Abstract
The U2 snRNP auxiliary factor (U2AF) is an essential splicing factor composed of two subunits, a large, 65-kDa subunit (U2AF65) and a small subunit, U2AF35. U2AF65 binds to the polypyrimidine tract upstream from the 3′ splice site and promotes U2 snRNP binding to the pre-mRNA. Based on in vitro studies, it has been proposed that U2AF35 plays a role in assisting U2AF65 recruitment to nonconsensus polypyrimidine tracts. Here we have analyzed in vivo the roles of the two subunits of U2AF in the selection between alternative 3′ splice sites associated with polypyrimidine tracts of different strengths. Our results reveal a feedback mechanism by which RNA interference (RNAi)-mediated depletion of U2AF65 triggers the downregulation of U2AF35. We further show that the knockdown of each U2AF subunit inhibits weak 3′ splice site recognition, while overexpression of U2AF65 alone is sufficient to activate the selection of this splice site. A variant of U2AF65 lacking the interaction domain with U2AF35 shows a reduced ability to promote this splicing event, suggesting that recognition of the weak 3′ splice site involves the U2AF heterodimer. Furthermore, our data suggest that, rather than being required for splicing of all pre-mRNA substrates containing a weak polypyrimidine tract, U2AF35 regulates the selection of weak 3′ splice sites in a specific subset of cellular transcripts.
The removal of introns from precursor mRNAs via splicing requires accurate recognition of splice sites by the spliceosome, an assembly of small nuclear ribonucleoprotein particles (snRNPs) and extrinsic (non-snRNP) protein splicing factors (reviewed in reference 13). Early recognition of the 3′ ends of introns is achieved in higher eukaryotes by the U2 snRNP auxiliary factor (U2AF), which is composed of a large, 65-kDa subunit (U2AF65) and a small, 35-kDa subunit (U2AF35) that interact to form a stable heterodimer (17, 37). U2AF65 binds to the polypyrimidine-rich tract that precedes the 3′ splice site, while U2AF35 interacts with the AG dinucleotide at the intron-exon boundary (20, 35, 38, 40). In contrast to U2AF65, which is essential for splicing, U2AF35 does not appear to be required for the splicing of introns containing strong polypyrimidine tracts, the so-called AG-independent introns (25). However, U2AF35 is essential in vitro for the splicing of introns that contain short or weak polypyrimidine tracts (11, 35).
The mechanism by which the two subunits of U2AF act to promote recognition of a weak 3′ splice site is still unclear. According to the current model, deviation from the consensus recognition sequences results in a decreased affinity of U2AF65 for the pre-mRNA (30). In this circumstance, the binding of U2AF35 to the AG can increase the affinity of U2AF for the pre-mRNA, either because the U2AF65/35 heterodimer makes one additional protein-RNA contact compared to U2AF65 alone (20, 27, 35) or because the heterodimeric complex provides an increased number of interaction surfaces for other proteins of the spliceosome (10).
The 65-kDa subunit of U2AF contains an N-terminal arginine-serine (RS)-rich domain, a U2AF35 interaction domain, and three RRM-type RNA-binding domains (38). The 35-kDa subunit contains a region with weak homology to an RRM-type RNA-binding domain (39) and a carboxy-terminal RS-rich domain, which mediates protein-protein interactions with similar RS domains in members of the serine-arginine (SR) family of splicing factors (34, 41). Many introns that contain nonconsensus splice sites depend on additional RNA sequence elements, termed splicing enhancers, for efficient splicing, and several studies indicate that SR proteins bound to splicing enhancers interact with U2AF35, thereby recruiting U2AF65 to the weak polypyrimidine tract (reviewed in references 2 and 9). U2AF35 is encoded by a conserved gene that has been duplicated during evolution, giving rise to a number of U2AF35-related proteins, which are predicted to maintain the ability to interact with U2AF65 (18, 29, 31, 32). Furthermore, we have recently described an alternatively spliced protein isoform of U2AF35 that interacts with U2AF65 (21). Thus, it is conceivable that a variety of U2AF heterodimeric complexes may form between U2AF65 and either U2AF35 or U2AF35-related proteins.
Based on previous studies indicating that U2AF35, in addition to U2AF65, was required to restore the in vitro splicing of introns with nonconsensus polypyrimidine tracts (11), here we have analyzed the roles of the two subunits of U2AF in promoting the recognition of a weak 3′ splice site in vivo.
MATERIALS AND METHODS
Minigene design and constructs.
