Abstract
Outer dense fiber 2 (Odf2) was initially identified as a major component of sperm tail cytoskeleton and later was suggested to be a widespread component of centrosomal scaffold that preferentially associates with the appendages of the mother centrioles in somatic cells. Here we report the identification of two Odf2-related centrosomal components, hCenexin1 and hCenexin1 variant 1, that possess a unique C-terminal extension. Our results showed that hCenexin1 is the major isoform expressed in HeLa cells, whereas hOdf2 is not detectably expressed. Mammalian polo-like kinase 1 (Plk1) is critical for proper mitotic progression, and its association with the centrosome is important for microtubule nucleation and function. Interestingly, depletion of hCenexin1 by RNA interference (RNAi) delocalized Plk1 from the centrosomes and the C-terminal extension of hCenexin1 was crucial to recruit Plk1 to the centrosomes through a direct interaction with the polo-box domain of Plk1. Consistent with these findings, the hCenexin1 RNAi cells exhibited weakened γ-tubulin localization and chromosome segregation defects. We propose that hCenexin1 is a critical centrosomal component whose C-terminal extension is required for proper recruitment of Plk1 and other components crucial for normal mitosis. Our results further suggest that the anti-Odf2 immunoreactive centrosomal antigen previously detected in non-germ line cells is likely hCenexin1.
Found in diverse eukaryotic organisms studied, the polo-like protein kinases (collectively termed Plks) are a conserved subfamily of Ser/Thr protein kinases that play pivotal roles during cell cycle progression and proliferation. Among Plks, Plk1 has been the most extensively studied enzyme because of its role in regulating various mitotic events (see reviews in references 2 and 35). Studies of cultured mammalian cells revealed that Plk1 localizes to the centrosomes and kinetochores in late S or early G2 and remains at these structures until anaphase/telophase (1, 11, 20, 33). In anaphase, Plk1 localization to the centrosomes and kinetochores weakens, as Plk1 relocalizes to the spindle midzone (later it becomes midbody) (11, 20, 33). The polo-box domain (PBD) present in the C-terminal noncatalytic region of Plk1 is critical for the dynamic subcellular localization of Plk1. In a PBD-dependent manner, Plk1 has been shown to interact with the peripheral Golgi protein Nir2, mitotic kinesin-like proteins MKLP-1 and MKLP-2, and centrosomal/midbody protein Cep55, and these interactions appear to be important to promote Plk1 localization to the midzone/midbody (10, 21, 22, 26). Yet, the components critical for Plk1 localization to the centrosomes and kinetochores are largely unknown. It has been shown that Plk1 interacts with Cep170, a centrosomal protein that localizes to the mother centriole (the one that is inherited from the mother cell) by associating with subdistal appendages (13). Plk1 also interacts with and phosphorylates another mother centriole-associating ninein-like protein (Nlp) to displace the latter from mitotic centrosomes (5, 31). Interestingly, both Cep170 and Nlp localize to the interphase centrosomes but not to the mitotic centrosomes (5, 13). In addition, knockdown of Cep170 does not cause any significant defects in centrosome structure and microtubule nucleation activity and fails to delocalize centrosomal proteins such as Plk1 from the centrosomes (13). These findings suggest the existence of an additional unidentified protein(s) that is critical for Plk1 localization to the centrosomes.
The centrosome is the main microtubule organizing center of the cell and is composed of a pair of barrel-shaped centrioles. Pericentriolar material surrounds the centrosome and tethers various proteins including γ-tubulin and ninein that are critical for microtubule nucleation and anchoring, suggesting that the centrosome serves as a scaffold to recruit various proteins important for proper bipolar spindle formation (see reviews in references 8 and 32). Among the proteins recruited to the centrosome is Plk1, whose association with the centrosome has been shown to promote microtubule nucleation and mitotic spindle formation (5, 6). However, how Plk1 is recruited to the centrosomes and which component(s) at the centrosomes is critically required for Plk1 recruitment remain elusive.
The outer dense fibers (ODFs) are prominent sperm tail-specific cytoskeletal structures and are conserved across the animal phylogenetic tree. In humans, ODFs are composed of at least 10 major and 15 minor proteins (30). Human Odf2 (hOdf2) is closely related to those isolated from rat and mouse testis cDNA libraries (4, 15, 34) and appears to be a major form of sperm tail outer dense fibers with a molecular mass of about 70 to 73 kDa (the hOdf2/1 and hOdf2/2 genes encode proteins of 610 and 638 amino acids [aa], respectively). Recent studies showed that a centrosomal protein immunoreactive to an anti-Odf2 antibody localizes to the fibrous distal/subdistal appendages of centrosomes in cultured human and mouse cells (24). Knockout of ODF2 in mouse F9 embryonic carcinoma cells eliminates distal/subdistal appendages and prevents primary cilium formation. Loss of ODF2 also disrupts two mother centriole-specific ninein dots, while leaving one dot on the proximal end of mother and daughter centrioles (16). These results raised the possibility that an anti-Odf2-immunoreactive protein(s) (or possibly Odf2 itself) localizes to the centrosomes of somatic cells and plays an important role in proper centrosome function.
In this paper, we report the isolation of two Odf2 splicing variants, hCenexin1 and hCenexin1 variant 1, as Plk1-interacting proteins at the centrosomes. Our results demonstrated that hCenexin1, but not Odf2, is the major isoform present at the centrosomes of HeLa cells. Interestingly, knockdown of hCenexin1 impaired Plk1 localization to the centrosomes and resulted in weakened γ-tubulin localization and a chromosome segregation failure. We propose that hCenexin1 is a critical centrosomal component that is required for proper recruitment of Plk1 and other proteins important for proper mitotic progression.
MATERIALS AND METHODS
Reverse transcription-PCR and plasmid construction.
Total RNAs purified from the HeLa cervical carcinoma cell line were reverse transcribed to first-strand cDNAs using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) and the oligo(dT)18 primer. The resulting products were then used as templates for subsequent PCR with a forward primer (5′-ATGAAGGACCGCTCTTCAACTCCCCCCT-3′) and a reverse primer (5′-TCCGCTCCCGATCTCCTCCTGCCTGA-3′) that were designed to target the full-length hOdf2 isoform 1. The obtained PCR fragments were cloned into pCR2.1-TOPO (Invtrogen) for sequence analyses. Among 24 independent clones sequenced, 9 were identical to the hCENEXIN1 (hODF2 isoform 1) sequence, whereas 15 were identical to the hCENEXIN1 variant 1 sequence.
pKL74 (pGBT9-PLK1) and pKL94 [pGBT9-PLK1(K82M)] were constructed by inserting a 2.2-kb EcoRI fragment containing either wild-type PLK1 or a kinase-inactive PLK1(K82M) (20) into pGBT9 vector (Clontech, Palo Alto, CA) digested with EcoRI. Two-hybrid screening was performed using a 2μm-LEU2 plasmid (pACT2)-based GAL4 activation domain-fused human testis cDNA library (MATCHMAKER; Clontech). To create a green fluorescent protein gene (GFP)-fused hCENEXIN1 expression construct (pKL2472), a KpnI-EcoRI fragment bearing the full-length open reading frame was cloned into pShuttle-GFP vector (14) digested with the corresponding enzymes. Truncated forms of pShuttle-GFP-hCENEXIN1 (pKL2477 [T1] to pKL2484 [T8]) were generated by self-ligating the obtained fragments of pKL2472 digested with Bsu36I and EcoRV (pKL2475), StuI and EcoRV (pKL2478), SanDI and EcoRV (pKL2479), KpnI and EcoNI (pKL2480), PmlI (pKL2481), SanDI and BglII (pKL2482), KpnI and StuI (pKL2483), or HindIII and EcoRV (pKL2484), respectively. pKL2630 and pKL2632 were generated to express the full-length open reading frames of hCENEXIN1 and hCENEXIN1 variant 1, respectively (these constructs do not contain any epitope tags and were used to mark the positions of hCenexin1 and hCenexin1 variant 1 in sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]). To create a bacterial expression construct containing the full-length hCENEXIN1 (pKL3049), a 2.4-kb (KpnI-EcoRV) fragment was cloned into pGEX-4T-3 digested with KpnI (created) and SmaI. Truncated hCenexin1 constructs, pKL3052, pKL3051, and pKL3050, were generated by inserting an EcoRI-SalI fragment of hCENEXIN1(1-249), hCENEXIN1(250-632), or hCENEXIN1(647-805), respectively, into pGEX-KG digested with the corresponding enzymes. Baculoviral constructs pAC702-HA-PLK1 and pAC702-HA-PLK1ΔPB1 (lacking the internal 430-bp SmaI fragment) have been described previously (19). To generate lentivirus-based RNA interference (RNAi) transfer plasmid pLKO.1-puro-sh-hCen781 (pKM183), the oligonucleotides (forward, 5′-CCGGAGACTAATGGAGCAACAAGCTCGAGCTTGTTGCTCCATTAGTCTTTTTTG-3′, and reverse, 5′-AATTCAAAAAAGACTAATGGAGCAACAAGCTCGAGCTTGTTGCTCCATTAGTCT-3′) were annealed and then inserted into pLKO.1-puro vector (a gift of S. A. Stewart and P. A. Sharp, Massachusetts Institute of Technology, Cambridge) digested with AgeI and EcoRI. pHR′-CMV-SV-puro vector was generated by replacing the lacZ sequence of pHR′-CMV-lacZ (25) with a 1.1-kb SalI-KpnI fragment containing the simian virus 40 enhancer/promoter upstream of the puromycin resistance gene (C.-Z. Giam, unpublished data). To generate pHR′-hCMV*-1-SV-puro-hCENEXIN1 WT (pKM166) and pHR′-hCMV*-1-SV-puro-hcenexin1-sil (pKM167), which express hCenexin1 under the hCMV*-1 tetracycline-controllable promoter (12), pHR′-CMV-SV-puro vector was first digested with ClaI (end filled) and SalI to remove the human cytomegalovirus (hCMV) promoter and then ligated with EcoRI (end-filled)-SalI fragments containing either hCMV*-1-SV-puro-hCENEXIN1 WT or hCMV*-1-SV-puro-hcenexin1-sil. The hcenexin1-sil mutant, which bears three silent mutations, was generated by PCR-based mutagenesis with the following primers: forward, 5′-AAAGACTCATGGAACAGCAAG-3′, and reverse, 5′-CTTGCTGTTCCATGAGTCTTT-3′ (silent mutations are indicated in boldface type).
Cell culture, transfection, and virus generation and infection.
