Abstract
Vpr, the viral protein R of human immunodeficiency virus type 1, induces G2 cell cycle arrest and apoptosis in mammalian cells via ATR (for “ataxia-telangiectasia-mediated and Rad3-related”) checkpoint activation. The expression of Vpr induces the formation of the γ-histone 2A variant X (H2AX) and breast cancer susceptibility protein 1 (BRCA1) nuclear foci, and a C-terminal domain is required for Vpr-induced ATR activation and its nuclear localization. However, the cellular target of Vpr, as well as the mechanism of G2 checkpoint activation, was unknown. Here we report that Vpr induces checkpoint activation and G2 arrest by binding to the CUS1 domain of SAP145 and interfering with the functions of the SAP145 and SAP49 proteins, two subunits of the multimeric splicing factor 3b (SF3b). Vpr interacts with and colocalizes with SAP145 through its C-terminal domain in a speckled distribution. The depletion of either SAP145 or SAP49 leads to checkpoint-mediated G2 cell cycle arrest through the induction of nuclear foci containing γ-H2AX and BRCA1. In addition, the expression of Vpr excludes SAP49 from the nuclear speckles and inhibits the formation of the SAP145-SAP49 complex. To conclude, these results point out the unexpected roles of the SAP145-SAP49 splicing factors in cell cycle progression and suggest that cellular expression of Vpr induces checkpoint activation and G2 arrest by interfering with the function of SAP145-SAP49 complex in host cells.
Human immunodeficiency virus type 1 (HIV-1) has four accessory genes—Vif, Vpr, Vpu, and Nef—that are dispensable for viral replication in vitro (for a review, see reference 10). The protein product of Vpr, viral protein R (Vpr), is of particular interest because it induces multiple effects in host cells, including transactivation of the long terminal repeats, cell cycle arrest, nuclear migration, and apoptosis (15, 17, 30, 34, 35, 37, 38). Several lines of evidence indicate that the ability of Vpr to induce cell cycle arrest at G2 depends on intrinsic signaling events of the cell's normal response to DNA damage (15, 17, 30, 34). Recent data reveal that the host proteins ATR (for “ataxia-telangiectasia and Rad3-related”), Rad17, and Hus1 are required for Vpr-induced G2 arrest (21, 35, 49). Because ATR and Rad17 colocalize and activate each other, thereby signaling G2 checkpoint activation (50), it is likely that Vpr induces G2 arrest by activating the ATR-Rad17 checkpoint signaling pathway. Previous works have shown that activated ATR in Vpr-transfected cells phosphorylates the histone 2A variant X (H2AX) and Chk1 and, in effect, takes advantage of the cell's normal response system to DNA damage (6, 33, 45, 49, 50). Vpr not only induces G2 arrest in mammalian cells but also in budding and fission yeast (24, 48), suggesting a highly conserved mechanism of cell cycle regulation in eukaryotes.
In eukaryotes the splicing of precursor mRNA (pre-mRNA) is essential for the expression of most protein-coding genes and is mediated by the sequential assembly and rearrangement of small nuclear ribonucleoprotein complexes, or spliceosomes, on the pre-mRNA (for a review, see reference 19). Among the best characterized of these are the components of U2 snRNP (for reviews, see references 17, 31, and 47). In mammals, functional 17S U2 snRNP is assembled from 12S U2 snRNP and two essential splicing factors, SF3a and SF3b (4). The SF3a and SF3b subunits are found in preparations of assembled spliceosomes and can be cross-linked to regions near the pre-mRNA branch point (1, 13, 14, 40). SF3b is composed of the four splicing-associated proteins (SAPs) SAP49, SAP130, SAP145, and SAP155 (for a review, see reference 19), which are highly conserved among eukaryotes. Budding yeast CUS1p, the homologue of human SAP145, was identified genetically by its ability to suppress U2 snRNA mutations (9, 14, 46) and to interact with Hsh49p, the yeast homologue of human SAP49, which binds to its CUS1 domain (7, 16). Genetic studies have recently shown that this domain is also required for normal growth in yeast (29), thus suggesting a regulatory link between RNA splicing and cell cycle progression.
Several different genetic studies also support the notion of a link between splicing factors and cell cycle progression. This evidence is twofold. First, independent screens for splicing factors and cell cycle regulators identified in both budding and fission yeasts a common group of genes (2, 36). Second, when mammalian cells were screened with a genome-scale small interfering RNA (siRNA) library. many genes required for G2-M transition were also found to be components of the splicing machinery (18). In addition, it is known that many well-known genes are repressed during mitosis either by transcriptional downregulation, polyadenylation, or translation (for a review, see reference 12) or by inhibition of pre-mRNA splicing (39). The significance of this mechanism is illustrated by the prominent function the ASF/SF2 protein, a member of the SR family of splicing proteins, plays in the maintenance of genome stability. ASF/SF2 prevents the formation of R loops that form when nascent transcripts and DNA template strands rehybridize (22, 23). Genetic inactivation of ASF/SF2 in DT40 cells leads to DNA double-strand breaks and subsequent checkpoint activation and G2 cell cycle arrest. In addition, in ASF/SF2 depleted cells R loops accumulate, which further indicate that the absence of splicing factors triggers genome instability and induces cell cycle arrest and checkpoint activation. Taken together, these observations suggest a highly conserved mechanism that links and regulates pre-mRNA splicing and cell cycle progression.
The present study provides evidence in support of a regulatory link between splicing factors and cell cycle progression. The initial objective of these investigations was to study the mechanism by which Vpr localizes to the nucleus and induces G2 arrest. Two-hybrid screening showed that Vpr binds host cell SAP145 and that this interaction is required for the nuclear targeting of Vpr. When Vpr binds to SAP145, it effectively depletes the cell of functional SAP145, triggers checkpoint activation, and induces G2 arrest. To conclude, SAP145 is a novel cellular target of Vpr, which induces activation of the checkpoint signal cascade, and thus G2 arrest, by binding to and thereby inhibiting the function of splicing factors SAP145-SAP49.
MATERIALS AND METHODS
Yeast two-hybrid screen.
Full-length Vpr and Vpr-ΔC lacking the C-terminal domain (amino acids 1 to 63) were fused in frame to the GAL4 DNA-binding domain of the pGBKT7 vector (Clontech Laboratories, Inc., Palo Alto, CA). Because Vpr expression inhibits normal growth in yeast, AH109 strain, which expresses Vpr fusion protein weakly and can grow on SD plates lacking tryptophan, was established. A human keratinocyte foreskin library (Clontech) was then screened using this strain (43).
Plasmid construction.