pIgM(-INH) and pAdML have been described previously (11) and used as PCR templates for amplification of the 5′IgMPy and 3′AdPY inserts (where IgM is immuglobulin M, INH is splicing inhibitory sequence, Py is polypyrimidine, and Ad is adenovirus). 5′IgMPy was obtained by PCR from pIgM(-INH) using primers IgFor and IgRev, followed by digestion with Asp718. The 3′AdPY insert corresponds to the second exon of the adenovirus major late promoter (AdML) preceded by the last 70 intronic nucleotides (nt). It was obtained by PCR from pAdML using primers AdFor and AdRev, followed by digestion with NotI (for primer sequences, see Table S1 in the supplemental material). The pyPY minigene was obtained by joining the 5′IgMPy and the 3′AdPY inserts by blunt-end ligation of the EcoRV extremities and cloning into Asp718/NotI sites of pCMV56 (Clontech). PYPY and pypy* were prepared from pyPY by replacing the Asp718/EcoRV 5′IgMPy insert and the EcoRV/NotI 3′AdPY insert, respectively, with the corresponding fragments amplified from the pIgM and pAdML mutant derivatives pPyAdML-IgM and pPy*AdML (12). The final chimeric minigenes were confirmed by sequencing. The plasmid expressing hemagglutinin (HA)-tagged U2AF65 (8) has already been described. To construct pcDNA-HA-U2AF65Δ35, U2AF65Δ35 cDNA was obtained by PCR from pU2AF6584Δ150 (8) using primers 65Δ35For and 65Δ35Rev containing EcoRI and BamHI restriction sites. The digested PCR product was then inserted into EcoRI/BamHI sites of pEGFP (Clontech). The obtained plasmid (pEGFP-U2AF65Δ35) was digested with EcoRI and XbaI, and the insert was then cloned into EcoRI/XbaI-cut pcDNA3/HA-N (4). The bidirectional tetracycline-responsive plasmid pBI-U2AF was generated by the sequential insertion of U2AF65-FLAG and U2AF35-HA constructs into the pBI Tet vector (Clontech). A C-terminal FLAG-tagged version of U2AF65 was initially made by PCR using pEGFP-U2AF65 (7) as the template and primers 65FlagFor and 65FlagRev containing SacI and EcoRI restriction sites. After digestion, the amplified product was introduced into the pEGFP-U2AF65 vector previously digested with SacI and EcoRI restriction enzymes to generate pEGFP-U2AF65-FLAG. This construct was confirmed by DNA sequencing. The cDNA of U2AF65-FLAG was then obtained by digestion of pEGFP-U2AF65-FLAG with Eco52I and SalI. The 5′ end of the obtained fragment was filled in with the Klenow fragment, and the cDNA was inserted into the blunt-ended NotI/SalI-cut pBI vector (Clontech), generating pBI-U2AF65-FLAG. The U2AF35-HA insert was obtained from pTRE-U2AF35-HA by digestion with BamHI, followed by fill-in and digestion with NheI. The obtained product was cloned into either blunt-ended MluI/NheI-digested pBI (pBI-U2AF35-HA) or pBI-U2AF65-FLAG (pBI-U2AF). The commercially available plasmid pTet-On vector (Clontech) was used to express the transactivating rtTA receptor.
Cell culture, RNA interference (RNAi), and transfection procedures.
Human HeLa cells (ECACC 93021013) were grown as monolayers in Dulbecco's minimum essential medium with Earle's salts, supplemented with 10% (vol/vol) fetal calf serum and 1% (vol/vol) nonessential amino acids (Gibco, Invitrogen). When pBI constructs were used, 2 μg/ml doxycycline (Sigma) was added to the medium at the time of transfection. Small interfering RNA (siRNA) duplexes were synthesized as 21-mers with 3′ dTdT overhangs (EUROGENTEC S.A.) (6). The sequences of the oligonucleotides used for targeting the U2AF65 and U2AF35 isoforms were as follows: h65, 5′-GCA CGG UGG ACU GAU UCG UdTdT-3′ (GenBank accession number NM_007279; nt 1271 to nt 1289); h35a, 5′-CCA UUG CCC UCU UGA ACA UdTdT-3′ (GenBank accession number NM_006758; nt 218 to nt 238); h35b, 5′-CCA UCU UGA UUC AAA ACA UdTdT-3′ (GenBank accession number AJ627978; nt 164 to nt 182) and h35ab, 5′-GGC UGU GAU UGA CUU GAA UdTdT-3′ (GenBank accession number NM_006758, nt 459 to nt 479). For siRNA transient transfection, 35-mm petri dishes were seeded with 6 × 104 cells prior to the day of transfection. A 150 nM concentration of each siRNA duplex was transfected using Lipofectin (Invitrogen) according to the supplier's recommendations. Cells were then incubated for 36 to 48 h before being transfected with 200 ng of the reporter plasmid using FuGENE6 reagent (Roche Diagnostics) and were analyzed 16 to 24 h after the second transfection. For transient-overexpression experiments, subconfluent HeLa cells were transfected with 1.5 μg of the total plasmid DNA using FuGENE6 reagent (Roche Diagnostics). Cells were analyzed 16 to 24 after transfection.
Western blot analysis.
Western blot analysis of transfected cells was performed using whole-cell extracts that were prepared in sodium dodecyl sulfate sample buffer as described previously (22). The lysates were boiled for 5 min and then fractionated by electrophoresis in either a 10% or 12% sodium dodecyl sulfate-polyacrylamide gel and transferred to nitrocellulose membranes by electroblotting. Western blotting was carried out by standard immunoblotting procedures. The following primary antibodies were used: mouse monoclonal antibodies directed against U2AF65 (MC3) (8), β-actin (clone AC-15; Sigma), and the FLAG epitope (M2; Sigma) and rabbit polyclonal sera directed against U2AF35 (kindly provided by Angus Lamond [3]) and the HA epitope (Y-11; Santa Cruz Biotechnology). Immunoblots were developed using horseradish peroxidase-coupled secondary antibodies and detected by enhanced chemoluminescence (ECL; Amersham Biosciences).
RT-PCR and real-time quantitative PCR.
Total RNA was extracted using the TRIzol reagent (Invitrogen) and treated with RNase-free DNase I (Roche Diagnostics). Reverse transcription-PCR (RT-PCR) mixtures were randomly primed, and cDNA was produced using Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. The PCR products were separated by gel electrophoresis and detected by ethidium bromide staining. Real-time quantitative RT-PCRs were performed with an ABI7000 sequence detector (Applied Biosystems, Foster City, CA), using SYBR green PCR master mix (Applied Biosystems). The relative expression levels of different isoforms of the same gene were calculated using the 2−ΔΔCt method as described previously (23). The PCR products were confirmed by restriction analysis and sequencing. The primer sequences are presented in Table S1 of the supplemental material.
Bioinformatic analysis.
