Abstract
We recently identified polynucleotide phosphorylase (PNPase) as a potential binding partner for the TCL1 oncoprotein. Mammalian PNPase exhibits exoribonuclease and poly(A) polymerase activities, and PNPase overexpression inhibits cell growth, induces apoptosis, and stimulates proinflammatory cytokine production. A physiologic connection for these anticancer effects and overexpression is difficult to reconcile with the presumed mitochondrial matrix localization for endogenous PNPase, prompting this study. Here we show that basal and interferon-β-induced PNPase was efficiently imported into energized mitochondria with coupled processing of the N-terminal targeting sequence. Once imported, PNPase localized to the intermembrane space (IMS) as a peripheral membrane protein in a multimeric complex. Apoptotic stimuli caused PNPase mobilization following cytochrome c release, which supported an IMS localization and provided a potential route for interactions with cytosolic TCL1. Consistent with its IMS localization, PNPase knockdown with RNA interference did not affect mitochondrial RNA levels. However, PNPase reduction impaired mitochondrial electrochemical membrane potential, decreased respiratory chain activity, and was correlated with altered mitochondrial morphology. This resulted in FoF1-ATP synthase instability, impaired ATP generation, lactate accumulation, and AMP kinase phosphorylation with reduced cell proliferation. Combined, the data demonstrate an unexpected IMS localization and a key role for PNPase in maintaining mitochondrial homeostasis.
TCL1 is a 14-kDa nonenzymatic oncoprotein whose mechanism of action in promoting B- and T-cell malignancies is not resolved (reviewed in reference 46). To identify potential TCL1-interacting proteins, TCL1 immunoprecipitation followed by mass spectrometry analysis from human pre-B cells was performed, and polynucleotide phosphorylase (PNPase) was identified as a potential binding partner (10). Immediately, our focus shifted to studies of PNPase to determine if this interaction was possible, because TCL1 was reportedly excluded from mitochondria under standard growth conditions (11), whereas PNPase is a nucleus-encoded mitochondrial exoribonuclease (35).
PNPases comprise an evolutionarily conserved enzyme family whose members regulate RNA levels in bacteria and plant chloroplasts. In Escherichia coli, PNPase associates with an RNA helicase and enolase in a degradosome complex, where it functions as an exoribonuclease and poly(A) polymerase to control RNA stability (32). In plants, PNPase localizes to the chloroplast stroma, where it functions as an exoribonuclease and poly(A) polymerase to mediate chloroplast RNA turnover (3, 29, 30, 53). In both bacteria and plants, poly(A) addition to RNA provides a signal for PNPase-mediated RNA degradation.
Recently, mammalian PNPase was identified as an up-regulated gene in melanoma cells induced to terminally differentiate and in senescent progeroid fibroblasts, potentially implicating increased PNPase in growth inhibition (26). PNPase is an 85-kDa protein that functions in vitro as a phosphate-dependent exoribonuclease (10, 26, 41) and poly(A) polymerase (33). By analogy with bacterial and chloroplastic PNPases, mammalian PNPase could be expected to regulate RNA levels in the mitochondrial matrix. However, robust PNPase overexpression inhibits cell growth (26) and induces apoptosis with down-regulation of MYC and BCL-xL expression (41), suggesting that PNPase may function outside the mitochondria to regulate transcript levels. Clearly, these observations deviate from a predicted role for PNPase in regulating mitochondrial RNA (mtRNA) levels. A possible explanation for this disparity includes aberrant localization of exogenous PNPase to nonmitochondrial compartments. Alternatively, PNPase might reside within or outside mitochondria, depending on cellular conditions, with a precedent for this notion provided by proteins involved in regulating apoptosis that include cytochrome c (cyto c), apoptosis-inducing factor (AIF), Smac/DIABLO, HtrA2/Omi, Bit1, and endonuclease G (8, 20, 28, 43, 50, 51). In the case of cytochrome c, for example, the intermembrane space (IMS) provides a location for its mitochondrial activity in respiration, whereas conditions that cause outer membrane (OM) permeability foster its release for cytosolic activity during apoptosis. A third possibility is that PNPase regulates growth and survival from within mitochondria, in addition to or instead of regulating nonmitochondrial MYC and BCL-xL expression levels. In fact, marked PNPase overexpression results in increased reactive oxygen species (ROS) production, which increases NF-κB activation and the expression of NF-κB target proinflammatory cytokine genes (40). Increased ROS production suggests a potential role for PNPase in respiration, since most ROS is produced within the mitochondria during oxidative phosphorylation (OX-PHOS) (1, 9).
To survive and grow, cancer cells employ strategies to evade cell death, terminal differentiation, and replicative senescence, which normally hold nonmalignant cells in check (18). Overexpressed PNPase seems to provide antigrowth, proapoptotic, and immune-activating functions that would be predicted to oppose cancerous transformation (26, 40, 41). However, these findings may or may not represent the function(s) of endogenously localized and physiologically expressed PNPase within cells. This distinction may be critical and, therefore, the current study aimed to determine the localization and role of endogenous PNPase in order to further understand its function and potential as an interacting partner for the TCL1 oncoprotein.
MATERIALS AND METHODS
Cells and Abs.
Cells were maintained in growth medium supplemented with 10% fetal bovine serum. Antibodies (Abs) used were as follows: phycoerythrin (PE)-conjugated anti-rabbit immunoglobulin G (IgG) and horseradish peroxidase-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch Labs); horseradish peroxidase-conjugated horse anti-mouse IgG, AMP kinase alpha (AMPKα), and phospho-AMPKα (Cell Signaling Technology); cytochrome c (BD Biosciences Pharmingen); myc-tag (Santa Cruz); OX-PHOS complex IV subunit II (Cox II) and complex V subunit α (Molecular Probes); and β-actin and β-tubulin (Sigma). Timm13 and Hsp60 Abs were generated as described previously (39). Rhodanese antiserum was generated in rabbits from injected bovine rhodanese (Sigma). ADP/ATP carrier (AAC), cytochrome b2, and α-ketoglutarate dehydrogenase (KDH) antisera were generated in rabbits from yeast (Saccharomyces cerevisiae)-purified proteins. Rabbits were vaccinated with keyhole limpet hemocyanin-linked acetylated-CGRDPADGRMRLSRKVLQA-amide to generate human PNPase antiserum and a peptide-purified Ab (Biosource International).
Plasmids and retroviruses.
Human PNPase cDNA was cloned by reverse transcription-PCR from a Nalm-6 pre-B-cell cDNA library with 5′-ccggtattatccATGGCGGCCTGCAGGTAC-3′ and 5′-ccgcacgtcgacTCACTGAGAATTAGATG-3′ primers (lowercase indicates linker sequence for cloning). PNPase was subcloned into pSP65 (Promega) for transcription/translation and into pcDNA3.1myc-His (Invitrogen) and pCMV-tag (Stratagene) myc-tagged expression vectors. For yeast expression, PNPase was subcloned into pYEX-BX (2μm ori, URA3; Clontech), and induction was controlled by the copper-inducible CUP1 promoter. The pYEX-BX-PNPase vector was transformed into GA74-1A yeast (25), and mitochondria and postmitochondrial supernatant were purified as described previously (24). PNPase RNA interference (RNAi) oligonucleotide hairpins were generated with DNA oligomers GATCCCCGAGAGGTTGGTACTTCTGATTCAAGAGATCAGAAGTACCAACCTCTCTtttttggaaac and tcgagtttccaaaaaAGAGAGGTTGGTACTTCTGATCTCTTGAATCAGAAGTACCAACCTCTCggg (gi 31657165). This sequence was cloned into pQSUPER, an H1 promoter-driven RNAi retroviral vector derived from pQCXIP that coexpresses green fluorescent protein (GFP) (a kind gift of B. Cobb, C. Li, and S. Smale). The parent vector, a scramble RNAi sequence, and a GFP-specific RNAi construct were used as controls. GFP-encoding sequences were removed from these vectors for experiments in which the electrochemical membrane potential (Δψ) was measured, and MitoTracker straining was used. For PNPase knockdown, HEK293 cells were transfected with 12 μg RNAi construct and 4 μg pHIT60 and 2 μg vesicular stomatitis virus G retroviral packaging vectors (kind gifts of P. Mischel) for 24 h using Lipofectamine 2000 transfection reagent (Invitrogen). Cells were selected with 1 μg/ml puromycin for 96 h. Cells were then grown in the absence of puromycin and used as polyclonal populations in all experiments.
Immunolocalization of PNPase.
MeWo melanoma cells were grown on poly-l-lysine coverslips, incubated with MitoTracker Green FM dye (Molecular Probes), fixed with methanol, and washed with 0.18% bovine serum albumin-phosphate-buffered saline (PBS), 1 mM CaCl2, and 1 mM MgCl2 (wash buffer). Cells were blocked with 10% goat serum-wash buffer, incubated with 0.46 μg of PNPase peptide Ab or 0.25 μg of rabbit IgG control Ab, and subsequently incubated with PE-conjugated anti-rabbit IgG. Microscopy was performed using a Zeiss Axioskop 2 Plus microscope with a Plan-APOCHROMAT 63×/1.0 oil objective. Images were acquired using a Zeiss Axiocam camera and Axiovision version 3.01 software.
Mitochondrial localization and import experiments.
