Abstract
The effects of transcriptional activation on the chromatin structure of the Saccharomyces cerevisiae HIS3 gene were addressed by mapping the precise positions of nucleosomes in uninduced and induced chromatin. In the absence of the Gcn4p activator, the HIS3 gene is organized into a predominant nucleosomal array. In wild-type chromatin, this array is disrupted, and several alternative overlapping nucleosomal arrays are formed. The disruption of the predominant array also requires the SWI/SNF remodeling machine, indicating that the SWI/SNF complex plays an important role in nucleosome mobilization over the entire HIS3 gene. The Isw1 remodeling complex plays a more subtle role in determining nucleosome positions on HIS3, favoring positions different from those preferred by the SWI/SNF complex. Both the SWI/SNF and Isw1 complexes are constitutively present in HIS3 chromatin, although Isw1 tends to be excluded from the HIS3 promoter. Despite the apparent disorder of HIS3 chromatin generated by the formation of multiple nucleosomal arrays, nucleosome density profiles indicate that some long-range order is always present. We propose that Gcn4p stimulates nucleosome mobilization over the entire HIS3 gene by the SWI/SNF complex. We suggest that the net effect of interplay among remodeling machines at HIS3 is to create a highly dynamic chromatin structure.
The central role of chromatin structure in gene regulation is currently the subject of intense interest. Much has been learned about the ATP-dependent chromatin remodeling machines, which use the free energy of ATP hydrolysis to move nucleosomes along DNA and alter nucleosome conformation (31), and the histone-modifying enzymes and their complexes. There is also a great deal of information available concerning changes in chromatin structure occurring at various gene promoters (3, 20). Because most of the obvious changes in chromatin structure occur at gene promoters, emphasis has been placed on these events, which are clearly of major importance. However, in our high-resolution studies of the chromatin structure of the CUP1 and HIS3 genes of Saccharomyces cerevisiae, we have obtained evidence pointing to more widespread changes in chromatin structure resulting from induction; these changes involve the entire gene and flanking sequences (9, 29). Our observations indicate that, at least for these genes, chromatin remodeling is not confined to the promoter but occurs on the scale of a chromatin domain.
It is now apparent that ATP-dependent chromatin remodeling machines can be grouped into several functional classes (2). One class includes the SWI/SNF and RSC complexes, which are capable of mobilizing nucleosomes and driving a conformational change in the nucleosome to create the remodeled state (24, 31). A second class is defined by the ISWI group of complexes, exemplified in budding yeast by the Isw1 and Isw2 complexes, which are capable of mobilizing nucleosomes to create arrays with characteristic spacing but cannot remodel nucleosome structure (19, 39). A third class of complex is defined by the Swr1 complex, which remodels chromatin by catalyzing replacement of H2A with a variant form, H2AZ (see reference 11 for a review). It is becoming clear that the regulation of at least some and perhaps all yeast genes, including HIS3, involves more than one remodeling complex (35).
HIS3 encodes an enzyme required for the biosynthesis of histidine (32). The transcription of HIS3 is activated in response to amino acid starvation via the Gcn4p activator (5, 6, 23). There is a single high-affinity binding site for Gcn4p in the HIS3 promoter. The activation of HIS3 by Gcn4p involves contributions from the SWI/SNF ATP-dependent chromatin remodeling machine (9, 22), the mediator complex (23), and the Gcn5p (13, 14) and Esa1p (25) histone acetyltransferase complexes. The HIS3 promoter also contains a poly(dA-dT) element, which stimulates Gcn4p-activated transcription by virtue of its intrinsic DNA structure (8, 17), perhaps by increasing the accessibility of nucleosomal DNA target sites (1).
Inspired by the work of Thoma and colleagues (37), we have developed a model system for purifying yeast genes as plasmid chromatin (10, 29). In a study of the chromatin structure of a purified TRP1ARS1 plasmid containing the HIS3 gene, we demonstrated that the plasmid chromatin exists in two alternative structural states which, for simplicity, are referred to as remodeled and unremodeled chromatin (9). HIS3 plasmid chromatin from uninduced cells is predominantly composed of fully supercoiled chromatin that is generally protected from cleavage by restriction enzymes, indicating that it has a canonical chromatin structure, with a minor component corresponding to remodeled chromatin. In contrast, induced chromatin is predominantly composed of remodeled chromatin, characterized by a much reduced level of negative supercoiling, decreased compaction, and increased sensitivity to restriction enzymes, indicating a highly accessible chromatin structure. Since the formation of remodeled chromatin requires both Gcn4p and the SWI/SNF remodeling machine, it has been suggested that Gcn4p recruits the SWI/SNF complex to the HIS3 promoter, where it directs the remodeling of a chromatin domain defined by the HIS3 gene, facilitating transcription (9).
We are interested in examining the structural consequences of interplay among remodeling machines. To address this interest, we have begun to study the effects of remodelers on HIS3 chromatin. Here, we have compared the positions of nucleosomes on the HIS3 gene in uninduced and induced wild-type, gcn4Δ, snf2Δ, and isw1Δ chromatin. Our studies reveal that extensive nucleosome movements occur on gene activation and that these require both the Gcn4p activator and the SWI/SNF complex. The Isw1 complex plays a more subtle role in determining nucleosome positions on HIS3. We found evidence for long-range order in the chromatin structure, even though it is composed of multiple alternative nucleosomal arrays. We propose that the net effect of the interplay among remodeling machines at HIS3 is to create a highly dynamic chromatin structure.
MATERIALS AND METHODS
Plasmids and yeast strains.
The TRP1ARS1HIS3 (TA-HIS3) strains (wild type, gcn4Δ, and snf2Δ) have been described previously (9). YTT186 (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5+isw1Δ::ADE2) (40) was transformed with TA-HIS3 as described previously (9). Strains used for chromatin immunoprecipitation (ChIP) experiments were based on the untagged wild-type strain W1588-4C (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5+) (40). W1588-4C, YTT1448 (ISW1-3FLAG::KanMX), and YTT1722 (SNF2-3FLAG::KanMX) were converted to his+ by the repair of the chromosomal HIS3 locus using the 982-bp EcoRI fragment containing HIS3 from pGEM-TA-HIS3B (9); transformants were selected on plates lacking histidine. The repair of the HIS3 locus was confirmed by Southern blotting. These strains were designated YDC111, YDC112, and YDC113, respectively. YDC112 and YDC113 were transformed to gcn4Δ::URA3 by using an XhoI digest of p385 to obtain YDC172 and YDC173, respectively. p385 was constructed by inserting the 1,162-bp SmaI-PmeI URA3 fragment from pNEB-URA3 (29) at the MscI site inside the GCN4 coding region in pBS-GCN4. This plasmid contained GCN4 as an NdeI-XhoI insert obtained by PCR using yeast genomic DNA with CATATGTCCGAATATCAGand CTCGAGGCGTTCGCCAAC as primers in pBluescript II SK(+). YDC188 (ISW1-3FLAG snf2Δ::URA3) was obtained by transforming YDC112 using the 1,648-bp snf2Δ::URA3 XhoI fragment from p435. The snf2Δ::URA3 fragment was made by PCR using pLY21 (15) as a template with CTCGAGTCGCGATATGTTAAAACCGCG and CTCGAGTCCGCTTCATCTGTG as primers and TA cloned into pCRIITOPO (Invitrogen) to give p435. YDC180 (SNF2-3FLAG isw1Δ::ADE2) was obtained by transforming YDC113 with a 3.1-kb fragment containing isw1Δ::ADE2 derived by PCR using genomic DNA from YTT186 with GGCGCGGGTACCGTGCACCGTATCCTCCATAGC and GGCGCGGGTACCGGACCAAGAAATCCAAAGCCTG as primers. For ChIP for Gcn4p, YDC172 (SNF2-3FLAG gcn4Δ::URA3) was transformed either with pRS415, a centromeric LEU2 vector (Stratagene), to obtain YDC197 or with p448 (pRS415 carrying GCN4 with 13 C-terminal myc tags) to obtain YDC196. p448 was constructed by transferring the 2.4-kb SalI-PvuII fragment containing the 13-myc-tagged GCN4 gene from pSK1 (35) to pRS415 cut with SalI and SmaI. A strain with a single base deletion in the Gcn4p binding site (YDC246, SNF2-3FLAG gcn4Δ::URA3 his3::ADE2 pRS-Gcn4-myc [p448]) was constructed by the transformation of YDC196 with an EcoRI digest of p522, resulting in the disruption of HIS3 by the insertion of ADE2. p522 was constructed by the insertion of an AvrII fragment containing ADE2, which was derived by PCR using yeast genomic DNA and GGCGCGCCTAGGTTCTTGAATAATACATAACTTTTCTTAAAAG and GGCGCGCCTAGGGATCTTATGTATGAAATTCTTAAAAAAGG as primers, into the AvrII site within the HIS3 open reading frame (ORF) in p517, with ADE2 and HIS3 in opposing orientations. p517 is pGEM-TAHIS3B with a deletion of the T residue in the Gcn4p binding site in the HIS3 promoter (ATGACTC [the deleted residue is underlined]) described previously (9). Transformants were selected on plates containing synthetic complete medium lacking leucine and adenine; the his− phenotype was confirmed. The presence of the point deletion in the HIS3 promoter was confirmed by showing the loss of the FokI site (Southern blotting) and by FokI digestion of a HIS3 promoter fragment obtained by PCR from genomic DNA. YDC247 is identical to YDC246, except that the HIS3 promoter is wild type. All strains were confirmed by Southern blotting.
Plasmid chromatin purification and monomer extension.
Plasmid chromatin was purified from late-log-phase cells grown in synthetic complete medium lacking tryptophan (uninduced) or both tryptophan and histidine (induced) as described previously (9). Micrococcal nuclease (MNase) digestions of TA-HIS3 chromatin were performed as described previously (4). The monomer extension protocol has been described previously (10). Briefly, 20 ng of purified TA-HIS3 chromatin was digested to nucleosome core particles in 10 mM HEPES-KOH (pH 7.9), 125 mM KCl, 2.5 mM MgCl2, 1 mM dithiothreitol, 2.5 mM CaCl2 for 5 min at 30°C with 0.05 to 0.1 U/μl of MNase (Worthington). The digestion was stopped by the addition of EDTA to 20 mM and sarcosyl to 0.5%. DNA was extracted, and core particle DNA (147 to 160 bp) was purified from a 3% (wt/vol) agarose gel. Core DNA was end labeled using T4 polynucleotide kinase and [γ-32P]ATP, denatured in 0.2 M NaOH, neutralized with 3 M sodium acetate, and precipitated. Labeled core DNA was annealed to single-stranded pGEM-TAHIS3B (9) acting as template and extended with Superscript II RNase H− reverse transcriptase (Invitrogen), in the presence or absence of an appropriate restriction enzyme. With the plus strand of pGEM-TAHIS3B as a template, EcoRV, XbaI, PmlI, MscI, or BglI was used for mapping. With the minus strand of pGEM-TAHIS3B strand as a template, DraIII, BglI, NdeI, PmlI, or NaeI was used. Extension products were resolved in 6% denaturing polyacrylamide gels.