The pcDNA3 N-terminal hemagglutinin (HA)-tagged Vpr expression vector, generously provided by T. Yamaguchi, was subcloned into the mammalian and bacterial expression vectors C1 EGFP (Clontech) and pGEX4T3 (Amersham Pharmacia Biosciences, Piscataway, NJ), respectively. Vpr-ΔC was subcloned into the pcDNA3-HA, C1-EGFP, and pGEX2TK vectors (Amersham Pharmacia) using the BamHI and EcoRI restriction sites. For Vpr-ΔN, base pairs 103 to 288 were amplified by PCR and subcloned into the pGEX2TK vector. Base pairs 709 to 2622 (Δ1), 1 to 708 (Δ2), and 1276 to 1926 (Δ4) of SAP145 (GenBank accession no. U41371) and base pairs 4 to 1275 of SAP49 (GenBank accession no. AAH90883) were amplified by PCR. The forward and reverse primer sequences used to amplify SAP145 Δ1 were 5′-GGATCCAAGCTTATGGATGACCCCTCTGTGGGCCCCAAGATCCCC and 3′-CTCGAGCTAAAACTTGAACTCCTTATATTTCTTGCT. The amplified SAP145 Δ1 product was subcloned into the HindIII and XhoI restriction sites of the pCMV-tag2B (Flag) vector (Stratagene, La Jolla, CA). The primer sequences used to amplify SAP145 Δ2 were 5′-GGATCCGATGGCGCCTGGGGCTGCCCAGGAG and 3′-AAGCTTTACTCTCTGTTCTCATCTCCAGGGGGC. The amplified SAP145 Δ2 product was subcloned into the BamHI and HindIII restriction sites of the pRSET-B vector (Invitrogen, Carlsbad, CA). The forward and reverse primers used to amplify SAP145 Δ4 were 5′-GGTACCGCTGCCGGGCCGATCTCCGAGCGGAATCAG and 3′-CTCGAGCTAAAACTTGAACTCCTTATATTTCTTGCT. The amplified SAP145 Δ4 product was subcloned into the BamHI and KpnI restriction sites of the pRSET-C vector. To generate the SAP145 Δ3 construct, pCMV-2B SAP145 Δ1 was cut with BstXI, and blunt ends were generated. After digestion with the BamHI restriction enzyme, the fragment was subcloned into the BamHI and PvuII restriction sites of the pRSET-B vector. To construct SAP145 Δ5, pTOPO SAP145 Δ1 was subcloned into the KpnI and EcoRI restriction sites of the pRSET-A vector. The amplified SAP49 was subcloned into the KpnI and XbaI restriction sites of the pCMV-N-terminal HA tag vector. Transfection was performed by using Lipofectamine 2000 (Invitrogen).
Antibody staining and microscopy.
The following antibodies were used: monoclonal anti-Flag M2 and anti-β-actin monoclonal antibodies (Sigma), polyclonal and monoclonal mouse anti-HA and polyclonal anti-glutathione S-transferase (GST; Santa Cruz, Santa Cruz, CA, and Babco, Richmond, CA), rabbit polyclonal anti-γH2AX (Bethyl Laboratories, Montgomery, TX), and mouse monoclonal anti-His6 antibody (Santa Cruz). Rabbit polyclonal anti-SAP145 and goat polyclonal breast cancer susceptibility protein 1 (BRCA1) antibodies were generously provided by R. Reed and F. J. Couch, respectively. Cells grown on glass coverslips were fixed at room temperature for 10 min with 4% paraformaldehyde, followed by incubation with 0.5% Triton-X for 5 min. For immunostaining, fixed cells were incubated for 30 min at 30°C with combinations of primary and secondary antibodies as specified in the figure legends. For the Vpr nuclear retention experiments, transfected cells were extracted with 0.5% Triton X-100 for 5 min and fixed for 5 min with 4% paraformaldehyde as described previously (21). Cells were imaged at 5-min intervals on a Nikon TE300.
Immunoblotting.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer, and the insoluble fraction was pelleted for 5 min in a microcentrifuge. The lysates were boiled for 5 min and then loaded onto a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE) gel. Histone proteins were extracted from the pellets with 0.1 M HCl, boiled in SDS gel sample buffer, and loaded onto a SDS-15% PAGE gel. The separated proteins were transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Bedford, MA). To reduce nonspecific binding, the membrane was blocked for 30 min by incubation in a 3% bovine serum albumin (BSA) solution for 30 min. Transferred proteins were detected after incubation of the membrane for 3 h with anti-HA, anti-β-actin, anti-His6, anti-GST, anti-SAP145, and anti-γ-H2AX antibodies. The blots were then washed in TBST (10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 0.2% Tween 20). Antigen-antibody complexes were detected with horseradish peroxidase chemiluminescence using the SuperSignal kit (Pierce, Rockford, IL).
Protein-binding assays.
Recombinant proteins were expressed in 1-liter cultures of Escherichia coli BL21 and solubilized by freeze-thawing in liquid N2, followed by sonication in 10 ml of RIPA buffer (20 mM Tris [pH 7.5], 500 mM NaCl, 5 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate) containing a full set of protease inhibitors (Boehringer Mannheim) and lysozyme (Sigma, St. Louis, MO). Soluble proteins were cleared by centrifugation, purified with 400 μl of glutathione-Sepharose (Amersham Pharmacia) or Ni-nitrilotriacetic acid (NTA) beads (QIAGEN, Valencia, CA), and washed in RIPA buffer. To determine the protein concentration, SDS-PAGE gels were stained with Coomassie blue. For in vivo protein-binding assays HeLa cells were transfected with HA-tagged Vpr expression plasmids. Expressed HA-tagged Vpr protein was immunoprecipitated from nuclear extracts by using protein G-Sepharose. Nuclear extracts were rotated for 3 h as previously described (9). The protein G-Sepharose beads were washed extensively with 250 mM NaCl-20 mM Tris (pH 7.8), and proteins were eluted from the beads by boiling them in SDS-containing sample buffer. To identify the SAP145-binding domain of Vpr, Flag-tagged SAP145 Δ1 proteins were synthesized in vitro by using the TNT T7/T3-coupled reticulocyte lysate kit (Promega, Madison, WI). In vitro-synthesized proteins were incubated for 1 h at 4°C in binding buffer (25 mM HEPES [pH 7.9], 150 mM NaCl, 0.1% NP-40, 5% glycerol, 0.5 mM dithiothreitol, 0.4 mM phenylmethylsulfonyl fluoride) with the full-length GST-Vpr fusion protein, ΔN, ΔC, or GST alone. Glutathione-Sepharose (20 μl; Amersham Pharmacia) was added, and incubation was continued at 4°C for an additional hour. Complexes were washed four times with binding buffer and eluted in SDS-PAGE gel loading buffer. For identification of the Vpr-binding domain of SAP145, His6-tagged SAP145 Δ2, Δ3, Δ4, or Δ5 proteins were incubated with GST-Vpr fusion protein as described above. Ni-NTA beads (20 μl; QIAGEN) were added, and the mixture was incubated for 1 h at 4°C. The precipitates were resolved by SDS-10% PAGE and analyzed by immunoblotting.
Cell line establishment.
HeLa cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. HA-tagged SAP49 expression plasmids were transfected into HeLa cells by using Lipofectamine (Invitrogen), and transfected cells were selected in G418 over a 2-week period (41). G418 was added to the growth medium 48 h after transfection, and the medium regularly changed. Several G418-resistent colonies were picked and expanded for immunostaining.