Mouse liver and yeast mitochondria were prepared as described previously (15, 39). Protein import assays with radiolabeled precursors, osmotic shock experiments, and alkali extraction in yeast mitochondria were described previously (25). PNPase (1:1,000), AAC (1:5,000), cytochrome b2 (1:2,000), and KDH (1:2,000) antisera were used in Western analyses. The protein concentration of crude and Percoll gradient-purified mitochondria was determined with a bicinchoninic acid assay (Pierce). Osmotic shock experiments with mouse liver mitochondria were performed similarly to those using yeast mitochondria except that sucrose was substituted for sorbitol. For immunodetection, 8% gels were immunoblotted for PNPase (1:1,000), while 15% gels were immunoblotted for Timm13 (1:500), Hsp60 (1:1,000), and rhodanese (1:1,000) (17).
Blue native gels and complex V catalytic staining reaction.
Cells (3 × 107) were pelleted and resuspended in 0.8 ml of isolation buffer (70 mM sucrose, 220 mM mannitol, 2 mM HEPES, pH 7.4) with a protease inhibitor cocktail (Sigma). Lysates were prepared with a glass Dounce homogenizer (Kontes) and spun at 700 × g for 10 min once to remove unbroken cells and nuclei at 4°C. The supernatant was spun at 10,000 × g for 25 min at 4°C and the resulting mitochondrial pellet resuspended at 2.5 mg/ml in 1% digitonin in resuspension buffer (50 mM NaCl, 10% glycerol, 20 mM HEPES-KOH, pH 7.4, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride [PMSF]). Mitochondria were solubilized as described previously with modifications (25). Solubilized mitochondria were resuspended in sample buffer (0.5% Coomassie brilliant blue G-250 [Serva], 50 mM 6-aminocaproic acid, 10 mM BisTris-HCl [pH 7.0], and 0.1 mM PMSF), and 300 μg protein was separated by electrophoresis with a continuous 5% to 13% gradient blue native polyacrylamide gel electrophoresis (BN-PAGE) gel. For the catalytic staining reaction of complex V, the BN-PAGE gel was incubated in 34 mM Tris, 270 mM glycine, 14 mM MgSO4, 0.2% Pb(NO3)2, and 8 mM ATP, pH 7.8, overnight at room temperature (49). For immunoblotting, proteins on the BN-PAGE gel were transferred onto a polyvinylidene difluoride membrane and blotted using standard procedures.
tBID-mediated mobilization assay.
Mouse liver mitochondria were purified by centrifugation in a Percoll gradient at 40,000 × g for 1 h at 4°C. The mitochondrial band was washed twice at 4°C. Purified mouse liver mitochondria (2.5 mg/ml) were incubated in buffer R (20 mM HEPES-KOH at pH 7.4, 220 mM mannitol, 68 mM sucrose, 10 mM KCl, 1.5 mM MgCl2, 1 mM Na-EDTA, 1 mM Na-EGTA, 1 mM dithiothreitol, 0.1 mM PMSF, and protease inhibitor cocktail [Sigma]) containing 100 mM KCl, 10 mM sodium succinate, and 1 mM ADP, with or without 12.5 ng/μl truncated BH3-interacting domain death agonist (tBID) at 37°C. Aliquots were taken at 15, 30, 60, and 120 min and subjected to centrifugation at 10,000 × g for 15 min at 4°C. The pellet and supernatant proteins were resolved by sodium dodecyl sulfate (SDS)-PAGE, and 8% gels were immunoblotted for PNPase (1:1,000), while 15% gels were immunoblotted for cytochrome c (1:1,000). Mouse liver mitochondria incubated in 1% Triton X-100 for 120 min at 37°C were used as a control (data not shown).
Staurosporine-mediated mobilization assay.
HeLa cells were plated on glass coverslips in Dulbecco's modification of Eagle’s medium and were transfected with the OCT-CFP plasmid by use of Lipofectamine 2000 (Invitrogen). Sixteen hours posttransfection, cells were treated with 1 μM staurosporine (LC Laboratories) and 10 μM zVAD-fmk (MP Biomedicals) for 3 h and then rinsed in PBS, fixed with 4% paraformaldehyde-PBS for 15 min, permeabilized with 0.2% Triton X-100-PBS for 4 min, and blocked with 5% bovine serum albumin, 5% fetal bovine serum, 0.02% Triton X-100 in PBS for 30 min at room temperature. Then, cells were incubated for 2 h with rabbit polyclonal PNPase and mouse cytochrome c Abs and then for 30 min with goat anti-rabbit Alexa Fluor 514 and goat anti-mouse Alexa Fluor 647 secondary antibodies (Molecular Probes). Cells were visualized with an Olympus 100× oil immersion objective, numerical aperture 1.4, at 1 airy unit, on an Olympus IX80 laser scanning confocal microscope operated by FV1000 software v1.4a.
Mitochondrial visualization, quantification, and mtRNA qPCR.
Cells were plated at 2 × 105 cells/well onto poly-l-lysine-coated four-well chambered coverslips. The next day, cells were stained with 100 nM MitoTracker Green FM dye for 30 min and visualized with a Zeiss Axiovert 200 M microscope, and images were captured with a Hamamatsu Orca-ER camera and analyzed with Zeiss Axiovision 4.1 software. For quantification of mitochondria, cells were stained with 50 nM MitoTracker Green FM at 5 × 105 cells/ml for 30 min and analyzed by flow cytometry. mtRNA levels were determined using an Applied Biosystems 7700 sequence detector. Briefly, total RNA was isolated using TRIzol reagent (Invitrogen). One microgram total RNA was reverse transcribed with oligo(dT) by use of a Protoscript first-strand cDNA synthesis kit (New England Biolabs) according to the manufacturer's protocol. The resulting cDNA was diluted 10-fold. Each real-time quantitative PCR (qPCR) reaction mixture contained 5 μl of the diluted cDNA stock, 300 nM of both forward and reverse primers, and 10 μl of a SYBR green probe master mix (Diagenode) in a total volume of 20 μl. PCR thermocycling parameters were as follows: 95°C for 15 min and 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. The quantitative expression values were extrapolated from standard curves for 36B4 gene expression, and all samples were analyzed in triplicate and compared in parallel to the expression of 36B4 (22). Replicated samples were then averaged, and induction (n-fold) was determined. Probe and primer sequences are available upon request.
Electrochemical membrane potential measurement.
Changes in Δψ were determined using a BD MitoScreen flow cytometry mitochondrial membrane potential detection kit (BD Biosciences Pharmingen) according to the manufacturer's instructions. Briefly, 1 × 106 cells were washed, pelleted, and resuspended in 500 μl of JC-1 (5,5′-6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazol-carbocyanine iodide) working solution. Cells were incubated at 37°C for 15 min. Following staining, cells were washed twice in assay buffer, and then they were resuspended in 500 μl of assay buffer and immediately analyzed using a BD FACSCalibur flow cytometer. Live cells were gated and analyzed.
Spectrophotometric activity assays.
Mitochondria were harvested as described above with the following modifications. After homogenization, cells were spun four times at 700 × g. The supernatant was spun at 12,000 × g for 15 min at 4°C. The resulting mitochondrial pellet was washed once with isolation buffer and protein concentration determined. Mitochondria were freeze-thawed three times at 1 mg/ml and used immediately for complex I and III, complex II and III, and complex IV spectrophotometric activity assays. Alternatively, mitochondria were snap-frozen in liquid nitrogen at 5 mg/ml and stored at −80°C for later analysis. At analysis, mitochondria were thawed, diluted to 1 mg/ml in isolation buffer, and freeze-thawed again twice. For complex V activity assays, freshly isolated mitochondria were used. Rotenone-sensitive complex I and III-linked activity was evaluated by incubating 12.5 μg or 25 μg mitochondria in 1 ml of buffer containing 40 mM sodium phosphate, pH 7.4, 20 μM NADH, 500 μM EDTA, 250 μM KCN with or without 5 μg rotenone for 15 min at 30°C. Then, 50 μM horse heart cytochrome c (Sigma) was added, and the reduction of ferricytochrome c was followed at 550 minus 540 nm at 30°C for 3.5 min with a spectrophotometer. Complex II and III-linked activity was determined similarly but by replacing NADH with 20 mM sodium succinate and omitting rotenone (47a). Complex IV activity was determined according to the method of Trounce et al. (47). Briefly, reduced cytochrome c was adjusted to 20 μM in 10 mM potassium phosphate, pH 7.4, and the stability of the absorbance was determined at 550 minus 540 nm for 1 min. Five or 10 μg of mitochondria was added, and the oxidation of ferrocytochrome c was monitored at 550 minus 540 nm at 30°C for 1 min. Oligomycin-sensitive complex V ATP hydrolysis activity was determined by incubating 30 μg mitochondria in 1 ml 100 mM Tris buffer, pH 8.0, containing 50 mM KCl, 2 mM MgCl2, 0.2 mM EDTA, 200 μM NADH, 1 mM phosphoenolpyruvate, 2 U/ml lactate dehydrogenase, 4 U/ml pyruvate kinase, 2.5 mM Mg-ATP, 4 μg/ml rotenone, 300 nM antimycin A, and 2 mM KCN in the presence or absence of 5 μg/ml oligomycin. The oxidation of NADH was followed at 340 minus 400 nm for 7 min at 30°C (2). Enzyme activities are shown as nmol/min/mg protein.
Lactate quantification.
PNPase RNAi and control HEK293 cells were plated at 1 × 105 cells/ml on day 0, and medium was collected each day for 6 days and subjected to lactate quantification using a Beckman LX20 automated chemistry analyzer. Lactate levels were divided by the number of viable cells, as determined by trypan blue exclusion.
ATP quantification.