HeLa, HCT116, U-2 OS, and CHO cells were cultured as subconfluent monolayers under the conditions recommended by the American Type Culture Collection (Manassas, VA). Sf9 insect cells were cultured in Grace's insect medium (Invitrogen) at 28°C. To enrich HeLa cells in G1, S, G2, or M, cells were treated with 0.3 mM of mimosine, 2.5 mM of thymidine, 0.5 μM of etoposide, or 0.1 μM of nocodazole, respectively. Plasmid transfection was carried out by using Lipofectamine 2000 (Invitrogen). For short interfering RNA (siRNA) transfection, indicated cells were transfected with 100 nM of siRNA oligonucleotides using Oligofectamine (Invitrogen). siRNAs against hCenexin1 (si-hCen781, 5′-AGACUAAUGGAGCAACAAG-3′; si-hCen1218, 5′-GGAUCUUUAUGUCGCUGAA-3′; si-hCen2066, 5′-GGCAGUUGGAGAGUGCCAU-3′; the numbers indicate the nucleotide positions of each si-hCen in relation to the start codon), Plk1 (si-Plk1, 5′-AGAUUGUGCCUAAGUCUCU-3′), and control luciferase GL2 (si-Luc, 5′-CGUACGCGGAAUACUUCGA-3′) were purchased from Dharmacon, Inc. (Lafayette, CO). Recombinant adenoviruses (GFP-hCenexin1 and GFP-T1-GFP-T7) were generated as described previously (14) using pKL2472, pKL2475, and pKL2478-pKL2484 constructs described above. To carry out Plk1 relocalization experiments, HeLa cells transfected with si-hCen781 were infected with adenoviruses expressing various hCenexin1 constructs. Viruses expressing the T1 or the T6 to T8 forms were used at a multiplicity of infection of 2. However, viruses expressing full-length GFP-hCenexin1 or the GFP-T2-GFP-T5 forms that contain the si-hCen781 target sequence were used at a multiplicity of infection of 10, a dose that was sufficient to overcome the effect of si-hCen781. For lentivirus production, pHR′-CMVΔR8.2Δvpr and pHR′-CMV-VSV-G (protein G of vesicular stomatitis virus) were cotransfected with either pLKO.1-puro-sh-hCen781 to generate shRNA virus or pHR′-hCMV*-1-SV-puro-hCENEXIN1 to generate hCenexin1 expression virus under the tetracycline-off hCMV*-1 promoter control (12). HeLa cells infected with the indicated viruses were selected with 2 μg/ml of puromycin before further experiments were conducted.
Antibody production and purification.
Bacterially expressed glutathione S-transferase (GST)-fused hCenexin1(250-632) [“(250-632)” indicates amino acids 250 to 632] was purified using glutathione agarose and then injected into rabbits to raise polyclonal anti-hCenexin antisera. To affinity purify the anti-hCenexin antibody, immunized sera were first depleted of anti-GST antibody using a GST column (Amersham Biosciences, Piscataway, NJ). The resulting sera were further purified using GST-hCenexin1(250-632) immobilized with Affigel-10 (Bio-Rad Laboratories, Hercules, CA).
Flow cytometry analyses.
To analyze cell cycle progression, flow cytometry analyses were carried out using a FACScan cell sorter (Becton Dickinson, San Jose, CA), and data were analyzed by the Flowjo program (Tree Star, San Carlos, CA).
Immunoprecipitation, in vitro protein-protein interaction, and immunoblotting analyses.
Immunoprecipitation was carried out as described previously (20). Briefly, cells were lysed in TBSN buffer (20 mM Tris-Cl [pH 8.0], 150 mM NaCl, 0.5% NP-40, 5 mM EGTA, 1.5 mM EDTA, 0.5 mM Na3VO4, and 20 mM p-nitrophenyl phosphate). The resulting lysates were clarified by centrifugation at 15,000 × g for 20 min at 4°C before immunoprecipitation with the specified antibody. Immunoprecipitated proteins were separated by 10% SDS-PAGE and then immunoblotted.
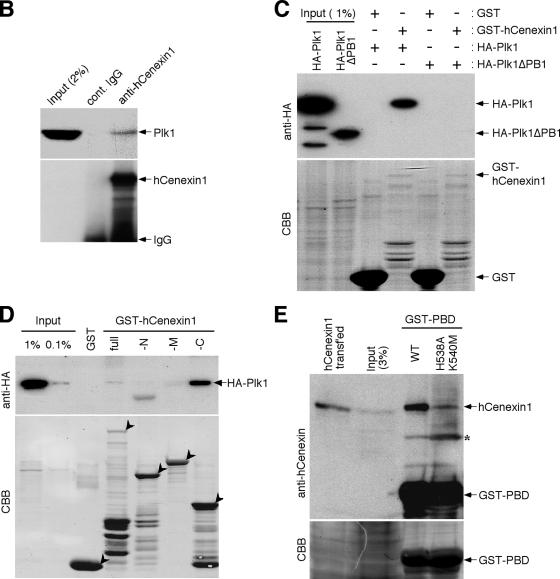
To investigate the interaction between hCenexin1 and Plk1, total cellular lysates (1 mg) prepared from Sf9 cells expressing HA-Plk1 or HA-Plk1ΔPB1 (lacking PB1 of the PBD) (19) were added to either bead-bound control GST (10 μg) or bead-bound GST-hCenexin1 fusion proteins (0.5 to 3 μg of the full-length or various truncated forms; see the Coomassie brilliant blue [CBB] gel in Fig. 7) and then incubated in a binding buffer (50 mM Tris HCl [pH 8.0], 300 mM NaCl, 0.5% NP-40, 5 mM EGTA, 1.5 mM EDTA, 0.5% deoxycholate) for 2 h at 4°C. After incubation, beads were washed extensively with the binding buffer and boiled in SDS-PAGE sample buffer to elute the coprecipitated proteins. Samples were separated by 10% SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and then detected by immunoblotting with anti-HA.11 antibody (Covance, Richmond, CA) using the enhanced chemiluminescence detection system (Pierce, Rockford, IL). The same membrane was stained with Coomassie blue to detect GST and GST-hCenexin1 ligands. To determine whether hCenexin1 interacts with the phosphate pincer cleft of the PBD, 6 μg of bead-bound GST-PBD or GST-PBD(H538A, K540M) (9) was incubated with mitotic HeLa lysates (2 mg), and the resulting precipitates were subjected to immunoblotting with anti-hCenexin antibody. Other immunoblotting analyses were also conducted similarly as described above.
FIG.7.
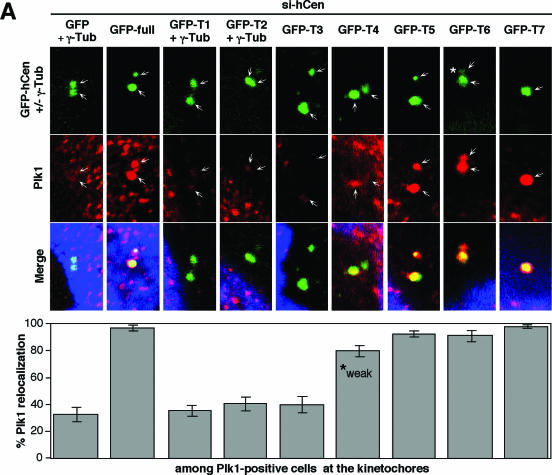
hCenexin1 promotes Plk1 localization to the centrosomes through the interaction between the C-terminal extension of hCenexin1 and the PBD of Plk1. (A) To examine the ability of hCenexin1 to relocalize Plk1 to the centrosomes, HeLa cells were first depleted of hCenexin1 and then infected with adenoviruses expressing either the full length or various truncations of GFP-hCenexin1 before being subjected to immunostaining analyses with anti-Plk1 antibody. Cells expressing GFP vector, GFP-T1, or GFP-T2 were costained with γ-tubulin antibody to mark the positions of centrosomes. The full-length GFP-hCenexin1 and GFP-T3-GFP-T7 constructs efficiently localized to the centrosomes by themselves. GFP-T4 was able to recruit Plk1 to the centrosomes, but the recruited Plk1 signals (marked “weak”) were significantly lower than those of Plk1 recruited by GFP-T5, GFP-T6, or GFP-T7. A nascent GFP-T6 signal (*) localized at the daughter centriole appeared to be sufficient to recruit Plk1 to that site. Arrows indicate centrosomes. Approximately 120 cells were counted in each of two independent experiments. Error bars indicate standard deviations. (B) Total cellular lysates prepared from nocodazole-arrested HeLa cells were subjected to immunoprecipitation with either control rabbit immunoglobulin G (IgG) or anti-hCenexin antibody. Coprecipitated Plk1 was detected with a mouse anti-Plk1 antibody. (C and D) To carry out in vitro binding analyses, the indicated bead-bound GST or GST-fused ligands (0.5 to 3 μg; see the CBB stain) were incubated with Sf9 cell lysates (1 mg) expressing either HA-Plk1 or HA-Plk1ΔPB1 (C) or HA-Plk1 (D). After pull-down, proteins were separated by 10% SDS-PAGE and then subjected to immunoblotting analyses. Afterwards, the same membranes were stained with CBB to visualize the ligand proteins. Arrowheads indicate ligands used for each binding. (E) Mitotic HeLa lysates (2 mg) were incubated with bead-bound GST-PBD or the corresponding GST-PBD(H538A, K540M) mutant (6 μg each), and the resulting precipitates were immunoblotted with anti-hCenexin antibody (top panel) followed by CBB staining (bottom panel). Because anti-hCenexin antibody was generated using GST-hCenexin1 fragment as an immunogen, it also detected the GST-PBD ligand. The asterisk indicates a cross-reacting band.
Indirect immunofluorescence microscopy and quantification.
Indirect immunostaining was carried out as described previously (33). Anti-hCenexin antibody was used at 4 μg/ml. Other primary antibodies used were rabbit anti-Cep135 antibody (A5-CEP) (27), rabbit anti-Plk1 antibody (Santa Cruz Biotechnologies, Santa Cruz, CA), mouse anti-HA.11 antibody (Covance), rabbit antininein antibody (a gift of J. B. Rattner, University of Calgary, Calgary, Canada), mouse anticentrin antibody (a gift of J. L. Salisbury, Mayo Clinic, Rochester, MN), rabbit antipericentrin antibody (Covance), mouse anti-CTR antibody (a gift of M. Bornens, Curie Institute, Paris, France), rabbit anti-Nek2 antibody (AbCam, Cambridge, MA), anti-α-tubulin antibody (Sigma, St. Louis, MO), and anti-γ-tubulin antibody (Santa Cruz Biotechnologies). All the appropriate secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). To stain chromosomes, cells were treated with PBST containing 0.1 μg/ml of 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma). Digital images were collected with a Zeiss LSM510 confocal microscope. For the quantification of the fluorescence signal intensities, images of unsaturated fluorescence signals were acquired with the same laser intensity at 1,024 × 1,024 pixels and 16-bit resolution. For each identifiable centrosomal signal (γ-tubulin, Plk1, and hCenexin1), fluorescence intensities were determined after subtraction of the background signal intensities using Zeiss AIM confocal software. The intercentriolar distance of centrin pairs was also measured using the same Zeiss software. Obtained data points were plotted with the Sigma Plot 9 program (Systat Software, Inc., Point Richmond, CA).