Indirect end-labeling analysis.
Nuclei were prepared from yeast cells grown to mid-log phase in synthetic medium and digested with MNase as described previously (4). The DNA was purified, digested with BamHI, and electrophoresed in long 1.5% (wt/vol) agarose gels. The gels were blotted and probed with a DED1 fragment corresponding to nucleotides +53 to +328 with respect to the DED1 start codon and labeled by random priming. The markers were prepared by mixing separate restriction digests of a 1,757-bp PCR fragment corresponding almost exactly to the genomic BamHI fragment containing HIS3 (the restriction enzymes used were HaeII, BsaBI, MscI, BglI, NheI, PstI, NsiI, XhoI, and AflII).
Chromatin immunoprecipitation.
ChIP experiments were based on the method of McConnell et al. (18). One liter of cells was grown to an optical density at 600 nm of about 0.5. For the induction with 3-aminotriazole (3-AT; Aldrich), cells at an initial A600 of 0.25 were grown for 2 to 4 h at 30°C. For the induction of strains YDC246 and YDC247 (which are his−), cells were grown in synthetic complete medium containing histidine to an optical density of about 0.5, collected by rapid filtration, resuspended in medium lacking histidine for 1 h, and then induced with 3-AT for 20 min. Cells were fixed with 1% formaldehyde for 20 min at room temperature on a shaker at 125 rpm by the addition of a one-tenth volume of 11% formaldehyde in 0.1 M NaCl, 50 mM HEPES-K, pH 7.6, 1 mM Na-EDTA. The reaction was stopped with 200 ml 2.5 M glycine and shaken for 5 min. Fixed cells were collected by vacuum filter, washed with 20 mM Tris-HCl, pH 8.0, 0.15 M NaCl, and stored in 2 aliquots at −80°C. For the preparation of cross-linked chromatin, fixed cells from a 500-ml culture were resuspended in 1 ml 0.1 M Tris-HCl, pH 8.0, 20% glycerol with protease inhibitors lacking EDTA (Roche), mixed with an equal volume of glass beads (acid washed; 425 to 600 μm; catalog no. G-8772; Sigma), and shaken vigorously for 40 min at 4°C. The lysate was diluted with 2 ml cold FA buffer (50 mM HEPES-K, pH 7.6, 0.15 M NaCl, 1 mM Na-EDTA, 1% Triton X-100, and 0.1% sodium deoxycholate with protease inhibitors as described above) and spun (1 min, 14,000 rpm, 4°C). The pellet was resuspended in 2 ml FA buffer and spun again; this wash was repeated. The pellet was resuspended in 1 ml FA buffer in a round-bottomed 14-ml tube placed in ice water for sonication using a Misonix Sonicator 3000 (14 pulses of 30 s separated by 30-s pauses for cooling; the power output setting was 2.5 for 6 W). The sample was diluted with 1.8 ml FA buffer and spun (14,000 rpm, 1 h, and 4°C). The supernatant containing the fragmented fixed chromatin was aliquoted and stored at −80°C. DNA concentrations were measured using the Hoechst assay. The average size of the DNA was 400 to 500 bp. For the IP reactions, two batches of protein G magnetic beads (catalog no. 100.04; Dynal), each with 20 μl suspension per IP sample were washed three times with 1 ml 5 mg/ml bovine serum albumin (BSA) (immunoglobulin G-free; catalog no. A-9085; Sigma) in phosphate-buffered saline (PBS) and incubated overnight with or without (mock) antibody on a rotator at 4°C. The monoclonal antibodies used were M2-anti-FLAG (5 μg/sample; catalog no. F-3165; Sigma) and anti-myc 9E10 (1 μg antibody/sample; catalog no. sc-40; Santa Cruz). The beads were washed twice with BSA-PBS and resuspended in 30 μl BSA-PBS per sample. Equal amounts of chromatin (11 μg DNA) were adjusted to 55 μg DNA/ml (600 μl) with FA buffer, spun (14,000 rpm, 15 min, and 4°C), and filtered (0.45-μm Ultra-free MC low binding filter; Millipore) to remove insoluble material. Input samples (60 μl) were removed; the cross-links were reversed by incubation at 65°C overnight in 5 mM Na-EDTA, 0.5% sodium dodecyl sulfate, followed by digestion with proteinase K at 50 μg/ml for 4 h at 55°C; the DNA was purified. IP samples (220 μl) were mixed with 30 μl beads with or without (mock) antibody on a rotator (90 min at room temperature). The beads were subjected to a series of 1-ml washes (5 min each on a rotator at room temperature): three washes with FA buffer, three with FA buffer containing 0.5 M NaCl, and two with radioimmunoprecipitation assay buffer (10 mM Tris-HCl, pH 8.0, 0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM Na-EDTA, and protease inhibitors). FLAG samples were eluted with 3-FLAG peptide (catalog no. F-4799; Sigma) at 5 mg/ml in 0.1 M HEPES-K, pH 7.6, 20% glycerol, 0.1 M KCl, 0.6 mM Na-EDTA, 2 mM MgCl2, 0.02% NP-40; 10 μl peptide was added to the drained beads and mixed; and 40 μl radioimmunoprecipitation assay buffer was added, and the elutions were placed in an Eppendorf Thermomixer (1,200 rpm for 30 min at 22°C). The elution was repeated, the eluates were pooled, the cross-links were reversed, and the purified DNA was dissolved in 25 μl of 10 mM Tris-HCl, pH 8.0, 0.1 mM Na-EDTA. Myc-antibody IPs were eluted with 120 μl of 50 mM Tris-HCl, pH 8.0, 5 mM Na-EDTA, 0.5% sodium dodecyl sulfate. For the quantification of DNA in the IP samples by PCR with internal control, 2.5 μl DNA was mixed with 22.5 μl of 2 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 0.2 μM each radiolabeled primer, and 0.5 U Pt Taq polymerase (Invitrogen) in Pt Taq buffer (all components were mixed together before DNA was added). Dilutions (1:100 and 1:500) of input DNA were assayed. Primers for PCR were the HIS3 promoter (CTTGGCCTCCTCTAGTACACTC and CATTTGTAATACGCTTTACTAGGGC, yielding a 238-bp fragment), HIS3 ORF (GACCATCACACCACTGAAGACT and AAAGTGCCTCATCCAAAGGCG; 120 bp), POL1 ORF (GCTCTGGTAGGCTGATATGTGA and GGGCCATTGTCATACTATTTACATC; 180 bp), and ARG1 promoter (ACGGCTCTCCAGTCATTTAT and GCAGTCATCAATCTGATCCA; 163 bp) (35). Different HIS3 promoter primers (CTTAGCGATTGGCATTATCACATAATG and GCCTTCGTTTATCTTGCCTGC; 102 bp), with Int-V (12) as internal control (CTTCCTGGCTGTCAGAATATGGG and CACCCCGAAGCTGCTTTCACAATAC; 153 bp), were also used (see Fig. 5E). One primer of each pair was end labeled using T4 kinase (USB). The PCRs were stopped with an equal volume of 20 mM Tris-HCl, pH 8.0, 10 mM Na-EDTA, 20% glycerol containing 0.5 μg of an MspI digest of pBR322 as carrier and analyzed in a denaturing 6% (19:1) polyacrylamide gel. The data were quantified using a phosphorimager.
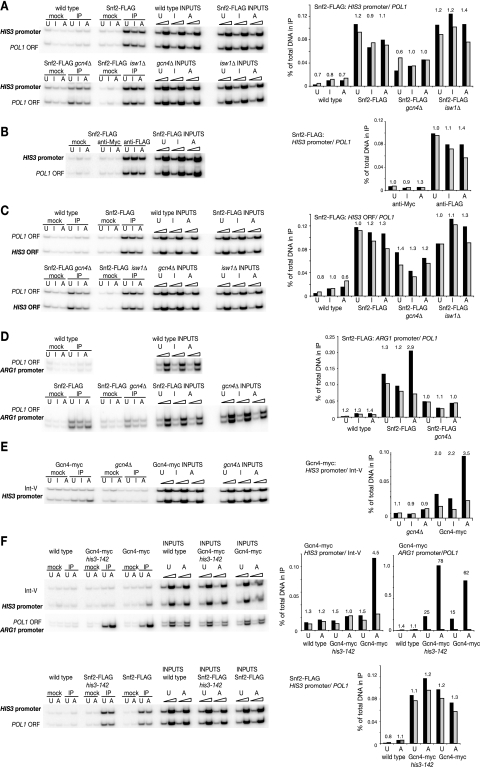
FIG. 5.
SWI/SNF remodeling complex is constitutively present on the chromosomal HIS3 gene. Results from ChIP experiments are shown. (A) The SWI/SNF complex is present at the HIS3 promoter independently of the Gcn4p activator. The Snf2p ATPase subunit of the SWI/SNF complex was tagged with three FLAG epitopes (SNF2-FLAG) in otherwise wild-type cells and in gcn4Δ and isw1Δ cells; the wild type has no tag. IP, DNA immunoprecipitated using anti-FLAG antibody; mock, no antibody. The amounts of DNA in the immunoprecipitates were measured using multiplex PCR with end-labeled PCR primers. The HIS3 promoter was a 238-bp fragment, and the POL1 ORF internal control was 180 bp. Samples were as follows: U, uninduced; I, partially induced; and A, fully induced with 3-AT. Two input DNA dilutions (1 in 100 and 1 in 500) were measured. The percentages of DNA in the IPs relative to the input for the HIS3 promoter (black bars) and the POL1 ORF (gray bars) were measured using a phosphorimager and are graphed at right. The HIS3 promoter/POL1 ORF ratios are given above each pair of bars. For clarity, the quantification of the mock samples is not shown but, in all cases, the mock samples were 0.01% or less of input DNA. (B) Immunoprecipitation using an unrelated monoclonal antibody (anti-Myc 9E10) using the SNF2-FLAG strain described in panel A. (C) The SWI/SNF complex is present on the HIS3 ORF. The experiment was carried out as described for panel A, except that primers for the HIS3 ORF were used (yielding a 120-bp fragment), together with the POL1 primers (180 bp). (D) Gcn4p recruits more SWI/SNF complex to the ARG1 promoter. The same samples were used as in panels A and C, except that primers for the ARG1 promoter (which, like HIS3, is induced by 3-AT and regulated by Gcn4p) were used (yielding a 163-bp fragment), together with the POL1 primers (180 bp). (E) Gcn4p is present at the HIS3 promoter. A gcn4Δ strain was transformed with a centromeric (low-copy) plasmid with or without an inserted GCN4 gene carrying 13 myc tags encoded at the C terminus. ChIP experiments used anti-myc antibody (IP) or no antibody (mock). Different primers from those above were used for the HIS3 promoter, resulting in a 102-bp fragment; the control was a noncoding region of chromosome V (Int-V), yielding a 153-bp fragment. (F) Gcn4p does not recruit significant amounts of SWI/SNF to the HIS3 promoter. Strains with or without a point deletion in the binding site for Gcn4p in the HIS3 promoter (the his3-142 mutation [34]) were used. These strains expressed both Gcn4-myc and Snf2-FLAG. Top, ChIP using anti-Myc antibody to detect Gcn4-myc at the mutated and wild-type HIS3 promoter and at the ARG1 promoter as a control. Note the different scales for the HIS3 and ARG1 promoters. Bottom, ChIP using anti-FLAG antibody to detect Snf2-FLAG at the mutated and wild-type HIS3 promoter.