Cellular ATP was quantified using an ATP determination kit (Molecular Probes) according to the manufacturer's instructions. Briefly, PNPase RNAi and control HEK293 cells plated at equal densities were lysed in passive lysis buffer (Promega). Equal volumes of cell lysate were added to the standard reaction solution, and luminescence was measured and normalized to protein amounts in each lysate. Values used fell in the linear range of the assay as determined by a standard curve.
Cell proliferation assay.
Cells were used at 105 cells/ml. MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; ATCC] assays were performed according to the manufacturer's instructions at 0, 24, 48, and 72 h following plating. Trypan blue exclusion assays were performed to assess the number of viable cells up to 6 days following plating.
RESULTS
Mitochondrial localization of PNPase.
N-terminally tagged PNPase localizes to the cytosol (26), whereas highly overexpressed PNPase localizes to both the cytosol and the mitochondria in HO-1 melanoma cells (42). Sequence inspection of the N terminus suggests that endogenous PNPase resides in the mitochondria (36). In agreement with this, both endogenous PNPase and C-terminally tagged PNPase localize to mitochondria in HeLa cells (35). These data suggest that the subcellular localization of PNPase is influenced by an unblocked N terminus, the level of exogenous overexpression, or the specific cell type examined. To distinguish between these possibilities, N- and C-terminally tagged PNPase expression constructs were generated and stably transfected into HEK293T cells. An N-terminal myc tag resulted in PNPase that is larger than the endogenous form (Fig. 1A), potentially by blocking the processing of a 37-amino-acid mitochondrial targeting sequence (37). In contrast, a C-terminal myc tag did not hinder mitochondrial import and removal of the targeting sequence to generate mature, processed PNPase (Fig. 1B and data not shown). The level of C-terminally myc-tagged PNPase overexpression was modest, and total PNPase was only slightly elevated, consistent with PNPase abundance remaining stable despite increased PNPase transcripts (reference 13 and data not shown).
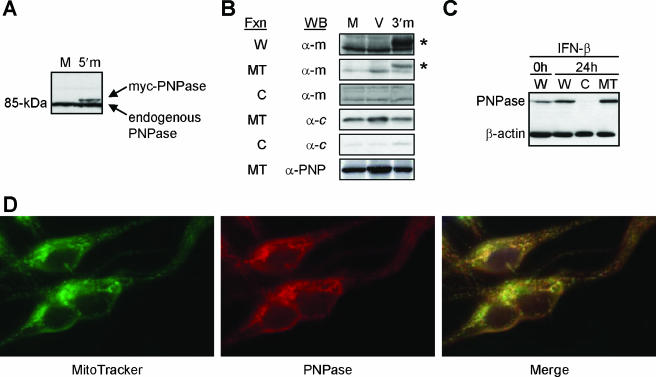
FIG. 1.
Mitochondrial localization of PNPase. (A) Recombinant human PNPase expression and detection. Immunoblots of whole-cell lysates from HEK293T cells mock (M) transfected or transfected with a 5′-myc-PNPase (5′m) expression construct are shown. (B) Exogenous PNPase localizes to mitochondria. HEK293T cells were mock (M) or empty vector (V) transfected or transfected with a PNPase-3′-myc (3′m) expression construct, and whole-cell lysates (W) and cytosolic (C) and mitochondrial (MT) fractions (Fxn) were probed for the myc epitope tag (α-m), for cytochrome c (α-c), or for PNPase (α-PNP). Asterisks indicate PNPase; additional bands are nonspecific. (C) Preexisting and IFN-β-induced PNPase localizes to P3HR-1 Burkitt lymphoma cell mitochondria. Cells were left untreated or were incubated with 1,000 U/ml IFN-β for 24 h, and whole-cell lysates (W) and cytosolic (C) and mitochondrial (MT) fractions were probed for PNPase and β-actin. (D) PNPase immunolocalizes to the mitochondria. MeWo melanoma cells were incubated with MitoTracker Green FM dye and PNPase peptide-purified rabbit antibody, followed by PE-conjugated anti-rabbit IgG. The right panel shows the images merged.
Induced endogenous PNPase might localize to the cytosol, as does robustly overexpressed exogenous PNPase (42). Type I interferons, such as beta interferon (IFN-β), induce PNPase gene expression (26, 27), although PNPase protein levels may change more modestly than exogenous overexpression (13). To test for the localization of induced PNPase, P3HR-1 Burkitt lymphoma cells were treated with 1,000 U/ml IFN-β for 24 h. PNPase protein was increased about twofold over unstimulated levels, with both preexisting and induced PNPase localized efficiently to mitochondria (Fig. 1C). The immunofluorescent colocalization of PNPase and mitochondria also visually confirmed the mitochondrial location of endogenous PNPase (Fig. 1D). Combined, these and prior results (26) indicated that the processed N-terminal sequence targeted PNPase to mitochondria and, when blocked, most, if not all, PNPase remained in the cytoplasm. The data also indicated that N-terminally unblocked PNPase was processed and localized to mitochondria in a variety of mammalian cell types, including HEK293T epithelial cells (Fig. 1B), P3HR-1 and Nalm-6 lymphoid cells (Fig. 1C and data not shown), and MeWo neural crest-derived melanoma cells (Fig. 1D). Endogenous PNPase levels appear tightly regulated (13) and suggest that forced overexpression could mislocalize abundant PNPase to the cytosol.
PNPase sublocalization in the mitochondrial IMS.
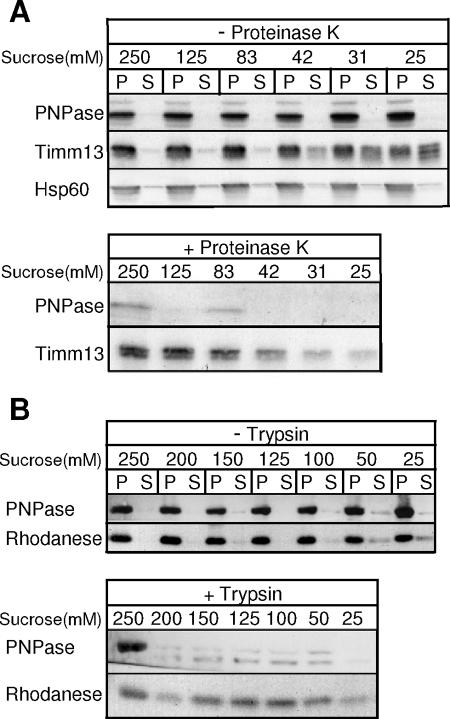
We next used a variety of approaches to localize PNPase within mitochondria. First, PNPase localization in mouse liver mitochondria was determined by incubation with decreasing concentrations of sucrose to selectively rupture the OM by osmotic shock (Fig. 2A). When the OM was disrupted and mitoplasts containing the inner membrane (IM), fragmented OM, and matrix components were separated from the soluble IMS components by centrifugation, PNPase remained in the pellet fraction, whereas IMS-localized Timm13 was partially released into the supernatant. However, when protease was added, PNPase proved very sensitive and degraded rapidly, supporting its localization in the IMS. By comparison, the matrix marker rhodanese was protected from protease treatment, until very low sucrose concentrations (50 mM and 25 mM) were reached, when rhodanese began to leak from the matrix (Fig. 2B). A control in which the matrix was opened with detergent showed that rhodanese was sensitive to and rapidly degraded by protease (data not shown).
FIG. 2.
PNPase localizes to mouse liver mitochondrial IMS. (A) Mouse liver mitochondria were subjected to osmotic shock with sucrose in the presence and absence of proteinase K and separated by centrifugation. PNPase, IMS marker Timm13, and matrix marker Hsp60 were detected by immunoblotting. Upper panel, mitoplasts (P) are separated from the soluble IMS (S); lower panel, mitoplasts are recovered by centrifugation. (B) Upper and lower panels are similar to those for panel A with immunoblotting for PNPase and the matrix marker rhodanese in the presence or absence of trypsin.
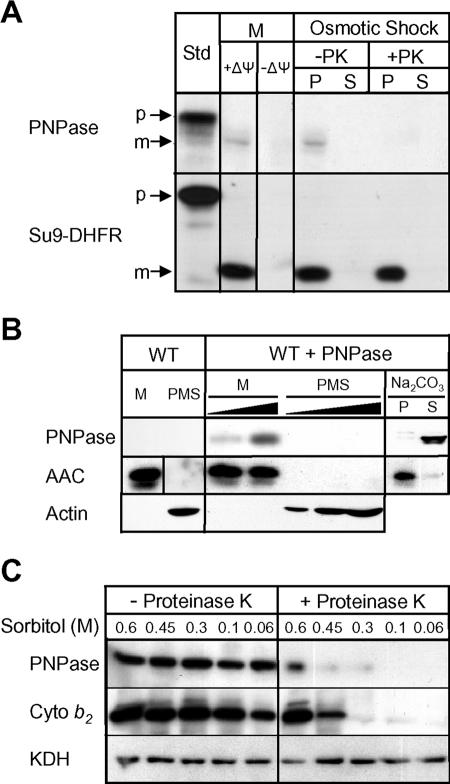
Since mitochondrial protein import is highly conserved from yeast to mammals (39), we utilized in organello import assays with purified yeast mitochondria and radiolabeled precursor PNPase for confirmatory studies. PNPase was imported in the presence of Δψ and coupled by cleavage of the PNPase targeting sequence. In control reactions, a synthetic protein consisting of the Neurospora crassa subunit 9 ATPase targeting sequence fused to dihydrofolate reductase (Su9-DHFR) and cytochrome c1 were also imported. Su9-DHFR was imported into the matrix, whereas cytochrome c1 was imported via the stop-transfer mechanism into the IMS (14). PNPase localization was analyzed after in vitro import by osmotic shock to selectively disrupt the OM; again, PNPase localized to the IMS because it was sensitive to added protease (Fig. 3A).
