Abstract
Mechanisms controlling nuclear hormone receptors are a central question to mammalian developmental and disease processes. Herein, we show that a subtle increase in O-GlcNAc levels inhibits activation of nuclear hormone receptors. In vivo, increased levels of O-GlcNAc impair estrogen receptor activation and cause a decrease in mammary ductal side-branching morphogenesis associated with loss of progesterone receptors. Increased O-GlcNAc levels suppress transcriptional expression of coactivators and of the nuclear hormone receptors themselves. Surprisingly, increased O-GlcNAc levels are also associated with increased transcription of genes encoding corepressor proteins NCoR and SMRT. The association of the enzyme O-GlcNAc transferase with these corepressors contributes to specific regulation of nuclear hormone receptors by O-GlcNAc. Overall, transcriptional inhibition is related to the integrated effect of O-GlcNAc by direct modification of critical elements of the transcriptome and indirectly through O-GlcNAc modification of the proteasome.
Nuclear hormone receptors are a superfamily of transcription factors that associate with conserved regions on gene promoters to regulate transcription of specific genes. Nuclear hormone receptors cycle on and off the chromatin both in the presence and in the absence of ligand stimulation (17). Ligand-bound receptors recruit coactivator complexes to target hormone response elements on gene promoters (13). In the case of estrogen receptor α (ERα), ligand binding enables a conformational change for stabilization of an ERα dimer that recruits coactivator proteins to activate transcription (5). Additionally, ligand binding appears to enable increased occupancy by nuclear hormone receptors with hormone response elements on gene promoters (19).
In contrast, corepressor transcription factors associate with nuclear hormone receptors at hormone response elements on gene promoters in the absence of an activating signal in order to maintain transcriptional repression (17). Corepressors work in concert with histone deacetylases (HDACs) to impair transcriptional activation by reducing DNA availability. Transcriptional repression is also linked to proteasome activity. Turnover of nuclear receptors in contact with specific promoters correlates with their proteolytic degradation. For example, proteasome-dependent degradation of ERα decreases responsiveness of the receptor to the activating signal (19). In addition, recycling of hormone-associated transcriptional cofactors is often required for gene activation (9).
The addition or removal of O-linked N-acetylglucosamine (O-GlcNAc) on proteins has been shown to function in parallel with HDACs to repress transcription of estrogen-responsive genes (32). Many transcription factors as well as the proteasome are directly modified by O-GlcNAc to affect their activity. In the case of transcription factor modification, O-GlcNAc addition to one of the activation domains of Sp1 or to the tail of RNA polymerase II impairs their function (8, 32), although not the capacity of Sp1 to associate with DNA (32). O-GlcNAc modification of the proteasome on the RPT2 subunit blocks all proteasomal ATPase activity, specifically prevents chymotryptic activity of the proteasome, and is linked to the stability of transcription factors such as Sp1 (33). Thus, O-GlcNAc can potentially play a role in transcription both by directly modifying the transcriptome and by preventing the recycling of transcription factors via inhibition of the proteasome.
O-GlcNAc modification occurs ubiquitously and involves the β-linkage of N-acetylglucosamine to the hydroxyls of serine or threonine residues of a protein by the enzyme O-GlcNAc transferase (OGT) (28). OGT has been shown to associate with the transcriptional corepressor mSin3a on repressed promoters. A domain containing the tetratricopeptide repeat domain, hydrophobic structural motifs thought to favor protein-protein interactions, is located at the N terminus of OGT and is responsible for OGT interaction with mSin3a. This interaction appears to target OGT, and thus O-GlcNAc modification, to sites of transcriptional repression (32). The removal of O-GlcNAc residues is catalyzed by the enzyme NCOAT (nuclear and cytoplasmic O-GlcNAcase and acetyltransferase; meningioma 5 or MGEA5; O-GlcNAcase), which is the only known protein bearing O-GlcNAcase (OGN) activity, in addition to containing a functional histone acetyltransferase (HAT) domain (25, 26). Thus, OGN activity by NCOAT is essential to relieve the effects of O-GlcNAc modification on proteins.
Although O-GlcNAc modification is associated with numerous intracellular proteins which take part in a wide variety of signaling pathways, a specific function for O-GlcNAc during organogenesis remains unclear. Herein, we describe that posttranslational modification by O-GlcNAc can regulate nuclear hormone receptor activation in vivo in the model system of the developing mouse mammary gland. To evaluate the complex function of O-GlcNAc in a relevant in vivo system, we utilized a novel mouse model in which O-GlcNAc levels could be inducibly controlled. We established that overexpression of a nonfunctional isoform of NCOAT protein operates in a dominant-negative fashion to increase O-GlcNAc levels. Increased O-GlcNAc levels impair transcriptional expression of nuclear hormone receptors and are associated with reduced ductal side branching during mammary gland development in our transgenic mouse model. This phenotype appears to result specifically from transcriptional inhibition of progesterone receptors by increased O-GlcNAc levels, although activation of estrogen receptors was also impaired. Inhibition of nuclear hormone receptors is a consequence of the dual role of O-GlcNAc in inhibiting transcription factor expression directly and in regulating transcription factors indirectly by inhibition of the proteasome. In this example, O-GlcNAc modification acts as a master switch for hormone-dependent gene expression and development. The coordinate modulation of transcription and of the proteasome by O-GlcNAc puts this protein modification at the center of developmental control.
MATERIALS AND METHODS
Plasmids.
