Abstract
The mechanism of client protein activation by Hsp90 is enigmatic, and it is uncertain whether Hsp90 employs a common route for all proteins. Using a mutational analysis approach, we investigated the activation of two types of client proteins, glucocorticoid receptor (GR) and the kinase v-Src by the middle domain of Hsp90 (Hsp90M) in vivo. Remarkably, the overall cellular activity of v-Src was highly elevated in a W300A mutant yeast strain due to a 10-fold increase in cellular protein levels of the kinase. In contrast, the cellular activity of GR remained almost unaffected by the W300A mutation but was dramatically sensitive to S485Y and T525I exchanges. In addition, we show that mutations S485Y and T525I in Hsp90M reduce the ATP hydrolysis rate, suggesting that Hsp90 ATPase is more tightly regulated than assumed previously. Therefore, the activation of GR and v-Src has various demands on Hsp90 biochemistry and is dependent on separate functional regions of Hsp90M. Thus, Hsp90M seems to discriminate between different substrate types and to adjust the molecular chaperone for proper substrate activation.
Heat shock protein 90 (Hsp90) is a highly conserved, abundant and constitutively expressed homodimeric molecular chaperone of the eukaryotic cytosol. It is specifically involved in the folding and conformational regulation of a limited subset of client proteins. Many natural substrates of Hsp90 are medically relevant signal transduction molecules, e.g., the nuclear receptors for steroid hormones and several kinases, some of them with oncogenic potential (19, 23, 24). To fulfill its biological function, Hsp90 cooperates with different cochaperones, such as Hop, p50, p23, Aha1, the immunophilins, and others, and acts as part of a multichaperone machine together with Hsp70.
Hsp90 is composed of a N-terminal nucleotide binding domain (Hsp90N), a middle domain (Hsp90M), and a C-terminal domain (Hsp90C) that mediates the dimerization of the protein. A hallmark of the Hsp90 reaction cycle is binding and hydrolysis of ATP (20, 21, 26, 34). Although the catalytic center for this reaction has been identified within the N-terminal domain of the protein, the interplay between this part and the other domains of Hsp90 during substrate activation is poorly understood. Emerging evidence suggests that the middle domain of Hsp90 plays an important role in this process. For example, it has been shown that Hsp90M interacts with Aha1, a cochaperone that stimulates Hsp90's rate of ATP hydrolysis and increases the efficiency of client protein activity (8, 11, 22). Moreover, communication between the middle and N-terminal domains of Hsp90 is essential in vivo (13), probably due to the role of a Hsp90M segment in the proper orientation of the γ-phosphate group of ATP for hydrolysis by the N-terminal catalytic domain (15). Furthermore, a peptide spanning 14 amino acid residues within Hsp90M has been suggested as the binding site for a natural client protein (30). Several point mutations within Hsp90M that exhibit temperature-sensitive growth defects in Saccharomyces cerevisiae have been identified by yeast genetic methods (2, 9, 19) or proposed and generated based on the recent crystal structure of this domain (15). Hence, a comprehensive analysis of these point mutations with regard to ATP hydrolysis rate, binding of cochaperones, and activation of natural Hsp90 client proteins in vivo should in turn yield information on the “normal” function of the middle domain of Hsp90 and the sites therein affected by these mutations.
We created site-directed exchanges W300A, E381K, E431K, S485Y, T525I, and the triple mutant F329A/L331A/F332A in Hsp90. The phenotypes of the respective yeast strains and the underlying biochemistry of mutant Hsp90 proteins were compared to mutations T22I and T101T that are located in the catalytic N-terminal domain and cause a general reduction of client protein activity due to their hyper- or hypoactive ATP hydrolysis rate. As a result, we find that v-Src and glucocorticoid receptor (GR), representing two major classes of Hsp90-dependent substrate proteins, are affected in different ways by mutations in the middle domain of Hsp90. This result suggests a role for Hsp90M in discriminating between various types of client proteins during their processes of activation. Moreover, we observed that the ATPase activity of the molecular chaperone Hsp90 is sensitive to segments within the middle domain of Hsp90 that are essential for GR activation but not involved in communication between Hsp90N and Hsp90M domains.
MATERIALS AND METHODS
Plasmid construction and yeast two-hybrid experiments.
Yeast Hsp90 was amplified from plasmid pTGPD/P82 (19), and the yeast cochaperones Aha1, p23 (Sba1), Hop (Sti1), and p50 (Cdc37) were amplified from wild-type yeast DNA. PCR products were inserted into the bacterial expression vector pProExHTa to generate an amino-terminal His6 sequence followed by a tobacco etch virus cleavage site. For expression in yeast cells, cochaperones were cloned into pYX122. The pYX122 vector is an autonomously replicating low-copy plasmid (1 to 5 copies per cell) for the expression of proteins under the control of the strong triose phosphate isomerase promoter. Point mutations T22I, T101I, W300A, E381K, E431K, S485Y, T525I, and the triple mutation F329A/L331A/F332A were inserted into the yeast vector pTGPD/P82 or into Hsp90 in the bacterial pProExHTa expression vector by using a QuikChange site-directed mutagenesis kit (Stratagene) with missense oligonucleotides. Desired exchanges were confirmed by DNA sequencing.
Protein purification and protein interaction assays.
Expression constructs in pProExHTa were transformed into Escherichia coli BL21(DE3)pLysS cells. Bacteria were grown at 18°C in LB medium supplemented with 100 mg/liter ampicillin and 34 mg/liter chloramphenicol to an optical density at 600 nm (OD600) of 1, and protein expression was induced for 5 h with 0.25 mM IPTG (isopropyl-β-d-thiogalactopyranoside). After harvesting, proteins were enriched from cell pellets by nickel-nitrilotriacetic acid chromatography at pH 8.0 essentially as described previously (20). Proteins were further purified by ion-exchange fast-performance liquid chromatography using ResourceQ columns or by gel filtration using Superose12 (Amersham Biosciences). For gel filtration analysis, 500-μl samples containing the indicated combinations of yeast protein Hsp90, Aha1, p23, Hop, or p50 (each at 5 μM) in 40 mM HEPES-KOH (pH 7.4), 100 mM KCl, and 2 mM MgCl2 were incubated for 10 min and separated at 4, 25, or 37°C as indicated in Fig. 3 with a Superose12 column equilibrated in the same buffer and using an ÁKTA chromatography system (Amersham Biosciences). When p23 was analyzed for interaction with Hsp90, 2 mM ATPγS was added (8). Fractions of 500 μl were collected for analysis beginning at a 6-ml elution volume. To quantify cochaperone binding, Coomassie-stained sodium dodecyl sulfate (SDS) gels were analyzed with a Bio-Rad ChemiDoc XRS system, and the binding of p23 or Aha1 to Hsp90 was determined as the percent ratio between cochaperone and Hsp90 in fractions 6 to 11.
