Abstract
The ING (inhibitor of growth) protein family includes a group of homologous nuclear proteins that share a highly conserved plant homeodomain (PHD) finger domain at their carboxyl termini. Members of this family are found in multiprotein complexes that posttranslationally modify histones, suggesting that these proteins serve a general role in permitting various enzymatic activities to interact with nucleosomes. There are three members of the ING family in Saccharomyces cerevisiae: Yng1p, Yng2p, and Pho23p. Yng1p is a component of the NuA3 histone acetyltransferase complex and is required for the interaction of NuA3 with chromatin. To gain insight into the function of the ING proteins, we made use of a genetic strategy to identify genes required for the binding of Yng1p to histones. Using the toxicity of YNG1 overexpression as a tool, we showed that Yng1p interacts with the amino-terminal tail of histone H3 and that this interaction can be disrupted by loss of lysine 4 methylation within this tail. Additionally, we mapped the region of Yng1p required for overexpression of toxicity to the PHD finger, showed that this region capable of binding lysine 4-methylated histone H3 in vitro, and demonstrated that mutations of the PHD finger that abolish binding in vitro are no longer toxic in vivo. These results identify a novel function for the Yng1p PHD finger in promoting stabilization of the NuA3 complex at chromatin through recognition of histone H3 lysine 4 methylation.
Chromatin structure can be regulated by the addition of posttranslational modifications to histones, including acetylation, methylation, phosphorylation, and ubiquitination. These modifications are thought to function by creating docking sites for factors which alter chromatin structure (27, 58). Consistent with this, several protein motifs have been identified that mediate the selective interaction of transcription-regulating complexes with specific histone modifications. The first such identified motif is the bromodomain, which preferentially interacts with acetylated histones (7, 10, 24, 49, 50, 65). Bromodomains are found in a number of protein complexes, including components of the basal transcription machinery and chromatin remodeling complexes (20, 41), which may explain, in part, how histone acetylation can facilitate transcription. Bromodomains are also found in a number of histone methyltransferases, suggesting that histone acetylation can mediate histone methylation (6). The ability of one histone modification to recruit another histone-modifying enzyme is a common recurring theme in chromatin research.
A second motif which has been shown to bind modified histones is the chromodomain, which preferentially interacts with methylated lysines (19, 52). Chromodomains are found in proteins involved in both the activation and silencing of transcription, consistent with the role of histone methylation in both events (31). Similar to bromodomains, chromodomains are found in chromatin remodeling complexes, histone acetyltransferases, and histone methyltransferases (6). As opposed to recognizing methylated lysine, some chromodomains act as DNA or RNA binding modules, and thus, not all chromodomain-containing proteins are methyl-histone binding proteins. Similarly, not all methyl-histone binding proteins contain a chromodomain. Recent studies have shown that proteins containing WD40 repeats, tudor, and MBT domains can also specifically interact with methylated histones (23, 30, 63). Additionally, other protein complexes which lack these domains have been shown to bind specifically to methylated histones, but the subunits which mediate these interactions are not yet known. These include the Isw1p chromatin remodeling complex and the NuA3 histone acetyltransferase complex (40, 55). Thus, it is highly likely that there are unidentified motifs which can mediate the interaction of chromatin modifying factors with posttranslationally modified histones.
The ING (inhibitor of growth) protein family includes a group of homologous nuclear proteins found in most eukaryotes (21). The first member of the ING family, human ING1, was discovered in a screen designed to identify genes whose expression was suppressed in cancer cells (16). It was subsequently shown that overexpression of ING1 inhibits cell growth and induces apoptosis, while expression of an antisense construct promotes transformation of cell lines and tumor formation in vivo (15, 17). Expression analyses of several tumor types show that ING1 is either mutated or down-regulated in many forms of cancer (18). Since the initial characterization of ING1, four other human homologs, ING2 to 5, have been identified through sequence homologies, with the greatest homology occurring within a PHD (plant homeodomain) finger domain at the carboxyl termini of the proteins (21). PHD fingers are found in many proteins involved in chromatin-mediated gene regulation, including the CBP/p300 acetyltransferases (3), the ACF chromatin remodeling complex (26), and the Drosophila melanogaster polycomblike protein (48). Consistent with this, all five human ING proteins have been reported to associate with a diverse group of histone acetyltransferases (HATs) and/or histone deacetylase complexes (HDACs). ING1 and ING2 are components of two related Sin3/HDAC1/2 complexes (9, 34, 57) while ING3, ING4, and ING5 are found in MYST-type HAT complexes (9, 10).
There are three members of the ING family in Saccharomyces cerevisiae: Yng1p, Yng2p, and Pho23p (37). Expression of human ING1 in yeast suppresses phenotypes associated with loss of YNG2, suggesting that YNG2 and ING1 have a conserved function (37). Like their mammalian counterparts, the yeast ING proteins are associated with multiprotein complexes that posttranslationally modify histones. Yng1p and Yng2p are associated with the NuA3 and NuA4 HAT complexes, respectively (22, 37, 46), while Pho23p is found associated with the Rpd3L HDAC complex (4, 38). Although none of these ING proteins are required for the structural integrity of their respective complexes, data suggest that both Yng1p and Yng2p are required for the ability of their complexes to acetylate nucleosomal substrates (5, 22, 46). Moreover, Yng1p is required for interaction of the NuA3 complex with nucleosomes in vitro (22). These data support the hypothesis that members of the ING protein family serve general roles in permitting various enzymatic activities to interact with nucleosomes and facilitate histone posttranslational modifications.
To understand the function of ING proteins in the posttranslational modification of histones, we made use of a genetic strategy to identify genes required for the binding of Yng1p to histones. Using an assay based on the toxicity of YNG1 overexpression, we showed that Yng1p interacts with the amino-terminal tail of histone H3 and that this interaction can be disrupted by loss of lysine 4 methylation within this tail. The toxicity of YNG1 overexpression is dependent on the Yng1p PHD finger, which we demonstrated can directly bind lysine 4-methylated histone H3 tails in vitro. We have previously shown that the binding of NuA3 to chromatin is regulated by methylation of histone H3 lysine 4 but were unable to confirm that this interaction was direct, as opposed to mediated by an auxiliary factor. These results confirm that the interaction of NuA3 with MeH3K4 is mediated by the PHD finger of Yng1p, suggesting a novel function for this motif.
MATERIALS AND METHODS
Yeast strains, plasmids, and genetic methods.