FIG. 3.
PNPase is actively imported and processed and localizes to the mitochondrial IMS. (A) Radiolabeled PNPase was imported into isolated yeast mitochondria (M) at 25°C in the presence and absence of Δψ. The OM was ruptured by osmotic shock to generate mitoplasts in the presence and absence of proteinase K (PK). Centrifugation separated mitoplasts (P) from the soluble IMS fraction (S). As a control, matrix-localized Su9-DHFR was also imported. Samples were analyzed by SDS-PAGE and fluorography. Standard (Std) refers to 10% of the radioactive precursor added to each assay. p, precursor; m, mature. (B) PNPase was expressed in S. cerevisiae under the control of the regulated CUP1 promoter (WT + PNPase). Cells were fractionated and mitochondria (M) were separated from the postmitochondrial supernatant (PMS) by centrifugation. Mitochondria were incubated with 0.1 M Na2CO3 (pH 11.5), and integral membrane proteins (P) were separated from soluble proteins (S) by centrifugation. Fifty- and 100-μg portions of M and increasing amounts of PMS were loaded in adjacent lanes. PNPase, the IM marker AAC protein, and the cytosolic marker β-actin were detected by immunoblotting. WT, wild type. (C) Yeast mitochondria containing PNPase as described for panel B were subjected to osmotic shock (incubation in solution with decreasing sorbitol concentration) in the presence and absence of proteinase K. Mitoplasts were recovered by centrifugation and separated by SDS-PAGE. PNPase, IMS marker cytochrome b2, and matrix marker KDH were detected by immunoblotting.
We further confirmed this import and localization with heterologous expression of PNPase in S. cerevisiae under control of the CUP1 gene copper-inducible promoter. PNPase and the mitochondrial IM marker, AAC, were localized exclusively to the mitochondrion (Fig. 3B). In contrast to the integral IM protein AAC, PNPase is a peripheral membrane protein because it was recovered in the supernatant after carbonate extraction (12). To further substantiate the IMS localization, mitochondria from yeast expressing PNPase were incubated in a hypo-osmotic buffer to selectively disrupt the OM, followed by centrifugation to separate the soluble IMS from the insoluble matrix, IM, and fragmented OM (23). When the OM was disrupted, PNPase remained associated with the membrane (Fig. 3C) and was not released with the soluble IMS components, such as cytochrome b2 (data not shown). However, when protease was added, PNPase and the resident IMS protein cytochrome b2 were degraded as the OM was disrupted (0.45 M sorbitol). The matrix control KDH was protected from protease, indicating that the IM remained intact. Together, the data from mammalian mitochondria, as well as import and localization studies with yeast, indicate that PNPase resides as a peripheral membrane protein in the mitochondrial IMS.
IMS-localized PNPase assembles in a multiprotein complex.
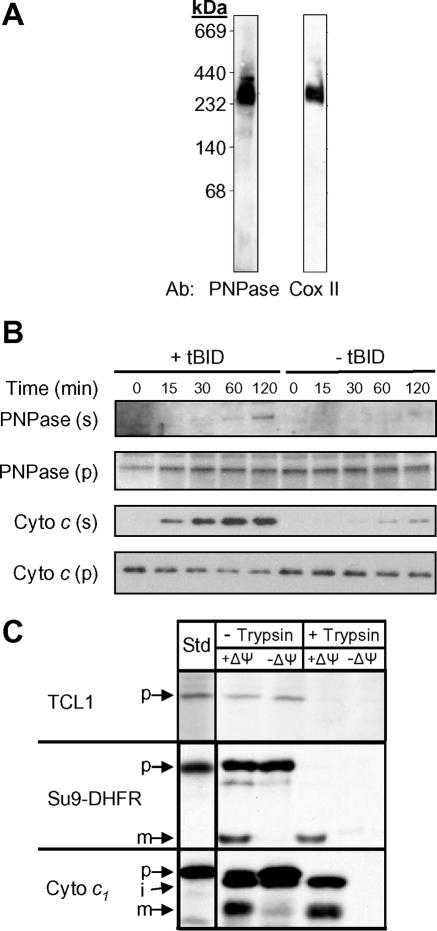
Structural studies show that bacterial PNPase forms a homotrimer of PNPase subunits, with a central cavity comprising the probable site of catalytic activity (44). A stacked PNPase trimer-dimer, creating a cylinder with an elongated central cavity, most likely exists in plants (4, 45). Human PNPase shares 34% amino acid sequence identity with bacterial PNPase and an even higher degree of sequence conservation in all of the key functional domains, suggesting the possibility for a similar trimer or trimer-dimer conformation (36, 45). To determine if IMS-localized PNPase exists as an 85-kDa monomer or in a multimeric complex, mouse liver mitochondrial proteins were size fractionated by BN-PAGE and immunoblotted for PNPase. Complexes containing PNPase were detected between ∼200 and 400 kDa (Fig. 4A); these may represent PNPase trimers, because PNPase immunoprecipitation did not identify heteromeric PNPase-associated proteins from mitochondrial extracts (data not shown).
FIG. 4.
PNPase exists in a multimeric IMS complex and is mobilized with tBID treatment. (A) Mouse liver mitochondria were size fractionated by BN-PAGE and immunoblotted for PNPase or respiratory complex IV. (B) PNPase is partially mobilized from mitochondria. Isolated mouse liver mitochondria were treated with or without tBID for 15, 30, 60, and 120 min and subjected to centrifugation. Supernatants (s) and mitochondrial pellets (p) were harvested and analyzed by immunoblotting. PNPase, cytochrome c, and Timm13 (data not shown) were released from mitochondria. Data are representative of three independent experiments. (C) Radiolabeled TCL1 was not imported into isolated yeast mitochondria at 25°C in the presence or absence of Δψ. After import, samples were treated with trypsin to remove nonimported precursor, and treatment was stopped with soybean trypsin inhibitor. Import-competent controls were matrix-localized Su9-DHFR and IMS-localized cytochrome c1. Samples were analyzed by SDS-PAGE and fluorography. Standard (Std) refers to 10% of the radioactive precursor added to each assay. p, precursor; i, intermediate; m, mature.
IMS mobilization of PNPase follows cytochrome c release with apoptotic stimuli.
The presence of PNPase in the IMS was puzzling because this compartment is not known to contain RNA substrates for PNPase exoribonuclease or poly(A) polymerase activities (10, 26, 33, 41). OM permeabilization and PNPase mobilization could expose cytosolic RNA to PNPase processing activity and provide a conduit for interactions with TCL1. A competing possibility is that PNPase could also be exposed passively to cytosolic proteins and RNAs while still attached to the IM following OM disruption (38). Since we identified PNPase through an interaction with TCL1 (10), and exogenous PNPase could possibly degrade cytosolic RNA to regulate transcript levels (40-42), PNPase mobilization from the IMS was determined.
Purified mouse liver mitochondria were incubated with or without tBID to permeabilize the OM, and the released proteins were subsequently separated by use of centrifugation. In the presence of tBID, PNPase was partially mobilized from the IMS over time (Fig. 4B). This mobilization was similar to the partial release of proapoptotic Bit1 with induction of anoikis (20). In the same assay, robust cytochrome c release preceded the mobilization of PNPase. Interestingly, TCL1, which lacks an identifiable mitochondrial targeting sequence, was not imported into energized mitochondria in a standard import assay (Fig. 4C), suggesting that a potential PNPase-TCL1 interaction required opening of the OM.
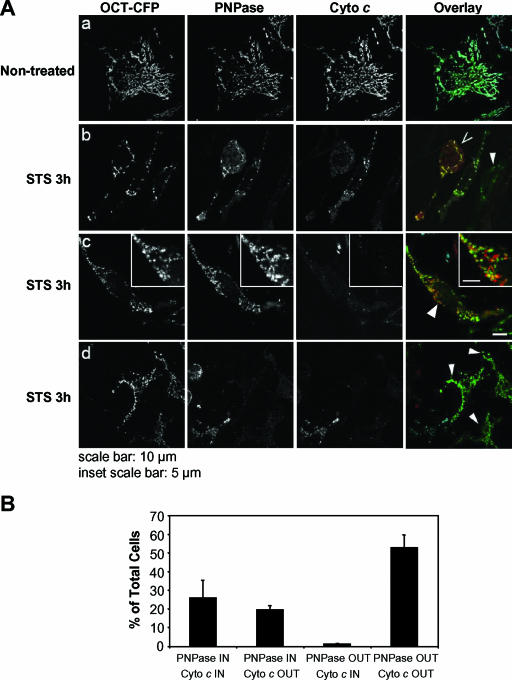
Given that PNPase exists in a membrane-associated multiprotein complex in the IMS (Fig. 3B and 4A), cell fractionation-based detection may underestimate its mobilization by nonspecific association with membranes or by aggregation and pelleting. Therefore, to further establish the mobilization of PNPase under conditions that cause cytochrome c release, staurosporine- or Fas (data not shown)-treated HeLa cells were evaluated by fluorescent microscopy for a mitochondrial matrix marker (OCT-CFP) (34) and by immunofluorescence staining for cytochrome c and PNPase (Fig. 5A). Untreated HeLa cells showed an almost complete overlapping staining pattern for CFP, cytochrome c, and PNPase for all cells evaluated. By contrast, with 3 h of staurosporine (or 6 h of Fas) treatment, CFP remained in a mitochondrial pattern, while both PNPase and cytochrome c staining distributions changed. PNPase and cytochrome c staining was classified into four patterns based upon overlap staining with the CFP matrix marker. In three independent experiments, the proportion of cells with cytochrome c released and PNPase retained (PNPase IN/cyto c OUT) exceeded the negligible number of cells that appeared to have PNPase mobilized and cytochrome c retained (PNPase OUT/cyto c IN) with apoptotic stimuli (Fig. 5A, panel b, and B). The mobilization of PNPase was also visually distinct from cytochrome c, as PNPase-containing aggregates that remained near the mitochondria but no longer overlapped completely with the stationary CFP matrix marker were not infrequently seen (Fig. 5A, panel c). Our interpretation of the data was that IMS-localized PNPase was mobilized after tBID treatment of isolated mitochondria (Fig. 4B) or staurosporine (and Fas) treatment of intact HeLa cells (Fig. 5 and data not shown). An alternative interpretation was that cytochrome c was released and IMS-localized PNPase degraded, although this interpretation was inconsistent with the appearance of diffuse or punctate PNPase staining in the cytosol following OM permeabilization. The retention of the CFP matrix marker and the loss of both cytochrome c and PNPase mitochondrial staining strongly supported the localization of PNPase in the IMS, as does our accompanying paper, which describes the PNPase import pathway into the IMS (37). Finally, mobilization or cytosolic exposure of IMS-localized PNPase provides a pathway for interactions with cytosolic TCL1.
