Abstract
Inhibition of the mutationally activated Wnt cascade in colorectal cancer cell lines induces a rapid G1 arrest and subsequent differentiation. This arrest can be overcome by maintaining expression of a single Tcf4 target gene, the proto-oncogene c-Myc. Since colorectal cancer cells share many molecular characteristics with proliferative crypt progenitors, we have assessed the physiological role of c-Myc in adult crypts by conditional gene deletion. c-Myc-deficient crypts are lost within weeks and replaced by c-Myc-proficient crypts through a fission process of crypts that have escaped gene deletion. Although c-Myc−/− crypt cells remain in the cell cycle, they are on average much smaller than wild-type cells, cycle slower, and divide at a smaller cell size. c-Myc appears essential for crypt progenitor cells to provide the necessary biosynthetic capacity to successfully progress through the cell cycle.
The mouse adult intestine is covered with a simple epithelium that bears finger-like extrusions and invaginations called villi and crypts of Lieberkühn. Proliferation of intestinal progenitor cells is restricted to the crypt area, while the villi are populated with differentiated cells. Self-renewal of the intestinal epithelium is a continuous process in which complete epithelial turnover along the crypt-villus axis takes 4 to 5 days (18). This process is driven by a rapidly dividing transit-amplifying population that originates from slowly dividing stem cells residing near the crypt bottom. Multiple studies have attributed a crucial role to the canonical Wnt pathway in normal epithelial homeostasis (24, 25, 31) as well as in colorectal tumorigenesis which is initiated by activating mutations in the Wnt cascade (23, 28, 32).
The transcription factor c-Myc has been identified as a target of the Wnt pathway in colorectal cancer (CRC) cells in vitro (17) and in normal crypts in vivo (42), as well as in intestinal epithelial cells acutely transformed upon in vivo deletion of the APC gene (37). A role for c-Myc in the proliferation of CRC cells was further suggested from in vitro cell culture experiments. CRC cell lines in which the Wnt pathway is blocked by means of a dominant-negative form of Tcf4 arrest in the G1 phase of the cell cycle and begin to differentiate (42). c-Myc is rapidly downregulated in this process. Moreover, when the expression of c-Myc is artificially maintained during Wnt pathway inhibition, the growth arrest phenotype does not ensue (42). This experiment implied that c-Myc plays a crucial role downstream of the Wnt cascade in maintaining the proliferative status of CRC cells.
Since the molecular characteristics of CRC cells and normal crypt progenitor cells are comparable (34), a similar role would be predicted for c-Myc in crypt proliferation. In this light, it was unexpected that conditional deletion of c-Myc in adult intestinal epithelium did not induce an overt phenotype (3). That study utilized a Cre-estrogen receptor fusion transgene driven by the intestine-specific villin promoter, inducible by tamoxifen injection. In the current study, we have pursued the induced deletion of c-Myc in adult crypts using a different Cre transgene that is inducible in the intestine.
MATERIALS AND METHODS
Mouse maintenance and Cre induction.
Mice were maintained on an outbred background segregating for C57BL6, Ola129, and C3H genomes. Mice were genotyped according to published protocols for the Mycfl allele (8), the Rosa26R allele (39), and the AhCre transgene (37). Cre activity was induced by daily intraperitoneal exposure to 80 mg of β-napthoflavone/kg of body weight as described by Sansom et al. (37).
β-Galactosidase analysis.
To determine the pattern of recombination at the Rosa26R reporter locus, both whole mounts and sectioned material were analyzed. Intestinal whole mounts were prepared on wax plates as previously described (44) and were fixed in ice-cold 2% formaldehyde-0.2% glutaraldehyde in phosphate-buffered saline (PBS; pH 7.4) for 1 h. For the generation of sections, 1- to 2-cm lengths of intestine were fixed on ice in 4% paraformaldehyde (PFA) for 4 to 6 h, subsequently immersed in 20% sucrose-PBS for 48 to 72 h at 0 to 4°C, and then snap-frozen and mounted in cryomountant (OCT) prior to cryo-sectioning. Air-dried sections and intestinal whole mounts were stained for β-galactosidase using 0.08% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) substrate [in 2 mM MgCl2, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6 · 6H2O, 0.1 M PBS, pH 7.4, and prepared from a 2% stock of X-Gal in dimethyl-formamide] overnight at 37°C and room temperature, respectively.
Histology, immunohistochemistry, BrdU labeling, and in situ hybridization.
Tissues were fixed in 10% formalin or 4% paraformaldehyde, paraffin embedded, and sectioned at 3 to 6 μm for hematoxylin and eosin (H&E) staining, argyrophilic nucleolar organizer region (AgNor) staining, or immunostaining as described elsewhere (37). The primary antibodies used were rabbit anti-lysozyme (1:500; DAKO), mouse anti-Ki67 (1:100; Novocastra), mouse anti-bromodeoxyuridine (BrdU; 1:500; Becton Dickinson), rabbit anti-synaptophysin (1:100; DAKO), rabbit anti-c-Myc (1:500; Upstate Biotechnology), and mouse anti-B23 (1:2,000; Sigma). The peroxidase-conjugated secondary antibodies used were mouse or rabbit EnVision+ (DAKO). For BrdU labeling, mice were injected with 100 μg of BrdU (Sigma) per kg body weight and killed after 2, 12, 24, 36, or 48 h. AgNor staining was performed as described elsewhere (40). For in situ hybridization, paraffin-embedded tissue was sectioned at 10 μm and processed for hybridization as previously described (14).
Cell size measuring.
All morphological parameters were quantified using a macro, written in the Pascal-based macro language of Scion Image, a Windows-based version of NIH Image (Scion, Frederick, Md.). Color images (RGB) were imported into the program. The second image plane (eight-bit gray) was selected and calibrated for magnification. Cell boundaries were marked with the selection tool using a digitizer board (Wacom Co., Ltd., Japan). All parameters (i.e., cell surface areas) were quantified with the measure option of the program. On average, 800 cells from 60 crypts per mouse were analyzed, with at least three mice representing the given genotype.