The search for human transcripts containing alternative 3′ splice sites of different strengths was carried out with the aid of the UCSC Genome Browser (http://genome.ucsc.edu/ [16] for the human genome assembly hg17, May 2004, NCBI Build 35 [19]). The gene region was defined by BLAT mapping (15) of the available RefSeq transcript (RNA) sequences (http://www.ncbi.nlm.nih.gov/projects/RefSeq/ [24]) for a particular gene. Using the UCSC Table Browser (14), the tables for the BLAT mappings of RefSeq and spliced mRNA and expressed sequence tags were obtained for this gene region. Making allowances only for the GT-AG, GC-AG, or AT-AC splice site consensus and excluding isoforms with extensive intron retentions, the nonredundant set of the longest isoforms and their corresponding accession numbers were determined. From this set of isoforms, we obtained a list of candidates containing a weak proximal 3′ splice site, a strong distal 3′ splice site, and a potential exonic splicing enhancer in the sequence between the alternate 3′ sites.
RESULTS AND DISCUSSION
RNAi-mediated knockdown of U2AF65 reduces the level of U2AF35.
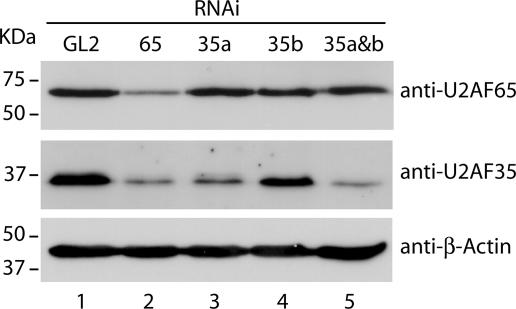
In order to analyze the role of the heterodimeric splicing factor U2AF in 3′ splice site recognition in vivo, we designed siRNA duplexes to knock down the expression of each subunit in HeLa cells. Since alternative splicing generates two different protein isoforms of U2AF35 (21), we used siRNA duplexes to specifically target the U2AF35a and U2AF35b mRNAs. Analysis by BLAST search confirmed that the selected sequences were unique to each target. As a negative control, we used the GL2 siRNA, which targets the firefly luciferase gene (5). Western blot analysis of HeLa total cell extracts shows that the U2AF35 protein is detected as two closely migrating bands: the major, lower band corresponds to the most abundant protein isoform, a, whereas the minor, upper band corresponds to the less abundant protein isoform, b (Fig. 1, gel labeled anti-U2AF35). Compared with cells treated with the GL2 control siRNA duplex (Fig. 1, lane 1), U2AF65-targeting siRNA caused a significant knockdown of both U2AF65 and U2AF35 protein levels (Fig. 1, lane 2). Treatment with siRNAs directed against U2AF35a decreased predominantly the lower U2AF35a band (Fig. 1, lane 3), whereas siRNAs directed against U2AF35b abolished detection of the minor upper band without affecting the U2AF35a lower band (Fig. 1, lane 4). Simultaneous treatment with siRNAs targeted to U2AF35a and U2AF35b effectively reduced both protein isoforms (Fig. 1, lane 5). Figure 1 further shows that RNAi-mediated knockdown of U2AF35a (either alone or in conjunction with U2AF35b) did not change the level of U2AF65 (lanes 3 to 5) compared with the level in cells treated with the GL2 control (lane 1).
FIG. 1.
RNAi-mediated knockdown of U2AF subunits. Western blot analysis of HeLa cell lysates prepared 48 h after transfection with siRNAs against luciferase (GL2) (lane 1), U2AF65 (lane 2), U2AF35a (lane 3), U2AF35b (lane 4), and U2AF35a simultaneously with U2AF35b (lane 5). The blot was probed with antibodies against U2AF65, U2AF35, and β-actin, as indicated. Molecular mass markers are shown on the left.
These results reveal a feedback mechanism by which depletion of the U2AF large subunit triggers the downregulation of the small subunit. Such a feedback loop is consistent with earlier studies indicating that U2AF heterodimer formation is essential for Drosophila viability (28) and with the more recent observation that several introns, the splicing of which is reduced by the genetic inactivation of the large subunit of U2AF in Schizosaccharomyces pombe, are also affected by inactivation of the small subunit (33). Furthermore, the finding that depletion of the small subunit caused no change in the level of the large subunit is consistent with previous in vitro data indicating that U2AF65 can perform its role in splicing independently of U2AF35 (11, 36, 38).
Roles of U2AF35 and U2AF65 in the recognition of a weak 3′ splice site in vivo.
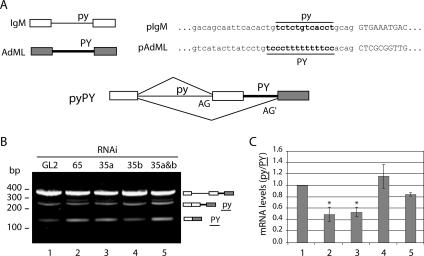
Previous biochemical complementation experiments performed with extracts chromatographically depleted of U2AF have indicated that U2AF35 is dispensable for in vitro splicing of pre-mRNAs that contain consensus 3′ splice site sequences but is required for the splicing of introns with weak 3′ splice sites (10, 11). In order to assess the role of the 3′ splice site sequence in U2AF35-dependent splicing in vivo, we combined RNAi against U2AF with the analysis of reporter minigenes. We constructed a reporter minigene chimera based on pre-mRNA sequences from AdML, which can be efficiently spliced in vitro in the absence of U2AF35 (36, 38), and the IgM pre-mRNA, which requires the presence of U2AF35 for efficient splicing in vitro (10-12). This reporter minigene is termed pyPY and consists of the sequence of IgM(−INH) containing the full-length M1 exon, the intronic sequence between exons M1 and M2, and a truncated form of exon M2 containing the purine-rich enhancer but lacking the splicing inhibitor sequence (11), fused to the 3′ half of the intron and exon 2 of the AdML sequence (Fig. 2A). When transfected into HeLa cells, the pyPY minigene gives rise to a primary transcript containing two alternative 3′ splice sites associated with a weak or a strong polypyrimidine tract (Fig. 2A). To analyze the effect of each U2AF subunit on the splicing of the pyPY minigene, HeLa cells were first treated with siRNAs directed against U2AF65, U2AF35a, U2AF35b, and both U2AF35 isoforms simultaneously. The cells were transfected 24 h later with the reporter plasmid, and transcripts derived from the pyPY minigene were analyzed 16 h posttransfection by RT-PCR and quantitative real-time PCR (Fig. 2B and C). The RT-PCR analysis of the control cells shows three major bands of approximately 375, 257, and 129 bp (Fig. 2B, lane 1). The 375-bp band corresponds to the unspliced primary transcript, the 257-bp band results from the use of the proximal 3′ splice site associated with the weak Py (py), and the 129-bp band corresponds to the use of the strong distal 3′ splice site (PY). The ratio between the intensities of the bands corresponding to the py and PY isoforms (py/PY) in GL2-treated cells is taken as the reference (Fig. 2C, bar 1). Knockdown of either U2AF65 or U2AF35a (alone or in combination with U2AF35b) significantly reduced the py/PY ratio (Fig. 2B and C, bars 2, 3, and 5), while in cells depleted of U2AF35b alone, the py/PY ratio was similar to that observed in cells treated with the GL2 control siRNA (Fig. 2B and C, bars 4 and 1). Thus, isoform b, which is 9- to 18-fold less abundant than U2AF35a in mammalian tissues (21), appears dispensable for the splicing of the weak 3′ splice site. The finding that depletion of U2AF65 caused an effect similar to that of depletion of U2AF35 is not surprising, taking into account that knockdown of the large subunit induced a downregulation of the small subunit (Fig. 1).