RESULTS
Isolation of hCenexin as a Plk1-interacting protein.
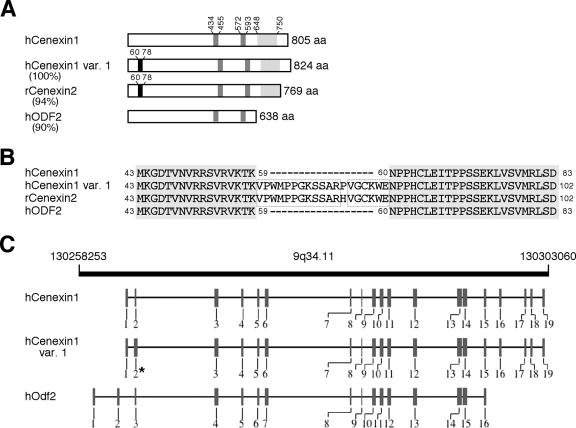
In an effort to identify novel Plk1-interacting proteins through a Saccharomyces cerevisiae two-hybrid screen, we used a GAL4 DNA-binding domain-fused kinase-inactive Plk1(K82M) (20) as a bait and screened a GAL4 activation domain-fused human testis cDNA library (MATCHMAKER; Clontech). One locus isolated multiple times contained the nucleotide sequences identical to the two overlapped C-terminal regions (aa 586 to 805 and aa 647 to 805) of the human outer dense fiber of sperm tail 2 (hOdf2) isoform 1 (GenBank accession number NM_002540) that was identified from a large-scale characterization of phosphoproteins from HeLa cells (3). For the reasons below together with a unique function of the C-terminal extension found in hOdf2 isoform 1, we renamed this gene-encoded product hCenexin1 (GenBank accession number DQ444714) for its functional clarity. To isolate other variants, we carried out reverse transcription-PCR using oligonucleotides complementary to the full-length hCenexin1 and total RNAs from asynchronously growing HeLa cells as template. These analyses resulted in the identification of an hCenexin1-like transcript (named hCenexin1 variant 1; GenBank accession number DQ444713) that bears a small insertion of 19 aa in the N-terminal exon2 (Fig. 1A to C). The insertion of a similar 19-aa sequence (aa 60 to 78) was previously shown to exist in a rat Cenexin homolog, rCenexin2 (Fig. 1B), and mouse Odf2/1 (30). Sequence alignment analyses showed that hOdf2, hCenexin1, and hCenexin1 variant 1 are splicing variants of the same genomic locus (Fig. 1C). Interestingly, however, the interspecies homology between hCenexin1 and rCenexin2 (94% identity; also 96% identity between hCenexin1 variant 1 and rCenexin2) was greater than the intraspecies homology between hCenexin1 and hOdf2 (90% identity) (Fig. 1A; also see Fig. S1 in the supplemental material). This finding suggests that hCenexin1 and its 19-aa insertion form, hCenexin1 variant 1, belong to a subfamily of rCenexin2-like proteins. Notably, both hCenexin1 and hCenexin1 variant 1, but not hOdf2, possess a unique C-terminal extension (aa 648 to 805 in hCenexin1) whose amino acid sequence is closely related to that of rCenexin2 (85% identity). Furthermore, the molecular sizes of hCenexin1 and hCenexin1 variant 1 (calculated sizes of 93 kDa and 95 kDa, respectively) are similar to that of the originally described 96-kDa Cenexin detected from purified centriolar preparations from lamb thymus (18). The existence of various Odf2 isoforms in one organism suggests that the expression and function of these proteins are likely to be highly regulated.
FIG. 1.
(A) Schematic comparison of primary amino acid sequences between hCenexin1 and its related proteins. hCenexin1 (accession no. DQ444714), hCenexin1 variant 1 (var. 1) (accession no. DQ444713), rCenexin2 (accession no. AF162756), and hOdf2 (accession no. AF012549) are shown. Percentages in parentheses indicate percent identity between hCenexin1 and the respective proteins at the amino acid level. Black boxes denote the 19-aa insertions found in hCenexin1 variant 1 and rCenexin2, whereas gray boxes indicate two putative leucine zipper motifs. Dotted boxes in the C-terminal extension are found only in the Cenexin subfamily and exhibit 85% identity. (B) Sequence alignment between hCenexin1 and its related sequences in the N-terminal insertion region. Gray boxes indicate the identical sequences among all four sequences, whereas the outlined boxes indicate the identical sequences within the 19-amino-acid insertion. (C) Schematic diagram of chromosome 9q34.11 region (from nucleotide 130258253 to nucleotide 130303060) (adapted from NCBI database). Vertical bars with numbers indicate exons for hCenexin1, hCenexin1 variant 1, and hOdf2. Note that the exon 2 for hCenexin1 variant 1 (*) is thicker than the corresponding exon for hCenexin1 because of the 19-aa insertion.
hCenexin1 is the major isoform that is highly expressed at the late stages of the cell cycle in HeLa cells.
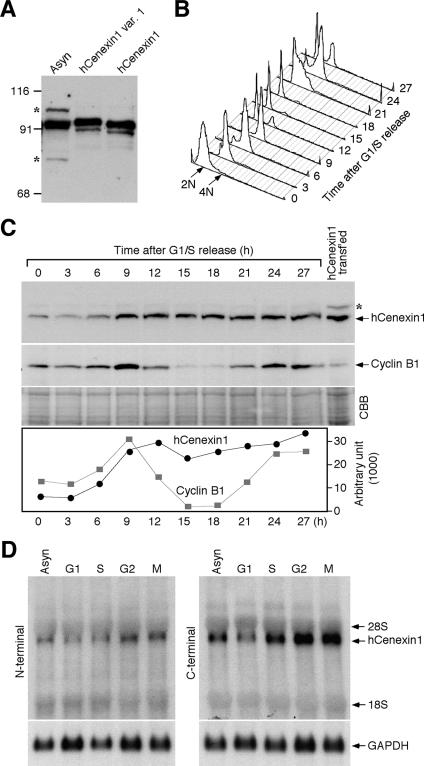
To investigate the expression of hCenexin proteins in cultured cells, we raised a polyclonal anti-hCenexin antibody against aa 250 to 632 of hCenexin1 (this sequence corresponds to aa 269 to 651 of hCenexin1 variant 1 and aa 255 to 623 of hOdf2). We next examined the expression levels of various isoforms in asynchronously growing HeLa cells. For comparison, HeLa lysates transfected with either the full-length hCenexin1 or hCenexin1 variant 1 construct were also included (untagged hCenexin1 and hCenexin1 variant 1 were expressed to mark the precise position of their migration in SDS-PAGE). Immunoblotting analysis with affinity-purified anti-hCenexin antibody detected a 93-kDa protein that comigrated with hCenexin1 (Fig. 2A). Proteins corresponding to the size of hCenexin1 variant 1 or hOdf2 were not detected (see also Fig. 5B), even though both hCenexin1 variant 1 and hOdf2 bear the hCenexin1 region that was used to generate the hCenexin antibody. These observations together with the result shown in Fig. 5 indicate that hCenexin1 is the major isoform expressed in HeLa cells. A 93-kDa protein was also detected in U-2 OS osteosarcoma cells (data not shown), hinting that hCenexin1 is ubiquitously expressed in human somatic cells.
FIG. 2.
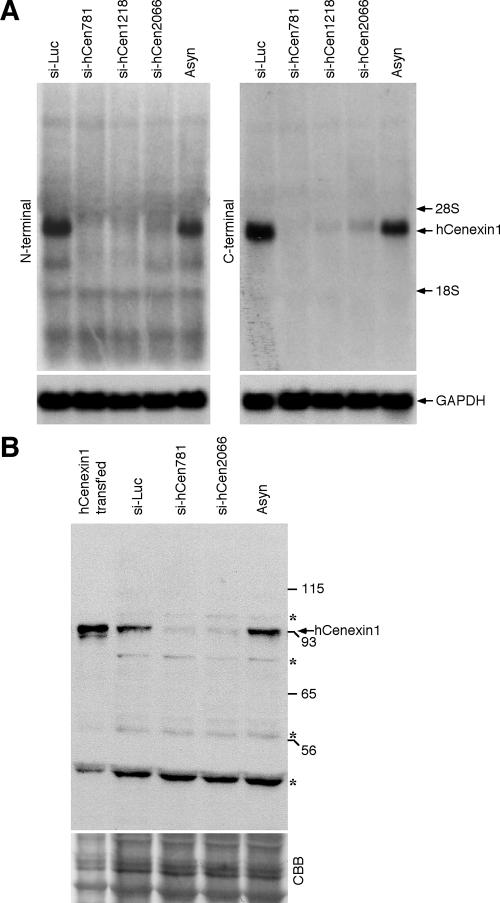
hCenexin1 is the major Odf2-related isoform abundantly expressed at the late stages of the cell cycle. (A) An anti-hCenexin antibody detects a 95-kDa protein from asynchronously growing HeLa cells (Asyn). For comparison, lysates from HeLa cells transfected with either the full-length hCENEXIN1 or the full-length hCENEXIN1 variant 1 (var. 1) were also separated after being diluted ∼20-fold (as a result, the endogenous 95-kDa protein was not detected). Asterisks indicate nonspecific proteins cross-reacting with anti-hCenexin antibody. Numbers at left are molecular masses in kilodaltons. (B and C) HeLa cells were arrested at the G1/S boundary by double thymidine block and then released into fresh medium. Samples harvested at the indicated time points were subjected to flow cytometry analysis (B) and immunoblotting analyses with the indicated antibodies (C). The same membrane was stained with CBB for loading controls. Asterisk, nonspecific cross-reacting protein. The levels of hCenexin1 and cyclin B1 were quantified using the Image J program (graph). (D) To synchronize the HeLa cells at specific stages of the cell cycle, cells were treated with either mimosine (G1), thymidine (S), etoposide (G2), or nocodazole (M) as described in Materials and Methods. Total RNAs were prepared for Northern blot analyses using the PCR fragments bearing either the N-terminal region (aa 120 to 253 of hCenexin1) (left panel) or the C-terminal extension region (aa 652 to 784 of hCenexin1) (right panel) as a probe. The same membrane was used to detect glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a loading control.
FIG.5.