Analysis of cell cycle kinetics.
After BrdU labeling, paraffin sections from mouse small intestines were prepared and stained with anti-BrdU antibody (Ab). Cells were scored per entire crypt and villus unit. At least 60 crypts and corresponding villi were analyzed per time point per mouse. Each genotype was represented with three animals. BrdU-labeled cells were normalized to total cell number per crypt or villus. The resulting percentage was then plotted against the induction time.
RESULTS AND DISCUSSION
Conditional deletion of c-Myc in adult crypts.
Deletion of c-Myc by gene targeting in mice causes mid-gestation lethality (7). Therefore, assessment of the role of c-Myc in specific tissue types in the adult mouse requires a conditional genetic approach. In order to address the role of c-Myc in the adult small intestine, we utilized a recently developed transgenic line in which expression of the Cre-recombinase is under the control of the Cyp1a promoter (19). Conditional deletion of c-Myc was achieved by crossing these mice with a loxP-flanked c-Myc allele (2). Using this approach, inducible Cre-mediated excision of the c-Myc allele in Cre+ Mycfl/fl mice occurs following intraperitoneal injection of β-naphthoflavone, which induces Cre activity within the crypts of the small intestine, including in the stem cells (19). When using a regimen which yields maximal recombination (either three injections in a single day or four daily injections of β-naphthoflavone), this results in near 100% recombination when scored using a surrogate marker of recombination (e.g., Rosa-26) (20, 39). This level of recombination remains stable in the intestinal epithelium for at least 1 year following induction, unless the recombination event (such as loss of Brca2 or β-catenin) is deleterious to the cell (16, 19).
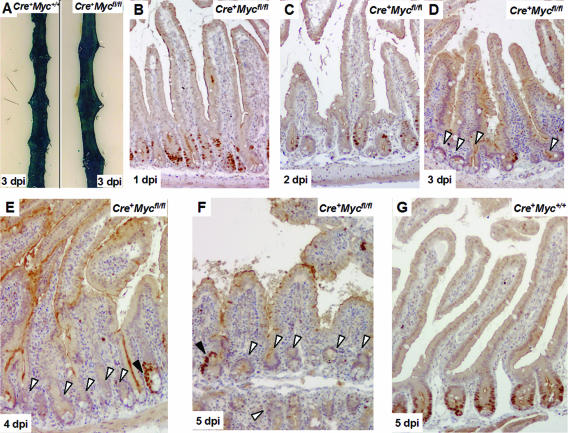
Mice 10 weeks of age were injected three times with β-naphthoflavone on the same day, and their intestines were analyzed on days 1, 2, 3, 4, 5, 7, and 14 postinduction (p.i.) by in situ hybridization (ISH) and immunohistochemistry (IHC) for c-Myc RNA and protein. During the first 2 days, levels of c-Myc RNA and protein gradually decreased compared to controls (Fig. 1B and C; see also Fig. S1A and B in the supplemental material). On serial sections, IHC results correlated well with those of ISH (see Fig. S1 in the supplemental material). At days 3 to 5, c-Myc RNA and protein became undetectable in virtually all crypts (Fig. 1D to F; see also Fig. S1C and D in the supplemental material). Occasional crypts showed clusters of c-Myc-positive cells, most notably at the latest time points (Fig. 1E and F). The near-complete c-Myc gene deletion at day 3 compared well to the highly efficient recombination frequency at the surrogate marker allele Rosa26R at day 3 in the same experiment (Fig. 1A).
FIG. 1.
Assessment of c-Myc deletion in β-napthoflavone-induced Cre+ Mycfl/fl intestines over a 5-day time course. (A) β-Galactosidase staining on intestinal whole mounts 3 days p.i. [dpi], indicating near-100% recombination at the Rosa26 reporter allele. Left, Cre+ Myc+/+; right, Cre+ Mycfl/fl. (B and C) c-Myc staining at days 1 and 2 p.i., showing reduced levels of c-Myc protein but still significant numbers of c-Myc-positive cells decorating the crypts. (D to F) Large areas of c-Myc-deficient epithelium at later time points (white arrows). Black arrows in panels E and F depict occasional c-Myc-proficient cells. Note that at later time points c-Myc-proficient cells clustered. (G) Representative section of Cre+ Myc+/+ control mice, where c-Myc protein was uniformly expressed at the lower half of the crypt region.
Histological analysis revealed no major alterations in crypt-villus architecture during the first 5 days (see Fig. S2A to J in the supplemental material). Moreover, c-Myc-deficient cells remained in cycle, as indicated by IHC for the proliferation marker Ki67 (see Fig. S2B and G in the supplemental material).
c-Myc-deficient crypts are replaced by c-Myc-proficient crypts through crypt fission.