Cloning of NCOAT variants was previously described (25-27). The Gal4-tagged NCOAT was previously described (29). For gene expression experiments, NCOAT and mutant NCOAT proteins were cloned into the mRFP-C1 vector. This vector was based on the backbone for EGFP-C1 (Clontech) in which the green fluorescent protein (GFP) sequence was replaced by a monomeric red fluorescent protein (mRFP) (a gift from R.S. Tsien, UCSD) in pEGFP-C1 parent vector. RNA interference (RNAi) was conducted using pSUPER (4) or pSUPER-retro-based vectors (OligoEngine, Seattle, WA), with both vectors targeting the same sequences. For tracking RNAi transfection efficiency during immunohistochemistry (IHC), mRFP-C1 was cotransfected at 1/10 the amount of pSUPER vector. Two small interfering RNAs that target sequence 5′-GGACAGATTCAAATAACAA or 5′-TGGCATCGACCTCAAAGCA of the human OGT coding region were interchangeably used to suppress OGT expression. Two small interfering RNAs that correspond to target sequence 5′-CATGAACGGAGTGAGGAAG or 5′-ACGCAAATTGGACCAGCTC of human NCOAT were interchangeably used to suppress NCOAT levels. Either a scrambled sequence that has no homology with any genes (5′-GACATAGCGTAAGCCTATC) or a sequence directed against enhanced green fluorescent protein (EGFP; Dharmacon) was used as a control. All transfections were conducted using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions.
Cell culture.
MCF10AT cells (Karmanos Cancer Institute, Detroit, MI) were cultured in Dulbecco modified Eagle medium-F12 (with l-glutamine, 15 mM HEPES, and sodium pyruvate; BioWhittaker, Cambrex, Walkersville, MD) supplemented with 0.1 μg/ml cholera toxin (Calbiochem, San Diego, CA), 10 μg/ml insulin (Sigma, St. Louis, MO), 0.5 μg/ml hydrocortisone (Sigma), 0.02 μg/ml epidermal growth factor (EGF; Upstate Biotechnology, Lake Placid, NY), and 5% horse serum (Invitrogen). Testing of RNAi constructs was conducted in BSC40 cells. These were cultured in Dulbecco modified Eagle medium with 10% fetal bovine serum (Invitrogen).
Enzyme activity assays.
The O-GlcNAcase activity assay was performed as previously described (25). Histone acetyltransferase activity was measured using the previously described “filter paper method” (26).
Mice.
The mouse mammary tumor virus (MMTV) β-globin transgene was previously used to make an MMTV-mEGFtr transgene (30). The 1-kb reverse tetracycline transactivator (rtTA) sequence (Clontech, Burlingame, CA) was inserted into β-globin exon 3. The complete 3.7-kb XhoI transgene fragment was isolated and microinjected into BL/6xSJL mouse zygotes. Founders were identified by PCR analysis (36 cycles of 94°C for 30 s, 55°C for 45 s, and 72°C for 45 s), using oligonucleotides designed to anneal to the MMTV sense sequence, 5′-TGCAACAGTCCTAACATTCA-3′, and rtTA antisense sequence, 5′-TGAATGTTAGGACTGTTGCA-3′.
A TRE-NCOATGK transgenic mouse line was constructed as a chimera of rat mutant NCOAT sequence (nucleotides 350 to 1110) inserted into the corresponding mouse sequence (nucleotides 350 to 1400) as previously described (29). Founders were identified by PCR.
MMTV-rtTA mice were crossed with mice bearing the TRE-NCOATGK transgene. This produced mice bearing one of either of the two possible transgenes or bitransgenic mice bearing an MMTV-rtTA/TRE-NCOATGK genotype. Doxycycline (DOX) was administered via drinking water (2 mg/ml) that was supplemented with saccharine sugar substitute. DOX was replaced every 48 h for the duration of the experiments. Approximately 20 to 30 mice were examined from each genetic category.
Histopathology.
Mammary tissue was dissected from adult mice, fixed in 4% paraformaldehyde, and embedded in paraffin. Sections were cut at 5 μm and stained with hematoxylin and eosin for histopathological evaluation.
Whole-mount analysis.
Analysis closely followed the protocol already outlined (2). Briefly, the right and left inguinal mammary glands were dissected and spread onto a glass slide. The glands were fixed for 24 h in Carnoy's fixative, washed for 15 min in 70% ethanol, washed for 5 min in distilled water, and stained in 0.2% carmine-0.5% aluminum potassium sulfate overnight. The glands were dehydrated sequentially in 70, 90, and 100% ethanol for 15 min each, cleared in xylene, and mounted with Permount (Sigma, St. Louis, MO). Side-branching morphogenesis was established by counting mean values of high-order branching (>4 branch points) and standard deviations for 25 mice from control or MMTV-rtTA/TRE-NCOATGK genetic categories. Student's t test was used to determine significance.
In situ hybridization analysis.
Mammary glands were embedded in Tissue-Tek optimal cutting temperature compound and frozen, and 5-μm sections were cut and affixed to glass slides. A biotin-labeled cDNA probe specific for full-length chimeric m/rNCOATGK was prepared using a biotin nick translation mix kit (Roche). Tissues were pretreated with 3% H2O2 and denatured in 70% formamide in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (pH 7.0, 74°C, 2 min). The remaining procedures essentially matched those previously described (21), which were modified from the Roche Nonradioactive In Situ Hybridization Application Manual. Briefly, biotinylated probe was denatured at 74°C for 10 min and applied to mammary sections for overnight hybridization at 37°C. Streptavidin-linked horseradish peroxidase and cyanine 3 (Cy3)-tyramide was employed following the manufacturer's instructions (NEN Life Sciences/PerkinElmer). Streptavidin was applied on tissues for 2 h at 37°C, followed by deposition of Cy3 for 10 min at room temperature. Sections were counterstained with DAPI (4′,6′-diamidino-2-phenylindole; Molecular Probes) for 5 min and mounted using antifade medium (0.2% N-propyl gallate; Sigma). For this and subsequent fluorescence microscopy, a Leica DMRB microscope coupled with a Leica DFC480 camera and standard filter set was used.
Protein expression and antibodies.