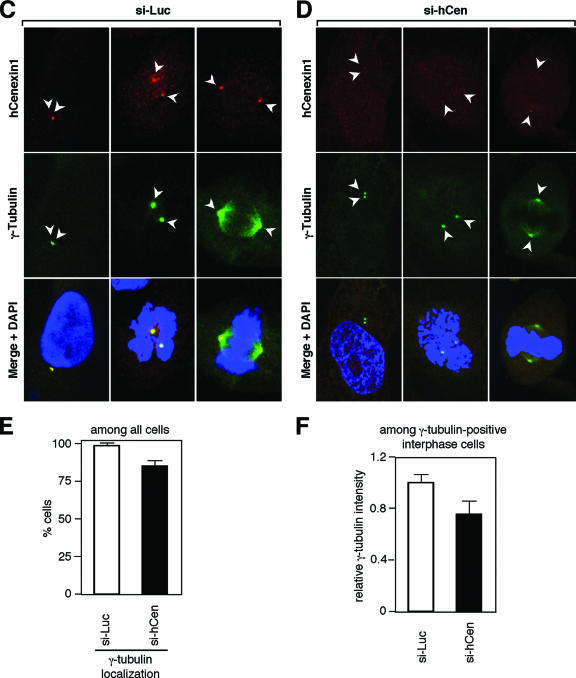
hCenexin1 is the major Odf2-related isoform expressed in HeLa cells. (A) Cells were transfected with either control luciferase siRNA (si-Luc) or the indicated siRNAs against hCenexin1. Total RNAs prepared from the cells 3 days after transfection were subjected to Northern blot analyses using either N-terminal (aa 120 to 253 of hCenexin1) or C-terminal (aa 652 to 784 of hCenexin1) probes. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) signals serve as loading controls for each lane. (B) Total lysates were prepared from HeLa cells transfected with the indicated siRNAs for 3 days and then subjected to immunoblotting analyses with anti-hCenexin antibody. Asynchronous cells (Asyn) and cells transfected with the full-length hCENEXIN1 were also loaded for comparison. Asterisks indicate proteins cross-reacting with the hCenexin1 antibody that were insensitive to si-hCen. CBB, CBB staining of the same membrane. Numbers at right are molecular masses in kilodaltons. (C to F) HeLa cells transfected with either si-Luc (C) or si-hCen781 (si-hCen) (D) were stained with anti-hCenexin and anti-γ-tubulin antibodies. Arrowheads mark the position of centrosomes. (E) These cells were then counted to determine the efficiency of γ-tubulin localization to the centrosomes. Approximately 250 cells were counted in each of two independent experiments. Error bars show standard deviations. (F) The intensities of γ-tubulin fluorescence for each sample were measured from more than 50 interphase cells with separated γ-tubulin signals chosen at random. Images were acquired with the same settings and then analyzed using Zeiss LSM 510 software. The level for the si-Luc cells was normalized to 1. Error bars indicate standard deviations.
To determine how hCenexin1 is regulated during cell cycle progression, we arrested the HeLa cells at the G1/S boundary by double thymidine block and released them into fresh medium to monitor the level of hCenexin1 expression (Fig. 2B). hCenexin1 was expressed at a low level at the G1/S boundary and S but became abundant at the later stages of the cell cycle (Fig. 2C). However, unlike the level of cyclin B1, which declined after mitotic exit and reaccumulated in the next G2 phase, the level of hCenexin1 largely remained high even in the next G1 and S phases (Fig. 2C). These observations suggest that hCenexin1 is a stable protein with a low turnover rate.
We next examined the levels of the hCenexin1 transcript after treating the HeLa cells with mimosine, thymidine, etoposide, or nocodazole to enrich the cell population in G1, S, G2, or M, respectively. Northern blot analyses using probes derived from either the N terminus (aa 120 to 253) or the C-terminally extended region (aa 652 to 784) of hCenexin1 detected a single transcript of ∼4.0 kb (Fig. 2D), whose abundance closely correlated with that of hCenexin1 protein at specific stages of the cell cycle. Thus, hCenexin1 expression is regulated at the transcriptional level. Interestingly, the C-terminal probe that does not bear a significant homology with hODF2 at the nucleotide sequence level also efficiently detected the 4.0-kb transcript, further supporting the argument that hCENEXIN1 is the major ODF2-related transcript in HeLa cells (see also Fig. 5A).
Complexity of hCenexin1 localization to the mother centrioles.
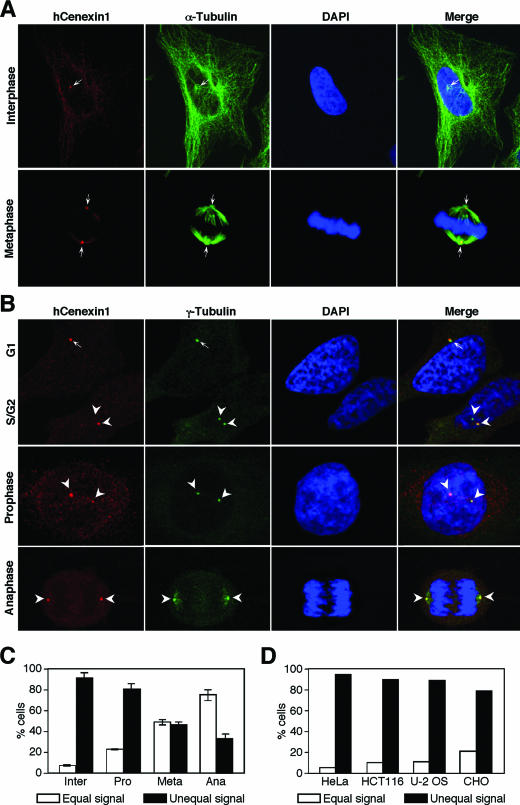
Next, we examined the subcellular localization of hCenexin1 in HeLa cells by carrying out indirect immunofluorescence studies using affinity-purified anti-hCenexin antibody. In coimmunostaining analysis with anti-α-tubulin antibody, we found that hCenexin1 localized at the microtubule organizing centers in interphase (Fig. 3A, upper panel) and spindle poles in mitosis (Fig. 3A, bottom panel), indicating that hCenexin1 is a centrosomal protein. Additional immunostainings with anti-γ-tubulin antibody showed that hCenexin1 localized to the unduplicated G1 centrosomes (Fig. 3B, top panel, arrow) but remained as a single dot even after centrosome duplication (Fig. 3B, top panel, two arrowheads). However, as cells proceeded through the cell cycle, hCenexin1 progressively localized to the other centrosome (Fig. 3B, middle panel) and exhibited approximately equal levels of signal intensities during mitotic progression (Fig. 3B, bottom panel, and Fig. 3C). Several cultured cell lines that we examined also exhibited unequal hCenexin1 localization similar to that observed in HeLa cells (Fig. 3D).
FIG. 3.
hCenexin1 preferentially localizes to one of the two centrosomes. (A and B) Asynchronously growing HeLa cells were subjected to coimmunostaining analyses with anti-hCenexin and anti-α-tubulin antibodies (A) or anti-hCenexin and anti-γ-tubulin antibodies (B). Arrows (G1) and arrowheads (S/G2) indicate centrosomes. (C) The percentages of cells with unequal hCenexin1 signals (estimated by eye) were determined by comparing hCenexin1 signals with two γ-tubulin signals within the same cells. More than 150 cells were counted in each of three independent experiments. Error bars indicate standard deviations. (D) Indicated cultured cells were immunostained with anti-hCenexin and anti-γ-tubulin antibodies. Among interphase centrosomes, the percentages of unequal hCenexin1 signals were determined as in panel C. More than 200 cells were counted for each cell line.
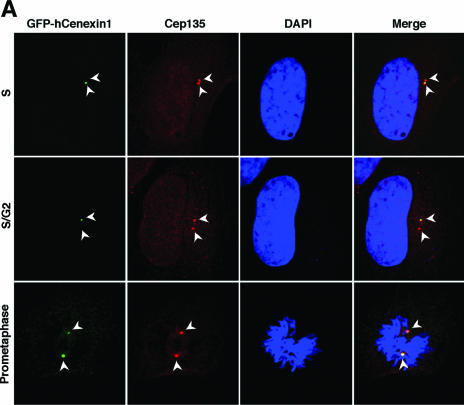
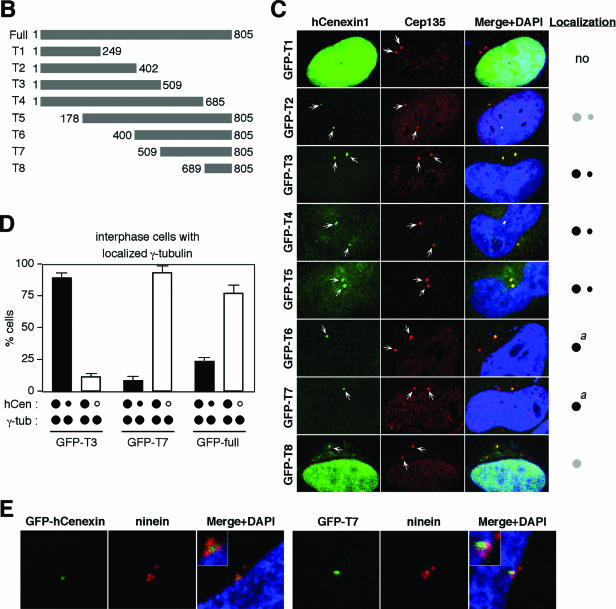
We then investigated the nature of hCenexin1 localization by transfecting a GFP-fused full-length hCenexin1 and its various truncation mutants (Fig. 4A and B) into HeLa cells and then examining the ability of each construct to localize to the centrosomes. A pericentrosomal protein, Cep135 (27), was used as a centrosome marker. The full-length GFP-hCenexin1 primarily localized to one of the two centrosomes in interphase and both centrosomes in mitosis (Fig. 4A and D), thus confirming the above endogenous hCenexin1 localization revealed by anti-hCenexin immunostaining. Under the same conditions, the N-terminal GFP-T1 (aa 1 to 249) accumulated in the nucleus but failed to localize to the centrosomes. The N-terminal region of GFP-hCenexin1 variant 1 (aa 1 to 268) bearing the 19-aa insertion or the full-length GFP-hCenexin1 variant 1 localized similarly to the respective GFP-T1 or GFP-hCenexin1 (data not shown), suggesting that the 19-aa insertion does not influence their subcellular localizations. GFP-T2 (aa 1 to 402) containing the N-terminal half of hCenexin1 inefficiently (∼34%) localized to both centrosomes with very weak signals (Fig. 4C). However, the GFP-T3, GFP-T4, and GFP-T5 forms efficiently localized (greater than 98% of GFP-positive cells) to both of the centrosomes but with characteristically uneven signal intensities (Fig. 4C and D). In contrast, GFP-T6 and GFP-T7, lacking the N-terminal half of hCenexin1, exhibited a strong tendency to localize to one of the two centrosomes (Fig. 4C and D), although a nascent fluorescent signal at the second centrosome was at times detectable at a high magnification (see Fig. 7A, GFP-T6 construct). The C-terminal-most GFP-T8 construct exhibited a localization pattern similar to those of GFP-T6 and GFP-T7 but frequently with strong nuclear and aggregated cytoplasmic signals (Fig. 4C).
FIG.4.