All strains used in this study (Table 1) are isogenic to S288C. Yeast culture and genetic manipulations were carried out using standard protocols (2, 39, 53). Genomic deletions or epitope tag integrations were verified by PCR analysis. The strains carrying the histone H3 tail deletion and the lysine 4- and 36-to-arginine mutations were derived by plasmid shuffle from FY2162 (11). This strain carries deletions of the HHT1-HHF1 and HHT2-HHF2 loci and carries HHT2-HHF2 on a URA3 plasmid (pHHT2). Due to the fact that this strain and all strains derived from it carry a wild-type version of HHF2 on a plasmid, for simplicity, the genotypes of these strains will be referred to as hht1Δ hht2Δ in the figures and figure legends. The plasmid pHHT2 was constructed by ligation of the SpeI fragment from pDM18 (11) into the SpeI site of pRS414. phht2Δ3-29 was generated by ligating the annealed phosphorylated oligonucleotides (5′-GATCCAAGCAAACACTCCACAATGGCCAGACCATCTA-3′ and 5′-CCGGTAGATGGTCTGGCCATTGTGGAGTGTTTGCTTG-3′) into the BamHI and AgeI sites of pHHT2. All other mutants were generated by PCR-mediated site-directed mutagenesis. The plasmids pGALYNG1 and pGALYNG1ΔPHD were generated by cloning the YNG1 and YNG1ΔPHD open reading frames into the BamHI sites of pGAL (44). The pGALYNG1HA, pGALYNG1ΔPHDHA, pGALYNG1Y157AHA, pGALYNG1D172AHA, and pGALYNG1W180AHA plasmids were constructed by first incorporating a triple hemagglutinin (HA) tag along with the CYC1 terminator into the pGAL vector using KpnI and SalI restriction sites, followed by ligation of a wild-type or mutant YNG1 open reading frame into the BamHI and SalI sites. For recombinant expression of the Yng1p PHD finger as a glutathione S-transferase (GST) fusion, the sequence corresponding to the YNG1 PHD finger was ligated into pGEX and transformed into BL21(DE3) cells for induction with isopropyl-β-d-thiogalactopyranoside (IPTG) and purification with glutathione resin (Amersham Biosciences).
TABLE 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| FY2162 | MATahis3Δ200 leu2Δ1 lys2-128Δ ura3-52 trp1Δ63 (hht1-hhf1)::LEU2 (hht2-hhf2)::HIS3 Ty912δ 35::his4 |
| Y2454 | MATα mfa1Δ::MFA1pr-HIS3 can1Δ ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 |
| YDM126 | MATahis3Δ200 leu2Δ1 lys2-128Δura3-52 trp1Δ63 SAS3-3HA::HISMX6 |
| YDM127 | MATahis3Δ200 leu2Δ1 lys2-128Δ ura3-52 trp1Δ63 SAS3-3HA::HISMX6 HTB1TAP::TRP |
| YDM137 | MATahis3Δ200 leu2Δ1 lys2-128Δ ura3-52 trp1Δ63 SAS3-3HA::HISMX6 HTB1TAP::TRP yng1ΔPHD::KAN |
| YDM138 | MATahis3Δ200 leu2Δ1 lys2-128Δ ura3-52 trp1Δ63 SAS3-3HA::HISMX6 HTB1TAP::TRP yng1::KAN |
| YDM153 | MATahis3Δ200 leu2Δ1 lys2-128Δ ura3-52 trp1Δ63 SAS3-3HA::HISMX6 HTB1TAP::URA set2::KAN |
| YLH101 | MATahis3Δ200 leu2Δ1 lys2-128Δ ura3-52 trp1Δ63 |
| YLH203 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 csp30::KAN |
| YLH204 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 csp50::KAN |
| YLH205 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 dot1::KAN |
| YLH206 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rad6::KAN |
| YLH209 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 lge1::KAN |
| YLH211 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ylr177::KAN |
| YLH220 | MATahis3Δ200 leu2Δ1 lys2-128Δ ura3-52 trp1Δ63 set1::HISMX6 |
| YLH298 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 set2::KAN |
| YLH326 | MATahis3Δ200 leu2Δ1 lys2-128Δ ura3-52 trp1Δ63 gcn5::TRP |
| YLH363 | MATahis3Δ200 leu2Δ1 lys2-128Δ ura3-52 trp1Δ63 SAS3-3HA::HISMX6 HTB1TAP::URA set2::TRP yng1ΔPHD::KAN |
Synthetic dosage resistance genome-wide screen.
The synthetic genetic array (SGA) starting strain Y2454 (MATα mfa1Δ::MFA1pr-HIS3 can1Δ ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0) (61) was transformed with either pGALYNG1 or pGAL (vector control). The resulting query strains were mated to the MATa deletion mutant array, and SGA methodology, previously described for plasmid-based synthetic dosage lethal SGA screen (42), was used with the following modifications: (i) medium lacking uracil was used to maintain the plasmids and (ii) the screen was performed at 25°C. The genome-wide synthetic dosage resistance screen was performed once, and all deletion mutant array strains that suppressed YNG1 toxicity were confirmed by reintroducing plasmids into each strain by means of traditional yeast transformation methods.
Chromatin pull-down assay.
The chromatin pull-down assay was performed using yeast whole-cell extracts prepared from mid-log cells in IPP150 buffer (10 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride, 2 μg/ml pepstatin A) with 2 mM CaCl2. Input protein amounts were normalized by Bradford protein assay, and 30 mg of protein was incubated with 25 μl of calmodulin affinity resin (Stratagene) for 2 h at 4°C with agitation. The resin was washed three times with 1 ml of calmodulin binding buffer (10 mM Tris-Cl, pH 8.0, 150 mM NaCl, 10 mM β-mercaptoethanol, 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2, 0.1% NP-40), and the tandem affinity purification (TAP)-tagged Htb1p was eluted with four washes with calmodulin elution buffer (10 mM Tris-Cl, pH 8.0, 150 mM NaCl, 10 mM β-mercaptoethanol, 1 mM magnesium acetate, 1 mM imidazole, 2 mM EGTA, 0.1% NP-40) for 10 min each at 4°C. The resulting eluates were pooled and incubated with 10 μl of immunoglobulin G Sepharose 6 Fast Flow resin (Amersham Biosciences). Samples were rotated at 4°C for 2 h and the resin washed three times with 1 ml of IPP150 buffer. Bound proteins were eluted by boiling in 50 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer and analyzed by immunoblotting with anti-HA-peroxidase (Roche Diagnostics) or peroxidase anti-peroxidase (Sigma-Aldrich) antibodies.
