Abstract
DDB1, a component of a Cul4A ubiquitin ligase complex, promotes nucleotide excision repair (NER) and regulates DNA replication. We have investigated the role of human DDB1 in maintaining genome stability. DDB1-depleted cells accumulate DNA double-strand breaks in widely dispersed regions throughout the genome and have activated ATM and ATR cell cycle checkpoints. Depletion of Cul4A yields similar phenotypes, indicating that an E3 ligase function of DDB1 is important for genome maintenance. In contrast, depletion of DDB2, XPA, or XPC does not cause activation of DNA damage checkpoints, indicating that defects in NER are not involved. One substrate of DDB1-Cul4A that is crucial for preventing genome instability is Cdt1. DDB1-depleted cells exhibit increased levels of Cdt1 protein and rereplication, despite containing other Cdt1 regulatory mechanisms. The rereplication, accumulation of DNA damage, and activation of checkpoint responses in DDB1-depleted cells require entry into S phase and are partially, but not completely, suppressed by codepletion of Cdt1. Therefore, DDB1 prevents DNA lesions from accumulating in replicating human cells, in part by regulating Cdt1 degradation.
Damaged DNA binding protein 1 (DDB1) was identified as part of a heterodimer that tightly associates with DNA following UV damage (14). This heterodimer of DDB1 and damaged DNA binding protein 2 (DDB2) was subsequently discovered to have a role in nucleotide excision repair (NER). There are two subpathways of NER: global genomic repair (GGR) removes lesions throughout the genome, while transcription-coupled repair (TCR) removes lesions more readily from the transcribed strand of active genes. Defects in the chromatin binding property of the DDB1-DDB2 complex reduce the GGR capacity of cells by approximately 50% (28, 61). Genetic defects in NER give rise to the autosomal recessive disorder xeroderma pigmentosum (XP), which is characterized by extreme sun sensitivity, premature aging, and an increased incidence of skin cancer. Eight complementation groups have been defined for XP, with seven resulting from unique defects in proteins required for NER (15, 58). Mutations in DDB2 are classified as XP complementation group E (XP-E) (30, 47).
Further studies revealed that it is not simply a heterodimer of DDB1-DDB2 that binds DNA and stimulates GGR following UV damage but a larger complex of proteins that possess an active ubiquitin ligase activity. In addition to DDB1 and DDB2, the soluble complex contains the E3 protein cullin 4A (Cul4A), the Roc1 RING subunit that is required for Cul4A activity, and a cullin regulatory complex termed the COP9 signalosome (CSN) (21). Following chromatin association, this active ubiquitin ligase targets XPC, a damage sensor for the GGR pathway. Ubiquitination of XPC does not result in proteosomal degradation but instead enhances the ability of XPC to remain associated with chromatin (56). Another target of this chromatin-bound complex is the DDB2 protein itself. DDB2 is rapidly degraded after UV damage and is ubiquitinated in vitro by DDB1-Cul4A (13, 39, 44). The ubiquitination of DDB2 reduces the DNA binding ability of DDB1-Cul4A and is necessary for efficient repair of UV-induced lesions (56). Recently, the monoubiquitination of histone H2A at sites of UV-induced damage was also shown to require DDB1-Cul4 (29). This modification may have an important role in facilitating GGR as well. Interestingly, a complex identical to that of DDB1-DDB2-Cul4A-Roc1-CSN was identified in which DDB2 is replaced by CSA, a protein that is defective in patients with Cockayne syndrome. Unlike the DDB2 complex, the CSA complex has a role in TCR (21).
DDB1 has important roles outside of NER as an adaptor molecule for the Cul4A ubiquitin ligase complex. Studies of Schizosaccharomyces pombe revealed that DDB1 and Pcu4, the Cul4 homologue in yeast, promote the ubiquitin-mediated degradation of an inhibitor of ribonucleotide reductase (RNR) (8, 25, 35). This inhibitor prevents the association of the two RNR subunits, an event that is necessary for the catalytic activity that converts nucleoside triphosphates to deoxynucleoside triphosphates for DNA synthesis and repair (12, 35). Additionally, the DDB1-Cul4A complex promotes Cdt1 degradation in human cells after ionizing radiation (IR) and UV damage and also has a role in the replication-dependent destruction of Cdt1 (2, 24, 26, 27, 45, 52). Cdt1 is a component of the prereplication complex, which assembles in an ordered fashion during G1 to license origins of replication for initiation of DNA synthesis. Regulation of Cdt1 is a critical means by which human cells prevent rereplication. Rereplication occurs when a replication origin fires more than once in a single cell division cycle (7, 17). This protein is targeted for proteasome-mediated degradation by two ubiquitin ligase complexes, SCFSkp2 and DDB1-Cul4A, and is also functionally inhibited by the binding of geminin (2, 24, 26, 34, 36, 45, 59, 71). These multiple levels of Cdt1 regulation emphasize the importance of restraining rereplication and highlight the need to understand how these distinct processes cooperate to maintain the integrity of the genome.
In addition to DDB1-DDB2, several other protein complexes are recruited to and activated by UV radiation-induced lesions. Principal among these UV response proteins is ATR. ATM- and Rad3-related kinase (ATR) is a member of the phosphatidylinositol 3-kinase-related kinase family, which also includes the damage response protein ataxia-telangiectasia mutated (ATM). These proteins are situated at the apex of the DNA damage response pathways and function to protect the stability of the genome by initiating signal transduction cascades that result in the inhibition of cell cycle progression and the coordination of DNA repair (1, 50). While ATM responds primarily to the formation of DNA double-strand breaks, ATR is activated by a wide variety of lesions and stresses, including UV radiation, DNA alkylation, and replication inhibitors, as well as double-strand breaks. Downstream targets of these active kinases that are important in mediating cell cycle arrest and facilitating DNA repair include the checkpoint kinases Chk1 and Chk2, p53, and the histone variant H2AX (4, 11, 22, 37, 40, 46, 63, 69). Since ATR is responsible for initiating the checkpoint response to UV radiation, and both DDB1 and ATR are recruited to UV lesions, we examined whether DDB1 functions in the ATR-mediated DNA damage response to this genotoxic stress. While depletion of DDB1 did not impair ATR-dependent responses to UV, surprisingly, we found that depletion of DDB1 caused DNA double-strand breaks and activation of checkpoint responses. This phenotype is due, in part, to the misregulation of DNA replication.
MATERIALS AND METHODS
Cell culture.
U2OS and HeLa cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 7.5% fetal bovine serum at 37°C in 5% CO2. The RPE-hTERT cell line was maintained in Dulbecco's modified Eagle's-F-12 medium supplemented with 10% fetal bovine serum and 0.2% sodium bicarbonate at 37°C in 5% CO2.
DNA constructs and transfections.
The DDB1 construct resistant to degradation by small interfering RNA (siRNA)-mediated silencing was created using site-directed mutagenesis. The following primer and its complement were used to introduce selected wobble base pair mutations: 5′-GGAGAGCAAGGATCTACTCTTTATCTTGACAGC-3′. Calcium phosphate transfections of the Phoenix amphotropic packaging cell line were performed to produce retroviruses. Following infection of U2OS cells with the viral medium, stable cell lines were selected using puromycin. C-terminal hemagglutinin (HA)-tagged Cdt1 expression constructs were generated by PCR in the pLPCX vector. Mutagenesis was performed using the following primers: 5′-TATGAAGCTTATGGAGGCTCGCCGCGCTACCGACGCAGCTGCGCGCCGCCGC-3′ and 5′-CAAGCTGGCCTGCCGGGCCCCCAGC-3′.
siRNA.
All siRNA oligonucleotides were purchased from Dharmacon, Inc. The siRNA duplexes were as follows: control siRNA sense strand, 5′-AUGAACGUGAAUUGCUCAAdTdT; DDB1 siRNA sense strand, 5′-GCAAGGACCUGCUGUUUAUUU; Cul4A siRNA sense strand, 5′-GAACCCAUAUUAUUAGUGAUU; DDB2 siRNA sense strand, 5′-GAUAUCAUGCUCUGGAAUUUU; XPC siRNA sense strand, 5′-GCAAAUGGCUUCUAUCGAAUU; XPA siRNA sense strand, 5′-GGAGACGAUUGUUCAUCAAUU; Cdt1 siRNA sense strand, 5′-GCGCAAUGUUGGCCAGAUCUU. The Skp2 depletions were achieved using a SMARTpool. Transfections were performed with 200 nM of siRNA using Oligofectamine reagent according to the manufacturer's instructions (Invitrogen).
Cell lysis.
Cells were lysed in Igepal lysis buffer (20 mM Tris, pH 7.5, 0.1 M NaCl, 1 mM EDTA, 0.5% Igepal CA 630, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 1 mM NaF, 20 mM μ-glycerolphosphate, 1 mM sodium vanadate, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonate).
Antibodies.
The DDB1 and bromodeoxyuridine (BrdU) antibodies were obtained from BD Biosciences. The ATM and XPC antibodies were purchased from Novus Biologicals. The replication protein A (RPA) p34 (9H8) and XPA antibodies were purchased from Neomarkers. The anti-p45 Skp2 and anti-HA.11 antibodies were obtained from Zymed and Covance, respectively. The Chk1 (G-4), p53 (DO-1), DDB2 (H-127), and Cdt1 (H-300) antibodies were purchased from Santa Cruz Biotechnology. The Mre11 (12D7) and p21 (EA10) antibodies were obtained from GeneTex, Inc., and Calbiochem, respectively. The Chk2 and ATRIP antibodies were described previously (16, 40). The phosphopeptide-specific antibody to pS1981 of ATM was purchased from Rockland, Inc. The phosphopeptide-specific antibodies to pT68 of Chk2, pS345 of Chk1, and pS15 of p53 were obtained from Cell Signaling Technology. The Cul4A antibody was a gift from Yue Xiong.