FIG. 2.
Effect of U2AF downregulation on alternative splicing of the pyPY reporter minigene. (A) Schematic representations of the reporter minigene chimeras of IgM and AdML splicing substrates; sequences derived from IgM are depicted as open boxes (exons) and thin lines (introns), and sequences derived from AdML are depicted as dark boxes (exons) and thick lines (introns). The minigene pyPY contains two alternative 3′ splice sites (AG and AG′) associated with polypyrimidine (Py) tracts with different strengths. The weak Py tract of IgM pre-mRNA is represented as “py,” and “PY” indicates a strong Py tract that matches the sequence of the AdML Py tract. (B) RT-PCR analysis of pyPY alternative splicing. Total RNA was isolated from HeLa cells doubly transfected with the indicated siRNAs and the reporter plasmid, and RT-PCR was carried out with the primers Rep.For and Rep.Rev (see Table S1 in the supplemental material). The predicted alternative splicing products are illustrated on the right by diagrams in which lines and boxes are as described for panel A. Molecular size markers are indicated on the left. (C) Quantitative real-time PCR analysis of the relative expression levels of the py and PY isoforms. The py isoform was amplified with primers Rep.bothF and Rep.proxR, and the PY isoform was amplified with primers Rep.bothF and Rep.distR (see Table S1 in the supplemental material). The ratios of the isoform levels (py/PY) were calculated from the formula 2−ΔΔCt (see Materials and Methods) using GL2-siRNA-treated cells as the calibrator. Results are presented as means ± standard deviations from at least four independent experiments. *, P < 0.05 relative to the results for the GL2-treated cells (Mann-Whitney U test).
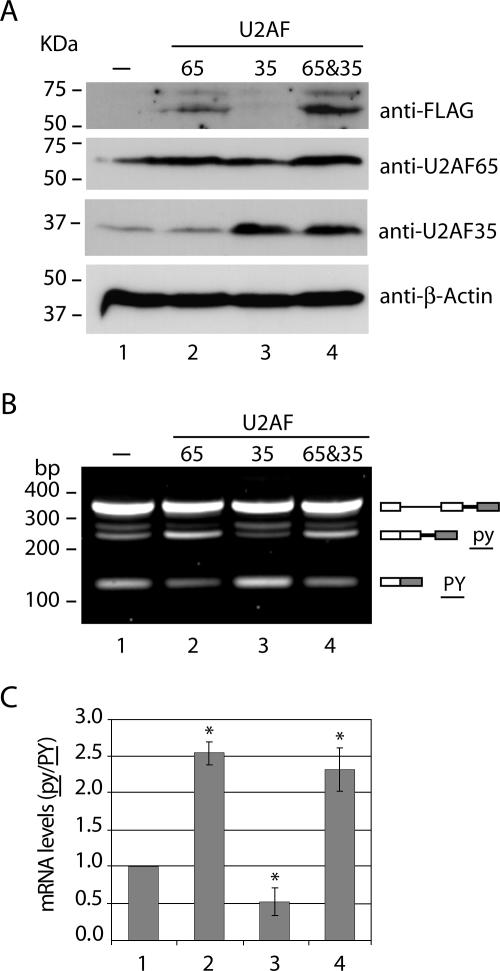
To test whether recognition of the proximal 3′ splice site associated with the weak polypyrimidine tract (py) is sensitive to both decreases and increases in the U2AF protein levels, we used expression constructs to transiently overexpress the two U2AF subunits in conjunction with the pyPY reporter minigene. The large subunit of U2AF was tagged with the FLAG epitope, and the small subunit with the HA epitope. The cells were transfected and analyzed 16 h posttransfection (Fig. 3). Western blot analysis using anti-FLAG antibody shows expression of the exogenous protein in cells transfected with plasmid constructs encoding only U2AF65 (Fig. 3A, lane 2) or both U2AF65 and U2AF35 (Fig. 3A, lane 4). Western blot analysis using anti-U2AF35 antibody further shows that U2AF35 is overexpressed in cells transfected with constructs encoding only U2AF35 (Fig. 3A, lane 3) or both U2AF65 and U2AF35 (Fig. 3A, lane 4). Notably, the level of endogenous U2AF35 remained unaltered in cells overexpressing U2AF65 (Fig. 3A, lane 2). Semiquantitative RT-PCR analysis reveals that exogenous expression of the U2AF65 protein, either alone or in conjunction with U2AF35, enhanced the splicing of the weak 3′ splice site (Fig. 3B, lanes 2 and 4) relative to that in control cells transfected with the pyPY minigene only (Fig. 3B, lane 1), causing an approximately 2.5-fold increase in the py/PY ratio (Fig. 3C, compare bars 1 and 2). In contrast, overexpression of U2AF35 alone caused a reduction in the py/PY ratio (Fig. 3B and C, bar 3).