FIG. 3.
Quantification of Hsp90 middle domain mutants binding to Aha1 and p23 at 4, 25, and 37°C. Protein complexes of p23 or Aha1 with Hsp90 wild-type and mutant proteins were formed and fractionated by gel filtration chromatography at different temperatures as indicated. Quantification of Coomassie-stained SDS gels was performed using a Bio-Rad ChemiDoc XRS system with the Quantity One software package. Binding activity was determined as the percent ratio of bound p23 or Aha1 versus Hsp90 protein in fractions 6 to 11. One representative gel per experiment is shown. For quantification, experiments were repeated three times, and the error bars show the standard deviations for the three experiments. (A) Quantification of p23 binding to Hsp90 wild type (WT) and Hsp90 mutants. (B) Quantification of Aha1 binding to Hsp90 wild type and Hsp90 mutants.
Assay of Hsp90 ATPase activity.
Recombinant wild-type or mutant Hsp90 proteins (5 μM) were incubated at 25 and at 37°C with or without the addition of Aha1 (5 μM) in 40 mM HEPES-KOH (pH 7.4) and 2 mM MgCl2 supplemented with 2 mM [α-32P]ATP (4 μCi/mM; Amersham Biosciences) for 15 min in a final volume of 20 μl. Aliquots of each reaction were stopped at time points by the addition of 5 mM EDTA and freezing in liquid nitrogen (20). The separation of ADP from ATP was achieved by thin-layer chromatography on polyethyleneimine-cellulose sheets (Merck) in 0.5 M LiCl and 0.5 M formic acid. ATPase activity was monitored by quantitation of [α-32P]ADP by using a liquid scintillation counter. Steady-state ATPase rates were calculated from the linear range of the reactions, and the ATP hydrolysis activity of 5 μM wild-type Hsp90 at 25°C was set to 100%. Inhibition of the ATPase activity by geldanamycin ensured the specificity of the measurement (20).
Yeast methods and manipulation.
Yeast strain ΔPCLDa/α (19) was used throughout this study, and standard methods for growth and transformation were employed. Cells were cultured on yeast extract-peptone-dextrose (YPD) medium or on SD, SRaf, or SGal selective minimal medium (0.67% yeast nitrogen base supplemented with 2% glucose, raffinose or galactose, respectively, and with nucleotides and amino acids depending on auxotrophy). Intrinsic wild-type Hsp90 of ΔPCLDa/α on the URA3-containing pKAT6 plasmid was replaced by a Hsp90 mutant gene or by wild-type Hsp90 on the expression vector pTGPD/P82 (19) by the plasmid shuffling technique as described previously (20, 32).
Expression and detection of v-Src in yeast cells.
After plasmid shuffling, yeast strains were transformed with Y316v-Src. Cells were selected on SD medium lacking Ura (SD-Ura medium), grown overnight in SRaf-Ura medium, diluted to an OD600 of 0.2, and grown on SGal-Ura medium for 6 h at 25°C to induce v-Src expression. For preparation of cell lysates, cultures were collected by centrifugation, resuspended in lysis buffer (8 M urea, 5% [wt/vol] sodium dodecyl sulfate, 40 mM Tris-HCl [pH 7.5], 0.1 mM EDTA) (28) and treated by ultrasonification for 5 s on ice. For immunoblotting, antibodies 4G10 (Upstate) and EC10 (Upstate) were used to detect phosphotyrosine residues and v-Src protein, respectively. Duplicates of samples were run on Coomassie-stained gels and served as loading controls. An ECL reagent kit (Amersham Biosciences) was used to visualize immunocomplexes. Quantification of blots and gels was performed with a Bio-Rad ChemiDoc XRS system using the Quantity One software package, and immunosignals were normalized to the protein content of the loading control.
Determination of GR activity in yeast cells.
For analysis of GR activity, yeast cells were transformed with the GR-expressing plasmid p2HGal/GR/CYC and the reporter plasmid pSX26.1 expressing β-galactosidase under the control of GR response elements (19). Cells were grown on SRaf-His-Ura medium, and GR synthesis was induced by shifting cultures to SGal-His-Ura medium. Receptors were activated at an OD600 of 0.2 by the addition of 10 μΜ deoxycorticosterone (DOC) for 1 h, cells were collected by centrifugation, and β-galactosidase activity was measured using an GalactoStar kit (Tropix) and normalized to the protein concentration of cell lysate that had been determined by the Bio-Rad protein assay kit. The mouse monoclonal antibody BuGR2 (Affinity Bioreagents) was used to detect levels of GR in yeast cell lysates, and quantification was carried out as described for v-Src.
RESULTS
Point mutations in the middle domain of Hsp90 confer growth defects.
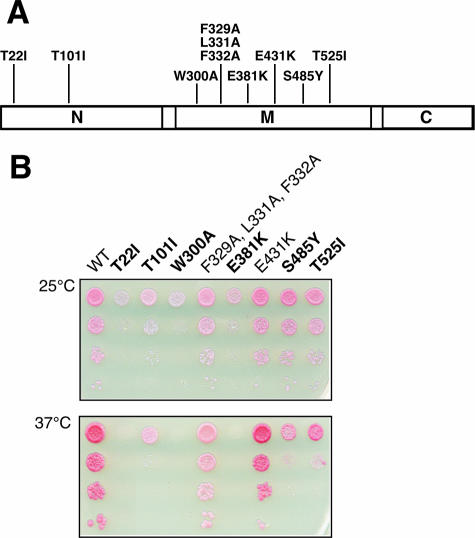
Missense mutations W300A, E381K, E431K, S485Y, and T525I in the middle domain of Hsp90 (Fig. 1A) have been identified by mutational analysis or deduced from structural data (2, 9, 15, 19) and show temperature-sensitive growth defects. Moreover, we generated the triple point mutation F329A/L331A/F332A because the potential of a hydrophobic patch consisting of amino acids F329, L331, and F332 has been recently discussed in the literature with regard to its impact on client protein activity (1, 15). For E431K and T525I, variations in the ability to activate different subclasses of steroid receptors (glucocorticoid receptor, progesterone receptor, estrogen receptor, and mineralocorticoid receptor) have been reported (2). However, with the exception of E381K (19), the consequences of the variety of Hsp90M mutations in handling different types or families of client proteins, such as v-Src, a kinase, or GR, a steroid receptor, have not been comparatively analyzed. Interestingly, GR itself, when bound as a substrate to Hsp90, can stimulate the ATPase activity of the molecular chaperone (14).