FIG. 5.
Staurosporine-mediated PNPase mobilization. (A) HeLa cells transfected with the mitochondrial matrix marker OCT-CFP were left untreated (a) or were treated for 3 h with 1 μM staurosporine (STS) in the presence of 10 μM zVAD-fmk (b, c, and d). Following fixation, the distribution of OCT-CFP was compared to those of endogenous PNPase and cytochrome c, as determined by indirect immunofluorescence. Black-and-white images of OCT-CFP, PNPase, and cytochrome c highlight the mobilization of PNPase and cytochrome c release. The colocalization of PNPase (red) with OCT-CFP (green) and cytochrome c (cyan) is revealed in the overlay in the rightmost panels. Symbols: >, cells showing mobilization of cytochrome c but not PNPase; arrowheads, cells showing mobilization of both cytochrome c and PNPase. (B) Evaluation of staining patterns shown in panel A. Approximately 250 cells were scored for PNPase and cytochrome c staining after 3 h of staurosporine treatment and placed into one of four categories relative to the OCT-CFP matrix marker. IN refers to PNPase and cytochrome c staining that overlaps with the stationary CFP marker, while OUT refers to dispersed cytosolic staining of low intensity and/or punctate staining that does not colocalize with CFP. The percentages of cells staining for each of the four patterns, totaling 100%, are shown. Almost all untreated HeLa cells had a PNPase IN/cyto c IN staining pattern (data not shown). Data shown are for duplicate cell scoring in one experiment, which is representative of three independent experiments.
PNPase deficiency does not alter mtRNA levels.
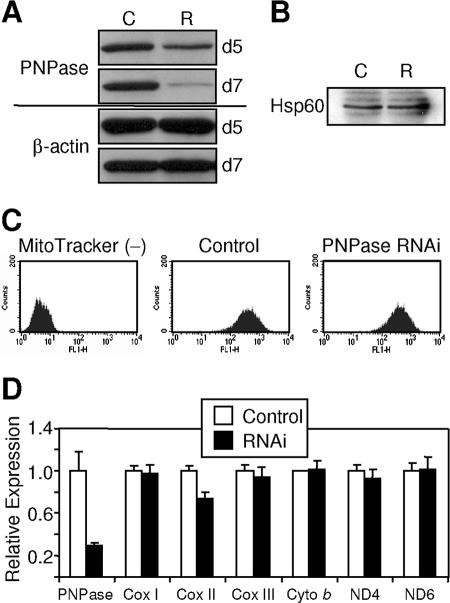
PNPase was evaluated for a role in regulating mtRNA levels, because an undetectable fraction of functional PNPase could possibly localize to the mitochondrial matrix. PNPase knockdown with RNAi was determined over time for HEK293 cells, with protein expression reduced 65 to 90% compared to that for control cells at 7 to 8 days posttransfection in numerous independent experiments (Fig. 6A). For this reason, day 8 or later PNPase knockdown cells were used in all experiments. PNPase knockdown and control cells showed similar amounts of Hsp60 (Fig. 6B), a mitochondrial matrix protein, and similar numbers of mitochondria (Fig. 6C). A recent study showed that PNPase knockdown in HeLa cells did not affect mtRNA levels, although the extent of knockdown was not provided (33). Our results with robust PNPase knockdown were consistent with this prior report in that PNPase knockdown exhibited no statistical effect on multiple mtRNAs, with the exception of a 20 to 30% decrease in Cox II mtRNA for unknown reasons (Fig. 6D). The lack of pan-mtRNA alterations with PNPase knockdown contrasts sharply with the panreduction of mtRNA in mitochondrial poly(A) polymerase (mtPAP) knockdown cells, which may represent the activity of a matrix-localized RNA-processing enzyme (33). We interpret these data as further evidence supporting PNPase localization in the IMS and a lack of function in regulating mtRNA in mammalian cells.
FIG. 6.
PNPase nadirs at or beyond day 7 post-RNAi transfection without altering mtRNA levels. (A) RNAi-mediated PNPase knockdown. HEK293 cells transfected with control (C) or PNPase RNAi knockdown (R) retroviral vectors were analyzed by immunoblotting for PNPase and β-actin at days 5 and 7 posttransfection. (B) PNPase knockdown and control cells contain similar amounts of the mitochondrial marker Hsp60. Western analysis was performed on whole-cell lysates for Hsp60. (C) Equivalent numbers of mitochondria in control and PNPase knockdown cells. Unstained (left), MitoTracker Green FM-stained control (middle), and PNPase knockdown (right) HEK293 cells are shown. MitoTracker Green FM stains mitochondria regardless of mitochondrial membrane potential. (D) PNPase deficiency does not alter mtRNA levels. A 75% reduction in PNPase has no differential effect on multiple mtRNA levels, with the exception of a 25 to 30% decrease in Cox II expression, as assessed by qPCR. Expression is normalized to a 36B4 control sequence (22). Data are representative of two independent experiments.
PNPase deficiency alters mitochondrial morphology.
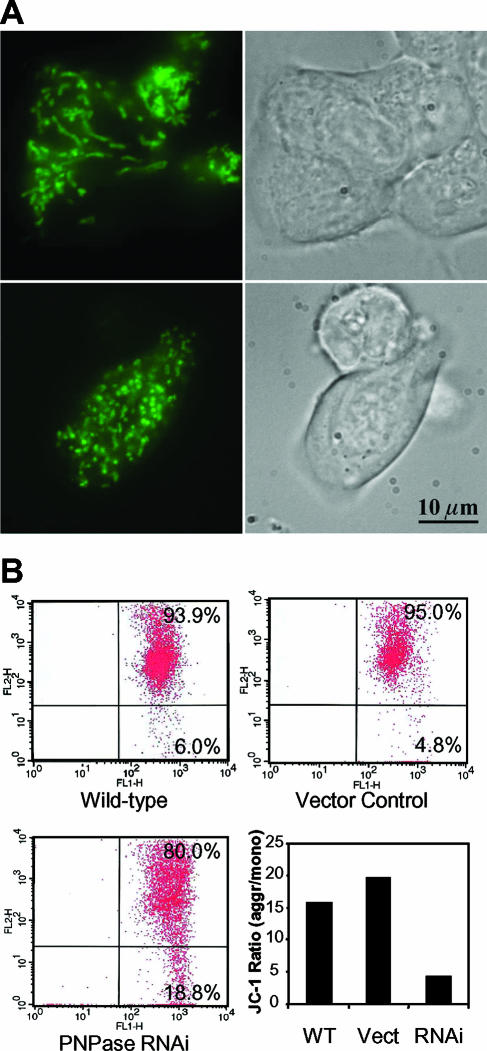
The localization of PNPase in the IMS prompted an assessment of mitochondrial morphology in PNPase knockdown cells stained with MitoTracker Green FM dye. Control HEK293 cells displayed a well-developed mitochondrial filamentous network, whereas PNPase knockdown cells featured punctately stained mitochondria due to the fragmentation of the network (Fig. 7A). This result suggests that PNPase knockdown tips the balance in favor of mitochondrial fission over fusion (6). Interestingly, the level of the OPA1 dynamin GTPase, a protein required for mitochondrial fusion (7), was unaltered by PNPase deficiency (data not shown). Furthermore, the data predict a potential defect in mitochondrial function from decreased PNPase levels and indicate a dependence of mitochondrial structure on IMS-localized PNPase.
FIG. 7.
PNPase knockdown alters mitochondrial morphology and compromises Δψ. (A) Fragmentation of the filamentous mitochondrial network with PNPase knockdown. MitoTracker Green FM-stained HEK293 control (top panels) and PNPase RNAi (bottom panels) cells show a change in mitochondrial morphology from filamentous and elongated to granularly shaped structures. (B) PNPase reduction compromises Δψ. Cells were stained with the mitochondrial membrane-sensitive dye JC-1; the live-gated cells are shown. When Δψ is high, the normally green dye aggregates (upper right quadrant in each flow cytometry profile) and stains red. The ratios of red aggregates (aggr) to green monomers (mono) (lower right quadrant in each flow cytometry profile) are indicated as a measure of Δψ. A result representative of five independent experiments is shown. WT, wild type; Vect, vector.
