Abstract
Stem cells are maintained in an undifferentiated state by interacting with a microenvironment known as the “niche,” which is comprised of various secreted and membrane proteins. Our goal was to identify niche molecules participating in stem cell-stem cell and/or stem cell-supporting cell interactions. Here, we isolated genes encoding secreted and membrane proteins from purified male germ stem cells using a signal sequence trap approach. Among the genes identified, we focused on the junctional adhesion molecule 4 (JAM4), an immunoglobulin type cell adhesion molecule. JAM4 protein was actually localized to the plasma membrane in male germ cells. JAM4 expression was downregulated as cells differentiated in both germ cell and hematopoietic cell lineages. To analyze function in vivo, we generated JAM4-deficient mice. Histological analysis of testes from homozygous nulls did not show obvious abnormalities, nor did liver and kidney tissues, both of which strongly express JAM4. The numbers of hematopoietic stem cells in bone marrow were indistinguishable between wild-type and mutant mice, as was male germ cell development. These results suggest that JAM4 is expressed in stem cells and progenitor cells but that other cell adhesion molecules may substitute for JAM4 function in JAM4-deficient mice both in male germ cell and hematopoietic lineages.
The most robust and regenerative stem cells are defined by their ability to permanently reconstitute full tissues from a single cell. Stem cells mediate tissue formation, maintenance, and repair based on complex interactions of cell-autonomous and nonautonomous mechanisms. An example of nonautonomous regulation is the highly specialized microenvironment called the niche, which commonly regulates stem cells in several tissues (16, 20). The niche is an anatomically defined, specifically constituted environment for stem cells, and the observation that some factors expressed there are expressed in different tissues suggests that common mechanisms maintain stem cells (2, 10, 29). For example, studies of gonadal tissue from Drosophila melanogaster have identified ancillary niche cells, molecular pathways governing interactions between stem cells and the local environment, and cell-cell interactions mediated by Drosophila epithelial cadherin (31, 34). Likewise, mammalian hematopoietic stem cells interact with osteoblasts, and both of these cell types express neural cadherin, the orthologue of Drosophila epithelial cadherin (4, 36).
Here, we used a signal sequence trap approach to identify membrane proteins mediating cell-cell interactions in the niche of male germ stem cells (spermatogonia) and hematopoietic stem cells (HSCs). Among the genes identified was that encoding the junctional adhesion molecule 4 (JAM4), which can be upregulated by retinoic acid and was previously shown to localize at tight junctions (TJs) (12, 19). JAM4 is a member of a cortical thymocyte marker of the Xenopus (CTX) protein family, including JAM-A (17), JAM-B (27), JAM-C (1), ESAM (22), and CAR (5). We show that JAM4 protein is localized at the cell surface of spermatogonia and in the lineage-negative, Kit-positive, and Sca-1-positive (Kit+ Sca-1+ lineage− [KSL]) population of hematopoietic cells. To understand the functional role of JAM4, we generated JAM4-deficient mice. JAM4-deficient mice showed no obvious phenotype in either the germ line or in hematopoiesis, suggesting that other family members may substitute for JAM4 function in the male germ line and in hematopoiesis.
MATERIALS AND METHODS
Mice.
C57BL/6 and ICR mice were purchased from Japan SLC (Shizuoka, Japan). Animal care was in accordance with the guidlines of Keio University for animal and recombinant DNA experiments.
Isolation of JAM4.
The signal sequence trap was performed as described previously (15). Briefly, total RNA was extracted from postnatal 1.5 (P1.5) gonocytes purified from Oct-4/enhanced green fluorescent protein (EGFP) transgenic mice (23, 24, 35). cDNA was synthesized from total RNA using random hexamers, separated through a SizeSep 400 Spin Column (Pharmacia, Uppsala, Sweden), and ligated to BstX1 adapters (Invitrogen, Carlsbad, CA). After ligation of cDNA into the pMX-SST vector, DNA was electroporated into DH10B-competent cells using a Gene Pulser (Bio-Rad, Hercules, CA) to make a cDNA library for the signal sequence trap.
Retroviruses representing the cDNA library were produced using the packaging cell line BOSC23. BA/F3 cells, an interleukin-3-dependent murine pro-B-cell line, were infected with retroviruses. Genomic DNA isolated from factor-independent BA/F3 clones was subjected to PCR to rescue integrated cDNAs as described previously (15). Rescued fragments were subcloned into pGEM-T vectors and sequenced using the Taq DyeTerminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA) and an automated sequencer (310 Genetic Analyzer; Applied Biosystems).
Antibodies and flow cytometry.
Monoclonal antibodies (MAbs) used for cell staining were the following: c-Kit, 2B8; Sca-1, E13-161.7; CD4, L3T4; CD8, 53-6.72; B220, RA3-6B2; TER-119, Gr-1, RB6-8C5; CD34, RAM34; and Mac-1, M1/70. All purchased from Pharmingen (San Diego, CA). Mouse hematopoietic stem cells were purified from bone marrow cells of 3-month-old mice. In brief, bone marrow mononuclear cells were flushed from femurs and tibias. Low-density cells were isolated from the flushed cells on Lymphoprep (1.086 g/ml; AXIS-SHIELD, Kimbolton, United Kingdom). After being washed, cells were incubated with anti-mouse CD16/CD32(FcγIII/II receptor) MAb (2.4G2) (eBioscience, San Diego, CA) at 4°C for 30 min. Cells were then simultaneously incubated with perCP-Cy5.5-conjugated lineage marker antibodies against CD4, CD8 B220, TER-119, and Gr-1 as well as phycoerythrin-conjugated anti-CD34 MAb, allophycocyanin-conjugated anti-c-Kit MAb, and fluorescein isothiocyanate-conjugated anti-Sca-1 MAb for 30 min at 4°C. Antibodies were used at 0.2 μg/1 × 106 cells. After being washed, samples were analyzed by a FACSCalibur (Becton Dickinson, San Jose, CA).