Complexity of hCenexin1 localization to the mother centrioles. (A) HeLa cells transfected with GFP-fused hCENEXIN1 were fixed and stained with anti-Cep135 antibody to determine the ability of hCenexin1 to localize to the centrosomes (arrowheads). (B and C) Various truncated forms of hCenexin1 depicted in panel B were transfected into HeLa cells to determine the ability of each construct to localize to the centrosomes (arrows) (C). Typical localization patterns of each construct are depicted as one or two dots (right). Big dots indicate strong signals for localization to the mother centrioles, whereas small dots indicate weak signals for localization to the daughter centrioles. Gray dots depict inefficient and weak localization. a, the GFP-T6 and GFP-T7 constructs frequently exhibited a very weak daughter centriole localization (see text). (D) Cells expressing GFP-T3, GFP-T7, or the full-length GFP-hCenexin1 were stained with anti-γ-tubulin antibody to determine their patterns of localization to the centrosomes. Only the interphase cells with two distinct γ-tubulin signals were subjected to counting. Black dots, localized signals; open dots, no localized signals. Approximately 250 cells were counted in each of two independent experiments. Error bars indicate standard deviations. (E) HeLa cells transfected with either the full-length GFP-hCenexin1 or the GFP-T7 were subjected to immunostaining with antininein antibody. Inset, a high magnification of the mother centriole area.
To confirm the unequal localization nature of hCenexin1 truncations, we then chose three of the constructs—namely, the full-length GFP-hCenexin1, GFP-T3, and GFP-T7, and quantitated their localization patterns in interphase HeLa cells. Among the cells with two distinct γ-tubulin signals, a large fraction (89%) of GFP-T3 localized to both centrosomes. In contrast, approximately 92% of GFP-T7 exhibited a preferential localization to one of the two centrosomes, while the full-length GFP-hCenexin1 displayed a somewhat reduced level (77%) of single-centrosome localization (Fig. 4D). Further immunostaining analyses with antininein antibody revealed that the singly localized full-length GFP-hCenexin1 and GFP-T7 dots colocalized with three ninein dots that associated with the mother centriole (23) (Fig. 4E). These findings suggest that the C-terminal region of hCenexin1 is important for mother centriole localization, whereas the N-terminal region confers the ability of hCenexin1 to localize to daughter centrosomes as well.
Depletion of hCenexin1 by siRNAs targeting either the N-terminal or the C-terminal extension region.
To investigate the defects associated with the depletion of hCenexin1, we used siRNA duplexes against hCENEXIN1. Three different siRNA oligonucleotides (si-hCen781, si-hCen1218, and si-hCen2066; the numbers indicate positions relative to the first nucleotide of the hCenexin1 start codon) efficiently downregulated the ∼4-kb hCENEXIN1 transcript that was detected by either the N-terminal (aa 120 to 253; common to both hCENEXIN1 and hODF2) or the C-terminal (aa 652 to 784; specific to hCENEXIN1) extension probe (Fig. 5A). It is notable that si-hCen2066, which targets the C-terminal extension of hCENEXIN1, but not hODF2, also drastically diminished the level of the 4-kb transcript detectable by the N-terminal probe (Fig. 5A). Since the nucleotide sequence of the N-terminal probe of hCENEXIN1 is identical to that of hODF2 and therefore should be able to detect the hODF2 transcript if present, these observations strongly suggest that hODF2 is not expressed or is expressed at an undetectable level in HeLa cells. In line with these observations, transfection of HeLa cells with either si-hCen781 or si-hCen2066 also efficiently depleted a single 93-kDa protein comigrating with the transfected hCenexin1 (Fig. 5B). These findings together with the results shown in Fig. 2 strongly support the notion that hCenexin1 is the major isoform expressed in HeLa cells.
Immunofluorescence studies revealed that cells transfected with either si-hCen781 or si-hCen2066 exhibited greatly diminished hCenexin1 signals, although ∼28% of the total population still possessed weakened but localized hCenexin1 signals (Fig. 5C and D; see Fig. 6B). These cells also displayed γ-tubulin signals that appeared to be weakened in most of the population (Fig. 5C and D) and absent in a small fraction (∼15%) of the cells (Fig. 5E). Measurement of the γ-tubulin fluorescence intensities among the interphase cells with two separated centrosomes revealed that the si-hCen cells possessed approximately 75% of the level of the γ-tubulin signals in the si-Luc cells (Fig. 5F). Thus, hCenexin1 is required for normal assembly of centrosome components important for γ-tubulin recruitment. Since hCen781 depleted hCenexin1 more efficiently than si-hCen2066 at both mRNA and protein levels (Fig. 5A; see also Fig. S2 in the supplemental material), hCen781 (hereafter hCen for simplicity) was used for the following experiments.
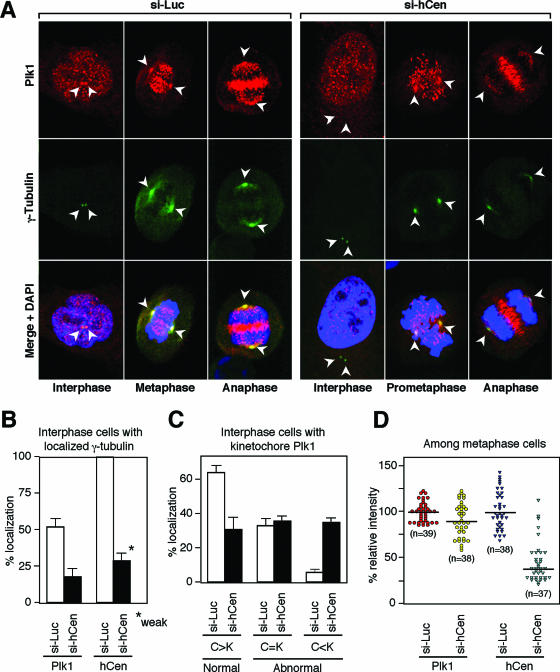
FIG.6.
Mutual requirement of hCenexin1 and Plk1 for proper localization to the centrosomes. (A) HeLa cells transfected with either control si-Luc or si-hCen781 (si-hCen) were subjected to immunostaining analyses with anti-Plk1 and anti-γ-tubulin antibodies. The γ-tubulin signals (arrowheads) serve as markers for centrosomes. (B) Quantitation of cells with the centrosomal Plk1 or the hCenexin1 signals was carried out using the samples costained with anti-γ-tubulin antibody. Since Plk1 signals are not readily detectable in early stages of the cell cycle, only G2 cells (Plk1 localizes to the centrosomes and kinetochores at this stage) with two distinct γ-tubulin signals were counted. Transfection of si-hCen greatly diminished, but did not eliminate, hCenexin1 signals (marked “weak”) at the centrosomes. More than 300 cells were counted in each of three independent experiments. Error bars show standard deviations. (C) Among the G2 cells with detectable Plk1 signals at the kinetochores (>300 cells), relative Plk1 signal intensities between the centrosomes and the kinetochores were visually compared and then scored in two independent experiments. C>K indicates that the centrosomal Plk1 signals are greater than the kinetochore Plk1 signals, whereas C<K indicates that the Plk1 signals from the kinetochores are greater than those from the centrosomes. (D) To quantify the level of Plk1 and hCenexin1 signal intensities among metaphase cells, acquired images (“n” indicates the number of cells) were analyzed using Zeiss confocal software and then plotted as described in Materials and Methods. The black horizontal lines indicate the average intensities in each group. (E) To determine whether Plk1 depletion influences the localization of hCenexin1 to the centrosomes, HeLa cells transfected with si-Plk1 were costained with anti-hCenexin and anti-γ-tubulin antibodies. Arrowheads indicate the positions of centrosomes. (F) Approximately 250 cells were counted in two independent experiments to determine the patterns of hCenexin1 and γ-tubulin localization. Error bars indicate standard deviations.
hCenexin1 and Plk1 are mutually required for proper localization to the centrosomes.
It has been shown that Plk1 is required for proper centrosome maturation and microtubule nucleation (6, 17). Since hCenexin1 interacted with Plk1 in a yeast two-hybrid assay and depletion of hCenexin1 resulted in a diminished level of γ-tubulin signals (Fig. 5E and F), we examined whether the depletion of hCenexin1 alters Plk1 localization to the centrosome among the cells with distinctly localized γ-tubulin signals (although γ-tubulin signals were diminished in si-hCen cells, they were strong enough to mark the centrosome locations in most of the cell population). The level of Plk1 signals at the kinetochores of the same cell was also examined for comparison. In control si-Luc cells, Plk1 was detectable in ∼53% of the interphase centrosomes (Fig. 6A and B) and the signals at the centrosomes were significantly greater than those at the kinetochores (Fig. 6C). In contrast, only 18% of the interphase si-hCen cells exhibited proper Plk1 localization and a large fraction of the cells exhibited undetectable or greatly weakened Plk1 signals at the centrosomes (Fig. 6A and B), even though hCenexin1 was not completely depleted from the cells (Fig. 6B, right graphs). Quantitation of Plk1 localization among the Plk1-positive cells (cells that display Plk1 dots at the kinetochores) further confirmed the idea that Plk1 localization to the centrosomes, but not to the kinetochores, was impaired in si-hCen cells (Fig. 6C). Thus, hCenexin1 is critical for proper localization of Plk1 to the interphase centrosomes. Since Plk1 function is required for proper centrosomal maturation and γ-tubulin recruitment (17), this observation is largely consistent with the weakened γ-tubulin signals observed in si-hCen cells (Fig. 5E and F). However, unlike interphase centrosomes, depletion of hCenexin1 modestly (∼83% of the control Plk1 intensity) delocalized Plk1 from the mitotic centrosomes (Fig. 6D). Perhaps, as the centrosomes mature before mitotic entry, they may recruit additional components capable of interacting with Plk1 during mitosis.
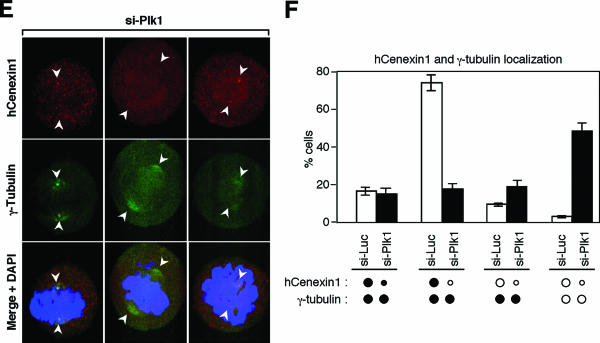
Next, we examined whether Plk1 is required for proper localization of hCenexin1 at the centrosomes. To this end, cells treated with either si-Luc or si-Plk1 were immunostained with anti-hCenexin and anti-γ-tubulin antibodies. Because of the lethality associated with the downregulation of Plk1, only the cells with normal DAPI morphologies (i.e., excluding the cells with apoptotic DNA morphology) were examined. In control si-Luc cells, approximately 75% of the cells exhibited only one centrosomally localized hCenexin1 and ∼16% of the cells exhibited two distinct hCenexin1 signals (Fig. 6F). Depletion of Plk1 induced delocalization of both hCenexin1 and γ-tubulin (Fig. 6E)—only a small fraction (∼17%) of cells displayed singly localized hCenexin1 signals and a large fraction (∼49%) possessed no detectable hCenexin1 and γ-tubulin signals (Fig. 6F). These results suggest that depletion of Plk1 leads to the delocalization of hCenexin1 from the centrosomes either directly or indirectly.
hCenexin1 promotes Plk1 localization to the centrosomes through direct binding.