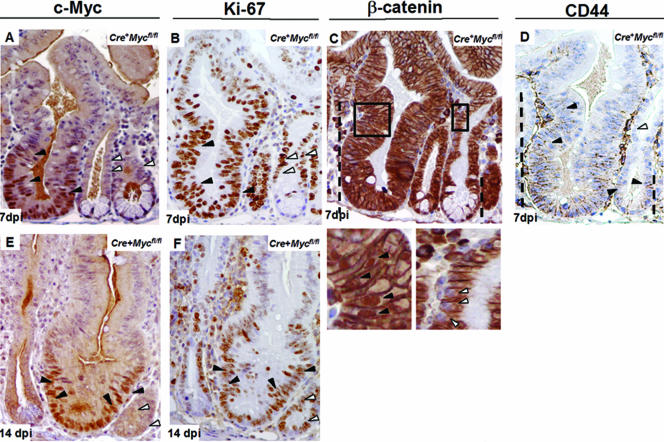
The observation of clusters of cells with high levels of c-Myc at days 4 and 5 postinduction indicated the possible expansion of rare unrecombined Mycfl/fl cells within an otherwise-c-Myc-deficient environment. This notion was confirmed upon histological analysis at days 7 and 14. H&E staining revealed the presence of a subset of crypts that were hyperplastic, highly reminiscent of repopulating crypts observed following exposure to DNA damage (5, 33). These crypts appeared to generate new crypts by fission as seen post-DNA damage (4). Virtually all enlarged crypts showed high levels of c-Myc expression (Fig. 2A and E). Somewhat unexpectedly, cells in c-Myc-deficient as well as in c-Myc-proficient crypts were cycling, as indicated by Ki67 positivity (Fig. 2B and F). Since Wnt signaling represents a dominant mitogen in the adult intestinal epithelium (31), we investigated the presence of nuclear β-catenin as an indicator of active Wnt signaling in the enlarged, c-Myc-proficient crypts (Fig. 2C). These crypts displayed nuclear β-catenin throughout the crypt region (Fig. 2C), while nuclear β-catenin is normally only obvious at crypt bottoms (42). This observation suggests that the mechanism underlying the intestinal repopulation involved increased activity of the Wnt pathway, a notion further supported by the increased expression domain of the Wnt target gene CD44 (42) in c-Myc-proficient crypts (Fig. 2D).
FIG. 2.
c-Myc-proficient cells in Cre+ Mycfl/fl mice are dominant over c-Myc-deficient cells and form enlarged crypts. (A and E) Representative sections from Cre+ Mycfl/fl mice at 7 and 14 days p.i. [dpi], respectively, stained with anti c-Myc antibody. Enlarged crypts are composed exclusively of c-Myc-proficient cells (black arrows), while the significantly smaller crypts contain cells expressing no c-Myc protein (white arrows). (B and F) Ki-67 staining showed cell proliferation in both c-Myc-proficient and -deficient crypts in Cre+ Mycfl/fl mice (black arrows versus white arrows). Note that c-Myc staining and Ki-67 staining were performed on consecutive sections. (C) Consecutive paraffin section of Cre+ Mycfl/fl mice at 7 days p.i. stained with antibody to β-catenin. The dashed lines depict the position until nuclear β-catenin translocation could be observed. A much higher cell position with nuclear β-catenin from a c-Myc-proficient crypt is also presented as a magnification of the squared area (left lower panel). In contrast, similar cell positions in a Myc-deficient crypt have no nuclear β-catenin (right lower panel). (D) Serial paraffin sections of Cre+ Mycfl/fl mice at 7 days postdeletion stained with antibody to CD44. The CD44 staining pattern follows that of nuclear β-catenin (dashed lines). Expanded CD44 expression in c-Myc-proficient crypts is marked with black arrows. Cells at equivalent positions in c-Myc-deficient crypts have no CD44 expression (white arrows).
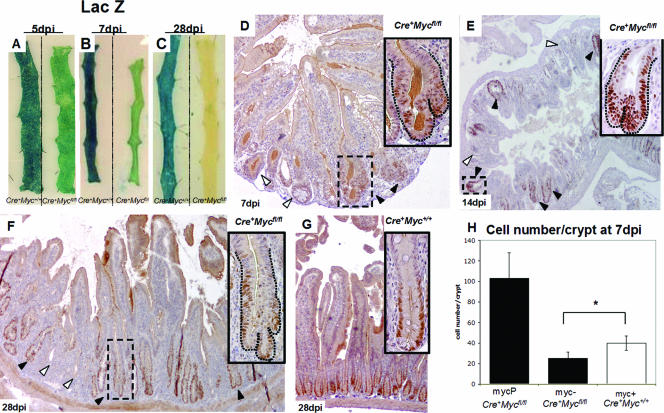
To determine the extent of competition between c-Myc-deficient and -proficient cells, we analyzed the pattern of recombination at a surrogate marker of Cre-mediated recombination by using mice additionally transgenic for the Rosa26R reporter line (39) at days 5, 7, and 28 p.i. LacZ whole-mount stains showed that recombined crypts were lost in the Cre+ Mycfl/fl mice after 28 days p.i., indicating a strong selection against c-Myc deficiency (Fig. 3A to C, tissue samples on right). By contrast, the pattern of Cre-mediated recombination as reported with Rosa26R remained unchanged over the course of 28 days in Cre+ Myc+/+ mice (Fig. 3A to C, tissue samples on left), implying that the expression of active Cre enzyme per se did not affect crypt homeostasis. The process of crypt fission by which c-Myc-deficient crypts are replaced is shown in Fig. 3D to F. Quantification of the frequency of crypt fission is given in Table 1.
FIG. 3.
Mycfl/fl crypts are lost in Cre+ Mycfl/fl mice after 4 weeks of induction and are replaced by Myc+/+ crypts through crypt fission. (A to C) LacZ-stained whole-mount preparations from Cre+ Mycfl/fl and control mice at day 5 (A), day 7 (B), and day 28 (C) p.i., indicating preferential repopulation with unrecombined cells in the induced Cre+ Mycfl/fl mice (right intestinal strip in each panel) which was almost complete at 28 days p.i. (D, E, and F) c-Myc staining at 7, 14, and 28 days p.i., respectively, in sections from Cre+ Mycfl/fl mice, indicating repopulation from c-Myc-proficient, hyperplastic crypts through crypt fission (black arrows). (G) Cre+ Myc+/+ control mice. (H) Graph showing reduced cell numbers per crypt in c-Myc-deficient crypts (myc-) from Cre+ Mycfl/fl mice compared to c-Myc-expressing crypts (myc+) of control Cre+ Myc+/+ mice at 7 days p.i. (Cre+ Mycfl/fl, 25 ± 6 [n = 2]; Cre+ Myc+/+, 40 ± 7 [n = 3]; *, P = 0.025 by Mann-Whitney U test). Data are means ± standard deviations; n is the number of mice analyzed. The first bar in the graph represents numbers of cells in c-Myc-proficient crypts (MycP) in Cre+ Mycfl/fl mice (103 ± 24; n = 2). A total of 100 crypts was analyzed per mouse.
