Abstract
The transcription factors of the Fos family have long been associated with the control of cell proliferation, although the molecular and cellular mechanisms that mediate this function are poorly understood. We investigated the contributions of Fos to the cell cycle and cell growth control using Drosophila imaginal discs as a genetically accessible system. The RNA interference-mediated inhibition of Fos in proliferating cells of the wing and eye discs resulted in a specific defect in the G2-to-M-phase transition, while cell growth remained unimpaired, resulting in a marked reduction in organ size. Consistent with the conclusion that Fos is required for mitosis, we identified cyclin B as a direct transcriptional target of Fos in Drosophila melanogaster, with Fos binding to a region upstream of the cyclin B gene in vivo and cyclin B mRNA being specifically reduced under Fos loss-of-function conditions.
The Fos family of leucine zipper transcription factors in mammals is comprised of four members, the proto-oncogene product c-Fos and FosB, Fra1, and Fra2. A variety of partly redundant functions have been ascribed to these different proteins, and as a family, they have been implicated in the regulation of multiple cellular processes ranging from growth and proliferation to stress response and apoptosis (9, 10, 26, 30). The ability of v-fos, the c-fos-derived retroviral oncogene, to transform avian and murine cells and the finding that the stimulation of quiescent cells with growth factors can rapidly induce c-fos gene expression have led to the suggestion that Fos proteins might regulate cell proliferation (5, 18). This notion was further supported by the finding that the microinjection of anti-Fos family antibodies into proliferating cells interfered with normal cell cycle progression (13). Another line of evidence in support of the idea that Fos might control cell proliferation came from genetic studies of mammalian systems. Murine fibroblasts deficient for both the c-fos and the fosB loci, but not the corresponding single mutants, showed impaired cell cycle progression (2). In addition to a potential role for Fos in the regulation of cell proliferation, this finding suggests functional redundancy among different mammalian Fos family members.
When c-Fos was ectopically expressed in transgenic mice, defects in bone morphogenesis ensued, and osteosarcomas formed due to the transformation of osteoblasts (7, 24, 25). Recent studies confirmed that AP-1 transcription factors are also associated with osteosarcomas in humans (21). In spite of the many indirect lines of evidence that link Fos to the control of cell proliferation and implicate it in tumorigenesis, studies of vertebrate systems have left many questions about the primary targets of Fos in this context unanswered.
The functional complexity and redundancy that characterize the mammalian Fos family have motivated a search for simpler and genetically more accessible systems of studying the molecular and biological functions of Fos. In Drosophila melanogaster, D-Fos (also known as D-FRA or Kayak) is the only known Drosophila homolog of Fos (23, 32). D-Fos has been implicated in a number of developmental processes: dorsal closure in the embryo, larval gut development, photoreceptor differentiation, wing vein differentiation, and thorax closure during pupal development (4, 12, 16, 22, 31). Employing Drosophila genetics and taking advantage of the fact that the Fos family has only one member in the fly, we generated evidence for direct roles for D-Fos in the regulation of cyclin B expression and the G2-to-M transition of the cell cycle.
MATERIALS AND METHODS
Fly strains.
Transgenic fly lines expressing wild-type D-Fos were generated by cloning the kayak cDNA into a pUAST transformation vector and subsequent P-element-mediated germ line transformation. The D-fos RNA interference (fosRNAi) gene was constructed by cloning a 976-bp-long fragment from D-fos cDNA in an inverted repeat manner into the pUAST vector. The cDNA fragment extends from positions +244 to +1220, with +1 referring to the ATG initiating codon. With the exception of the experiment in which adult wing size was measured in a kaysro-1 heterozygous mutant background (FI47), D-fos loss-of-function experiments employed a fly line carrying two copies of upstream activation sequence (UAS) fosRNAi on the third chromosome (FI35/19). The GMRGal4 fly line was a gift from M. Mlodzik. apterous Gal4 and scalloped Gal4 flies were provided by S. Cohen. The following fly lines were obtained from the Bloomington stock center: (i) T80Gal4; (ii) enGal4; (iii) eyelessGal4; (iv) hsFLP, Act5C>CD2>Gal4; (v) tubGal80TS; (vi) UAS p35; UAS CD8GFP; (vii) kaysro-1; and (viii) Oregon R.
Immunohistochemistry.
Immunostaining of wing and eye imaginal discs from third-instar larvae was performed as described by Sullivan et al. (27). The following antibodies were used: rabbit anti-D-Fos (diluted 1:1,000) (4), rabbit anti-cleaved caspase 3 (diluted 1:100; Cell Signaling), mouse anti-phospho histone 3 (diluted 1:50; Cell Signaling), rat anti-Elav (diluted 1:100; Hybridoma Bank), mouse anti-cyclin B (F2F4, diluted 1:20; Hybridoma Bank), and tetramethyl rhodamine isocyanate- and cyanine 5-conjugated secondary immunoglobulin G antibodies (Jackson ImmunoResearch Laboratories).
Clonal analysis.
Random flippase (FLP) recombinase target/FLP-induced clones were generated in wing imaginal discs as described in reference 19. Flies carrying hsFLP; Act5C>CD2>Gal4, UAS EGFP were crossed with either Oregon R or UAS fosRNAi flies. Eggs were collected for 4 h, and 60 h after eggs were laid, larvae were heat treated for 2 h at 37°C in a water bath. Wing discs were dissected and fixed 110 h after egg deposition, and enhanced green fluorescent protein (EGFP)-positive clones in 19 wing discs of each genotype were counted using a Leica TCS SP2 confocal microscope.
Flow cytometry.
Fly stocks carrying UAS fosRNAi or Oregon R flies were crossed with either enGal4, UAS EGFP flies or a line carrying T80Gal4, UAS EGFP; tubGal80TS. Cell dissociation and sorting of 18 wing discs per each genotype from third-instar larvae were carried out as described in reference 19. Experiments using a Gal4/Gal80 TARGET system (17) for the transient induction of fosRNAi were performed as follows. Eggs were collected, and larvae were raised at 22°C. Transgene expression was induced by incubation in a 37°C water bath for 1.5 h. After heat shock, larvae were kept at 27°C for 6 h before wing discs were dissected and dissociated into cell suspensions for flow cytometry (Becton Dickinson Vantage fluorescence-activated cell sorter, CellQuest software).
In situ hybridization.
The expression of cyclin B mRNA in eye imaginal discs from third-instar larvae was visualized using digoxigenin-labeled antisense mRNA probes as described in reference 28.