Since hCenexin1 is required for proper Plk1 localization, expression of a localization-competent hCenexin1 may promote Plk1 localization to the centrosomes. To examine this possibility, cells were first depleted of hCenexin1 by si-hCen to delocalize Plk1 from the centrosomes and then infected with adenoviruses expressing either the full-length or the various truncated forms of GFP-hCenexin1. In si-hCen cells expressing control GFP, approximately 68% of the Plk1-positive population (a fraction of cells with detectable kinetochore Plk1 signals) displayed a much-weakened or undetectable level of Plk1 signals at the centrosomes; only ∼32% of this population exhibited proper Plk1 localization to the centrosomes. Expression of the full-length GFP-hCenexin1 induced relocalization of Plk1 to the centrosomes in ∼96% of the Plk1-positive population (Fig. 7A). Similarly, expression of the centrosome-localizing GFP-T5, GFP-T6, and GFP-T7 forms also efficiently targeted Plk1 to the centrosomes as they localized to the same locations. In contrast, expression of localization-competent GFP-T4 bearing only a part of the C-terminal extension restored Plk1 signals in approximately 80% of the population, but the intensity of the Plk1 signal was about half of those induced by the GFP-T5, GFP-T6, and GFP-T7 forms (Fig. 7A; marked “weak” for GFP-T4). In addition, expression of GFP-T3 lacking the whole C-terminal extension failed to relocalize Plk1, even though GFP-T3 itself is fully capable of localizing to the centrosomes. As expected, localization-defective GFP-T1 and GFP-T2 were not able to target Plk1 to the centrosomes under the same conditions (Fig. 7A). The T8 construct was not included in this analysis, because of its unusually strong localization to both nucleus and cytoplasm. These observations suggest that the C-terminal extension of hCenexin1 is important for proper recruitment of Plk1 to the centrosomes.
To determine whether the C-terminal extension of hCenexin1 directly interacts with Plk1, we first investigated whether immunoprecipitation of endogenous hCenexin1 coprecipitates Plk1. Using cellular lysates prepared from nocodazole-treated HeLa cells (the condition that induces strong colocalization of hCenexin1 and Plk1 at the mitotic spindle poles), we found that hCenexin1, but not the control immunoglobulin G, coprecipitated endogenous Plk1 (Fig. 7B). In vitro binding studies showed that precipitation of bacterially expressed GST-hCenexin1 coprecipitated HA-Plk1 but not HA-Plk1ΔPB1 lacking the PB1 of the PBD (Fig. 7C), suggesting that an intact PBD is required for the hCenexin1-Plk1 interaction. In addition, the C-terminal extension (aa 647 to 805) of hCenexin1 (-C), which was isolated from the yeast two-hybrid screen, also interacted with Plk1. Neither the N-terminal (-N) nor the middle (-M) region of hCenexin1 interacted with Plk1 under the same conditions (Fig. 7D). In a second experiment, we found that wild-type PBD precipitated hCenexin1 from the mitotic HeLa lysates severalfold more efficiently than the corresponding phosphate pincer PBD(H538A, K540M) mutant (Fig. 7E). Taken together, these data support the hypothesis that the PBD of Plk1 interacts with the phosphorylated C-terminal extension of hCenexin1, although the kinase responsible for this phosphorylation event has yet to be identified.
hCenexin1 is required for proper localization of ninein and centrin.
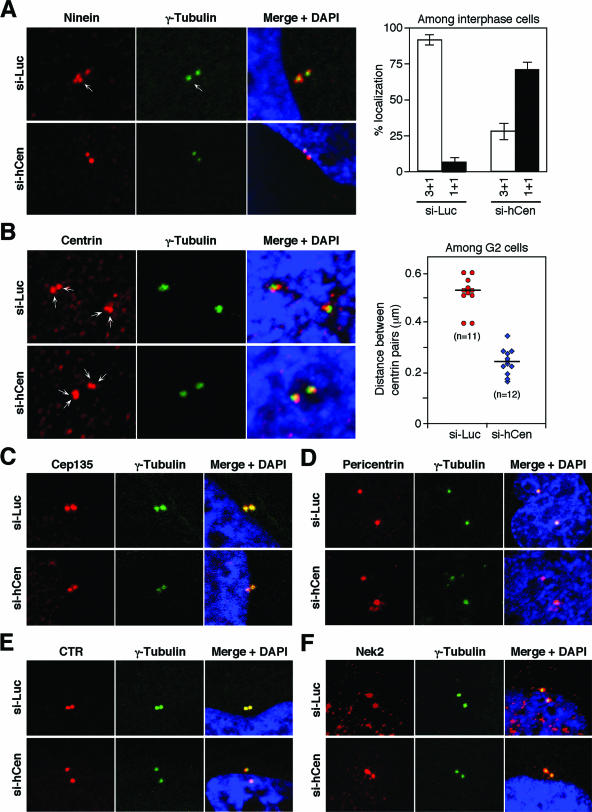
Odf2 had been suggested as a scaffold component of the centrosome matrix that is likely important for recruiting other centrosomal proteins (24). Thus, we investigated the effect of hCenexin1 knockdown on the localization of various centrosomal components. Ninein localizes to the appendages of mature centrioles during interphase, which are implicated in microtubule anchoring. In control cells, three ninein-positive dots and one ninein-positive dot were detected on each mother and daughter γ-tubulin-positive centriole, respectively. Similarly to a previous report (16), depletion of hCenexin1 by RNAi greatly diminished the fraction of cells with the normal 3 + 1 ninein configuration (three mother centrioles plus one daughter centriole) (Fig. 8A). Closely correlating with the efficiency of hCenexin1 depletion shown in Fig. 6B, ∼72% of the si-hCen cells exhibited the 1 + 1 configuration (one mother centriole plus one daughter centriole) as opposed to 29% of the si-Luc interphase cells. These results suggest that hCenexin1 is required for normal ninein localization to the mother centrioles.
FIG. 8.
Knockdown of hCenexin disrupts proper localization of ninein and centrin. HeLa cells transfected with either control si-Luc or si-hCen781 (si-hCen) for 3 days were subjected to immunostainings with the indicated antibodies. (A) Knockdown of hCenexin1 drastically altered the localization patterns of ninein. More than 200 cells were counted in each of two independent experiments. Error bars indicate standard deviations. (B) Knockdown of hCenexin1 diminishes the intercentriolar distance between the centrin pairs. Confocal images were acquired and analyzed as described in Materials and Methods. The resulting data points were plotted using the Sigma Plot 9 program. “n” denotes the number of cells analyzed. (C to F) Under the same conditions, the localization of Cep135 (C) was mildly weakened (see text), whereas localizations of pericentrin (D), CTR (E), and Nek2 (F) were largely unchanged.
We also examined the localization patterns of other centrosomal components. Centrin localizes to one or two pairs of dots within a given interphase centrosome and to one pair of dots at each pole of the spindle during mitosis (29). We found that control si-Luc cells exhibited an average intercentriolar distance of 0.53 μm between pairs of centrin signals, whereas si-hCen cells displayed a significantly diminished distance of 0.23 μm (Fig. 8B). These results suggest that hCenexin1 is required for the formation and/or maintenance of normal centrin assembly. In addition, depletion of hCenexin1 also diminished the Cep135 fluorescence to ∼77% of the level in the si-Luc cells (Fig. 8C and data not shown). However, under the same conditions, depletion of hCenexin1 did not appear to significantly influence localization of other centrosome-localizing proteins such as pericentrin, CTR, and Nek2 (Fig. 8D to F). Taken together, these observations suggest that hCenexin1 is required for the localization of a set of centrosomal proteins.
Knockdown of hCenexin induces severe mitotic defects and apoptosis.
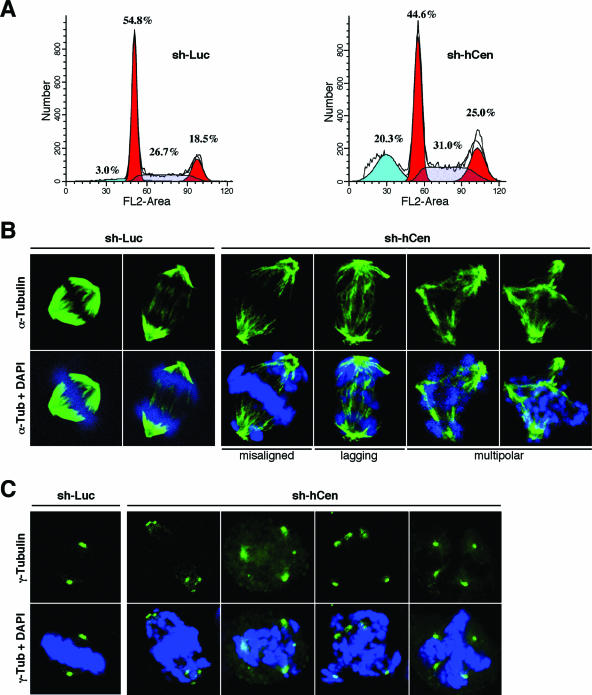
Since knockdown of hCenexin results in improper localization of a set of centrosomal proteins including Plk1, we examined whether depletion of hCenexin1 results in any defect during cell cycle progression. As a result of a low turnover rate of hCenexin1, silencing of hCenexin1 expression by the transfection of si-hCen did not permit us to achieve prolonged hCenexin1 depletion and to reliably examine the phenotype. Thus, to stably maintain effective hCenexin1 depletion for a longer period of time (5 to 7 days), we generated lentiviruses expressing either control sh-Luciferase (sh-Luc) or sh-hCenexin (sh-hCen; nucleotide sequence used for the lentivirus construct is identical to that for si-hCen781), infected HeLa cells, and then selected the sh-hCen population with puromycin before further examination. As expected, cells infected with sh-hCen-expressing viruses exhibited a significant level of hCenexin1 depletion in most of the population (hCenexin1 was depleted effectively 4 days after the sh-hCen virus infection, and the phenotype was manifest 5 to 7 days after infection). Flow cytometry analyses of these cells 6 days after infection showed that, in comparison to sh-Luc cells, sh-hCen cells exhibited significant increases in both G2/M (6.5%) and apoptotic (17.3%) populations (Fig. 9A). Viral infection itself did not cause a significant alteration in the cell cycle (data not shown). Close inspection of the mitotic cells revealed that the sh-hCen cells largely possessed much-weakened spindle structures (Fig. 9B). In addition, 31% of the sh-hCen cells displayed misaligned chromosomes, whereas 12% of them exhibited lagging chromosomes. Approximately 8% of them also exhibited multipolar spindles, as evidenced by the presence of multiple γ-tubulin signals within a single cell (Fig. 9C and D). Cells with misaligned chromosomes also frequently exhibited multiply clustered centrosomes (Fig. 9C). As expected if these mitotic defects were due to the lack of the hCenexin1 function, the defects were more pronounced in the cells with undetectable levels of hCenexin1 at the centrosomes (see Fig. S3 in the supplemental material). Under these conditions, the control sh-Luc cells exhibited only a low level of mitotic defects (Fig. 9D). Thus, depletion of the hCenexin1 population from the centrosomes results in improper bipolar spindle formation and chromosome missegregation that ultimately lead to apoptotic cell death.
