamine, penicillin/streptomycin, and 50 μM β-mercaptoethanol. Primary tumor cells were plated at three concentrations in a six-well plate: 5 × 106, 1 ×107, and 2 × 107 cells per well. Tumor cells were left undisturbed for 1 week and subsequently fed three times weekly by careful aspiration and addition of fresh medium. When nonadherent cells became confluent, they were transitioned to a 60-mm dish and finally to a 10-cm dish.
IPCR and Southern blot analysis.
Genomic DNA was isolated from MoMLV-infected mut tal1/scl (R188G;R189G) tumors. One microgram of DNA was digested overnight with PstI, heat inactivated, and ligated in a total volume of 600 μl overnight at 16°C. The ligation was ethanol precipitated and digested with ClaI. Following the ClaI digest, inverse PCR (IPCR) was performed using the Long Template Expand kit (Roche, Indianapolis, IN) and primers to MoMLV long terminal repeat (5′CTTGTGGTCTCGCTGTTCCTT3′) and to the region adjacent to the PstI site within MoMLV (5′TTAAGCTAGCTTGCCAAACCTACAGGT3′). PCR products were electrophoresed on a 1% agarose gel, extracted, cloned into pCR4-TOPO (Invitrogen, Carlsbad, CA), and sequenced.
Genomic DNA was extracted from MoMLV-infected tal1/scl tumors and uninfected tal1/scl tumors. Fifteen micrograms was digested overnight with EcoRV (notch1 cluster region I) or Asp718 (notch1 cluster region II), electrophoresed on a 0.8% agarose-1× Tris-borate-EDTA gel, and transferred to a nylon membrane. The membrane was then probed with a cDNA probe corresponding to Notch1 cluster region I or cluster region II as in reference 13.
Western blot analysis.
Protein was isolated from MoMLV-infected tumors and wild-type thymocytes using radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitor tablets (Roche). Fifty micrograms of total protein was resolved on a 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, transferred to a nitrocellulose membrane, and probed with polyclonal antibodies against intracellular Notch1 or with an antibody specific for Notch1 activated by cleavage at Val 1744 (#2421; Cell Signaling Technology). Blots were stripped and reprobed with β-actin (A5441; Sigma, St. Louis, MO) to control for equal loading.
Flow cytometry.
The NotchIC T-ALL 3404 cell line was cultured with doxycycline (2 μg/ml) for the time periods indicated in the text; stained with CD25-phycoerythrin (PE) conjugate, CD4-fluorescein isothiocyanate (FITC), or CD8-PE (Pharmingen); and analyzed by flow cytometry. For cell cycle analysis, the cells were fixed in 70% ethanol, stained with propidium iodide (PI), and analyzed for DNA content.
Microarray analysis.
The Dox-regulated NotchIC T-ALL cell line was treated with doxycycline (2 μg/ml) for 24 h, and RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA), purified using a QIAGEN RNeasy Mini kit (QIAGEN, Valencia, CA), and hybridized to Affymetrix mouse genome 430A2.0 arrays (Affymetrix, Santa Clara, CA). The experiment was performed in triplicate to minimize artifacts. The normalization was performed using the D-chip, and the expression level was modeled using the perfect-match-only model. The probe sets absent in 50% of the arrays of either group were not included in the analysis. The set of differentially expressed genes was determined on the basis of an unpaired Student's t test. All genes with a >1. 5-fold change and a P value of <0.05 were considered to show significant differential expression. For the hierarchical clustering, the top probes showing more than a twofold change and P value of <0.05 were used. A total of 59 unique genes were identified from 81 probe sets. The interaction networks between the genes showing significant differential expression (Fc > 1.5 and P < 0.05) were generated using the web-based interactive package Ingenuity Systems. This analysis results in identification of networks that are differentially perturbed between the two groups.
RNA analyses.
RNA was extracted from MoMLV-infected tal1/scl tumors, tal1/scl tumors, wild-type thymocytes, and tal1/scl thymocytes using Trizol. cDNA was synthesized using the Superscript first-strand synthesis system (Invitrogen). deltex, hes1, and pre-Tα expression was assayed using primers as described in reference 9. Specific c-myc primers were designed using Primer Express software (Applied Biosystems): forward, 5′-CTGTTTGAAGGCTGGATTTCCT-3′; reverse, 5′-CAGCACCGACAGACGCC-3′. To determine c-myc expression levels, cDNA was serially diluted and quantitated using the SYBR green kit (QIAGEN) containing the c-myc-specific primers and β-actin-specific primers. The copy number for c-myc was normalized to the copy number for β-actin.
ChIP assay.
Approximately 108 cells were resuspended in cold phosphate-buffered saline solution containing 1% formaldehyde and incubated for 10 min at 4°C with continuous shaking. After 10 min, the cross-linking was reversed by adding glycine to a final concentration of 0.125 M and incubating the cells at 4°C for 5 min with continuous shaking. Cells were then harvested and washed twice with ice-cold phosphate-buffered saline. Nuclei were isolated by incubating the cells in nucleus isolation buffer {5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 8.0, 85 mM KCl, 0. 5% NP40} for 20 to 30 min on ice. The nuclei were harvested at 4°C by centrifuging the cell suspension at 6,000 rpm for 6 min. The nuclei were resuspended in 1. 5 ml of RIPA buffer (150 mM NaCl, 1% [vol/vol] Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris [pH 8. 0], 5 mM EDTA) containing protease inhibitors. Nuclear lysates were sonicated to yield chromatin fragments of approximately100 to 700 bp as assessed by agarose gel electrophoresis. Nuclear debris was removed by centrifuging the lysates at 4°C for 15 min at 14,000 rpm. Nuclear extracts were precleared by incubation with the protein A Sepharose CL-4B beads (Pharmacia) for 30 min with rocking at 4°C. After centrifugation for 2 min at 3,000 rpm at 4°C, the supernatants were transferred to a new tube. Immunoprecipitation was performed by rocking the extracts overnight at 4°C with the respective antibody. Protein A Sepharose CL-4B beads were added, and the immunocomplexes were allowed to bind to the beads for 2 h at 4°C with rocking. Immunocomplexes bound to beads were then washed once each with RIPA buffer, RIPA buffer containing 500 mM NaCl, and immunoprecipitation wash buffer (10 mM Tris Cl [pH 8.0], 250 mM LiCl, 0. 5% NP40, 0. 5% deoxycholate, 1 mM EDTA) and once with Tris-EDTA. Between washes, samples were rocked end over end for 5 min. Beads were resuspended in 300 μl of elution buffer (50 mM Tris Cl, pH 8.0, 10 mM EDTA, 1% SDS) followed by overnight incubation at 55°C. DNA was phenol-chloroform extracted, ethanol precipitated, allowed to air dry, and finally dissolved in sterile water. Immunoprecipitated DNAs for acetylated histones H3 and H4 were diluted sixfold and threefold, respectively, to keep the PCR in the linear range of amplification. The following set of primers was used to amplify different regions of the genes indicated: for the promoter region of c-myc, 5′-GGAAACTGGGAAATTAATGTA-3′ (forward) and 5′-TTCCCAGAAAGGGGGAGGAGTGA-3′ (reverse); for the TATA region of c-myc, 5′-ACTAGCGCGCGAGCAAGAGAAAA-3′ (forward) and 5′-GGGATTAGCCAGAGAATCTCTCT-3′ (reverse); and for the hes1 promoter region, 5′-GACCTTGTGCCTAGCGGCCA-3′ (forward) and 5′-TCTGTCCCCTAAGGCGACAA-3′ (reverse).