FIG. 3.
Effect of U2AF overexpression on alternative splicing of pyPY. (A) HeLa cells were cotransfected with the reporter minigene and the pTet-On plasmid alone (lane 1) or together with Tet-responsive expression plasmids (pBI) coding for U2AF65-FLAG (lane 2), for U2AF35-HA (lane 3), and for both U2AF65-FLAG and U2AF35-HA (lane 4) and grown in the presence of 2 μg/ml doxycycline. Total cell extracts were prepared after 24 h, and the proteins were analyzed by Western blotting with the indicated antibodies. (B) RT-PCR analysis of pyPY alternative splicing. RNA was isolated from HeLa cells cotransfected for 24 h with the indicated plasmids and the reporter minigene. The predicted alternatively spliced products are illustrated on the right by diagrams in which lines and boxes are as described in the legend for Fig. 2A. Molecular size markers are indicated on the left. (C) Quantitative real-time PCR analysis of the relative expression levels of py and PY isoforms. The ratios between the isoform levels (py/PY) were calculated from the formula 2−ΔΔCt relative to the ratio present in cells transfected with the empty vector (bar 1). Results are presented as means ± standard deviations from at least four independent experiments. *, P < 0.05 relative to the results for the control (bar 1) (Mann-Whitney U test).
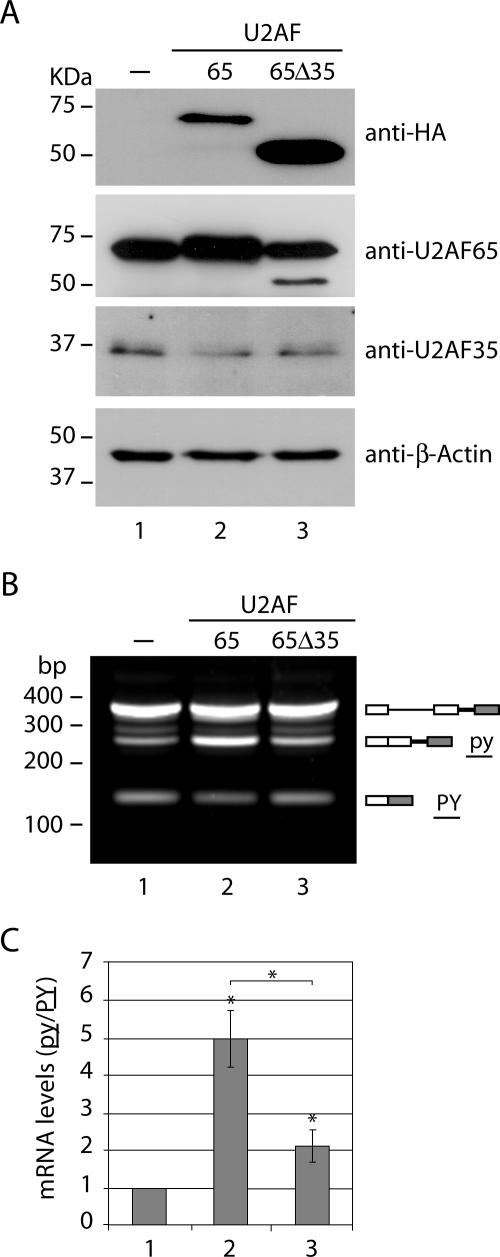
Thus, as expected, increasing the levels of the U2AF heterodimer by simultaneous overexpression of both subunits enhanced the splicing of the weak 3′ splice site. Moreover, overexpression of U2AF35 alone failed to enhance the recognition of the weak 3′ splice site, in agreement with the view that the splicing activity of this protein requires heterodimerization with U2AF65 (12). But why is overexpression of U2AF65 alone sufficient for stimulating the splicing of the weak polypyrimidine tract? One possibility is that the highest concentration of U2AF65 in the cell can compensate for the low affinity of the binding of this protein to a weak polypyrimidine tract (10). Alternatively, exogenously expressed U2AF65 may form functional heterodimers with different partners present in cells, namely, the alternatively spliced isoform U2AF35b (21) and the U2AF35-related proteins (29, 31, 32). To address this hypothesis, we cotransfected the reporter pyPY minigene with a U2AF65 mutant construct carrying a partial deletion of the region implicated in the interaction with U2AF35. The mutant is called U2AF65Δ35 and harbors a deletion of amino acid residues 84 to 150 (8). Western blot analysis using anti-HA antibody confirms the expression of exogenous full-length and mutant forms of U2AF65. As shown before, the levels of endogenous U2AF35 were not altered in cells overexpressing the large subunit (Fig. 4A). Semiquantitative RT-PCR analysis reveals that exogenous expression of the full-length U2AF65 protein enhanced splicing of the weak 3′ splice site (Fig. 4B, lane 2) relative to that in control cells transfected with the plasmid reporter only (Fig. 4B, lane 1), causing an approximately fivefold increase in the py/PY ratio (Fig. 4C). Most probably, the strongest effect on splicing observed in this experiment compared to the results depicted in Fig. 3 correlates with the higher level of expression of the exogenous protein due to the use of different expression systems. In contrast to the overexpression of full-length U2AF65, which caused a fivefold increase in splicing of the weak splice site (Fig. 4C, bar 2), overexpression of the mutant U2AF65Δ35 induced only a twofold increase in splicing (Fig. 4C, bar 3). Importantly, splicing of the strong 3′ splice site was not inhibited by the expression of U2AF65Δ35 (Fig. 4B, compare bands for PY in lanes 1 and 3), arguing that the reduced effect of this mutant on the weak site is not a consequence of its inability to support splicing. Taken together, our results support the view that the large subunit of the U2AF heterodimer cooperates with the small subunit to facilitate recognition of a weak 3′ splice site in vivo.