RT-PCR.
Gonocytes and spermatogonia were purified from Oct-4/EGFP transgenic mice after staining with allophycocyanin-conjugated anti-c-Kit MAb (clone 2B8). HSCs were isolated with the indicated markers by a FACS Vantage (Becton Dickinson, San Jose, CA) as described previously (14). Total RNA was purified using an RNeasy mini kit (QIAGEN, Valencia, CA). Reverse transcription-PCR (RT-PCR) was preformed using an Advantage RT-for-PCR kit (Becton Dickinson, San Jose, CA) according to instructions. JAM4 primers were 5′-TAACCAAATGGTGGTGCTGA-3′ and 5′-CCACCATGACAGACACTTGG-3′. Glyceraldehyde 3-phosphate dehydrogenase (G3PDH) primers were 5′-GGAAAGCTGTGGCGTGATG-3′ and 5′-CTGTTGCTGTAGCCGTATTC-3′. PCR was performed under the following conditions: 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s.
Northern blot hybridization.
Northern blot analysis was performed using Multiple-Tissue Northern blots (mouse) purchased from Clontech (Mountain View, CA). RNA was hybridized with a 32P-radiolabeled probe derived from the full-length JAM4 coding sequence. To check for JAM4 expression in mutants, total liver RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA). Seven micrograms of total RNA was separated on 1.0% agarose, 2.4% formaldehyde gels and transferred to Hybond-N+ nylon membranes (GE Healthcare, Piscataway, NJ) in 10× SSC overnight (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Blots were hybridized with a 32P-radiolabeled probe derived from the entire coding sequence of JAM4 or G3PDH.
Generation of JAM4−/− mice.
A 10-kb mouse genomic DNA EcoRI fragment from a bacterial artificial chromosome containing the JAM4 exon 1 was cloned into pBluescriptII KS+ (Stratagene, La Jolla, CA). To generate the targeting vector, a loxP site was inserted into a BamHI site in the 5′-untranslated region. The PGK-neo cassette flanked by loxP sites was inserted into a KpnI site downstream of exon 1 (see Fig. 3a). The vector was linearized with NotI and electroporated into TT2 mouse embryonic stem (ES) cells, and cells were selected in 250 μg/ml (active weight) neomycin (G418) (Invitrogen, Carlsbad, CA) for 10 days. Neomycin-resistant colonies were screened for homologous recombination by PCR with the 5′ primer 5′-GGTCCCTGCTCCCTGAAACAATAT-3′ and the 3′ primer 5′-CTCCAGACTGCCTTGGGAAAAGTA-3′. PCR was performed under the following conditions: 40 cycles of 94°C for 30 s, 60°C for 1 min, and 72°C for 3 min. Positive colonies were propagated and confirmed by Southern blot analysis. Heterozygously targeted clones were transfected with a vector expressing Cre recombinase and the puromycin resistance gene (Cre-pac) (33) and selected in puromycin (Sigma, St. Louis, MO) (0.5 μg/ml) for 78 h. Genomic DNA from individual puromycin-resistant colonies was digested and checked for deletion of PGK-neo and exon 1. ES cells were aggregated with ICR mouse morulae to generate chimeric mice. Male chimeras were mated with C57BL/6 females to yield heterozygotes, and heterozygotes were bred to produce homozygotes. Genotypes were determined by EcoRI-digested or XbaI-digested tail genomic DNA using probe A or probe B, respectively, as shown in Fig. 3. Genotypes of knockout mice were determined by genomic PCR using three primers: a forward primer for both wild-type and targeted alleles (5′-ATACTCTGAGAGAGCACTGCTCTG-3′), a reverse primer for the wild-type allele (5′-CAATGGCAGGAATGAGAGCTACTG-3′), and a reverse primer for the targeted allele (5′-CACACCTTTTCCAGACCATCCTGTT-3′). Genomic PCR was performed under the following conditions: 25 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s.