RESULTS
The notch1 locus is a common retroviral insertion site in a tal1 transgenic mouse model of T-ALL.
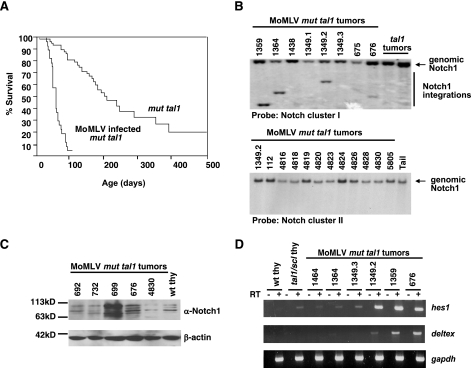
Transgenic mice expressing Tal1 or a DNA-binding mutant Tal1 (mut tal1) develop T-cell acute lymphoblastic leukemia after a long latency, indicating that additional genetic events are required to induce leukemia (21, 31). Both tal1 models induce T-cell leukemia primarily by interfering with E47/HEB function (31, 33). To identify cooperating oncogenes in this T-cell leukemia mouse model, we performed RIM. Wild-type or mut tal1 transgenic neonates were injected with MoMLV. As expected, infection with MoMLV greatly accelerates the rate of disease, with a median survival latency of 81 days for the infected mut tal1 transgenic mice compared to 215 days for uninfected mut tal1 mice (Fig. 1A). In addition, MoMLV-infected mut tal1 mice develop leukemia with complete penetrance. As expected, MoMLV-infected wild-type FVB/N mice also develop disease, but with a significantly longer latency than MoMLV-infected mut tal1 transgenic mice (data not shown). We utilized IPCR to clone the region adjacent to the MoMLV integration in the genome. We determined that the MoMLV provirus integrated into intron 28 of the notch1 gene, which likely results in expression of a constitutively active intracellular Notch1 protein. To determine the frequency of notch1 integrations in MoMLV-infected mut tal1 tumors, we performed Southern blot screening on the remaining MoMLV-infected tumors. We digested genomic DNA from MoMLV-infected mut tal1 tumors with EcoRV, transferred it to a membrane, and probed it with notch1 cDNA probes. The probe contains notch1 cDNA exons 22 to 26 or cluster region I, which has previously been shown to be a common site of integration in MMTVD/myc and E2A/Pbx1 transgenic mice (13, 17). Thirty-two percent (8/25) of the MoMLV-infected mut tal1 tumors contained integrations in the notch1 locus (data not shown and Fig. 1B). Surprisingly, additional Southern blot screening failed to detect MoMLV integrations in the 3′ cluster region II, which encompasses the regulatory PEST domain (Fig. 1B). Previous studies have shown that Notch1 can cooperate with the expression of dominant-negative isoforms of Ikaros, and these forms of Ikaros are also detected in human T-ALL (3, 6, 40).
FIG. 1.
Notch1 activation in MoMLV-infected mut tal1 tumors. (A) Disease acceleration in MoMLV-infected mut tal1 transgenic mice. Shown is a Kaplan-Meier survival plot of MoMLV-infected tal1/scl transgenic mice. The cohort of tal1/scl transgenic mice consisted of 62 mice, and the cohort of infected mice consisted of 27 mice. (B) The notch1 locus is a common integration site in infected tumors. Genomic DNA from MoMLV-infected tumors (1359, 1364, 1438, 1349.1, 1349.2, 1349.3, 675, and 676) and from two uninfected tal1 tumors was digested with EcoRV and hybridized with a probe to cluster region I of Notch1 (13). (C) Notch1 activation in MoMLV-infected tumors. Lysates from tumors with MoMLV integrations in notch1 (699, 676, and 4830) were probed with an anti-Notch1 (α-Notch1) antibody and compared to lysates from infected tal1 tumors with no detectable Notch1 integrations (692 and 732) or from wild-type thymus (wt thy). (D) Hes1 and Deltex are expressed in tumors with MoMLV insertions in Notch1. hes1 and deltex expression was examined in wild-type and preleukemic tal1 thymocytes and MoMLV-infected tumors with insertions in Notch1 using RT-PCR. Glyceraldehyde-3-phosphate dehydrogenase (gapdh) was used as an internal control.
To determine if the integration of MoMLV into the notch1 allele increased intracellular Notch1 levels, we performed Western blot analysis of MoMLV-infected mut tal1 tumors (Fig. 1C). Tumors that contain notch1 integrations (no. 699 and 676) display increased levels of Notch1IC compared to wild-type thymocytes or MoMLV-infected tumors without Notch1 integrations (no. 692 and 732). To further determine if the Notch1 pathway was activated in these tumors, we examined expression of known Notch1 target genes, hes1 and deltex, using reverse transcription-PCR (RT-PCR). Compared to wild-type thymocytes, MoMLV-infected mut tal1 tumors exhibit increased levels of hes1 and deltex expression, suggesting that the Notch1 pathway is constitutively active in the infected mut tal1 tumors (Fig. 1D). Insertions were also frequently observed in the c-myc locus (6/25) (Table 1). Of the 14 tumors with insertions in notch1 or c-myc, only one tumor (no. 4830) contained proviral insertions in both notch1 and c-myc. The remaining 13/14 tumors contained retroviral insertions in either notch1 or c-myc, suggesting that Notch1 or c-Myc activation can cooperate with tal1 to induce leukemia in mice (Table 1).
TABLE 1.
MoMLV-infected tal1 tumors harbor proviral integrations in notch1 or c-myc
| MoMLV tumor | Presence of integration ina:
|
|
|---|---|---|
| Notch | c-Myc | |
| 1349.2 | + | − |
| 1349.3 | + | − |
| 1359 | + | − |
| 1364 | + | − |
| 676 | + | − |
| 731 | + | − |
| 699 | + | − |
| 4830 | + | + |
| 702 | − | + |
| 105 | − | + |
| 4819 | − | + |
| 4820 | − | + |
| 4823 | − | + |
| 4829 | − | + |
+, integration present; −, integration absent.
Effects of GSI treatment on mouse tal1 leukemic cell lines and primary tal1 tumor tissue.
The high frequency of MoMLV integrations in notch1 in the tumors derived from MoMLV-infected tal1 mice prompted us to test whether activating notch1 mutations could be detected in the DNA isolated from spontaneous, primary mouse tal1 tumors. As the RIM screen predicted, we found a high frequency (20/27 [74%]) of mutations that create frameshifts or introduce premature stop codons, resulting in truncations in the PEST domain of Notch1, whereas mutations in the heterodimerization domains of Notch1 were rarely detected (32).