PNPase supports Δψ and OX-PHOS.
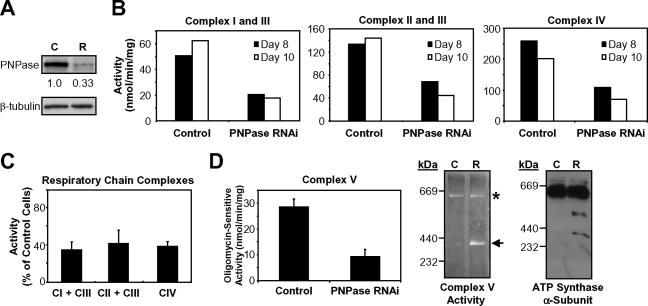
In addition to the altered morphology, a respiratory dysfunction was observed for mitochondria with PNPase knockdown. Consistent with this notion, viable PNPase knockdown cells showed a three- to fourfold loss in Δψ compared to live-gated control cells, implicating PNPase in maintaining Δψ under normal growth conditions (Fig. 7B). Loss of Δψ is associated with crippled OX-PHOS (16), which prompted us to examine the function of the respiratory chain. Coupled respiratory complexes I and III, coupled complexes II and III, and individual complexes IV and V were each functionally impaired in PNPase knockdown HEK293 mitochondria compared to those in control mitochondria, as determined by enzymatic analysis (Fig. 8B to D). Furthermore, BN-PAGE coupled with an in-gel ATP hydrolysis activity assay showed that the loss of PNPase led to structural instability of complex V. Increased dissociation of the F1- ATP synthase subcomplex from the fully assembled FoF1-ATP synthase was observed for PNPase knockdown mitochondria. This was confirmed by immunoblotting that showed enhanced dissociation of ATP synthase α-subunits (Fig. 8D). Oxygen consumption in HEK293T PNPase knockdown cells was also compromised compared with that in control cells (data not shown). The specificity of these OX-PHOS defects to the loss of PNPase was confirmed by the observation of similar but less dramatic OX-PHOS defects in HEK293 cells transfected with a second PNPase RNAi construct that reduced PNPase protein expression 20 to 40% at its nadir (data not shown). Combined, the data indicate that PNPase is required to support OX-PHOS and that PNPase knockdown cells are associated with an altered mitochondrial morphology, disrupted Δψ, and an impaired respiratory chain.
FIG. 8.
PNPase reduction impairs respiratory chain function. (A) Representative immunoblot of PNPase knockdown (R) and control (C) cells used for the experiments described below. PNPase was reduced to 33% of control HEK293 cells on day 8 after RNAi transfection by densitometry. (B) Respiratory chain complex activities in mitochondria from control and HEK293 PNPase knockdown cells measured spectrophotometrically. Activities are expressed as nmol cytochrome c reduction or oxidation/min/mg of mitochondrial protein. Data are from day 8 and day 10 of two independent PNPase knockdown experiments. (C) Summary of the data in panel B. Complex (CI, CII, CIII, and CIV) activities are expressed as percentages of the activity of control cells (normalized to 100%). (D) Reduced activity and partial dissociation of the ATP synthase in PNPase knockdown cells. Oligomycin-sensitive ATP hydrolysis activity was determined spectrophotometrically (left panel). Mitochondria from control and PNPase knockdown cells were analyzed by BN-PAGE followed by in-gel ATP hydrolysis activity staining (middle panel) or immunoblotting for the α-subunit of the F1-ATP synthase subcomplex (right panel). Symbols: *, fully assembled FoF1-ATP synthase complex (theoretical molecular mass, ∼600 kDa); ←, F1 subcomplex (theoretical molecular mass, 371 kDa).
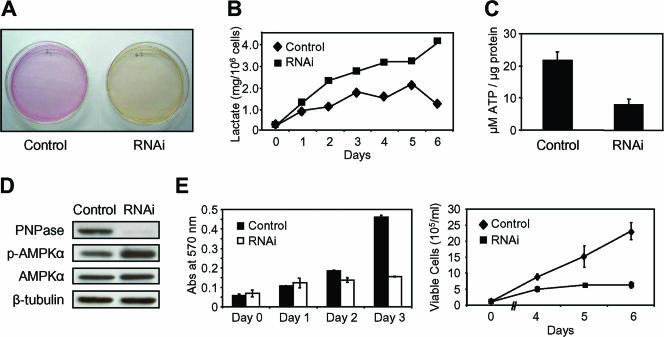
ATP reduction from PNPase deficiency increased AMPK phosphorylation and slowed cell growth.
Consistently, the growth medium for PNPase-deficient HEK293, HeLa, and Jurkat T cells turned yellow more rapidly than that for control cells (Fig. 9A and data not shown). Because exogenous PNPase overexpression inhibits cell growth (26), one possibility was that the yellowing was from increased growth of PNPase knockdown cells. Alternatively, the enhanced yellowing may have stemmed from accumulated lactate in the medium secondary to impaired cell respiration. Therefore, energy charge status and cell growth were evaluated to distinguish between these possibilities. Indeed, lactate reproducibly accumulated in the medium of PNPase knockdown HEK293, Jurkat T, and HeLa cells more rapidly on a per live cell basis than control cell medium over time (Fig. 9B and data not shown). PNPase knockdown cells also exhibited a >50% decrease in steady-state ATP levels (Fig. 9C) and showed a robust induction of AMPK phosphorylation (Fig. 9D), which served as an indicator of an increased AMP/ATP ratio and reduced cell energy charge. Consistent with a defect in OX-PHOS, the rate of cell proliferation in PNPase-deficient cells was reduced in MTT and trypan blue exclusion assays (Fig. 9E), whereas there was no significant difference in cell death levels between PNPase knockdown and control cells (data not shown). Combined, the data indicate that PNPase deficiency causes protean cellular defects, primarily affecting OX-PHOS and mitochondrial morphology, resulting in secondary changes, such as reduced ATP and lactate accumulation, leading to enhanced AMPK phosphorylation and diminished cell growth (Fig. 10).
FIG. 9.
Reduced cell energy charge and slowed cell growth with PNPase reduction. (A) Growth medium color change for continuously growing HEK293 cells (and HeLa and Jurkat T cells [data not shown]) with PNPase knockdown (RNAi). (B) PNPase knockdown cells have increased lactate accumulation over time. Cells were incubated in medium with 10% serum, and lactate was measured each day for 6 days. Day 0 equals day 8 following PNPase RNAi knockdown. (C) PNPase knockdown cells have impaired ATP production. ATP levels were measured in both control and PNPase knockdown cells. Three independent experiments showed similar results. (D) Increase in AMPK phosphorylation with PNPase knockdown as assessed by immunoblotting. (E) Cells with reduced PNPase have slowed growth. Control and PNPase RNAi cells were assessed in triplicate each day for 3 days by MTT assay (left panel) or for up to 6 days by trypan blue exclusion assay (right panel) for changes in cell growth. Data are representative of three independent experiments.
FIG. 10.
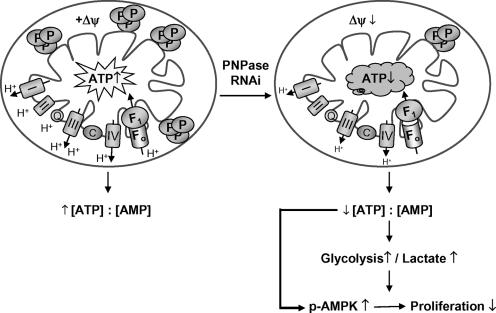
Model for the effect of reduced PNPase activity. Loss of PNPase (P) leads to reduced respiratory chain complex activity, decreased Δψ, and impaired ATP production. See Discussion for details. Symbols: ↑, increase; ↓, decrease; +Δψ, presence of normal electrochemical membrane potential.
DISCUSSION
In this study, the subcellular localization and physiologic requirement for endogenous mammalian PNPase were determined. Our data showed that PNPase localized to mitochondria in multiple cell types, specifically to the IMS in a peripheral membrane protein complex. With a classical mitochondrial targeting sequence and prokaryotic and chloroplastic stroma counterparts, PNPase was expected to regulate mtRNA levels from within the matrix (35, 52). However, PNPase was surprisingly localized to the IMS, and mtRNAs were almost completely unaffected by PNPase reduction at its nadir. Mobilization of PNPase followed cytochrome c release from mitochondria treated with proapoptotic stimuli, which strongly supported an IMS localization for PNPase and also suggested a potential route for interactions with cytosolic TCL1, which was how we initially identified PNPase (10). An IMS location also could provide endogenous PNPase with RNA substrates under conditions that cause mitochondrial OM permeabilization (26, 41). In addition to its surprising IMS localization, PNPase exoribonuclease was unexpectedly required to maintain the filamentous structure of mitochondria, and its reduction caused a loss in Δψ with impairment of the respiratory chain, leading to a metabolic alteration that reduced cellular energy charge and caused decreased cell growth without increased cell death. Interestingly, more than 30 years ago an obscure RNase activity was isolated from the IMS of rat liver mitochondria (5). Our identification of PNPase in the IMS may provide a source for this previously isolated RNase activity.