RNA interference in HeLa cells.
cDNA fragments corresponding to bp 1025 to 1332 of the human SAP145 or bp 4 to 178 of human SAP49 were inserted into the pTOPO vector (Invitrogen). SAP145 or SAP49 cDNA flanked by T7-promoter binding sites was amplified by PCR using the M13F and T7-TOPO primers (5′-GCGTAATACGACTCACTATAGGTAACGGCCGCCAGTGTGCTG) and the pTOPO-SAP145 or SAP49 plasmids as templates, respectively. RNA strands were synthesized from PCR-derived linear template carrying a T7 promoter at both ends by using the MEGAscript kit (Ambion, Inc., Austin, TX). siRNAs for SAP145, SAP49, and luciferase as a control were prepared as described previously (44).
Cell cycle analysis.
HeLa cells were transfected with plasmids or siRNAs. At different time points the cells were treated with trypsin and fixed at −20°C with 70% EtOH. After being washed with phosphate-buffered saline, the cells were incubated for 30 min with 10 μg of RNase A/ml in citrate buffer and stained with 25 μg of propidium iodide/ml for an additional 30 min at 37°C. Cell cycle analysis was carried out on a FACScan (BD Biosciences, Bedford, MA) as described previously (42).
RESULTS
Identification of SAP145 as a Vpr-interacting protein.
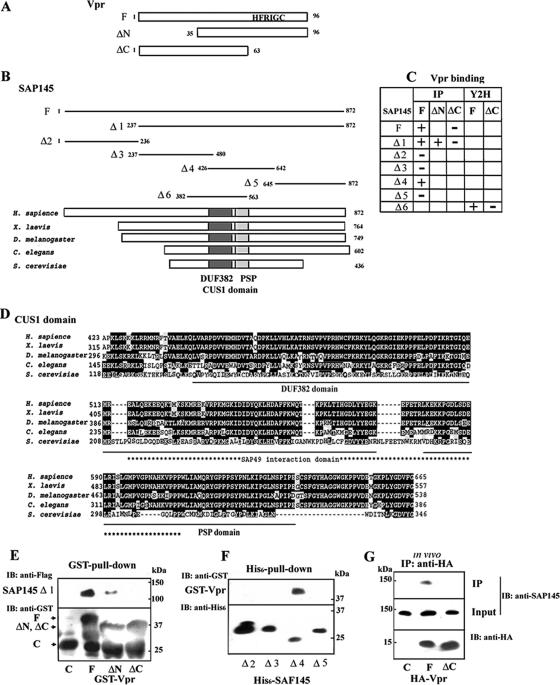
Recently, it has been reported that Vpr localizes to nuclear speckles through its C-terminal domain (21). Given the essential role the C-terminal domain of Vpr (Fig. 1A) plays in G2 cell cycle arrest and nuclear localization (3, 24, 27), it has been predicted that this domain might interact with other proteins that are associated with nuclear speckles and are involved in cell cycle regulation. To identify cellular proteins interacting with Vpr, the two-hybrid system was used to screen a human keratinocyte foreskin cDNA library with full-length Vpr as bait. This screen produced 28 positive clones from a total of 2 × 107 transformants. Clones requiring the C-terminal domain of Vpr for interaction were then identified from these 28 positive clones by using Vpr-ΔC as a bait. Among these 28 clones, two that reacted with full-length Vpr, but not the C-terminal deletion mutant Vpr-ΔC, were isolated. Both carried the same gene (Fig. 1B and C). DNA sequencing of the cDNA insert in both clones revealed that it (Fig. 1B; Δ6, SAP145382-563) corresponded to the CUS1 domain of the SAP145 subunit of the multimeric splicing factor 3b (SF3b) (Fig. 1B) (7), a protein present in the 17S form of the human U2 snRNP. The direct interaction between Vpr and SAP145 was independently confirmed in an in vitro binding assay. GST-tagged Vpr, Vpr-ΔN, or Vpr-ΔC lacking the N-terminal or C-terminal domains was expressed in E. coli, bound to glutathione-Sepharose, and incubated with SAP145 Δ1 protein synthesized in vitro through translation of recombinant SAP145 Δ1 in the reticulocyte lysate system. Under these conditions, GST-Vpr (Fig. 1E, lane F) and Vpr-ΔN (lane ΔN), but not Vpr-ΔC (lane ΔC) or GST alone (lane C), bound to SAP145237-872 Δ1, indicating that binding of the Vpr protein to SAP145 occurs through its C-terminal domain. The SAP145 domain required for interaction with Vpr was identified in in vitro binding assays by using His6-tagged SAP145 mutant proteins and GST-Vpr. His6-tagged SAP145 mutant proteins were incubated with GST-Vpr, and the complex was pulled down with nickel beads. These experiments showed that the GST-Vpr fusion protein bound selectively to SAP145426-642 that includes the CUS1 domain (Fig. 1F, lane Δ4) but not SAP1451-236 (lane Δ2), SAP145237-480 (lane Δ3), and SAP145645-872 (lane Δ5). To confirm whether Vpr associates with SAP145 in vivo, HA-tagged Vpr or Vpr-ΔC expression plasmids were transfected into HeLa cells, and expressed Vpr proteins were immunoprecipitated from nuclear cell extracts with anti-HA antibodies. The immunoprecipitated proteins were analyzed by immunoblotting with an anti-SAP145 antibody. The immunoblots demonstrated that SAP145 coimmunoprecipitated with the Vpr wild-type protein but not with the Vpr-ΔC deletion mutant, a finding consistent with the in vitro binding results. Because Vpr directly binds to chromatin (8, 21), the in vivo interaction between Vpr and SAP145 may be mediated through chromatin. To test this possibility, the effect of treatment with micrococcal nuclease was investigated. Digestion of chromosomal DNA with micrococcal nuclease had no effect on the interaction between Vpr and SAP145, indicating that it is not through a direct interaction with chromosomal DNA (data not shown). Figure 1C summarizes the interaction of the CUS1 domain of SAP145 with Vpr. Based on these results it can be concluded that the CUS1 domain, which spans amino acids 426 to 563, is sufficient for the interaction of SAP145 with Vpr. In addition, another subunit of SF3b, SAP49, binds to CUS1 through its N-terminal RNA recognition motif (RRM1) (7, 16). Because Vpr can arrest both mammalian and yeast cells in G2 (3, 21, 24, 25, 49), it is very likely that the Vpr protein targets molecules that are conserved among species and are part of the same pathways. Indeed, the CUS1 and RRM1 domains in SAP145 and SAP49, respectively, are highly conserved in species from yeasts to humans (Fig. 1B and C). In addition, genetic results have revealed that the CUS1 domain is required for normal growth in yeast (16, 29).
FIG. 1.