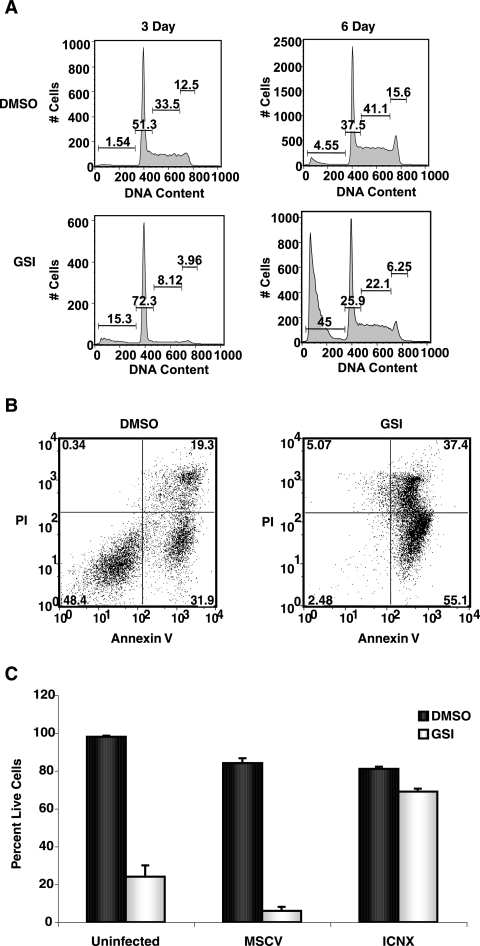
In vitro studies using GSI to inhibit Notch1 have shown that human T-ALL cell lines remain dependent on Notch1 signaling for their growth and survival (43, 44). Interestingly, γ-secretase inhibitor (GSI) treatment of human T-ALL cell lines results in predominantly G1 arrest, and a minority of human T-ALL lines exhibit apoptosis (43). In contrast, treatment of mouse tal1 tumor lines with GSI predominantly results in increases in sub-G1 cells without evidence of G1 arrest (32) (Fig. 2A). The reason for the difference in responses to GSI treatment between human and mouse T-ALL cell lines is unclear. One possibility might be that G1 arrest precedes accumulation in sub-G1. To examine this, we treated mouse T-ALL lines having Notch1 mutations with GSI and examined the cell cycle profiles at earlier time points. GSI treatment of mouse tal1 lines 720 and 130 for 3 days reveals induction of G1 arrest and an increase in sub-G1 cells (Fig. 2A) (data not shown). GSI treatment of mouse leukemic line 130 results in a 21% increase in the percentage of leukemic cells in G0/G1 phase. A concomitant 14% increase in sub-G1 cells was also observed in the GSI-treated cultures (Fig. 2A). By 6 days of GSI treatment, there was a 10-fold increase in sub-G1 cells, with 45% of the cells in the sub-G1 fraction. Similar results were obtained with tal1 leukemic line 720 (data not shown). These studies reveal that in the tal1 cell lines examined, GSI treatment results in an initial G1 arrest followed by an accumulation of sub-G1 cells, suggesting that Notch1 is required for leukemic growth.
FIG. 2.
GSI treatment induces G0/G1 arrest followed by apoptosis, and leukemic growth is rescued by expression of intracellular Notch1. (A) G0/G1 arrest precedes the accumulation of sub-G1 cells. Mouse T-ALL cell line 130 with Notch1 PEST truncation was treated with DMSO or 1 μM N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT) for 3 or 6 days, and cells were assayed for DNA content by staining with PI. (B). GSI treatment of primary tal1 tumor tissue induces apoptosis. Primary thymic masses isolated directly from the animal were left untreated or were treated with 1 μM GSI. Three days later, cells were stained with FITC-Annexin V/PI and analyzed by flow cytometry. (C) GSI-induced effects in mouse T-ALL cells are rescued by human intracellular NOTCH1 (ICNX) expression. Mouse T-ALL line 720 was left uninfected or was infected with vector only or with a retrovirus that expresses human intracellular NOTCH1 (ICNX) and then was left untreated or was treated with 1 μM GSI (MRK-003) for 3 days, and the percentage of viable cells was determined by trypan blue exclusion. The rescue assay was repeated a minimum of three times. A representative experiment is shown.
These in vitro studies suggest that GSI may have efficacy in vivo; however, cell line sensitivity may not reflect primary tumor response in vivo, and therefore it is important to demonstrate GSI efficacy against primary tumor tissue. Primary thymic masses were isolated directly from diseased tal1 transgenic mice and exposed to carrier (dimethyl sulfoxide [DMSO]) or GSI for 3 days. The tumor tissue exhibited variable amounts of background apoptosis upon placement in culture. The addition of GSI to these primary cultures resulted in significant increase in apoptotic cells, as detected by FITC-annexin V/PI staining (Fig. 2B; average increase of 51% apoptotic cells in the GSI-treated primary cultures).
The GSI-induced apoptosis of primary tal1 thymic masses suggests that GSI may have efficacy in vivo in mouse models of T-ALL and, most importantly, in patients. However, the possibility exists that the apoptosis observed in GSI-treated mouse tal1 tumor cell lines and tumor tissue may be in part mediated by inhibition of other γ-secretase-dependent pathways. To rule out this possibility and demonstrate that T-cell leukemic lines and tumor tissue were dependent on Notch1 for growth, we tested whether intracellular Notch1 expression can rescue the GSI-induced apoptosis in multiple mouse tal1 tumor lines. Mouse tal1 tumor lines were left uninfected or were infected with murine stem cell virus (MSCV) retroviral vector only or with a retrovirus that expresses intracellular NOTCH1 (ICNX), and the cultures were treated with either DMSO or GSI. Following 3 days of culture with the GSI, 80% of the uninfected or MSCV-infected cultures had undergone apoptosis (Fig. 2C). Retroviral expression of ICNX prevented the GSI-induced apoptosis, demonstrating that inhibition of the Notch1 pathway is responsible for the effects on leukemic growth.
Development of conditional Notch1IC T-ALL line.
The critical signal(s) Notch1 activation provides during leukemogenesis remains unknown. To elucidate how Notch1 contributes to leukemogenesis and to specifically regulate Notch1IC expression, we developed a Dox-regulated Notch1IC leukemic cell line. To develop this system, we isolated tumors arising in Eμ/tTA/NotchIC transgenic mice (4) and adapted them to culture. Of 14 tumors isolated, only three tumor lines successfully converted to culture, and one of the three remained responsive to doxycycline. Western blot analysis of the Dox-regulated NotchIC T-ALL line demonstrates that following Dox treatment for 24 h, Notch1IC levels decrease to levels seen in wild-type thymocytes (Fig. 3A). To confirm the Notch1IC signaling pathway was intact in this cell line and functioning in a Dox-dependent manner, we performed RT-PCR and assayed hes1 and deltex expression levels following Dox treatment. We observed a significant reduction in hes1 and deltex expression levels following 18 h of Dox treatment and failed to detect any hes1 or deltex expression following 24 h of Dox treatment (Fig. 3B). Decreases in pre-Tα expression were also observed following Dox treatment or Notch1IC repression, indicating that the expression of known Notch1 target genes in T cells was also affected.