Immunofluorescence.
For γH2AX and Mre11 immunostaining, cells grown on glass coverslips were fixed and permeabilized with 100% methanol at −20°C for 15 min. After being rinsed twice with phosphate-buffered saline (PBS), cells were incubated in 100% acetone at −20°C for 30 seconds. The cells were then air dried for 1 min, rinsed six times with PBS, and blocked for 15 min at room temperature with 5% bovine serum albumin (BSA) in PBS. Primary antibodies recognizing γH2AX or Mre11 were diluted in 1% BSA-PBS and incubated on cells for 20 min at 37°C with 5% CO2. After being washed three times with PBS, cells were incubated in secondary antibodies, fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit immunoglobulin G or rhodamine red-X-conjugated donkey anti-mouse immunoglobulin G (Jackson ImmunoResearch Laboratories, Inc.), diluted in 1% BSA-PBS for 20 min at 37°C with 5% CO2. Cells were washed and counterstained with 4′,6′-diamidino-2-phenylindole (DAPI). For RPA34 immunostaining, cells were fixed with 3% paraformaldehyde at room temperature for 10 min and permeabilized with 0.5% Triton X-100 for 10 min on ice. After being blocked at room temperature for 15 min with 5% BSA-PBS, the RPA34 antibody was diluted in 1% BSA-PBS and incubated on cells for 20 min at 37°C with 5% CO2. Following three washes with PBS, cells were incubated in FITC-conjugated goat anti-mouse secondary antibody, diluted in 1% BSA-PBS, for 20 min at 37°C with 5% CO2. The cells were washed and counterstained with DAPI. All images were obtained with a Zeiss Axioplan microscope equipped with a Zeiss camera and software.
Fragile site analysis.
Fragile sites were induced by treatment of U2OS cells with 0.1 μM aphidicolin for 24 h. Metaphase cells were enriched by treating with Demecolcine solution (Sigma) for 1 h at 37°C. Cells were then incubated in a hypotonic solution (3:1, 0.566% KCl-0.8% sodium citrate) for 15 min at 37°C, fixed by multiple washes with Carnoy fixative (3:1 methanol-acetic acid), and dropped onto slides. Slides were baked at 90°C for 30 min and stained with Giemsa stain. Metaphase spreads were scored for chromosomal gaps and breaks, and common fragile sites were identified based on the idiogram in the work of Richards (48).
RESULTS
Depletion of DDB1 activates cell cycle checkpoints.
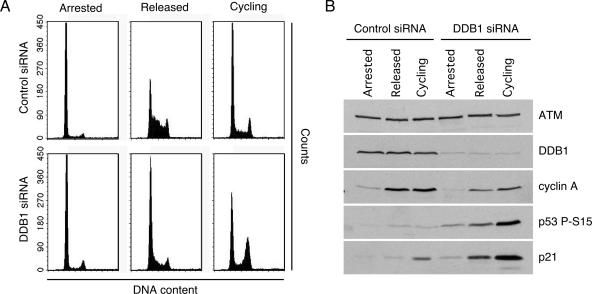
Both DDB1 and ATR are activated by UV radiation and are recruited to sites of UV-induced DNA damage. In S. pombe both these proteins are required for the proteolysis of the replication inhibitor Spd1. The damage inducibility of DDB1, along with its functional relationship to the ATR homologue Mec1 in yeast, led us to test whether DDB1 has a role in the UV response initiated by ATR in human cells. We did not observe any defects in ATR activation in response to DNA damage in DDB1-depleted cells (data not shown). However, during the course of testing this hypothesis we examined the cell cycle distribution of DDB1-depleted cells to ensure that depletion of DDB1 was not having an adverse effect on cell cycle progression. Surprisingly, we found a striking change in the cell cycle distribution of undamaged, DDB1-depleted cells (Fig. 1A). This population shows a considerable increase in cells with 4n DNA content compared to that observed following treatment with a control siRNA. To determine if this accumulation is occurring in G2 or mitosis, cells were stained with an antibody to phosphorylated histone H3 on serine 10. This phosphorylation event occurs during chromosome condensation and is routinely used as a marker of mitotic cells (23). DDB1 depletion does not increase the percentage of mitotic cells relative to that observed in control cells (Fig. 1A). In fact, there was a significant decrease in the percentage of mitotic cells in the DDB1-depleted population. Similar results were obtained with two independent DDB1 siRNA oligonucleotides in both U2OS and HeLa cell lines (data not shown). This observation suggests that depletion of DDB1 causes a cell cycle arrest in G2.
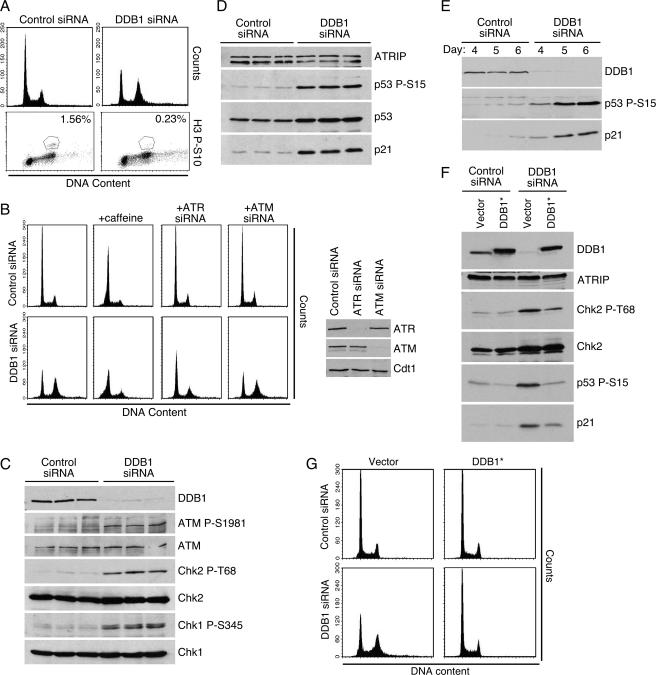
FIG. 1.
Depletion of DDB1 activates cell cycle checkpoints. (A) The cell cycle distribution of HeLa cells transfected with control or DDB1 siRNA oligonucleotides was analyzed by flow cytometry detection of DNA content following propidium iodide staining 3 days after transfection. The percentage of mitotic cells in each population was determined by immunostaining with a phosphopeptide-specific antibody to histone H3 S10. (B) HeLa cells transfected with DDB1 or control siRNA oligonucleotides were processed for flow cytometry 4 days after transfection. Where indicated, the cells were cotransfected with ATM- or ATR-specific siRNA or treated with 8 mM caffeine for 24 h prior to analysis. The depletion efficiency of the ATR and ATM siRNA was monitored by immunoblot analysis. (C to E) Checkpoint signaling was examined in HeLa (C), U2OS (D), and RPE-hTERT (E) cells 3 days after transfection with control or DDB1 siRNA oligonucleotides. Cell lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted with the indicated antibodies. (F and G) U2OS cells were infected with retroviruses encoding an empty vector or an siRNA-immune Flag-DDB1 cDNA (DDB1*). Following selection the cells were transfected with control or DDB1 siRNA oligonucleotides. Checkpoint signaling was examined 3 days after transfection by immunoblotting with the indicated antibodies (F) or monitoring DNA content by flow cytometry of propidium iodide-stained cells (G).
An arrest in G2 may indicate that cell cycle progression is halted due to the activation of a cell cycle checkpoint. Treatment with caffeine abrogates the G2/M checkpoint (32, 51); therefore, if the G2 arrest in DDB1-depleted cells results from checkpoint activation, the addition of caffeine should overcome this accumulation. Treatment with caffeine for 24 h does reverse the G2 accumulation of DDB1-depleted cells but also induces apoptosis (as measured by cells with <2n DNA content [Fig. 1B]). Since caffeine has numerous cellular effects, we also directly inhibited two of the checkpoint proteins that initiate the DNA damage response signaling cascade, ATR and ATM. Codepletion of DDB1 with ATR also suppresses the G2 accumulation that is observed following DDB1 depletion (Fig. 1B). Interestingly, codepletion of DDB1 with ATM cannot prevent the accumulation of cells in G2 (Fig. 1B). The sensitivity of the 4n accumulation to treatment with caffeine and ATR siRNA provides further evidence that the arrest is in G2 and that it is dependent on the activation of cell cycle checkpoints.
To determine which checkpoint pathways are induced after DDB1 depletion, we immunoblotted cells with phosphopeptide-specific antibodies that detect activated checkpoint proteins. Immunoblotting of HeLa extracts revealed that ATM S1981, Chk2 T68, and Chk1 S345 are all phosphorylated after depletion of DDB1 (Fig. 1C). A similar observation was made in U2OS cells, where p53 S15 is phosphorylated and p21 is induced specifically after DDB1 depletion (Fig. 1D). To confirm that these results are not limited to transformed cells, the experiment was repeated in an untransformed, telomerase-immortalized epithelial cell line (RPE-hTERT). The results again show p53 S15 phosphorylation and p21 induction specifically after DDB1 depletion (Fig. 1E). These data indicate that depletion of DDB1 causes activation of both ATM- and ATR-dependent cell cycle checkpoints, since Chk2 T68 is predominantly an ATM phosphorylation site and Chk1 S345 is an ATR phosphorylation site (9, 37, 40, 72). Even though both ATM and ATR signaling pathways are activated in the absence of DDB1, ATR siRNA but not ATM siRNA relieved the G2 checkpoint despite efficient depletion of both proteins (Fig. 1B). The dependency of the G2 arrest on ATR, but not ATM, may be explained by the role that each of these proteins plays in the G2/M checkpoint. While ATM is important for initiation of the G2/M checkpoint response, maintenance of the cell cycle arrest largely relies on the activity of ATR (10).