IHC was conducted following the protocol outlined by Transduction Laboratories (Burlingame, WI). Mammary sections were deparaffinized and subjected to antigen retrieval using antigen retrieval solution (Transduction Laboratories). For all monoclonal antibodies, a MOM (mouse on mouse) kit from Transduction Laboratories was employed. IHC was conducted using RL2 (α-O-GlcNAc, monoclonal antibody produced in our laboratory), α-ERα (MC20), and α-PRA/B (C20) (each a rabbit polyclonal antibody from Santa Cruz Biotechnology, Santa Cruz, CA), anti-EGFR (AB-20; LabVision/Neomarkers, Fremont, CA), or Ki67 (MM1; Novocastra). For terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) analysis, an in situ cell death detection kit was used (Roche). Sections were counterstained with DAPI (Molecular Probes) and mounted using antifade mounting solution (Vector Laboratories).
Immunoprecipitation and Western blot analysis were performed following the general protocol from Santa Cruz Biotechnology. Lysates and immunoprecipitates were treated with 1× EDTA-free complete protease inhibitors (Roche, Burlingame, WI). Antibodies against Gal4-DBD (N19), NCoR (N19), SMRT (1212), mSin3a (K20), AIB1 (NCoA3, F2), SRC1 (M20), ERα (MC20), and OGT (V18) were obtained from Santa Cruz. Anti-Sp1 (rabbit polyclonal) was produced in our laboratory.
Quantification of IHC data was conducted using ImageJ. Single green-channel images (for RL2, ERα, or PRA/B IHC) were converted to TIFF format. Three positive transfectants (containing mRFP signal) and three negative transfectants were sampled and a mean value ± standard deviation was calculated for each set of samples. Student's t test was used to determine significance between control and experimental samples. Quantification of Western blots was conducted using ImageJ for the depicted experiment. Each value represents the densitometry of a band divided by the densitometry for the corresponding β-actin.
Quantitative RT-PCR analysis.
RNA was purified using an RNeasy kit (QIAGEN). cDNA was created using a cDNA archive kit (Applied Biosystems), and relative RNA levels were evaluated by quantitative real-time reverse transcriptase (RT)-PCR technology with an ABI Prism 7700 detection system and SYBR green reagent (Applied Biosystems). PCRs contained 1× SYBR green master mix, 66 nM primers, and cDNA equivalent to 10 ng total RNA in a 15-μl volume. Target mRNA levels were normalized against the GAPDH mRNA level from a standard curve established from the same total RNA sample. Primers for ERα, ERβ, progesterone receptor A (PRA), PRB, SMRT, NCoR, and β-actin are available upon request.
Proteasome assays.
Proteasome analysis using GFP-degron or Suc-LLVY-AMC was performed following the previously established protocols (33).
RESULTS
Overexpression of mutant NCOAT causes an increase in the levels of intracellular O-GlcNAc.
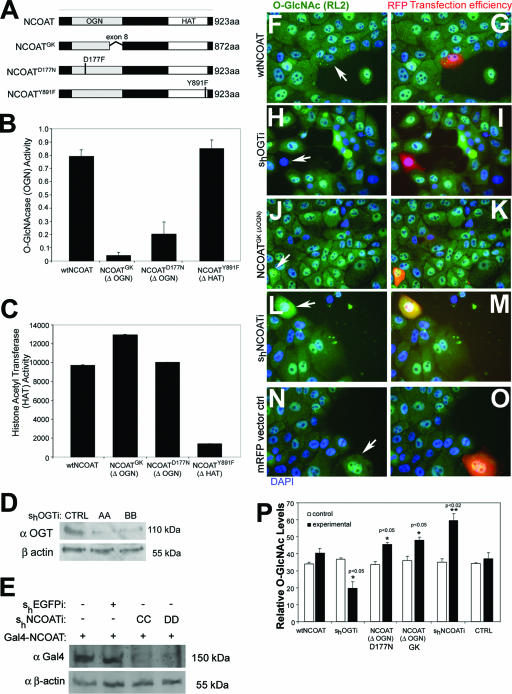
We first evaluated whether overexpression of NCOAT splice variants that do not possess OGN activity could function in a dominant-negative capacity to cause an increase in O-GlcNAc levels. To accomplish this, we designed several constructs to control the enzymes which govern O-GlcNAc levels. A splice variant lacking exon 8 of the NCOAT protein was identified from Goto-Kakizaki rats (NCOATGK) (27) as well as a point mutant (NCOATD177N) (Fig. 1A) (26) and was cloned into expression vectors. These two variants of NCOAT were shown to lack OGN activity but retain HAT activity (Fig. 1B and C). Another point mutation in the HAT domain (NCOATY891F) was also identified (Fig. 1A) (26). This mutant retains OGN activity but lacks HAT activity (Fig. 1B and C).
FIG. 1.
Overexpression of mutant NCOAT increases intracellular O-GlcNAc levels. (A) Cartoon depiction of wild-type NCOAT, NCOATGK splice variant, NCOATD177N, and NCOATY891F. (B) O-GlcNAcase activity for the indicated NCOAT constructs was measured using an in vitro pNP-GlcNAc assay. Error bars represent standard errors of the means. (C) Results of an in vitro HAT assay for the indicated NCOAT constructs using [3H]acetyl coenzyme A to modify immobilized histone tails. Error bars represent standard errors of the means. (D) Western blot showing levels of endogenous OGT in MCF10AT whole-cell lysates in the absence or presence of short hairpin RNAi constructs (AA and BB) against OGT. (E) Western blot of Gal4(DBD) in cells transfected with cytomegalovirus-Gal4(DBD)-tagged NCOAT to test short hairpin RNAi constructs (CC and DD) against NCOAT. (F to O) Immunohistochemical detection of O-GlcNAc levels (green channel) in response to 18 h of transfection with the indicated constructs. Positive transfectants were tracked with mRFP, and DAPI (blue) was used for nuclear staining. For overexpression of NCOAT variants lacking OGN activity (ΔOGN), only the NCOATGK image is shown (J and K). (P) O-GlcNAc levels were quantified by densitometry of three separate transfectants with the indicated constructs, and arbitrary densitometric values ± standard deviations are shown. The surrounding cells were used to establish control levels for O-GlcNAc. Student's t test was used to establish significant differences, which are indicated as P values of <0.05 (*) and <0.02 (**). wt, wild type.