FIG. 3.
A conditional Notch1IC mouse T-ALL cell line. (A) Dox administration represses Notch1IC expression. Fifty micrograms of protein isolated from wild-type thymocytes and doxycycline-regulated T-ALL cell line 3404 was left untreated or was treated with 2 μg/ml Dox. Cell lysates were separated on a 7. 5% SDS-PAGE gel and then probed with anti-Notch1 (α-Notch1) antibody. (B) Decreased hes1, deltex, and pre-Tα expression upon the addition of Dox. Expression of hes1, deltex, and pre-Tα in the Dox-regulated NotchIC T-ALL line was examined by RT-PCR following mock treatment or doxycycline (2 μg/ml) treatment for the time periods indicated. Glyceraldehyde-3-phosphate dehydrogenase (gapdh) was used as an internal control. (C) Dox treatment or Notch1IC inhibition induces G0/G1 arrest followed by an increase in sub-G1 cells. Following 48 or 72 h of doxycycline treatment, cells were assayed for DNA content by staining with PI. Similar to GSI treatment of mouse T-ALL lines, Dox treatment of 3404 cells induces G0/G1 arrest followed by an increase in sub-G1 cells.
G1 arrest and apoptosis upon Notch1 inhibition.
To test whether Notch1IC provides an essential proliferative/survival signal(s), we performed cell cycle analysis in the presence and absence of doxycycline. Forty-eight hours following Dox treatment, there was an increase in cells in G1 phase (35.6% to 48%) but no significant change in the percentage of sub-G1 cells (Fig. 3C). However, following 72 h of Dox treatment, there was a statistically significant increase in cells in the sub-G1 phase of the cell cycle (15% to 45.3%), indicating an increase in cells undergoing apoptosis (P = 0.0009) (Fig. 3C). In addition, when the sub-G1 cells were removed, a 22% increase in cells in G0/G1 phase of the cell cycle was observed (P < 0.002; data not shown), demonstrating that Notch1IC in this cell system provides an essential proliferative signal, similar to what has been observed when GSI are added to human and mouse T-ALL cell cultures (32, 43).
Notch1 target genes in T-ALL.
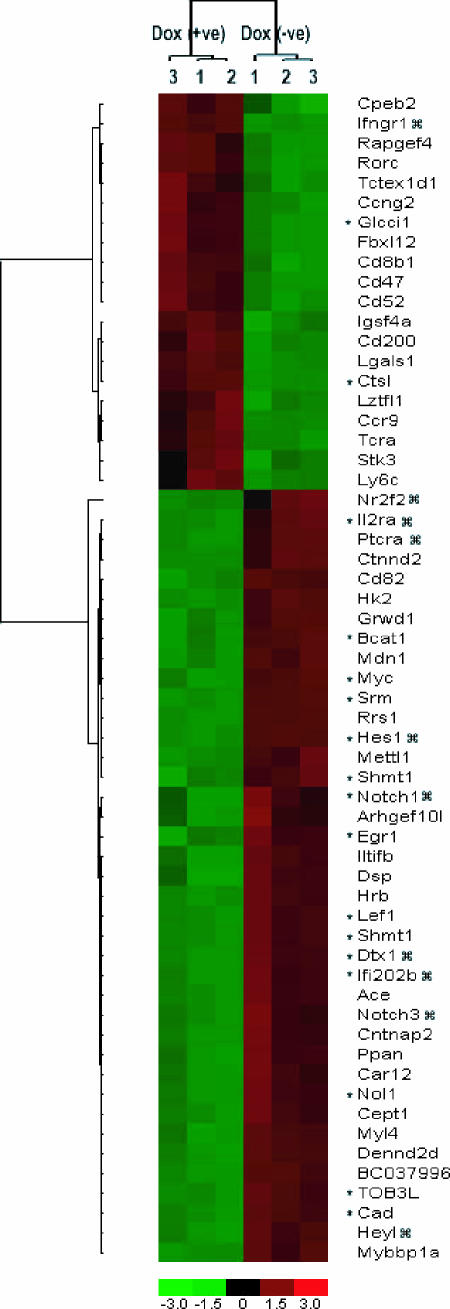
To determine how NotchIC stimulates T-cell proliferation during leukemogenesis, we performed an Affymetrix microarray experiment comparing gene expression profiles in the Notch1IC T-ALL line in the absence of Dox and following a 24-h Dox treatment to suppress Notch1. The systematic analysis of the gene expression data (as explained in Materials and Methods) resulted in the identification of a set of differentially expressed genes (Fc > 1.5 and P < 0.05). As expected, in the presence of continuous expression of Notch1IC, many known Notch1 target genes were induced, including hes1, deltex, il-2rα, and pre-Tα (1, 9) (Fig. 4). In addition, multiple genes implicated in Notch1 signaling were also found to be upregulated in the absence of doxycycline: for example, notch3, adam19 (meltrin β), and members of the interferon 200 gene family (ifi202b) (Fig. 4) (1, 9, 20). The array data show an increase in interleukin-10 (IL-10) levels; IL-10 is known to be upregulated following Notch1IC activation by Jagged1 (2).
FIG. 4.
Notch1-regulated genes in mouse T-ALL. Shown is a heat map of clustered samples in columns and clustered genes in rows of expression data of mouse leukemic cell line 3404 cultured in the presence or absence of doxycycline or Notch1IC. The pseudocolor representation of gene expression ratios is shown with the scale below. The hierarchical clustering was performed for genes having a change of more than twofold (P value of 0.005). NotchIC target genes are indicated by  ; c-myc target genes are indicated by an asterisk.
; c-myc target genes are indicated by an asterisk.
Expression of Notch1IC in this Dox-regulated T-ALL cell line appears to induce multiple genes implicated in regulation of Notch1 signaling. For example, deltex is highly expressed in the absence of doxycycline and has been proposed to antagonize or promote Notch1 function, depending on the model system (2, 27). In addition, Notch1IC induces expression of both nr2f2 (nuclear receptor subfamily 2, group F, member 2) and nrarp (Notch-regulated ankyrin repeat protein). Nrarp has been shown to inhibit Notch function in a CSL-dependent manner and is also capable of inhibiting thymocyte development in vivo (23, 45). As suggested by previous studies, Notch1IC induces the expression of genes that negatively regulate its transcriptional activity or function.
Notch1 directly regulates c-myc expression.
To interpret the gene expression data, a network-based analysis of the differentially expressed genes (360 genes) was performed using the trial version of Ingenuity Systems. The analysis identified many networks with significant interactions, including the gamma interferon (IFN-γ) pathway. Out of these, a network depicting c-myc as a potential Notch1-regulated gene was found to be primarily stimulated (Fig. 4). Real-time PCR analysis confirmed that c-myc expression was doxycycline or Notch1 dependent in the conditional NotchIC leukemic cell line. In the absence of Dox, an average 3.2-fold increase in c-myc expression was observed. Moreover, the expression of 35 known c-Myc target genes was also affected (Fig. 4; genes noted with asterisks).