Chromatin immunoprecipitation.
Chromatin from continuously dividing S2 cells was collected according to the standard protocol of a commercial chromatin immunoprecipitation assay kit (Upstate). Rabbit anti-D-Fos antibody (4) was used for immunoprecipitation. Promoter regions with or without AP-1 sites of cyclin B, puckered (puc), and hsp26 were detected in the precipitated material by PCR using primers 5′-TTGTCGCGCACATGTTCTG-3′ and 5′-TTGTGGCTGCACGACGAA-3′ for cyclin B. Primers for puc and hsp26 were described by Lee et al. (14).
RESULTS
fosRNAi efficiently suppresses endogenous D-Fos expression in imaginal discs.
Larval imaginal discs of Drosophila melanogaster have become a favorite genetic system for studying the control of cell and organ growth in an intact tissue context. We decided to investigate the function of D-Fos in this system by using a loss-of-function approach. Existing alleles of D-fos are poorly characterized and are not well suited for this type of experiment. Some are too weak (as in the case of kay2), while the strongest known allele (kay1) is cell lethal and represents a deficiency that removes several genes (32). A number of recently reported fos mutants, such as those with shroud alleles (6), have intermediate phenotypes, but their mutations may affect only certain splice variants produced from the expansive fos locus. Dominant negative forms of D-Fos have been used but may affect other leucine zipper proteins, such as the Fos dimerization partner Jun, making the interpretation of their effects potentially difficult (1, 4). Thus, we chose to generate a specific loss-of-function situation for Fos by an RNAi-based strategy. The transgene lines used in our study direct the expression of an inverted repeat containing 976 nucleotides of the D-fos transcript (subsequently referred to as fosRNAi). The transcription of this product is inducible by the yeast transcription factor Gal4 and can thus be regulated in a tissue- and time-specific manner using appropriate driver lines. Semiquantitative reverse transcription (RT)-PCR analysis showed that endogenous D-fos mRNA levels were significantly decreased when fosRNAi was ubiquitously expressed in third-instar larvae (see Fig. S1A in the supplemental material). Further characterization of the fosRNAi showed that it is effective and specific: fosRNAi expression can reduce the endogenous levels of D-Fos protein in imaginal discs and recapitulate the described fos mutant phenotypes (see Fig. S1B and S1C in the supplemental material). Such phenotypes can be rescued by the co-overexpression of fos cDNA (see Fig. S1D to S1G in the supplemental material).
D-Fos is required for normal growth.
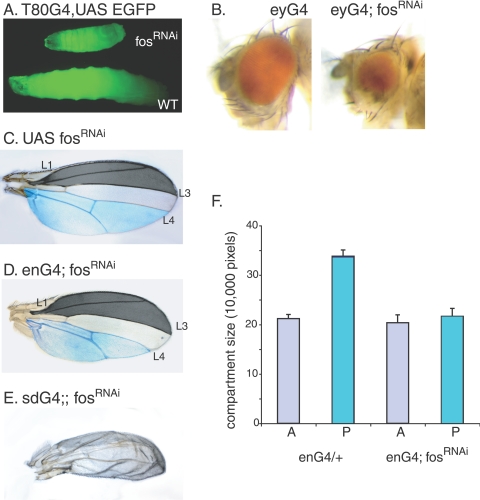
To investigate the function of D-Fos in growth control, we ubiquitously expressed fosRNAi under the direction of the T80Gal4 driver throughout the larval stages (29). The resulting larvae showed severely delayed overall growth and died at various stages of development, never surviving to adult flies (Fig. 1A).
FIG. 1.
Reduction of D-Fos activity interferes with normal growth. (A) Ubiquitous knockdown of D-fos mRNA during larval development results in smaller body size. Wild-type (WT) flies and flies carrying fosRNAi were crossed with flies expressing T80Gal4 and UAS EGFP. Eggs from each genotype were collected for 4 h, and larvae were raised on apple juice agar with yeast paste at 27°C. The image shows larvae of the same age. (B) Eye-specific expression of fosRNAi under the control of eyGal4 (eyG4) resulted in defective eye development with various levels of penetrance. Phenotypes range from a complete loss of the eye to an almost wild-type appearance. (C and D) The loss of D-fos in a tissue-specific manner caused a size decrease in the affected area. In the absence of a Gal4 driver, flies carrying UAS fosRNAi show no developmental defects. When fosRNAi was expressed in the wing imaginal disc under the control of the enGal4 (enG4) driver at 22°C, the emerging adult flies showed a reduced size of the posterior compartment where this driver is active (compare areas highlighted in blue). (E) When expression levels of D-Fos were suppressed throughout wing primordium development by the expression of fosRNAi under the control of sdGal4, the emerging adult wings became severely hypotrophic. (F) Compartment sizes of adult wings shown in panels C and D were measured by counting pixels in digital images by using Adobe Photoshop. Twenty adult wings per genotype were analyzed. The sections of the anterior compartment that were measured for the quantitative analyses are demarcated by L1 and L3 (marked in dark gray). The size of the area below vein L4, highlighted in blue, was calculated as a measure for posterior compartment size.
To examine the contribution of D-Fos to the development of adult tissues, we expressed fosRNAi in eye imaginal discs during larval development under the control of the eyeless Gal4 driver (eyGal4). In this experiment, eyGal4 directs high levels of RNAi expression from two UAS-linked transgenes encoding FosRNAi to ensure the effective suppression of fos mRNA in the entire eye imaginal disc starting in early larval development (8). This expression of fosRNAi resulted in rough eyes that are smaller than those of wild-type animals (Fig. 1B). The phenotype had various levels of penetrance and, in some cases, even entailed the complete ablation of the eye. At an elevated temperature (29°C), Gal4-driven transgenes are more highly expressed than at the standard cultivation temperature of 25°C, and in the genotype used above, D-fos mRNA levels should be more efficiently suppressed. Under such conditions, eye imaginal discs in the third-instar larvae remained extremely small and were very fragile (data not shown), suggesting that D-Fos is necessary for normal disc growth and development.