FIG.9.
Depletion of hCenexin results in chromosome segregation failures and apoptosis. (A) HeLa cells were infected with lentiviruses expressing either control sh-luciferase (sh-Luc) or sh-hCen781 (sh-hCen) and then selected with puromycin for 5 days. Cells were harvested 6 days after infection for flow cytometry analyses. The obtained data were analyzed by using the ModFit program. (B to D) The same samples obtained in panel A were subjected to immunostaining analyses with anti-α-tubulin antibody (B) or anti-γ-tubulin antibody (C), and the defects in chromosome segregation were quantified (>250 cells) in each of three independent experiments (D). (E to G) HeLa cells were infected with lentiviruses expressing either control sh-Luc or sh-hCen. After the cells were selected for 5 days, they were additionally infected with lentiviruses expressing either hCenexin1-WT or the hCenexin1-sil mutant insensitive to sh-hCen. Samples were harvested as shown in panel E. Growth rates of either sh-Luc cells or sh-hCen cells were determined in the absence (Dox+) (F) or presence (Dox−) (G) of hCenexin1 expression. To determine the level of hCenexin1 expression, indicated samples were subjected to immunoblotting analyses with anti-hCenexin antibody. Asterisks indicate a nonspecific protein cross-reacting with anti-hCenexin antibody. (H) HeLa cells preparedas in panel E were subjected to immunostaining analyses to determine whether expression of hCenexin-sil complements the mitotic defect associated with the depletion of hCenexin1. Cells transfected with an hCenexin1-sil construct (last lane; hCen-sil transf'ed) were loaded to mark the position of hCenexin1 migrating in the gel. More than 250 cells were counted in each of three independent experiments. Error bars indicate standard deviations.
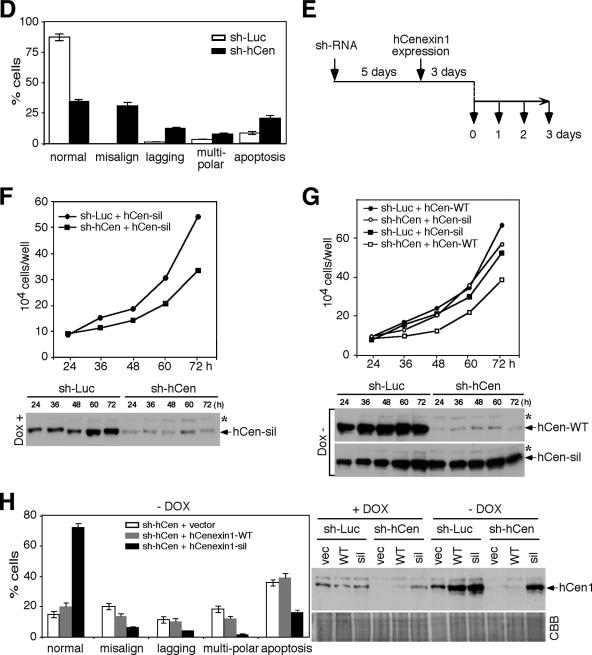
To investigate whether the observed mitotic defect can be rescued by the expression of hCenexin1, HeLa cells were first depleted of hCenexin1 by infecting the cells with the sh-hCen-expressing lentivirus. After puromycin selection, cells were verified for the hCenexin1 depletion and then additionally infected with lentiviruses expressing either full-length hCenexin1-WT or hCenexin1-sil (bearing the sh-hCen-insensitive silent mutations; see Materials and Methods) under doxycycline (Dox) control (Fig. 9E). As expected from the mitotic defect associated with the depletion of hCenexin1 (Fig. 9B to D), sh-hCen cells exhibited a significantly diminished growth rate in the absence of hCen-sil expression (Dox+) (Fig. 9F). Expression of the hCenexin1-sil mutant, but not the corresponding hCenexin1-WT, under Dox− conditions, rescued the sh-hCen growth defect (Fig. 9G). Consistent with these observations, immunostaining analyses revealed that sh-hCen cells expressing the hCenexin1-sil mutant dramatically increased the hCenexin1-positive population (data not shown) and greatly diminished both mitotic defects and apoptosis (Fig. 9H). Under the same conditions, either control vector or hCenexin1-WT did not yield detectable levels of hCenexin1 expression and, as a result, failed to rescue the defects associated with the hCenexin1 depletion (Fig. 9H). These data indicate that hCenexin1 is required for normal spindle formation and mitotic progression.
DISCUSSION
hCenexin1, but not hOdf2, is the major isoform expressed in HeLa cells.
Odf2 was originally identified as a major component of the sperm tail cytoskeleton and thought to be expressed exclusively in spermatogenic cells (4, 15, 30, 34). Later studies showed that a monoclonal antibody, mAb101, generated against purified chicken centrosomes specifically recognizes an ODF2-encoded protein from a λgt11 expression library and detects a ∼90-kDa protein in lysates prepared from cultured mammalian cells (24). Immunostaining analyses with mAb101 antibody showed that it decorates preferentially the distal/subdistal appendages of the mother centriole in G1 and the daughter centriole at the late stages of the cell cycle (16, 24). Based on these observations, Odf2 has been proposed to function as a centrosomal component in nonsperm cells. However, whether the centrosomal antigen immunoreactive to the anti-mAb101 antibody is Odf2 itself or an Odf2-related protein(s) has not been clarified. Interestingly, previous reports showed that ODF2 sequences from higher eukaryotic organisms encode proteins of a much smaller calculated size (610 aa and 638 aa for hOdf2/1 and hOdf2/2, respectively; 638 aa for both mOdf2 and rOdf2; and 659 aa for cOdf2) than the actual protein size detected with mAb101 or other anti-Odf2 antibodies (4, 15, 24, 30). In addition, although the cDNA sequence of this protein is yet to be isolated, Lange and Gull reported the detection of a 96-kDa mother centriole-specific protein named Cenexin using an antibody generated against a centriolar preparation from lamb thymus (18). These findings raise the possibility that the above 90- to 96-kDa protein detected in non-germ line cells could be an isoform(s) of Odf2 that is immunoreactive to mAb101 or other anti-Odf2 antibodies. In support of this argument, we demonstrated the existence of two ODF2-related hCENEXIN genes (hCENEXIN1 and the gene for its 19-aa insertion form hCENEXIN1 variant 1) in HeLa cells that encode proteins of 805 aa and 824 aa, respectively. Database analyses showed that hCenexin1 and hOdf2 are splicing variants of the same genomic locus and are identical at the nucleotide level except for both the N and C termini of hCenexin1. Interestingly, both hCenexin1 and hCenexin1 variant 1 possess a unique C-terminal extension (∼160 amino acids long) that is closely related to the analogous C-terminal extension found in rCenexin2. These observations suggest that the members of the Cenexin family constitute a distinct subfamily of hOdf2 isoforms. However, whether hOdf2 is still expressed in non-germ line cells has not been clearly determined. Several lines of evidence provided here indicate that hCenexin1, but not hOdf2, is the major isoform expressed in HeLa cells. First, anti-hCenexin antibody detected a 93-kDa protein that comigrated with the transfected full-length hCenexin1. Second, a C-terminal probe that specifically recognizes hCENEXIN1 mRNA, but not the hODF2 mRNA, detected a 4-kb transcript as efficiently as the N-terminal common region probe. Third, an si-RNA duplex directed against the C-terminal extension of hCenexin1, but not hOdf2, efficiently depleted both the ∼4-kb mRNA and the 93-kDa protein. Since the cell cycle-dependent subcellular localization of hCenexin1 closely correlates with that of the mAb101-immunoreactive antigen, we speculate that the mother centriole-specific 90-kDa protein detected in somatic cells by mAb101 is most likely hCenexin1.
hCenexin1 is required for proper localization of Plk1 to the centrosomes.
Plk1 localizes to the centrosomes as it is expressed during the late stages of the cell cycle, and this localization appears to be critical for Plk1's role in regulating microtubule nucleation (5, 6). However, how Plk1 localizes to the centrosomes remains elusive. Earlier studies have shown that Plk1 binds to centrosomal proteins such as Nlp and Cep170 (5, 13). However, these proteins do not localize to the mitotic centrosomes where Plk1 abundantly localizes. Moreover, depletion of Nlp or Cep170 does not appear to alter Plk1 localization to the centrosomes, suggesting that either these proteins are not critical for Plk1 localization to the centrosomes or they are not the only proteins regulating Plk1 localization. We observed that depletion of hCenexin1 greatly impaired Plk1 localization to the interphase centrosomes. This defect was rescued by the expression of the localization-competent hCenexin1 constructs bearing the C-terminal extension but not by the C-terminal deletion mutants. Furthermore, the C-terminal extension region was sufficient to bind to the PBD of Plk1 in vitro. These findings suggest that the C-terminal extension of hCenexin1 directly interacts with the PBD of Plk1 and that this step is important to recruit Plk1 to the centrosomes. Previous studies showed that the PBD of Plk1 interacts with a phosphorylated epitope that is frequently generated by Cdc2 or by Plk1 itself (5, 9, 21, 36). Whether these kinases are responsible for generating the phospho-epitope for the PBD binding remains to be further investigated.
It is noteworthy that a nascent hCenexin1 signal at the daughter centrosomes appears to be sufficient to recruit Plk1 to this location (Fig. 7A, an asterisk for GFP-T6), suggesting that a small fraction of hCenexin1 is sufficient to target Plk1 to the centrosomes. This opinion helps explain why Plk1 exhibits relatively even signals at both mother and daughter centrosomes, even though its interacting proteins such as hCenexin1, ninein, and Cep170 are all mother centriole oriented. It is also possible that hCenexin1 has a broader role as a scaffold to properly maintain the centrosome structure, thus enabling other Plk1-binding proteins to recruit Plk1 to the centrosomes. Interestingly, the C-terminal domain of hCenexin1 almost exclusively localizes to the mother centriole, whereas the N-terminal domain of hCenexin1 has a much looser preference for the mother centrioles than the full-length hCenexin1. This differential localization pattern could be achievable if different regions of hCenexin1 interact with different sets of proteins that are distinct between mother and daughter centrioles. Identification of additional hCenexin1-interacting proteins will likely be critical to better understand the nature of hCenexin1 localization.
Requirement of hCenexin1 for proper mitotic progression.