Interaction of Vpr with SAP145. (A) Schematic representation of the Vpr deletion mutants used in (E, and G). The wild-type and mutant Vpr constructs were N-terminally fused to the GST or HA tag. A C-terminal domain containing HFRIG motif is required for Vpr-induced G2 arrest. (B) Schematic representation of the SAP145 deletion mutants used in panels E and F. (C) The Vpr and SAP145 interactions occurring in in vitro binding (IP) and yeast two-hybrid (Y2H) assays are summarized. (D) Comparison of the structures of the metazoan members of the Cus1/SAP145 family from metazoans: human SAP145 (Swiss-Prot accession no. Q13435), Xenopus laevis (GenBank accession no. BC094200), Caenorhabditis elegans (GenBank accession no. AAB69931), Drosophila melanogaster (GenBank accession no. NM134895), and Saccharomyces cerevisiae Cus1p (Swiss-Prot accession no. Q02554). The CUS1 domain including the DUF382 and PSP regions resides in similar positions in the human, X. laevis, D. melanogaster, C. elegans, and S. cerevisiae homologues. The positions of the DUF381 and PSP regions are schematically shown by black and hatched boxes, respectively. An asterisk indicates the position of the SAP49/Hsh49p interaction domain. Underlining shows the DUF382 and PSP domains, respectively. (E) GST alone (C), GST-Vpr (F), or mutants (ΔN and ΔC) were incubated with Flag-tagged SAP145 Δ1 protein expressed in the reticulocyte system and precipitated with glutathione-Sepharose 4B beads. The bound proteins were analyzed by immunoblotting with anti-Flag and GST antibodies. (F) Identification of the Vpr-interacting region in the SAP145 protein. Four N-terminal His6-tagged SAP145 constructs (see panel B) were tested for interaction with GST-Vpr (lane F) by using an in vitro His6-pull-down assay. The bound proteins were precipitated with nickel-beads and analyzed by immunoblotting with anti-GST and anti-His6 antibodies. (G) Vector alone (lane C) or the expression vectors HA-Vpr (lane F) or Vpr-ΔC (lane ΔC) were transfected into HeLa cells. After 48 h, the cells were harvested and then immunoprecipitated with a monoclonal anti-HA antibody. Bound proteins were analyzed by immunoblotting with the polyclonal anti-HA and SAP145 antibodies.
Vpr colocalizes with SAP145 in a speckled distribution.
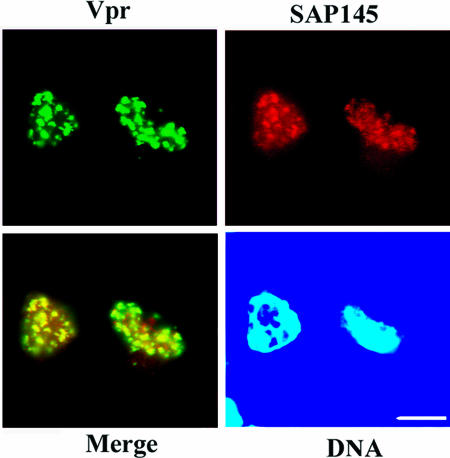
Consistent with previous results (21), Vpr was found for the most part at the nuclear speckles after transfection of HeLa cells with a recombinant GFP-Vpr fusion construct (see Fig. S1A, middle, in the supplemental material). After in situ extraction of cells, a subpopulation of Vpr was found to be retained in the nucleus to give a characteristic clumped pattern. In contrast, this nuclear retention of Vpr cannot be observed in the cells expressing the Vpr-ΔC deletion mutant. Vpr-ΔC still localizes to the nucleus, but extraction-resistant nuclear staining at the nuclear speckles is significantly diminished (Fig. S1A, bottom). These data suggest that Vpr localizes at nuclear speckles through its C-terminal domain (Fig. S1B). To examine the subcellular distribution of SAP145 with Vpr, HeLa cells were cotransfected with the recombinant GFP-Vpr fusion and Flag-tagged SAP145 Δ1 plasmids, and subsequently immunostained with an anti-Flag antibody. The nuclear distributions of SAP145 (Fig. 2) and SAP49 (see below) were speckled. Since nuclear speckle localization was not observed in cells transfected with SAP145 siRNA, the possibility that this observation is due to nonspecific binding of the Flag antibody could be excluded (data not shown). These cotransfection experiments showed that Vpr colocalizes with SAP145 in a speckled distribution similar to that seen for other snRNP proteins (Fig. 2) (5, 26).
FIG. 2.
Vpr colocalizes with SAP145 in a speckled distribution. Subcellular localization of SAP145 and Vpr. HeLa cells were cotransfected with GFP-Vpr and Flag-SAP145 Δ1 expression vectors. Cells were briefly extracted with detergent, fixed, and immunostained with anti-Flag antibody. For nuclear retention experiments, transfected cells were subjected to 0.5% Triton X-100 extraction for 5 min followed by 4% paraformaldehyde fixation for 5 min.
Depletion of either SAP145 or SAP49 induces G2 arrest.
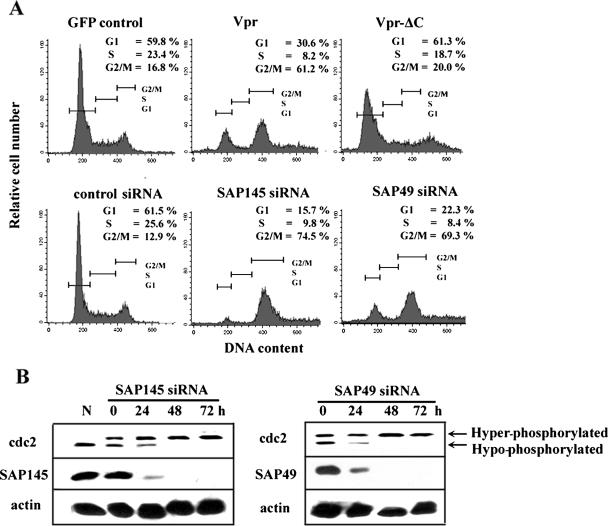
The C-terminal domain of Vpr was implicated in arresting cells in G2 (3, 24, 26, 27). Consistent with these results, wild-type Vpr induced G2 arrest in HeLa cells, as was demonstrated by fluorescence-activated cell sorting (FACS) analysis of cells expressing wild-type Vpr (Fig. 3A, upper panel). The same effect was not observed with the deletion mutant Vpr-ΔC. Furthermore, recent studies have also demonstrated that inactivation of the ASF/SF2 splicing factor in DT40 cells led to G2 arrest (22). To examine whether depletion of SAP145 causes cell cycle arrest in G2, SAP145 was depleted by RNA interference, and the effects of this depletion on cell cycle progression were analyzed by FACS analysis. In cells depleted of SAP145, but not in control cells depleted of luciferase, a significant shift toward cells arrested in G2 was observed (Fig. 3A, bottom). This finding is comparable to what was observed in Vpr-expressing cells. To investigate whether SAP49, which forms a complex with SAP145, has a similar effect on cell cycle progression, the effect of SAP49 depletion was tested. As shown in Fig. 3A, these experiments gave results virtually identical to those obtained with SAP145-depeleted cells. To determine whether the cells were arrested in the G2 or M phase, the phosphorylation status of the cdc2 kinase was analyzed by immunoblot analysis with an anti-cdc2 antibody (Fig. 3B). Although the cdc2 protein levels remain constant throughout the cell cycle, its activity is regulated by phosphorylation and dephosphorylation. At the G2/M boundary, cdc2 is activated by cdc25C-mediated dephosphorylation, which triggers entry into mitosis (for a review, see reference 28). Immunoblots of whole-cell lysates showed that neither SAP145 nor SAP49 siRNA affected the total concentration of cdc2 (Fig. 3B). However, the proportion of the phosphorylated inactive form of cdc2 was increased in cells depleted of either SAP145 or SAP49. In contrast, transfection of control luciferase siRNA did not affect the phosphorylation status of cdc2 (data not shown). This indicates that depletion of either SAP145 or SAP49 induces G2 cell cycle arrest and that SAP145-SAP49 complex formation may be required for G2-M progression.