Growth defects were also manifested when D-Fos levels were reduced during wing development. The expression of fosRNAi (from two UAS-linked transgenes) in the posterior compartment of the larval wing imaginal disc under the control of enGal4 at 25°C resulted in complete pupal lethality. At a lower temperature, however, a small portion of pupae hatched to adult flies. The posterior wing compartment in such escapers was smaller than the corresponding area in control flies (Fig. 1C and D, highlighted in blue). Quantitative analysis of the compartment sizes revealed that the posterior part of wings from fly lines expressing fosRNAi became significantly smaller than that of controls, while the anterior compartment, in which enGal4 is not active and therefore fosRNAi was not expressed, did not show a change in size (Fig. 1F). When the levels of endogenous D-Fos were decreased in the entire wing anlage (using the scalloped Gal4 driver [sdGal4]), the resulting adult wing was small and malformed (Fig. 1 E). The observation that both ubiquitous and tissue-specific knock-down of D-Fos during larval development results in decreased organ size indicates a universal role for D-Fos in normal growth.
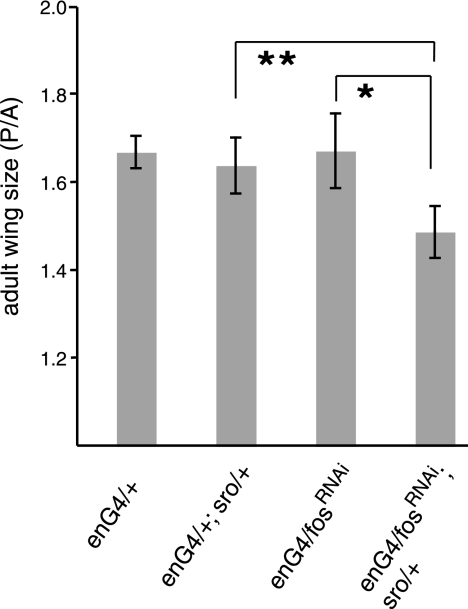
To validate the specificity of the RNAi approach, we combined UAS-driven fosRNAi expression with existing fos mutant alleles. At 22°C, neither the expression of a single copy of fosRNAi under the control of enGal4 nor heterozygosity for the recessive kaysro-1 allele (6) had a measurable effect on adult wing size (Fig. 2 and data not shown). However, when those two genotypes were combined and the weak RNAi for fos was expressed in a kaysro-1 heterozygous background, the posterior compartment of the adult wings became significantly smaller than that of control flies (Fig. 2). This finding indicates that the observed phenotype is caused by a loss of D-Fos function.
FIG. 2.
The Fos RNAi-mediated suppression of wing growth is enhanced by heterozygosity for a fos loss-of-function allele. A moderate reduction in D-fos by weak RNAi expression (using a single UAS fosRNAi transgene, FI47) under the control of enGal4 (enG4) causes no detectable change in wing size relative to that of controls. The same RNAi expression in a kaysro-1 heterozygous mutant background, however, results in a significant reduction in the size of the posterior compartment where enGal4 is active. Growth effects were quantitated by measuring the ratio between the sizes of the posterior and the anterior (P/A) compartments. Eighteen to 21 adult wings were analyzed for each genotype. *, P < 0.0002; **, P < 0.0002, two-tailed t test.
In what follows, we describe experiments to delineate whether the wing size decrease resulting from D-Fos suppression is a consequence of reduced cell growth or proliferation or whether it might be caused by apoptotic mechanisms.
The loss of D-Fos causes a growth disadvantage in proliferating tissues.
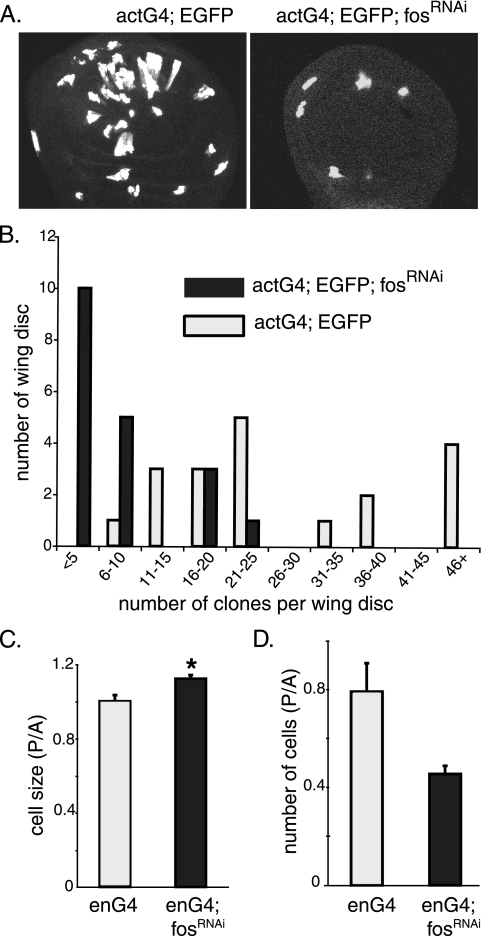
The wing imaginal disc in the early instar stages of larval development consists mainly of asynchronously dividing cells, which makes it a suitable system for clonal analysis of proliferating cell populations. We started to investigate the role of D-Fos in tissue/organ growth at the cellular level by generating random clones expressing fosRNAi in such discs. The number of clones that emerged when fosRNAi was expressed, along with EGFP, in the clonal tissue was much smaller than that of control clones which expressed EGFP alone (Fig. 3A and B). The low yield of clones expressing fosRNAi is consistent with inefficient growth and cell cycle progression followed by the resulting loss of the clonal tissue caused by cell competition with the surrounding wild-type cells.
FIG. 3.
Loss of D-Fos results in a growth disadvantage in proliferating tissue. (A and B) Random clones expressing EGFP alone (left panel) or fosRNAi along with EGFP (right panel) were generated in wing imaginal discs (see Materials and Methods), and the number of recovered clones per disc was counted. The expression of fosRNAi reduced the yield of clones significantly (panel A shows representative images, and panel B shows a quantification based on 19 wing discs from each genotype). (C) Cell sizes of wild-type and fosRNAi-expressing cells were measured and compared. fosRNAi was expressed together with EGFP in the posterior wing compartment under the control of enGal4 (enG4). Cell sizes were assessed by measuring FSC in a flow cytometry assay. The histogram displays the ratio of the FSC of posterior cells to that of anterior cells (P/A). Cells with reduced levels of D-Fos became bigger than control cells (*, P < 0.0003, two-tailed t test). (D) Posterior compartments in which fosRNAi was expressed contributed fewer cells to the wing imaginal disc than the corresponding area of control discs, in which only EGFP was expressed. Cell numbers were counted by flow cytometry.