FIG. 1.
Point mutations generated along the Hsp90 sequence and resulting phenotypes for S. cerevisiae. (A) Schematic representation of the domain organization of Hsp90. Positions of amino acid substitutions refer to the sequence of yeast Hsc82. (B) Viability of mutant yeast strains at 25 and 37°C. Shuffled cells were grown overnight in YPD medium, adjusted to 1 × 108 cells per ml, and 10-fold serially diluted. Two-microliter portions were spotted onto YPD agar plates and incubated for 72 h at 25°C or for 48 h at 37°C. Boldface type indicates cells with temperature-sensitive growth defects. WT, wild type.
Thus, we decided to compare exchanges in Hsp90M to wild-type Hsp90 and to T22I and T101I, which are mutated in the N-terminal catalytic domain of Hsp90, exhibit a temperature-sensitive phenotype, and are reduced in the activation of heterologous client proteins v-Src and GR (19), probably due to their hyper- or hypoactive intrinsic ATP hydrolysis rate (25). Point mutations in Hsp90 were purified as recombinant proteins for biochemical analysis or introduced into yeast cells by plasmid shuffling to generate a system for in vivo analysis. Phenotypes of the resulting strains are depicted at 25 and 37°C and cells with temperature-sensitive growth defects are indicated in Fig. 1B.
With respect to cell growth, client protein activation, or ATPase activity, the F329A/L331A/F332A triple mutant behaved like the F332Q exchange that had been created to introduce a polar amino acid into that hydrophobic patch (15). Hence, it is unlikely that this hydrophobic patch plays a central role in substrate binding, in agreement with previous results (15).
Intrinsic ATP hydrolysis rate of Hsp90 point mutants and stimulation of their ATPase activity by Aha1.
Defects of T22I and T101I mutations in the N-terminal catalytic domain of Hsp90 result from its ATPase hyper- and hypoactivity (Table 1) (25, 33). In addition, R380A and Q384A point mutations that affect residues required for orientation of the γ-phosphate of ATP for nucleophilic attack by the catalytic center in the N-terminal domain of Hsp90 were identified in the middle domain of Hsp90 (15). Therefore, mutations of residues of the catalytic domain or those involved in communication between the Hsp90N and the Hsp90M domain led to decreased ATPase activity and compromised client protein activation (15). However, other segments of Hsp90M are not known to affect the ATPase activity of the molecular chaperone (15). To analyze Hsp90M further, we purified the Hsp90 point mutants indicated above and tested their intrinsic ATP hydrolysis rate as well as their susceptibility to Aha1 stimulation at 25 and at 37°C (Table 1). T22I or T101I, serving as a control, showed increased or decreased intrinsic ATPase activities, respectively (Table 1) (25, 33), and the T22I mutant displayed a reduced response to Aha1-dependent ATPase stimulation, as reported recently (33). In contrast, W300A, F329A/L331A/F332A, and E381K exhibited no striking difference from wild-type Hsp90, with ATPase stimulation by Aha1 being slightly decreased for the W300A and E381K mutants (Table 1) (11). The mutations E431K, S485Y, and T525I that map outside the contact region between the N-terminal and the middle domain of Hsp90 showed compromised ATP hydrolysis rate at 25 and at 37°C, an observation that had not been expected (15). The mutant E431K was stimulated by Aha1 to wild-type ATPase activity at 25 and at 37°C (Table 1). In contrast, Aha1 stimulated S485Y and T525I mutants up to the level of nonstimulated wild-type Hsp90 at 25°C. At 37°C, however, Aha1 exerted no stimulation on the low intrinsic ATPase activity of S485Y and T525I mutants (Table 1).
TABLE 1.
Relative ATPase activity of Hsp90 mutantsa
| Construct | ATPase activity at 25°C
|
ATPase activity at 37°C
|
||||
|---|---|---|---|---|---|---|
| Unstimulated | Aha1 stimulated | Fold stimulation | Unstimulated | Aha1 stimulated | Fold stimulation | |
| Wild type | 100 | 557 | 5.6 | 369 | 1,456 | 3.9 |
| T22I | 702 | 1,552 | 2.2 | 1,436 | 1,998 | 1.4 |
| T101I | 30 | 124 | 4.1 | 56 | 147 | 2.6 |
| W300A | 105 | 403 | 3.8 | 365 | 1,002 | 2.7 |
| F329A/L331A/F332A | 89 | 514 | 5.8 | 398 | 1,374 | 3.5 |
| E381K | 97 | 428 | 4.4 | 312 | 855 | 2.7 |
| E431K | 42 | 619 | 14.7 | 129 | 1,450 | 11.2 |
| S485Y | 31 | 122 | 3.9 | 121 | 104 | 0.9 |
| T525I | 43 | 133 | 3.1 | 132 | 176 | 1.3 |
Values are given as percentages of results for the wild type.
Mutations W300A, S485Y, and T525I in the middle domain of Hsp90 impair its interactions with the cochaperone p23 or Aha1.