RESULTS
The TRP1ARS1HIS3 minichromosome.
The TRP1ARS1HIS3 (TA-HIS3) plasmid (9) was constructed by the insertion of the HIS3 gene at the EcoRI site of TRP1ARS1, a well-studied, high-copy yeast plasmid (37, 45) (Fig. 1A). The TRP1 gene encodes an enzyme required for the biosynthesis of tryptophan and serves as a selection marker; cells were grown in synthetic medium lacking tryptophan to maintain the plasmid. In the following experiments using TA-HIS3, two different growth conditions were employed: (i) HIS3 expressed at basal (uninduced) levels (synthetic medium lacking tryptophan but including histidine) and (ii) induced HIS3 (synthetic medium lacking both tryptophan and histidine). The induction of HIS3 under these conditions is weak (only about twofold), but the changes in chromatin structure associated with this induction are quite profound (9).
FIG. 1.
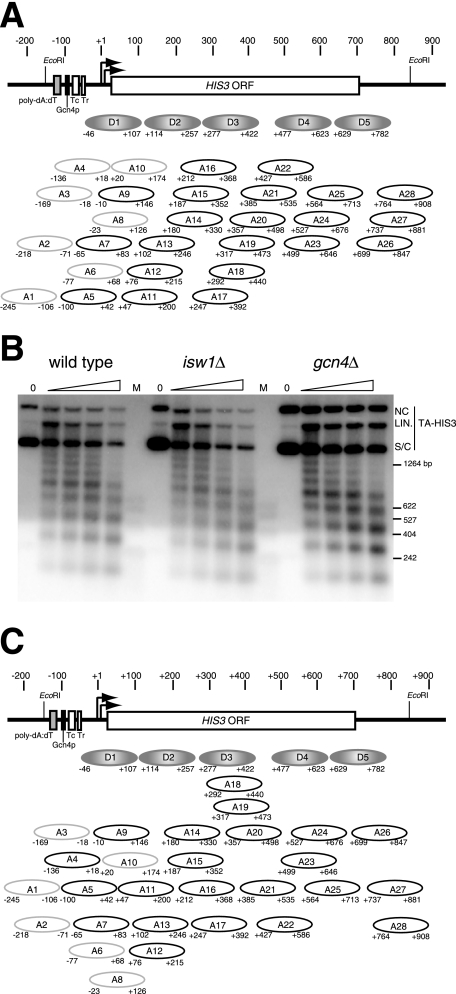
Monomer extension mapping of nucleosomes positioned on HIS3 in TA-HIS3 chromatin purified from wild-type and gcn4Δ cells. (A) Map of the TRP1ARS1HIS3(TA-HIS3) plasmid (9). HIS3 was inserted at the EcoRI site of the TRP1ARS1 plasmid (37). TA-HIS3 has only 2,435 bp and contains no bacterial sequences. TRP1 is expressed at basal levels because the upstream activation sequence for TRP1 is not present in TRP1ARS1. The unique XbaI site used in the mapping experiments shown here is located in the TRP1 ORF. (B) Schematic diagram of the monomer extension method for mapping the positions of nucleosomes (41). The approach is to obtain DNA from nucleosome core particles (mononucleosomes or monomers) and use this as primer in a primer extension reaction; since both strands of nucleosomal DNA can act as primers, a single-stranded template must be used (otherwise two sets of extension products will be obtained). pGEM-TAHIS3 contains the entire TA-HIS3 sequence as an insert. Note that this method can resolve overlapping positions. (C) High-resolution monomer extension mapping of TA-HIS3 chromatin purified from uninduced (unind.) and induced wild-type cells and from gcn4Δ cells. Nucleosome positions were mapped with respect to the XbaI site in the TRP1 ORF (see panel A). The products of monomer extension reactions using nucleosomal DNA were analyzed with (+) or without (−) XbaI digestion in long sequencing gels. The purpose of monomer extension without XbaI cleavage was to identify bands resulting from premature termination by reverse transcriptase (lanes 4, 6, and 8). In the samples digested with XbaI, each band represents the distance of the downstream border of a nucleosome from the XbaI site. The bands which are quantitatively above background are labeled (nucleosomes D1 to D5 and A4 to A28); some are quantitatively so minor that they have been ignored. Some bands have been grouped (indicated by black bars at right) because they are less than 15 bp apart and so might represent incomplete trimming of the same nucleosome. The HIS3 transcription unit (with transcription start site at nucleotide +1) is shown at right, together with a series of gray ovals indicating the positions of the predominant (D) nucleosomes; these nucleosomes are present in wild-type and gcn4Δ chromatin but are clearly predominant in gcn4Δ chromatin. End-labeled markers: 100 bp ladder (New England Biolabs) (lane 1), a DdeI digest of λ DNA (lane 2), and a HinfI digest of λ DNA (lane 3). A phosphorimage is shown. The position information is summarized in Fig. 2A. (D) Scans of the monomer extension maps shown in panel C. The lanes corresponding to gcn4Δ chromatin (lane 9), induced wild-type chromatin (lane 7), and uninduced wild-type chromatin (lane 5) were scanned using a phosphorimager and normalized to the major peak at the top of the gel.
Attempts to increase the level of induction using 3-AT, a competitive inhibitor of the enzyme encoded by HIS3, were only marginally successful (about twofold). In contrast, a fivefold induction of chromosomal HIS3 by 3-AT was observed in cells lacking the TA-HIS3 plasmid (not shown). The amount of HIS3 mRNA in TA-HIS3 strains was about 10-fold higher than that in cells carrying only chromosomal HIS3, presumably reflecting the fact that the TA-HIS3 strains carry multiple copies of HIS3 (we estimated that each cell contains 10 to 20 TA-HIS3 molecules on average) (data not shown). It is therefore likely that 3-AT was ineffective in TA-HIS3-containing cells because it could not inhibit the excess HIS3 enzyme efficiently. This explanation is consistent with our observation that the growth of TA-HIS3 strains was only modestly affected by 3-AT, whereas the growth of strains with chromosomal HIS3 was virtually halted by 3-AT, which was clearly quite poisonous (not shown). Thus, as discussed in our previous study (9), the effects of induction on HIS3 chromatin structure were best determined by comparing the chromatin structures of TA-HIS3 isolated from induced wild-type cells and from gcn4Δ cells, which lack the Gcn4p activator and so provide basally expressing chromatin. There is an approximately fivefold induction of HIS3 in synthetic medium lacking histidine relative to that of gcn4Δ cells (9).
Mapping nucleosome positions using the monomer extension technique.
An advantage of the high-copy plasmid system is that plasmid chromatin can be purified in sufficient quantities to perform high-resolution monomer extension mapping of the precise locations of the nucleosomes. The monomer extension method (41) is based on the protection of 147 bp of DNA in the nucleosome core particle from digestion by MNase. Since this is the operational definition of the nucleosome, the method provides information about nucleosome positions. This contrasts with the standard technique, mapping by indirect end labeling, which requires relatively early MNase digestion times and therefore measures relative protection rather than nucleosome positions since, under these conditions, nucleosomes are not trimmed to their borders (further discussed below).
The monomer extension method involves the complete digestion of purified plasmid chromatin to nucleosome core particles using MNase (Fig. 1B). Briefly, the nucleosomal DNA is used as a primer and single-stranded target plasmid DNA is used as a template in a primer extension reaction. The product DNA is digested with a restriction enzyme having a unique site in the plasmid, and a map is produced from the sizes of the product bands, which represent the distance of the far border of each nucleosome to the restriction site. More specifically, nucleosomal DNA (145 to 160 bp) is purified from a gel and end labeled with T4 polynucleotide kinase. Labeled core DNA is annealed to single-stranded template plasmid and used as a primer for extension using a DNA polymerase. The template plasmid in our case (pGEM-TAHIS3) contains the entire TA-HIS3 plasmid sequence as an insert. Single-stranded DNA must be used as the template because both strands of core particle DNA would anneal if the template were double stranded; this would result in extension in both directions, confounding interpretation. The replicated DNA is then digested with a restriction enzyme which has a unique site in the insert to be mapped (i.e., TA-HIS3). The DNA fragments produced are resolved and measured accurately in long sequencing gels. Long stretches of chromatin can be mapped in great detail in one reaction, and nucleosome positions can be quantified relative to one another.
To map HIS3 chromatin, the product DNA was digested with XbaI, which has a single site in pGEM-TAHIS3, located within the TRP1 gene (Fig. 1A). The XbaI site was chosen because almost all of HIS3 can be mapped in a single reaction. The lengths of the resulting DNA fragments were determined accurately in sequencing gels; each band represents a nucleosome border, defining the distance from the far border of the nucleosome to the XbaI site. A control reaction designed to detect any sequence-dependent pausing/termination by reverse transcriptase involved an identical reaction except for the omission of XbaI; there should be no bands in the control other than the long extension products.
The monomer extension technique is not yet widely used, so it is worthwhile to discuss some technical points. A slight underdigestion of chromatin by MNase results in core particles that are not completely trimmed to 147 bp. Consequently, bands within about 15 bp of one another might represent different degrees of trimming of the same positioned nucleosome. For this reason, clusters of bands within 15 bp were counted as the same nucleosome in our analysis. If core particles are heavily overdigested by MNase, nicks begin to appear. Labeled core DNA was routinely checked in denaturing gels: the size range was typically 140 to 160 bp with very little nicking. In any case, nicking would not affect the result because kinase does not label nicks and end-labeled nicked DNA strands liberated on denaturation of nucleosomal DNA would give the correct result on extension. Proteins other than nucleosomes which might be bound to the minichromosome will not interfere with nucleosome mapping unless they protect 140 to 160 bp of DNA against extensive digestion by MNase because nucleosomal DNA is subsequently gel purified. Major contributions from nonhistone proteins would appear as nucleosome-free gaps in the map.
The transcriptional activator Gcn4p directs the disruption of a predominant nucleosome array on HIS3.
HIS3 plasmid chromatin was purified from uninduced and induced wild-type cells and from gcn4Δ cells grown in the absence of histidine. Their chromatin structures were compared by monomer extension mapping relative to the XbaI site inside the TRP1 open reading frame. In uninduced and induced wild-type cells, a very complex chromatin structure was observed (Fig. 1C, lanes 5 and 7). Many bands of various intensities were observed; each band indicates the downstream border of a nucleosome with respect to the XbaI site. These bands were not the result of problems with elongation by the reverse transcriptase because no bands were observed without XbaI digestion (Fig. 1C, lanes 4, 6, and 8). Theoretically, the HIS3 gene could accommodate a maximum of about six uniquely positioned nucleosomes if packed close together. However, there were many more than six bands in uninduced and induced chromatin and neighboring bands were much less than 147 bp apart (the size of a nucleosome). Thus, the monomer extension map revealed the presence of many overlapping nucleosome positions on the HIS3 gene. Since nucleosomes cannot physically overlap, these positions represent alternative positions and the chromatin structure observed must represent the superimposition of several alternative chromatin structures. It is concluded that the HIS3 gene can exist in any of several overlapping nucleosomal arrays in wild-type cells.