To confirm that activation of cell cycle checkpoints is due specifically to the depletion of DDB1, we created a DDB1 construct using site-directed mutagenesis of wobble base pairs that is resistant to RNA inhibition by one of the siRNA oligonucleotides. U2OS cell lines were generated that stably express an empty vector or a Flag-tagged siRNA-immune DDB1 mRNA (DDB1*). Depletion of DDB1 from the cell line stably expressing an empty vector results in phosphorylation of Chk2 T68 and p53 S15, as well as p21 induction (Fig. 1F). However, depletion of DDB1 from U2OS cells expressing the siRNA-resistant DDB1 mRNA produces significantly less checkpoint protein phosphorylation and p21 induction (Fig. 1F). The slight amount of residual checkpoint signaling observed in some experiments is likely due to variability in the expression of the DDB1* cDNA in the polyclonal population. Analysis of cell cycle profiles by flow cytometry also revealed that expression of the siRNA-resistant DDB1 mRNA can suppress the G2 accumulation of cells that is observed after DDB1 depletion (Fig. 1G). The findings that checkpoint activation and cell cycle arrest can be complemented by expression of an siRNA-resistant DDB1 construct confirm that the observed phenotype is due specifically to depletion of DDB1.
Depletion of DDB1 results in γH2AX, Mre11, and RPA focus formation.
The phosphorylation of Chk2 T68 and Chk1 S345 indicates that both the ATM and ATR damage response pathways are activated. In principle, activation of these kinases could be due to the elimination of a negative regulatory mechanism or the presence of genotoxic stress. To differentiate between these hypotheses, we examined whether phosphorylation of H2AX by checkpoint kinases is restricted to foci or distributed diffusely throughout the genome. If the depletion of DDB1 relieves an inhibitory mechanism restraining the activity of ATM or ATR, then these kinases will be generally active without the presence of DNA damage and H2AX phosphorylation is expected to occur throughout the chromatin. A similar phenomenon is observed when the ATR-activating fragment of TopBP1 is overexpressed in cells (31). Under these circumstances ATR is activated and H2AX is phosphorylated, but the phosphorylation is pan-nuclear rather than occurring in distinct foci because there is no actual DNA damage created by overexpression of the TopBP1 fragment (D. Cortez, unpublished data). In contrast, if DDB1 depletion causes the accumulation of DNA damage, then phosphorylated H2AX should be restricted to discrete intranuclear foci (49). We monitored phosphorylated H2AX (γH2AX) in U2OS cells by indirect immunofluorescence using a phosphopeptide-specific antibody to Ser-139. In cells treated with control siRNA there is little γH2AX focus formation (Fig. 2A and 2B). Approximately 5% of the undamaged control cells contain γH2AX foci. Irradiation of control cells with 8 Gy of IR, to induce the formation of DNA double-strand breaks, results in a corresponding increase in cells with γH2AX foci to 75% (Fig. 2A and 2B). Depletion of DDB1 also induces γH2AX focus formation, with 67% of DDB1-depleted cells displaying intranuclear foci (Fig. 2A and 2B). Irradiation of DDB1-depleted cells further enhances the percentage with γH2AX foci to 84% (Fig. 2B). The formation of γH2AX foci in the majority of cells following depletion of DDB1 suggests that the loss of this protein generates DNA damage.
FIG. 2.
DDB1 depletion results in γH2AX, Mre11, and RPA focus formation. U2OS cells were transfected with control or DDB1 siRNA oligonucleotides. Cells transfected with control siRNA were also irradiated with 8 Gy of IR 2 hours prior to analysis where indicated. (A) Three days after siRNA transfection the cells were fixed and stained with antibodies to H2AX phospho-S139 and Mre11, followed by the appropriate FITC and rhodamine red-X secondary antibodies. The nucleus was visualized with DAPI. (B) The percentage of U2OS cells displaying five or more intranuclear γH2AX foci 3 days after transfection with control or DDB1 siRNA oligonucleotides was quantified. (C) The number of γH2AX foci per cell was quantitated. (D) Cells were fixed and stained with an antibody to RPA34. DAPI staining was used to visualize the nucleus.
To further support the finding that depletion of DDB1 results in DNA damage, U2OS cells transfected with control or DDB1 siRNA were also examined for Mre11 focus formation. Mre11 is one component of the MRN (Mre11-Rad50-Nbs1) complex, which localizes to sites of double-strand breaks and facilitates repair of the damaged DNA (42). Immunostaining with γH2AX and Mre11 reveals that the γH2AX foci colocalize with Mre11 foci in the irradiated cells, as well as in the DDB1-depleted cells (Fig. 2A). This suggests that the γH2AX foci do indeed represent sites of DNA damage, as proteins necessary for the repair of double-strand breaks are also recruited to these sites within the cell.
The extent of DNA damage was characterized by examining the number of γH2AX foci generated per cell in control, irradiated, and DDB1-depleted cells. While 80% of control cells contained no γH2AX foci, 90% of DDB1-depleted cells displayed γH2AX foci ranging in numbers from 3 to 86, with an average of 22 γH2AX foci per cell (Fig. 2C). In comparison, control cells display an average of one focus per cell, while those irradiated with the lethal dose of 8 Gy of IR display an average of 66 γH2AX foci per cell (Fig. 2C).
In addition to monitoring γH2AX and Mre11 focus formation as markers of DNA damage, we also examined RPA focus formation. RPA is a three-subunit protein complex that binds single-stranded DNA in eukaryotic cells and, like γH2AX and Mre11, forms intranuclear foci after DNA damage (43). Immunostaining for the RPA34 subunit of this heterotrimeric protein complex indicates that similarly to irradiated cells, DDB1-depleted cells display numerous RPA34 foci (Fig. 2D). These results provide additional evidence that depletion of DDB1 results in DNA damage.
DDB1 maintains genome integrity as part of a Cul4A ubiquitin ligase complex.
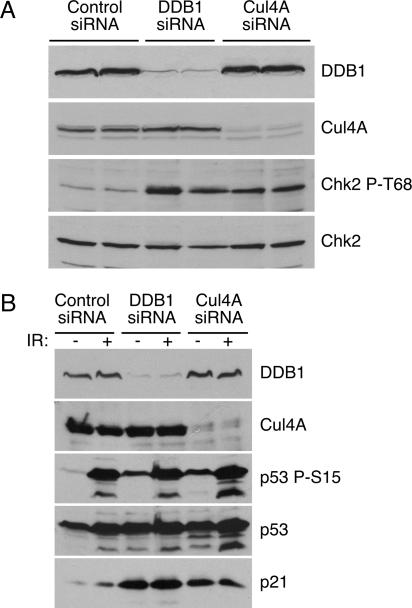
All currently known roles of DDB1 involve its functioning as part of a multiprotein complex containing Cul4A that targets substrates for ubiquitination (2, 26, 29, 54, 56, 64, 65, 70). To determine if defective ubiquitination contributes to the phenotype observed after DDB1 depletion, we used siRNA to deplete Cul4A. Depletion of Cul4A from HeLa cells results in the phosphorylation of Chk2 T68, analogous to what is observed following depletion of DDB1 (Fig. 3A). Similar phenotypes are also observed in the untransformed RPE-hTERT cells. Checkpoint activation is absent from undamaged RPE-hTERT cells treated with control siRNA and is induced following irradiation, as indicated by p53 S15 phosphorylation (Fig. 3B). The individual depletions of DDB1 and Cul4A result in p53 S15 phosphorylation and p21 induction in the absence of exogenous DNA damage, and further increases in p53 phosphorylation are observed after ionizing radiation (Fig. 3B). Additionally, depletion of Cul4A produces a modest increase in cells with 4n DNA content (see Fig. S1 in the supplemental material). This phenotype is reminiscent of that observed following DDB1 depletion and correlates with the phosphorylation of checkpoint proteins; however, the cell cycle arrest following depletion of Cul4A is less striking than the arrest typically observed following depletion of DDB1. This difference may be due to differences in the silencing efficiency of the siRNA oligonucleotides or may indicate that DDB1 is limiting in the DDB1-Cul4A ubiquitin ligase complex. The activation of checkpoint pathways following depletion of Cul4A is consistent with the hypothesis that the DNA damage observed following depletion of DDB1 results from defective ubiquitination of a DDB1-Cul4A substrate(s).
FIG. 3.
Depletion of Cul4A yields a DNA damage phenotype similar to that of DDB1 depletion. (A) HeLa cells were transfected with control, DDB1, or Cul4A siRNA oligonucleotides. Three days after transfection cell lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted with the indicated antibodies. (B) RPE-hTERT cells were transfected with control, DDB1, or Cul4A siRNA oligonucleotides. Three days after transfection the indicated samples were irradiated with 8 Gy of IR and incubated 2 hours prior to analysis. Checkpoint signaling was monitored by immunoblotting with the indicated antibodies.
Defective NER is not responsible for the DNA damage observed after DDB1 depletion.