We additionally utilized pSUPER vector-based short hairpin (sh) RNAi constructs (4) to regulate endogenous protein expression levels of OGT (Fig. 1D) and NCOAT (Fig. 1E). For identification of shNCOATi, it was necessary to observe knockdown of overexpressed NCOAT tagged with the Gal4 DNA binding domain (DBD) (Fig. 1E), as no antibody is currently available against endogenous NCOAT protein.
The effectiveness of these constructs was measured by IHC analysis of O-GlcNAc modification levels in transfected MCF10AT cells. Knockdown of OGT levels using shOGT interference (shOGTi) causes a decrease in O-GlcNAc levels, as we have previously shown (Fig. 1H, I, and P) (33). In contrast, RNAi knockdown of NCOAT caused an increase in O-GlcNAc levels (Fig. 1L, M, and P). Likewise, expression of NCOAT lacking OGN activity (NCOATGK or NCOATD177N; only GK splice variant transfection IHC is shown) also caused an increase in O-GlcNAc levels (Fig. 1J, K, and P). We therefore reasoned that these NCOAT variants which lack OGN activity can function in a dominant-negative capacity similar to the knockdown of NCOAT protein by RNAi. Overexpression of the NCOATY891F point mutant lacking HAT activity but retaining OGN function did not affect O-GlcNAc levels (data not shown).
Increased O-GlcNAc levels impair nuclear hormone receptor expression.
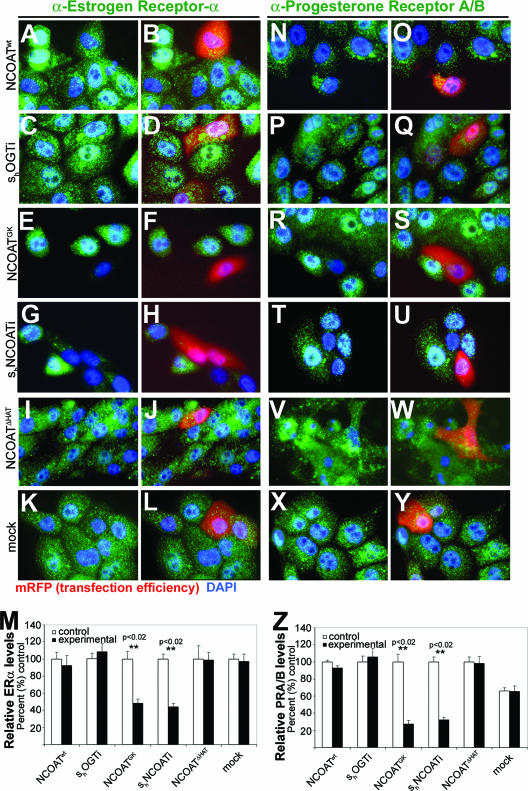
We previously observed that increased O-GlcNAc modifications were associated with repressed gene promoters that are downstream of nuclear hormone receptor activation (32). In the current study, we assessed the effect of O-GlcNAc levels on the expression of nuclear hormone receptors themselves in MCF10AT cells. These “premalignant” human breast cells retain nuclear hormone receptor responsiveness to both estrogen and progesterone and express ERα and PRB but not the PRA hormone receptor isoforms (23). We relied upon high reproducibility of our plasmid constructs shown in Fig. 1 to correlate levels of O-GlcNAc with nuclear hormone receptor expression (Fig. 2A to Z) and quantified those data using densitometry (Fig. 2M and Z). Lowering O-GlcNAc levels with shOGTi had no significant affect on nuclear hormone expression levels at the protein level (Fig. 2C, D, M, P, Q, and Z). However, increased O-GlcNAc levels in response to overexpression mutant NCOAT lacking OGN activity or to shNCOATi consistently caused a 50% and 60% reduction in ERα (Fig. 2E to H and M) and PRA/B expression (Fig. R to U and Z), respectively. This reduction in nuclear hormone receptor expression levels was statistically significant compared to the controls for both ERα (Fig. 2M) and PRA/B (Fig. 2Z). Overexpression of wild-type NCOAT (Fig. 2A, B, N and O), the NCOAT HAT point mutant (Fig. 2I, J, V, and W), or cells mock transfected with mRFP only (Fig. 2K, L, X, and Y) showed no significant differences in nuclear hormone receptor expression between control and experimental groups (Fig. 2M and Z). At the time of immunohistochemical testing, neither the level of apoptosis as assessed by TUNEL staining nor IHC analysis of the general proliferation marker Ki67 showed any changes induced by transfection of the various constructs (data not shown). Cell cycle was not affected by increased O-GlcNAcylation 18 h after transfection (data not shown).
FIG. 2.
Increased levels of O-GlcNAc inhibit nuclear hormone receptor expression in MCF10AT cells. (A to L) Immunohistochemical detection of ERα expression in response to transfection of the construct indicated (at left). Positive transfectants were tracked with mRFP, and DAPI was used for nuclear staining. (M) Densitometry analysis for ERα expression in response to transfection of the indicated constructs expressed as a percent change from the control values. (N to Y) Immunohistochemical detection of PRA/B expression in response to transfection of the indicated constructs. (Z) Densitometry analysis for PRA/B expression in response to transfection of the indicated constructs expressed as a percent change from the control values. For all IHC panels, primary antibody appears in green, positive transfectants were tracked with mRFP (red), and DAPI (blue) was used for detection of the nucleus. ImageJ was used to calculate densitometry, and Student's t test was used to evaluate significant differences between controls and experimental transfectants. P values of <0.02 (**) are indicated above significantly different groups, and error bars represent standard deviations.
O-GlcNAc regulates hormone-dependent mammary gland development in mice.