It should be noted that the structure of uninduced wild-type chromatin was quite variable (unlike that of induced chromatin); in most experiments, multiple overlapping positions were observed as shown here (Fig. 1C, lane 5) but, in some experiments, a more ordered structure, similar to that of gcn4Δ chromatin (see below), was observed (not shown). This variability was probably due to variable degrees of HIS3 induction in uninduced cells (9). It is important to understand that, although the monomer extension maps were very complex, the same bands were observed in all mapping experiments; it was their intensities that varied, indicating that nucleosomes were redistributed among a large but fixed number of possible alternative positions.
The chromatin structure of HIS3 expressed at basal levels was analyzed by mapping TA-HIS3 chromatin derived from gcn4Δ cells. This had a more ordered structure over most of the HIS3 gene (Fig. 1C, lane 9): an array of predominant positions was apparent (labeled D1 to D5). Many relatively minor alternative positions were also detected, representing a significant fraction of the total nucleosomes (labeled A1 to A28). D1, D2, and, to a lesser extent, D3 were clearly stronger than their neighboring overlapping positions (Fig. 1D), but there was more ambiguity in the region of the D4 and D5 nucleosomes; D4 was only marginally stronger than the nearby A22 nucleosome, and D5 was weaker than A25. D4 and D5 were included in the predominant array rather than A22 and A25 because the latter pair of positions overlap one another and A25 overlaps D4. Also, in some mapping experiments (e.g., see Fig. 3), D5 was stronger than A25. In uninduced and induced wild-type cells, the D positions were less prominent and some of the A positions were enhanced such that they were of intensities similar to those of the D positions (e.g., A5, A7, A9, A11, A12, and A15) (Fig. 1C, lanes 5 and 7). Thus, in wild-type cells, the predominant array was only one of several possible alternative nucleosome arrays. The nucleosome positions corresponding to the bands observed are shown in detail in Fig. 2A.
FIG. 3.
Nucleosome mobilization in HIS3 chromatin requires the Gcn4p activator and the SWI/SNF remodeling complex. (A) Monomer extension mapping of HIS3 in TA-HIS3 chromatin purified from induced wild-type, gcn4Δ, snf2Δ, and isw1Δ cells. Nucleosomes were mapped with respect to the XbaI site in the TRP1 ORF (see Fig. 1A); monomer extension without XbaI cleavage was to identify bands due to premature termination by the DNA polymerase. The bands corresponding to nucleosomes D1 to D5 and A4 to A28 are indicated (Fig. 2A shows a summary map). The asterisk indicates a nucleosome-free gap apparent in isw1Δ chromatin (see the text). Markers (lanes 1, 2, and 3) were as described in the legend for Fig. 1C. A phosphorimage is shown. (B) Phosphorimager scans of the monomer extension maps shown in panel A. The scans were normalized to the major peak at the top of the gel.
FIG. 2.
HIS3 chromatin is organized into overlapping nucleosomal arrays. (A) Arrays of positioned nucleosomes on HIS3. The precise positions of the nucleosomes mapped by monomer extension (as in Fig. 1) are given with downstream borders (relative to the HIS3 promoter) mapped with respect to the XbaI site in the TRP1 ORF (Fig. 1) and upstream borders mapped in independent experiments using other restriction enzymes (data not shown). HIS3 coordinates are given with respect to the transcription start sites at nucleotides +1 and +13 (33) (indicated by the arrows). The most important sequence elements in the HIS3 promoter are indicated by labeled boxes: the poly(dA-dT) element, the binding site for the Gcn4p activator, and the two TATA elements (TR and TC) (32). TA-HIS3 chromatin from gcn4Δ cells has a predominant array of nucleosomes over the HIS3 gene (Fig. 1), shown here as gray ovals labeled D1 to D5. This D array is not predominant in wild-type cells; the A nucleosomes, which overlap the D nucleosomes, are much more prominent in wild-type cells (A1 to A28). Quantitatively minor positions (A1 to A4, A6, A8, and A10) are shown as faint ovals. Nucleosomes on the HIS3 promoter elements (A1 to A3) were mapped in separate monomer extension experiments (data not shown). (B) Nucleosomal organization of the TA-HIS3 minichromosome in wild-type, isw1Δ, and gcn4Δ cells. Nuclei containing TA-HIS3 chromatin were digested with different amounts of MNase, and the resulting DNA fragments were separated in an agarose gel, which was then blotted and probed with TA-HIS3 DNA labeled by random priming. Some of the chromatin was resistant to digestion, perhaps due to clumping of nuclei or unlysed spheroplasts. The supercoiled (S/C), nicked circular (NC), and linear (LIN.) forms of TA-HIS3 DNA are indicated. (C) Possible arrangements of the nucleosomes shown in panel A into alternative arrays with predicted short repeat lengths (165 to 175 bp). Some nucleosomes could be alternatives for the same array (e.g., A18 and A19 could substitute for D3 and A3-A9-A14 could be replaced with A4-A10-A15). In this diagram, each nucleosome is depicted only once, but more arrays could be drawn if the same nucleosomes can occur in more than one array.
The appearance of so many alternative nucleosome positions in wild-type chromatin suggested that the predominant (D) nucleosomal array observed in gcn4Δ chromatin was “scrambled” in wild-type chromatin, with D nucleosomes displaced to alternative A positions. However, a strong nucleosomal repeat was preserved in TA-HIS3 chromatin from wild-type and gcn4Δ cells (Fig. 2B), indicating that the chromatin was still highly ordered. The repeat lengths of wild-type, gcn4Δ, and isw1Δ TA-HIS3 chromatin were about 165 bp, as expected (38). Taken together, the repeat length and the monomer extension mapping data suggested that nucleosomes are arranged in alternative arrays, with an average spacing of about 165 bp. Consistent with this interpretation, the A nucleosomes can be arranged into several alternative arrays of nonoverlapping positions with predicted repeats of about 170 bp (Fig. 2C). The arrangements shown are not the only possibilities; some nucleosomes could be alternatives in other arrays, and more arrays can be drawn if a nucleosome is allowed to appear in more than one array.
Nucleosomes on the HIS3 promoter were mapped in separate experiments using different restriction enzymes (not shown). Although some nucleosomes were detected on the HIS3 promoter (A1 to A4), these were all quantitatively relatively minor, indicating that nucleosome positions containing the TATA boxes, the Gcn4p binding site, and/or the poly(dA-dT) site were not usually occupied (see Discussion). The predominant nucleosome on the HIS3 promoter, D1, covered the transcription start sites but not the TATA boxes or the Gcn4p binding site; there was no predominant position covering these regulatory elements, leaving the Gcn4p binding site relatively nucleosome free.
We conclude that, in the absence of the Gcn4p transcriptional activator, the HIS3 gene was organized into a predominant array of nucleosomes superimposed on a significant background of multiple alternative positions. In wild-type cells, the D positions were still present but no longer predominant; the chromatin structure was much less ordered, presumably because Gcn4p is part of a system for mobilizing nucleosomes from the D positions to the alternative A positions.
The mobilization of nucleosomes on the HIS3 gene requires the SWI/SNF remodeling complex.
It seemed likely that the mobilization of nucleosomes from the D to the A positions was due to the recruitment of a remodeling machine to the HIS3 promoter by Gcn4p. The remodeling machine involved would presumably be capable of mobilizing nucleosomes on DNA in vitro; candidates included the SWI/SNF, Isw1, Isw2, and RSC remodeling complexes. We tested the roles of the SWI/SNF and Isw1 remodeling complexes in nucleosome mobilization on HIS3 using mutants in their ATPase subunits.
The chromatin structures of TA-HIS3 purified from gcn4Δ, snf2Δ, and isw1Δ cells grown under inducing conditions were compared with that of induced wild-type chromatin by monomer extension mapping (Fig. 3). The D nucleosome array was predominant in gcn4Δ and snf2Δ chromatin but not in isw1Δ chromatin. The structures of gcn4Δ and snf2Δ chromatin were almost identical (Fig. 3B), except that the A7 and A25 positions were more prominent in snf2Δ chromatin than in gcn4Δ chromatin. Nucleosomes D2, D3, and D4, which occupy most of the HIS3 coding region, gave consistently weaker signals relative to nucleosomes D1 and D5 (this effect is dependent on the Isw2 remodeling complex [Y. Kim, unpublished observations]). Thus, both Gcn4p and the Snf2p subunit of the SWI/SNF complex were required for the mobilization of HIS3 nucleosomes from the D to the A positions. In contrast, Isw1p was not required for this mobilization.
The Isw1p remodeling complex was particularly interesting because it has been shown to organize nucleosomes into arrays with approximately 175-bp spacing in vitro (40). However, the nucleosomal spacing was unaffected in isw1Δ cells (Fig. 2B), so it is presumably not the only complex capable of making such arrays. Although isw1Δ chromatin resembled wild-type chromatin, the two were not identical (Fig. 3). Nucleosomes A25 and A28 were prominent in isw1Δ chromatin, and there were some more minor quantitative differences, such as increased occupancy of the A15 and A19 positions, perhaps indicating that Isw1p promotes the formation of some A arrays over others. A nucleosome-free gap between the A25 and A28 nucleosomes was present in the map for isw1Δ chromatin, corresponding to a major weakening of the D5, A26, and A27 positions. This region corresponds to the 3′ end of HIS3 where transcript termination probably occurs; the gap of about 50 bp lies between nucleotides +713 and +764 (relative to the transcription start site at +1 and the stop codon at +685) (Fig. 2A). Thus, Isw1 is required for the movement of nucleosomes into positions A26, A27, and D5 at the 3′ end of HIS3, presumably at the expense of A25 and A28. The biological significance of this change in chromatin structure is unclear since the isw1Δ mutation did not have a significant effect on HIS3 transcription (not shown). However, it is clear that the SWI/SNF and Isw1 complexes tend to direct nucleosomes to different destinations on HIS3.
Chromatin structure of the chromosomal HIS3 locus.
It was important to determine whether chromosomal HIS3 chromatin undergoes structural transitions similar to those of HIS3 plasmid chromatin. However, the monomer extension method cannot be applied to single-copy genes in the context of the entire genome because the huge excess of nucleosomes derived from the rest of the genome results in nonspecific annealing to template DNA, contributing too much background (41). Plasmid-borne HIS3 can be purified away from the rest of the genome, and the signal is improved by the high copy number of TA-HIS3. Therefore, we had to use the standard indirect end-labeling method to probe the chromatin structure of the chromosomal HIS3 gene. This method relies on a relatively mild digestion of chromatin in nuclei using MNase or DNase I, which cleaves the relatively accessible linker DNA between nucleosomes.