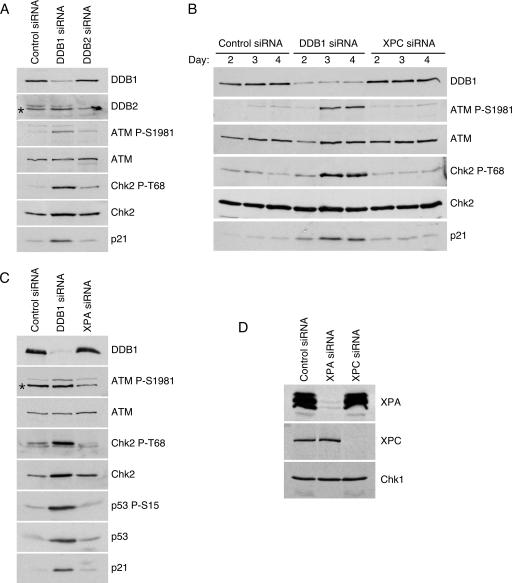
One way in which DDB1 may exert its genome protective function is through its role in NER (28, 29, 56, 61). Since the chromatin binding activity of the DDB1-DDB2-Cul4A ubiquitin ligase complex is important for the GGR subpathway of NER, we determined whether a defect in this repair process was the cause of the phenotype observed following DDB1 depletion (21, 30). To examine this possibility, we disrupted GGR by depleting DDB2. Depletion of DDB2 using siRNA oligonucleotides does not resemble the phenotype observed following depletion of DDB1 (Fig. 4A). ATM S1981 and Chk2 T68 phosphorylation, as well as increased p21 levels, is observed after depletion of DDB1 but is minimal after depletion of DDB2. Also, depletion of DDB2 does not cause a G2 accumulation (see Fig. S2 in the supplemental material). We further examined the potential role that impaired GGR activity may have in contributing to the damage phenotype by depleting XPC, a damage sensor protein required for the GGR subpathway of NER (6, 55, 68). Loss of XPC function is more detrimental to GGR than the loss of DDB1-DDB2-Cul4A ubiquitin ligase activity (38). However, despite an effective reduction in protein levels (Fig. 4D), depletion of XPC by siRNA again does not produce the phenotypic characteristics of DDB1 depletion. ATM S1981 and Chk2 T68 phosphorylation is not observed following depletion of XPC from cells, nor is there a significant increase in p21 levels (Fig. 4B). Depletion of XPC also does not result in a G2 accumulation of cells (see Fig. S2 in the supplemental material). These results suggest that defects in GGR are unlikely to be the cause of the DNA damage observed after DDB1 depletion.
FIG. 4.
Disruption of NER does not cause activation of cell cycle checkpoint signaling. (A) Cells were transfected with control, DDB1, or DDB2 siRNA oligonucleotides. Three days after transfection cell lysates were harvested, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and immunoblotted with the indicated antibodies. (B) Checkpoint activation was monitored on days 2, 3, and 4 after transfection with control, DDB1, and XPC siRNA oligonucleotides by immunoblotting with the indicated antibodies. (C) Cells were transfected with control, DDB1, or XPA siRNA oligonucleotides. Three days after transfection, checkpoint activation was examined by immunoblotting with the indicated antibodies. (D) The efficiency of depletion by the XPA and XPC siRNA oligonucleotides was monitored by immunoblot analysis. The asterisks denote nonspecific, cross-reacting proteins.
In addition to functioning in the GGR subpathway of NER, DDB1 also has a role in TCR (21). The DDB1-Cul4A ubiquitin ligase complex functions independently of DDB2 in this capacity. To determine whether defective TCR contributes to the phenotype observed after DDB1 depletion, we depleted XPA using siRNA oligonucleotides. Like DDB1, XPA is required for both GGR and TCR, with the loss of this protein reducing the repair capacity of cells to approximately 10% (6, 38, 55). Depletion of XPA from cells does not produce a phenotype reminiscent of DDB1 depletion. There is no phosphorylation of checkpoint proteins, no increase in p21 levels, and no aberration in the cell cycle distribution of these cells (Fig. 4C; see also Fig. S2 in the supplemental material). Immunoblotting assays for XPA demonstrate that the siRNA oligonucleotides effectively reduce XPA protein levels in these experiments (Fig. 4D). These data indicate that the phenotype observed after depletion of DDB1 is not due to defective GGR or TCR.
Misregulation of Cdt1 contributes to the DNA damage observed following depletion of DDB1.
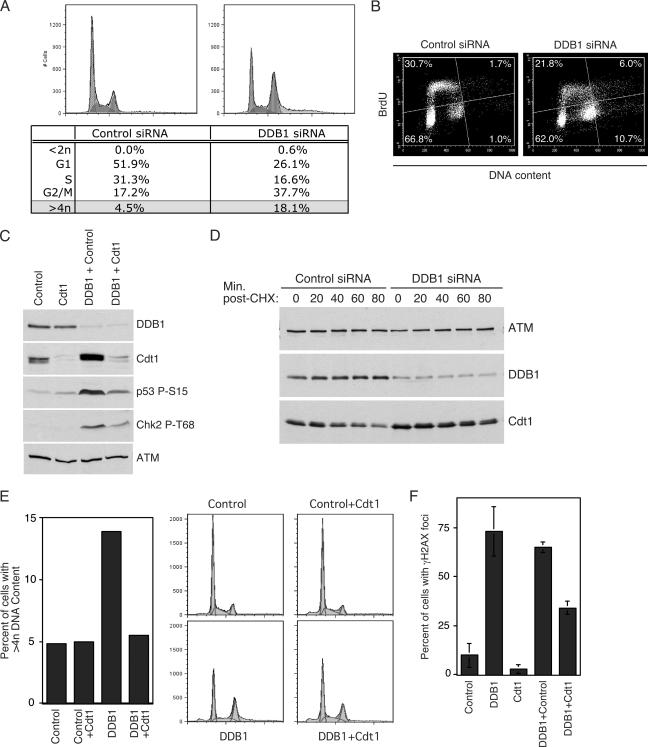
Aside from its role in NER, another function ascribed to DDB1 is the regulation of Cdt1 by DDB1-Cul4A. The DDB1-Cul4A ubiquitin ligase complex targets this replication licensing factor for degradation following UV and IR damage (24, 26). It also regulates Cdt1 degradation during normal cell cycle progression in combination with an SCF ubiquitin ligase complex (2, 27, 45, 52). Regulation of Cdt1 is critical, as overexpression of this protein leads to rereplication of genomic DNA. During the course of characterizing the phenotype resulting from DDB1 depletion, it was apparent in the DNA content profiles generated by flow cytometry that depletion of DDB1 increases the percentage of cells with more than 4n DNA content (Fig. 5A). While only 4.5% of control cells reside in this population, 18.1% of DDB1-depleted cells display a DNA content greater than 4n. The existing link between DDB1-Cul4A and Cdt1 degradation suggests that the increase in the percentage of cells with more than 4n DNA content after DDB1 depletion may result from rereplication due to the misregulation of Cdt1. To more definitively determine whether DDB1 depletion causes rereplication, we monitored incorporation of the thymidine analogue BrdU by flow cytometry. Depletion of DDB1 from U2OS cells again enhanced the percentage of cells with greater than 4n DNA content (Fig. 5B). Furthermore, nearly 36% of DDB1-depleted cells with greater than 4n DNA content are actively undergoing DNA synthesis and incorporating BrdU (6% of the total DDB1-depleted population). DNA synthesis in cells with greater than 4n DNA content suggests that this population does arise from rereplication. The presence of active cell cycle checkpoints has likely limited the extent of BrdU incorporation in the DDB1-depleted cells displaying greater than 4n DNA content, as well as in some S-phase cells, as evidenced by the diminished BrdU incorporation in cells possessing a DNA content between 2n and 4n. The cells with greater than 4n DNA content after DDB1 depletion are unlikely to result from defects in cytokinesis, since we found no enhancement in the number of multinucleated cells in the DDB1-depleted population (see Fig. S3 in the supplemental material).
FIG. 5.
Misregulation of Cdt1 contributes to the damage observed following DDB1 depletion. (A) The DNA content of HeLa cells was analyzed by flow cytometry following propidium iodide staining 4 days after transfection with control or DDB1 siRNA oligonucleotides. Cell cycle distributions were quantified using FlowJo fluorescence-activated cell sorting analysis software. (B) U2OS cells were transfected with control or DDB1 siRNA oligonucleotides. Four days after transfection the cells were labeled with BrdU. After fixation, the cells were immunostained with an anti-BrdU antibody and DNA content was monitored by flow cytometry using propidium iodide staining. (C) U2OS cells were transfected with control, Cdt1, and DDB1 siRNA oligonucleotides alone or in combination, as indicated. Cdt1 abundance and checkpoint activation were examined 4 days after transfection by immunoblotting with the indicated antibodies. (D) Cells stably expressing an HA-tagged Cdt1 protein were transfected with control or DDB1 siRNA oligonucleotides. Two days after transfection cells were treated with 100 μM cycloheximide (CHX) and harvested at the times indicated. Depletion of DDB1 and the stability of Cdt1 were monitored by immunoblot analyses. (E and F) U2OS cells were transfected with control, Cdt1, and DDB1 siRNA oligonucleotides alone or in combination, as indicated. (E) The extent of rereplication and G2 accumulation were monitored by flow cytometric analysis of DNA content. Quantitation was performed using the FlowJo software analysis program. (F) Intranuclear γH2AX focus formation was examined by indirect immunofluorescence. The graph depicts the percentage of cells in each population displaying γH2AX foci.
The presence of rereplication in DDB1-depleted cells suggests that regulation of the replication licensing factor Cdt1, a known substrate of the DDB1-Cul4A ubiquitin ligase complex, may be disrupted. Consistent with this interpretation, we observed an increase in the abundance of Cdt1 protein in the DDB1-depleted cell population (Fig. 5C). Furthermore, the Cdt1 protein is more stable in the DDB1-depleted cells (approximate half-life of 120 min) than in control cells (approximate half-life of 65 min) (Fig. 5D). Thus, Cdt1 turnover is misregulated in DDB1-depleted cells.