A variety of in vitro and in vivo systems in mammalian and yeast cells demonstrated and confirmed the IMS localization and membrane association of PNPase within mitochondria. Detailed import studies also showed that PNPase utilized a novel import pathway to reach its destination in the IMS (37). Import required energized mitochondria and was coupled to the N-terminal processing of a 37-amino-acid mitochondrial targeting sequence (reference 37 and data not shown). We determined that basal and IFN-β-induced PNPase were imported into mitochondria, and appreciable nonmitochondrial PNPase was not detected under normal growth conditions. These results contrast with studies that show exogenous overexpression of PNPase decreases MYC and BCL-xL transcript levels and induces growth arrest and apoptosis, presumably from a cytosolic localization of PNPase (41, 42). Interestingly, high-level adenovirus-mediated PNPase overexpression (100 PFU) in these prior studies results in apoptosis, whereas a lower but still robust level of PNPase overexpression (25 PFU) does not produce enhanced apoptosis over control infections. We interpret these data with caution, because when combined with our current studies of endogenous PNPase localization they seem to suggest that growth arrest and apoptosis from exogenous PNPase overexpression could result from aberrant localization of PNPase, causing nonphysiologic activity in the cytosol. A cytosolic accumulation of PNPase could resemble a physiologic mobilization of PNPase from mitochondria, as demonstrated here for tBID-treated isolated mouse liver mitochondria and staurosporine- and Fas (data not shown)-treated HeLa cells. However, this mobilization follows cytochrome c release, suggesting that physiologic PNPase mobilization does not induce apoptosis but rather may play a role in the processing of cytosolic RNAs during the clearance phase of cell death in vivo.
PNPase knockdown was correlated with mitochondrial dysfunction. Mitochondria became fragmented, which is a hallmark of mitochondrial dysfunction (7). This result contrasted with a study that showed no change in the appearance or respiratory function of HeLa mitochondria with reduced PNPase expression (33). A possible explanation for these discrepant results is that not enough time may have passed to permit the PNPase knockdown to become effective in these prior studies. We interpret the global dysfunction in respiration from PNPase deficiency as a consequence of the role for PNPase in mitochondrial homeostasis.
Based on our results, we propose a model for the range of cellular effects arising from a reduction in endogenous PNPase (Fig. 10). Under physiologic growth conditions, PNPase localizes to the mitochondrial IMS as a peripheral membrane protein in a multimeric complex. A PNPase reduction of >65% results in a morphological change in the mitochondrial filamentous network, causing fragmentation and mitochondrial fission. This change is correlated with a loss of Δψ and a functional impairment of OX-PHOS, as observed with the reduction of the matrix-associated mtPAP RNA-processing protein (33). However, reduced mtPAP decreases the steady-state level of multiple mtRNAs, suggesting that impaired OX-PHOS directly results from reduced expression of essential mtRNAs, which does not appear to be the source for OX-PHOS defects in PNPase-deficient mitochondria. Interestingly, an OX-PHOS impairment is likewise reported for the knockdown of AIF, a 57-kDa redox flavoprotein that also localizes as a peripheral IM protein in the IMS and might be expected, unlike PNPase, to affect respiration (43). AIF is homologous to bacterial oxidoreductases, and knockout and RNAi knockdown reveal a dysfunction in complex I of the electron transport chain, resulting in elevated lactate and decreased ATP levels (48). These metabolism endpoints are similar to the current findings for PNPase-deficient cells. However, PNPase deficiency does not alter the level of multiple mtRNAs, unlike mtPAP, nor does it appear to cause a defect in a specific respiratory chain complex, unlike AIF.
Mitochondrial dysfunction resulting from PNPase deficiency led to secondary effects including reduced ATP production. In addition to reduced ATP production from defective OX-PHOS, a complementary mechanism for further reducing ATP might include increased ATP catabolism as an adaptive response, through the ATP synthase acting as an ATP hydrolase, which would help stabilize Δψ and inhibit the onset of apoptosis (2). Reduced cell energy charge, in turn, would enhance the phosphorylation of AMPK, which could slow cell growth through a potential link to p53-mediated inhibition of the cyclin E cell cycle checkpoint protein (21, 31). However, PNPase regulation of this cell growth pathway, of an alternative TOR-TSC2 growth pathway (19), or of as-yet-undefined growth pathways is likely secondary to its primary role in mitochondrial homeostasis from its location within the IMS. Because of the complications imparted by delayed kinetics of PNPase knockdown, the generation of PNPase knockout cells will be critical for helping to determine the intricacies by which PNPase maintains mitochondrial integrity and respiratory function.
Acknowledgments
We thank Paul D. Boyer (UCLA) for insightful discussions and Roberta Gottlieb (Scripps Research Institute) and Sabine Rospert (University of Freiburg) for reagents.
This work was supported by NIH grants T32CA09056, F31HD041889, T32CA009120, R01GM061721, R01GM073981, R01CA90571, and R01CA107300; a CIHR Fellowship Award; Muscular Dystrophy Association grant 022398; and CMISE, a NASA URETI Institute (NCC 2-1364). C.M.K. is an Established Investigator of the American Heart Association, and M.A.T. is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Published ahead of print on 11 September 2006.
REFERENCES
- 1.Agarwal, S., and R. S. Sohal. 1995. Differential oxidative damage to mitochondrial proteins during aging. Mech. Ageing Dev. 85:55-63. [DOI] [PubMed] [Google Scholar]
- 2.Appleby, R. D., W. K. Porteous, G. Hughes, A. M. James, D. Shannon, Y. H. Wei, and M. P. Murphy. 1999. Quantitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. Eur. J. Biochem. 262:108-116. [DOI] [PubMed] [Google Scholar]
- 3.Baginsky, S., and W. Gruissem. 2002. Endonucleolytic activation directs dark-induced chloroplast mRNA degradation. Nucleic Acids Res. 30:4527-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baginsky, S., A. Shteiman-Kotler, V. Liveanu, S. Yehudai-Resheff, M. Bellaoui, R. E. Settlage, J. Shabanowitz, D. F. Hunt, G. Schuster, and W. Gruissem. 2001. Chloroplast PNPase exists as a homo-multimer enzyme complex that is distinct from the Escherichia coli degradosome. RNA 7:1464-1475. [PMC free article] [PubMed] [Google Scholar]
- 5.Baudhuin, P., C. Peeters-Joris, and J. Bartholeyns. 1975. Hepatic nuclease. 2. Association of polyadenylase, alkaline ribonuclease and deoxyribonuclease with rat-liver mitochondria. Eur. J. Biochem. 57:213-220. [DOI] [PubMed] [Google Scholar]
- 6.Chan, D. C. 11 May 2006, posting data. Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. [Online.] doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed]
- 7.Chen, H., A. Chomyn, and D. C. Chan. 2005. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 280:26185-26192. [DOI] [PubMed] [Google Scholar]
- 8.Du, C., M. Fang, Y. Li, L. Li, and X. Wang. 2000. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102:33-42. [DOI] [PubMed] [Google Scholar]
- 9.Finkel, T., and N. J. Holbrook. 2000. Oxidants, oxidative stress and the biology of ageing. Nature 408:239-247. [DOI] [PubMed] [Google Scholar]
- 10.French, S. W., D. W. Dawson, H.-W. Chen, R. N. Rainey, S. A. Sievers, C. E. Balatoni, L. Wong, J. J. Troke, M. T. N. Nguyen, C. M. Koehler, and M. A. Teitell. 28 August 2006, posting date. The TCL1 oncoprotein binds the RNase PH domains of the PNPase exoribonuclease without affecting its RNA degrading activity. Cancer Lett. [Online.] doi: 10.1016/j.canlet.2006.07.06. [DOI] [PubMed]
- 11.Fu, T. B., L. Virgilio, M. G. Narducci, A. Facchiano, G. Russo, and C. M. Croce. 1994. Characterization and localization of the TCL-1 oncogene product. Cancer Res. 54:6297-6301. [PubMed] [Google Scholar]
- 12.Fujiki, Y., A. L. Hubbard, S. Fowler, and P. B. Lazarow. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93:97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gewartowski, K., R. Tomecki, L. Muchowski, A. Dmochow Ska, A. Dzwonek, M. Malecki, H. Skurzak, J. Ostrowski, and P. P. Stepien. 2006. Up-regulation of human PNPase mRNA by beta-interferon has no effect on protein level in melanoma cell lines. Acta Biochim. Pol. 53:179-188. [PubMed] [Google Scholar]
- 14.Glick, B. S., A. Brandt, K. Cunningham, S. Muller, R. L. Hallberg, and G. Schatz. 1992. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell 69:809-822. [DOI] [PubMed] [Google Scholar]
- 15.Glick, B. S., and L. A. Pon. 1995. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 260:213-223. [DOI] [PubMed] [Google Scholar]
- 16.Gottlieb, R. A. 2001. Mitochondria and apoptosis. Biol. Signals Recept. 10:147-161. [DOI] [PubMed] [Google Scholar]