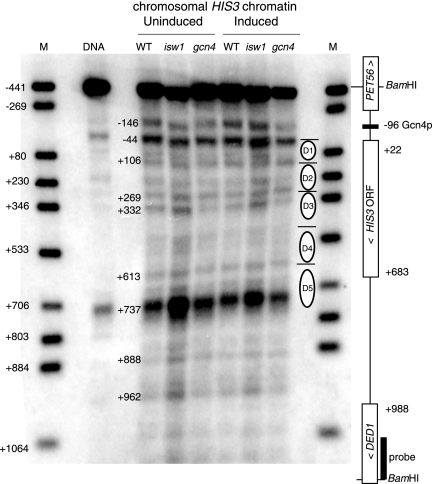
The 1,767-bp BamHI fragment encompassing the entire chromosomal HIS3 gene and parts of the upstream PET56 gene and the downstream DED1 gene was mapped (Fig. 4). The probe used abutted the BamHI site in the DED1 coding region. The indirect end-labeling pattern of protein-free genomic DNA was dominated by two MNase hypersensitive sites mapping to nucleotides −44 and +737. The coordinates are given with respect to the upstream transcription start site of HIS3, designated nucleotide +1 (33). The strong hypersensitive site at nucleotide +737 was located just downstream of the HIS3 stop codon (at nucleotide +683). MNase has a strong preference for cleaving DNA at CATA and CTA sequences (7). The hypersensitive site at nucleotide +737 coincided with a cluster of six CATA sequences located between nucleotides +682 and +780, which probably accounts for the observed hypersensitivity. In fact, many (but not all) of the bands observed in the protein-free DNA coincide with CATA sequences, partly accounting for the cleavage pattern (not shown).
FIG. 4.
Analysis of the chromatin structure of chromosomal HIS3 by indirect end labeling. Nuclei from uninduced and induced wild-type (WT), isw1Δ, and gcn4Δ cells were digested with MNase. Purified DNA was digested with BamHI and electrophoresed in an agarose gel. A Southern blot was probed with the DED1 fragment indicated. M, marker corresponding to a mixture of restriction digests of the 1,767-bp BamHI HIS3 fragment obtained by PCR; the bands are labeled relative to the transcription start site of HIS3 at nucleotide +1. The positions of the nucleosomes in the D array identified by monomer extension (Fig. 1 and 2) are shown. Note that the major MNase cleavage sites identified by Losa et al. (16) were at nucleotides −156, −47, +107, +285, +608, +770, and +811. Those identified by Sekinger et al. (26) were at nucleotides −168, −48, +117, and +306 (only the 5′ half of HIS3 was mapped). These coordinates were adjusted to the first transcription start site at nucleotide +1 (33) for comparison.
Chromatin samples from uninduced and induced wild-type, isw1Δ, and gcn4Δ cells were compared (Fig. 4). An examination of HIS3 chromatin structure in uninduced wild-type cells revealed many bands, particularly in the 5′ half of HIS3. These bands were mostly significantly less than 147 bp apart and were therefore too close together to be attributed to uniquely positioned nucleosomes. HIS3 chromatin in gcn4Δ cells yielded almost the same set of bands as did wild-type chromatin, but their relative intensities were different, with a subset of relatively strong bands having the spacing expected of nucleosomes, suggesting the presence of a predominant nucleosomal array. The weaker bands between the stronger bands in the 5′ half of HIS3 can be accounted for by the presence of an alternative array(s). The more complex structure of wild-type HIS3 chromatin is consistent with the presence of more than one nucleosomal array.
The predominant array in gcn4Δ cells coincided reasonably well with the D array observed by monomer extension (Fig. 4), with the exception of the D5 nucleosome, which overlapped the MNase hypersensitive site at the 3′ end of HIS3 (discussed below). The chromatin structure of HIS3 in isw1Δ cells was very similar to that of the wild type, except that the bands corresponding to cut sites located within the D nucleosomes were somewhat more prominent in isw1Δ chromatin and the 3′ HIS3 hypersensitive site was much more pronounced. The latter could reflect the fact that the 3′ hypersensitive site at nucleotide +737 lies between nucleosomes A25 (nucleotides +564 to +713) and A28 (nucleotides +764 to +908) in the nucleosome-free gap observed in the monomer extension map for isw1Δ chromatin. Induction had little effect on the chromatin structure of HIS3 in wild-type, isw1Δ, and gcn4Δ cells (the latter was expected because HIS3 cannot be induced in the absence of Gcn4p). In the HIS3 promoter, there was a MNase cut site at nucleotide −146, but the distance between this site and the 5′ hypersensitive site at nucleotide −44 is too small to accommodate a nucleosome.
To compare the indirect end-labeling data with the monomer extension data, the differences between the methods must be discussed. Indirect end labeling requires a relatively low level of digestion by MNase, whereas monomer extension requires a limit digest to obtain nucleosome core particles. MNase has both endonuclease and exonuclease activities; initially, it cuts DNA rapidly at the preferred sequences CATA and CTA (7) and then it trims the DNA ends relatively slowly. Further digestion by MNase generates smaller nucleosomal oligomers; these are eventually reduced to mononucleosomes and trimmed to nucleosome core particles, which are stable to MNase for an extended period. Thus, the digestion conditions required for indirect end labeling correspond to initial cleavages at favored sites in the linkers with very little trimming, without which the borders and position of a nucleosome cannot be determined with accuracy. This problem probably accounts for the large variation in nucleosome sizes reported using this method.Indirect end labeling works best with strongly ordered nucleosomal arrays and cannot resolve complex chromatin structures (such as those detected using monomer extension), which would appear more like protein-free DNA.
In the indirect end label map of wild-type HIS3 chromatin, some weak MNase cut sites were apparent near the centers of the D1 and D2 nucleosomes (Fig. 4). These cut sites were more prominent in isw1Δ chromatin (particularly in induced isw1Δ chromatin). These bands are consistent with the presence of the alternative overlapping arrays identified by monomer extension. We argue that MNase cuts at its favored sites when they are in the linker DNA, but not when they are in the nucleosome. Thus, the degree of cleavage at each preferred site is an indicator of the probability of that site being present in the linker; a particular site could be in the linker in one array and be cut but be nucleosomal in another array and be protected. A case in point is that of nucleosome D5, which overlaps the strong 3′ hypersensitive site (see above); the fact that the D5 nucleosome was present in the nucleosome population used for monomer extension proves that the hypersensitive site is not cut when it is inside this nucleosome. Thus, the band was strong primarily because it is a strongly favored cut site in protein-free DNA; its presence in chromatin indicates that this site is in the linker in a significant fraction of wild-type HIS3 chromatin. This site probably reflected cleavage at any of a cluster of six CATA sequences spread over 100 bp; the probability of one or more of these CATA sequences being in a linker would be high.
It is concluded that chromosomal HIS3 has a more ordered chromatin structure in the absence of Gcn4p, consistent with the formation of a predominant nucleosomal array. This predominant array is similar to that detected using monomer extension with HIS3 plasmid chromatin.
The SWI/SNF remodeling complex is constitutively present on the HIS3 gene.
It seemed likely that the Gcn4p activator recruits the SWI/SNF complex to the HIS3 promoter (22, 36). Accordingly, the ChIP assay was employed to determine whether the SWI/SNF complex was present at the HIS3 promoter and whether its presence depended on the Gcn4p activator. Strains having a wild-type chromosomal HIS3 locus were used instead of the TA-HIS3 plasmid strains. This was to avoid complications in the ChIP analysis due to differential recovery and fragmentation of plasmid chromatin relative to chromosomal chromatin (which we had observed in preliminary experiments). Strains carrying SNF2 with three FLAG tags were grown to mid-log phase and cross-linked with formaldehyde under three different growth conditions: noninduced (with histidine), induced (no histidine), and highly induced (no histidine and treated with 10 mM 3-AT for 3 h). DNA purified from cross-linked chromatin immunoprecipitated with anti-FLAG antibody was analyzed by quantitative multiplex PCR, using part of the coding region of the POL1 gene as an internal control (Fig. 5). The POL1 gene is often used as a control because it is relatively long and so there should be no contributing signal from the POL1 promoter.
In cells carrying SNF2-FLAG, the IP signals observed for the HIS3 promoter were at least 10 times those obtained either in the absence of antibody (mock) or for a strain carrying untagged SNF2 (Fig. 5A). However, the signal at POL1 was also positive and the HIS3/POL1 ratio was not significantly different from that of input, indicating that SWI/SNF was just as likely to be in the middle of the POL1 gene as at the HIS3 promoter. In addition, there was no effect of induction on the presence of SWI/SNF at HIS3. Several other control regions were tested in addition to the POL1 coding region with identical results (not shown). The experiment was also repeated using both hemagglutinin-tagged and myc-tagged SNF2 strains, with the same result (not shown). Thus, the SWI/SNF complex was constitutively present at the HIS3 promoter.
In gcn4Δ cells carrying SNF2-FLAG, there was an approximately twofold decrease in the amount of Snf2-FLAG at the HIS3 promoter relative to wild-type cells, but there was also a proportionate decrease at POL1. This result suggested that there was a general reduction in the amounts of chromatin-bound SWI/SNF complex in gcn4Δ cells, probably the result of an indirect effect (see below). It should be noted that gcn4Δ cells grew quite poorly relative to the wild type and that they almost stopped growing in the presence of 3-AT. The absence of Isw1p had no effect on the ChIP signals for Snf2-FLAG (Fig. 5A). Essentially the same results were obtained for the SWI/SNF complex on the HIS3 open reading frame (Fig. 5C). Thus, there was no evidence here for the recruitment of SWI/SNF to the HIS3 promoter by Gcn4p. Instead, the SWI/SNF complex appeared to be present constitutively at the HIS3 promoter and on the HIS3 gene.
As a positive test for the recruitment of the SWI/SNF complex by Gcn4p, the ARG1 promoter was examined by using the same IP samples. ARG1 is also induced by 3-AT via starvation for histidine and consequent activation of GCN4 mRNA for translation (5). There is good evidence for Gcn4p-dependent recruitment of the SWI/SNF complex to the ARG1 promoter (43). This observation was confirmed: induction with 3-AT resulted in a threefold increase in Snf2-FLAG levels at the ARG1 promoter relative to POL1 (Fig. 5D). It was also clear that the SWI/SNF complex was present at the ARG1 promoter in uninduced cells at levels similar to those observed at the HIS3 promoter (Fig. 5A). The 3-AT-induced increase in the binding of the SWI/SNF complex at the ARG1 promoter was abolished in the gcn4Δ mutant, showing that it was dependent on Gcn4p. We conclude that Gcn4p recruited additional SWI/SNF complex to the ARG1 promoter in 3-AT-treated cells but not to the HIS3 promoter.