The increased stability of Cdt1 and the rereplication observed following DDB1 depletion suggest that misregulation of Cdt1 may be contributing to the phenotype observed in DDB1-depleted cells. To examine this possibility, we used siRNA to codeplete Cdt1 and DDB1 from U2OS cells. If deregulation of Cdt1 is promoting rereplication in DDB1-depleted cells, then depletion of Cdt1 with siRNA should reduce the percentage of cells with greater than 4n DNA content. The level of Cdt1 depletion that was achieved in combination with a control siRNA did not alter the cell cycle profile, the percentage of replicating cells, or the percentage of cells with more than 4n DNA content (Fig. 5E and data not shown). In contrast, codepletion of Cdt1 with DDB1 reduces the percentage of cells with more than 4n DNA content from 14% to 5%, a level similar to that observed in control cells (Fig. 5E). This confirms that the population of cells with greater than 4n DNA content after DDB1 depletion does represent cells in which rereplication has occurred due to Cdt1 misregulation.
Since codepletion of Cdt1 with DDB1 can reduce the percentage of rereplicating cells, we next asked whether the codepletion of Cdt1 with DDB1 can alleviate the DNA damage and checkpoint activation caused by DDB1 depletion. As observed previously, depletion of DDB1 significantly enhances the percentage of cells displaying γH2AX foci. The codepletion of DDB1 with Cdt1 considerably reduces the amount of DNA damage generated by the loss of DDB1, as the percentage of cells with γH2AX foci was reduced by nearly 50% (Fig. 5F). A similar alleviation in the DNA damage phenotype was observed by immunoblot analysis. As noted previously, depletion of DDB1 results in phosphorylation of p53 S15 and Chk2 T68 (Fig. 5C). These events are not observed in control or Cdt1-depleted cells. When DDB1 is depleted in combination with Cdt1, the levels of phosphorylated p53 S15 and Chk2 T68 are significantly reduced (Fig. 5C). Collectively, these results indicate that misregulation of Cdt1, and the subsequent rereplication, contributes to the formation of DNA damage and the activation of checkpoint responses after DDB1 depletion. It should also be noted that reducing the level of Cdt1 below that of control cells only partially alleviates the DNA damage associated with DDB1 depletion. Thus, misregulation of Cdt1 may be only one of the factors that cause DNA damage in DDB1-depleted cells.
Disruption of SCFSkp2-mediated Cdt1 regulation does not cause DNA damage.
Multiple mechanisms of Cdt1 regulation exist, and our results suggest that disruption of a single mechanism is sufficient to cause deregulation of Cdt1, rereplication, and DNA damage. To more precisely examine the ubiquitin-dependent mechanisms of Cdt1 regulation and their potential to generate DNA damage when disrupted, we transiently expressed wild-type Cdt1 or Cdt1 mutants where one or both of the ubiquitin-mediated degradation pathways were abrogated. Disruption of the DDB1-Cul4A-dependent ubiquitination was accomplished by mutating residues of Cdt1 that are critical for its interaction with the DNA polymerase processivity factor PCNA (Q3A, V6A, F9A, F10A = ΔPIP). Binding of Cdt1 to PCNA is required for degradation of Cdt1 by DDB1-Cul4A (2, 27, 45, 52). We also disrupted the SCFSkp2 ubiquitination pathway for Cdt1 by removing a cyclin-dependent kinase (CDK) phosphorylation site in Cdt1 at threonine 29 (T29A). The binding and degradation of Cdt1 by SCFSkp2 require the prior phosphorylation of Cdt1 on T29 by CDKs (34, 36, 57, 60). Introduction of an empty vector into HeLa cells does not cause DNA damage; however, overexpression of a wild-type Cdt1 protein induces DNA damage signaling (Fig. 6A), as previously reported (33, 41, 66, 74). A similar level of p53 S15 phosphorylation is observed upon disruption of the SCFSkp2 degradation pathway (T29A), indicating that the loss of this regulatory mechanism does not cause any further damage than overexpression of wild-type Cdt1 does. Notably, disruption of DDB1-Cul4A-dependent Cdt1 degradation (ΔPIP) resulted in greater p53 S15 phosphorylation than did overexpression of either wild-type Cdt1 or the T29A mutant. This suggests that the two ubiquitin-dependent mechanisms of Cdt1 degradation are not redundant but that disruption specifically of the DDB1-Cul4A-mediated Cdt1 degradation pathway has a greater potential to generate DNA damage.
FIG. 6.
Disruption of SCFSkp2-dependent Cdt1 degradation does not generate DNA damage. (A) HeLa cells were transfected with an empty vector or vectors encoding wild-type (wt) Cdt1; Cdt1 Q3A, V6A, F9A, and F10A (ΔPIP); or Cdt1 T29A. Checkpoint activation was examined in these cells 2 days after transfection by immunoblotting. (B) HeLa cells were transfected with control, DDB1, or Skp2 siRNA oligonucleotides. Three days after transfection cell lysates were harvested and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and checkpoint activation was monitored by immunoblotting with the indicated antibodies.
To further examine the ability of these ubiquitin ligase complexes to maintain genome stability, we assessed checkpoint activation following depletion of DDB1 or Skp2 by siRNA. While depletion of DDB1 results in the phosphorylation of Chk1 S345, Chk2 T68, and p53 S15, these markers of DNA damage signaling are not observed following depletion of Skp2 (Fig. 6B). Interestingly, depletion of DDB1 elevates Cdt1 levels to a greater extent than that observed following depletion of Skp2, suggesting that disruption of the DDB1-Cul4A ubiquitin ligase complex is more detrimental to the proper regulation of Cdt1 protein levels than loss of the SCFSkp2 ubiquitin ligase complex. Together, these results support our conclusion that misregulation of Cdt1 generates DNA damage and suggests that disruption of DDB1-Cul4A-mediated Cdt1 degradation is more detrimental to genome stability than disruption of the SCFSkp2-mediated degradation pathway.
Accumulation of DNA damage following DDB1 depletion requires cell cycle progression.
To further test the model that rereplication resulting from misregulation of Cdt1 is contributing to the phenotype observed after DDB1 depletion, we examined the cell cycle dependency of the phenotype. If our model is correct, then the accumulation of DNA damage should require progression into S phase. RPE-hTERT cells were treated with control or DDB1 siRNA and grown to confluence as a means of causing contact inhibition and a G0 arrest. Cell cycle analysis by propidium iodide staining shows a significant accumulation of cells with a 2n DNA content following the arrest by contact inhibition (Fig. 7A). Approximately 93% of control cells and 88% of DDB1-depleted cells arrested by this method display a DNA content of 2n. G0-arrested cells were also released into the cell cycle and harvested at a point in time when cells were advancing into S phase. This population allows us to examine the extent of DNA damage that occurs during progression through a single S phase. It should be noted, however, that DDB1-depleted cells take more time to be released from a G0 arrest and do not release as synchronously as control cells (Fig. 7A). Finally, control and DDB1-depleted cells were allowed to cycle unperturbed throughout the course of the experiment. The extent of DNA damage signaling in these samples was then examined by immunoblotting. DDB1-depleted cells that are arrested by contact inhibition show very little p53 S15 phosphorylation or p21 induction (Fig. 7B). The entrance of DDB1-depleted cells into S phase corresponds with an increase in the levels of p53 S15 phosphorylation and p21 induction, whereas the analogous control cells showed no increase in these markers of DNA damage. While it is unclear why DDB1-depleted cells are released slowly from G0, these data indicate that the damage observed after DDB1 depletion can occur during a single cell cycle and that the damage requires progression into S phase. Cells depleted of DDB1 that are allowed to cycle during the course of the experiment show further increases in p53 S15 phosphorylation and p21 induction, suggesting that the damage is accumulating and further activating checkpoint responses over time. Cyclin A immunoblotting of these samples was used as a marker of S and G2 phases of the cell cycle and corresponds with the flow cytometry data (Fig. 7B).
FIG. 7.
Cell cycle progression is required for the accumulation of DNA damage in DDB1-depleted cells. RPE-hTERT cells were transfected with control or DDB1 siRNA oligonucleotides. Cells were arrested by growth to confluence, arrested, and released back into the cell cycle by splitting them into subconfluent densities or maintained at subconfluent densities during the course of the experiment. All samples were harvested on the fourth day after siRNA transfection, and DNA content was measured by flow cytometry of propidium iodide-stained cells (A) or cell lysates were prepared and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and checkpoint activation was monitored by immunoblotting (B).
Analysis of the location of chromosomal breaks in DDB1-depleted cells.