Next, we carried out flow cytometry experiments to investigate whether D-Fos might affect cell growth (i.e., the acquisition of cell mass) or cell division, two processes that can be separately controlled (20). Thus, we expressed EGFP in the posterior part of the wing imaginal disc with or without fosRNAi using enGal4. The EGFP-negative anterior compartment served as an internal control. Wing discs of the respective genotypes were dissected from third-instar larvae and dissociated into cell suspensions by trypsin digestion. Cell numbers, sizes, and cell cycle profiles of EGFP-positive and -negative cell populations were then determined using flow cytometry. D-fos-deficient cells were consistently larger than control cells, as shown by a comparison of graphs of the forward light scatter (FSC) of cells of the posterior and the anterior compartment (Fig. 3C). This result indicates that D-Fos might not be directly required for cell growth in proliferating cells. Instead, it appears that cells lacking Fos are compromised in cell cycle progression and continue to grow when wild-type cells would already have undergone mitosis. Consistent with this interpretation, the suppression of D-Fos activity caused a decreased compartment size (Fig. 1F) and a reduction in the total cell number in the compartment (Fig. 3D). Taken together, these data indicate that D-Fos is required for efficient cell proliferation but is not limiting for cell growth.
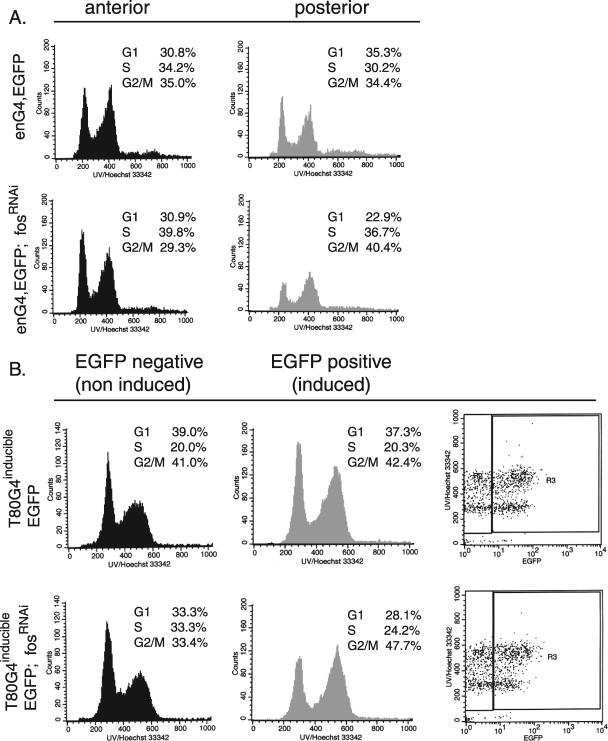
To investigate further whether D-Fos might have a function in the regulation of cell cycle progression, as suggested based on the findings presented above, we asked whether cells devoid of D-Fos would display a change in cell cycle profile relative to that of control cells. First, we suppressed D-Fos in the posterior compartments of wing imaginal discs using enGal4-driven fosRNAi expression and monitored the effects on cell cycle distribution (Fig. 4A). Cells of the anterior compartments in which no RNAi was expressed served as an internal control. The comparative analysis of EGFP-positive and -negative cell populations revealed that the reduction of D-Fos increased the fraction of cells at the G2/M phase of the cell cycle, whereas the portion of cells in G1 was reduced relative to the same fraction in the wild-type control. This finding supports the notion that the lack of D-fos causes an accumulation of cells at or around the G2-to-M transition.
FIG. 4.
D-Fos is required for cell cycle progression. (A) The expression of fosRNAi and EGFP was induced in the posterior compartment of wing imaginal discs under the control of the enGal4 (eng4) driver. Dissected wing discs were dissociated into cell suspensions and subjected to flow cytometry. Histograms show the DNA contents of EGFP-negative and -positive cells derived from either compartment. Cells lacking the D-Fos function accumulated in the G2/M phase of the cell cycle to a higher level than that in the control cells. (B) fosRNAi expression was transiently induced throughout the third instar larval stage by using the T80Gal4/tubGal80TS TARGET system. The top two panels show the results of a control experiment with discs expressing EGFP alone. The histograms show DNA contents and cell cycle profiles in the wing disc cells 6 h after transgene induction. Transgene activity was monitored by EGFP expression, shown in the far-right plots. After the transient reduction of D-fos, cells in G2/M were more prevalent than in the control. The quantitation is based on at least three independent experiments for each genotype.
In the experiment shown in Fig. 4A, fosRNAi was expressed throughout larval development. To rule out the possibility that the resulting phenotypes might be an indirect consequence of this long-term suppression of D-Fos activity, we used an inducible TARGET system to transiently express fosRNAi in third-instar larvae wing discs (17). The activity of the inducible system was monitored by EGFP expression. The cell cycle profile recorded at 6 h after the induction of fosRNAi expression was consistent with the results of the previous experiment measuring the effects of long-term suppression of D-fos. When D-Fos function was thus restricted for a short period of time only, the fraction of G1/S cells decreased, while more cells accumulated in the G2/M phase of the cell cycle (Fig. 4B, lower panels). Cells of the EGFP-negative (Fig. 4B, left [noninduced]panels) population in the same wing discs served as internal controls in this experiment. Cell cycle profiles from wing discs in which no fosRNAi was expressed did not display changes of cell cycle distribution (Fig. 4B, upper panel). The accumulation of Fos-depleted cells in the G2/M pool is consistent with a delay in mitosis and with the bigger average size of D-Fos-depleted cells, as measured by forward scatter (Fig. 3C).
The loss of Fos results in increased apoptosis.