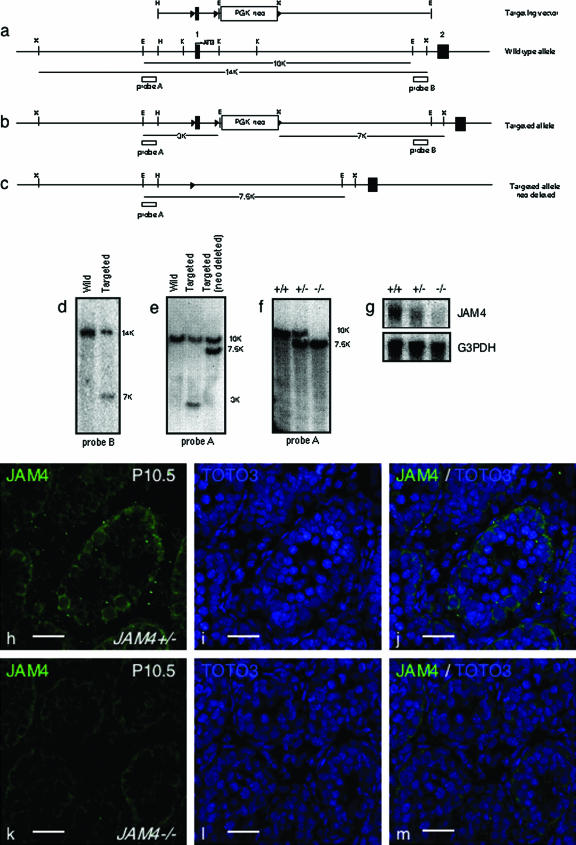
FIG. 3.
Targeted disruption of mouse JAM4 gene. (a) Structure of the targeting vector and JAM4 gene locus. Filled rectangles represent exons, and open triangles represent loxP sites. Probe A and probe B used in Southern blot analysis are shown as boxes. E, EcoRI; X, XbaI; H, HindIII; and K, KpnI; these are restriction sites. (b) Targeted allele before Cre-mediated recombination of loxP sites. (c) Targeted allele after Cre-mediated recombination resulting in deletion of the gene fragment containing JAM4 exon 1 and the PGK-neo cassette. (d) Southern blot analysis of wild-type ES cells and the targeted clone. We obtained seven clones that showed successful recombination at this step. The representative data (clone no. 190f) is shown. The 14-kb wild-type allele and the 7-kb targeted allele XbaI fragments identified by external probe B are shown. (e) Southern blot analysis of the wild-type allele, the targeted allele before Cre-mediated recombination, and the targeted allele after Cre-mediated recombination. We chose two individual clones to treat with Cre. After the Cre treatment, we obtained two clones from each parent clone that was successfully deleted of exon 1 and a part of intron 1. The representative data (clone no. 190f20) are shown. The DNA samples were digested with EcoRI. The 10-kb wild-type and the 3-kb targeted allele before Cre-mediated recombination as well as the 7.5-kb targeted allele after Cre-mediated recombination identified by external probe A are shown. (f) JAM4-deficient mice from clone 190f20d were used for further analysis. Southern blot analysis of tail genomic DNA from wild-type (+/+), heterozygous (+/−), and homozygous (−/−) mice using probe A. (g) Northern blot analysis of mice of the indicated genotypes. JAM4 expression was completely abolished in JAM4−/− mice. (h to m) Immunohistological analysis of JAM4 in P10.5 testis from JAM4 heterozygotes (+/−) and homozygotes (−/−). JAM4 (h and k), TOTO3 (i and l), and merged images (j and m) are shown. Scale bars, 25 μm (h to m).
Histological and immunohistological analysis.
For histology, testis, kidney, and liver tissues were fixed with Bouin's fixative overnight, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (HE). For immunohistological analysis, testes were perfused with PLP fixative (0.01 M NaIO4, 2% paraformaldehyde in 0.075 M phosphate buffer) at 4°C for 1 h. After being washed, testes were sequentially immersed in 10%, 20%, and 30% (wt/vol) sucrose in phosphate-buffered saline (PBS) and embedded in Tissue-Tek O.C.T. compound (Sakura Finetechnical, Tokyo, Japan). Sectioned specimens were used for immunohistochemistry. Briefly, specimens were blocked with 1% bovine serum albumin in PBS for 1 h and incubated with polyclonal rabbit anti-mouse JAM4 antibodies at 4°C overnight (1:4,000) (12). After the specimens were washed three times with PBS, antibodies were detected with the Alexa 488-conjugated goat anti-rabbit immunoglobulin G (IgG) (A-21441; Invitrogen, Carlsbad, CA) at room temperature for 2 h (1:200). Images were obtained with a confocal microscope (Olympus FV1000; Olympus, Tokyo, Japan).
Blood counts.
Blood samples were obtained by retro-orbital sinus puncture, treated with heparin, and analyzed using an automated system (Celltac α; Nihon Koden, Tokyo, Japan).
RESULTS
Isolation of JAM4.
Green fluorescent protein (GFP)-positive gonocytes from P1.5 newborn testes were purified from Oct-4/GFP transgenic mice as described previously (24, 35). A cDNA library generated from purified gonocytes was constructed and used for the signal sequence trap method developed by Kojima and Kitamura (15). cDNA was ligated to the 5′ end of the coding sequence of the retroviral vector pMX-SST, which lacks the original signal sequence. Retroviruses harboring the library were produced and used to infect BA/F3 cells, an interleukin-3-dependent murine pro-B-cell line. Cells transduced with constitutively active granulocyte-macrophage colony-stimulating factor receptor with a signal sequence became factor independent and were analyzed by PCR. Among the rescued fragments were known membrane proteins such as the mannose-6-phosphate receptor, minopontin, and complement 1qb. As a second screen, we chose genes expressed in hematopoietic stem cells (HSCs). One of the candidates identified in stem cell and progenitor cell populations of both male germ cell and hematopoietic cell lineages was the gene encoding the cell adhesion protein JAM4. JAM4 protein shares structural features with CTX family molecules, such as two Ig-like domains, a single transmembrane domain, and a cytoplasmic tail exhibiting a canonical PDZ domain binding motif (7).