Peptide pull-down assays.
Biotinylated histone peptides were from Upstate Biotechnology or were synthesized at the Stanford PAN facility. To verify the peptide concentrations, 0.5 μg of each peptide was spotted onto an Amersham Hybond-C Extra membrane and allowed to dry overnight at room temperature in the dark. The membrane was then probed with an anti-biotin antibody. For pull-down assays, 1 μg of GST-Yng1PHD was incubated with 0.5 μg of biotinylated histone peptides in binding buffer (50 mM Tris-HCl, pH 7.5, 300 mM NaCl, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride, plus protease inhibitors) for 4 h to overnight at 4°C with rotation. After a 1-h incubation with streptavidin beads (Amersham Biosciences) and extensive washing, bound proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting.
RESULTS
Yng1p interacts with the histone H3 tail in vivo.
Yng1p is a member of the ING protein family and a component of the NuA3 histone acetyltransferase complex. Previously published biochemical and genetic analyses demonstrated that, while Yng1p is not required for the structural integrity of the NuA3 complex, it is important for NuA3 function (22). These data, together with the fact that other ING proteins are also components of histone-modifying complexes (9, 22, 34, 37, 38, 47, 57), led us to speculate that ING proteins mediate the interaction of various chromatin-modifying complexes with histones. Consistent with this hypothesis, NuA3 purified from a yng1Δ strain shows reduced interaction with chromatin reconstituted in vitro compared to NuA3 from a wild-type strain (22). However, whether YNG1 is required for the interaction of NuA3 with chromatin in vivo was unknown. We attempted to address this issue using chromatin immunoprecipitation analysis, but to date, we have been unable to reproducibly chromatin immunoprecipitate any component of NuA3 to a specific genomic locus. However, using a chromatin pull-down assay, we were able to show that Sas3p, the catalytic subunit of NuA3, interacts with chromatin in vivo, and that this interaction is dependent on the amino-terminal tail of histone H3 (40). To determine whether this interaction is also dependent on Yng1p, we fused a TAP tag to the carboxyl terminus of H2B (Htb1p) to enable us to purify native chromatin and associated proteins using a modified TAP protocol. Using wild-type and yng1Δ strains that also expressed a triple-HA-tagged Sas3p, we purified chromatin and performed anti-HA Western blot analysis for Sas3p. Immunodetection for TAP-tagged histone H2B (Htb1TAP) was used as a loading control to ensure that we were pulling down equal amounts of chromatin in each sample. Consistent with previously published data (40), precipitation of bulk chromatin coprecipitated Sas3p, indicating that this protein is indeed capable of binding chromatin (Fig. 1A, compare lanes 1 and 2). However, when a similar experiment was performed using a strain with a deletion of YNG1, there was significantly less Sas3p in the precipitate (Fig. 1A, compare lanes 2 and 3). The fact that the interaction of Sas3p with chromatin is severely compromised in a yng1Δ strain indicates that Yng1p is required for the interaction of the NuA3 complex with chromatin in vivo.
FIG. 1.
Yng1p interacts with the histone H3 tail in vivo. (A) Chromatin pull-down assays were performed on strains YDM126, YDM127, and YDM138, and the resulting samples were subjected to Western blotting with peroxidase antiperoxidase (Htb1TAP) or anti-HA antibodies (Sas3HA). (B and C) Tenfold serial dilutions of yeast strains YLH101 (B) or FY2162 transformed with pHHT2 or phht2Δ3-29 (C) containing the indicated plasmids were plated on synthetic complete medium lacking uracil containing either dextrose or galactose as a carbon source and incubated at 30°C for 3 days. (D) Normalized amounts of whole-cell extracts from the indicated strains (FY2162 transformed with pHHT2 or phht2Δ3-29) transformed with pGALYNG1HA and grown on galactose were subjected to Western blotting with immunodetection for HA. +, present; −, absent.
To provide further support that Yng1p is capable of directly interacting with histones, we developed a genetic approach. We rationalized that if Yng1p is capable of interacting with histones in vivo, then an overabundance of Yng1p may be detrimental to the cell due to the fact that it may interfere with the normal regulation of chromatin structure. To test this possibility, we fused the YNG1 open reading frame (ORF) to the GAL1 promoter on a URA3-based centromeric plasmid (pGALYNG1). The plasmid was introduced into wild-type yeast cells and the transformants selected by growth on uracil dropout media with dextrose. A plasmid carrying the GAL1 promoter but lacking the YNG1 ORF was used as a negative control (pGAL). Cells carrying the pGAL and pGALYNG1 plasmids were spotted onto uracil dropout media with either dextrose or galactose as a carbon source. Cells containing the control plasmid grew well on both media, but cells containing the pGALYNG1 plasmid failed to grow on galactose, indicating that overexpression of YNG1 is toxic to the cell (Fig. 1B). Deletion of SAS3 did not rescue the Yng1p toxicity, indicating that this phenotype is due to an overabundance of Yng1p alone and not aberrant targeting of NuA3 HAT activity (data not shown).
We have previously shown that the interaction of NuA3 with chromatin is dependent on the amino-terminal tail of histone H3 (40). If the Yng1p toxicity is due to the interaction of overabundant Yng1p with the H3 tail, then deletion of this tail should rescue the toxicity. To test this, we introduced the pGAL and pGALYNG1 plasmids into strains which carried a plasmid-borne copy of HHT2 as the only source of histone H3 expression in the cell. Cells with a plasmid expressing full-length histone H3 failed to grow on galactose when carrying the pGALYNG1 plasmid. However, cells expressing histone H3 with a truncation of amino acids 3 to 29 were resistant to YNG1 overexpression, suggesting that the Yng1p toxicity is dependent on the H3 tail (Fig. 1C). To confirm that the rescue of the Yng1p toxicity was not due to a GAL1 transcription defect in the hht2Δ3-29 strain, we took advantage of the fact that the addition of an HA tag to the carboxyl terminus of Yng1p suppresses the toxicity of this protein. This enabled us to measure the relative levels of Yng1p in wild-type and mutant strains. Figure 1D shows that deletion of the H3 tail had no impact of the levels of Yng1p, suggesting that the rescue of the Yng1p toxicity seen upon deletion of the H3 tail was not due to a GAL1 transcription defect. Thus, the facts that YNG1 is toxic when overexpressed and that this toxicity can be rescued by deletion of the H3 tail suggest that Yng1p is directly interacting with the H3 tail in vivo.