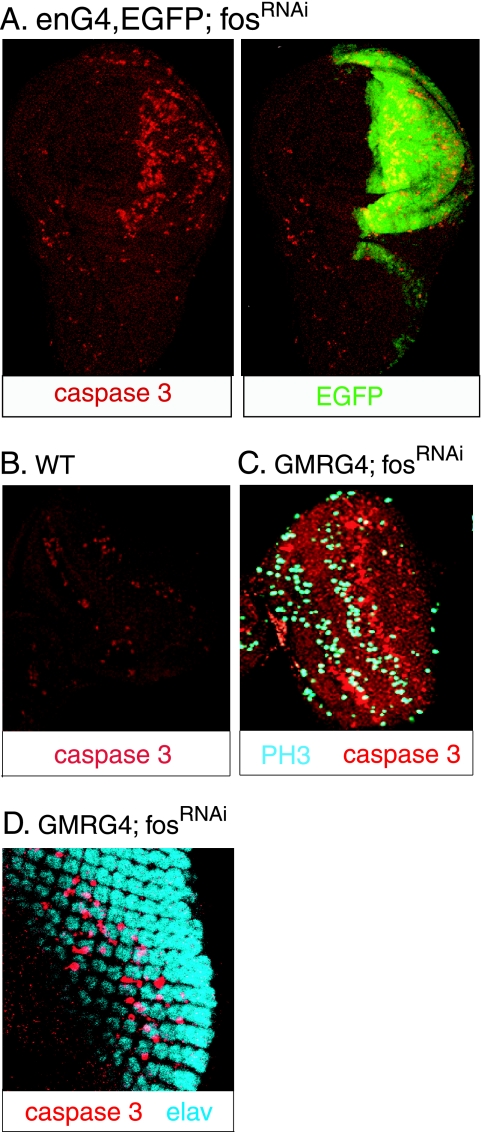
The results described above indicate that D-Fos-deficient cells are compromised in their ability to progress through the cell cycle. Such a defect may be expected to lead to mitotic problems and the elimination of the affected cells by apoptosis to avoid the accumulation of damaged or dysfunctional cells. We examined whether the loss of D-Fos might have such an effect in vivo. When D-Fos levels were reduced by the expression of fosRNAi in the posterior compartments of the wing imaginal discs, many cells in the affected area underwent apoptosis (Fig. 5A). Similarly, we found that D-Fos deficiency can also cause apoptosis in the developing eye. D-Fos function was suppressed in the eye imaginal disc by expressing fosRNAi under the control of the GMRGal4 driver. This driver directs transgene expression to the posterior part of the eye disc, which includes cells partaking in the second mitotic wave (SMW). The SMW follows the morphogenetic furrow and sweeps across the eye imaginal-disc epithelium in a posterior-to-anterior direction. In the SMW, undifferentiated cells go through one last synchronized cell cycle before cell division ceases altogether and the adult eye is formed by a subsequent series of cell recruitment and differentiation steps. Eye discs expressing fosRNAi under the control of GMRGal4 showed a distinctive stripe of apoptotic cell death immediately posterior to the SMW compared to the wild-type eye discs (Fig. 5B and C). Apoptosis occurred in nonneuronal cells, suggesting that the cells that are affected are the same cells that had, shortly before, initiated the SMW (Fig. 5D). The defined location of the stripe of apoptotic cells in the wake of the SMW suggests that cell death occurs after the initiation of mitosis and might be a response to a defect in mitotic progression.
FIG. 5.
Loss of D-Fos in proliferating cells causes apoptosis. (A) D-Fos expression was reduced in the posterior compartment of wing imaginal discs by enGal4 (enG4)-driven fosRNAi expression, and apoptotic cells were visualized by immunostaining with anti-activated caspase 3 antibody (shown in red). Many apoptotic cells were detected in the enGal4 domain (marked by EGFP expression [green]) but not in the anterior compartment, in which no fosRNAi was expressed. (B) In wild-type (WT) third-instar eye imaginal discs, apoptotic cells are scarce. (C and D) The expression of fosRNAi under the control of GMRGal4 (GMRG4) in the posterior eye imaginal disc, encompassing differentiating cells and the cells of the second mitotic wave (marked by anti-phospho histone 3 [PH3] staining [blue]), caused massive cell death in nonneuronal cells (Elav negative [Elav is in blue]). A distinct row of apoptotic cells emerges at a short distance behind the second mitotic wave.
When the SMW sweeps across the eye imaginal disc, it produces enough cells to complete the ommatidial clusters, which also include nonneuronal accessory cells like pigment cells. The experiments described above suggest that the depletion of D-Fos interferes with the SMW so that insufficient numbers of precursor cells are generated for the differentiation of the full complement of accessory cells. In support of this hypothesis, eyes emerging from larvae expressing fosRNAi under the control of GMRGal4 are distinctly smaller than wild-type eyes (see Fig. S1D and S1F in the supplemental material). The analysis of sections prepared from adult eyes supports this interpretation further. Eyes developing under loss-of-Fos conditions (expressing fosRNAi under the control of GMRGal4) lose the regular ommatidial pattern observed in the wild type. Many ommatidia are fused, a phenotype that is caused by the distinct shortage of pigment cells (Fig. 6A and C).
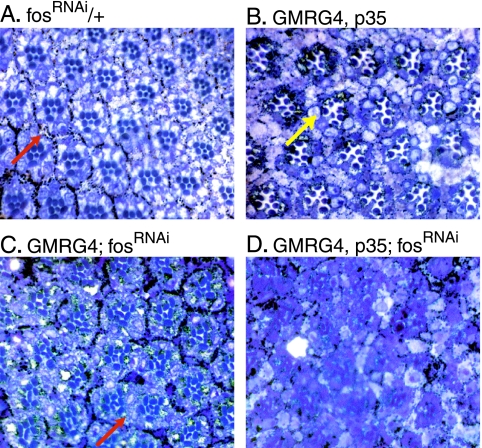
FIG. 6.
Prevention of apoptosis enhances the fosRNAi-mediated adult eye phenotype. (A) Tangential sections of wild-type adult eyes (UAS fosRNAi is not active due to the absence of a Gal4 driver) display a normal, stereotyped hexagonal pattern throughout the section. Note that pigment cells (red arrow, pigment granules appear black) completely surround each ommatidium. (B) Ectopic expression of p35 results in increased space between ommatidia due to the presence of superfluous cells (yellow arrow) which were not eliminated as a part of normal eye development. (C) When D-Fos expression is reduced, many ommatidia become fused as insufficient numbers of pigment cells are present to separate the ommatidia (red arrow). (D) Coexpression of p35 and fosRNAi under the control of GMRGal4 (GMRG4) greatly exacerbates the disturbance in the ommatidial structure observed in panel C.
The lack of pigment cells observed in GMRGal4; fosRNAi eyes is consistent with a failed second mitotic wave and, as a consequence of that, the loss of precursor cells. To test this interpretation and to rule out the possibility that Fos acts as a general survival factor in this situation, fosRNAi was coexpressed with baculovirus p35 (an inhibitor of apoptosis) in the eye imaginal disc, and the resulting adult eye structures were analyzed. The coexpression of p35 did not rescue the defect caused by the loss of Fos (Fig. 6D). Rather, the phenotype became much more severe, and the ommatidial organization was completely disrupted. We conclude that the apoptosis observed in eye discs in which Fos function is reduced is a secondary consequence of a cell cycle problem. The apoptotic removal of cells thus affected evidently prevents a more serious phenotype that might be caused by the presence of cells with mitotic aberrations.