ChIP experiments were used to confirm that Gcn4p binds at the HIS3 promoter on induction. A gcn4Δ strain was transformed with a centromeric plasmid carrying GCN4 tagged with 13 myc epitopes at its C terminus or with empty vector as a control (Fig. 5E). These strains also carried the SNF2-FLAG gene. In 3-AT-treated cells, there was a 3.5-fold increase in the binding of Gcn4-myc relative to Int-V as internal control (the same result was obtained using POL1 as internal control [data not shown]). In the uninduced and partially induced conditions (i.e., no 3-AT), Gcn4p was detected at the HIS3 promoter but at lower levels (twofold). The binding of Snf2-FLAG at the HIS3 promoter was also constitutive in these strains (not shown).
All Gcn4p-dependent genes are affected in a gcn4Δ strain, resulting in pleiotropic effects. This problem was circumvented by constructing a strain with a mutated Gcn4p binding site and expressing both SNF2-FLAG and GCN4-myc. The mutation was a single-base deletion (of the first T in the TGACTC sequence recognized by Gcn4p; the his3-142 mutation [34]). In order to introduce this mutation into the chromosomal HIS3 gene, it was necessary to insert a selection marker (ADE2) into the HIS3 ORF, rendering the strain unable to grow in the absence of histidine. Consequently, the induction procedure was altered: cells were grown in medium containing histidine, rapidly harvested, resuspended for an hour in medium lacking histidine, and then induced with 3-AT for 20 min prior to cross-linking. ChIP experiments were performed to confirm the absence of Gcn4p at the mutated HIS3 promoter (Fig. 5F). Gcn4p was not detected at the mutated HIS3 promoter in uninduced or in 3-AT-induced cells. However, Gcn4p was observed at the wild-type promoter after a 3-AT-induction of an otherwise identical strain (4.5-fold over POL1), as expected. As a positive control, the presence of Gcn4p at the ARG1 promoter was confirmed in both strains: Gcn4p cross-linked very well in uninduced cells (15- to 25-fold) and extremely well in 3-AT-induced cells (60- to 80-fold). The much higher degree of cross-linking of Gcn4p at the ARG1 promoter relative to the HIS3 promoter in wild-type cells might reflect more efficient cross-linking, but such a large difference might indicate that Gcn4p bound more tightly at ARG1 than at HIS3. There was no significant difference in the amount of SWI/SNF at the HIS3 promoter (or at POL1) in the presence or absence of bound Gcn4p (Fig. 5F). This experiment provides further evidence that Gcn4p does not recruit additional SWI/SNF to the HIS3 promoter.
Isw1p is constitutively present on the HIS3 gene, but excluded from the HIS3 promoter.
We tested whether Isw1p is present at HIS3 and whether its presence at HIS3 is dependent on the Gcn4p activator or on the SWI/SNF remodeling machine by using a strain with FLAG-tagged ISW1 (Fig. 6). In cells with FLAG-tagged Isw1p, the IP signals observed for the POL1 control were at least 10 times those obtained either in the absence of antibody (mock) or with a strain lacking the FLAG tag (wild type), indicating that Isw1p was present in the middle of the POL1 gene. In uninduced cells, Isw1p was modestly depleted at the HIS3 promoter relative to POL1 (about twofold) (Fig. 6A). Induction resulted in further depletion of Isw1p from the HIS3 promoter (about threefold relative to POL1). The induction-dependent depletion of Isw1p was partly dependent on the SWI/SNF complex since it was significantly weaker in the snf2Δ strain (Fig. 6A), suggesting that the SWI/SNF complex might play a role in depleting the HIS3 promoter of Isw1p. In the absence of the Gcn4p activator, there was also a depletion of Isw1p relative to POL1 (three- to fivefold) independent of growth condition. This result suggested that Gcn4p might recruit Isw1p to the HIS3 promoter. However, Isw1p was detected at high levels on the HIS3 coding region, independent of growth condition, Gcn4p, and the SWI/SNF complex (Fig. 6B). This observation indicates that Isw1p is excluded from the promoter upon HIS3 induction rather than recruited to it.
FIG. 6.
Isw1 remodeling complex is constitutively present on the chromosomal HIS3 gene but tends to be excluded from the HIS3 promoter. (A) The Isw1 complex tends to be excluded from the HIS3 promoter. Shown are results from ChIP experiments. The Isw1p ATPase subunit of the Isw1 complex was tagged with 3 FLAG epitopes (ISW1-FLAG) in otherwise wild-type cells and in gcn4Δ and snf2Δ cells; wild type has no tag. IP, DNA immunoprecipitated using anti-FLAG antibody; mock, no antibody. The amounts of DNA in the immunoprecipitates were measured using multiplex PCR with end-labeled PCR primers. The HIS3 promoter was a 238-bp fragment, and the POL1 ORF internal control was 180 bp. Samples were as follows: U, uninduced; I, partially induced; A, fully induced with 3-AT. Two input DNA dilutions (1 in 100 and 1 in 500) were measured. The percentage of DNA in the IPs relative to the input for the HIS3 promoter (black bars) and the POL1 ORF (gray bars) were measured using a phosphorimager and are graphed at the right. The HIS3 promoter/POL1 ORF ratios are given above each pair of bars. For clarity, quantification of the mock samples is not shown but, in all cases, the mock samples were less than 0.02% of the input DNA. (B) The Isw1 complex is present on the HIS3 ORF. The experiment was carried out as described for panel A, except that primers for the HIS3 ORF were used (yielding a 120-bp fragment), together with the POL1 primers (180 bp). The percentage of DNA in the IPs relative to the input for the HIS3 ORF (black bars) and the POL1 ORF (gray bars) were measured using a phosphorimager and are graphed at the right. The HIS3 ORF/POL1 ORF ratios are given above each pair of bars. For clarity, the quantification of the mock samples is not shown but, in all cases, the mock samples were less than 0.02% of the input DNA.
In conclusion, Isw1p tended to be excluded from the HIS3 promoter but was present on the HIS3 coding region under all conditions tested. Although the cause of Isw1p exclusion from the promoter is not known, one possibility is a physical block, such as the formation of a large complex at the promoter. This could, in principle, account for the exclusion of Isw1p under the opposite conditions of nonactivated/basal expression (gcn4Δ) and highly induced (wild type treated with 3-AT); under these conditions, different blocking complexes might be present at the promoter. The SWI/SNF complex appears to be a contributing factor, although it did not affect the presence of Isw1p on the coding region even though both complexes were present there.
DISCUSSION
Chromatin structure of HIS3.
Several studies have determined the locations of nucleosomes on the HIS3 gene in wild-type cells using a variety of methods (16, 26, 44). The nucleosome positions on chromosomal HIS3 that we have identified using indirect end labeling are in good agreement with those reported for various HIS3 plasmids (16) and for the chromosomal HIS3 gene (26). They correspond reasonably well with the D nucleosomal array detected by monomer extension. The MNase bands in between the bands flanking nucleosomes D1 and D2 were significantly stronger in our wild-type chromatin than in the other studies (16, 26), suggesting that the A arrays were more prominent in our study; the reason for this is unclear. There is also agreement that the HIS3 promoter is substantially less likely to be nucleosomal than the coding region; nucleosomes were detected over the regulatory sites [i.e., the TATA boxes, Gcn4p binding site, and poly(dA-dT) sequence], but these positions have low occupancy (the bands corresponding to positions A1 to A3 are very weak [data not shown]). Consistent with this, we have shown previously that 60 to 80% of HIS3 promoters are accessible to a restriction enzyme that cuts at the Gcn4p binding site (9).
An important issue is whether the chromatin structure of HIS3 is typical of yeast genes. We have studied nucleosome positions on only one other gene, CUP1 (28-30), with similar results. However, since we were unable to identify the remodeling complexes acting at CUP1, we turned to HIS3 for further studies, because it was known to be affected by the SWI/SNF complex. Most of the work described here necessarily utilized plasmid rather than chromosomal chromatin, but our indirect end-labeling study strongly suggests that HIS3 plasmid chromatin is a good model for chromosomal HIS3 chromatin.
We conclude that there is good agreement on the nucleosome positions constituting the predominant array on HIS3, both in the plasmids and in the chromosome. However, we observed additional, alternative nucleosomal arrays using the monomer extension method. This reflects the fact that the indirect end-labeling method cannot detect more than one nucleosomal array because it depends on identifying nucleosomes by finding nucleosome-sized gaps in the MNase cleavage pattern. Indirect end-labeling experiments often provide data with series of major bands too close together to be assigned to positioned nucleosomes, and it is usually concluded that there are no positioned nucleosomes. We suggest that these patterns might actually reflect the presence of multiple alternative arrays of positioned nucleosomes with no predominant arrays.
The nucleosome density profile of HIS3.
Recently, both nucleosome scanning and histone ChIP were used to measure the nucleosome densities of HIS3 and other yeast genes (26). The nucleosome scanning method involves the preparation of mononucleosomal DNA for use as template in a series of PCRs utilizing primer pairs spaced about 30 bp apart to measure the relative amount of DNA in nucleosomes along a gene of interest. Another recent study (44) used nucleosomal DNA in a similar way to probe microarrays to measure nucleosome density on a genomic scale. Both studies reported a sinusoidal nucleosome density profile along part of the HIS3 gene, with peaks and troughs that coincide quite well with the approximate nucleosome positions derived by indirect end labeling. Although these reported positions also agree quite well with our D array, our data indicate that the D array is predominant in only gcn4Δ and snf2Δ chromatin. In wild-type cells, our monomer extension data revealed the presence of multiple overlapping arrays (Fig. 2A). However, the monomer extension method determines nucleosome positions, whereas indirect end-labeling can reveal only a predominant array, and the ChIP and nucleosome scanning methods report histone or nucleosome density, which are not necessarily the same as nucleosome position.