Functional defects might be an intrinsic property of the respective Hsp90 mutant or may result from compromised interaction between Hsp90 and a particular cochaperone. To investigate whether mutations in the middle domain impair binding of Hsp90 to cochaperones, we used purified Hsp90 and the cochaperones p23, Aha1, Hop, and p50 to test their interaction by gel filtration chromatography (8). Hop and p50 use the C-terminal or N-terminal domain of Hsp90 for binding and, hence, no difference was detected compared to the wild type when their interaction with the various Hsp90M mutants was tested (data not shown). Binding sites for p23 and Aha1 overlap on the middle domain of the Hsp90 chaperone, as shown recently by crystal structure analysis of full-length Hsp90 and by nuclear magnetic resonance analysis of Hsp90M (1, 12). When we probed p23 for interaction with Hsp90M mutants, binding to W300A appeared almost like that of the wild type. The affinity of S485Y and T525I to p23 was strongly reduced compared to other mutants or to wild-type Hsp90 (Fig. 2). This observation is supported by the finding that immobilized p23 cannot pull out Hsp90 from S485Y and T525I mutant strains (5). When the interaction to Aha1 was tested, W300A, S485Y, and T525I exhibited reduced binding capacity (Fig. 2). To analyze more rigorously the effects of Hsp90 mutants on cochaperone binding, we performed a series of gel filtration experiments at 4, 25, and 37°C. As a measure for binding activity, the percent ratio of p23 and Aha1 shifted with Hsp90 in fractions 6 to 11 was quantified at each temperature (Fig. 3). Compared to wild-type Hsp90, binding of p23 was reduced to about 4/5 for W300A and to about 1/10 for T525I, and binding was negligible for S485Y (Fig. 3A) at each temperature. Binding of Aha1 to W300A was modestly reduced compared to wild-type Hsp90 at 25 and 37°C, consistent with structural predictions (16), probably accounting for the lower stimulation of W300A ATPase activity by Aha1 (Fig. 3B). The S485Y and T525I exchanges further reduced binding of Aha1 to Hsp90 to about 1/5 to 1/10 of that of the wild type, depending on the mutation (Fig. 3B), which may explain the decreased stimulation of ATPase activity of these mutants by Aha1 at 25°C (Table 1). However, binding of Aha1 to S485Y and T525I mutants is not further compromised by raising the temperature to 37°C (Fig. 3B). Thus, failure of ATPase stimulation most probably results from a temperature-dependent defect of the Hsp90 protein itself at 37°C rather than from impaired binding of Aha1. Despite the lower cochaperone binding strength of S485Y and T525I yeast strains, their growth was not impaired at 25°C, and growth defects could be detected only at the elevated temperature of 37°C. W300A showed a dramatic growth defect even at 25°C and was not viable at 37°C (Fig. 1B). As ATPase activity and stimulation by Aha1 is only mildly affected, impaired cochaperone binding seems to be only a minor contribution to the defects of this mutant but does not account for its major defects. Therefore, proper binding of p23 and Aha1 appears to be not a general requirement for cell viability but might be important for specific functions of Hsp90. This view is supported by the fact that deletions of p23, Aha1, and Hop are tolerated by yeast cells under normal conditions (5, 7, 11).
FIG. 2.
Interaction of p23 and Aha1 with middle domain mutants of Hsp90. Purified p23 (left panel) and Aha1 (right panel) as well as wild-type (WT) and mutant Hsp90 proteins were incubated as indicated and fractionated by gel filtration chromatography on a Superose12 column. Fractions were analyzed by SDS-polyacrylamide gel electrophoresis. Marker proteins are shown on top (thyroglobulin, 669 kDa; bovine serum albumin, 67 kDa). In the case of p23, a negative control in the absence of p23 (w/o) is included.
Cellular activity of the heterologous client protein v-Src is highly sensitive to the W300A mutation in the middle domain of Hsp90.
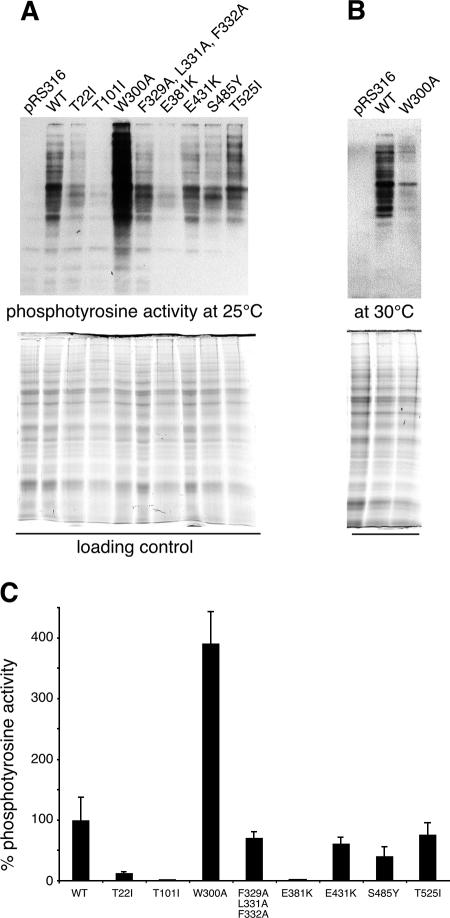
Mutations T22I and T101I in Hsp90N interfere with the ATPase cycle of the molecular chaperone, resulting in a general loss of Hsp90-dependent client protein activity that affects GR and v-Src equally (19, 25). For some point mutations mapping to the middle domain of Hsp90, the activity of either GR or v-Src has been analyzed (2, 9, 19). However, no systematic approach has been applied to elucidate whether these mutations affect both client proteins equally or whether differences between GR and v-Src activation arise. Thus, we tested W300A, F329A/L331A/F332A, E381K, E431K, S485Y, and T525I at 25°C for their ability to activate v-Src and GR and to compare their defects in activating both kinds of client proteins. v-Src activity was low in T22I and T101I strains and in the E381K mutant as reported previously (19), whereas the v-Src activity of the triple mutant F329A/L331A/F332A, as well as of E431K, S485Y, and T525I, was only moderately affected in vivo (Fig. 4). Surprisingly, the W300A mutant was hyperactive in v-Src activation (Fig. 4A). In contrast, the W300A mutation led to minor activity of the client protein v-Src when grown at 30°C (Fig. 4B) (15). Hence, the W300A exchange results in a temperature-sensitive functional frailty of a segment of the Hsp90 middle domain that is very critical for v-Src activation.
FIG. 4.
Point mutations in the sequence of the middle domain of Hsp90 interfere with the activation of the Hsp90-dependent client proteins v-Src. (A) Cell lysates were prepared from normal or mutant ΔPCLDa/α strains expressing v-Src or from empty vector control cells (pRS316) grown at 25°C as described in Materials and Methods. Proteins were separated by polyacrylamide gel electrophoresis on 7.5% gels and transferred to nitrocellulose membranes. Phosphotyrosine activity was monitored by Western blot analysis with antibody 4G10. Quantification of protein load was performed with a Bio-Rad ChemiDoc XRS system as described in Materials and Methods and used for normalization of phosphotyrosine activity. One representative blot and gel out of three independent experiments are shown. (B) Phosphotyrosine activity of control cells (pRS316) and of wild-type and W300A cells grown at 30°C. (C) Quantification of v-Src-dependent phosphotyrosine levels from the Western blots shown in panel A. Error bars show the standard deviations from the means for the three experiments. The activity of wild-type (WT) cells was set to 100%.
Cellular activity of heterologous Hsp90 client proteins GR and v-Src responds differently to mutations in the middle domain of the molecular chaperone.