Yng1p toxicity is dependent on lysine 4 methylation of histone H3.
Both biochemical and genetic experiments suggest that Yng1p serves as a histone binding protein which binds the amino-terminal tail of histone H3. The histone amino-terminal tails are subjected to numerous posttranslational modifications, and these modifications are thought to regulate the binding of chromatin-modifying factors to histones (27, 58). We have previously shown that the binding of the NuA3 complex to chromatin is dependent on either the methylation of histone H3 lysine 4 by the Set1p methyltransferase or methylation of H3 lysine 36 by the Set2p methyltransferase (40). However, the absence of a chromo, WD40-repeat, tudor, or MBT domain in any of the NuA3 subunits suggests that these interactions may be mediated by an unidentified auxiliary factor. To determine how the binding of Yng1p to chromatin is regulated, we performed a synthetic dosage resistance screen for mutants that are resistant to YNG1 overexpression. This screen used SGA methodology to introduce the pGALYNG1 plasmid into the set of 4,700 viable haploid deletion mutants. YNG1 overexpression was induced on galactose-containing medium, and a synthetic dosage resistance phenotype was scored by comparing growth of the deletion mutant overexpressing YNG1 to the vector control plasmid pGAL. Mutant strains which tested positive for resistance were retransformed with the pGAL and pGALYNG1 plasmids and retested using standard plating (Fig. 2A). Two genes were identified that, when deleted, conferred resistance to YNG1 overexpression: YLR177w and LGE1. The function of YLR177w is unknown, but LGE1 is required for RAD6-dependent ubiquitination of histone H2B and is also involved in regulating gene expression in a pathway independent of RAD6 (25, 33, 66). To determine whether the requirement of LGE1 for Yng1p toxicity was dependent on H2B ubiquitination, we sought to determine whether rad6Δ strains also showed resistance. Figure 2B shows that, similar to a lge1Δ mutant, the growth of a rad6Δ mutant is not inhibited by aberrant expression of YNG1, suggesting that it is loss of H2B ubiquitination that confers resistance to YNG1 overexpression.
FIG. 2.
Yng1p toxicity is rescued by loss of the COMPASS histone methyltransferase complex. Tenfold serial dilutions of yeast strains YLH101, YLH211, and YLH209 (A) and YLH101, YLH206, YLH220, YLH203, YLH204, YLH298, and YLH205 (B) containing the indicated plasmids were plated on synthetic complete medium lacking uracil containing either dextrose or galactose as a carbon source and incubated at 30°C for 3 days.
Ubiquitination of H2B is required for di- and trimethylation of lysine 4 of histone H3 by the SET1-dependent COMPASS complex (8, 43). This, when taken together with the fact that Yng1p toxicity is dependent on the H3 tail, may suggest that the resistance to Yng1p toxicity seen in a rad6Δ mutant is an indirect effect of loss of H3 K4 methylation. To address this, we tested the susceptibility of set1Δ mutants to Yng1p toxicity as well as strains carrying deletions of CSP30 and CSP50, other genes which encode subunits of the COMPASS complex. Figure 2B demonstrates that loss of any gene required for H3 K4 methylation provides resistance to Yng1p toxicity, suggesting that Yng1p requires this methylation mark to bind chromatin. In contrast, the growth of set2Δ, dot1Δ, or gcn5Δ strains (Fig. 2B and data not shown), which lack methylation of H3 K36 (59), methylation of H3 K79 (14, 35, 45, 62), or acetylation of H3 K9, 18, 23, and 27, respectively (32, 60), is inhibited by overexpression of Yng1p, suggesting that Yng1p can still bind chromatin in the absence of these modifications. As a final confirmation that H3 K4 methylation is required for Yng1p to bind chromatin, we examined Yng1p toxicity in a strain in which lysine 4 of histone H3 had been mutated to an arginine. Figure 3A demonstrates that, similar to mutations that disrupt the COMPASS complex, mutation of lysine 4 confers resistance to Yng1p toxicity, further confirming the importance of this residue for the binding of Yng1p to chromatin. In contrast, mutation of histone H3 lysine 36 to arginine did not rescue the growth of cells overexpressing Yng1p, indicating that this residue does not interact with Yng1p. Once again, we were able to confirm that the growth of the lysine 4 mutant on galactose was not due to a transcription defect of the GAL1 promoter by demonstrating that there are equal levels of Yng1HA in the wild type versus the K4R mutant (Fig. 3B).
FIG. 3.
The interaction of Yng1p with histone H3 is dependent on lysine 4. (A) Tenfold serial dilutions of yeast strain FY2162 transformed with pHHT2, phht2K4R, or phht2K36R containing the indicated plasmids were plated on synthetic complete medium lacking uracil containing either dextrose or galactose as a carbon source and incubated at 30°C for 3 days. (B) Normalized amounts of whole-cell extracts from the indicated strains (FY2162 transformed with pHHT2 or phht2K4R) transformed with pGALYNG1HA and grown on galactose were subjected to Western blotting with immunodetection for HA.
The Yng1p PHD finger is required for histone binding.
Members of the ING family contain a PHD finger domain within their carboxyl termini (21). This zinc finger-like motif is found in various proteins involved in chromatin-mediated gene regulation (1), and several studies have implicated PHD fingers in mediating the interaction of chromatin-modifying factors with histones (12, 54). Indeed, the Yng2p PHD finger is required for full activation of NuA4-dependent genes, suggesting that this motif is required for some aspect of HAT complex function (46). Despite the intriguing possibility that the Yng1p PHD finger mediates the interaction of NuA3 with chromatin, we have previously demonstrated that Yng1p lacking the PHD domain is still able to rescue phenotypes associated with loss of YNG1, suggesting that this region of the protein is not required for the function of the NuA3 complex (22). Consistent with this, a chromatin pull-down assay performed on YNG1 and yng1ΔPHD strains showed no significant requirement for the Yng1p PHD finger for the interaction of Sas3HA with Htb1TAP (Fig. 4A, compare lanes 2 and 3). However, we have also previously shown that NuA3 can interact with chromatin that is methylated at lysines 4 or 36, and loss of both methyl marks is required to abolish the binding of NuA3 to chromatin (40). Thus, the fact that loss of the Yng1p PHD finger alone does not disrupt NuA3 function does not rule out the possibility that this domain mediates interaction with methylated histone H3 lysine 4, since a failure to bind methylated lysine 4 would be compensated for by binding of NuA3 to methylated lysine 36. To provide support for this hypothesis, we repeated the chromatin pull-down assays using strains lacking Set2p, the histone H3 lysine 36 methyltransferase. Figure 4A shows that, in the absence of SET2, the PHD finger of Yng1p is crucial for the binding of Sas3HA to chromatin (compare lanes 4 and 5). This result suggests that the PHD finger of Yng1p does mediate the interaction of NuA3 with histones but is redundant with the interaction of an independent component of NuA3 with methylated histone H3 lysine 36. Additionally, the fact that the Yng1p PHD finger is only required for chromatin binding in a set2Δ strain, whereas loss of the entire YNG1 ORF disrupts chromatin binding in a SET2 strain, suggests that Yng1p has two independent domains: a carboxyl-terminal PHD finger which binds lysine 4-methylated histone H3 and an amino-terminal domain which is required for some aspect of NuA3-chromatin interaction, which cannot be compensated for by interaction of NuA3 with histone H3 methylated at lysine 36.