These studies suggest that Notch1 promotes leukemic growth by sustaining c-myc mRNA levels. Consistently, we found c-myc mRNA levels increased between 9- and 23-fold in primary tal1 tumors that harbor mutations in Notch1 (Fig. 5A). To determine whether increased c-myc mRNA levels reflect increased Notch1 activity, we quantified c-myc mRNA levels in the presence/absence of GSI. GSI treatment resulted in three- to fourfold decreases in c-myc mRNA levels, yet c-myc mRNA levels were maintained in leukemic cells expressing human intracellular Notch1, even in the presence of GSI (Fig. 5B). Collectively, these data demonstrate that Notch1 activity regulates c-myc expression in mouse leukemic T cells. Interestingly, we found hes1, deltex, and c-myc mRNA levels all increased in preleukemic tal1 thymocytes (39), suggesting that Notch1 activation and c-myc deregulation may be early events in T-cell leukemogenesis.
FIG. 5.
Leukemic growth is Notch1 and c-myc dependent. (A) c-myc expression is increased in primary tal1 tumors that contain mutations in Notch1. c-myc RNA levels were examined using real-time PCR in three primary tal1 tumors with insertions/deletions in the PEST region of Notch1. (B) GSI treatment results in downregulation of c-myc mRNA levels, whereas c-myc levels are maintained in leukemic cells infected with an intracellular Notch1 retrovirus. c-Myc RNA levels were measured using real-time PCR. RNA was harvested from the leukemic cell line 720 treated with DMSO or 1 μM N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT) and from 720 cells that were infected with MSCV or with a human ICNX retrovirus treated with DMSO or 1 μM DAPT. c-Myc levels were quantified using a real-time PCR assay. (C) Leukemic growth is rescued by c-Myc or intracellular Notch1 expression. Multiple mouse leukemic cell lines were left uninfected or were infected with MSCV or with retroviruses expressing either human intracellular NOTCH1 (ICNX) or mouse c-myc. Cultures were treated with either DMSO or DAPT (1 μm) for 6 days, and viable cells were determined by trypan blue staining. The experiment was repeated at least three times, and one representative experiment is shown.
To test whether enforced c-myc expression rescues the GSI-induced growth arrest and apoptosis, we infected two Notch1-dependent mouse tal1 cell lines, 720 and 130, that express mutated forms of Notch1 lacking an intact PEST domain. Leukemic cell lines were left uninfected or were infected with MSCV or with retroviruses expressing either intracellular NOTCH1 (ICNX) or c-myc. Following 6 days of GSI treatment, 80% of 720 cells had undergone apoptosis (Fig. 2A and 5B). Retroviral expression of c-myc prevented the GSI-induced apoptosis and promoted growth to a similar extent as the expression of intracellular Notch1 (ICNX), suggesting that sustained activation of c-myc is one major target of Notch1 in T-ALL.
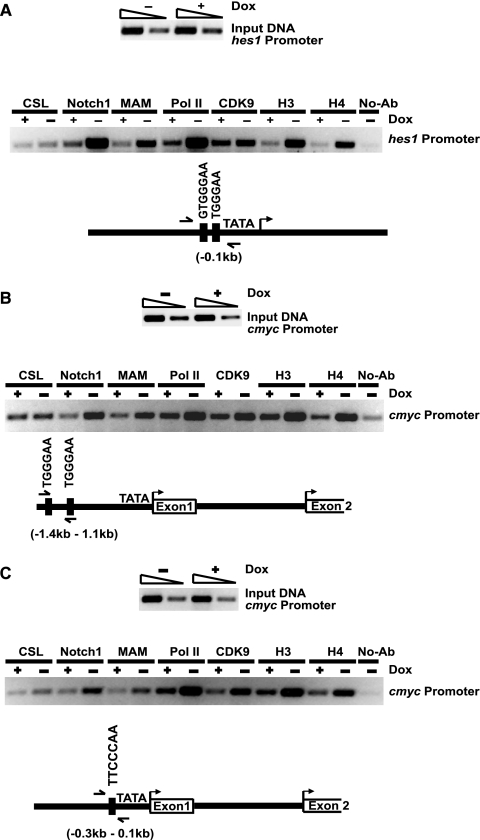
The kinetics of c-myc expression in the Notch1-regulated leukemic cell line and the presence of conserved, tandem CSL binding sites in the regulatory regions of mouse and human c-myc suggest that c-myc may be directly regulated by Notch1. To address whether Notch1 regulates c-myc expression directly, we examined Notch1IC recruitment to c-myc regulatory regions using a ChIP assay. Previous work has demonstrated Notch1 recruitment to the hes1 promoter (16); we therefore used hes1 as a positive control for our ChIP assays. Consistent with these published studies, we observed Dox-dependent recruitment of intracellular Notch1 and its coactivator, MAM, to the mouse hes1 promoter region in our leukemic cell line (Fig. 6A). Consistent with the induction of hes1 expression in the absence of Dox (presence of Notch1IC), we observed recruitment of RNA polymerase II/CDK9 and increases in histone H3 and H4 acetylation (Fig. 6A). Similarly, in the absence of Dox, we found intracellular Notch1 recruited to the mouse c-myc promoter region (Fig. 6B and C). To test whether Notch1IC recruitment correlated with changes in c-myc transcriptional activity, we compared the acetylated histone H3 and H4 binding as well as RNA polymerase II recruitment to the mouse c-myc promoter in the presence and absence of Dox. In the absence of Dox and the presence of intracellular Notch1, increases in acetylated histones H3 and H4 and RNA polymerase II recruitment were consistently observed, indicating that c-myc can be directly regulated by intracellular Notch1. Whereas when the expression of Notch1IC was inhibited by Dox, decreased binding of Notch1IC, MAM, RNA polymerase II, and CDK9 was observed. The Dox-mediated suppression of Notch1IC expression was also accompanied by decreases in histone H3 and H4 acetylation at the c-myc regulatory regions (Fig. 6B and C). None of these interactions were observed in the absence of specific antisera, and none of these proteins bound mouse c-myc in a 3′ intergenic region downstream of the polyadenylation site (data not shown). In sum, the Dox-induced growth arrest and apoptosis were coincident with the depletion of the Notch1IC/MAM transcriptional complex from c-myc regulatory regions and with decreases in c-myc expression. Considered together, these studies strongly suggest that Notch1 contributes to T-cell leukemogenesis by directly regulating the expression of c-myc.
FIG. 6.
Dox-dependent recruitment of Notch1IC and MAM to the hes1 and c-myc promoters. (A) ChIP assay for CSL, Notch1IC, MAM, RNA polymerase II (Pol II), CDK9, and acetylated histones H3 and H4. Briefly, the Dox-dependent 3404 leukemic cell line was grown in RPMI medium with 10% fetal bovine serum at 37°C and treated with doxycycline (2 μg/ml) for 48 h (+Dox) or left untreated (−Dox). Immunoprecipitations (IPs) were carried out using the respective monoclonal or polyclonal antibodies or without antibody (No-ab) as a control. Immunoprecipitated and input DNAs were amplified by PCR using primers specific for the mouse hes1 promoter (A) or the mouse c-myc promoter (B and C). PCR-amplified products were analyzed on 2% agarose gels and stained with ethidium bromide.