Depletion of hCenexin1 delocalizes centrosomal Plk1 and alters localization of other components important for normal microtubule nucleation and function. To determine the functional significance of the hCenexin1 during mitosis, we depleted hCenexin1 by lentivirus-based RNAi and selected the sh-hCen cells with puromycin before further examination. Since cells were maintained under selection, we were able to prevent growth in undepleted cells. As with the impaired Plk1 localization and diminished γ-tubulin recruitment to the centrosomes, the sh-hCen cells exhibited severe mitotic defects such as chromosome alignment and segregation. A significant fraction of these cells also showed multipolar chromosomes and apoptotic cell death. Ectopic expression of sh-hCen-insensitive hCenexin1-sil mutant, but not the hCenexin1-WT, effectively rescued these mitotic defects. These findings demonstrated that silencing of hCenexin1 ultimately leads to chromosome missegregation and apoptosis. However, whether these defects are solely attributable to the impaired Plk1 localization to the centrosomes remains to be further investigated. A recent report showed that, similarly to the hCenexin1-depleted cells as shown in Fig. 8, mouse F9 odf2−/− cells lack two of the three ninein dots on the mother centriole. In contrast to our findings, however, these cells did not exhibit any apparent defects in microtubule nucleation and cell cycle progression (16), even though ninein has been implicated for proper assembly of centrosomal proteins and also microtubule nucleation by docking the γ-tubulin ring complex (7, 28). How this discrepancy has arisen is not clear at present. One possibility is that the odf2−/− cells may have been adapted for the complete loss of ODF2 during selection, thus compromising the defects that the odf2−/− cells originally had.
Our data suggest that hCenexin is a unique form of Odf2 that plays an important role in targeting Plk1 to the centrosomes. The importance of the C-terminal extension of hCenexin1 in recruiting Plk1 predicts that Cenexin proteins, but not Odf2, are required for normal centrosome functions and mitotic processes. Whether Cenexin is ubiquitously expressed in all cell types for proper centrosome assembly and mitosis and whether the role of Odf2 is confined to spermatogenesis as originally thought remain to be further investigated. In this regard, it will be interesting to examine whether the expression of Cenexin proteins and Odf2 is differentially regulated among different cell types and tissues. Generation of Cenexin- and Odf2-specific antibodies and further studies in various experimental systems will be required to address these questions.
Supplementary Material
Acknowledgments
We are grateful to Susan Garfield for critical reading of the manuscript and assistance with confocal microscopy. We also thank Ki-Young Lee for sharing data prior to publication; Michael B. Yaffe for GST-PBD constructs; and Sheila A. Stewart, Phillip A. Sharp, and Irvin S. Y. Chen for lentivirus constructs.
This work was supported in part by National Cancer Institute intramural grants (K.S.L. and T.M.), Korean Ministry of Health and Welfare grant 02-PJ1-PG10-21304-0003 (S.R.K.), NIH grants R01 CA115884 and RO1CA 75638 (C.-Z.G.), and NIH grant NIH GM55735 (R.K.).
Footnotes
Published ahead of print on 11 September 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arnaud, L., J. Pines, and E. A. Nigg. 1998. GFP tagging reveals human Polo-like kinase 1 at the kinetochore/centromere region of mitotic chromosomes. Chromosoma 107:424-429. [DOI] [PubMed] [Google Scholar]
- 2.Barr, F. A., H. H. Sillje, and E. A. Nigg. 2004. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5:429-440. [DOI] [PubMed] [Google Scholar]
- 3.Beausoleil, S. A., M. Jedrychowski, D. Schwartz, J. E. Elias, J. Villen, J. Li, M. A. Cohn, L. C. Cantley, and S. P. Gygi. 2004. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA 101:12130-12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brohmann, H., S. Pinnecke, and S. Hoyer-Fender. 1997. Identification and characterization of new cDNAs encoding outer dense fiber proteins of rat sperm. J. Biol. Chem. 272:10327-10332. [DOI] [PubMed] [Google Scholar]
- 5.Casenghi, M., P. Meraldi, U. Weinhart, P. I. Duncan, R. Korner, and E. A. Nigg. 2003. Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev. Cell 5:113-125. [DOI] [PubMed] [Google Scholar]
- 6.de Cárcer, G., M. do Carmo Avides, M. J. Lallena, D. M. Glover, and C. González. 2001. Requirement of Hsp90 for centrosomal function reflects its regulation of Polo kinase stability. EMBO J. 20:2878-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delgehyr, N., J. Sillibourne, and M. Bornens. 2005. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J. Cell Sci. 118:1565-1575. [DOI] [PubMed] [Google Scholar]
- 8.Doxsey, S., W. Zimmerman, and K. Mikule. 2005. Centrosome control of the cell cycle. Trends Cell Biol. 15:303-311. [DOI] [PubMed] [Google Scholar]
- 9.Elia, A. E., P. Rellos, L. F. Haire, J. W. Chao, F. J. Ivins, K. Hoepker, D. Mohammad, L. C. Cantley, S. J. Smerdon, and M. B. Yaffe. 2003. The molecular basis for phospho-dependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 115:83-95. [DOI] [PubMed] [Google Scholar]
- 10.Fabbro, M., B. B. Zhou, M. Takahashi, B. Sarcevic, P. Lal, M. E. Graham, B. G. Gabrielli, P. J. Robinson, E. A. Nigg, Y. Ono, and K. K. Khanna. 2005. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev. Cell 9:477-488. [DOI] [PubMed] [Google Scholar]
- 11.Golsteyn, R. M., K. E. Mundt, A. M. Fry, and E. A. Nigg. 1995. Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J. Cell Biol. 129:1617-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guarguaglini, G., P. I. Duncan, Y. D. Stierhof, T. Holmstrom, S. Duensing, and E. A. Nigg. 2005. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol. Biol. Cell 16:1095-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoyer-Fender, S., C. Petersen, H. Brohmann, K. Rhee, and D. J. Wolgemuth. 1998. Mouse Odf2 cDNAs consist of evolutionary conserved as well as highly variable sequences and encode outer dense fiber proteins of the sperm tail. Mol. Reprod. Dev. 51:167-175. [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa, H., A. Kubo, S. Tsukita, and S. Tsukita. 2005. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 7:517-524. [DOI] [PubMed] [Google Scholar]
- 17.Lane, H. A., and E. A. Nigg. 1996. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 135:1701-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange, B. M., and K. Gull. 1995. A molecular marker for centriole maturation in the mammalian cell cycle. J. Cell Biol. 130:919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, K. S., and R. L. Erikson. 1997. Plk is a functional homolog of Saccharomyces cerevisiae Cdc5, and elevated Plk activity induces multiple septation structures. Mol. Cell. Biol. 17:3408-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, K. S., Y.-L. Yuan, R. Kuriyama, and R. L. Erikson. 1995. Plk is an M-phase-specific protein kinase and interacts with a kinesin-like protein, CHO1/MKLP-1. Mol. Cell. Biol. 15:7143-7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Litvak, V., R. Argov, N. Dahan, S. Ramachandran, R. Amarilio, A. Shainskaya, and S. Lev. 2004. Mitotic phosphorylation of the peripheral Golgi protein Nir2 by Cdk1 provides a docking mechanism for Plk1 and affects cytokinesis completion. Mol. Cell 14:319-330. [DOI] [PubMed] [Google Scholar]
- 22.Liu, X., T. Zhou, R. Kuriyama, and R. L. Erikson. 2004. Molecular interactions of Polo-like-kinase 1 with the mitotic kinesin-like protein CHO1/MKLP-1. J. Cell Sci. 117:3233-3246. [DOI] [PubMed] [Google Scholar]
- 23.Mogensen, M. M., A. Malik, M. Piel, V. Bouckson-Castaing, and M. Bornens. 2000. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J. Cell Sci. 113:3013-3023. [DOI] [PubMed] [Google Scholar]
- 24.Nakagawa, Y., Y. Yamane, T. Okanoue, S. Tsukita, and S. Tsukita. 2001. Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol. Biol. Cell 12:1687-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 26.Neef, R., C. Preisinger, J. Sutcliffe, R. Kopajtich, E. A. Nigg, T. U. Mayer, and F. A. Barr. 2003. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J. Cell Biol. 162:863-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohta, T., R. Essner, J. H. Ryu, R. E. Palazzo, Y. Uetake, and R. Kuriyama. 2002. Characterization of Cep135, a novel coiled-coil centrosomal protein involved in microtubule organization in mammalian cells. J. Cell Biol. 156:87-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ou, Y. Y., G. J. Mack, M. Zhang, and J. B. Rattner. 2002. CEP110 and ninein are located in a specific domain of the centrosome associated with centrosome maturation. J. Cell Sci. 115:1825-1835. [DOI] [PubMed] [Google Scholar]
- 29.Paoletti, A., M. Moudjou, M. Paintrand, J. L. Salisbury, and M. Bornens. 1996. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 109:3089-3102. [DOI] [PubMed] [Google Scholar]
- 30.Petersen, C., L. Fuzesi, and S. Hoyer-Fender. 1999. Outer dense fibre proteins from human sperm tail: molecular cloning and expression analyses of two cDNA transcripts encoding proteins of approximately 70 kDa. Mol. Hum. Reprod. 5:627-635. [DOI] [PubMed] [Google Scholar]
- 31.Rapley, J., J. E. Baxter, J. Blot, S. L. Wattam, M. Casenghi, P. Meraldi, E. A. Nigg, and A. M. Fry. 2005. Coordinate regulation of the mother centriole component Nlp by Nek2 and Plk1 protein kinases. Mol. Cell. Biol. 25:1309-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rieder, C. L., S. Faruki, and A. Khodjakov. 2001. The centrosome in vertebrates: more than a microtubule-organizing center. Trends Cell Biol. 11:413-419. [DOI] [PubMed] [Google Scholar]
- 33.Seong, Y. S., K. Kamijo, J. S. Lee, E. Fernandez, R. Kuriyama, T. Miki, and K. S. Lee. 2002. A spindle checkpoint arrest and a cytokinesis failure by the dominant-negative polo-box domain of Plk1 in U-2 OS cells. J. Biol. Chem. 277:32282-32293. [DOI] [PubMed] [Google Scholar]
- 34.Shao, X., H. A. Tarnasky, U. Schalles, R. Oko, and F. A. van der Hoorn. 1997. Interactional cloning of the 84-kDa major outer dense fiber protein Odf84. Leucine zippers mediate associations of Odf84 and Odf27. J. Biol. Chem. 272:6105-6113. [DOI] [PubMed] [Google Scholar]
- 35.van Vugt, M. A., and R. H. Medema. 2005. Getting in and out of mitosis with Polo-like kinase-1. Oncogene 24:2844-2859. [DOI] [PubMed] [Google Scholar]
- 36.Yuan, J., F. Eckerdt, J. Bereiter-Hahn, E. Kurunci-Csacsko, M. Kaufmann, and K. Strebhardt. 2002. Cooperative phosphorylation including the activity of polo-like kinase 1 regulates the subcellular localization of cyclin B1. Oncogene 21:8282-8292. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.