TABLE 1.
Crypt fission frequency
| Genotype | Crypt fission frequency (%)a
|
||||
|---|---|---|---|---|---|
| 1 dpi | 4 dpi | 7 dpi | 14 dpi | 28 dpi | |
| Cre+Mycfl/fl | 0, 0 | 0, 0 | 4, 3 | 12, 8 | 8, 6 |
| Cre+Myc+/+ | 0, 0 | 0, 0 | 0, 0 | 0, 0 | 0, 0 |
A total of 100 crypts were analyzed in each of two mice at each of the indicated days p.i. (dpi). Crypt fission was first detected at 7 dpi and affected up to 12% of the crypts at various time points.
Loss of c-Myc leads to decreased cell numbers per crypt.
To address the difference in size between c-Myc-deficient and c-Myc-proficient crypts, we determined cell numbers in c-Myc-deficient versus c-Myc-proficient crypts from induced Cre+ Mycfl/fl mice. As a control, we included crypts from induced Cre+ Myc+/+ control mice. Cell number per crypt was analyzed by counting nuclei from c-Myc-stained sections at day 7, which contained both c-Myc-deficient crypts and repopulating c-Myc-proficient crypts, facilitating a direct comparison within the same animal. It was thus observed that c-Myc-deficient crypts contained fourfold-fewer cells per crypt than c-Myc-proficient, hyperplastic crypts in the same animal (Fig. 3H) (Mann-Whitney U test, P < 0.01). c-Myc-deficient crypts from the Cre+ Mycfl/fl mice also contained fewer cells than Cre+ Myc+/+ control mice (Fig. 3H) (Mann-Whitney U test, P = 0.025), while hyperplastic crypts in Cre+ Mycfl/fl mice contained two- to threefold more cells than crypts in control mice.
c-Myc deletion does not affect apoptotic rates in the intestinal epithelium.
Numerous studies in diverse model systems have linked either upregulated or downregulated c-Myc expression to apoptosis (30). We therefore performed IHC against the cleaved form of caspase-3, as a marker for cellular apoptosis, in our time course experiment. Activated caspase-3-positive cells were seen at the tip of the villus, and occasional positive cells were observed in the crypt area in both Cre+ Mycfl/fl and Cre+ Myc+/+ mice at all time points (see Fig. S3A to D in the supplemental material). Counting of apoptotic cells in Cre+ Myc+/+ and Cre+ Mycfl/fl epithelium at 3, 5, and 7 days revealed no significant differences between c-Myc-proficient and -deficient crypts, and indeed the apoptotic index was not elevated over basal levels (see Fig. S3E in the supplemental material) (Mann-Whitney U test, P > 0.2). Apparently, loss of c-Myc in the small intestine did not affect the rate of apoptosis.
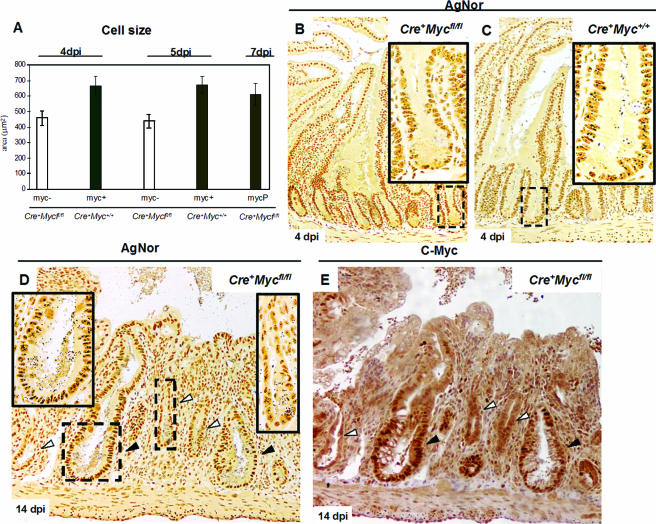
c-Myc-deficient crypt cells are smaller than c-Myc-proficient crypt cells and enter mitosis with decreased cell size.
As we observed that c-Myc-proficient crypts were larger than their c-Myc-deficient counterparts, we determined cell size in serial paraffin sections stained for both c-Myc and β-catenin. This double labeling allowed us to identify cell boundaries (β-catenin), as well as identify c-Myc-proficient and -deficient crypts. c-Myc-deficient cells were smaller in longitudinal crypt sections than control c-Myc-proficient cells from Cre+ Myc+/+ mice (Fig. 4A) (459.5 μm2 [n = 4] and 661.9 μm2 [n = 3], respectively; Mann-Whitney U test, P = 0.02) at 4 day p.i. There was no difference in cell size between c-Myc-proficient cells from Cre+ Mycfl/fl mice (Fig. 4A) and from Cre+ Myc+/+ mice (611 μm [n = 3] versus 661.9 μm2 and 672 μm2; Mann-Whitney U test, P = 0.66). These differences were also apparent if scored in transverse sections: c-Myc-deficient cells were smaller than c-Myc-proficient cells from Cre+ Myc+/+ mice (Fig. 4B) (437 μm2 [n = 4] and 672 μm2 [n = 4], respectively; Mann-Whitney U test, P = 0.01) at 5 days p.i.
FIG. 4.