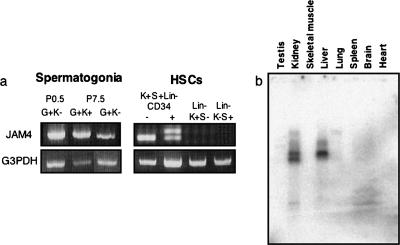
Next, we confirmed that JAM4 is expressed in spermatogonia and HSCs (Fig. 1a). We previously reported that in P7.5 spermatogonia, Kit expression can be used to differentiate stem cell activity in an Oct-4-positive progenitor population: spermatogonia showing strong stem cell activity are concentrated in an Oct-4-positive/Kit-negative population (23). In spermatogonia, JAM4 was expressed in both Oct-4-positive/Kit-negative and Oct-4-positive/Kit-positive populations (Fig. 1a). In HSCs, stem cells are highly purified in a KSL (Kit+, Sca-1+, and lineage marker−)/CD34-negative fraction rather than in the KSL/CD34-positive fraction (25). Neither K+ S− L− nor K− S+ L− populations in bone marrow show stem cell activities like that of lineage-positive cells (26). PCR analysis showed that JAM4 was expressed in KSL/CD34-positive and -negative populations but not in K+ S− L− or K− S+ L− cells in bone marrow. Therefore, we concluded that JAM4 is expressed on both stem and progenitor cells in both testis and the bone marrow (Fig. 1a). Northern blot analysis showed that JAM4 is also expressed in adult kidney and liver (Fig. 1b).
FIG. 1.
Expression of JAM4 mRNA in neonatal testes and bone marrow. GFP-positive gonocytes and spermatogonia were purified from P0.5 and P7.5 testes, respectively, from Oct-4/EGFP transgenic mice. All P0.5 gonocytes are Kit negative. P7.5 spermatogonia exhibit both Kit-negative and Kit-positive populations. Each population was sorted, and total RNA was isolated. (a) RT-PCR analysis of JAM4 in spermatogonia (left) and HSCs (right). G, Oct-4/EGFP; K, Kit; S, Sca-1; Lin, lineage markers. (b) Northern blot analysis of JAM4 in adult tissues. JAM4 is expressed in adult kidney and liver tissue.
Immunohistological analysis of JAM4 in testis.
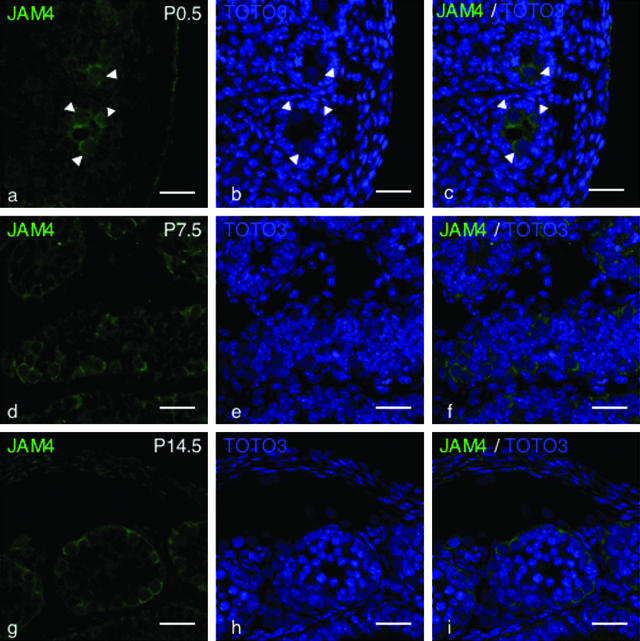
To analyze expression and localization of JAM4 in neonate testis, immunohistological analysis using an anti-JAM4 antibody was performed. At P0.5, JAM4 was expressed on gonocytes (Fig. 2a to c). Expression in germ cell membranes was observed in P7.5 and P14.5 testes. JAM4 protein was not highly concentrated at cell-cell borders, different from Occludin, a typical tight-junction (TJ) protein (21). JAM4 protein was equally distributed and did not show polarity on the surface of spermatogonia at P7.5 and P14.5. In addition, Sertoli cells were weakly positive for JAM4 at P0.5, P7.5, and P14.5 (Fig. 2a to c). The expression of JAM4 in gonocytes, spermatogonia, and Sertoli cells was also confirmed by in situ hybridization analysis (data not shown).
FIG. 2.
Immunohistological analysis of JAM4 in developing testes. Sections of testis at the indicated periods were stained with anti-JAM4 antibody and TOTO3. Bar, 25 μm. JAM4 (a, d, and g), TOTO3 (b, e, and h), and merged images (c, f, and i) of testis sections at the indicated days are shown (P0.5, P7.5, and P14.5). JAM4 is expressed on gonocytes localized at the center of testis cords (a) (arrowheads). Spermatogonia attached to the basement membrane in both P7.5 (d) and P14.5 (g) testes express JAM4.
Targeted disruption of JAM4.