- 17.Hammen, P. K., D. G. Gorenstein, and H. Weiner. 1994. Structure of the signal sequences for two mitochondrial matrix proteins that are not proteolytically processed upon import. Biochemistry 33:8610-8617. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 19.Inoki, K., T. Zhu, and K. L. Guan. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115:577-590. [DOI] [PubMed] [Google Scholar]
- 20.Jan, Y., M. Matter, J. T. Pai, Y. L. Chen, J. Pilch, M. Komatsu, E. Ong, M. Fukuda, and E. Ruoslahti. 2004. A mitochondrial protein, Bit1, mediates apoptosis regulated by integrins and Groucho/TLE corepressors. Cell 116:751-762. [DOI] [PubMed] [Google Scholar]
- 21.Jones, R. G., D. R. Plas, S. Kubek, M. Buzzai, J. Mu, Y. Xu, M. J. Birnbaum, and C. B. Thompson. 2005. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 18:283-293. [DOI] [PubMed] [Google Scholar]
- 22.Joseph, S. B., M. N. Bradley, A. Castrillo, K. W. Bruhn, P. A. Mak, L. Pei, J. Hogenesch, M. O'Connell, R., G. Cheng, E. Saez, J. F. Miller, and P. Tontonoz. 2004. LXR-dependent gene expression is important for macrophage survival and the innate immune response. Cell 119:299-309. [DOI] [PubMed] [Google Scholar]
- 23.Koehler, C. M., E. Jarosch, K. Tokatlidis, K. Schmid, R. J. Schweyen, and G. Schatz. 1998. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science 279:369-373. [DOI] [PubMed] [Google Scholar]
- 24.Koehler, C. M., D. Leuenberger, S. Merchant, A. Renold, T. Junne, and G. Schatz. 1999. Human deafness dystonia syndrome is a mitochondrial disease. Proc. Natl. Acad. Sci. USA 96:2141-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koehler, C. M., S. Merchant, W. Oppliger, K. Schmid, E. Jarosch, L. Dolfini, T. Junne, G. Schatz, and K. Tokatlidis. 1998. Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J. 17:6477-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leszczyniecka, M., D. C. Kang, D. Sarkar, Z. Z. Su, M. Holmes, K. Valerie, and P. B. Fisher. 2002. Identification and cloning of human polynucleotide phosphorylase, hPNPase old-35, in the context of terminal differentiation and cellular senescence. Proc. Natl. Acad. Sci. USA 99:16636-16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leszczyniecka, M., Z. Z. Su, D. C. Kang, D. Sarkar, and P. B. Fisher. 2003. Expression regulation and genomic organization of human polynucleotide phosphorylase, hPNPase(old-35), a type I interferon inducible early response gene. Gene 316:143-156. [DOI] [PubMed] [Google Scholar]
- 28.Li, L. Y., X. Luo, and X. Wang. 2001. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 412:95-99. [DOI] [PubMed] [Google Scholar]
- 29.Li, Q. S., J. D. Gupta, and A. G. Hunt. 1998. Polynucleotide phosphorylase is a component of a novel plant poly(A) polymerase. J. Biol. Chem. 273:17539-17543. [DOI] [PubMed] [Google Scholar]
- 30.Lisitsky, I., A. Kotler, and G. Schuster. 1997. The mechanism of preferential degradation of polyadenylated RNA in the chloroplast. The exoribonuclease 100RNP/polynucleotide phosphorylase displays high binding affinity for poly(A) sequence. J. Biol. Chem. 272:17648-17653. [DOI] [PubMed] [Google Scholar]
- 31.Mandal, S., P. Guptan, E. Owusu-Ansah, and U. Banerjee. 2005. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev. Cell 9:843-854. [DOI] [PubMed] [Google Scholar]
- 32.Mohanty, B. K., and S. R. Kushner. 2000. Polynucleotide phosphorylase functions both as a 3′ → 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:11966-11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagaike, T., T. Suzuki, T. Katoh, and T. Ueda. 2005. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J. Biol. Chem. 280:19721-19727. [DOI] [PubMed] [Google Scholar]
- 34.Neuspiel, M., R. Zunino, S. Gangaraju, P. Rippstein, and H. McBride. 2005. Activated mitofusin 2 signals mitochondrial fusion, interferes with Bax activation, and reduces susceptibility to radical induced depolarization. J. Biol. Chem. 280:25060-25070. [DOI] [PubMed] [Google Scholar]
- 35.Piwowarski, J., P. Grzechnik, A. Dziembowski, A. Dmochowska, M. Minczuk, and P. P. Stepien. 2003. Human polynucleotide phosphorylase, hPNPase, is localized in mitochondria. J. Mol. Biol. 329:853-857. [DOI] [PubMed] [Google Scholar]
- 36.Raijmakers, R., W. V. Egberts, W. J. van Venrooij, and G. J. Pruijn. 2002. Protein-protein interactions between human exosome components support the assembly of RNase PH-type subunits into a six-membered PNPase-like ring. J. Mol. Biol. 323:653-663. [DOI] [PubMed] [Google Scholar]
- 37.Rainey, R. N., J. D. Galvin, H.-W. Chen, S. W. French, M. A. Teitell, and C. M. Koehler. 2006. A new function in translocation for the mitochondrial i-AAA protease Yme1: import of polynucleotide phosphorylase into the intermembrane space. Mol. Cell. Biol. 26:8488-8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ricci, J. E., C. Munoz-Pinedo, P. Fitzgerald, B. Bailly-Maitre, G. A. Perkins, N. Yadava, I. E. Scheffler, M. H. Ellisman, and D. R. Green. 2004. Disruption of mitochondrial function during apoptosis is mediated by caspase cleavage of the p75 subunit of complex I of the electron transport chain. Cell 117:773-786. [DOI] [PubMed] [Google Scholar]
- 39.Roesch, K., S. P. Curran, L. Tranebjaerg, and C. M. Koehler. 2002. Human deafness dystonia syndrome is caused by a defect in assembly of the DDP1/TIMM8a-TIMM13 complex. Hum. Mol. Genet. 11:477-486. [DOI] [PubMed] [Google Scholar]
- 40.Sarkar, D., I. V. Lebedeva, L. Emdad, D. C. Kang, A. S. Baldwin, Jr., and P. B. Fisher. 2004. Human polynucleotide phosphorylase (hPNPaseold-35): a potential link between aging and inflammation. Cancer Res. 64:7473-7478. [DOI] [PubMed] [Google Scholar]
- 41.Sarkar, D., M. Leszczyniecka, D. C. Kang, I. V. Lebedeva, K. Valerie, S. Dhar, T. K. Pandita, and P. B. Fisher. 2003. Down-regulation of Myc as a potential target for growth arrest induced by human polynucleotide phosphorylase (hPNPaseold-35) in human melanoma cells. J. Biol. Chem. 278:24542-24551. [DOI] [PubMed] [Google Scholar]
- 42.Sarkar, D., E. S. Park, L. Emdad, A. Randolph, K. Valerie, and P. B. Fisher. 2005. Defining the domains of human polynucleotide phosphorylase (hPNPaseOLD-35) mediating cellular senescence. Mol. Cell. Biol. 25:7333-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Susin, S. A., N. Zamzami, and G. Kroemer. 1998. Mitochondria as regulators of apoptosis: doubt no more. Biochim. Biophys. Acta 1366:151-165. [DOI] [PubMed] [Google Scholar]
- 44.Symmons, M. F., G. H. Jones, and B. F. Luisi. 2000. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Struct. Fold Des. 8:1215-1226. [DOI] [PubMed] [Google Scholar]
- 45.Symmons, M. F., M. G. Williams, B. F. Luisi, G. H. Jones, and A. J. Carpousis. 2002. Running rings around RNA: a superfamily of phosphate-dependent RNases. Trends Biochem. Sci. 27:11-18. [DOI] [PubMed] [Google Scholar]
- 46.Teitell, M. A. 2005. The TCL1 family of oncoproteins: co-activators of transformation. Nat. Rev. Cancer 5:640-648. [DOI] [PubMed] [Google Scholar]
- 47.Trounce, I. A., Y. L. Kim, A. S. Jun, and D. C. Wallace. 1996. Assessment of mitochondrial oxidative phosphorylation in patient biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 264:484-509. [DOI] [PubMed] [Google Scholar]
- 47a.Trumpower, B. L., and C. A. Edwards. 1979. Purification of a reconstitutively active iron-sulfur protein (oxidation factor) from succinate cytochrome c reductase complex of bovine heart mitochondria. J. Biol. Chem. 254:8697-8706. [PubMed] [Google Scholar]
- 48.Vahsen, N., C. Cande, J. J. Briere, P. Benit, N. Joza, N. Larochette, P. G. Mastroberardino, M. O. Pequignot, N. Casares, V. Lazar, O. Feraud, N. Debili, S. Wissing, S. Engelhardt, F. Madeo, M. Piacentini, J. M. Penninger, H. Schagger, P. Rustin, and G. Kroemer. 2004. AIF deficiency compromises oxidative phosphorylation. EMBO J. 23:4679-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Coster, R., J. Smet, E. George, L. De Meirleir, S. Seneca, J. Van Hove, G. Sebire, H. Verhelst, J. De Bleecker, B. Van Vlem, P. Verloo, and J. Leroy. 2001. Blue native polyacrylamide gel electrophoresis: a powerful tool in diagnosis of oxidative phosphorylation defects. Pediatr. Res. 50:658-665. [DOI] [PubMed] [Google Scholar]
- 50.van Loo, G., M. van Gurp, B. Depuydt, S. M. Srinivasula, I. Rodriguez, E. S. Alnemri, K. Gevaert, J. Vandekerckhove, W. Declercq, and P. Vandenabeele. 2002. The serine protease Omi/HtrA2 is released from mitochondria during apoptosis. Omi interacts with caspase-inhibitor XIAP and induces enhanced caspase activity. Cell Death Differ. 9:20-26. [DOI] [PubMed] [Google Scholar]
- 51.Verhagen, A. M., P. G. Ekert, M. Pakusch, J. Silke, L. M. Connolly, G. E. Reid, R. L. Moritz, R. J. Simpson, and D. L. Vaux. 2000. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102:43-53. [DOI] [PubMed] [Google Scholar]
- 52.von Heijne, G. 1992. Cleavage-site motifs in protein targeting sequences. Genet. Eng. 14:1-11. [DOI] [PubMed] [Google Scholar]
- 53.Yehudai-Resheff, S., M. Hirsh, and G. Schuster. 2001. Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell. Biol. 21:5408-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]