D-Fos regulates cyclin B expression.
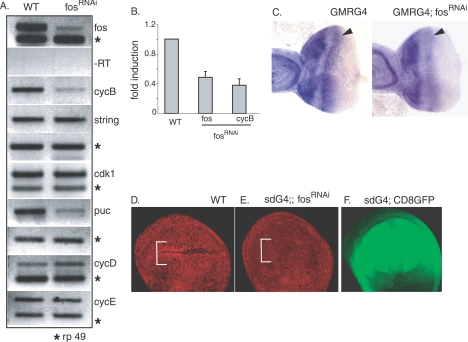
The cell cycle defect observed under conditions of depleted Fos function prompted us to investigate whether D-Fos might transcriptionally regulate genes associated with cell cycle control. We transiently induced the expression of fosRNAi in third-instar larvae and prepared cDNA from total larval lysates. Among several cell cycle genes tested for induction by semiquantitative RT-PCR, we found that the expression of cyclin B was reproducibly and substantially reduced (Fig. 7A). Cyclin B is the regulatory subunit of the cyclin B/CDK1 complex, one of the principal regulators of the G2-to-M transition of the mitotic cell cycle. The string gene, the Drosophila homolog of cdc25, and the cdk1 gene are required for progression through mitosis and for mitotic exit. The expression levels of those genes were not affected by the transient depletion of D-fos. Conversely, the puc gene, an established D-Fos target, was considerably down-regulated, providing a positive control for this experiment. Experiments using quantitative real time RT-PCR confirmed the conclusions of the conventional RT-PCR analysis and showed a significant suppression of cyclin B mRNA levels under conditions of reduced D-Fos activity (Fig. 7B). The Fos dependence of cyclin B expression was further confirmed by in situ hybridization and immunostaining. We directed fosRNAi expression in a third-instar eye imaginal disc under the control of GMRGal4 and monitored cyclin B mRNA levels. Compared to what occurred in the control eye disc, the cyclin B signal in the region of the SMW was significantly reduced when fosRNAi was expressed in the eye disc (Fig. 7C).
FIG. 7.
D-Fos regulates cyclin B expression. (A) fosRNAi was ubiquitously expressed in third-instar larvae using the T80Gal4/TubGal80TS TARGET system. Seventeen hours after RNAi induction, total mRNA from third-instar larval lysates was prepared for analysis by semiquantitative RT-PCR (RT). The transient reduction of D-fos levels decreased cyclin B (cycB) mRNA expression but did not affect other genes encoding cell cycle regulators, including string, cdk1, cyclin D (cycD), and cyclin E (cycE). Expression levels of puc (a known D-Fos target gene) were determined as a positive control for the efficiency of the D-fos knockdown. WT, wild type; rp 49, ribosomal protein 49. (B) Real-time RT-PCR was conducted to quantitatively measure the reduction of cyclin B mRNA levels when the expression of D-fos was perturbed, using the same mRNA source as in panel A. (C) In situ hybridization with antisense cyclin B mRNA probe was performed on third-instar eye imaginal discs. The GMRGal4 (GMRG4) driver directed the expression of fosRNAi in the posterior part of an eye imaginal disc, including the SMW (arrowheads). Compared to that in the control eye disc and anterior part of the same disc, the intensity of cyclin B expression levels was significantly decreased in the SMW in the eye disc where D-fos expression was reduced. (D and E) The discernible accumulation of cyclin B protein in the ZNC (bracket) in the anterior compartment of wild-type late third-instar wing imaginal disc (D) was abolished when D-fos expression was knocked down by the expression of fosRNAi under the control of sdGal4 (E). An anti-cyclin B antibody was used to detect cyclin B protein levels. (F) The expression domain of sdGal4 is visualized by means of UAS-linked CD8 GFP expression. Anterior is to the left in all imaginal-disc figures.
Toward the end of the third instar stage, a subset of cells along the dorsoventral boundary in the wing imaginal disc became arrested in the G1 or G2 stage of the cell cycle and formed a distinct “zone of nonproliferating cells” (ZNC) (11). As opposed to the G1-arrested cells in the ZNC of the posterior compartment, a group of ZNC cells in the anterior compartment of the wing disc arrest at the G2 phase and are characterized by a pronounced cyclin B accumulation (Fig. 7D). When D-Fos expression was disrupted in the wing disc by sdGal4-driven fosRNAi, the elevated levels of cyclin B expression in the anterior ZNC were no longer detectable at the protein level, as shown by anti-cyclin B antibody staining of the affected area (Fig. 7E). Insufficient levels of cyclin B expression, as discerned in the above-described experiments, provide an explanation for the G2/M-specific cell cycle defect observed as a consequence of reduced D-Fos function.
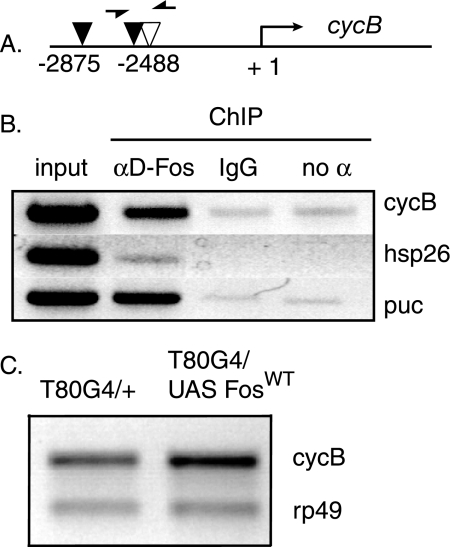
After establishing a role for D-Fos in the expression of cyclin B in mitotically active tissues, we wondered whether this might reflect a direct transcriptional regulation of the cyclin B gene by Drosophila Fos. To address this question, we performed chromatin immunoprecipitation experiments. We searched the promoter regions of the cyclin B gene and identified two perfect AP-1 binding sites and one site with a single nucleotide mismatch within 3 kb of the translation start site (Fig. 8A). Chromatin immunoprecipitation was conducted in S2 cells using an anti-D-Fos antibody to detect binding of endogenous D-Fos protein to the putative response elements in the cyclin B locus. PCR amplification of the precipitated DNA confirms that D-Fos binds to the promoter region of cyclin B (Fig. 8B) but not to the promoter region of hsp26, which served as a negative control for anti-D-Fos antibody (14). puc, as an established Fos target gene (15), was used as a positive control.