To analyze JAM4 function in vivo, we generated JAM4-deficient mice by using a conditional strategy (Fig. 3a and b; see Materials and Methods) in which Cre activity deleted exon 1 and part of intron 1 (Fig. 3b and c). Southern blot hybridization analysis of ES cells showed successful homologous recombination (Fig. 3d) and deletion (Fig. 3e), and correctly targeted ES cell clones transmitted the mutant allele through the germ line of aggregation chimeras. Heterozygous (JAM4+/−) F1 mice were viable and fertile. F1 heterozygous intercrosses produced viable offspring with a Mendelian distribution of each genotype (Table 1). JAM4−/− mice were indistinguishable from littermate controls in terms of growth and development. Proper targeting was confirmed by Southern and Northern blotting methods (Fig. 3f and g). Immunohistological analysis of JAM4−/− testis sections showed a decrease in JAM4 positivity (Fig. 3h to j) compared to JAM4 with our without testis sections (Fig. 3k to m). JAM4 positivity was weakly observed in the testis of JAM4−/− mice, indicating either cross-reactivity of the polyclonal antibody with related proteins or incomplete disruption of JAM4. Since Northern blot analysis showed no evidence of JAM4 expression, we favor the former possibility.
TABLE 1.
Summary of genotyping from intercrossing of F1 mice
| Gender | Wild type (+/+) | Heterozygote (+/−) | Homozygote (−/−) | Total |
|---|---|---|---|---|
| Male | 15 | 53 | 23 | 91 |
| Female | 16 | 35 | 21 | 72 |
| Total | 31 | 88 | 44 | 163 |
Normal differentiation of JAM4-deficient testicular cells.
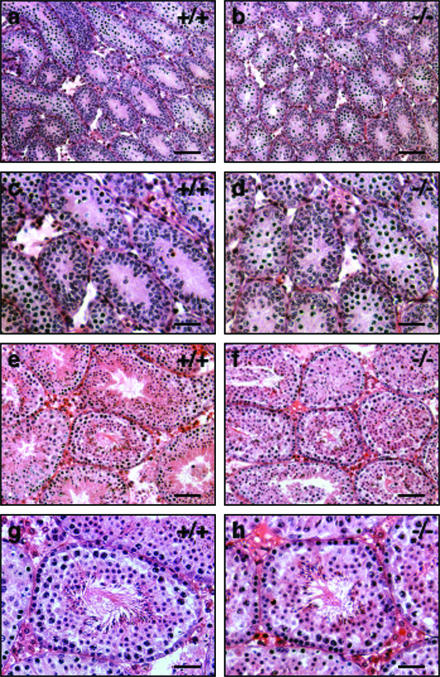
We investigated male germ cell development and differentiation in JAM4−/− mice at postnatal stages. Testis tissue from P10.5 mice was sectioned, and the sections were stained with HE. Tissues from JAM4−/− mice showed normal morphology (Fig. 4a and b). All seminiferous tubules contained spermatogenic cells in both JAM4+/+ and JAM4−/− mice. In adult stages (12 weeks), the time of spermatogenesis reported to cycle the complete differentiation progression at each tubule, there was no obvious difference between mutants and wild-type mice in appearance of testicular cells (Fig. 4c and d). These results indicate that JAM4 deficiency does not grossly affect male germ cell development.
FIG. 4.
Histological analysis of neonatal testis and adult testis. HE-stained sections of JAM4+/+ (a, c, e, and g) and JAM4−/− (b, d, f, and h) testes at P10.5 (a to d) as well as at 12 weeks of age (e to h) are shown. Panels c, g, d, and h show high-magnification images of panels a, e, b, and f, respectively. Scale bars are 100 μm (a, b, e, and f) and 50 μm (c, d, g, and h).
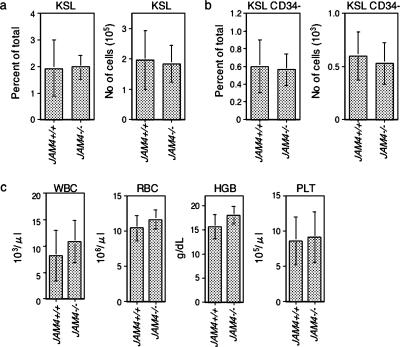
JAM4−/− mice exhibit normal numbers of HSCs.
JAM4 was specifically expressed in both CD34-negative and -positive HSCs in bone marrow mononuclear cells. Therefore, we asked whether the proportions of these types of HSCs were altered in JAM4−/− mice. Bone marrow mononuclear cells were isolated and stained for cell surface markers. JAM4−/− mice showed similar proportions and numbers of KSL cells, which include both short- and long-term repopulating cells (Fig. 5a). JAM4−/− mice also have equivalent populations of CD34-negative HSCs compared to wild-type mice (Fig. 5b). In addition, blood counts were also similar in mutant and wild-type mice (Fig. 5c). The hemoglobin concentration was indistinguishable between the wild-type and knockout mice, as were counts of white blood cells, red blood cells, and platelets. These results indicate that JAM4−/− HSCs likely have the potential to produce daughter cells with the proper number and function of peripheral blood components.
FIG. 5.