FIG. 3.
Depletion of either SAP145 or SAP49 induces G2 cell cycle arrest. (A) The cell cycle profiles of GFP- (control), wild-type GFP-Vpr-, or GFP-Vpr-ΔC-transfected HeLa cells were analyzed by FACS at 48 h after transfection (top). Cell cycle phenotypes of SAP145- or SAP49-depleted cells (bottom). HeLa cells with or without stably expressed HA-tagged SAP49 were transfected with either luciferase siRNA (control) or SAP145- and SAP49-specific siRNAs. The cell cycle distribution was analyzed by FACS 48 h after transfection. The G1, S, and the G2/M phases are indicated. (B) Immunoblot analysis with whole-cell lysates from HeLa cells transfected for 0, 24, 48, or 72 with either SAP145 or SAP49 siRNAs. Blots were probed with anti-cdc2, anti-SAP145, or anti-HA (SAP49) antibodies. (N) sample from nocodazole-synchronized cells. The positions of hypo- and hyperphosphorylated cdc2 are indicated.
Depletion of either SAP145 or SAP49 induces focus formation of γ-H2AX and BRCA1.
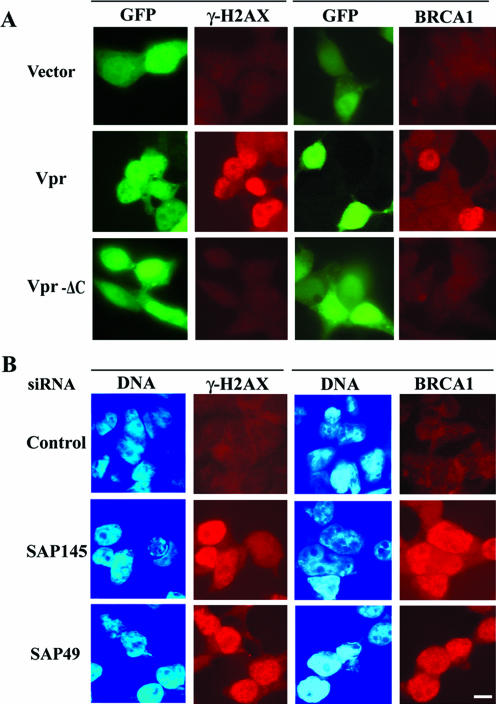
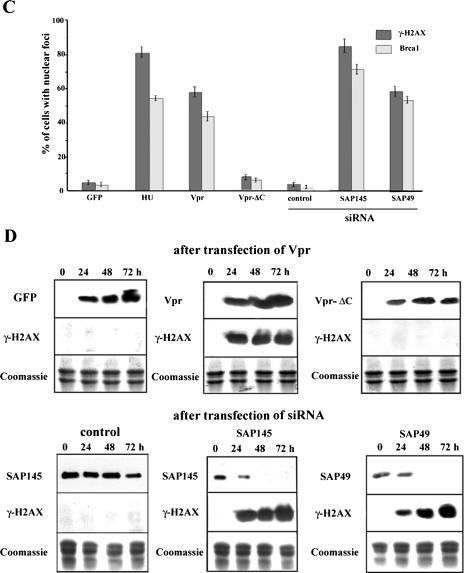
Recent reports suggest that the ATR-mediated signaling pathway is required for Vpr-induced G2 arrest (49). H2AX, a histone protein variant that is randomly incorporated in nucleosomes (6, 50), is phosphorylated as an immediate response to DNA damage and will form nuclear foci (11, 32). Indeed, Vpr through ATR activation induces phosphorylated H2AX (also referred to as γ-H2AX) and BRCA1 focus formation (21, 49). Depletion of ASF/SF2 will also result in phosphorylation of H2AX (23). These results raised the question of whether depletion of SAP145 or SAP49 can induce the focus formation of γ-H2AX and BRCA1. Consistent with earlier results (21, 49), immunostaining experiments showed that in cells transfected with a Vpr expression plasmid, the expression of the γ-H2AX protein was increased (Fig. 4A). However, the same increase was not observed in cells transfected with an expression plasmid of the deletion mutant Vpr-ΔC. Likewise, depletion of either SAP145 or SAP49 also induced the phosphorylation of H2AX (Fig. 4B). Quantification of cells displaying intense γ-H2AX immunostaining showed more than 85% of cells with SAP145 siRNA and 62% of cells with SAP49 siRNA positive for γ-H2AX foci, whereas 58% of Vpr-expressing cells and less than 4% of cells transfected with the GFP control were found to be positive for γ-H2AX foci (Fig. 4C). As a positive control, 82% of the cells treated for 2 h with hydroxyurea were found to have positive γ foci. As shown in Fig. 4B and C, these experiments gave results virtually identical to those obtained with immunostaining analysis with BRCA1 antibody (Fig. 4A, B, and C). These observations were further verified by immunoblotting (Fig. 4D). Detectable levels of γ-H2AX protein appeared as early as 24 h posttransfection of either SAP145 or SAP49 siRNAs and increased as a function of the time-dependent depletion of each gene (Fig. 4D). These results indicate that in SAP145- or SAP49-depleted cells the checkpoint signal cascade can be activated and will lead to G2 cell cycle arrest.
FIG.4.
Depletion of either SAP145 or SAP49 induces formation of γ-H2AX and BRCA1 foci. (A) HeLa cells were transfected with vector (GFP), GFP-Vpr, or GFP-Vpr-ΔC. At 48 h after transfection, the cells were stained with γ-H2AX- or BRCA1-specific antibodies (red). (B) Cells were transfected with either luciferase (control), SAP145, or SAP49 siRNAs. At 48 h after transfection, cells were stained with γ-H2AX- or BRCA1-specific antibodies (red). (C) γ-H2AX- and BRCA1-positive and γ-H2AX- and BRCA1-negative cells were visually counted. As a positive control, cells were treated with 10 mM hydroxyurea for 2 h. Cells were transfected with several vectors or siRNAs as shown in panels A and B. The results were averaged over three independent experiments (>200 cells each); bars indicate the standard deviations. (D) Immunoblot analysis with acid-soluble extracts (middle and bottom) or whole-cell extract (top) from HeLa cells transfected with either GFP (control), GFP-Vpr, or GFP-Vpr-ΔC plasmids or HeLa cells stably expressing HA-tagged SAP49, transfected with either luciferase (control), SAP145, or SAP49 siRNAs for 0, 24, 48, or 72 h. Blots were probed with anti-γ-H2AX (top), anti-GFP (Vpr), anti-SAP145, or anti-HA (SAP49) antibodies. Proteins were stained with Coomassie blue (bottom).