FIG. 4.
Recognition of the weak 3′ splice site of the pyPY minigene requires U2AF heterodimer formation. (A) HeLa cells were either cotransfected with the reporter plasmid and empty vector (lane 1) or cotransfected with the reporter minigene and cytomegalovirus-driven expression plasmids coding for full-length (lane 2) and mutant (lane 3) forms of U2AF65 tagged with the HA epitope. Total cell extracts were prepared after 24 h, and the proteins were analyzed by Western blotting with the indicated antibodies. (B) RT-PCR analysis of pyPY alternative splicing. Total RNA was isolated from HeLa cells cotransfected with the indicated plasmids and the reporter minigene. The predicted alternatively spliced products are illustrated on the right by diagrams in which lines and boxes are as described in the legend for Fig. 2A. Molecular size markers are indicated on the left. (C) Quantitative real-time PCR analysis of the relative expression levels of py and PY isoforms. The ratios between the isoform levels (py/PY) were calculated from the formula 2−ΔΔCt, relative to the ratio present in cells transfected with the empty vector (bar 1). Results are presented as means ± standard deviations from at least four independent experiments;. *, P < 0.05 relative to the results for the control (bar 1) (Mann-Whitney U test).
U2AF35-independent recognition of a weak 3′ splice site.
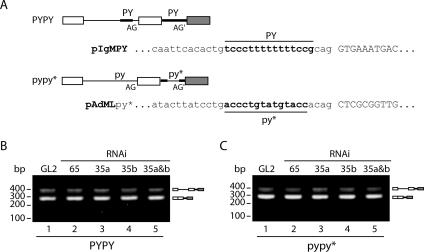
Having shown that knockdown of U2AF35 reduces recognition of a weak 3′ splice site located upstream of a strong 3′ splice site, we next tested the effect of U2AF35 depletion on the splicing of alternative 3′ splice sites associated with polypyrimidine tracts that are similar in strength. Two additional reporter minigenes were constructed (Fig. 5A). In one of them, both alternative 3′ splice sites are associated with the strong AdML polypyrimidine tract (PYPY). In the other, the two alternative 3′ splice sites are associated with weak polypyrimidine tracts, due to the replacement of all uridine residues in the strong AdML polypyrimidine tract by adenosines and guanidines (pypy*). The results obtained by semiquantitative RT-PCR analysis show that in GL2-treated cells, one major mRNA band corresponding to splicing of the proximal site in both reporters is observed (Fig. 5B and C, lane 1). Thus, in the two transcripts containing alternative 3′ splice sites associated with polypyrimidine tracts of similar strengths, there is a strong preference for splicing of the proximal site irrespective of its being weak or strong. This is in agreement with previous studies showing that in model human β-globin pre-mRNAs containing a single 5′ splice site and tandemly duplicated 3′ splice sites, the proximal 3′ site is preferentially selected (26).
FIG. 5.
Effect of U2AF downregulation on the splicing of PYPY and pypy* reporter minigenes. (A) Schematic representation of the reporter minigene chimeras PYPY and pypy*. Sequences derived from IgM are depicted as open boxes (exons) and thin lines (introns); sequences derived from AdML are depicted as dark boxes (exons) and thick lines (introns). PY denotes a strong Py tract and matches the sequence of the AdML Py tract; py* corresponds to the AdML Py tract in which five U's were replaced by A and G. (B and C) RT-PCR analysis of PYPY and pypy* alternative splicing. Total RNA was isolated from HeLa cells doubly transfected with the indicated siRNAs and the reporter plasmids (PYPY or pypy*). The relative positions of unspliced and spliced products are indicated on the right by diagrams in which lines and boxes are as described for panel A, and molecular size markers are shown on the left.
In clear contrast with the results obtained with the pyPY reporter minigene, depletion of either U2AF65 or U2AF35a (alone or in combination with U2AF35b) had no detectable effect on the splicing pattern of the reporters PYPY and pypy* (Fig. 5B and C). This shows that the requirement for U2AF35 can be completely relieved in vivo by improving the polypyrimidine tract of the IgM pre-mRNA reporter, as previously reported for in vitro data (12). Additionally, our results indicate that an intron with a weak polypyrimidine tract can be efficiently spliced under conditions of reduced expression of both U2AF65 and U2AF35, arguing that in vivo, not all pre-mRNA substrates containing a weak polypyrimidine tract are equally dependent on U2AF35 for splicing.
U2AF-dependent and U2AF-independent recognition of alternative 3′ splice sites in endogenous pre-mRNAs.