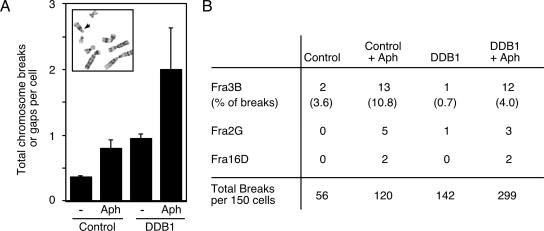
Our data indicate that lack of DDB1 function causes DNA damage, partially through deregulation of Cdt1 that allows rereplication to occur. Since the DNA damage is associated with replication, we hypothesized that the double-strand breaks may preferentially occur at chromosomal fragile sites. These chromosome regions are particularly vulnerable to breakage, especially when cells experience replication stress such as that caused by low doses of aphidicolin (18, 48). Therefore, we examined whether the DNA double-strand breaks that form following DDB1 depletion occur preferentially at fragile sites. Metaphase spreads from U2OS cells treated with a control siRNA or DDB1 siRNA were analyzed for chromosomal gaps and breaks by trypsin-Giemsa banding. Depletion of DDB1 from U2OS cells enhances the occurrence of chromosomal gaps and breaks in mitotic cells more than twofold over the number of events observed in control cells (Fig. 8A). This increase is similar to what we observed upon treating cells with aphidicolin. Interestingly, treatment of DDB1-depleted cells with aphidicolin merely caused an additive effect. Further characterization of the location of these chromosomal breaks reveals that while the addition of aphidicolin enhances breakage events at common fragile sites, such as FRA3B, depletion of DDB1 alone causes breakage events to occur at a wide variety of loci throughout the genome, with no enhancement in the number of breaks at any known fragile site (Fig. 8B). For example, over 10% of breaks in aphidicolin-treated cells occur at a single fragile site (FRA3B), but only 0.7% of breaks caused by DDB1 depletion were at this site. These results were replicated in the untransformed RPE-hTERT cells, where depletion of DDB1 again enhanced the number of chromosome breakage events observed relative to control cells (data not shown). These data indicate that the cause of the double-strand breaks in DDB1-depleted cells is distinct from the replication problems created by aphidicolin and suggest that rereplication does not cause DNA damage due to the stalling of replication forks at fragile sites.
FIG. 8.
Depletion of DDB1 enhances chromosome breaks. (A) U2OS cells were transfected with control or DDB1 siRNA oligonucleotides and processed for metaphase spread analysis 3 days after transfection. Fragile sites were induced by the addition of 0.1 μM aphidicolin (Aph) 24 h prior to harvest. Identification of chromosome gaps and breaks was facilitated by trypsin-Giemsa banding, as shown in the inset. The arrowhead indicates the location of a chromosome break. The graph depicts the average number of chromosome gaps or breaks per cell. (B) The chromosome location of the gaps and breaks was recorded based on banding patterns. FRA3B, FRA2G, and FRA16D breakages were scored as described previously (48).
DISCUSSION
Our data indicate that DDB1 is required for maintaining genome stability in human cells. Silencing of DDB1 expression results in an accumulation of DNA damage and activation of cell cycle checkpoints. DDB1 protects the integrity of the genome as part of an E3 ubiquitin ligase complex containing Cul4A. One target of this complex that must be degraded to preserve genome integrity is the replication licensing factor Cdt1. Multiple levels of Cdt1 regulation exist in human cells. These mechanisms possess unique and necessary roles in the regulation of Cdt1, to properly restrain this protein and prevent the adverse cellular consequences associated with its misregulation.
A role for DDB1 in maintaining genome stability has previously been noted in other organisms. Shimanouchi and colleagues found that depletion of DDB1 in Drosophila melanogaster promoted the loss of heterozygosity in somatic cells (53). Holmberg et al. also found that deletion of DDB1 in S. pombe enhances the mutation rate more than 20-fold (25). These data correlate well with our data from human cells. Depletion or deletion of DDB1 results in genome instability, as assessed by the generation of DNA double-strand breaks in human cells (this study) or an enhanced mutation rate in yeast, and in neither organism is this instability the result of defective NER. In S. pombe, the genetic instability associated with deletion of DDB1 could be partially suppressed by removing Spd1, an inhibitor of RNR (25). DDB1 and Pcu4 (the Cul4 homologue in yeast) promote the ubiquitin-mediated degradation of Spd1 to allow association of the RNR subunits and production of deoxynucleoside triphosphates for DNA synthesis and repair (8, 12, 25, 35). A human homologue of Spd1 has not been identified; therefore, it is unclear whether this function of DDB1 is conserved in higher eukaryotes. However, removal of Spd1 could not completely alleviate the genetic instability created by the loss of DDB1 in S. pombe. This suggests that DDB1 is required for other aspects of mutation avoidance in addition to regulating DNA replication through the degradation of Spd1. Likewise, we suggest that human DDB1 mediates genome stability, in part by controlling DNA replication through the degradation of Cdt1. However, the inability of Cdt1 depletion to completely alleviate the phenotype observed following DDB1 depletion indicates that additional functions of DDB1 are critical for its role in the maintenance of genome integrity.
The DDB1-Cul4A ubiquitin ligase complex regulates DNA replication in multicellular eukaryotes by mediating the degradation of Cdt1. Exposure to exogenous DNA-damaging agents induces the destruction of Cdt1 specifically by DDB1-Cul4A (24, 26). Our data indicate that DDB1 also has an important role in the regulation of Cdt1 in the absence of exogenous DNA damage. In agreement with our proposal, recent reports have implicated DDB1-Cul4A in the replication-dependent degradation of Cdt1 and identified PCNA as a critical mediator of the DDB1-Cul4A-dependent Cdt1 ubiquitination (2, 27, 45, 52). The destruction of Cdt1 by the DDB1-Cul4A ubiquitin ligase complex is one of three mechanisms known in human cells to regulate Cdt1. Additionally, the replication-dependent degradation of this protein can be accomplished by an SCFSkp2 ubiquitin ligase complex, which is targeted to Cdt1 by a CDK-mediated phosphorylation event (34, 36, 57, 60). Cdt1 is also functionally inhibited by the binding of geminin (59, 71). These multiple levels of Cdt1 regulation suggest that the proper restraint of this protein activity is crucial.
Disruption of Cdt1 regulation is detrimental to genome stability and cell viability (25, 41, 53). Highly elevated levels of Cdt1 expression lead to significant amounts of rereplication in human cells, Drosophila, and Xenopus laevis (3, 33, 62, 66). Rereplication induces activation of DNA damage response pathways in humans, Xenopus, and yeast (20, 33, 41, 66, 74). The phenotype observed after DDB1 depletion is consistent with the phenotype observed after rereplication. We found a significant increase in DNA damage and activation of both ATR- and ATM-mediated damage response pathways (Fig. 1 and 2). Notably, we see this enhancement of genome instability with only moderate increases in Cdt1 levels (Fig. 5C), in contrast to the significantly greater levels of Cdt1 overexpression that were used in previous studies. Furthermore, we observe direct evidence that rereplication is occurring in DDB1-depleted cells with the incorporation of BrdU by cells with greater than 4n DNA content (Fig. 5B). The rereplication observed after DDB1 depletion is not extensive, perhaps due in part to the restraints imposed by the checkpoint response (33). The elevated levels of Cdt1 protein, rereplication, and DNA damage suggest that misregulation of Cdt1 is contributing to the phenotype observed after DDB1 depletion. In support of this hypothesis, we found that there was indeed deregulation of Cdt1, as the degradation of Cdt1 proteins was significantly delayed following depletion of DDB1 (Fig. 5D). Additionally, reducing Cdt1 protein levels in DDB1-depleted cells could prevent rereplication and eliminate approximately half of the DNA damage and checkpoint activation (Fig. 5). Importantly, the level of Cdt1 reduction achieved in these experiments did not interfere with normal DNA replication. Therefore, the reduction in DNA damage by codepleting Cdt1 with DDB1 is unlikely to be an indirect consequence of slowing the cell cycle. Our results clearly demonstrate that depletion of DDB1 results in the misregulation of Cdt1, which stabilizes and elevates the cellular levels of this protein. The consequence of this misregulation is rereplication, which contributes to the DNA damage and checkpoint activation observed after DDB1 depletion.
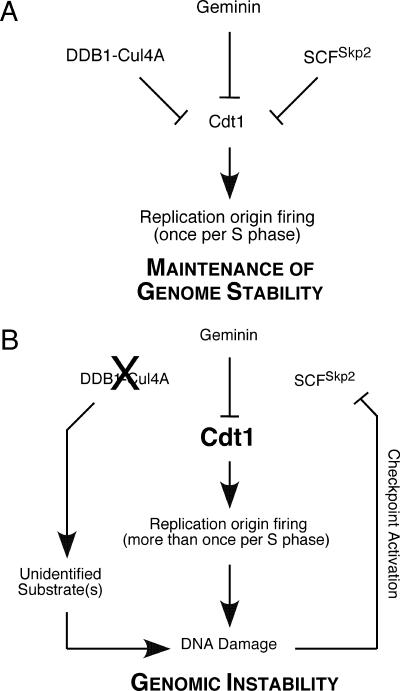
Our data indicate that rereplication occurs in DDB1-depleted cells despite the presence of other mechanisms that operate to suppress the refiring of replication origins. Previous data from human systems have suggested that the DDB1-Cul4 and SCFSkp2 mechanisms of Cdt1 destruction are redundant in the absence of exogenous DNA damage (45, 52). However, the disruption of a single mode of Cdt1 regulation in other organisms was shown to be sufficient to have adverse effects. In Xenopus, disruption of the DDB1-Cul4 degradation pathway stabilizes Cdt1 and induces significant levels of rereplication (2). Depletion of Cul4 from Caenorhabditis elegans also stabilizes Cdt1 and results in massive rereplication, with cells exhibiting up to 100n DNA content (73). The stabilization of Cdt1 and the presence of rereplication after DDB1 depletion argue against redundant roles for DDB1-Cul4A and SCFSkp2 in the destruction of Cdt1 in human cells. Additionally, this suggestion is supported by our observation that expression of a Cdt1 mutant insensitive to DDB1-Cul4A degradation results in greater DNA damage than expression of either wild-type Cdt1 or a mutant that is insensitive to the SCFSkp2 destruction pathway (Fig. 6A). We propose that the DDB1-dependent degradation of Cdt1 is particularly important because the loss of DDB1 creates a situation in which one other mechanism of Cdt1 regulation is also inactivated. Depletion of DDB1 generates DNA damage that activates cell cycle checkpoints, which in turn function to inactivate CDK complexes (50). Since the SCFSkp2-mediated destruction of Cdt1 requires a CDK-mediated phosphorylation event, this pathway is inhibited by the presence of active checkpoints. Therefore, disruption of DDB1 eliminates both ubiquitin-dependent mechanisms of Cdt1 regulation, which will likely result in further rereplication, greater DNA damage, and amplification of genome instability (Fig. 9). It should be noted that cells generate DNA damage intrinsically as a consequence of respiration and DNA metabolism. Replication forks encounter DNA lesions and experience difficulty in replicating through specific regions of the genome during every round of DNA synthesis. The act of growing cells in culture can also increase cellular stresses. Thus, the distinction between DNA damage-dependent Cdt1 degradation and replication-dependent degradation is largely one of degree.