FIG. 4.
The Yng1p PHD finger is required for histone binding. (A) Chromatin pull-down assays were performed on strains YDM126, YDM127, YDM137, YDM153, and YLH363, and the resulting samples were subjected to Western blotting with peroxidase antiperoxidase (Htb1TAP) or anti-HA antibodies (Sas3HA). (B) Tenfold serial dilutions of yeast strain YLH101 containing the indicated plasmids were plated on synthetic complete medium lacking leucine containing either dextrose or galactose as a carbon source and incubated at 30°C for 3 days. (C) Normalized amounts of whole-cell extracts from wild-type yeast strains (YLH101) carrying a pGALYNG1HA (lane 1) or pGALYNG1ΔPHDHA (lane 2) plasmid and grown on galactose were subjected to Western blotting for HA. +, present; −, absent.
If the PHD finger of Yng1p mediates the interaction of this protein with histones, then an overabundance of Yng1p lacking the PHD finger should not result in growth inhibition. Figure 4B demonstrates that overexpression of YNG1ΔPHD is not toxic, further supporting the fact that the PHD finger is required for Yng1p to bind nucleosomes. To show that Yng1p lacking the PHD finger is stably expressed, we used immunoblot analysis to demonstrate that HA-tagged Yng1p and Yng1ΔPHD are present in similar amounts in yeast whole-cell extracts (Fig. 4C, compare lanes 1 and 2). These data, when taken together with the fact that histone H3 K4 methylation is required for Yng1p to bind chromatin, suggests that the PHD finger of Yng1p may be a methyl-histone binding module that specifically recognizes H3 K4 methylation.
The Yng1p PHD finger binds lysine 4-methylated histone H3.
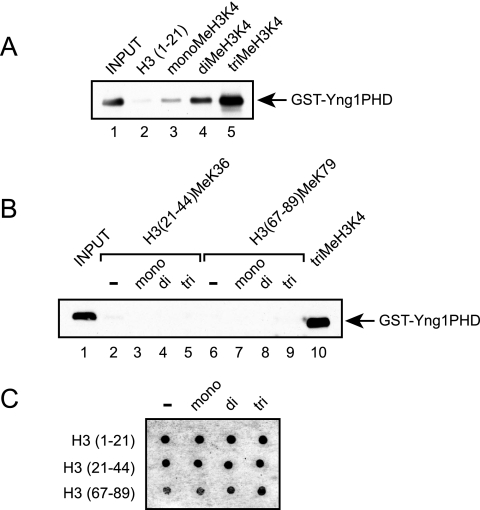
To determine whether the Yng1p PHD finger is capable of directly interacting with lysine 4-methylated histone H3, we expressed the Yng1p PHD finger as a GST fusion in bacteria and examined the binding of this protein to unmodified and methylated histone H3 peptides (H3 residues 1 to 21) bound to beads. Figure 5A shows that while the Yng1p PHD finger failed to interact with unmodified peptides, it was able to interact with peptides that were mono-methylated at lysine 4 (Fig. 5A, compare lanes 2 and 3). This interaction was further enhanced when the peptides were di- or trimethylated (compare lane 3 with lanes 4 and 5). To determine whether the Yng1p PHD finger specifically recognizes methylated lysine 4, as opposed to other methyl marks, we examined the binding of the Yng1p PHD finger to peptides corresponding to histone H3 methylated at lysine 36 (H3 residues 21 to 44) or 79 (H3 residues 67 to 89). Figure 5B shows that the Yng1p PHD finger failed to interact with these peptides regardless of the level of methylation, suggesting that the Yng1p PHD finger specifically recognizes H3 K4 methylation. Figure 5C shows the relative levels of each peptide used as determined by dot immunoblotting with anti-biotin antibodies. Unfortunately, due to the reduced charge of the H3 (67 to 89) peptides, these peptides do not bind the membrane as well as the H3 (1 to 21) and H3 (21 to 44) peptides, resulting in a weaker signal. To further verify the amount of peptide used, the concentrations were also assayed using a Bradford protein assay.
FIG. 5.
The Yng1p PHD finger binds lysine 4-methylated histone H3. (A and B) Histone peptide binding assays were performed with the indicated biotinylated peptides and purified GST-Yng1PHD. Shown are Western blots of peptide-bound GST-Yng1PHD protein with GST antibodies. Input lanes represent 10% of the GST protein used in the pull-down assays. (C) Five hundred nanograms of biotinylated histone peptides was spotted onto membranes and immunodetected with antibiotin antibodies. −, absent.
As a final confirmation that Yng1p specifically binds methylated H3 K4 in vivo, we sought to determine whether mutations which disrupt the Yng1p-triMeH3K4 peptide interaction in vitro alleviate YNG1 toxicity. ING2, a mammalian homolog of Yng1p, has also been shown to interact with triMeH3K4 peptides, and residues Y215, D230, and W238 of this protein are important for peptide binding (51, 56). We created alanine substitutions of the corresponding residues in Yng1p (Fig. 6A) and tested the mutants for binding to triMeH3K4 peptides and for inhibition of growth when overexpressed. Mutation of D172 and W180 to alanine totally abolished the binding of the Yng1p PHD finger to triMeH3K4 peptides, while residual binding was still observed in the Y157A mutant (Fig. 6B). Consistent with this, overexpression of the D172A and W180A mutants in yeast had no effect on cell growth compared to vector alone, and mutation of Y157 to alanine resulted in minor growth inhibition (Fig. 6C). Immunoblot analysis of yeast whole-cell extracts demonstrated that the wild type and mutant versions were present at comparable levels in the cell (Fig. 6D). The close correlation between the effect of the various Yng1p mutations on triMeH3K4 peptide binding in vitro and overexpression toxicity indicates that the Yng1p PHD finger is binding lysine 4-methylated histone H3 in vivo. This is consistent with previously published data that H3 K4 methylation is required for NuA3 function (40) and suggests a novel function for the Yng1p PHD finger.