We next examined whether increasing O-GlcNAc levels would affect hormone-dependent development in vivo. The observation that increased levels of O-GlcNAc were associated with aberrant expression of nuclear hormone receptors in MCF10AT cells suggested that increased O-GlcNAc levels may play a role in regulating hormone-dependent mammary gland development. Major development for the mammary gland occurs postnatally. During puberty, the rudimentary ductal system extends from the nipple region to fill the mammary fat pad. Ductal side branching increases as a function of estrous cycles and reaches its maximum potential during pregnancy (20). Studies have demonstrated that pubertal development of the mammary ductal system in rodents is regulated by the concerted actions of estrogen and progesterone (15). In mice, tissue recombination experiments have shown that the absence of both alleles of the estrogen receptor (ER) impairs ductal elongation. Specifically, expression of ERα was found to be critical in both the stromal and the epithelial compartments of the mammary gland. Knockout experiments demonstrate that epithelial progesterone receptors (PRs) are responsible for ductal side-branching morphogenesis (3, 11). Stromal expression of both ER and PR appears to be necessary for normal ductal outgrowth at puberty (7, 12).
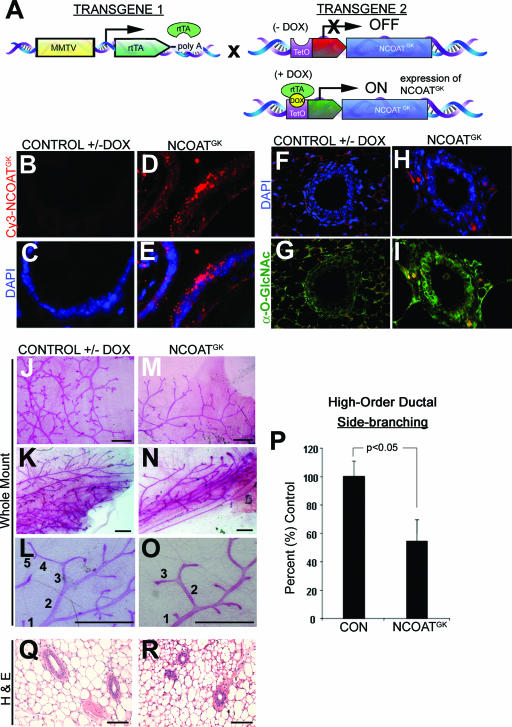
Previous studies suggest that impaired activity of NCOAT is detrimental to cellular viability (10) and may impair cell cycle progression (24); we sought to establish an inducible bitransgenic mouse model in which expression of mutant NCOAT (NCOATGK) could increase intracellular O-GlcNAc levels. The first line of transgenic mice expressed the MMTV-rtTA promoter transgene. This promoter is specific to the epithelial compartment of the mammary gland (6). The second line expressed the NCOATGK splice variant that lacks O-GlcNAcase activity (Fig. 1). This transgene was created as a chimera of mouse NCOAT with the exon 8 flanking regions of rat NCOAT. These two transgenic mouse lines were then crossed to create MMTV-rtTA/TRE-NCOATGK bitransgenic mice (Fig. 3A). In the presence of doxycycline, the MMTV transactivator binds to the TRE-NCOATGK transgene to induce NCOATGK expression in the mammary epithelial compartment, as we previously showed by Northern blotting (29) and as assessed herein by tyramide-amplified in situ analysis (Fig. 3D and E). For biochemical evaluation of the mammary gland during development, we cut paraffin sections for IHC analysis to examine the in vivo levels of O-GlcNAc in mammary ductal epithelia by IHC with RL2. A striking increase in O-GlcNAc levels was observed in the mammary ductal epithelium in response to overexpression of the NCOATGK transgene (Fig. 3F to I). Notably, these results were similar to our observation that overexpression of an inactive NCOAT could function as a dominant-negative protein to increase the levels of cellular O-GlcNAc (Fig. 1).
FIG. 3.
Transgenic expression in the mammary epithelium of mutant NCOAT that lacks OGN activity causes increased O-GlcNAc levels and reduced mammary ductal side-branching morphogenesis. (A) Mice bearing an MMTV-rtTA transgene (transgene 1) were crossed with mice bearing a tetracycline response element (TRE)-NCOATGK transgene (transgene 2). Administration of DOX to MMTV-rtTA/TRE-NCOATGK bitransgenic mice induced expression of the NCOATGK transgene. (B to E) Detection of the NCOATGK transcript in the mammary epithelium using fluorescence in situ hybridization analysis with tyramide amplification in mammary gland sections from bitransgenic mice with or without DOX. (F to I) Immunohistochemical detection of O-GlcNAc levels (RL2) in sections from mammary gland ducts taken from bitransgenic mice with or without DOX. (J to O) Whole-mount analysis of ductal side-branching morphogenesis in 8-week-old bitransgenic mice with or without administration of DOX. Panels L and O demonstrate counting methodology for high-order ductal side branches (>4 branch points from the primary duct). (P) Graphical representation of differences in side-branching morphogenesis between control bitransgenic animals (−DOX) and bitransgenic mice expressing the NCOATGK transgene (+DOX). Expression of the NCOATGK transgene was associated with a 40% decrease in high-order ductal side-branching morphogenesis (P < 0.05, Student's t test). Error bars represent standard deviations from the sample means. (Q and R) Hematoxylin- and eosin-stained (H & E) sections of mammary glands in mice bearing the indicated genotype. Bars on images represent 1 mm. For all experiments, approximately 25 animals from each category were examined. Bitransgenic mice without induction of the transgene are also representative of mice bearing a single transgene with or without DOX (data not shown).
We next examined the mammary glands from MMTV-TRE/rtTA-NCOATGK bitransgenic mice by whole-mount analysis for developmental abnormalities during pubertal development. For these experiments, we initiated transgene expression at 2 to 3 weeks of age, at which point postpubertal mammary development begins (20). Our analysis revealed a 40% decrease in high-order ductal side-branching morphogenesis in response to the expression of NCOATGK in 8-week-old mice (Fig. 3J to P). However, lobuloalveolar development was not affected and the ducts themselves appeared histologically normal by pathological evaluation of hematoxylin- and eosin-stained sections (Fig. 3Q and R). Thus, although the ducts grew throughout the fat pad and formed histologically normal lobuloalveolar structures, ductal side-branching morphogenesis was impaired.