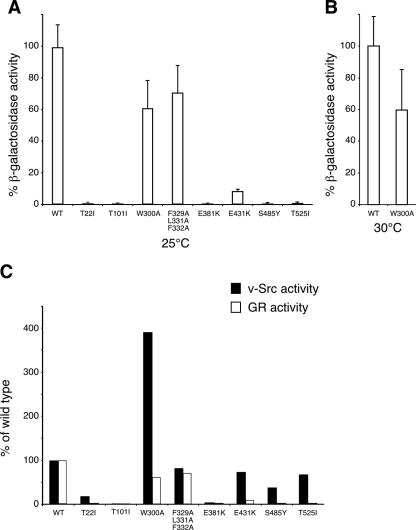
When GR was tested, a dramatic loss of activity was observed for T22I and T101I, similar to the effect of these mutants on v-Src activity (Fig. 5A). In contrast to its effect on v-Src activity, the W300A exchange had only a minor effect on GR at 25 and at 30°C and still exhibited ∼60% of wild-type activity (Fig. 5A and B) at both temperatures. The triple mutant F329A/L331A/F332A affected neither v-Src activity nor GR activity significantly. This result suggests that loss of hydrophobicity in this patch has no major impact on client protein activity and, as substrate proteins enter Hsp90 in a prefolded, close-to-native state, they might not primarily bind to exposed hydrophobic side chains of Hsp90 (35). In contrast, the E381K mutation resulted in negligible activity for kinase and hormone receptor activity (Fig. 5C). E431K, S485Y, and T525I mutations are outside the contact region between Hsp90N and Hsp90M (15). The three mutations have a moderate effect on v-Src activity, lowering it to ∼50 to 75% of that of the wild type (Fig. 4C and 5C), but show a massive impairment of GR activity (Fig. 5A and C). Taken together, these results indicate that W300A strongly affects cellular activity of v-Src kinase but compromises GR hormone receptor activity only moderately (Fig. 5C). The E431K, S485Y, and T525I mutants, however, showed minor effects on v-Src activity but a dramatic loss of GR activity (Fig. 5C). From these results, it is reasonable to conclude that substrate activation of the two types of Hsp90-dependent client proteins, v-Src and GR, has different demands on the biochemistry of the molecular chaperone. Thus, Hsp90 may use individual mechanistic routes to activate the various kinds of client proteins, as demonstrated for the examples of v-Src and GR in this study.
FIG. 5.
GR and v-Src activity in wild-type and mutant Hsp90 yeast strains. Mutant and wild-type strains were cotransformed with GR and the reporter plasmid pSX26.1 containing β-galactosidase under the control of GR response elements. GR was activated by the addition of 10 μM DOC as described in Materials and Methods. GR-dependent β-galactosidase activity was measured using a GalactoStar kit (Tropix) and normalized to the protein concentration of the lysate. Activities are averages from at least five independent experiments, and error bars are indicated. The background activity of expressed but not DOC-activated GR was subtracted, resulting in lower specific activities than those reported in previous studies (19). (A) GR activity in wild-type and indicated mutant strains grown at 25°C. (B) GR activity in wild-type and W300A strains measured at 30°C. (C) v-Src and GR activity measured for wild-type and mutant yeast strains at 25°C. Activity of the wild-type (WT) strain was set to 100%.
Mutations in the middle domain of Hsp90 affect steady-state levels of v-Src and GR differently.
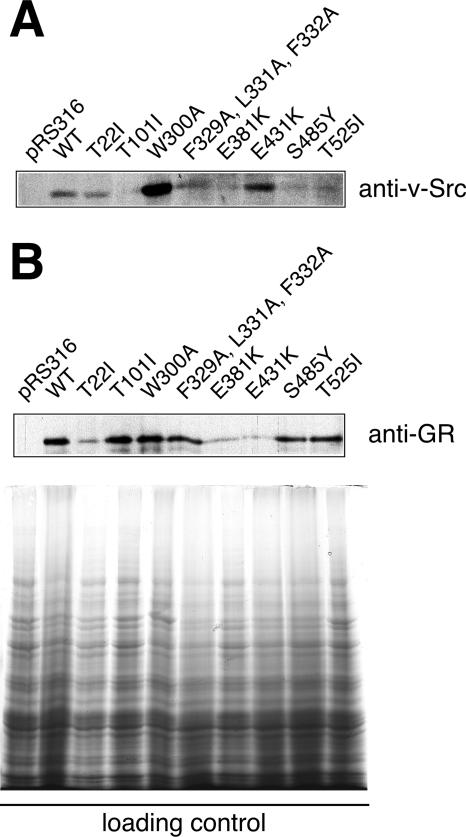
In order to examine different client protein handling by Hsp90 further, steady-state levels of client proteins v-Src and GR were monitored and quantified in yeast cells by Western blotting using specific antibodies (Fig. 6; Table 2) (19, 28). When v-Src accumulation was analyzed, only F329A/L331A/F332A exhibited near wild-type levels, while in most other mutants, v-Src was reduced (Fig. 6A). However, a dramatic increase of v-Src levels was detected in the W300A mutant, which seems to account for some of the properties of this strain (Fig. 6A). The accumulation of GR reached wild-type levels for most mutants, with the exception of T22I (19) and the middle domain mutants E381K and E431K (Fig. 6B). Interestingly, most Hsp90M mutations affected steady-state levels of v-Src and GR differently. For example, v-Src accumulation in T101I, E381K, S485Y, and T525I strains was lower than in wild-type cells but tremendously elevated in the W300A strain at 25°C (Fig. 6A). Conversely, GR accumulation was low in T22I, E381K, and E431K strains (Fig. 6B). To evaluate whether cellular protein levels or specific activity of the substrate are mainly affected by the various Hsp90 mutants, we determined specific client protein activities as a quotient of cellular activity (as measured by phosphotyrosine or β-galactosidase activity) and cellular levels of protein accumulation (Table 2). T101 and E381K reduce cellular activity as well as specific activity of v-Src and GR to similar levels, suggesting that v-Src and GR are equally dependent on general Hsp90 function (Table 2). The specific activities of GR and v-Src respond similarly to W300A, whereas this mutant leads to a 10-fold accumulation of v-Src protein compared to wild-type cells and hence enhanced cellular kinase activity. Obviously, W300A increases cellular activity of v-Src basically by enhancing the accumulation of the protein, while it does not affect steady-state protein levels of GR. E431K is reduced by GR accumulation but confers wild-type-like specific activity to GR. S485Y and T525I mutants affect v-Src and GR differently. Although the accumulation of v-Src is decreased, the specific activity of v-Src is even higher in both strains than in the wild type. On the other hand, S485Y and T525I mutations lead to GR accumulation at almost wild-type levels, but the receptor fails to attain activity. This suggests that v-Src and GR are dependent on different specific biochemical properties located along the middle domain of Hsp90. Moreover, our study shows that E431K on one hand and S485Y and T525I exchanges on the other hand have different consequences on the biochemistry of Hsp90, a result which can explain the observation that E431K specifically impairs GR activity, whereas S485Y and T525I affect all steroid receptors (2). Hence, it is tempting to speculate that the basic biochemical requirements for hormone receptor activation are located within an area covered from S485 through T525 on the Hsp90 protein.