FIG. 6.
The toxicity of YNG1 overexpression correlates with the level of Yng1p-triMeH3K4 binding. (A) Schematic representation of Yng1p (open bar), the Yng1 PHD finger (black bar), the coordinating cysteines and histidine residues of the PHD finger (underlined), and the residues subjected to mutation (highlighted). (B) Histone peptide binding assays were performed with the indicated biotinylated peptides and purified wild-type and mutant versions of GST-Yng1PHD. Shown are Western blots of peptide-bound GST-Yng1PHD proteins with GST antibodies. Input lanes represent 10% of the GST protein used in the pull-down assays. (C) Tenfold serial dilutions of yeast strain YLH101 containing the indicated plasmids were plated on synthetic complete medium lacking uracil containing either dextrose or galactose as a carbon source and incubated at 30°C for 3 days. (D) Normalized amounts of whole-cell extract from a wild-type yeast strain (YLH101) carrying plasmids expressing the indicated HA-tagged versions of Yng1p from a GAL1 promoter and grown on galactose were subjected to Western blotting for HA.
DISCUSSION
We have previously shown that NuA3 requires either the methylation of lysine 4 or 36 of histone H3 to bind chromatin (22, 40). However, the NuA3 subunit or subunits that mediate these interactions was unknown, and the possibility existed that the interaction of NuA3 with methylated histone H3 is mediated by an auxiliary factor. The results of this study demonstrate that Yng1p, a subunit of NuA3, is capable of directly interacting with lysine 4-methylated histone H3. Overexpression of YNG1 inhibits cell growth, and this toxicity is dependent on methylation of lysine 4 on the histone H3 tail. Furthermore, mutations within Yng1p that disrupt the interaction of this protein with methylated histone H3 peptides in vitro alleviate the toxicity of YNG1 overexpression in vivo. These results confirm that NuA3 directly interacts with lysine 4-methylated histone H3 via the Yng1p subunit.
The ability of Yng1p to act as a methyl histone binding protein is unusual in that this protein does not contain chromo, WD40-repeat, tudor, or MBT domains, which are found in other proteins that bind methylated histones (6). The toxicity of YNG1 overexpression is dependent on the PHD finger of Yng1p, suggesting that it is this motif which is responsible for binding lysine 4-methylated histone H3. Moreover, the PHD finger of Yng1p shows preferential interaction in vitro with histone H3 peptides that have been methylated on lysine 4, confirming that the Yng1p PHD finger is a methyl-histone binding module. PHD fingers are found in multiple complexes that regulate chromatin structure; however, the function of the majority of these domains is unknown. The PHD fingers of the p300 histone acetyltransferase and ACF1, a subunit of an ATP-dependent nucleosome remodeling complex, are required for these proteins to bind histones (12, 54), but whether this binding is dependent on the methylation state of the histones was not investigated. However, the fact that the binding of the ACF1 PHD fingers to histones was not dependent on the presence of the histone tails argues against this possibility for ACF1 (12). The ability of the Yng1p PHD finger to bind lysine 4-methylated histone H3 therefore represents a novel function for this motif.
The high conservation of the PHD fingers within the ING family proteins has led many to speculate that these domains share a common ligand, and thus, it is possible that other members of the ING family also specifically interact with lysine 4-methylated histone H3. Recent work has shown that the NuA4 HAT complex requires H3 lysine 4 and lysine 36 methylation for interaction with the MET16 and RPS11B promoters (43). NuA4 contains Eaf3p, a chromodomain protein which has been shown to interact with methylated lysine 36 (4, 13, 28, 29); however, the subunit that mediates interaction with methylated lysine 4 is not known. The results of this study suggest that this interaction may be mediated by the ING protein, Yng2p. This has been confirmed by parallel studies showing that the PHD fingers of all ING proteins bind lysine 4-methylated histone H3 (51, 56). Since ING proteins are found in both HAT and HDAC complexes, histone H3 lysine 4 methylation could conceivably allow both HAT and HDAC activities to bind chromatin. Alternatively, lysine 4 methylation may act in a combinatorial fashion with other histone modifications to recruit or retain different chromatin-modifying complexes to different regions of the genome. For example, maximum binding of the NuA3 HAT complex to chromatin requires methylation of lysines 4 and 36. Interestingly, another subunit of NuA3, Nto1p, also contains a PHD finger, suggesting the possibility that histone H3 lysine 36 methylation is recognized by the Nto1p PHD finger. Recent studies have also shown that the PHD finger of the NURF (nucleosome remodeling factor) mediates the interaction of this complex with lysine-methylated histone H3 tails (36, 64). Whether other PHD fingers bind methylated histones and whether they show specificity for methylation of H3 K4 or other methylation marks will be the subject of future study.
Acknowledgments
Support for this work was provided by grants to L.H. from the Natural Sciences and Engineering Research Council of Canada and the Canadian Institutes of Heath Research and to O.G. from the National Institute of Health (KO8AG19245). L.H. is a Canadian Institutes of Health Research New Investigator and a Scholar of the Michael Smith Foundation for Health Research. O.G. is a recipient of a Burroughs Wellcome Career Development Award in Biomedical Sciences. K.B. was supported by a Canadian Institutes of Health Research postdoctoral fellowship and a MSFHR postdoctoral fellowship and is a Canada Research Chair in Functional and Chemical Genomics.
We also gratefully acknowledge the valuable comments provided by Jacques Côté, Jerry Workman, and members of the Molecular Epigenetics Group of the Life Sciences Institute at the University of British Columbia. We are also grateful to Fred Winston for providing yeast strains and plasmids.
Footnotes
Published ahead of print on 21 August 2006.
REFERENCES
- 1.Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20:56-59. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M. 1987. Current protocols in molecular biology. Wiley-Interscience, New York, N.Y.