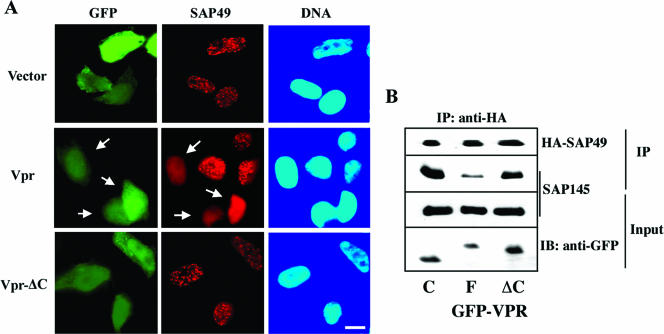
Expression of Vpr protein excludes SAP49 from nuclear speckles and inhibits formation of the SAP145-SAP49 complex.
The results presented above suggest that Vpr induced checkpoint activation and thereby G2 arrest through interference with the SAP145-SAP49 function by an unknown mechanism. Since Vpr binds to the common CUS1 domain, which is essential for interaction with SAP49, one possibility to explain the inhibition of the SAP145-SAP49 complex by Vpr might be through functional interference. To explore this possibility, a Vpr expression plasmid was transfected into cells stably expressing HA-tagged SAP49, and the subcellular distribution of SAP49 was examined. There was a clear difference in the localization of SAP 49 between control cells and cells expressing Vpr. SAP49 localized in control cells such as SAP145 at the nuclear speckles (Fig. 5A and Fig. 2), but it disappeared from the nuclear speckles and diffused broadly into the nucleoplasm in cells expressing Vpr but not Vpr-ΔC (Fig. 5A). SAP49 localized to nuclear speckles in only 35% of Vpr-expressing cells, whereas 84 or 71% of GFP-expressing (vector alone) cells or Vpr-ΔC-expressing cells, respectively, displayed nuclear speckle localization. To next verify the effect of Vpr on SAP145-SAP49 complex formation, nuclear extracts from HeLa cell expressing HA-tagged SAP49 were subjected to immunoprecipitation with anti-HA-antibody, and the resulting immunoprecipitates were subjected to immunoblot analysis with anti-HA and SAP145 antibodies (Fig. 5B). The SAP145 protein coimmunoprecipitated with the SAP49 protein in the nuclear extract from the GFP-expressing control cells, indicating that both proteins exit in a tight complex as described previously (7, 16), whereas it was dramatically inhibited by the expression of Vpr but not by the expression of Vpr-ΔC. These data are quite the opposite of previous results that showed colocalization of Vpr with SAP145 to the nuclear speckles (Fig. 2). These findings also indicate that Vpr induces atypical localization of SAP49 but not of SAP145. The data presented here suggest that the localization of SAP49 to nuclear speckle depends on SAP145 but not vice versa. To conclude, these observations raise the possibility that Vpr inhibits the function of the SAP145-SAP49 complex by interfering with the interaction between the two proteins.
FIG. 5.
Expression of Vpr excludes SAP49 from nuclear speckles and inhibits the formation of SAP145-SAP49 complex. (A) HeLa cells stably expressing HA-tagged SAP49 were transfected with GFP alone (vector), GFP-Vpr, or GFP-Vpr-ΔC. Cells were fixed and stained with an anti-HA antibody. In Vpr-expressing cells SAP49 disappeared from the nuclear speckles and diffused broadly into the nucleoplasm (open arrow). (B) Nuclear extracts from HeLa cell expressing HA-tagged SAP49 were subjected to immunoprecipitation with anti-HA-antibody, and the resulting immunoprecipitates were subjected to immunoblot analysis with anti-HA and SAP145 antibodies. Equal amounts of proteins prepared from HeLa cells transfected with GFP, GFP-Vpr, or GFP-Vpr-ΔC were blotted with β-actin or GFP antibodies. The SAP145 protein coimmunoprecipitated with the HA-tagged SAP49 protein in the nuclear extract from the GFP-expressing control cells. It was inhibited by expression of Vpr but not Vpr-ΔC.
DISCUSSION
This study describes how the HIV-1 Vpr protein localizes to nuclear speckles and induces checkpoint activation and G2 cell cycle arrest through functional interference with host cell proteins. The data presented herein link Vpr to the two splicing factors SAP145 and SAP49. The Vpr protein binds through its C-terminal domain to the CUS1 domain of SAP145 (Fig. 1A and D). Moreover, Vpr colocalizes with SAP145 in a speckled distribution (Fig. 2). These data suggest that SAP145 is a cellular target for Vpr anchoring to nuclear speckles. In addition, the C-terminal domain plays a crucial role in ATR-mediated G2 arrest but not in ATM-mediated arrest (21, 49), thus indicating that the C-terminal domain tightly links the nuclear localization of Vpr with its activation of the ATR pathway. ATM responds primarily to DNA double-strand breaks, whereas ATR is crucial to the cell's response to DNA replication stress and broader DNA lesions (6, 50). It is therefore most likely that Vpr induces replication stress, thus activating the ATR-mediated G2 checkpoint pathway. Indeed, Vpr has no effect on the formation of double-strand breaks that activate the ATM cascade (21) but induces instead foci formation of γ-H2AX and BRCA1 and probably the accumulation of chromatin-associated replication protein A through the ATR-mediated checkpoint cascade (21) (Fig. 4A and D). The results presented here show that depletion of SAP145 or SAP49 leads to rapid formation of γ-H2AX and BRCA1 foci (Fig. 4B, C, and D), clearly indicating that the ATM and/or ATR cascade is activated in SAP145- or SAP49-depleted cells. Furthermore, depletion of either SAP145 or SAP49 leads to G2 arrest (Fig. 3). Since Vpr expression and loss-of-function of SAP145 and SAP49 result in similar phenotypes, it has been predicted that in addition to the physical interaction between Vpr and the SAP145-SAP49 complex a functional relationship exists.