Size and biosynthetic capacity are reduced in c-Myc-deficient cells. (A) Graph summarizing cell size reduction in c-Myc-deficient cells (myc-). Data were analyzed at two time points after induction, and similar results were obtained (Cre+Mycfl/fl, 459 ± 46 μm2, 4 days p.i. [dpi]; Cre+ Myc+/+, 662 ± 61 μm2, 4 days p.i.; Cre+ Mycfl/fl, 437 ± 43 μm2, 5 days p.i.; Cre+ Myc+/+, 672 ± 53 μm2, 5 days p.i., n = 3). The last bar represents the cell size of c-Myc-proficient cells (MycP) of Cre+ Mycfl/fl mice at 7 days p.i. (611 ± 70 μm2; n = 2). Data are means ± standard deviations, and n is the number of mice analyzed per time point. (B) Underrepresentation of AgNor dots, regions of ribosomal gene synthesis, in c-Myc-deficient cells and crypts of Cre+ Mycfl/fl mice at 4 days p.i. (C) Corresponding control Cre+ Myc+/+ mice with multiple large AgNor dots per nucleus. Insets are magnifications of representative squared areas. (D) Sections from Cre+Mycfl/fl mice at 7 days p.i. and incubated with AgNor. Note that at this stage, smaller c-Myc-deficient crypts (white arrows, right-hand magnification inset) clearly have fewer and smaller AgNors compared in the same section with c-Myc-proficient crypts (black arrows, left-hand magnification inset). (E) Consecutive paraffin section stained with c-Myc Ab, showing smaller crypts with fewer AgNors being c-Myc negative (white arrows).
We next determined the size of the cells undergoing mitosis from Cre+ Mycfl/fl and Cre+ Myc+/+ mice. Mitotic cells are easily identifiable in H&E-stained sections because they migrate towards the crypt lumen and round up before initiating cell division. Serial paraffin sections of the intestines of mutant and control mice were stained with H&E and for c-Myc. The size of mitotic cells was measured directly from H&E-stained sections using Scion imaging software. Only cells from completely c-Myc-deficient crypts were scored. Dividing c-Myc-deficient cells were significantly smaller than their c-Myc-proficient counterparts (633 μm2 [n = 3] versus 747 μm2 [n = 4]; Mann-Whitney U test, P = 0.03).
Reduced biosynthetic activity in c-Myc-deficient crypt cells.
To assess the biosynthetic activity of c-Myc-deficient cells in our system, we used AgNor staining (40). AgNor proteins are argyrophillic (silver-stainable) acidic fibrillar proteins that surround interphase nucleolar organizer regions. No rRNA synthesis takes place in the absence of AgNor proteins, and rRNA biosynthesis is directly proportional to the AgNor protein concentration. Image analysis of the size and number of these regions can be used to distinguish metabolically active cell populations from less active ones (9, 38). AgNor staining of sections from induced Cre+ Mycfl/fl and Cre+ Myc+/+ mice revealed a dramatic decrease of AgNor+ regions in c-Myc-deficient cells and crypts. The difference was most directly apparent when adjacent c-Myc-proficient and -deficient crypts in Cre+ Mycfl/fl mice were examined (Fig. 4B to D).
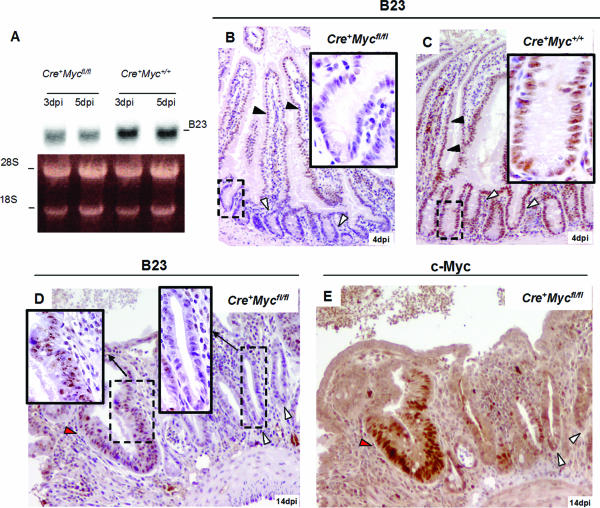
A major component of AgNors is nucleophosmin (B23) (35, 36), which is a direct c-Myc target gene product (13, 46; http://www.myc-cancer-gene.org/index.asp). Nucleophosmin is involved in the late steps of ribosomal particle organization. To investigate if nucleophosmin behaves as a c-Myc target gene in our model system, we performed Northern blot analysis on intestinal RNA samples and IHC and found that the expression of nucleophosmin was greatly reduced in the c-Myc-deficient crypts (Fig. 5A to D).
FIG. 5.
Nucleophosmin (B23) protein, involved in ribosomal gene synthesis and a component of AgNor dots, is downregulated in Cre+ Mycfl/fl mice. (A) Northern blot showing downregulation of B23 RNA in Cre+ Mycfl/fl mice (two left lanes, upper panel) compared with Cre+ Myc+/+ mice (two right lanes, upper panel). Lower panel: ethidium bromide-stained 1% denatured agarose gel. (B and C) IHC for B23 protein in mouse small intestine samples. Crypt cells from Cre+ Mycfl/fl mice express almost undetectable levels of B23 protein (white arrows in panel B), and expression in the villus is reduced (black arrows in panel B). Cre+ Myc+/+ control intestine samples show uniform B23 expression throughout crypt villus units (white arrows in panel C). (D) B23 IHC at 14 days p.i. of intestine from Cre+ Mycfl/fl mice, clearly underscoring the difference in B23 levels between c-Myc-deficient (black arrows, left inset) and -proficient crypts (white arrows, right inset). (E) IHC with c-Myc Ab on a consecutive paraffin section, confirming that B23-negative crypts are also c-Myc negative. dpi, days p.i.
Loss of c-Myc leads to reduced cell cycle kinetics.
The difference in AgNor staining and nucleophosmin expression suggested that reduced protein synthesis in c-Myc-deficient cells caused a reduced cellular growth rate, resulting in a smaller cell size (see above) and slower cell cycle kinetics. Ki67 is a nuclear antigen marking actively cycling cells in all phases of the cycle. As in c-Myc-proficient crypts, essentially all cells in c-Myc-deficient crypts were Ki67+, indicating that cells were actively in cycle. Of note, no expression differences were found in the putative Myc target genes cyclin D1, cyclin D2, and CDK4 (6, 29) between c-Myc-proficient and -deficient crypts (see Fig. S4 in the supplemental material). We have reported previously that c-Myc in colorectal cancer cells represses the expression of p21, which is also a good marker of terminally differentiated villus cells (42). The p21 expression domain was not extended into c-Myc-deficient crypts (see Fig. S4 in the supplemental material), which may be explained by the fact that cells in c-Myc-deficient crypts cycle, albeit at a slower pace, and do not differentiate.