FIG. 9.
Model highlighting the importance of DDB1 in Cdt1 regulation. (A) Geminin, DDB1-Cul4A, and SCFSkp2 cooperate to restrain the activity and levels of Cdt1, ensuring that origins of replication fire only once per S phase. (B) siRNA depletion of DDB1 disrupts the DDB1-Cul4A-dependent regulation of Cdt1 and causes DNA damage due to rereplication and impaired regulation of an additional unidentified substrate. DNA damage activates cell cycle checkpoints, which in turn function to inactivate CDK complexes. SCFSkp2-mediated degradation of Cdt1 requires a prior phosphorylation event by CDK; therefore, this mechanism of Cdt1 regulation is disrupted by the presence of active checkpoints. The deregulation results in increased Cdt1 levels that promote rereplication, leading to greater DNA damage, and amplification of genomic instability.
DDB1-dependent regulation of Cdt1 is not sufficient to explain all of the genome instability that arises from DDB1 depletion. Codepletion of Cdt1 with DDB1 eliminates rereplication; however, it does not completely prevent DNA damage or checkpoint activation. Additional genome maintenance functions of DDB1 likely have cell cycle dependency, since the loss of DDB1 from G0-arrested cells does not cause DNA damage (Fig. 7). Identifying other substrates of DDB1-Cul4A whose degradation is important in preventing genetic instability will be important. Another question that remains unanswered is the mechanism by which rereplication activates checkpoint pathways. Our analyses suggest that rereplication causes DNA double-strand breaks in a manner that is distinct from those arising at chromosome fragile sites during replication stress. It is unclear whether these breaks are at random locations or whether they may cluster near specific genomic regions that are prone to rereplication. The breaks do not appear to cluster near centromeres. This suggests that attachment of the mitotic spindle to a rereplicated centromere on a single chromatid, and the subsequent breakage of the chromosome during anaphase, is not a major mechanism contributing to these breaks. One possibility is that rereplication causes double-strand breaks as the second fork originating from a refired origin encounters the Okazaki fragments generated by the original replication fork. A second possibility is that the forks initiated at a refired origin are defective, stall, and eventually collapse, thus generating DNA damage.
Recent studies have highlighted a role for replication stress as an early event in the initiation of cancer (5, 19, 67). Aberrant DNA replication in precancerous lesions produces DNA damage and activates checkpoint pathways. Inactivation of these damage response pathways is associated with tumor progression due to genetic instability. Disruption of DDB1-dependent functions also has the ability to create replication stress and genetic instability, perhaps providing an avenue through which cancer progression can be facilitated. Changes in the activity of the DDB1-Cul4A ubiquitin ligase complex, or in mechanisms regulating replication origin firing, may therefore play important roles in the process of tumorigenesis.
Supplementary Material
Acknowledgments
This work was supported by a National Cancer Institute grant to D.C. D.C. is also supported by the Pew Scholars Program in the Biological Sciences, sponsored by the Pew Charitable Trusts. C.L. is supported by a Department of Defense predoctoral fellowship.
We thank Jeffrey Salisbury, Xiaohua Wu, and Yue Xiong for reagents.
Footnotes
Published ahead of print on 28 August 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abraham, R. T. 2004. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amsterdam) 3:883-887. [DOI] [PubMed] [Google Scholar]
- 2.Arias, E. E., and J. C. Walter. 2006. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 8:84-90. [DOI] [PubMed] [Google Scholar]
- 3.Arias, E. E., and J. C. Walter. 2005. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 19:114-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banin, S., L. Moyal, S. Shieh, Y. Taya, C. W. Anderson, L. Chessa, N. I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674-1677. [DOI] [PubMed] [Google Scholar]
- 5.Bartkova, J., Z. Horejsi, K. Koed, A. Kramer, F. Tort, K. Zieger, P. Guldberg, M. Sehested, J. M. Nesland, C. Lukas, T. Orntoft, J. Lukas, and J. Bartek. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434:864-870. [DOI] [PubMed] [Google Scholar]
- 6.Berg, R. J., H. J. Ruven, A. T. Sands, F. R. de Gruijl, and L. H. Mullenders. 1998. Defective global genome repair in XPC mice is associated with skin cancer susceptibility but not with sensitivity to UVB induced erythema and edema. J. Investig. Dermatol. 110:405-409. [DOI] [PubMed] [Google Scholar]
- 7.Blow, J. J., and A. Dutta. 2005. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 6:476-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bondar, T., A. Ponomarev, and P. Raychaudhuri. 2004. Ddb1 is required for the proteolysis of the Schizosaccharomyces pombe replication inhibitor Spd1 during S phase and after DNA damage. J. Biol. Chem. 279:9937-9943. [DOI] [PubMed] [Google Scholar]
- 9.Brown, A. L., C. H. Lee, J. K. Schwarz, N. Mitiku, H. Piwnica-Worms, and J. H. Chung. 1999. A human Cds1-related kinase that functions downstream of ATM protein in the cellular response to DNA damage. Proc. Natl. Acad. Sci. USA 96:3745-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, E. J., and D. Baltimore. 2003. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 17:615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677-1679. [DOI] [PubMed] [Google Scholar]
- 12.Chabes, A., and L. Thelander. 2000. Controlled protein degradation regulates ribonucleotide reductase activity in proliferating mammalian cells during the normal cell cycle and in response to DNA damage and replication blocks. J. Biol. Chem. 275:17747-17753. [DOI] [PubMed] [Google Scholar]
- 13.Chen, X., Y. Zhang, L. Douglas, and P. Zhou. 2001. UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J. Biol. Chem. 276:48175-48182. [DOI] [PubMed] [Google Scholar]
- 14.Chu, G., and E. Chang. 1988. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science 242:564-567. [DOI] [PubMed] [Google Scholar]
- 15.Cleaver, J. E. 2005. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat. Rev. Cancer 5:564-573. [DOI] [PubMed] [Google Scholar]
- 16.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge. 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294:1713-1716. [DOI] [PubMed] [Google Scholar]
- 17.Diffley, J. F. 2004. Regulation of early events in chromosome replication. Curr. Biol. 14:R778-R786. [DOI] [PubMed] [Google Scholar]
- 18.Glover, T. W., M. F. Arlt, A. M. Casper, and S. G. Durkin. 2005. Mechanisms of common fragile site instability. Hum. Mol. Genet. 14(Spec. No. 2):R197-R205. [DOI] [PubMed] [Google Scholar]
- 19.Gorgoulis, V. G., L. V. Vassiliou, P. Karakaidos, P. Zacharatos, A. Kotsinas, T. Liloglou, M. Venere, R. A. Ditullio, Jr., N. G. Kastrinakis, B. Levy, D. Kletsas, A. Yoneta, M. Herlyn, C. Kittas, and T. D. Halazonetis. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434:907-913. [DOI] [PubMed] [Google Scholar]
- 20.Green, B. M., and J. J. Li. 2005. Loss of rereplication control in Saccharomyces cerevisiae results in extensive DNA damage. Mol. Biol. Cell 16:421-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groisman, R., J. Polanowska, I. Kuraoka, J. Sawada, M. Saijo, R. Drapkin, A. F. Kisselev, K. Tanaka, and Y. Nakatani. 2003. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113:357-367. [DOI] [PubMed] [Google Scholar]
- 22.Guo, Z., A. Kumagai, S. X. Wang, and W. G. Dunphy. 2000. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 14:2745-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gurley, L. R., J. A. D'Anna, S. S. Barham, L. L. Deaven, and R. A. Tobey. 1978. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur. J. Biochem. 84:1-15. [DOI] [PubMed] [Google Scholar]
- 24.Higa, L. A., I. S. Mihaylov, D. P. Banks, J. Zheng, and H. Zhang. 2003. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 5:1008-1015. [DOI] [PubMed] [Google Scholar]
- 25.Holmberg, C., O. Fleck, H. A. Hansen, C. Liu, R. Slaaby, A. M. Carr, and O. Nielsen. 2005. Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev. 19:853-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu, J., C. M. McCall, T. Ohta, and Y. Xiong. 2004. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 6:1003-1009. [DOI] [PubMed] [Google Scholar]
- 27.Hu, J., and Y. Xiong. 2006. An evolutionarily conserved function of proliferating cell nuclear antigen for Cdt1 degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA damage. J. Biol. Chem. 281:3753-3756. [DOI] [PubMed] [Google Scholar]