DISCUSSION
Retroviral insertional mutagenesis revealed that the T-cell oncogene tal1 cooperates with activated Notch1 to induce T-ALL in mice. The notch1 proviral integrations occurred in cluster region I, which contains the LIN-12/Notch repeat (LNR) and epidermal growth factor-like repeat (EGFR) motifs. Conversely, no integrations were detected in cluster region II, located within the 3′ regulatory regions that contain the PEST domain. Previous RIM screens of both E2A-Pbx1 and MMTVD/myc transgenic mice detected MoMLV integrations in the 3′ region of notch1, and these were shown to cooperate with the initiating oncogene to cause disease (13, 17). It is unclear why spontaneous tal1 tumors appear to preferentially truncate the PEST domain (32), whereas analysis of MoMLV-infected tal1 tumors detected integrations in the midregion of notch1 and no 3′ integrations were detected. One possibility is that too few tumors were examined in the RIM screen (n = 25) and that with larger screens, 3′ integrations in notch1 would also be observed. Nonetheless, this screen revealed that increases in Notch1 signaling contribute to mouse T-ALL and prompted us to investigate the status of the Notch1 receptor in spontaneous Tal1 mouse tumors. That analysis revealed that mouse tal1 tumors harbor truncated versions of intracellular Notch1 that delete the PEST regulatory region (32).
To understand how Notch1 mutations contribute to T-cell leukemia, we developed a mouse leukemic line that expresses Notch1IC in a Dox-dependent manner. The conditional Notch1IC T-ALL line provides multiple advantages over other experimental systems to examine the function of Notch1 in leukemogenesis. Although GSI are commonly used to inhibit the cleavage and activation of Notch receptors (8), they do not specifically inhibit Notch1 but also inhibit cleavage of a number of other transmembrane proteins (7, 18, 25, 29). Moreover, the response of human and mouse leukemic cell lines to GSI is not uniform: sensitive cell lines undergo variable amounts of cell cycle arrest/apoptosis, and effects on cell growth occur anywhere from 3 to 7 days following GSI treatment (32, 43). In contrast, doxycycline specifically inhibits Notch1IC expression and allows temporal comparison of the recruitment of intracellular Notch1 and its coactivators to target gene loci.
Gene expression profiling of this Dox-regulated Notch1IC T-ALL line demonstrated upregulation of numerous known Notch1IC target genes, including nrarp, nr2f2, notch3, hes1, cd25, adam19 (or β-meltrin), deltex, il-10, irf-4, egr1, and pre-Tα (1, 9, 20). Ingenuity Systems software analysis revealed the c-Myc pathway as the central pathway (other than Notch1) induced in the absence of Dox or presence of active Notch1 signaling. c-Myc and 35 known c-Myc target genes (cad, odc1, nol1, bcat1, srm, and others denoted with an asterisk in Fig. 4) were induced/repressed in the absence of Dox or presence of intracellular Notch1. We demonstrate that Notch1IC mediates leukemic growth by inducing c-myc expression since retroviral expression of c-myc rescues the growth arrest/apoptosis induced upon GSI treatment of multiple mouse tal1 leukemic lines. In our RIM screen, proviral insertions were also observed in the c-myc locus (6/25 tumors), suggesting that a certain threshold level of c-myc expression is essential for induction/maintenance of T-ALL (achieved by either Notch1 activation or direct insertion near c-myc).
Preleukemic studies of mouse models of T-ALL have demonstrated that T-cell oncogenes like tal1 and/or lmo1/2 arrest thymocyte development (5, 19, 31), and thus activation of Notch1 and potentially c-myc may occur to rescue developmentally arrested, preleukemic thymocytes. Consistent with this model, we detected evidence of Notch1 activation and increased c-myc levels in preleukemic tal1 thymocytes (39), suggesting that Notch1 mutations are early events in T-cell leukemogenesis and may reflect a response to differentiation arrest. Evidence of early Notch1 activation has also been observed in SCL/LMO1 transgenic mice (26). Although c-myc levels clearly correlate with Notch1 activity in preleukemic or leukemic stages of disease and retroviral c-myc expression prevents GSI-induced cell cycle arrest (Fig. 5C), it was unclear whether c-myc activation was a direct or indirect effect of Notch1 activation. The kinetics of c-myc activation (as compared to the direct Notch1 target, hes1) suggests that Notch1IC/MAM complex may directly stimulate c-myc transcription. Consistent with the direct model, the promoter of the mouse c-myc gene contains three CSL binding sites (Fig. 6B and C). One of the three CSL binding sites found in the promoter region is also conserved in the human c-MYC promoter region, suggesting that this may also be Notch1 regulated in human T-ALL. Moreover, increases in hes1 or c-myc mRNA levels were accompanied by an enrichment of Notch1IC, Mastermind, and RNA polymerase II/CDK9 at the promoter regions containing the CSL binding sites. The increase in c-myc transcriptional activity was also associated with increases in histone acetylation. These Dox-dependent changes were specific to the promoter region of c-myc that contained conserved CSL binding sites and when 3′ regions of c-myc were interrogated by ChIP, no evidence of Notch1IC or MAM binding was observed.
c-myc has also been recently implicated as a Notch1 target gene in mouse mammary tumorigenesis (22). Mouse mammary tumor virus (MMTV) LTR-driven expression of Notch1IC induces mammary tumors in female mice, and an absence of c-myc delays tumor incidence and penetrance. Mammary tumors express increased c-myc mRNA levels, and a Notch1/Cbf1 complex was shown to bind the c-myc promoter (22). Thus, our study and that of Klinakis et al. reveal c-myc as a direct Notch1 target gene in diverse transformed cell types (i.e., mouse leukemic T cells and mammary tumor cells). Although Klinakis et al. provide genetic evidence that links Notch1 to c-myc, Notch1IC-induced mammary tumors do form, albeit with increased latency, in the absence of c-myc (22). Thus, it remains unclear whether Notch1 target genes other than c-myc also contribute to mammary tumorigenesis or whether Myc family members compensate for an absence of c-myc in the tumors that do form in the MMTV-Notch1IC “null” Myc mice. For potential therapeutic reasons, it will be important to test whether mammary tumor growth, like T-cell leukemic growth, remains Notch1 dependent.
In our study, we provide functional data that demonstrate that Notch1 supports leukemic growth by maintaining c-myc mRNA levels. Consistently, primary tumors isolated from tal1 transgenic mice with increased endogenous Notch1 activity due to deletions in the PEST region (32) exhibit 9- to 23-fold increases in c-myc mRNA levels (Fig. 5A). Notch1 inhibition results in three- to fourfold decreases in c-myc mRNA levels and induction of G1 arrest and apoptosis (Fig. 2 and 5). Thus, this work identifies c-myc as both a direct and critical target of Notch1 in T-cell leukemogenesis.