- 3.Bordoli, L., S. Husser, U. Luthi, M. Netsch, H. Osmani, and R. Eckner. 2001. Functional analysis of the p300 acetyltransferase domain: the PHD finger of p300 but not of CBP is dispensable for enzymatic activity. Nucleic Acids Res. 29:4462-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson, K. K. Lee, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581-592. [DOI] [PubMed] [Google Scholar]
- 5.Choy, J. S., B. T. Tobe, J. H. Huh, and S. J. Kron. 2001. Yng2p-dependent NuA4 histone H4 acetylation activity is required for mitotic and meiotic progression. J. Biol. Chem. 276:43653-43662. [DOI] [PubMed] [Google Scholar]
- 6.de la Cruz, X., S. Lois, S. Sanchez-Molina, and M. A. Martinez-Balbas. 2005. Do protein motifs read the histone code? Bioessays 27:164-175. [DOI] [PubMed] [Google Scholar]
- 7.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 8.Dover, J., J. Schneider, M. A. Tawiah-Boateng, A. Wood, K. Dean, M. Johnston, and A. Shilatifard. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277:28368-28371. [DOI] [PubMed] [Google Scholar]
- 9.Doyon, Y., C. Cayrou, M. Ullah, A. J. Landry, V. Cote, W. Selleck, W. S. Lane, S. Tan, X. J. Yang, and J. Cote. 2006. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 21:51-64. [DOI] [PubMed] [Google Scholar]
- 10.Doyon, Y., W. Selleck, W. S. Lane, S. Tan, and J. Cote. 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 24:1884-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duina, A. A., and F. Winston. 2004. Analysis of a mutant histone H3 that perturbs the association of Swi/Snf with chromatin. Mol. Cell. Biol. 24:561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eberharter, A., I. Vetter, R. Ferreira, and P. B. Becker. 2004. ACF1 improves the effectiveness of nucleosome mobilization by ISWI through PHD-histone contacts. EMBO J. 23:4029-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisen, A., R. T. Utley, A. Nourani, S. Allard, P. Schmidt, W. S. Lane, J. C. Lucchesi, and J. Cote. 2001. The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J. Biol. Chem. 276:3484-3491. [DOI] [PubMed] [Google Scholar]
- 14.Feng, Q., H. Wang, H. H. Ng, H. Erdjument-Bromage, P. Tempst, K. Struhl, and Y. Zhang. 2002. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12:1052-1058. [DOI] [PubMed] [Google Scholar]
- 15.Garkavtsev, I., I. A. Grigorian, V. S. Ossovskaya, M. V. Chernov, P. M. Chumakov, and A. V. Gudkov. 1998. The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control. Nature 391:295-298. [DOI] [PubMed] [Google Scholar]
- 16.Garkavtsev, I., A. Kazarov, A. Gudkov, and K. Riabowol. 1996. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat. Genet. 14:415-420. [DOI] [PubMed] [Google Scholar]
- 17.Garkavtsev, I., and K. Riabowol. 1997. Extension of the replicative life span of human diploid fibroblasts by inhibition of the p33ING1 candidate tumor suppressor. Mol. Cell. Biol. 17:2014-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong, W., K. Suzuki, M. Russell, and K. Riabowol. 2005. Function of the ING family of PHD proteins in cancer. Int. J. Biochem. Cell. Biol. 37:1054-1065. [DOI] [PubMed] [Google Scholar]
- 19.Grewal, S. I. 2000. Transcriptional silencing in fission yeast. J. Cell. Physiol. 184:311-318. [DOI] [PubMed] [Google Scholar]
- 20.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 21.He, G. H., C. C. Helbing, M. J. Wagner, C. W. Sensen, and K. Riabowol. 2005. Phylogenetic analysis of the ING family of PHD finger proteins. Mol. Biol. Evol. 22:104-116. [DOI] [PubMed] [Google Scholar]
- 22.Howe, L., T. Kusch, N. Muster, R. Chaterji, J. R. Yates III, and J. L. Workman. 2002. Yng1p modulates the activity of Sas3p as a component of the yeast NuA3 histone acetyltransferase complex. Mol. Cell. Biol. 22:5047-5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, Y., J. Fang, M. T. Bedford, Y. Zhang, and R. M. Xu. 2006. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 312:748-751. [DOI] [PubMed] [Google Scholar]
- 24.Hudson, B. P., M. A. Martinez-Yamout, H. J. Dyson, and P. E. Wright. 2000. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J. Mol. Biol. 304:355-370. [DOI] [PubMed] [Google Scholar]
- 25.Hwang, W. W., S. Venkatasubrahmanyam, A. G. Ianculescu, A. Tong, C. Boone, and H. D. Madhani. 2003. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11:261-266. [DOI] [PubMed] [Google Scholar]
- 26.Ito, T., M. E. Levenstein, D. V. Fyodorov, A. K. Kutach, R. Kobayashi, and J. T. Kadonaga. 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13:1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 28.Joshi, A. A., and K. Struhl. 2005. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell 20:971-978. [DOI] [PubMed] [Google Scholar]
- 29.Keogh, M. C., S. K. Kurdistani, S. A. Morris, S. H. Ahn, V. Podolny, S. R. Collins, M. Schuldiner, K. Chin, T. Punna, N. J. Thompson, C. Boone, A. Emili, J. S. Weissman, T. R. Hughes, B. D. Strahl, M. Grunstein, J. F. Greenblatt, S. Buratowski, and N. J. Krogan. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123:593-605. [DOI] [PubMed] [Google Scholar]
- 30.Kim, J., J. Daniel, A. Espejo, A. Lake, M. Krishna, L. Xia, Y. Zhang, and M. T. Bedford. 2006. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 7:397-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kouzarides, T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12:198-209. [DOI] [PubMed] [Google Scholar]
- 32.Kristjuhan, A., J. Walker, N. Suka, M. Grunstein, D. Roberts, B. R. Cairns, and J. Q. Svejstrup. 2002. Transcriptional inhibition of genes with severe histone h3 hypoacetylation in the coding region. Mol. Cell. 10:925-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krogan, N. J., M. Kim, A. Tong, A. Golshani, G. Cagney, V. Canadien, D. P. Richards, B. K. Beattie, A. Emili, C. Boone, A. Shilatifard, S. Buratowski, and J. Greenblatt. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23:4207-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuzmichev, A., Y. Zhang, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2002. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol. Cell. Biol. 22:835-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lacoste, N., R. T. Utley, J. M. Hunter, G. G. Poirier, and J. Cote. 2002. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 277:30421-30424. [DOI] [PubMed] [Google Scholar]