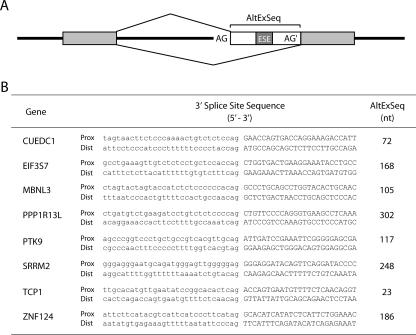
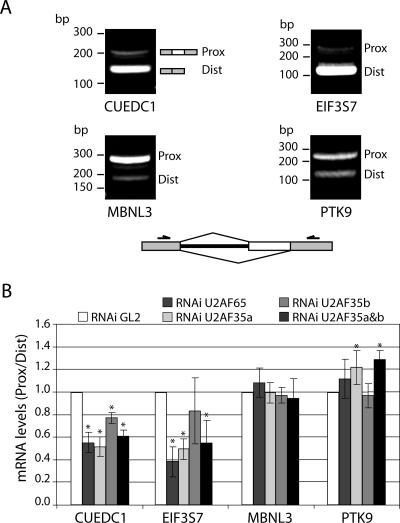
To determine whether U2AF plays a role in the alternative splicing of endogenous pre-mRNAs, we performed a bioinformatics search for human transcripts containing alternative 3′ splice sites of different strengths. First, we searched for genes that contain a proximal 3′ splice site associated with a weak polypyrimidine tract and a distal 3′ splice site associated with a strong polypyrimidine tract. A 3′ splice site was considered strong if it contained four consecutive T bases within the 30 nucleotides upstream of the splice site and weak if it contained no more than two consecutive T bases within that region. Because the reporter minigene harbors a purine-rich splicing enhancer in exon M2 35 nucleotides downstream of the proximal 3′ splice site, we further searched for candidates with a potential enhancer in the sequence between the two alternative 3′ splice sites. The selection of candidates was therefore restricted to pre-mRNAs containing at least one GAA motif (1) within 20 to ∼300 nucleotides downstream of the proximal 3′ splice site (Fig. 6A). Using these criteria, we identified 70 candidate pre-mRNAs (see Table S2 in the supplemental material) corresponding to diverse functional groups, such as cell proliferation and apoptosis genes, signal transduction and transcription regulators, or metabolic pathway components. From this candidate list, eight genes were randomly chosen for validation (Fig. 6B) and only four (CUEDC1, EIF3S7, MBNL3, and PTK9) were found to be alternatively spliced in HeLa cells producing the predicted isoforms (Fig. 7A). Following the knockdown of U2AF proteins, splicing of the weak proximal site was significantly reduced in the CUEDC1 and EIF3S7 mRNAs but not in the MBNL3 or PTK9 transcripts (Fig. 7B). Notably, for both the CUEDC1 and the EIF3S7 mRNA, the major isoform detected in nontreated cells corresponds to splicing of the stronger distal site, whereas for MBNL3 and PTK9, splicing of the proximal site predominates (Fig. 7A). We therefore speculate that the MBNL3 and PTK9 pre-mRNAs contain additional specific elements that stimulate splicing of the weaker 3′ site in a U2AF-independent manner.
FIG. 6.
Candidate endogenous genes with alternative 3′ splice sites. (A) Schematic structural representation of candidate genes. The candidates contain a proximal 3′ splice site (AG) associated with a weak polypyrimidine tract, a distal 3′ splice site (AG′) associated with a strong Py tract, and a putative exonic splicing enhancer (ESE) within the sequence between the alternative 3′ splice sites (AltExSeq). (B) Sequences of the proximal (Prox) and distal (Dist) 3′ splice sites of candidates chosen for further analysis. Intron sequences are in lowercase letters; exon sequences are in capital letters. The number of nucleotides between the two alternative 3′ sites (AltExSeq) is indicated.
FIG. 7.
Effect of U2AF downregulation on alternative splicing of endogenous genes. (A) RT-PCR analysis of transcripts from the CUEDC1, EIF3S7, MBNL3, and PTK9 genes. Total RNA was extracted from HeLa cells treated for 48 h with siRNAs against luciferase. The relative positions of mRNA isoforms resulting from the use of either the proximal (Prox) or distal (Dist) 3′ splice site are indicated by diagrams on the right, and molecular size markers are shown on the left. The darts above the schematic structural representation under the gels indicate primer pairs. For sequence details, see Tables S1 and S2 in the supplemental material. (B) Quantitative real-time PCR analysis of the relative levels of expression of Prox and Dist isoforms of the CUEDC1, EIF3S7, MBNL3, and PTK9 genes. Total RNA was extracted from cells treated with siRNAs against luciferase (GL2), U2AF65, U2AF35a, U2AF35b, and U2AF35a simultaneously with U2AF35b, as indicated. Specific and separate primer pairs were used for the amplification of each isoform (see Table S1 in the supplemental material). The ratios between the isoform levels (Prox/Dist) were calculated with the formula 2−ΔΔCt (see Materials and Methods) using GL2-siRNA-treated cells as the calibrator. Results are presented as means ± standard deviations from at least four independent experiments. *, P < 0.05 relative to the results for the GL2-treated cells (Mann-Whitney U test).
In conclusion, and in agreement with the results obtained with the reporter minigenes, the data strongly suggest that U2AF regulates the splicing of a subset of endogenous pre-mRNAs containing alternative 3′ splice sites associated with polypyrimidine tracts of different strengths.
Supplementary Material
Acknowledgments
We thank Juan Valcárcel and Angus Lamond for reagents and for stimulating discussions.
This work was supported by grants from Fundação para a Ciência e Tecnologia, Portugal (POCI/SAU-MMO/57700/2004), the Human Frontier Science Program Organization (RG0300/2000-M), and the European Commission (EURASNET). T. R. Pacheco was supported by an FCT fellowship (PRAXIS XXI/BD/18044/98).