Although c-Myc promotes entry into the cell cycle by increasing E2F1 levels, increased c-Myc and/or E2F1 levels have been shown to activate the Arf-p53 oncogene checkpoint (11, 46). Consistently, Eμ-myc transgenic mice exhibit apoptosis and acquire Ink4a/Arf deletions (46). INK4A/ARF losses are also frequent in human T-ALL and in mouse tal1 tumors (14, 39). Thus, Notch1 mutation may induce and sustain elevated c-myc levels and thereby provide selective pressure for INK4A/ARF loss.
A percentage of mouse T-ALL lines exhibit evidence of Notch1 activation but appear resistant to GSI, suggesting that additional members of the Notch1 signaling pathway may be mutated in these tumors. For example, inactivating mutations in the negative regulators Numb, Itch, or Fbw7/Sel10 or in the kinases such as CycC:CDK8 (16) which trigger Notch1 ubiquitination could lead to increased Notch1 activation and GSI resistance. Similarly, mutations in Notch1 coactivators Mastermind-like 1, 2, and 3 may also contribute to T-ALL. Although these in vitro studies suggest that GSI may have efficacy in 50% of human T-ALL patients that harbor mutations in the Notch1 receptor (43), the identification of GSI-resistant mouse and human T-ALL lines and the implication that Notch1 activation contributes to mammary tumorigenesis (22) urge the development of additional Notch1 pathway therapeutics perhaps aimed at displacing the intracellular Notch1/MAM complex from the CSL repressor or potentially directly targeting c-Myc.
Acknowledgments
We thank Gerry Weinmaster for rat notch1 cluster I and II cDNA probes, Jon Aster for the human NOTCH1 ICNX retroviral construct, and Warren Pear for the anti-Notch1 antibody. We thank Merck Research Laboratories, Inc., for supplying the GSI (MRK-003) used in some of the in vitro studies.
V.M.S. is a Lymphoma Research Foundation Fellow, and A.J.C. and M.A.K. are Scholars of the Leukemia and Lymphoma Society of America. This work was supported by grants from the ACS and NCI to A.J.C. and by CA-096899 from the NCI to M.K.
Footnotes
Published ahead of print on 5 September 2006.
REFERENCES
- 1.Adler, S. H., E. Chiffoleau, L. Xu, N. M. Dalton, J. M. Burg, A. D. Wells, M. S. Wolfe, L. A. Turka, and W. S. Pear. 2003. Notch signaling augments T cell responsiveness by enhancing CD25 expression. J. Immunol. 171:2896-2903. [DOI] [PubMed] [Google Scholar]
- 2.Bain, G., E. C. Maandag, D. J. Izon, D. Amsen, A. M. Kruisbeek, B. C. Weintraub, I. Krop, M. S. Schlissel, A. J. Feeney, and M. van Roon. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell 79:885-892. [DOI] [PubMed] [Google Scholar]
- 3.Beverly, L. J., and A. J. Capobianco. 2003. Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell 3:551-564. [DOI] [PubMed] [Google Scholar]
- 4.Beverly, L. J., D. W. Felsher, and A. J. Capobianco. 2005. Suppression of p53 by Notch in lymphomagenesis: implications for initiation and regression. Cancer Res. 65:7159-7168. [DOI] [PubMed] [Google Scholar]
- 5.Chervinsky, D. S., X.-F. Zhao, D. H. Lam, M. Ellsworth, K. W. Gross, and P. D. Aplan. 1999. Disordered T-cell development and T-cell malignancies in SCL LMO1 double-transgenic mice: parallels with E2A-deficient mice. Mol. Cell. Biol. 19:5025-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortes, M., E. Wong, J. Koipally, and K. Georgopoulos. 1999. Control of lymphocyte development by the Ikaros gene family. Curr. Opin. Immunol. 11:167-171. [DOI] [PubMed] [Google Scholar]
- 7.Cowan, J. W., X. Wang, R. Guan, K. He, J. Jiang, G. Baumann, R. A. Black, M. S. Wolfe, and S. J. Frank. 2005. Growth hormone receptor is a target for presenilin-dependent gamma-secretase cleavage. J. Biol. Chem. 280:19331-19342. [DOI] [PubMed] [Google Scholar]
- 8.Deftos, M. L., Y. W. He, E. W. Ojala, and M. J. Bevan. 1998. Correlating notch signaling with thymocyte maturation. Immunity 9:777-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deftos, M. L., E. Huang, E. W. Ojala, K. A. Forbush, and M. J. Bevan. 2000. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity 13:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Strooper, B., W. Annaert, P. Cupers, P. Saftig, K. Craessaerts, J. S. Mumm, E. H. Schroeter, V. Schrijvers, M. S. Wolfe, W. J. Ray, A. Goate, and R. Kopan. 1999. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398:518-522. [DOI] [PubMed] [Google Scholar]
- 11.Dimri, G. P., K. Itahana, M. Acosta, and J. Campisi. 2000. Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14ARF tumor suppressor. Mol. Cell. Biol. 20:273-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dumortier, A., R. Jeannet, P. Kirstetter, E. Kleinmann, M. Sellars, N. R. dos Santos, C. Thibault, J. Barths, J. Ghysdael, J. A. Punt, P. Kastner, and S. Chan. 2006. Notch activation is an early and critical event during T-cell leukemogenesis in Ikaros-deficient mice. Mol. Cell. Biol. 26:209-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feldman, B. J., T. Hampton, and M. L. Cleary. 2000. A carboxy-terminal deletion mutant of Notch1 accelerates lymphoid oncogenesis in E2A-PBX1 transgenic mice. Blood 96:1906-1913. [PubMed] [Google Scholar]
- 14.Ferrando, A. A., D. S. Neuberg, J. Staunton, M. L. Loh, C. Huard, S. C. Raimondi, F. G. Behm, C. H. Pui, J. R. Downing, D. G. Gilliland, E. S. Lander, T. R. Golub, and A. T. Look. 2002. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell 1:75-87. [DOI] [PubMed] [Google Scholar]
- 15.Fryer, C. J., E. Lamar, I. Turbachova, C. Kintner, and K. A. Jones. 2002. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 16:1397-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fryer, C. J., J. B. White, and K. A. Jones. 2004. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell 16:509-520. [DOI] [PubMed] [Google Scholar]
- 17.Girard, L., Z. Hanna, N. Beaulieu, C. D. Hoemann, C. Simard, C. A. Kozak, and P. Jolicoeur. 1996. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 10:1930-1944. [DOI] [PubMed] [Google Scholar]
- 18.Haas, I. G., M. Frank, N. Veron, and R. Kemler. 2005. Presenilin-dependent processing and nuclear function of gamma-protocadherins. J. Biol. Chem. 280:9313-9319. [DOI] [PubMed] [Google Scholar]
- 19.Herblot, S., A. M. Steff, P. Hugo, P. D. Aplan, and T. Hoang. 2000. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat. Immunol. 1:138-144. [DOI] [PubMed] [Google Scholar]