Hematopoietic cell populations and numbers in JAM4−/− mice. (a and b) The percentage (left graphs) and numbers (right graphs) of Kit+ Sca-1+ lineage− (KSL) (a) and KSL/CD34− (b) bone marrow mononuclear cells of JAM4+/+ mice and JAM4−/− mice are shown. (c) Peripheral blood components of wild-type and JAM4−/− mice. WBC, white blood cell; RBC, red blood cell; HGB, hemoglobin; PLT, platelet.
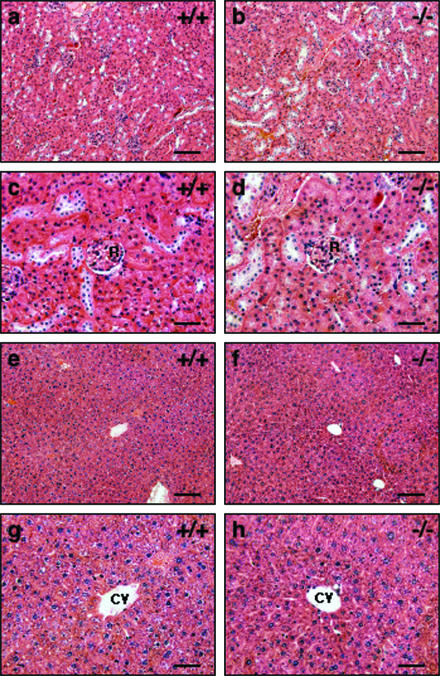
Histological analysis of adult kidney and liver in JAM4−/− mice.
As noted, JAM4 was expressed in adult kidney and liver. The function of JAM4 expression in liver is not known; however, in kidneys JAM4 reportedly functions as a tight-junction protein with MAGI-1 in podocytes and colocalizes with ZO-1 to constitute the podocyte slit membrane (12). When we examined tissue sections of adult kidney and liver for defects, sections of both tissues appeared normal compared with wild-type littermates on the light microscopic level (Fig. 6).
FIG. 6.
Histological analysis of kidney and liver of JAM4−/− mice. HE-stained sections of kidney (a to d) and liver (e to h) from JAM4+/+ (a, c, e, and g) and JAM4−/− (b, d, f, and h) mice at 12 weeks of age are shown. Panels c, g, d, and h are high-magnification images of panels a, e, b, and f, respectively. Scale bars are 100 μm (a, b, e, and f) and 50 μm (c, d, g, and h). R, renal glomerulus. CV, central vein.
DISCUSSION
Interaction with surrounding cells mediated by adhesion molecules is a common mechanism to sustain stem cells over numerous tissues and species (2, 20). Here we identified factors involved in interactions between stem cells and themselves or with supporting cells. Using a signal sequence trap strategy, we identified JAM4 from a cDNA library made from P1.5 gonocytes as a cell surface molecule expressed on both spermatogonia and HSCs.
Previously, JAM4 was identified as a novel MAGI-1 binding protein (11). MAGI-1 is a membrane-associated guanylate kinase (MAGUK) protein located at tight junctions in epithelial cells reported to interact with various molecules and to function as a scaffold protein at cell junctions (6, 13). Therefore, we predict that JAM4 provides adhesion machinery at cell junctions together with MAGI-1. In testis, TJs between Sertoli cells, which form at least around 2 weeks after birth (3), are called the blood-testis barrier. JAM4 protein did not colocalize with ZO-1 protein in the testis, suggesting that JAM4 functions as a cell adhesion molecule rather than a tight-junction protein in the testis at this stage. Therefore, JAM might participate in homophilic cell adhesion between spermatogonia-spermatogonia, spermatogonia-Sertoli cells, and Sertoli cells-Sertoli cells. In spermatogonia, JAM4 is expressed in both Kit-positive and -negative populations, but it is not detected in testicular cells at the meiotic phase. Kit-negative and Kit-positive spermatogonia are known as self-renewing stem cells and progenitor cells, respectively (23, 30). In bone marrow, JAM4 is specifically expressed in both CD34-negative and -positive populations of KSL cells, which correspond to stem cells and progenitor cells, respectively (25). In our study we found that JAM4−/− mice, which were born from heterozygote crosses with the expected Mendelian ratio, are viable and grow normally in the specific-pathogen-free condition. JAM4−/− mice also exhibited no abnormalities in hematopoiesis, suggesting that either JAM4 is not critical for normal HSC development and maintenance or that a functionally redundant molecule(s) compensates for its function in the knockout mouse. JAM4 is more closely related to ESAM and CAR than to other JAMs. The former conserve the type II PDZ domain binding motif, whereas the other JAMs conserve a type I motif. This difference may account for variations in factors interacting with the cytoplasmic domain (8). In the CTX family, some members are predicted to function in testis, namely, BT-IgSF (brain- and testis-specific immunoglobulin superfamily), CAR (coxsackie and adenovirus receptor), and JAM-C. BT-IgSF is preferentially expressed in brain and testis (32), while CAR is present in spermatids and spermatozoa (18). JAM-C mainly localizes in spermatids. Targeted disruption of JAM-C revealed a crucial role for this protein in spermatogenesis: JAM-C mutants were infertile due to lack of mature spermatozoa (9). Therefore, the biological function of JAM-4 might be masked by that of a JAM4-related molecule(s) at the spermatogonial level.
Similarly, we saw no obvious abnormalities in adult kidney and liver tissues of JAM4−/− mice, indicating that JAM4 is dispensable for normal kidney and liver development.