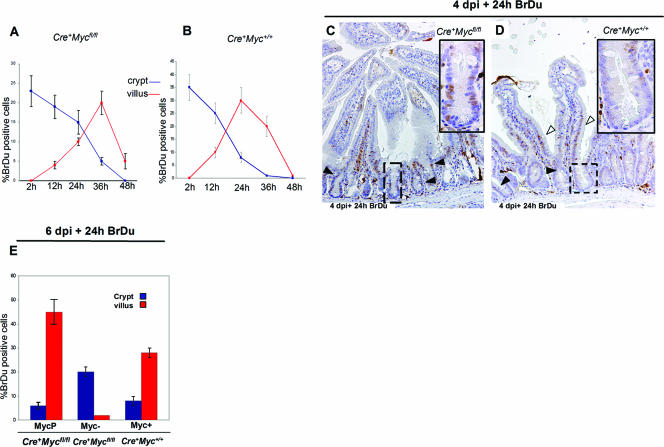
The rate at which crypt progenitor cells transit into the postmitotic villus compartment is directly correlated to the rate of cell division in the crypt (18). To investigate whether the loss of c-Myc would result in slowing down of the cell cycle, we pulsed c-Myc-proficient and c-Myc-deficient mice at 4 days postdeletion with BrdU to label S-phase cells in the crypts and followed the kinetics of the migrating crypt cells. As shown in Fig. 6, virtually all BrdU-labeled cells left the crypts within 24 h, while c-Myc-deficient cells were retained within the crypts for at least 50% longer. Thus, all crypts cell cycle in the absence of c-myc, yet c-Myc-deficient crypts contain fewer cells which are retained for a longer period of time within the crypt. Together, this implies that cell cycle kinetics are strongly reduced in the absence of c-Myc. This was further confirmed by BrdU pulsing at 6 days postinduction of Cre+ Mycfl/fl mice followed by analysis 24 h later (Fig. 6E and F).
FIG. 6.
c-Myc-deficient crypt cells from Cre+ Mycfl/fl mice cycle profoundly slower than c-myc-expressing cells from Cre+ Myc+/+ control mice. (A and B) BrdU time course. Mice at 4 days p.i. (dpi) were injected with BrdU and harvested 2, 12, 24, 36, and 48 h later. BrdU-positive cells were scored as a percentage where the number of positive cells was normalized with the total number of cells per crypt (blue lines) or villus (red lines). Villus BrdU+ cells reached maximum numbers at 36 h, while the maximum number for control Cre+Myc+/+ mice was already reached at 24 h. (C and D) IHC to BrdU antibody 24 h postinjection. Significant numbers of BrdU+ cells did not exit the crypt in Cre+ Mycfl/fl mice (C, black arrows), while only a few cells decorated the crypts in Cre+ Myc+/+ mice (D, black arrows), with the majority of cells present in the villus (D, white arrows). (E) Graph summarizing the percentage of BrdU-labeled cells in the crypt and villus in Cre+ Mycfl/fl and Cre+ Myc+/+ mice induced for 6 days and pulsed for 24 h. Data are means ± standard deviations, and a minimum of three mice were analyzed per genotype per BrdU time point.
The current study establishes a central role for c-Myc in the rate of self-renewal within the intestinal epithelium. Crypt progenitor cells that lack c-Myc are smaller, have less protein translation machinery, and appear to cycle slower than their wild-type counterparts. Consequently, c-Myc-deficient crypts are competed out by c-Myc-proficient crypts within weeks. The Wnt cascade is the primary driver of cell proliferation in intestinal crypts. Our study implies that c-Myc, as a target of Tcf4 (17), is a crucial component of the intestinal genomic program driven by the Wnt cascade.
A role for c-Myc in the accumulation of cell mass was initially suggested through studies of wing patterning in Drosophila melanogaster (22) and was subsequently also confirmed in several mammalian tissues in vivo, e.g., in B lymphocytes, in hepatocytes, and in epidermis (2, 21, 45). c-Myc is considered to exert this effect by regulating gene transcription by all three RNA polymerases (1, 11, 12, 15, 26), thus regulating many target genes directly involved in cellular metabolism and protein synthesis (10). The smaller cell size of c-Myc-deficient cells that we reported above is predicted to be a reflection of reduced protein synthesis and a consequent reduced rate of cell growth.
In contrast to the current study, loss of c-Myc had no effect on cell size in some other settings. An allelic series of c-Myc hypomorph/null mice demonstrated that c-Myc regulates body size in mice (41). Yet, this study uncovered no size difference in either hematopoietic cells or primary embryo fibroblasts. Rather, primary fibroblasts of embryos with decreased c-Myc levels displayed an elongated cell cycle. Another study reported a similar situation for c-Myc-deficient fibroblasts that exhibited significant prolongation of the cell cycle without any change in cell size. Notably, the rate of protein synthesis in these cells was two- to threefold lower than in the c-Myc+/+ parental cell line (27). One interpretation of these data is that cell division is coupled to cell size in these situations. Therefore, any defect in cell growth would inevitably result in fewer cells.
The difference between our study and that of Bettess and colleagues (3) most likely relates to deletion efficiency accomplished with the different Cre transgenes in the earliest crypt progenitors. We recently used the same two Cre transgenes within one study to inducibly delete the Notch pathway CSK/RBP-Jκ gene from intestinal epithelium (43). We found that the Cyp1a-Cre transgene deleted the CSK/RBP-Jκ gene highly efficiently throughout the small intestinal epithelium, while the villin Cre-ER transgene yielded patchy gene deletion in only a subset of the treated animals.