Footnotes
Published ahead of print on 28 August 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aznarez, I., E. M. Chan, J. Zielenski, B. J. Blencowe, and L. C. Tsui. 2003. Characterization of disease-associated mutations affecting an exonic splicing enhancer and two cryptic splice sites in exon 13 of the cystic fibrosis transmembrane conductance regulator gene. Hum. Mol. Genet. 12:2031-2040. [DOI] [PubMed] [Google Scholar]
- 2.Blencowe, B. J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25:106-110. [DOI] [PubMed] [Google Scholar]
- 3.Chusainow, J., P. M. Ajuh, L. Trinkle-Mulcahy, J. E. Sleeman, J. Ellenberg, and A. I. Lamond. 2005. FRET analyses of the U2AF complex localize the U2AF35/U2AF65 interaction in vivo and reveal a novel self-interaction of U2AF35. RNA 11:1201-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell 2:233-239. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 6.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gama-Carvalho, M., M. P. Carvalho, A. Kehlenbach, J. Valcarcel, and M. Carmo-Fonseca. 2001. Nucleocytoplasmic shuttling of heterodimeric splicing factor U2AF. J. Biol. Chem. 276:13104-13112. [DOI] [PubMed] [Google Scholar]
- 8.Gama-Carvalho, M., R. D. Krauss, L. Chiang, J. Valcarcel, M. R. Green, and M. Carmo-Fonseca. 1997. Targeting of U2AF65 to sites of active splicing in the nucleus. J. Cell Biol. 137:975-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graveley, B. R., K. J. Hertel, and T. Maniatis. 2001. The role of U2AF35 and U2AF65 in enhancer-dependent splicing. RNA 7:806-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guth, S., C. Martínez, R. K. Gaur, and J. Valcárcel. 1999. Evidence for substrate-specific requirement of the splicing factor U2AF35 and for its function after polypyrimidine tract recognition by U2AF65. Mol. Cell. Biol. 19:8263-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guth, S., T. Ø. Tange, E. Kellenberger, and J. Valcárcel. 2001. Dual function for U2AF35 in AG-dependent pre-mRNA splicing. Mol. Cell. Biol. 21:7673-7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5-14. [DOI] [PubMed] [Google Scholar]
- 14.Karolchik, D., A. S. Hinrichs, T. S. Furey, K. M. Roskin, C. W. Sugnet, D. Haussler, and W. J. Kent. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32:D493-D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kent, W. J. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kent, W. J., C. W. Sugnet, T. S. Furey, K. M. Roskin, T. H. Pringle, A. M. Zahler, and D. Haussler. 2002. The human genome browser at UCSC. Genome Res. 12:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kielkopf, C. L., N. A. Rodionova, M. R. Green, and S. K. Burley. 2001. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell 106:595-605. [DOI] [PubMed] [Google Scholar]
- 18.Kitagawa, K., X. Wang, I. Hatada, T. Yamaoka, H. Nojima, J. Inazawa, T. Abe, K. Mitsuya, M. Oshimura, A. Murata, et al. 1995. Isolation and mapping of human homologues of an imprinted mouse gene U2af1-rs1. Genomics 30:257-263. [DOI] [PubMed] [Google Scholar]
- 19.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 20.Merendino, L., S. Guth, D. Bilbao, C. Martinez, and J. Valcarcel. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402:838-841. [DOI] [PubMed] [Google Scholar]
- 21.Pacheco, T. R., A. Q. Gomes, N. L. Barbosa-Morais, V. Benes, W. Ansorge, M. Wollerton, C. W. Smith, J. Valcarcel, and M. Carmo-Fonseca. 2004. Diversity of vertebrate splicing factor U2AF35: identification of alternatively spliced U2AF1 mRNAS. J. Biol. Chem. 279:27039-27049. [DOI] [PubMed] [Google Scholar]
- 22.Pacheco, T. R., L. F. Moita, A. Q. Gomes, N. Hacohen, and M. Carmo-Fonseca. 19 July 2006. RNAi knockdown of hU2AF35 impairs cell cycle progression and modulates alternative splicing of Cdc25 transcripts. Mol. Biol. Cell [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 23.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pruitt, K. D., T. Tatusova, and D. R. Maglott. 2005. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33:D501-D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reed, R. 1989. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 3:2113-2123. [DOI] [PubMed] [Google Scholar]
- 26.Reed, R., and T. Maniatis. 1986. A role for exon sequences and splice-site proximity in splice-site selection. Cell 46:681-690. [DOI] [PubMed] [Google Scholar]
- 27.Rudner, D. Z., K. S. Breger, R. Kanaar, M. D. Adams, and D. C. Rio. 1998. RNA binding activity of heterodimeric splicing factor U2AF: at least one RS domain is required for high-affinity binding. Mol. Cell. Biol. 18:4004-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudner, D. Z., R. Kanaar, K. S. Breger, and D. C. Rio. 1998. Interaction between subunits of heterodimeric splicing factor U2AF is essential in vivo. Mol. Cell. Biol. 18:1765-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shepard, J., M. Reick, S. Olson, and B. R. Graveley. 2002. Characterization of U2AF26, a splicing factor related to U2AF35. Mol. Cell. Biol. 22:221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, C. W., and J. Valcarcel. 2000. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25:381-388. [DOI] [PubMed] [Google Scholar]
- 31.Tronchere, H., J. Wang, and X. D. Fu. 1997. A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature 388:397-400. [DOI] [PubMed] [Google Scholar]
- 32.Tupler, R., G. Perini, and M. R. Green. 2001. Expressing the human genome. Nature 409:832-833. [DOI] [PubMed] [Google Scholar]
- 33.Webb, C. J., S. Lakhe-Reddy, C. M. Romfo, and J. A. Wise. 2005. Analysis of mutant phenotypes and splicing defects demonstrates functional collaboration between the large and small subunits of the essential splicing factor U2AF in vivo. Mol. Biol. Cell 16:584-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, J. Y., and T. Maniatis. 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75:1061-1070. [DOI] [PubMed] [Google Scholar]
- 35.Wu, S., C. M. Romfo, T. W. Nilsen, and M. R. Green. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402:832-835. [DOI] [PubMed] [Google Scholar]
- 36.Zamore, P. D., and M. R. Green. 1991. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 10:207-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zamore, P. D., and M. R. Green. 1989. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc. Natl. Acad. Sci. USA 86:9243-9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zamore, P. D., J. G. Patton, and M. R. Green. 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355:609-614. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, M., P. D. Zamore, M. Carmo-Fonseca, A. I. Lamond, and M. R. Green. 1992. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc. Natl. Acad. Sci. USA 89:8769-8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zorio, D. A., and T. Blumenthal. 1999. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402:835-838. [DOI] [PubMed] [Google Scholar]
- 41.Zuo, P., and T. Maniatis. 1996. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 10:1356-1368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.