- 36.Li, H., S. Ilin, W. Wang, E. M. Duncan, J. Wysocka, C. D. Allis, and D. J. Patel. 2006. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442:91-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loewith, R., M. Meijer, S. P. Lees-Miller, K. Riabowol, and D. Young. 2000. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol. Cell. Biol. 20:3807-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loewith, R., J. S. Smith, M. Meijer, T. J. Williams, N. Bachman, J. D. Boeke, and D. Young. 2001. Pho23 is associated with the Rpd3 histone deacetylase and is required for its normal function in regulation of gene expression and silencing in Saccharomyces cerevisiae. J. Biol. Chem. 276:24068-24074. [DOI] [PubMed] [Google Scholar]
- 39.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 40.Martin, D. G. E., D. E. Grimes, K. Baetz, and L. Howe. 2006. Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin. Mol. Biol. Cell 26:3018-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matangkasombut, O., R. M. Buratowski, N. W. Swilling, and S. Buratowski. 2000. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 14:951-962. [PMC free article] [PubMed] [Google Scholar]
- 42.Measday, V., K. Baetz, J. Guzzo, K. Yuen, T. Kwok, B. Sheikh, H. Ding, R. Ueta, T. Hoac, B. Cheng, I. Pot, A. Tong, Y. Yamaguchi-Iwai, C. Boone, P. Hieter, and B. Andrews. 2005. Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation. Proc. Natl. Acad. Sci. USA 102:13956-13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morillon, A., N. Karabetsou, A. Nair, and J. Mellor. 2005. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell 18:723-734. [DOI] [PubMed] [Google Scholar]
- 44.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng, H. H., R. M. Xu, Y. Zhang, and K. Struhl. 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277:34655-34657. [DOI] [PubMed] [Google Scholar]
- 46.Nourani, A., Y. Doyon, R. T. Utley, S. Allard, W. S. Lane, and J. Cote. 2001. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell. Biol. 21:7629-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nourani, A., L. Howe, M. G. Pray-Grant, J. L. Workman, P. A. Grant, and J. Cote. 2003. Opposite role of yeast ING family members in p53-dependent transcriptional activation. J. Biol. Chem. 278:19171-19175. [DOI] [PubMed] [Google Scholar]
- 48.O'Connell, S., L. Wang, S. Robert, C. A. Jones, R. Saint, and R. S. Jones. 2001. Polycomblike PHD fingers mediate conserved interaction with enhancer of zeste protein. J. Biol. Chem. 276:43065-43073. [DOI] [PubMed] [Google Scholar]
- 49.Ornaghi, P., P. Ballario, A. M. Lena, A. Gonzalez, and P. Filetici. 1999. The bromodomain of Gcn5p interacts in vitro with specific residues in the N terminus of histone H4. J. Mol. Biol. 287:1-7. [DOI] [PubMed] [Google Scholar]
- 50.Owen, D. J., P. Ornaghi, J. C. Yang, N. Lowe, P. R. Evans, P. Ballario, D. Neuhaus, P. Filetici, and A. A. Travers. 2000. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 19:6141-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pena, P. V., F. Davrazou, X. Shi, K. L. Walter, V. V. Verkhusha, O. Gozani, R. Zhao, and T. G. Kutateladze. 2006. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442:100-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pray-Grant, M. G., J. A. Daniel, D. Schieltz, J. R. Yates III, and P. A. Grant. 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433:434-438. [DOI] [PubMed] [Google Scholar]
- 53.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 54.Ragvin, A., H. Valvatne, S. Erdal, V. Arskog, K. R. Tufteland, K. Breen, O. Y. AM, A. Eberharter, T. J. Gibson, P. B. Becker, and R. Aasland. 2004. Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. J. Mol. Biol. 337:773-788. [DOI] [PubMed] [Google Scholar]
- 55.Santos-Rosa, H., R. Schneider, B. E. Bernstein, N. Karabetsou, A. Morillon, C. Weise, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2003. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell 12:1325-1332. [DOI] [PubMed] [Google Scholar]
- 56.Shi, X., T. Hong, K. L. Walter, M. Ewalt, E. Michishita, T. Hung, D. Carney, P. Pena, F. Lan, M. R. Kaadige, N. Lacoste, C. Cayrou, F. Davrazou, A. Saha, B. R. Cairns, D. E. Ayer, T. G. Kutateladze, Y. Shi, J. Cote, K. F. Chua, and O. Gozani. 2006. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442:96-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skowyra, D., M. Zeremski, N. Neznanov, M. Li, Y. Choi, M. Uesugi, C. A. Hauser, W. Gu, A. V. Gudkov, and J. Qin. 2001. Differential association of products of alternative transcripts of the candidate tumor suppressor ING1 with the mSin3/HDAC1 transcriptional corepressor complex. J. Biol. Chem. 276:8734-8739. [DOI] [PubMed] [Google Scholar]
- 58.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 59.Strahl, B. D., P. A. Grant, S. D. Briggs, Z. W. Sun, J. R. Bone, J. A. Caldwell, S. Mollah, R. G. Cook, J. Shabanowitz, D. F. Hunt, and C. D. Allis. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suka, N., Y. Suka, A. A. Carmen, J. Wu, and M. Grunstein. 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8:473-479. [DOI] [PubMed] [Google Scholar]
- 61.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 62.van Leeuwen, F., P. R. Gafken, and D. E. Gottschling. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109:745-756. [DOI] [PubMed] [Google Scholar]
- 63.Wysocka, J., T. Swigut, T. A. Milne, Y. Dou, X. Zhang, A. L. Burlingame, R. G. Roeder, A. H. Brivanlou, and C. D. Allis. 2005. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121:859-872. [DOI] [PubMed] [Google Scholar]
- 64.Wysocka, J., T. Swigut, H. Xiao, T. A. Milne, S. Y. Kwon, J. Landry, M. Kauer, A. J. Tackett, B. T. Chait, P. Badenhorst, C. Wu, and C. D. Allis. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442:86-90. [DOI] [PubMed] [Google Scholar]
- 65.Zeng, L., and M. M. Zhou. 2002. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 513:124-128. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, X., A. Kolaczkowska, F. Devaux, S. L. Panwar, T. C. Hallstrom, C. Jacq, and W. S. Moye-Rowley. 2005. Transcriptional regulation by Lge1p requires a function independent of its role in histone H2B ubiquitination. J. Biol. Chem. 280:2759-2770. [DOI] [PubMed] [Google Scholar]