FIG. 8.
D-Fos binds to the promoter region of the cyclin B (cycB) gene and induces its expression in vivo. (A) Schematic view of putative AP-1 sites in the promoter region of the Drosophila cyclin B gene. The +1 denotes the translation start site. Red arrowheads mark two perfect AP-1 sites (TGACTCA), and the green arrowhead represents an imperfect AP-1 site with one nucleotide mismatch (TGATTCA). Arrows indicate the positions of the DNA primers used for PCR. (B) Chromatin immunoprecipitation (ChIP) was performed with chromatin extracted from S2 cells as described in Materials and Methods. PCR amplification shows that D-Fos binds to the AP-1 sites in the promoter region of the cyclin B gene. puc was used as a positive control for the binding of anti-D-Fos (αD-Fos) antibody to regions containing known AP-1 sites, and hsp26 was used as a negative control. IgG, immunoglobulin G; no α, no antibody. (C) Ectopic D-Fos expression increased endogenous cyclin B mRNA levels. Wild-type D-Fos (FosWT) was overexpressed under the control of T80Gal4 (T80G4) throughout larval development at 29°C, and mRNA from third-instar total larval lysate was prepared for semiquantitative RT-PCR analysis. Endogenous cyclin B mRNA levels were increased with wild-type D-Fos. rp49, ribosomal protein 49.
To explore whether Fos is not only necessary but also sufficient for cyclin B expression, we tested whether the overexpression of D-Fos would induce an increase in cyclin B transcript levels. Ubiquitous D-Fos expression was directed under the control of T80Gal4, and mRNA from total larval lysates was used to quantify endogenous cyclin B mRNA levels. Overexpression of D-Fos caused moderately but significantly increased levels of cyclin B mRNA, as detected by RT-PCR (Fig. 8C). Taken together, these findings indicate that D-Fos positively regulates cyclin B at the transcriptional level and provides a plausible mechanism by which the transcription factor could promote the G2/M transition of mitosis.
DISCUSSION
Since its discovery more than 20 years ago (5), Fos has been suggested to regulate cell cycle progression and/or cell growth. Definitive experiments to confirm this function and to elucidate molecular mechanisms by which Fos might promote cell proliferation have, in spite of a large body of research on the topic, been scarce. This is explained by a number of experimental impediments and the biological complexity of Fos function, especially in vertebrates.
We have conducted a loss-of-function analysis relying mostly on RNAi technology in the Drosophila imaginal-disc system to dissect the function of Fos in the growth control of an intact developing tissue. The data obtained in these studies suggest the following model. In continuously cycling cells, D-Fos is required for the propagation of the cell cycle. If D-Fos function is reduced, components that are limiting for the successful transition from the G2 phase to mitosis are not supplied in sufficient amounts. Consequently, cells cannot leave G2/M, which causes the accumulation of cells of 4N DNA content and an overall increase of cell size, as determined by forward scatter measurements. Such a block of cell cycle function in primary cells is expected to cause the activation of checkpoints leading ultimately to the apoptotic removal of the affected cells. The conclusion that the observed increased frequency of cell death results from defects in the cell cycle is supported by experiments with eye imaginal discs. The removal of D-Fos function from cells slated for transit through a developmentally defined mitotic wave results in apoptosis at a relatively sharp time point after cell division would normally have been completed. This indicates that apoptosis is a consequence of rather than a cause for the reported cell cycle defects. Analysis of eye discs in which Fos function has been depleted but apoptosis is inhibited by the expression of p35 supports this view: in such a genotype, defects of eye development are enhanced rather than suppressed, indicating that Fos is not required just for survival signaling. The identification of cyclin B as a transcriptional target of D-Fos in imaginal discs offers a molecular explanation for the G2/M phenotype observed under D-Fos loss-of-function conditions.
It is possible that Fos has functions in other stages of the cell cycle that do not become phenotypically apparent at the levels of Fos suppression achieved by the RNAi-based approach employed in our studies. It has, for example, been suggested that Fos has a function during the G1/S transition and regulates cyclin D transcription (2, 3). It is important to note, however, that, in contrast to our studies of continuously growing imaginal-disc cells, these experiments were conducted on cultured cells that entered the cell cycle from a quiescent state upon serum stimulation. For this G0-to-G1 transition, the de novo synthesis of cyclin D can be expected to be limiting and require higher levels of Fos activity than in asynchronously cycling cells.
The studies presented here show that Fos can control specific aspects of cell cycle progression, at least in Drosophila imaginal-disc cells. This observation, if it can be extended to higher organisms, might explain the oncogenic activities of Fos proteins. However, it is important to keep in mind that Fos is a protein with complex and pleiotropic functions that can interact with multiple other transcription factors and signaling pathways. Thus, efforts to unravel the contributions of Fos proteins to cancer and other pathologies will have to consider this complexity and integrate the contribution of Fos to processes other than growth control, such as the differentiation and control of cell mobility.
Supplementary Material
Acknowledgments
We thank Heinrich Jasper, Jiyong Zhao, Hucky Land, Mirka Uhlirova, and Gerasimos Sykiotis for thoughtful comments on the manuscript and Chris Sommers for the generation of transgenic animals. We are grateful to Jean-Antoine Lepesant for initiating the collaboration between the Paris and Rochester labs and to Ursula Weber for communicating unpublished information about fos alleles. Peter Keng and Tara Calcagni are acknowledged for their assistance in FACS analysis.
This work was supported by NIH grant R01 EY014624-01 to D.B.