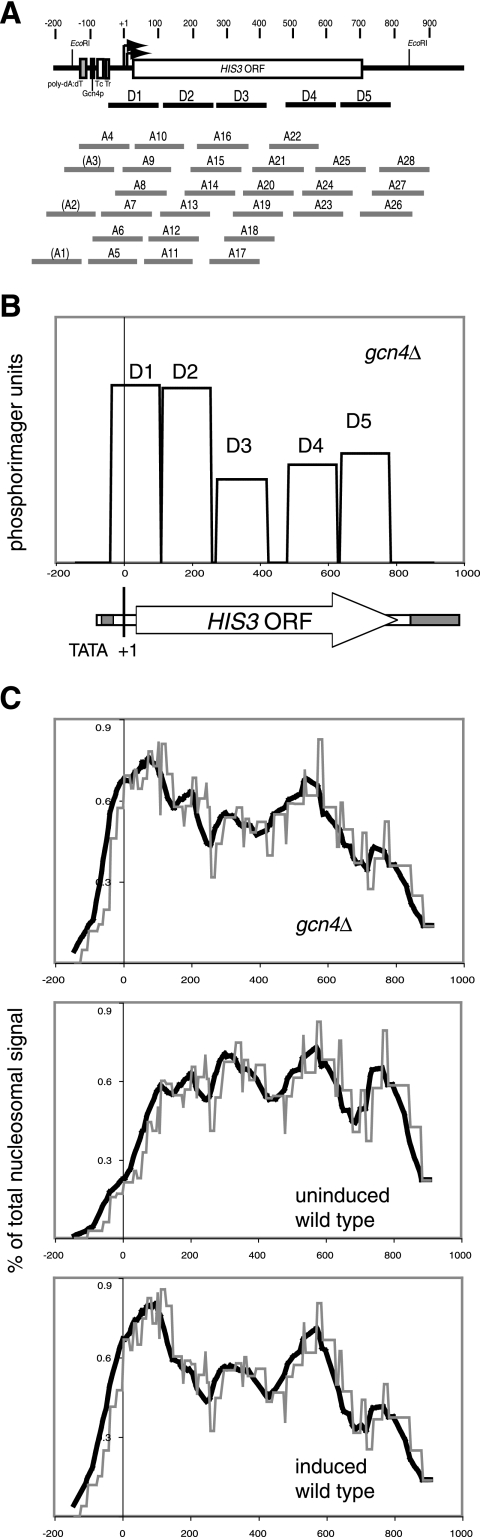
To compare our high-resolution positioning data with the published nucleosome density data, we converted our data to a nucleosome density function. This is possible because the relative amounts of each nucleosome are given by the scan of a single monomer extension lane (the data in Fig. 1 were used). The HIS3 gene was divided into 5-bp intervals, and each nucleosome was assumed to occupy 145 bp. Each nucleosome (i.e., D1 to D5 and A4 to A25) was given a score equal to its phosphorimager signal corrected for background; this score was placed in the 29 bins of 5 bp, corresponding to the 145 bp it occupies on HIS3 (Fig. 7A). The scores for each nucleosome at each 5-bp interval were summed across the HIS3 gene. The score was then plotted as a percentage of the total nucleosomal signal against HIS3 coordinate. For illustrative purposes, the signals for only the D array (in gcn4Δ chromatin) are shown in Fig. 7B. From this, it is obvious that a square waveform is predicted for a single nucleosomal array; the amplitude corresponding to each nucleosome should be the same in the case of a unique array (i.e., if nucleosomes are in identical positions on all copies of HIS3). Thus, the sinusoidal waveforms observed by others (26, 44) are not really consistent with a unique array of nucleosomes, although it might be argued that the square wave function is smoothed out at lower resolution (but see below).
FIG. 7.
Nucleosome density profiles for the HIS3 gene reveal long-range order in HIS3 chromatin. (A) Schematic depiction of nucleosomal D and A arrays (from Fig. 2A). Each nucleosome was assumed to occupy 145 bp using the downstream border determined by monomer extension analysis (given in Fig. 2A). A1 to A3 were not included in the nucleosome density analysis because they are too close to the XbaI site to be detected by monomer extension using this enzyme and so are not present on the maps in Fig. 1. (B) Illustration of the predicted square waveform for the nucleosome density of an array of uniquely positioned nucleosomes. The D array in gcn4Δ chromatin is shown; the relative heights reflect the phosphorimager signals. In the idealized case of a unique array, the amplitudes would be identical. A scaled map of the HIS3 gene is shown below. (C) Calculated nucleosome density functions for uninduced and induced wild-type chromatin and gcn4Δ chromatin. The relative amounts of each nucleosome (D1 to D5 and A4 to A28) were determined from the phosphorimager scans (Fig. 1D). Each nucleosome was assigned a score equal to its phosphorimager signal after correction for background. The HIS3 gene was divided into 5-bp intervals; each nucleosome occupied 145 bp, corresponding to 29 bins of 5 bp each. The scores were integrated for each bin across the HIS3 gene to obtain nucleosome density functions, which were then normalized by calculating the integrated signal in each 5-bp bin as a percentage of the total integrated signal. This is plotted as a function of the HIS3 coordinate (with the first transcription start site as nucleotide +1). The profiles were smoothed using a 50-bp (10 bins) running average to simulate a lower resolution.
When the scores for all of the nucleosomes in our monomer extension data are integrated across the HIS3 gene and converted to a percentage of the total integrated signal, a remarkable result is obtained: a sinusoidal pattern is apparent for all three chromatin samples, even for uninduced and induced wild-type chromatin. The sinusoidal pattern is more obvious when the data are smoothed to simulate lower resolution (Fig. 7C). In the case of gcn4Δ chromatin, there are five peaks, reflecting the contribution of the predominant D array. In wild-type chromatin, there are only four peaks, although the first peak has a shoulder, reflecting the contribution of the D2 nucleosome. These patterns are very similar to those observed by other researchers (26), who mapped about half of the HIS3 gene by nucleosome and H4 ChIP scanning; the main difference is that the other researchers resolved D1 and D2 clearly, whereas we did not. Otherwise, the positions of the peaks and troughs are the same (once the different HIS3 coordinates used in reference 26 are taken into account). In the microarray study (44), only the first nucleosome on HIS3 (equivalent to D1) is shown; its position is in agreement with our data and those of others (26). The patterns reported for many other genes (44) are strikingly similar to the nucleosome density profiles we report here (Fig. 7). Thus, our monomer extension data predict a nucleosome density pattern just like that observed for HIS3 using the nucleosome/ChIP scanning assays, even though multiple overlapping positions are present. Therefore, we suggest that the yeast genome might not be composed mostly of uniquely positioned nucleosomes as proposed (44). Instead, the microarray data could be interpreted to indicate that the genome is organized into multiple alternative nucleosomal arrays, similar to those we have observed on HIS3.
In conclusion, there is an underlying long-range order in the overlapping arrays of nucleosomes on HIS3, with maxima and minima in nucleosome density. This results from the summation of the square wave functions for each overlapping nucleosome array (as in Fig. 7B), yielding a “constructive interference” pattern corresponding to the nucleosome density. In physical terms, the pattern indicates that the alternative positions occur in clusters, resulting in maxima and minima.
The Gcn4p activator and the SWI/SNF complex direct nucleosome mobilization over the entire HIS3 gene.
Our observations suggest that, in wild-type cells, HIS3 chromatin is organized into one of several alternative nucleosomal arrays. In the absence of Gcn4p or the SWI/SNF complex, the D array is predominant, though it is not the only significant array. Thus, both Gcn4p and SWI/SNF are required for mobilizing nucleosomes from positions in the D array to positions in the alternative A arrays. It seems likely that the SWI/SNF complex is directly responsible for mobilizing the nucleosomes on the HIS3 gene since it is capable of mobilizing nucleosomes in vitro (24). Consistent with this model, our ChIP experiments indicate that the SWI/SNF complex is present at the HIS3 promoter and on the HIS3 gene. Our data indicate that the effects of the SWI/SNF complex are not limited to the chromatin structure of the promoter but include that of the entire HIS3 gene.
An alternative possibility might be that the nucleosome mobilization observed is due to the passage of RNA polymerase II rather than the direct consequence of SWI/SNF activity. Although it is difficult to rule this model out entirely, there is some evidence against it. Firstly, the D array is predominant in HIS3 chromatin from gcn4Δ and snf2Δ cells even though the levels of basal transcription are quite high (9). Secondly, the TATA box mutations we have described previously (9) do not affect nucleosome positions; the monomer extension map resembles wild-type chromatin, even though the levels of HIS3 transcription are very low (not shown).
It was expected that the role of Gcn4p in mobilization is to recruit the SWI/SNF complex to the HIS3 promoter. However, we found that SWI/SNF is present at HIS3 independently of Gcn4p. The simplest explanation for our observations is that SWI/SNF is not recruited to HIS3; it is present constitutively. Alternatively, some additional SWI/SNF might indeed be recruited by Gcn4p but rapidly departs from the HIS3 promoter, moving down the gene. Another possibility is that a Gcn4p-independent mechanism is responsible for the recruitment of SWI/SNF to HIS3, perhaps involving the general transcription factors (27). The recruitment of additional SWI/SNF by Gcn4p at ARG1 might reflect the contribution of other transcription factors not present at HIS3, such as the ArgR complex, which associates with the ARG1 promoter in a Gcn4p-dependent manner (42). In any case, the presence of the SWI/SNF complex on the HIS3 coding region is consistent with a role in gene-wide nucleosome mobilization.
The role of the Isw1 complex in remodeling HIS3 chromatin appears to be more subtle than that of SWI/SNF; it is involved in minor but significant changes in HIS3 chromatin structure. In the absence of Isw1p, a nucleosome-free gap of about 50 bp appears at the 3′ end of HIS3, where transcript termination probably occurs, suggesting that Isw1 favors the mobilization of nucleosomes to positions covering this gap. The observed accumulation of RNA polymerase II at the 3′ ends of yeast genes in an isw1 mutant (21) might contribute to the formation of this nucleosome-free gap. It has been proposed that the Isw1 complex coordinates transcript elongation and termination (21). The Isw1 complex also appears to influence the relative amounts of the various A and D positions, perhaps favoring the formation of some arrays over others. The constitutive presence of Isw1p on the HIS3 gene is consistent with these structural data. The level of Isw1p at the HIS3 promoter is lower than that on the coding region and is affected by the SWI/SNF complex. However, the marginal effects of Isw1p on HIS3 transcription suggest that these changes in chromatin structure are not very important for HIS3 expression. Like the SWI/SNF complex, the Isw1 complex also influences which positions are occupied by nucleosomes on HIS3 but it favors different positions from the SWI/SNF complex.
A working model for the activation of HIS3 chromatin.
We now have a very detailed picture of the activation of HIS3 chromatin. In the absence of Gcn4p or the SWI/SNF complex, the chromatin structure of HIS3 is relatively ordered, with the D array of nucleosomes predominant. In the presence of Gcn4p and the SWI/SNF complex, several overlapping arrays of nucleosomes are formed on the HIS3 gene, indicating that there is a net movement of nucleosomes from the D array to the alternative A arrays. Although this chromatin is more disordered than that formed in the absence of Gcn4p or the SWI/SNF complex, some long-range order persists, as indicated by the nucleosome density profiles, which suggest that the various arrays represent clusters of alternative nucleosome positions. Previously, we have presented evidence suggesting that nucleosomes on the HIS3 gene undergo major conformational changes that are dependent on Gcn4p and the SWI/SNF complex (9). These conformational changes do not substantially affect the protection of nucleosomal DNA from MNase because nucleosomal DNA was prepared in reasonable yield from induced chromatin for monomer extension studies. This is perhaps surprising given the accessibility to restriction enzymes; new experiments aimed at understanding the structural basis of the loss of supercoils from induced chromatin should cast light on this issue.
As a working model, we propose that the binding of Gcn4p at the promoter initiates the formation of a much more dynamic chromatin structure over the entire HIS3 gene, involving nucleosome movements and conformational changes (Fig. 8). In the presence of Gcn4p, increased activity of the SWI/SNF complex on HIS3 results in increased mobilization of nucleosomes from the D positions to the various A positions. The Isw1 complex contributes to this mobilization by mobilizing nucleosomes to different positions from those favored by SWI/SNF. We speculate that the increased dynamics of activated HIS3 chromatin structure might render the underlying DNA more transparent. The continuous movement of nucleosomes by the remodeling complexes envisaged in our model might create windows of opportunity for various factors to bind over the entire HIS3 gene, including the various elongation complexes, stimulating HIS3 transcription.