FIG. 6.
Steady-state expression levels of Hsp90-dependent client proteins v-Src and GR in wild-type and mutant yeast strains. Quantification was performed with a Bio-Rad ChemiDoc XRS system using the Quantity One software package and normalized to the protein content of loading controls. A typical blot for v-Src and GR levels is shown, and experiments were done in triplicate for quantification. Data were used to calculate specific activity of v-Src and GR as presented in Table 2. (A) Cell lysates from v-Src expressing yeast (same as in Fig. 4A, see that figure legend for loading control) were blotted for v-Src protein levels using the specific monoclonal antibody EC10. (B) Cell lysates from GR-expressing yeast were blotted for GR protein levels using the specific monoclonal antibody BuGR2. Loading controls are shown on the bottom. WT, wild type.
TABLE 2.
Activity and accumulation of v-Src and GR in mutant Hsp90 yeast strainsa
| Yeast strain | Result for:
|
|||||
|---|---|---|---|---|---|---|
| v-Src
|
GR
|
|||||
| Cellular activity | Accumulation | Sp act | Cellular activity | Accumulation | Sp act | |
| Wild-type | 100 | 100 | 100 | 100 | 100 | 100 |
| T22I | 11.6 | 83.5 ± 11.9 | 13.9 | 1.1 | 11.7 ± 1.0 | 9.4 |
| T101I | 0.3 | 23.8 ± 9.4 | 1.3 | 0.2 | 115.6 ± 21.0 | 0.2 |
| W300A | 389.9 | 926.1 ± 84.0 | 42.1 | 61.4 | 118.0 ± 26.2 | 52.0 |
| F329A/L331A/F332A | 67.7 | 94.9 ± 8.4 | 71.3 | 71.4 | 77.9 ± 29.7 | 91.7 |
| E381K | 1.6 | 20.5 ± 2.8 | 7.8 | 0.8 | 10.9 ± 2.8 | 7.3 |
| E431K | 60.8 | 149.7 ± 52.7 | 40.6 | 9.1 | 7.9 ± 2.6 | 115.2 |
| S485Y | 40.4 | 26.6 ± 4.8 | 151.9 | 0.3 | 77.3 ± 13.0 | 0.4 |
| T525I | 76.8 | 28.7 ± 18.9 | 267.6 | 1.6 | 102.7 ± 15.6 | 1.6 |
Values are given as percentages of results for the wild type.
Compromised GR activity in S485Y and T525I mutants can be rescued in part by overexpression of p23.
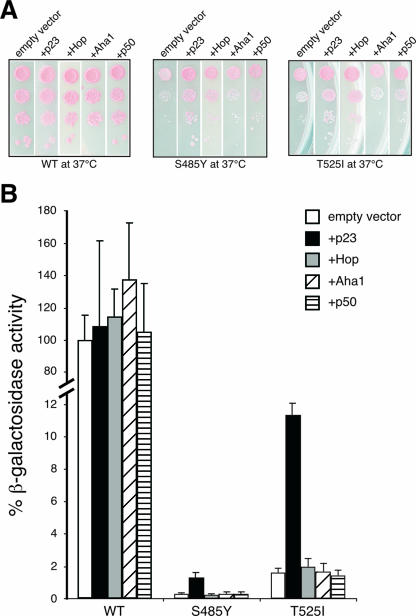
The activity of v-Src and GR is sensitive to Hsp90 mutations W300A or S485Y and T525I, respectively (Fig. 5C), and the yeast mutants exhibit temperature-dependent growth defects (Fig. 1B). We therefore asked whether these defects can be rescued by the overexpression of Hsp90 cochaperones. As none of the cochaperones, Aha1, Hop, p50, or p23, could rescue the W300A mutant strain (data not shown), the deficiencies caused by this exchange seem to originate from intrinsic failures. In contrast, the overexpression of p23 and Hop, but not of Aha1 or p50, in S485Y and T525I partially rescued their growth defects at 37°C (Fig. 7A). When these two mutants were assessed for rescue of GR activity, p23 but neither Hop, Aha1, nor p50 overexpression led to a severalfold increase of β-galactosidase reporter activity; GR activity was never fully restored but reached about 12% of wild-type levels in the T525I strain (Fig. 7B). This finding supports the view that S485Y and T525I compromise Hsp90 biochemistry in more than p23 binding and stresses the specific roles of the segment encompassing S485 to T525 and of the cochaperone p23 in the assembly of Hsp90 complexes with GR (4, 10, 17, 18).
FIG. 7.
Effect of cochaperone overexpression on the viability and client protein activity of Hsp90 mutant strains S485Y and T525I. (A) S485Y and T525I strains were cotransformed with empty vector serving as a control or p23, Hop, Aha1, and p50 expression plasmids. Pictures were taken from different plates that had been incubated together at 37°C. (B) GR activity measured for wild-type and mutant yeast strains cotransformed with empty vector, p23, Hop, Aha1, or p50 expression plasmids. Activity of the wild-type (WT) strain was set to 100%.
DISCUSSION
Hsp90 has been established as a nucleotide-dependent chaperone, and the mechanism of ATP hydrolysis on the molecular level has been resolved (15, 20, 21, 26, 29, 34). Yet, how energy created by the hydrolysis of ATP within the N-terminal domain of Hsp90 is used for a conformational change within the client protein to reach its native state remains enigmatic. Approaches using classical chaperone substrates like firefly luciferase, rhodanese, or citrate synthase suggested binding of the substrate protein to the N- and C-terminal domains of Hsp90 (31, 36). Recent studies, however, using natural substrate proteins of Hsp90 propose instead that the middle domain of the molecular chaperone plays a role in client protein binding (6, 30). Therefore, Hsp90M may have a central position in substrate recognition, distinguish between different kinds of substrates, and adjust the molecular chaperone to the demands for activation of a specific client protein. To gain insight into the function of Hsp90M and to figure out whether activation of all kinds of substrates follows a general route or whether different client proteins have distinct demands on specific Hsp90 functions, we used a series of Hsp90 point mutations. For analysis, we focused on the activity of Hsp90-dependent substrates v-Src and GR in vivo, as well as on underlying functional properties, such as ATPase activity and the interaction with Hsp90 cochaperones. Some constraints of Hsp90 biochemistry affected v-Src and GR activity equally. The mutation T101I in the N-terminal catalytic domain that interferes with the ATP hydrolysis cycle of Hsp90 (27) and the E381K mutation in the middle domain both result in a general impairment of client protein activity in the cell. Some other mutations in the middle domain of Hsp90, however, showed diverse biochemical deficits, had different capacities to activate v-Src and GR, and seem therefore to be capable of discriminating between Hsp90-dependent client proteins.