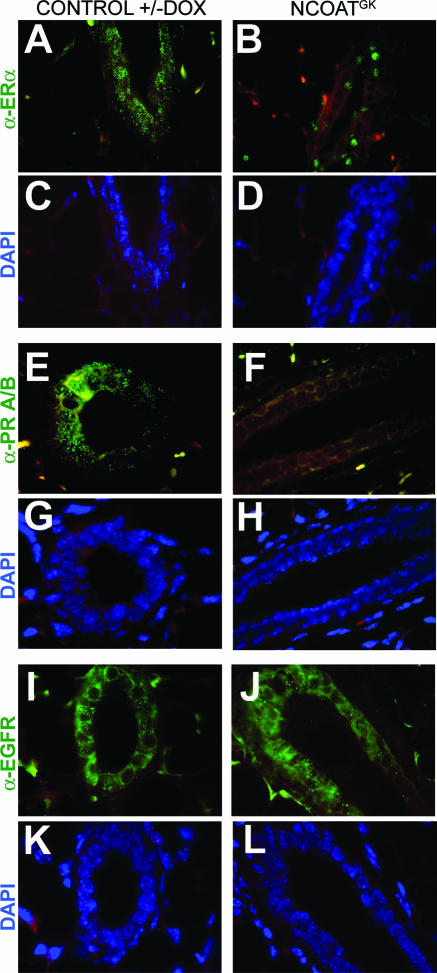
We have previously shown that expression of genes downstream of estrogen was impaired by expression of NCOATGK (29). Here, approximately 25 mammary glands from control or experimental mice were sectioned for examination of nuclear hormone receptor expression patterns by IHC. Using an antibody specific for ERα, we found that its levels were reduced markedly in response to expression of the NCOATGK transgene (Fig. 4A to D). In this case, increased O-GlcNAc did not ablate expression of ERα completely but instead relegated it to a spotty pattern of expression in which receptors appeared to localize to the nucleus. In contrast, PRA/B expression was not detected by an antibody which detects both PR subtypes (Fig. 4E to H). Expression of the epidermal growth factor receptor appeared unaffected by overexpression of the NCOATGK transgene (Fig. 4I to L), supporting the notion that O-GlcNAc regulates distinct signaling pathways in vivo.
FIG. 4.
Increased O-GlcNAc levels impair nuclear hormone receptor expression in vivo during pubertal mouse mammary gland development. (A to D) Immunohistochemical detection of ERα expression in sections from bitransgenic mouse mammary glands either with or without DOX induction of NCOATGK expression. (E to H) Immunohistochemical detection of PRA/B expression in sections from bitransgenic mouse mammary glands either with or without DOX induction of NCOATGK expression. (I to L) Immunohistochemical detection of epidermal growth factor receptor (EGFR) expression in sections from bitransgenic mouse mammary glands either with or without DOX induction of NCOATGK expression. For all panels, primary antibody is depicted in green and DAPI nuclear staining is depicted in blue, and Texas Red filter was overlaid on all panels to demonstrate background staining in yellow. Mammary sections are from 8- to 10-week-old mice. Bitransgenic mice without induction of the transgene are also representative of mice bearing a single transgene with or without DOX (data not shown).
We did not observe changes in proliferation by Ki67 IHC or apoptosis by TUNEL staining in response to expression of the NCOATGK transgene (data not shown).
Inhibition of the proteasome by O-GlcNAc contributes to inactivation of nuclear hormone receptors.
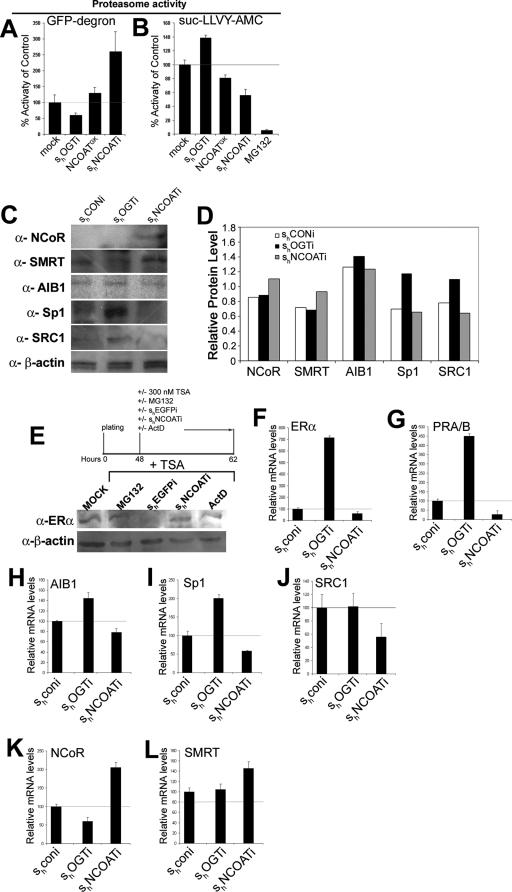
Inhibition of the proteasome causes localization of ERα to the nuclear envelope, where it is inactive (19). Likewise, we observed that ERα may localize to the nucleus in mice expressing NCOATGK in their mammary epithelium (Fig. 4A to D). Because our previous studies suggest that O-GlcNAc can inhibit proteasome activation (33), we evaluated whether modulating O-GlcNAc levels could affect proteasome activity in MCF10AT cells. We first utilized the GFP-degron construct, which produces GFP-labeled protein that is continually degraded by the proteasome (1), to analyze proteasome activity in MCF10AT cells in response to differing O-GlcNAc levels induced by our constructs. MCF10AT cells were cotransfected with GFP-degron and shOGTi, NCOATGK, or shNCOATi. Fluorimetric analysis of lysates demonstrated that reduction of O-GlcNAc by shOGTi induces an approximate 20% activation in proteasome activity (Fig. 5A). In contrast, increased O-GlcNAc levels in response to NCOATGK or shNCOATi marked a decrease in proteasome activity (Fig. 5A).