- 20.Huang, Y. H., D. Li, A. Winoto, and E. A. Robey. 2004. Distinct transcriptional programs in thymocytes responding to T cell receptor, Notch, and positive selection signals. Proc. Natl. Acad. Sci. USA 101:4936-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelliher, M. A., D. C. Seldin, and P. Leder. 1996. Tal-1 induces T cell acute lymphoblastic leukemia accelerated by casein kinase IIalpha. EMBO J. 15:5160-5166. [PMC free article] [PubMed] [Google Scholar]
- 22.Klinakis, A., M. Szabolcs, K. Politi, H. Kiaris, S. Artavanis-Tsakonas, and A. Efstratiadis. 2006. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc. Natl. Acad. Sci. USA 103:9262-9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krebs, L. T., M. L. Deftos, M. J. Bevan, and T. Gridley. 2001. The Nrarp gene encodes an ankyrin-repeat protein that is transcriptionally regulated by the notch signaling pathway. Dev. Biol. 238:110-119. [DOI] [PubMed] [Google Scholar]
- 24.Kurooka, H., and T. Honjo. 2000. Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 275:17211-17220. [DOI] [PubMed] [Google Scholar]
- 25.Lammich, S., M. Okochi, M. Takeda, C. Kaether, A. Capell, A. K. Zimmer, D. Edbauer, J. Walter, H. Steiner, and C. Haass. 2002. Presenilin-dependent intramembrane proteolysis of CD44 leads to the liberation of its intracellular domain and the secretion of an Abeta-like peptide. J. Biol. Chem. 277:44754-44759. [DOI] [PubMed] [Google Scholar]
- 26.Lin, Y. W., R. A. Nichols, J. J. Letterio, and P. D. Aplan. 2006. Notch1 mutations are important for leukemic transformation in murine models of precursor-T leukemia/lymphoma. Blood 107:2540-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuno, K., R. J. Diederich, M. J. Go, C. M. Blaumueller, and S. Artavanis-Tsakonas. 1995. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development 121:2633-2644. [DOI] [PubMed] [Google Scholar]
- 28.McGill, M. A., and C. J. McGlade. 2003. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 278:23196-23203. [DOI] [PubMed] [Google Scholar]
- 29.Ni, C. Y., M. P. Murphy, T. E. Golde, and G. Carpenter. 2001. Gamma-secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science 294:2179-2181. [DOI] [PubMed] [Google Scholar]
- 30.Noseda, M., L. Chang, G. McLean, J. E. Grim, B. E. Clurman, L. L. Smith, and A. Karsan. 2004. Notch activation induces endothelial cell cycle arrest and participates in contact inhibition: role of p21Cip1 repression. Mol. Cell. Biol. 24:8813-8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Neil, J., M. Billa, S. Oikemus, and M. Kelliher. 2001. The DNA binding activity of TAL-1 is not required to induce leukemia/lymphoma in mice. Oncogene 20:3897-3905. [DOI] [PubMed] [Google Scholar]
- 32.O'Neil, J., J. Calvo, K. McKenna, V. Krishnamoorthy, J. C. Aster, C. H. Bassing, F. W. Alt, M. Kelliher, and A. T. Look. 2006. Activating Notch1 mutations in mouse models of T-ALL. Blood 107:781-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Neil, J., J. Shank, N. Cusson, C. Murre, and M. Kelliher. 2004. TAL1/SCL induces leukemia by inhibiting the transcriptional activity of E47/HEB. Cancer Cell 5:587-596. [DOI] [PubMed] [Google Scholar]
- 34.Oswald, F., B. Tauber, T. Dobner, S. Bourteele, U. Kostezka, G. Adler, S. Liptay, and R. M. Schmid. 2001. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell. Biol. 21:7761-7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu, L., C. Joazeiro, N. Fang, H. Y. Wang, C. Elly, Y. Altman, D. Fang, T. Hunter, and Y. C. Liu. 2000. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J. Biol. Chem. 275:35734-35737. [DOI] [PubMed] [Google Scholar]
- 36.Reschly, E. J., C. Spaulding, T. Vilimas, W. V. Graham, R. L. Brumbaugh, I. Aifantis, W. S. Pear, and B. L. Kee. 2006. Notch1 promotes survival of E2A-deficient T cell lymphomas through pre-T cell receptor-dependent and -independent mechanisms. Blood 107:4115-4121. (First published 31 January 2006; doi: 10.1182/blood-2005-09-3551.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ronchini, C., and A. J. Capobianco. 2001. Induction of cyclin D1 transcription and CDK2 activity by Notchic: implication for cell cycle disruption in transformation by Notchic. Mol. Cell. Biol. 21:5925-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satoh, Y., I. Matsumura, H. Tanaka, S. Ezoe, H. Sugahara, M. Mizuki, H. Shibayama, E. Ishiko, J. Ishiko, K. Nakajima, and Y. Kanakura. 2004. Roles for c-Myc in self-renewal of hematopoietic stem cells. J. Biol. Chem. 279:24986-24993. [DOI] [PubMed] [Google Scholar]
- 39.Shank-Calvo, J. A., K. Draheim, M. Bhasin, and M. A. Kelliher. 2006. p16Ink4a or p19Arf loss contributes to Tal1-induced leukemogenesis in mice. Oncogene 25:3023-3031. [DOI] [PubMed] [Google Scholar]
- 40.Sun, L., M. L. Crotty, M. Sensel, H. Sather, C. Navara, J. Nachman, P. G. Steinherz, P. S. Gaynon, N. Seibel, C. Mao, A. Vassilev, G. H. Reaman, and F. M. Uckun. 1999. Expression of dominant-negative Ikaros isoforms in T-cell acute lymphoblastic leukemia. Clin. Cancer Res. 5:2112-2120. [PubMed] [Google Scholar]
- 41.Tetzlaff, M. T., W. Yu, M. Li, P. Zhang, M. Finegold, K. Mahon, J. W. Harper, R. J. Schwartz, and S. J. Elledge. 2004. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc. Natl. Acad. Sci. USA 101:3338-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallberg, A. E., K. Pedersen, U. Lendahl, and R. G. Roeder. 2002. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by Notch intracellular domains in vitro. Mol. Cell. Biol. 22:7812-7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weng, A. P., A. A. Ferrando, W. Lee, J. P. T. Morris, L. B. Silverman, C. Sanchez-Irizarry, S. C. Blacklow, A. T. Look, and J. C. Aster. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306:269-271. [DOI] [PubMed] [Google Scholar]
- 44.Weng, A. P., and A. Lau. 2005. Notch signaling in T-cell acute lymphoblastic leukemia. Fut. Oncol. 1:511-519. [DOI] [PubMed] [Google Scholar]
- 45.Yun, T. J., and M. J. Bevan. 2003. Notch-regulated ankyrin-repeat protein inhibits Notch1 signaling: multiple Notch1 signaling pathways involved in T cell development. J. Immunol. 170:5834-5841. [DOI] [PubMed] [Google Scholar]
- 46.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]