A recent report shows that the expression pattern of JAM4 in podocytes changes in proteinuric rat models (11), indicating that JAM4 may function in pathological conditions.
Acknowledgments
We thank Yuri Shimoada for excellent technical assistance.
This work was partly supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Science, and Technology of Japan.
Footnotes
Published ahead of print on 18 September 2006.
REFERENCES
- 1.Aurrand-Lions, M., L. Duncan, C. Ballestrem, and B. A. Imhof. 2001. JAM-2, a novel immunoglobulin superfamily molecule, expressed by endothelial and lymphatic cells. J. Biol. Chem. 276:2733-2741. [DOI] [PubMed] [Google Scholar]
- 2.Brakebusch, C., and R. Fassler. 2005. Beta 1 integrin function in vivo: adhesion, migration and more. Cancer Metastasis Rev. 24:403-411. [DOI] [PubMed] [Google Scholar]
- 3.Byers, S. G. R., H. N. Dai, and B. Hoxter. 1991. Development of Sertoli cell junctional specializations and the distribution of the tight-junction-associated protein ZO-1 in the mouse testis. Am. J. Anat. 191:35-47. [DOI] [PubMed] [Google Scholar]
- 4.Calvi, L. M., G. B. Adams, K. W. Weibrecht, J. M. Weber, D. P. Olson, M. C. Knight, R. P. Martin, E. Schipani, P. Divieti, F. R. Bringhurst, L. A. Milner, H. M. Kronenberg, and D. T. Scadden. 2003. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425:841-846. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, C. J., J. T. Shieh, R. J. Pickles, T. Okegawa, J. T. Hsieh, and J. M. Bergelson. 2001. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc. Natl. Acad. Sci. USA 98:15191-15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobrosotskaya, I., R. K. Guy, and G. L. James. 1997. MAGI-1, a membrane-associated guanylate kinase with a unique arrangement of protein-protein interaction domains. J. Biol. Chem. 272:31589-31597. [DOI] [PubMed] [Google Scholar]
- 7.Du Pasquier, L., I. Zucchetti, and R. De Santis. 2004. Immunoglobulin superfamily receptors in protochordates: before RAG time. Immunol. Rev. 198:233-248. [DOI] [PubMed] [Google Scholar]
- 8.Ebnet, K., A. Suzuki, S. Ohno, and D. Vestweber. 2004. Junctional adhesion molecules (JAMs): more molecules with dual functions? J. Cell Sci. 117:19-29. [DOI] [PubMed] [Google Scholar]
- 9.Gliki, G., K. Ebnet, M. Aurrand-Lions, B. A. Imhof, and R. H. Adams. 2004. Spermatid differentiation requires the assembly of a cell polarity complex downstream of junctional adhesion molecule-C. Nature 431:320-324. [DOI] [PubMed] [Google Scholar]
- 10.Hall, P. E., J. D. Lathia, N. G. Miller, M. A. Caldwell, and C. French-Constant. 2006. Integrins are markers of human neural stem cells. Stem Cells 24:2078-2084. [DOI] [PubMed]
- 11.Harita, Y., N. Miyauchi, T. Karasawa, K. Suzuki, G. D. Han, H. Koike, T. Igarashi, F. Shimizu, and H. Kawachi. 2006. Altered expression of junctional adhesion molecule 4 in injured podocytes. Am. J. Physiol. Renal Physiol. 290:F335-F344. [DOI] [PubMed] [Google Scholar]
- 12.Hirabayashi, S., M. Tajima, I. Yao, W. Nishimura, H. Mori, and Y. Hata. 2003. JAM4, a junctional cell adhesion molecule interacting with a tight junction protein, MAGI-1. Mol. Cell. Biol. 23:4267-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ide, N., Y. Hata, H. Nishioka, K. Hirao, I. Yao, M. Deguchi, A. Mizoguchi, H. Nishimori, T. Tokino, Y. Nakamura, and Y. Takai. 1999. Localization of membrane-associated guanylate kinase (MAGI)-1/BAI-associated protein (BAP) 1 at tight junctions of epithelial cells. Oncogene 18:7810-7815. [DOI] [PubMed] [Google Scholar]
- 14.Iwama, A., O. H. Negishi, M. Kato, Y. Morita, Y. Tsukui, H. Ema, H. Kamijo, T. Katoh-Fukui, Y. Koseki, H. van Lohuizen, and M. Nakauchi. 2004. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity 21:843-851. [DOI] [PubMed] [Google Scholar]
- 15.Kojima, T., and T. Kitamura. 1999. A signal sequence trap based on a constitutively active cytokine receptor. Nat. Biotechnol. 17:487-490. [DOI] [PubMed] [Google Scholar]
- 16.Li, L., and T. Xie. 2005. Stem cell niche: structure and function. Annu. Rev. Cell Dev. Biol. 21:605-631. [DOI] [PubMed] [Google Scholar]
- 17.Martin-Padura, I., S. Lostaglio, M. Schneemann, L. Williams, M. Romano, P. Fruscella, C. Panzeri, A. Stoppacciaro, L. Ruco, A. Villa, D. Simmons, and E. Dejana. 1998. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 142:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mirza, M., J. Hreinsson, M. L. Strand, O. Hovatta, O. Soder, L. Philipson, R. F. Pettersson, and K. Sollerbrant. 2006. Coxsackievirus and adenovirus receptor (CAR) is expressed in male germ cells and forms a complex with the differentiation factor JAM-C in mouse testis. Exp. Cell Res. 312:817-830. [DOI] [PubMed] [Google Scholar]