FIG. 5.
Proteasome inhibition and transcriptional regulation are integrated via O-GlcNAc (A) Degradation versus synthesis of GFP-degron in MCF10AT cells cotransfected with the indicated constructs. Increased O-GlcNAc levels were associated with increased GFP fluorescence. Degradation of the proteolytic GFP-degron plasmid was measured using a fluorimeter. (B) Degradation of Suc-LLVY-AMC chymotryptic proteolytic peptide substrate is impaired in the presence of increased O-GlcNAc levels. MCF10AT cells were transfected with the indicated constructs, and lysates were incubated with peptide. LLVY cleavage was measured using a fluorimeter. (C) Western blot of corepressors and coactivator proteins in MCF10AT cells retrovirally transfected with RNAi targeting OGT or NCOAT. (D) Densitometry was performed using ImageJ for the blots shown in panel C. (E) Western blot for ERα in MCF10AT cell lysates treated under the indicated conditions and with TSA to induce proteolytic clearance of ERα. (F to L) Results of real-time quantitative RT-PCR of nuclear hormone receptors ERα and PRA/B, coactivators SRC1 and SRC3/AIB1, and corepressor proteins NCoR and SMRT. Samples were normalized to GAPDH. For all experiments depicted, error bars represent standard errors of the means. CON, control.
Our previous data suggest that O-GlcNAc specifically inhibits chymotryptic activity of the proteasome (33). To demonstrate that differing O-GlcNAc levels can specifically regulate proteasome activity, we used the fluorigenic chymotryptic peptide Suc-LLVY-AMC for an in vitro measure of activity of the proteasome in MCF10AT cell culture. By decreasing O-GlcNAc levels, shOGTi increased proteasome activity by 20%, while expression of NCOATGK or shNCOATi reduced proteasome activity by 20% or 40%, respectively (Fig. 5B). In contrast, proteasome inhibitor MG132 lessened proteasome activity by 90 to 95% (Fig. 5B). Thus, NCOATGK or shNCOATi increased O-GlcNAc levels enough to cause a partial, but not total, inhibition of proteasome activity.
Proteasomal degradation of corepressor molecules plays a critical role in the switch from repression to activation. We therefore analyzed protein levels by Western blot and densitometry analyses for corepressors NCoR and SMRT, as well as coactivators SRC1, AIB1, and Sp1, in whole-cell lysates from MCF10AT cells in response to variation of O-GlcNAc levels. These molecules were selected based on prior investigation in our laboratory (29) and their general importance to mouse mammary gland morphogenesis. We observed that increased O-GlcNAc levels in response to shNCOATi were associated with reduced levels of coactivators, including SRC1, AIB1, and SRC3 (Fig. 5C and D). On the other hand, the expressions of NCoR and SMRT were still expressed in the presence of increased O-GlcNAc levels (Fig. 5C and D). In contrast, coactivators Sp1 and SRC displayed increased levels in response to reduction of O-GlcNAc by OGT RNAi (Fig. 5C and D). Thus, the levels of general inhibitory molecules remained increased in association with increased O-GlcNAc levels while coactivator proteins were at low levels under those same conditions.
It has been shown that the proteasome plays an essential role in the cycling of nuclear hormone receptors to maintain cells in a state of readiness for activation (16). In the case of ERα, the receptor is stabilized in a proteasome-dependent fashion in response to proteasome inhibitors or transcriptional inhibitors, such as actinomycin D. Receptors are localized to the nuclear envelope and their transactivation potential is reduced (19). Those prior studies by Reid et al. show that treatment of cells with trichostatin A (TSA), an HDAC inhibitor, causes clearance of ERα from cells due to proteolytic degradation (19). Thus, to evaluate whether O-GlcNAc plays a role in this process, we treated MCF10AT cells with a combination of TSA and MG132 proteasome inhibitor, shEGFPi, shNCOATi, or the transcription inhibitor actinomycin D. Western blot analysis demonstrated that ERα expression is maintained in the presence of TSA by MG132 or actinomycin D (Fig. 5E). While RNAi against the control, EGFP, allows for degradation of ERα in the presence of TSA, RNAi against NCOAT restored levels of ERα (Fig. 5E).
Increasing O-GlcNAc levels impairs transcription of nuclear hormone activators/coactivators and enhances transcription of corepressors.
O-GlcNAc modification inhibits the operation of essential elements of the transcriptome, such as Sp1. We therefore investigated whether O-GlcNAc could regulate expression of nuclear hormone receptors, as well as several major hormone-dependent coactivators and corepressors, at the transcriptional level. To accomplish this, RNA was isolated from MCF10AT cells transfected with retrovirally expressed RNAi against OGT or NCOAT. Quantitative RT-PCR was utilized to measure transcript levels, as indicated, and normalize them to GAPDH. We observed that decreased O-GlcNAc levels by shOGTi were correlated with enhanced expression of nuclear hormone receptors ERα and of PRA/B (Fig. 5F and G). Likewise, shOGTi enhanced expression of both Sp1 and SRC3/AIB1, although SRC1 appeared unaffected by reduced O-GlcNAc levels (Fig. 5H to J). On the other hand, increased O-GlcNAc levels by shNCOATi were associated with a decrease in transcription of nuclear hormone receptors and coactivators (Fig. 5F to L).
Decreased RNA levels of O-GlcNAc by shOGTi were associated with a decrease in the level of corepressor protein NCoR but not SMRT (Fig. 5K and L). Strikingly, however, levels of NCoR transcription were increased 2-fold and levels of SMRT transcription were increased 1.5-fold following an increase in O-GlcNAc by shNCOATi (Fig. 5K and L). These data are consistent with the model that O-GlcNAc modulates transcription by both direct mechanisms and regulation of the proteasome (31-33).