In principle, there are at least two possible explanations for the functional relationship between Vpr and SAP145-SAP49. The simplest explanation would be that as a direct consequence of the expression of Vpr defects in pre-mRNA splicing occur, which will lead to G2 cell cycle arrest. In this scenario, the functional inhibition of the SAP145-SAP49 complex as a result of expression of Vpr leads to impaired pre-mRNA processing of genes essential for the G2-M transition. Intriguingly, herpes simplex virus IE63 protein interacts with SAP145 and inhibits splicing function (5). Likewise, Vpr was also reported to inhibit the splicing of beta-globin pre-mRNA in vivo and in vitro (20). Thus, it is likely that Vpr inhibits the splicing function by interfering with SAP145. Unexpectedly, however, the expression of Vpr did not affect the splicing status of several genes such as cdk1, cyclin B1, cdc25C, and wee1, which are essential for the G2-M transition (data not shown). In addition, checkpoint regulators such as ATR, RAD17, and Hus1 are required for Vpr-mediated G2 arrest (49). These observations indicate Vpr does not induce cell cycle arrest by simply inhibiting the proper splicing of pre-mRNA. Alternatively, it is possible that Vpr activates the DNA damage checkpoint and thereby induces cell cycle arrest through inhibition of the function of the SAP145-SAP49 complex. Recent reports have shown that depletion of the ASF/SF2 splicing factor induces genomic instability (23). In ASF/SF2-depleted cells, R loop structures are formed by rehybridization of nascent transcripts and DNA template strands. In addition, frequently double-strand breaks and thus G2 checkpoint activation were observed. These data suggest that ASF/SF2 prevents R-loop formation by binding to nascent mRNA precursors, thereby preventing the transcripts from associating with template DNA (23). Since the phenotypes of cells depleted of SAP145 or SAP49 are indistinguishable from those in which ASF/SF2 has been depleted, it is conceivable that the SAP145-SAP49 complex plays an important role in maintaining genomic stability and that Vpr activates the G2 checkpoint cascade by interfering with the formation of the SAP145-SAP49 complex. Indeed, since the depletion of SAP145 or SAP49 induced the rapid nuclear focus formation of γ-H2AX (Fig. 4A, C, and D) and G2 arrest (Fig. 3), the expression of Vpr most likely interferes with the function of SAP145-SAP49 complex, leading to G2 checkpoint activation. However, we cannot exclude the possibility that Vpr induces the checkpoint activation by interfering with the splicing of several genes required for genomic stability.
Although the precise mechanism of Vpr interference with the function of the SAP145-SAP49 complex remains unknown, it is important to note that the CUS1 domain of SAP145, which is required for its interaction with Vpr, is highly conserved in species from yeasts to humans (28) (Fig. 1B and D). This domain also overlaps with the domain that is essential for the interaction with Hsh49p, a homologue of human SAP49 (16), which is indispensable for the assembly of SF3b and thus normal growth. Since the expression of Vpr excludes the SAP49 protein from nuclear speckles and inhibits the formation of SAP145-ASP49 complex (Fig. 5), it is most likely that Vpr inhibits the function of SAP145-SAP49 by interfering with the interaction between SAP145 and SAP49 in host cells. Because Vpr-induced G2 arrest is critical for efficient HIV-1 replication and cytopathicity, it is conceivable that pharmacological prevention of Vpr-induced G2 arrest may yield new approaches for therapeutic intervention. The Vpr-SAP145 interaction may be the obvious target for drug design in the future.
Supplementary Material
Acknowledgments
We are grateful to R. Kuriyama, S. Sturm, O. Niwa, and V. Planelles for helpful comments and to S. Skjolaas for helpful advice. We thank T. Yamaguchi for Vpr plasmids, R. Reed for SAP145 antibody, and F. J. Crouch for Brca1 antibody.
This study was supported by the Minnesota Medical Foundation and the Uehara Memorial Foundation.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 21 August 2006.
REFERENCES
- 1.Bennett, M., S. Pinol-Roma, D. Staknis, G. Dreyfuss, and R. Reed. 1992. Differential binding of heterogeneous nuclear ribonucleoproteins to mRNA precursors prior to spliceosome assembly in vitro. Mol. Cell. Biol. 12:3165-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Yehuda, S., I. Dix, C. S. Russell, M. McGarvey, J. D. Beggs, and M. Kupiec. 2000. Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics 156:1503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berglez, J. M., L. A. Castelli, S. A. Sankovich, S. C. Smith, C. C. Curtain, and I. G. Macreadie. 1999. Residues within the HFRIGC sequence of HIV-1 vpr involved in growth arrest activities. Biochem. Biophys. Res. Commun. 264:287-290. [DOI] [PubMed] [Google Scholar]
- 4.Brosi, R., H. P. Hauri, and A. Kramer. 1993. Separation of splicing factor SF3 into two components and purification of SF3a activity. J. Biol. Chem. 268:17640-17746. [PubMed] [Google Scholar]
- 5.Bryant, H. E., S. E. Wadd, A. I. Lamond, S. J. Silverstein, and J. B. Clements. 2001. Herpes simplex virus IE63 (ICP27) protein interacts with spliceosome-associated protein 145 and inhibits splicing prior to the first catalytic step. J. Virol. 75:4376-4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burma, S., B. P. Chen, M. Murphy, A. Kurimasa, and D. J. Chen. 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276:42462-42467. [DOI] [PubMed] [Google Scholar]
- 7.Champion-Arnaud, P., and R. Reed. 1994. The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes Dev. 15:1974-1983. [DOI] [PubMed] [Google Scholar]
- 8.Coeytaux, E., D. Coulaud, E. Le Cam, O. Danos, and A. Kichler. 2003. The cationic amphipathic alpha-helix of HIV-1 viral protein R (Vpr) binds to nucleic acids, permeabilizes membranes, and efficiently transfects cells. J. Biol. Chem. 278:18110-18116. [DOI] [PubMed] [Google Scholar]
- 9.Das, B. K., L. Xia, L. Palandjian, O. Gozani, Y. Chyung, and R. Reed. 1999. Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Mol. Cell. Biol. 19:6796-6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emerman, M., and M. H. Malim. 1998. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science 280:1880-1884. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez-Capetillo, O., A. Lee, M. Nussenzweig, and A. Nussenzweig. 2004. H2AX: the histone guardian of the genome. DNA Repair 3:959-967. [DOI] [PubMed] [Google Scholar]
- 12.Gottesfeld, J. M., and D. J. Forbes. 1997. Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 22:197-202. [DOI] [PubMed] [Google Scholar]
- 13.Gozani, O., J. G. Patton, and R. Reed. 1994. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 13:3356-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gozani, O., R. Feld, and R. Reed. 1996. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 15:233-243. [DOI] [PubMed] [Google Scholar]
- 15.He, J., S. Choe, R. Walker, P. Di Marzio, and D. O. Morgan, and N. R. Landau. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Igel, H., S. Wells, R. Perriman, and M. Ares, Jr. 1998. Conservation of structure and subunit interactions in yeast homologues of splicing factor 3b (SF3b) subunits. RNA 4:1-10. [PMC free article] [PubMed] [Google Scholar]