FIG. 8.
Working model for the transcriptional activation of HIS3 chromatin. The chromatin structure of the HIS3 gene expressed at basal levels (as in gcn4Δ chromatin) is characterized by a predominant nucleosomal array (D1 to D5), although A arrays are also present. In the presence of the Gcn4p activator, the activity of the SWI/SNF complex is stimulated, resulting in a net mobilization of nucleosomes from the D arrays to the A arrays. The Isw1 complex also affects the distribution of the nucleosomes, particularly at the 3′-end of HIS3. The inference is that HIS3 chromatin structure is highly dynamic. The nucleosomal flux created by the competing activities of the various remodeling complexes should facilitate access to the DNA for both transcript initiation and elongation complexes. Also note that we have previously shown that HIS3 nucleosomes apparently undergo a major conformational change requiring both Gcn4p and the SWI/SNF complex (9), which might increase the transparency of the chromatin still further.
We imagine that the D array is formed by an as-yet-unidentified remodeling complex capable of moving nucleosomes from A to D positions. If this is the case, the chromatin structure of HIS3 might represent a balance between the nucleosome-mobilizing activity of this putative remodeling complex and the activities of the SWI/SNF and Isw1 complexes, which favor the A arrays. Induction might alter this balance in favor of the SWI/SNF and Isw1 complexes. From our work and that of others, a picture is beginning to emerge in which several remodeling machines act on HIS3 chromatin. These include the SWI/SNF, Isw1, and Isw2 (Y. Kim, unpublished data) complexes, all of which appear to mobilize nucleosomes to different positions on HIS3. Apparently, each complex has a different specific task designed to circumvent different types of chromatin obstacles to transcription. This picture appears to be complex, but perhaps it is quite general. We propose that the various remodeling machines might perform their specific functions at most or all yeast genes, but their particular tasks are more or less critical at different genes, dependent on their specific chromatin structures, resulting in mutant phenotypes of varying severity.
Acknowledgments
We thank Chang-Hui Shen for plasmids, strains, and comments on the manuscript and Alan Hinnebusch for helpful discussion, comments on the manuscript, and pSK1. We thank Joe Reese for antibodies and discussion of ChIP experiments.
This research was supported by the Intramural Research Program of the NIH (NICHD) and by NIH grant GM58465 to T. Tsukiyama. T. T. is a Leukemia and Lymphoma Society Scholar.
Footnotes
Published ahead of print on 18 September 2006.
REFERENCES
- 1.Anderson, J. D., and J. Widom. 2001. Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol. Cell. Biol. 21:3830-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cairns, B. R. 2005. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr. Top. Genet. Dev. 15:185-190. [DOI] [PubMed] [Google Scholar]
- 3.Ercan, S., M. J. Carrozza, and J. L. Workman. 2004. Global nucleosome distribution and the regulation of transcription in yeast. Genome Biol. 5:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eriksson, P. R., G. Mendiratta, N. B. McLaughlin, T. G. Wolfsberg, L. Mariño-Ramírez, T. A. Pompa, M. Jainerin, D. Landsman, C.-H. Shen, and D. J. Clark. 2005. Global regulation by the yeast Spt10 protein is mediated through chromatin structure and the histone upstream activating sequence elements. Mol. Cell. Biol. 25:9127-9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinnebusch, A. G., and K. Natarajan. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 1:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hope, I. A., and K. Struhl. 1985. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell 43:177-188. [DOI] [PubMed] [Google Scholar]
- 7.Hörz, W., and W. Altenburger. 1981. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 9:2643-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer, V., and K. Struhl. 1995. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 14:2570-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim, Y., and D. J. Clark. 2002. SWI/SNF-dependent long-range remodeling of yeast HIS3 chromatin. Proc. Natl. Acad. Sci. USA 99:15381-15386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, Y., C.-H. Shen, and D. J. Clark. 2004. Purification and nucleosome mapping analysis of native yeast plasmid chromatin. Methods 33:59-67. [DOI] [PubMed] [Google Scholar]
- 11.Korber, P., and W. Hörz. 2004. Swred not shaken: mixing the histones. Cell 117:5-7. [DOI] [PubMed] [Google Scholar]
- 12.Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt, M. A. Boateng, O. W. Ryan, A. Golshani, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol. Cell 11:721-729. [DOI] [PubMed] [Google Scholar]
- 13.Kuo, M.-H., J. Zhou, P. Jambeck, M. E. A. Churchill, and C. D. Allis. 1998. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12:627-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo, M.-H., E. vom Bauer, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 15.Laurent, B. C., M. A. Treitel, and M. Carlson. 1991. Functional interdependence of the yeast SNF2, SNF5 and SNF6 proteins in transcriptional activation. Proc. Natl. Acad. Sci. USA 88:2687-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Losa, R., S. Omari, and F. Thoma. 1990. Poly(dA) · poly(dT) rich sequences are not sufficient to exclude nucleosome formation in a constitutive yeast promoter. Nucleic Acids Res. 18:3495-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mai, X., S. Chou, and K. Struhl. 2000. Preferential accessibility of the yeast HIS3 promoter is determined by a general property of the DNA sequence, not by specific elements. Mol. Cell. Biol. 20:6668-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McConnell, A. D., M. E. Gelbart, and T. Tsukiyama. 2004. Histone fold protein Dls1p is required for Isw2-dependent chromatin remodeling in vivo. Mol. Cell. Biol. 24:2605-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellor, J., and A. Morillon. 2004. ISWI complexes in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1677:100-112. [DOI] [PubMed] [Google Scholar]
- 20.Mellor, J. 2005. Dynamics of chromatin remodeling at promoters. Mol. Cell 19:147-157. [DOI] [PubMed] [Google Scholar]
- 21.Morillon, A., N. Karabetsou, J. O'Sullivan, N. Kent, N. Proudfoot, and J. Mellor. 2003. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell 115:425-435. [DOI] [PubMed] [Google Scholar]
- 22.Natarajan, K., B. M. Jackson, H. Zhou, F. Winston, and A. G. Hinnebusch. 1999. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol. Cell 4:657-664. [DOI] [PubMed] [Google Scholar]
- 23.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 25.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6:1297-1307. [DOI] [PubMed] [Google Scholar]
- 26.Sekinger, E. A., Z. Moqtaderi, and K. Struhl. 2005. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell 18:735-748. [DOI] [PubMed] [Google Scholar]
- 27.Sharma, V. M., B. Li, and J. C. Reese. 2003. SWI/SNF-dependent chromatin remodeling of RNR3 requires TAFIIs and the general transcription machinery. Genes Dev. 17:502-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen, C.-H., and D. J. Clark. 2001. DNA sequence plays a major role in determining nucleosome positions in yeast CUP1 chromatin. J. Biol. Chem. 276:35209-35216. [DOI] [PubMed] [Google Scholar]
- 29.Shen, C.-H., B. P. Leblanc, J. A. Alfieri, and D. J. Clark. 2001. Remodeling of yeast CUP1 chromatin involves activator-dependent repositioning of nucleosomes over the entire gene and flanking sequences. Mol. Cell. Biol. 21:534-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen, C.-H., B. P. Leblanc, C. Neal, R. Akhavan, and D. J. Clark. 2002. Targeted histone acetylation at the yeast CUP1 promoter requires the transcriptional activator, the TATA boxes and the putative histone acetylase encoded by SPT10. Mol. Cell. Biol. 22:6406-6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, C., and C. L. Peterson. 2005. ATP-dependent chromatin remodeling. Curr. Top. Dev. Biol. 65:115-148. [DOI] [PubMed] [Google Scholar]
- 32.Struhl, K. 1983. Promoter elements, regulatory elements and chromatin structure of the yeast his3 gene. Cold Spring Harbor Symp. Quant. Biol. 47:901-910. [DOI] [PubMed] [Google Scholar]
- 33.Struhl, K. 1985. Nucleotide sequence and transcriptional mapping of the yeast pet56-his3-ded1 gene region. Nucleic Acids Res. 13:8587-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Struhl, K., and D. E. Hill. 1987. Two related regulatory sequences are required for maximal induction of Saccharomyces cerevisiae his3 transcription. Mol. Cell. Biol. 7:104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swanson, M. J., H. Qiu, L. Sumibcay, A Krueger, S. Kim, K. Natarajan, S. Yoon, and A. G. Hinnebusch. 2003. A multiplicity of cofactors is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 23:2800-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Syntichaki, P., I. Topalidou, and G. Thireos. 2000. The Gcn5 bromodomain coordinates nucleosome remodelling. Nature 404:414-417. [DOI] [PubMed] [Google Scholar]
- 37.Thoma, F., L. W. Bergman, and R. T. Simpson. 1984. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J. Mol. Biol. 177:715-733. [DOI] [PubMed] [Google Scholar]
- 38.Thomas, J. O., and V. Furber. 1976. Yeast chromatin structure. FEBS Lett. 66:274-280. [DOI] [PubMed] [Google Scholar]
- 39.Tsukiyama, T. 2002. The in vivo functions of ATP-dependent chromatin remodelling factors. Nat. Rev. Mol. Cell. Biol. 3:422-429. [DOI] [PubMed] [Google Scholar]
- 40.Tsukiyama, T., J. Palmer, C. C. Landel, J. Shiloach, and C. Wu. 1999. Characterization of the Imitation Switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yenidunya, A., C. Davey, D. J. Clark, G. Felsenfeld, and J. Allan. 1994. Nucleosome positioning on chicken and human globin gene promoters in vitro. Novel mapping techniques. J. Mol. Biol. 237:401-414. [DOI] [PubMed] [Google Scholar]
- 42.Yoon, S., C. K. Govind, H. Qiu, S. Kim, J. Dong, and A. G. Hinnebusch. 2004. Recruitment of the ArgR/Mcm1p repressor is stimulated by the activator Gcn4p: a self-checking activation mechanism. Proc. Natl. Acad. Sci. USA 101:11713-11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon, S., H. Qiu, M. J. Swanson, and A. G. Hinnebusch. 2003. Recruitment of SWI/SNF by Gcn4p does not require Snf2p or Gcn5p but depends strongly on SWI/SNF integrity, SRB mediator, and SAGA. Mol. Cell. Biol. 23:8829-8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan, G., Y. Liu, M. F. Dion, M. D. Slack, L. F. Wu, S. J. Altschuler, and O. J. Rando. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309:626-630. [DOI] [PubMed] [Google Scholar]
- 45.Zakian, V. A., and J. F. Scott. 1982. Construction, replication and chromatin structure of TRP1 RI circle, a multiple-copy synthetic plasmid derived from Saccharomyces cerevisiae chromosomal DNA. Mol. Cell. Biol. 2:221-232. [DOI] [PMC free article] [PubMed] [Google Scholar]