W300A is located adjacent to a region (amino acids 327 to 340) that was proposed to serve as the binding region for kinase substrates (30) and forms part of a conformationally flexible loop with an amphipathic structure (15). On the atomic level, W300 makes a hydrophobic interaction with a “shallow hydrophobic recess on the outer face of Aha1” (16), which may explain the observation that the W300A exchange slightly decreases the affinity of Hsp90 to this cochaperone. Our results show that cellular v-Src activity is highly sensitive to this point mutation at 25 and at 30°C, whereas GR activity was only barely affected by the W300A exchange. Since ATPase activity as well as binding to and stimulation by Aha1 were only a little affected by the W300A mutation, other aspects of Hsp90 biochemistry that affect v-Src activation may be altered. When we analyzed steady-state levels of client proteins in the yeast cytosol, it turned out that v-Src but not GR accumulation was conspicuously elevated in W300A cells, while this mutation affected the specific activity of both client proteins similarly. As W300 is in close neighborhood to the loop for kinase binding but not a part of this segment (30), it seems that W300 might be critical for the control of v-Src loading on Hsp90 or for v-Src release from Hsp90. Although higher accumulation of v-Src results in a higher phosphotyrosine activity at 25°C in the W300A strain, such a charge may block the Hsp90 chaperone system for other substrate proteins (3). This effect might gain the upper hand at 30°C, decrease the already low vitality of W300A cells and, as a consequence, lead to a loss of v-Src activity.
Exchanges S485Y and T525I resulted in moderately abridged cellular v-Src activity but disturbed cellular GR activity almost completely, strongly reduced binding of Hsp90 to p23 and Aha1, and harmed the ATPase activity of the molecular chaperone. While the specific activity of GR in these mutant strains was reduced to ∼1%, the specific activity of v-Src was even higher than in the Hsp90 wild-type strain. S485Y and T525I map to the small αβα middle-segment domain encompassing residues 435 to 525 of Hsp90's middle domain that interacts via ion pairs with Aha1 (1, 15, 16). Although not directly involved in these interactions, mutations S485Y and T525I interfere with Hsp90 binding to Aha1 either by altering the local conformation of this segment or by adding hydrophobicity to the region and thus obstructing the formation of polar contacts. Ali et al. (1) report in their recent atomic structure of full-length Hsp90 that “the interface between the C domain and the small middle-segment domain flexes on going from the unconstrained M-C structure to the ATP-bound conformation, bringing the small middle domains about 10 Å closer together. This displaces the projecting helix-strand segments, which tilt downwards and become less well ordered.” Since cochaperone binding to Hsp90 and Hsp90 ATPase activity are affected by the S485Y and T525I mutations, the conformational change described above could be an additional mode to regulate Hsp90 activity. Therefore, our biochemical data are complementary to the structural data and identify the small αβα middle domain as part of a structural element with impact on the ATPase reaction cycle of the molecular chaperone. However, it remains to be elucidated by molecular dynamics studies how the movement of the αβα middle-segment domains mechanistically affects the ATPase reaction, which is controlled by the N-terminal domain together with an additional catalytic loop in the large middle segment (15).
We have shown that v-Src and GR have different demands on Hsp90 biochemistry and that their activity is affected differently by point mutations in the middle domain of Hsp90. Thus, it is plausible to suggest that the working mechanism of Hsp90 is not a unique process but is dependent on the type of substrate protein to be processed. As different kinds of cochaperones contribute to the efficiency of substrate activation, they might regulate the Hsp90 core chaperone for tuning different intrinsic activation programs for substrate proteins that can be executed by the molecular chaperone.
Acknowledgments
We thank S. Lindquist (Whitehead Institute, Cambridge, Mass.) for plasmids pTGPD/P82, p2HGal/GR/CYC, pSX26.1, and Y316v-Src.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 284/project Z3).
Footnotes
Published ahead of print on 18 September 2006.
REFERENCES
- 1.Ali, M. M. U., S. M. Roe, C. K. Vaughan, P. Meyer, B. Panaretou, P. W. Piper, C. Prodromou, and L. H. Pearl. 2006. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature 440:1013-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohen, S. P., and K. R. Yamamoto. 1993. Isolation of Hsp90 mutants by screening for decreased steroid receptor function. Proc. Natl. Acad. Sci. USA 90:11424-11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brychzy, A., T. Rein, K. F. Winklhofer, F. U. Hartl, J. C. Young, and W. M. J. Obermann. 2003. Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. EMBO J. 22:3613-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dittmar, K. D., D. R. Demady, L. F. Stancato, P. Krishan, and W. B. Pratt. 1997. Folding of the glucocorticoid receptor by heat shock protein (hsp) 90-based chaperone machinery. J. Biol. Chem. 272:21213-21220. [DOI] [PubMed] [Google Scholar]
- 5.Fang, Y. F., A. E. Fliss, J. Rao, and A. J. Caplan. 1998. Sba1 encodes a yeast Hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol. Cell. Biol. 18:3727-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fontana, J., D. Fulton, Y. Chen, T. A. Fairchild, T. J. McCabe, N. Fujita, T. Tsuruo, and W. C. Sessa. 2002. Domain mapping studies reveal that the M domain of hsp90 serves as a molecular scaffold to regulate Akt-dependent phosphorylation of endothelial nitric oxide synthase and NO release. Circ. Res. 90:866-873. [DOI] [PubMed] [Google Scholar]
- 7.Gerber, M. R., A. Farrell, R. J. Deshaies, I. Herskowitz, and D. O. Morgan. 1995. Cdc37 is required for association of the protein kinase Cdc28 with G1 and mitotic cyclins. Proc. Natl. Acad. Sci. USA 92:4651-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harst, A., H. Lin, and W. M. J. Obermann. 2005. Aha1 competes with Hop, p50 and p23 for binding to the molecular chaperone Hsp90 and contributes to kinase and hormone receptor activation. Biochem. J. 387:789-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimura, Y., S. Matsumoto, and I. Yahara. 1994. Temperature-sensitive mutants of hsp82 of the budding yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 242:517-527. [DOI] [PubMed] [Google Scholar]