- 17.Jowett, J. B., V. Planelles, B. Poon, N. P. Shah, M. L. Chen, and I. S. Chen. 1995. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2+M phase of the cell cycle. J. Virol. 69:6304-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kittler, R., G. Putz, L. Pelletier, I. Poser, A. K. Heninger, D. Drechsel, S. Fischer, I. Konstantinova, B. Habermann, H. Grabner, M. L. Yaspo, H. Himmelbauer, B. Korn, K. Neugebauer, M. T. Pisabarro, and F. Buchholz. 2004. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature 432:1036-1040. [DOI] [PubMed] [Google Scholar]
- 19.Kramer, A. 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65:367-409. [DOI] [PubMed] [Google Scholar]
- 20.Kuramitsu, M., C. Hashizume, N. Yamamoto, A. Azuma, M. Kamata, N. Yamamoto, Y. Tanaka, and Y. Aida. 2005. A novel role for Vpr of human immunodeficiency virus type 1 as a regulator of the splicing of cellular pre-mRNA. Microbes Infect. 7:1150-1160. [DOI] [PubMed] [Google Scholar]
- 21.Lai, M., E. S. Zimmerman, V. Planelles, and J. Chen. 2005. Activation of the ATR pathway by human immunodeficiency virus type 1 Vpr involves its direct binding to chromatin in vivo. J. Virol. 79:15443-15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, X., J. Wang, and J. L. Manley. 2005. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 19:2705-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, X., and J. L. Manley. 2005. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122:365-378. [DOI] [PubMed] [Google Scholar]
- 24.Macreadie, I. G., L. A. Castelli, D. R. Hewish, A. Kirkpatrick, A. C. Ward, and A. A. Azad. 1995. A domain of human immunodeficiency virus type 1 Vpr containing repeated H(S/F)RIG amino acid motifs causes cell growth arrest and structural defects. Proc. Natl. Acad. Sci. USA 92:2770-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masuda, M., Y. Nagai, N. Oshima, K. Tanaka, H. Murakami, H. Igarashi, and H. Okayama. 2000. Genetic studies with the fission yeast Schizosaccharomyces pombe suggest involvement of wee1, ppa2, and rad24 in induction of cell cycle arrest by human immunodeficiency virus type 1 Vpr. J. Virol. 74:2636-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Misteli, T., J. F. Caceres, and D. L. Spector. 1997. The dynamics of a pre-mRNA splicing factor in living cells. Nature 387:523-527. [DOI] [PubMed] [Google Scholar]
- 27.Mueller, S. M., and S. M. Lang. 2002. The first HxRxG motif in simian immunodeficiency virus mac239 Vpr is crucial for G2/M cell cycle arrest. J. Virol. 76:11704-11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nurse, P. 1990. Universal control mechanism regulating onset of M-phase. Nature 344:503-508. [DOI] [PubMed] [Google Scholar]
- 29.Pauling, M. H., D. S. McPheeters, and M. Ares, Jr. 2000. Functional Cus1p is found with Hsh155p in a multiprotein splicing factor associated with U2 snRNA. Mol. Cell. Biol. 20:2176-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Re, F., D. Braaten, E. K. Franke, and J. Luban. 1995. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J. Virol. 69:6859-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reed, R. 1996. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr. Opin. Genet. Dev. 6:215-220. [DOI] [PubMed] [Google Scholar]
- 32.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858-5868. [DOI] [PubMed] [Google Scholar]
- 33.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogel, M. E., L. I. Wu, and M. Emerman. 1995. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J. Virol. 2:882-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roshal, M., B. Kim, Y. Zhu, P. Nghiem, and V. Planelles. 2003. Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J. Biol. Chem. 278:25879-25886. [DOI] [PubMed] [Google Scholar]
- 36.Russell, C. S., S. Ben-Yehuda, I. Dix, M. Kupiec, and J. D. Beggs. 2000. Functional analyses of interacting factors involved in both pre-mRNA splicing and cell cycle progression in Saccharomyces cerevisiae. RNA 6:1565-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakai, K., J. Dimas, and M. J. Lenardo. 2006. The Vif and Vpr accessory proteins independently cause HIV-1-induced T-cell cytopathicity and cell cycle arrest. Proc. Natl. Acad. Sci. USA 109:3369-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherman, M. P., C. M. de Noronha, L. A. Eckstein, J. Hataye, P. Mundt, S. A. Williams, J. A. Neidleman, M. A. Goldsmith, and W. C. Greene. 2003. Nuclear export of Vpr is required for efficient replication of human immunodeficiency virus type 1 in tissue macrophages. J. Virol. 77:7582-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin, C., and J. L. Manley. 2002. The SR protein SRp38 represses splicing in M phase cells. Cell 111:407-417. [DOI] [PubMed] [Google Scholar]
- 40.Staknis, D., and R. Reed. 1994. Direct interactions between pre-mRNA and six U2 small nuclear ribonucleoproteins during spliceosome assembly. Mol. Cell. Biol. 14:2994-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terada, Y., M. Tatsuka, S. Jinno, and H. Okayama. 1995. Requirement for tyrosine phosphorylation of Cdk4 in G1 arrest induced by ultraviolet irradiation. Nature 376:358-362. [DOI] [PubMed] [Google Scholar]
- 42.Terada, Y., M. Tatsuka, F. Suzuki, Y. Yasuda, S. Fujita, and M. Otsu. 1998. AIM-1: a mammalian midbody-associated protein required for cytokinesis. EMBO J. 17:667-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terada, Y., Y. Uetake, and R. Kuriyama. 2003. Interaction of Aurora-A and centrosomin at the microtubule-nucleating site in Drosophila and mammalian cells. J. Cell Biol. 163:757-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terada, Y. 2006. Aurora-B/AIM-1 regulates dynamic behavior of HP1 alpha at G2-M transition. Mol. Biol. Cell 17:3232-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward, I., M. K. Minn, and J. Chen. 2004. UV-induced ataxia-telangiectasia-mutated and Rad3-related (ATR) activation requires replication stress. J. Biol. Chem. 279:9677-9680. [DOI] [PubMed] [Google Scholar]
- 46.Wells, S. E., M. Neville, M. Haynes, J. Wang, H. Igel, and M. Ares, Jr. 1996. CUS1, a suppressor of cold-sensitive U2 snRNA mutations, is a novel yeast splicing factor homologous to human SAP 145. Genes Dev. 10:220-232. [DOI] [PubMed] [Google Scholar]
- 47.Will, C., and R. Luhrman. 1997. Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol. 9:320-328. [DOI] [PubMed] [Google Scholar]
- 48.Zhao, Y., and R. T. Elder. 2000. HIV-1 VPR modulates cell cycle G2/M transition through an alternative cellular mechanism other than the classic mitotic checkpoints. Front. Biosci. 5:D905-916. [DOI] [PubMed] [Google Scholar]
- 49.Zimmerman, E. S., J. Chen, J. L. Andersen, O. Ardon, J. L. Dehart, J. Blackett, S. K. Choudhary, D. Camerini, P. Nghiem, and V. Planelles. 2004. Human immunodeficiency virus type 1 Vpr-mediated G2 arrest requires Rad17 and Hus1 and induces nuclear BRCA1 and gamma-H2AX focus formation. Mol. Cell. Biol. 24:9286-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zou, L., D. Cortez, and S. J. Elledge. 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16:198-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.