In conclusion, our data indicate that cell proliferation in the intestinal epithelium is reduced in the absence of c-Myc. The c-Myc deficiency leads to a reduction in cell size which appears linked to biosynthetic capacity. Apparently, intestinal progenitor cells are not subject to an absolute cell size checkpoint for entry into mitosis, a characteristic in which they may differ from other cell types, such as primary embryo fibroblasts. The failed perpetuation of c-Myc-negative crypts triggers a regenerative response in crypts that have escaped c-Myc gene deletion, which is reminiscent of postirradiation repair. Finally, c-Myc plays no role in the regulation of apoptosis under our experimental conditions.
Supplementary Material
Acknowledgments
This work was supported by CR-UK and by the Wales Gene Park.
Footnotes
Published ahead of print on 5 September 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arabi, A., S. Wu, K. Ridderstrale, H. Bierhoff, C. Shine, K. Fatyoul, S. Fahlen, P. Hydbring, O. Soderberg, I. Grummt, L. G. Larson, and A. P. Write. 2005. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 7:303-310. [DOI] [PubMed] [Google Scholar]
- 2.Baena, E., A. Gandarillas, M. Vallespinos, J. Zanet, O. Bachs, C. Redondo, I. Fabregat, C. Martinez-A, and I. M. de Alboran. 2005. c-Myc regulates cell size and ploidy but is not essential for postnatal proliferation in liver. Proc. Natl. Acad. Sci. USA 102:7286-7291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bettess, M. D., N. Dubois, M. J. Murphy, C. Dubey, C. Roger, S. Robine, and A. Trumpp. 2005. c-Myc is required for the formation of intestinal crypts but dispensable for homeostasis of the adult intestinal epithelium. Mol. Cell. Biol. 25:7868-7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booth, C., and C. S. Potten. 2000. Gut instincts: thoughts on intestinal epithelial stem cells. J. Investig. Dermatol. 114:667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booth, D., and C. S. Potten. 2001. Protection against mucosal injury by growth factors and cytokines. J. Natl. Cancer Inst. Monogr. 29:16-20. [DOI] [PubMed] [Google Scholar]
- 6.Bouchard, C., K. Thieke, A. Maier, R. Saffrich, J. Hanley-Hyde, W. Asorge, S. Reed, P. Sicinski, J. Bartek, and M. Eilers. 1999. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 18:5321-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, A. C., M. Wims, G. D. Spotts, S. R. Hann, and A. Bradley. 1993. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 7:671-682. [DOI] [PubMed] [Google Scholar]
- 8.de Alboran, I. M., R. C. O'Hagan, F. Gartner, B. Malynn, L. Davidson, R. Rickert, K. Rajewsky, R. A. DePinho, and F. W. Alt. 2001. Analysis of c-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14:45-55. [DOI] [PubMed] [Google Scholar]
- 9.Derenzini, M. 2000. The AgNORs. Micron 31:117-120. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez, P. C., S. R. Frank, L. Wang, M. Schroeder, S. Liu, J. Greene, A. Cocito, and B. Amati. 2003. Genomic targets of the human c-Myc protein. Genes Dev. 17:1115-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez-Roman, N., C. Grandori, R. N. Eisenman, and R. J. White. 2003. Direct activation of RNA polymerase III transcription by c-Myc. Nature 421:290-294. [DOI] [PubMed] [Google Scholar]
- 12.Grandori, C., N. Gomez-Roman, Z. A. Felton-Edkins, C. Ngouenet, D. A. Galloway, R. N. Eisenman, and R. J. White. 2005. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 7:311-318. [DOI] [PubMed] [Google Scholar]
- 13.Greasley, P. J., C. Bonnard, and B. Amati. 2000. Myc induces the nucleolin and BN51 genes: possible implication in ribosome biogenesis. Nucleic Acids Res. 28:446-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregorieff, A., R. Grosschedl, and H. Clevers. 2004. Hindgut defects and transformation of the gastro-intestinal tract in Tcf4−/− Tcf1−/− embryos. EMBO J. 23:1825-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grewal, S. S., L. Li, A. Orian, R. N. Eisenman, and B. A. Edgar. 2005. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell Biol. 7:295-302. [DOI] [PubMed] [Google Scholar]
- 16.Hay, T. J., T. Patrick, D. J. Winton, O. J. Sansom, and A. R. Clarke. 2005. Brca2-deficient cells in the murine small intestine are sensitised to p53-dependent apoptosis and are spontaneously deleted. Oncogene 24:3842-3846. [DOI] [PubMed] [Google Scholar]
- 17.He, T. C., A. B. Sparks, C. Rago, H. Hermeking, L. Zawel, L. T. da Costa, P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Identification of c-MYC as a target of the APC pathway. Science 281:1509-1512. [DOI] [PubMed] [Google Scholar]
- 18.Heath, J. P. 1996. Epithelial cell migration in the intestine. Cell Biol. Int. 20:139-146. [DOI] [PubMed] [Google Scholar]
- 19.Ireland, H., R. Kemp, C. Houghton, L. Howard, A. R. Clarke, O. J. Sansom, and D. J. Winton. 2004. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: effect of loss of beta-catenin. Gastroenterology 126:1236-1246. [DOI] [PubMed] [Google Scholar]
- 20.Ireland, H., C. Houghton, L. Howard, and D. J. Winton. 2005. Cellular inheritance of a Cre-activated reporter gene to determine Paneth cell longevity in the murine small intestine. Dev. Dyn. 233:1332-1336. [DOI] [PubMed] [Google Scholar]