- 10.Kovacs, J. J., P. J. Murphy, S. Gaillard, X. Zhao, J. T. Wu, C. V. Nicchitta, M. Yoshida, D. O. Toft, W. B. Pratt, and T. P. Yao. 2005. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol. Cell 18:601-607. [DOI] [PubMed] [Google Scholar]
- 11.Lotz, G. P., H. Lin, A. Harst, and W. M. J. Obermann. 2003. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation and stimulates the ATPase activity of the molecular chaperone. J. Biol. Chem. 278:17228-17235. [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Yamout, M. A., R. P. Venkitakrishnan, N. E. Preece, G. Kroon, P. E. Wright, and H. J. Dyson. 2006. Localization of sites of interaction between p23 and Hsp90 in solution. J. Biol. Chem. 281:14457-14464. [DOI] [PubMed] [Google Scholar]
- 13.Matsumoto, S., E. Tanaka, T. K. Nemoto, T. Ono, T. Takagi, J. Imai, Y. Kimura, I. Yahara, T. Kobayakawa, T. Ayuse, K. Oi, and A. Mizuno. 2002. Interaction between the N-terminal and middle regions is essential for the in vivo function of HSP90 molecular chaperone. J. Biol. Chem. 277:34959-34966. [DOI] [PubMed] [Google Scholar]
- 14.McLaughlin, S. H., H. W. Smith, and S. E. Jackson. 2002. Stimulation of the weak ATPase activity of human hsp90 by a client protein. J. Mol. Biol. 315:787-798. [DOI] [PubMed] [Google Scholar]
- 15.Meyer, P., C. Prodromou, B. Hu, C. Vaughan, S. M. Roe, B. Panaretou, P. W. Piper, and L. H. Pearl. 2003. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol. Cell 11:647-658. [DOI] [PubMed] [Google Scholar]
- 16.Meyer, P., C. Prodromou, C. Liao, B. Hu, S. M. Roe, C. K. Vaughan, I. Vlasic, B. Panaretou, P. W. Piper, and L. H. Pearl. 2004. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery EMBO J. 23:511-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morishima, Y., K. C. Kanelakis, P. J. M. Murphy, E. R. Lowe, G. J. Jenkins, Y. Osawa, R. K. Sunahara, and W. B. Pratt. 2003. The hsp90 cochaperone p23 is the limiting component of the multiprotein hsp90/hsp70-based chaperone system in vivo where it acts to stabilize the client protein · hsp90 complex. J. Biol. Chem. 278:48754-48763. [DOI] [PubMed] [Google Scholar]
- 18.Murphy, P. J., Y. Morishima, J. J. Kovacs, T. P. Yao, and W. B. Pratt. 2005. Regulation of the dynamics of hsp90 action on the glucocorticoid receptor by acetylation/deacetylation of the chaperone. J. Biol. Chem. 280:33792-33799. [DOI] [PubMed] [Google Scholar]
- 19.Nathan, D. F., and S. Lindquist. 1995. Mutational analysis of hsp90 function: interactions with a steroid receptor and a protein kinase. Mol. Cell. Biol. 15:3917-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obermann, W. M., H. Sondermann, A. A. Russo, N. P. Pavletich, and F. U. Hartl. 1998. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J. Cell Biol. 143:901-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panaretou, B., C. Prodromou, S. M. Roe, R. O'Brien, J. E. Ladbury, P. W. Piper, and L. H. Pearl. 1998. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 17:4829-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panaretou, B., G. Siligardi, P. Meyer, A. Maloney, J. K. Sullivan, S. Singh, S. H. Millson, P. A. Clarke, S. Naaby-Hansen, R. Stein, R. Cramer, M. Mollapour, P. Workman, P. W. Piper, L. H. Pearl, and C. Prodromou. 2002. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone Aha1. Mol. Cell 10:1307-1318. [DOI] [PubMed] [Google Scholar]
- 23.Pratt, W. B., and D. O. Toft. 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 228:111-133. [DOI] [PubMed] [Google Scholar]
- 24.Pratt, W. B., and D. O. Toft. 1997. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 18:306-360. [DOI] [PubMed] [Google Scholar]
- 25.Prodromou, C., B. Panaretou, S. Chohan, G. Siligardi, R. O'Brien, J. E. Ladbury, S. M. Roe, P. W. Piper, and L. H. Pearl. 2000. The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. EMBO J. 19:4383-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prodromou, C., S. M. Roe, R. O'Brien, J. E. Ladbury, P. W. Piper, and L. H. Pearl. 1997. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell 90:65-75. [DOI] [PubMed] [Google Scholar]
- 27.Prodromou, C., G. Siligardi, R. O'Brien, D. N. Woolfson, L. Regan, B. Panaretou, J. E. Ladbury, P. W. Piper, and L. H. Pearl. 1999. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 18:754-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riggs, D. L., P. J. Roberts, S. C. Chirillo, J. Cheung-Flynn, V. Prapapanich, T. Ratajczak, R. Gaber, D. Picard, and D. F. Smith. 2003. The Hsp90-binding peptidylprolyl isomerase FKBP52 potentiates glucocorticoid signaling in vivo. EMBO J. 22:1158-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roe, S. M., M. M. Ali, P. Meyer, C. K. Vaughan, B. Panaretou, P. W. Piper, C. Prodromou, and L. H. Pearl. 2004. The mechanism of Hsp90 regulation by the protein kinase-specific cochaperone p50cdc37. Cell 116:87-98. [DOI] [PubMed] [Google Scholar]
- 30.Sato, S., N. Fujita, and T. Tsuruo. 2000. Modulation of Akt kinase activity by binding to Hsp90. Proc. Natl. Acad. Sci. USA 97:10832-10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheibel, T., T. Weikl, and J. Buchner. 1998. Two chaperone sites in Hsp90 differing in substrate specificity and ATP dependence. Proc. Natl. Acad. Sci. USA 95:1495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sikorski, R., and J. D. Boeke. 1991. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 194:302-318. [DOI] [PubMed] [Google Scholar]
- 33.Siligardi, G., G. Hu, B. Panaretou, P. W. Piper, L. H. Pearl, and C. Prodromou. 2004. Co-chaperone regulation of conformational switching in the Hsp90 ATPase cycle. J. Biol. Chem. 279:51989-51998. [DOI] [PubMed] [Google Scholar]
- 34.Stebbins, C. E., A. A. Russo, C. Schneider, N. Rosen, F. U. Hartl, and N. P. Pavletich. 1997. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell 89:239-250. [DOI] [PubMed] [Google Scholar]
- 35.Young, J. C., I. Moarefi, and F. U. Hartl. 2001. Hsp90: a specialized but essential protein-folding tool. J. Cell Biol. 154:267-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young, J. C., C. Schneider, and F. U. Hartl. 1997. In vitro evidence that hsp90 contains two independent chaperone sites. FEBS Lett. 418:139-143. [DOI] [PubMed] [Google Scholar]