- 28.Hwang, B. J., J. M. Ford, P. C. Hanawalt, and G. Chu. 1999. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl. Acad. Sci. USA 96:424-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kapetanaki, M. G., J. Guerrero-Santoro, D. C. Bisi, C. L. Hsieh, V. Rapic-Otrin, and A. S. Levine. 2006. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. USA 103:2588-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keeney, S., A. P. Eker, T. Brody, W. Vermeulen, D. Bootsma, J. H. Hoeijmakers, and S. Linn. 1994. Correction of the DNA repair defect in xeroderma pigmentosum group E by injection of a DNA damage-binding protein. Proc. Natl. Acad. Sci. USA 91:4053-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumagai, A., J. Lee, H. Y. Yoo, and W. G. Dunphy. 2006. TopBP1 activates the ATR-ATRIP complex. Cell 124:943-955. [DOI] [PubMed] [Google Scholar]
- 32.Lau, C. C., and A. B. Pardee. 1982. Mechanism by which caffeine potentiates lethality of nitrogen mustard. Proc. Natl. Acad. Sci. USA 79:2942-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, A., and J. J. Blow. 2005. Cdt1 downregulation by proteolysis and geminin inhibition prevents DNA re-replication in Xenopus. EMBO J. 24:395-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, X., Q. Zhao, R. Liao, P. Sun, and X. Wu. 2003. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J. Biol. Chem. 278:30854-30858. [DOI] [PubMed] [Google Scholar]
- 35.Liu, C., K. A. Powell, K. Mundt, L. Wu, A. M. Carr, and T. Caspari. 2003. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev. 17:1130-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, E., X. Li, F. Yan, Q. Zhao, and X. Wu. 2004. Cyclin-dependent kinases phosphorylate human Cdt1 and induce its degradation. J. Biol. Chem. 279:17283-17288. [DOI] [PubMed] [Google Scholar]
- 37.Liu, Q., S. Guntuku, X. S. Cui, S. Matsuoka, D. Cortez, K. Tamai, G. Luo, S. Carattini-Rivera, F. DeMayo, A. Bradley, L. A. Donehower, and S. J. Elledge. 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 14:1448-1459. [PMC free article] [PubMed] [Google Scholar]
- 38.Magnaldo, T., and A. Sarasin. 2004. Xeroderma pigmentosum: from symptoms and genetics to gene-based skin therapy. Cells Tissues Organs 177:189-198. [DOI] [PubMed] [Google Scholar]
- 39.Matsuda, N., K. Azuma, M. Saijo, S. Iemura, Y. Hioki, T. Natsume, T. Chiba, K. Tanaka, and K. Tanaka. 2005. DDB2, the xeroderma pigmentosum group E gene product, is directly ubiquitylated by cullin 4A-based ubiquitin ligase complex. DNA Repair (Amsterdam) 4:537-545. [DOI] [PubMed] [Google Scholar]
- 40.Matsuoka, S., M. Huang, and S. J. Elledge. 1998. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 282:1893-1897. [DOI] [PubMed] [Google Scholar]
- 41.Melixetian, M., A. Ballabeni, L. Masiero, P. Gasparini, R. Zamponi, J. Bartek, J. Lukas, and K. Helin. 2004. Loss of geminin induces rereplication in the presence of functional p53. J. Cell Biol. 165:473-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mirzoeva, O. K., and J. H. Petrini. 2001. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 21:281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myers, J. S., and D. Cortez. 2006. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J. Biol. Chem. 281:9346-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nag, A., T. Bondar, S. Shiv, and P. Raychaudhuri. 2001. The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol. Cell. Biol. 21:6738-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishitani, H., N. Sugimoto, V. Roukos, Y. Nakanishi, M. Saijo, C. Obuse, T. Tsurimoto, K. I. Nakayama, K. Nakayama, M. Fujita, Z. Lygerou, and T. Nishimoto. 2006. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 25:1126-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886-895. [DOI] [PubMed] [Google Scholar]
- 47.Rapic-Otrin, V., V. Navazza, T. Nardo, E. Botta, M. McLenigan, D. C. Bisi, A. S. Levine, and M. Stefanini. 2003. True XP group E patients have a defective UV-damaged DNA binding protein complex and mutations in DDB2 which reveal the functional domains of its p48 product. Hum. Mol. Genet. 12:1507-1522. [DOI] [PubMed] [Google Scholar]
- 48.Richards, R. I. 2001. Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet. 17:339-345. [DOI] [PubMed] [Google Scholar]
- 49.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sancar, A., L. A. Lindsey-Boltz, K. Unsal-Kacmaz, and S. Linn. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73:39-85. [DOI] [PubMed] [Google Scholar]
- 51.Schlegel, R., and A. B. Pardee. 1986. Caffeine-induced uncoupling of mitosis from the completion of DNA replication in mammalian cells. Science 232:1264-1266. [DOI] [PubMed] [Google Scholar]
- 52.Senga, T., U. Sivaprasad, W. Zhu, J. H. Park, E. E. Arias, J. C. Walter, and A. Dutta. 2006. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J. Biol. Chem. 281:6246-6252. [DOI] [PubMed] [Google Scholar]
- 53.Shimanouchi, K., K. Takata, M. Yamaguchi, S. Murakami, G. Ishikawa, R. Takeuchi, Y. Kanai, T. Ruike, R. Nakamura, Y. Abe, and K. Sakaguchi. 2006. Drosophila damaged DNA binding protein 1 contributes to genome stability in somatic cells. J. Biochem. (Tokyo) 139:51-58. [DOI] [PubMed] [Google Scholar]
- 54.Shiyanov, P., A. Nag, and P. Raychaudhuri. 1999. Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J. Biol. Chem. 274:35309-35312. [DOI] [PubMed] [Google Scholar]
- 55.Sugasawa, K., J. M. Ng, C. Masutani, S. Iwai, P. J. van der Spek, A. P. Eker, F. Hanaoka, D. Bootsma, and J. H. Hoeijmakers. 1998. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell 2:223-232. [DOI] [PubMed] [Google Scholar]
- 56.Sugasawa, K., Y. Okuda, M. Saijo, R. Nishi, N. Matsuda, G. Chu, T. Mori, S. Iwai, K. Tanaka, K. Tanaka, and F. Hanaoka. 2005. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121:387-400. [DOI] [PubMed] [Google Scholar]
- 57.Sugimoto, N., Y. Tatsumi, T. Tsurumi, A. Matsukage, T. Kiyono, H. Nishitani, and M. Fujita. 2004. Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding. J. Biol. Chem. 279:19691-19697. [DOI] [PubMed] [Google Scholar]
- 58.Svoboda, D. L., L. P. Briley, and J. M. Vos. 1998. Defective bypass replication of a leading strand cyclobutane thymine dimer in xeroderma pigmentosum variant cell extracts. Cancer Res. 58:2445-2448. [PubMed] [Google Scholar]
- 59.Tada, S., A. Li, D. Maiorano, M. Mechali, and J. J. Blow. 2001. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 3:107-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takeda, D. Y., J. D. Parvin, and A. Dutta. 2005. Degradation of Cdt1 during S phase is Skp2-independent and is required for efficient progression of mammalian cells through S phase. J. Biol. Chem. 280:23416-23423. [DOI] [PubMed] [Google Scholar]
- 61.Tang, J. Y., B. J. Hwang, J. M. Ford, P. C. Hanawalt, and G. Chu. 2000. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol. Cell 5:737-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomer, M., N. R. May, B. D. Aggarwal, G. Kwok, and B. R. Calvi. 2004. Drosophila double-parked is sufficient to induce re-replication during development and is regulated by cyclin E/CDK2. Development 131:4807-4818. [DOI] [PubMed] [Google Scholar]
- 63.Tibbetts, R. S., K. M. Brumbaugh, J. M. Williams, J. N. Sarkaria, W. A. Cliby, S. Y. Shieh, Y. Taya, C. Prives, and R. T. Abraham. 1999. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 13:152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ulane, C. M., and C. M. Horvath. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304:160-166. [DOI] [PubMed] [Google Scholar]
- 65.Ulane, C. M., J. J. Rodriguez, J. P. Parisien, and C. M. Horvath. 2003. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J. Virol. 77:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vaziri, C., S. Saxena, Y. Jeon, C. Lee, K. Murata, Y. Machida, N. Wagle, D. S. Hwang, and A. Dutta. 2003. A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell 11:997-1008. [DOI] [PubMed] [Google Scholar]
- 67.Venkitaraman, A. R. 2005. Medicine: aborting the birth of cancer. Nature 434:829-830. [DOI] [PubMed] [Google Scholar]
- 68.Volker, M., M. J. Mone, P. Karmakar, A. van Hoffen, W. Schul, W. Vermeulen, J. H. Hoeijmakers, R. van Driel, A. A. van Zeeland, and L. H. Mullenders. 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8:213-224. [DOI] [PubMed] [Google Scholar]
- 69.Ward, I. M., and J. Chen. 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276:47759-47762. [DOI] [PubMed] [Google Scholar]
- 70.Wertz, I. E., K. M. O'Rourke, Z. Zhang, D. Dornan, D. Arnott, R. J. Deshaies, and V. M. Dixit. 2004. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303:1371-1374. [DOI] [PubMed] [Google Scholar]
- 71.Wohlschlegel, J. A., B. T. Dwyer, S. K. Dhar, C. Cvetic, J. C. Walter, and A. Dutta. 2000. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290:2309-2312. [DOI] [PubMed] [Google Scholar]
- 72.Zhao, H., and H. Piwnica-Worms. 2001. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 21:4129-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhong, W., H. Feng, F. E. Santiago, and E. T. Kipreos. 2003. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 423:885-889. [DOI] [PubMed] [Google Scholar]
- 74.Zhu, W., Y. Chen, and A. Dutta. 2004. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell. Biol. 24:7140-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.