- 21.Iritani, B. M., and R. N. Eisenman. 1999. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc. Natl. Acad. Sci. USA 96:13180-13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston, L. A., D. A. Prober, B. A. Edgar, R. N. Eisenman, and P. Gallant. 1999. Drosophila myc regulates cellular growth during development. Cell 98:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korinek, V., N. Barker, P. J. Morin, D. van Wichen, R. de Weger, K. W. Kinzler, B. Vogelstein, and H. Clevers. 1997. Constitutive transcriptional activation by a beta-catenin-Tcf complex in Apc−/− colon carcinoma. Science 275:1784-1787. [DOI] [PubMed] [Google Scholar]
- 24.Korinek, V., N. Barker, P. Moerer, E. van Donselaar, G. Huls, P. J. Peters, and H. Clevers. 1998. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 19:373-383. [DOI] [PubMed] [Google Scholar]
- 25.Kuhnert, F., C. R. Davis, H. T. Wang, P. Chu, M. Lee, J. Yan, R. Nusse, and C. J. Kuo. 2004. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc. Natl. Acad. Sci. USA 101:266-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levens, D. L. 2003. Reconstructing MYC. Genes Dev. 17:1071-1077. [DOI] [PubMed] [Google Scholar]
- 27.Mateyak, M. K., A. J. Obaya, S. Adachi, and J. M. Sedivy. 1997. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 8:1039-1048. [PubMed] [Google Scholar]
- 28.Morin, P. J., A. B. Sparks, V. Korinek, N. Barker, H. Clevers, B. Vogelstein, and K. W. Kinzler. 1997. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or Apc. Science 275:1787-1790. [DOI] [PubMed] [Google Scholar]
- 29.O'Connell, B. C., A. F. Cheung, C. P. Simkevich, W. Tam, X. Ren, M. K. Mateyak, and J. M. Sedivy. 2003. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J. Biol. Chem. 278:12563-12573. [DOI] [PubMed] [Google Scholar]
- 30.Pelengaris, S., B. Rodolf, and T. Littlewood. 2000. Action of Myc in vivo-proliferation and apoptosis. Curr. Opin. Genet. Dev. 10:100-105. [DOI] [PubMed] [Google Scholar]
- 31.Pinto, D., A. Gregorieff, H. Begthel, and H. Clevers. 2003. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 17:1709-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polakis, P. 1999. The oncogenic activation of beta-catenin. Curr. Opin. Genet. Dev. 9:15-21. [DOI] [PubMed] [Google Scholar]
- 33.Potten, C. S., G. Owen, and S. A. Roberts. 1990. The temporal and spatial changes in cell proliferation within the irradiated crypts of the murine small intestine. Int. J. Radiat. Biol. 57:185-199. [DOI] [PubMed] [Google Scholar]
- 34.Radtke, F., and H. Clevers. 2005. Self-renewal and cancer of the gut: two sides of a coin. Science 307:1904-1909. [DOI] [PubMed] [Google Scholar]
- 35.Roussel, P., P. Belenguer, F. Amalric, and D. Hernandez-Verdun. 1992. Nucleolin is an Ag-NOR protein; this property is determined by its amino-terminal domain independently of its phosphorylation state. Exp. Cell Res. 203:259-269. [DOI] [PubMed] [Google Scholar]
- 36.Roussel, P., and D. Hernandez-Verdun. 1994. Identification of Ag-NOR proteins, markers of proliferation related to ribosomal gene activity. Exp. Cell Res. 214:465-472. [DOI] [PubMed] [Google Scholar]
- 37.Sansom, O. J., K. R. Reed, A. J. Hayes, H. Ireland, N. Brinkmann, I. P. Newton, E. Batlle, P. Simon-Assmann, H. Clevers, I. S. Nathke, A. R. Clarke, and D. J. Winton. 2004. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 18:1385-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sirri, V., P. Roussel, M. C. Gendron, and D. Hernandez-Verdun. 1997. Amount of the two major Ag-NOR proteins, nucleolin, and protein B23 is cell-cycle dependent. Cytometry 28:147-156. [PubMed] [Google Scholar]
- 39.Soriano, P. 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21:70-71. [DOI] [PubMed] [Google Scholar]
- 40.Trere, D. 2000. AgNOR staining and quantification. Micron 31:127-131. [DOI] [PubMed] [Google Scholar]
- 41.Trumpp, A., Y. Refaeli, T. Oskarsson, S. Gasser, M. Murphy, G. R. Martin, and J. M. Bishop. 2001. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 414:768-773. [DOI] [PubMed] [Google Scholar]
- 42.van de Wetering, M., E. Sancho, C. Verweij, W. de Lau, I. Oving, A. Hurlstone, K. nav de Horn, E. Batlle, D. Coudreuse, A. P. Haramis, M. Tjon-Pon-Fong, P. Moerer, M. van den Born, G. Soete, S. Pals, M. Eilers, R. Medema, and H. Clevers. 2002. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111:241-250. [DOI] [PubMed] [Google Scholar]
- 43.van Es, J. H., M. E. van Gijn, O. Riccio, M. van den Born, M. Vooijs, H. Begthel, M. Cozijnsen, S. Robine, D. J. Winton, F. Radtke, and H. Clevers. 2005. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 453:959-963. [DOI] [PubMed] [Google Scholar]
- 44.Winton, D. J., and B. A. J. Ponder. 1990. Stem cell organisation in mouse small intestine. Proc. R. Soc. Lond. Ser. B 241:13-18. [DOI] [PubMed] [Google Scholar]
- 45.Zanet, J., S. Pibre, C. Jacquet, A. Ramirez, I. M. de Alboran, and A. Gandarillas. 2005. Endogenous Myc controls mammalian epidermal cell size, hyperproliferation, endoreplication and stem cell amplification. J. Cell Sci. 118:1693-1704. [DOI] [PubMed] [Google Scholar]
- 46.Zeller, K. I., T. J. Haggerty, J. F. Barret, Q. Guo, and C. V. Dang. 2001. Characterisation of nucleophosmin (B23) as a Myc target by chromatin immunoprecipitation. J. Biol. Chem. 276:48285-48291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.