- 19.Moog-Lutz, C., F. Cave-Riant, F. C. Guibal, M. A. Breau, Y. Di Gioia, P. O. Couraud, Y. E. Cayre, S. Bourdoulous, and P. G. Lutz. 2003. JAML, a novel protein with characteristics of a junctional adhesion molecule, is induced during differentiation of myeloid leukemia cells. Blood 102:3371-3378. [DOI] [PubMed] [Google Scholar]
- 20.Moore, K. A., and I. R. Lemischka. 2006. Stem cells and their niches. Science 311:1880-1885. [DOI] [PubMed] [Google Scholar]
- 21.Moroi, S., M. Saitou, K. Fujimoto, A. Sakakibara, M. Furuse, O. Yoshida, and S. Tsukita. 1998. Occludin is concentrated at tight junctions of mouse/rat but not human/guinea pig Sertoli cells in testes. Am. J. Physiol. 274:C1708-C1717. [DOI] [PubMed] [Google Scholar]
- 22.Nasdala, I., K. Wolburg-Buchholz, H. Wolburg, A. Kuhn, K. Ebnet, G. Brachtendorf, U. Samulowitz, B. Kuster, B. Engelhardt, D. Vestweber, and S. Butz. 2002. A transmembrane tight junction protein selectively expressed on endothelial cells and platelets. J. Biol. Chem. 277:16294-16303. [DOI] [PubMed] [Google Scholar]
- 23.Ohbo, K., S. Yoshida, M. Ohmura, O. Ohneda, T. Ogawa, H. Tsuchiya, T. Kuwana, J. Kehler, K. Abe, H. R. Scholer, and T. Suda. 2003. Identification and characterization of stem cells in prepubertal spermatogenesis in mice small star, filled. Dev. Biol. 258:209-225. [DOI] [PubMed] [Google Scholar]
- 24.Ohmura, M., S. Yoshida, Y. Ide, G. Nagamatsu, T. Suda, and K. Ohbo. 2004. Spatial analysis of germ stem cell development in Oct-4/EGFP transgenic mice. Arch. Histol. Cytol. 67:285-296. [DOI] [PubMed] [Google Scholar]
- 25.Osawa, M., K. Hanada, H. Hamada, and H. Nakauchi. 1996. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science 273:242-245. [DOI] [PubMed] [Google Scholar]
- 26.Osawa, M., K. Nakamura, N. Nishi, N. Takahasi, Y. Tokuomoto, H. Inoue, and H. Nakauchi. 1996. In vivo self-renewal of c-Kit+ Sca-1+ Lin(low/-) hemopoietic stem cells. J. Immunol. 156:3207-3214. [PubMed] [Google Scholar]
- 27.Palmeri, D., A. van Zante, C. C. Huang, S. Hemmerich, and S. D. Rosen. 2000. Vascular endothelial junction-associated molecule, a novel member of the immunoglobulin superfamily, is localized to intercellular boundaries of endothelial cells. J. Biol. Chem. 275:19139-19145. [DOI] [PubMed] [Google Scholar]
- 28.Reference deleted.
- 29.Shinohara, T., M. R. Avarbock, and R. L. Brinster. 1999. Beta1- and alpha6-integrin are surface markers on mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA 96:5504-5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shinohara, T., K. E. Orwig, M. R. Avarbock, and R. L. Brinster. 2000. Spermatogonial stem cell enrichment by multiparameter selection of mouse testis cells. Proc. Natl. Acad. Sci. USA 97:8346-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song, X., C. H. Zhu, C. Doan, and T. Xie. 2002. Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science 296:1855-1857. [DOI] [PubMed] [Google Scholar]
- 32.Suzu, S., Y. Hayashi, T. Harumi, K. Nomaguchi, M. Yamada, H. Hayasawa, and K. Motoyoshi. 2002. Molecular cloning of a novel immunoglobulin superfamily gene preferentially expressed by brain and testis. Biochem. Biophys. Res. Commun. 296:1215-1221. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi, M., M. Sanbo, S. Watanabe, I. Naruse, M. Mishina, and T. Yagi. 1998. Efficient production of Cre-mediated site-directed recombinants through the utilization of the puromycin resistance gene, pac: a transient gene-integration marker for ES cells. Nucleic Acids Res. 26:679-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamashita, Y. M., D. L. Jones, and M. T. Fuller. 2003. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301:1547-1550. [DOI] [PubMed] [Google Scholar]
- 35.Yoshimizu, T., N. Sugiyama, M. De Felice, Y. I. Yeom, K. Ohbo, K. Masuko, M. Obinata, K. Abe, H. R. Scholer, and Y. Matsui. 1999. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev. Growth Differ. 41:675-684. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, J., C. Niu, L. Ye, H. Huang, X. He, W. G. Tong, J. Ross, J. Haug, T. Johnson, J. Q. Feng, S. Harris, L. M. Wiedemann, Y. Mishina, and L. Li. 2003. Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425:836-841. [DOI] [PubMed] [Google Scholar]