Footnotes
Published ahead of print on 11 September 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bohmann, D., M. C. Ellis, L. M. Staszewski, and M. Mlodzik. 1994. Drosophila Jun mediates Ras-dependent photoreceptor determination. Cell 78:973-986. [DOI] [PubMed] [Google Scholar]
- 2.Brown, J. R., E. Nigh, R. J. Lee, H. Ye, M. A. Thompson, F. Saudou, R. G. Pestell, and M. E. Greenberg. 1998. Fos family members induce cell cycle entry by activating cyclin D1. Mol. Cell. Biol. 18:5609-5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burch, P. M., Z. Yuan, A. Loonen, and N. H. Heintz. 2004. An extracellular signal-regulated kinase 1- and 2-dependent program of chromatin trafficking of c-Fos and Fra-1 is required for cyclin D1 expression during cell cycle reentry. Mol. Cell. Biol. 24:4696-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciapponi, L., D. B. Jackson, M. Mlodzik, and D. Bohmann. 2001. Drosophila Fos mediates ERK and JNK signals via distinct phosphorylation sites. Genes Dev. 15:1540-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curran, T., and N. M. Teich. 1982. Candidate product of the FBJ murine osteosarcoma virus oncogene: characterisation of a 55,000 kDa phosphoprotein. Virology 116:221-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giesen, K., U. Lammel, D. Langehans, K. Krukkert, I. Bunse, and C. Klämbt. 2003. Regulation of glial cell number and differentiation by ecdysone and Fos signaling. Mech. Dev. 120:401-413. [DOI] [PubMed] [Google Scholar]
- 7.Grigoriadis, A. E., K. Schellander, Z. Q. Wang, and E. F. Wagner. 1993. Osteoblasts are target cells for transformation in c-fos transgenic mice. J. Cell Biol. 122:685-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazelett, D. J., M. Bourouis, U. Walldorf, and J. E. Treisman. 1998. decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development 125:3741-3751. [DOI] [PubMed] [Google Scholar]
- 9.Hess, J., P. Angel, and M. Schorpp-Kistner. 2004. AP-1 subunits: quarrel and harmony among siblings. J. Cell Sci. 117:5965-5973. [DOI] [PubMed] [Google Scholar]
- 10.Jochum, W., E. Passegue, and E. F. Wagner. 2001. AP-1 in mouse development and tumorigenesis. Oncogene 20:2401-2412. [DOI] [PubMed] [Google Scholar]
- 11.Johnston, L. A., and B. A. Edgar. 1998. Wingless and Notch regulate cell-cycle arrest in the developing Drosophila wing. Nature 394:82-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kockel, L., J. G. Homsy, and D. Bohmann. 2001. Drosophila AP-1: lessons from an invertebrate. Oncogene 20:2347-2364. [DOI] [PubMed] [Google Scholar]
- 13.Kovary, K., and R. Bravo. 1991. The Jun and Fos protein families are both required for cell cycle progression in fibroblasts. Mol. Cell. Biol. 11:4466-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, N., C. Maurange, L. Ringrose, and R. Paro. 2005. Suppression of Polycomb group proteins by JNK signalling induces transdetermination in Drosophila imaginal discs. Nature 438:234-237. [DOI] [PubMed] [Google Scholar]
- 15.Martin-Blanco, E., A. Gampel, J. Ring, K. Virdee, N. Kirov, A. M. Tolkovsky, and A. Martinez-Arias. 1998. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 12:557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin-Blanco, E., J. C. Pastor-Pareja, and A. Garcia-Bellido. 2000. JNK and decapentaplegic signaling control adhesiveness and cytoskeleton dynamics during thorax closure in Drosophila. Proc. Natl. Acad. Sci. USA 97:7888-7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGuire, S. E., P. T. Le, A. J. Osborn, K. Matsumoto, and R. L. Davis. 2003. Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302:1765-1768. [DOI] [PubMed] [Google Scholar]
- 18.Müller, R., R. Bravo, J. Burckhardt, and T. Curran. 1984. Induction of c-fos gene and protein by growth factors precedes activation of c-myc. Nature 312:716-720. [DOI] [PubMed] [Google Scholar]
- 19.Neufeld, T. P., A. F. de la Cruz, L. A. Johnston, and B. A. Edgar. 1998. Coordination of growth and cell division in the Drosophila wing. Cell 93:1183-1193. [DOI] [PubMed] [Google Scholar]
- 20.Neufeld, T. P., and B. A. Edgar. 1998. Connections between growth and the cell cycle. Curr. Opin. Cell Biol. 10:784-790. [DOI] [PubMed] [Google Scholar]
- 21.Papachristou, D. J., A. Batistatou, G. P. Sykiotis, I. Varakis, and A. G. Papavassiliou. 2003. Activation of the JNK-AP-1 signal transduction pathway is associated with pathogenesis and progression of human osteosarcomas. Bone 32:364-371. [DOI] [PubMed] [Google Scholar]
- 22.Riese, J., G. Tremml, and M. Bienz. 1997. D-Fos, a target gene of Decapentaplegic signaling required during Drosophila endoderm induction. Development 124:3353-3361. [DOI] [PubMed] [Google Scholar]
- 23.Riesgo-Escovar, J. R., and E. Hafen. 1997. Common and distinct roles of DFos and DJun during Drosophila development. Science 278:669-672. [DOI] [PubMed] [Google Scholar]
- 24.Rüther, U., C. Garber, D. Komitowski, R. Muller, and E. F. Wagner. 1987. Deregulated c-fos expression interferes with normal bone development in transgenic mice. Nature 325:412-416. [DOI] [PubMed] [Google Scholar]
- 25.Rüther, U., D. Komitowski, F. R. Schubert, and E. F. Wagner. 1989. c-fos expression induces bone tumors in transgenic mice. Oncogene 4:861-865. [PubMed] [Google Scholar]
- 26.Shaulian, E., and M. Karin. 2001. AP-1 in cell proliferation and survival. Oncogene 20:2390-2400. [DOI] [PubMed] [Google Scholar]
- 27.Sullivan, W., M. Ashburner, and R. S. Hawley. 2000. Drosophila protocols. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Tautz, D., and C. Pfeifle. 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98:81-85. [DOI] [PubMed] [Google Scholar]
- 29.Wilder, E. L., and N. Perrimon. 1995. Dual functions of wingless in the Drosophila leg imaginal disc. Development 121:477-488. [DOI] [PubMed] [Google Scholar]
- 30.Wisdom, R. 1999. AP-1: one switch for many signals. Exp. Cell Res. 253:180-185. [DOI] [PubMed] [Google Scholar]
- 31.Zeitlinger, J., and D. Bohmann. 1999. Thorax closure in Drosophila: involvement of Fos and the JNK pathway. Development 126:3947-3956. [DOI] [PubMed] [Google Scholar]
- 32.Zeitlinger, J., L. Kockel, F. A. Peverali, D. B. Jackson, M. Mlodzik, and D. Bohmann. 1997. Defective dorsal closure and loss of epidermal decapentaplegic expression in Drosophila fos mutants. EMBO J. 16:7393-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.