O-GlcNAc transferase associates with corepressor proteins NCoR and SMRT.
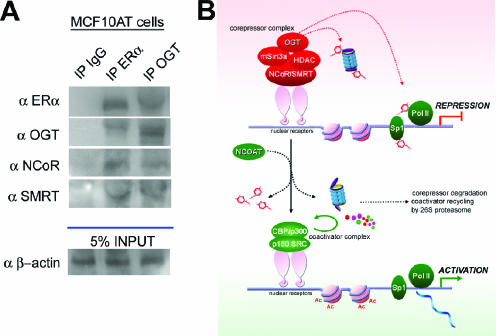
We previously showed that OGT may be targeted to promoters by its interaction with corepressor proteins, such as mSin3a (32). The nuclear corepressor proteins NCoR and SMRT are upregulated in response to increased O-GlcNAc levels (Fig. 5). Moreover, we previously observed that OGT may directly associate with either NCoR or SMRT in a pull-down assay and with NCoR under conditions in which hemagglutinin-tagged OGT was overexpressed (33). We therefore tested whether OGT could be targeted to sites of nuclear hormone transcriptional repression by interaction with NCoR and SMRT. Immunoprecipitation followed by Western blot analysis demonstrated that endogenous NCoR, SMRT, and ERα coprecipitate with OGT in MCF10AT cells (Fig. 6A). Thus, the upregulation of NCoR and SMRT in response to increased O-GlcNAc levels (Fig. 5) would enhance transcriptional repression in two ways: first, simply by their upregulation alone, and second, by their interaction with OGT, which may serve to target O-GlcNAc to sites of transcriptional repression.
FIG. 6.
OGT associates with NCoR and SMRT nuclear corepressor proteins to target O-GlcNAc modification to sites of repressed nuclear hormone receptors. (A) OGT coimmunoprecipitated with NCoR, SMRT, and ERα in reciprocal immunoprecipitation (IP) experiments. Ig, immunoglobulin. (B) Proposed model for O-GlcNAc as a regulator of nuclear hormone receptor signaling. During transcriptional repression, O-GlcNAc modification is targeted to sites of transcriptional repression by the interaction of OGT with corepressors on nuclear hormone receptors. O-GlcNAc modification of critical elements of the transcriptome and the proteasome contributes to transcriptional repression. These may include Sp1 and RNA polymerase II (Pol II). For transition from repression conditions to activation of the nuclear hormone receptors, NCOAT activity is required to remove O-GlcNAc residues from critical elements of the transcriptome and the proteasome. The removal of O-GlcNAc residues enables degradation of corepressors and proteolytic recycling of coactivators, culminating in activation of the transcriptional machinery. Generally, repressor elements are depicted in red and activating elements in green.
DISCUSSION
This paper provides a novel link between direct regulation of nuclear hormone receptor activation and regulation of transcription factor activity by the proteasome. The current study suggests that posttranslational modification by O-GlcNAc unifies these processes. Herein, we describe a role for O-GlcNAc in regulating nuclear hormone receptors in vivo during whole-tissue organogenesis. Strikingly, the experiments presented here suggest that O-GlcNAc is a specific and physiological inhibitor of nuclear hormone receptor signaling in vivo. This regulatory mechanism suggests that O-GlcNAc controls both transcription and the proteasome.
Overexpression of variants of NCOAT which lack O-GlcNAcase activity was sufficient to induce a subtle increase in O-GlcNAc levels and was associated with impaired expression of nuclear hormone receptors. Increased O-GlcNAc levels result from a dominant-negative effect of the NCOAT variants (Fig. 1). The ability of NCOATGK to prevent removal of O-GlcNAc from DNA-associated proteins was previously observed (29). Likewise, overexpression of NCOATGK and NCOATD177N produced an effect similar to knockdown of NCOAT by RNAi, allowing us to conclude that the O-GlcNAcase activity of NCOAT is critical to expression of the nuclear hormone receptors. O-GlcNAcase activity of NCOAT is particularly critical since overexpression of NCOATY891F, which retains O-GlcNAcase activity but lacks HAT activity, did not alter expression of the nuclear hormone receptors (Fig. 1 and 2). The HAT activity, but not the O-GlcNAcase activity, of NCOAT may be redundant.
By controlling O-GlcNAc through manipulation of the enzymes which regulate this posttranslational modification, we induce physiologically relevant changes in O-GlcNAc levels. The resultant phenotype observed in mice expressing NCOATGK in their mammary epithelia is associated with impaired PR expression that occurs in response to increased O-GlcNAc levels. This phenotype correlates with that observed in knockout of PR in the mammary epithelial compartment, which is associated with aberrant mammary ductal side-branching morphogenesis (7). In contrast, studies show that the total absence of ERα in the epithelial compartment severely impairs mammary ductal outgrowth (12). The singular presence of a mutant isoform of ERα that retains at least some transactivation potential in the epithelial compartment is sufficient for pubertal ductal outgrowth to occur (12). Our data suggest that ERα expression is not altogether lost in response to increased O-GlcNAc levels but is instead relegated to a restricted pattern of expression (Fig. 4A to D). A failure to knock out all ERαs may permit some ductal outgrowth, but the markedly reduced ERα signaling may play an indirect role in ductal side-branching morphogenesis. ERα can also act as a transcriptional coactivator of PR. The PR promoter contains critical Sp1 sites that flank an estrogen response element and that are essential to PR expression (18). Our laboratory has previously shown that O-GlcNAc modification of Sp1 impairs its ability to activate transcription (31). In addition, we show here that OGT can associate with ERα in the presence of transcriptional corepressors (Fig. 6A). Thus, it is likely that an existing ERα in the mammary epithelium associates with a corepression complex that contains OGT and that targets O-GlcNAc modification to critical Sp1 sites on the PR promoter. Based on this suggested model, we think that impaired mammary ductal side branching results from impairment of PR in direct or indirect response to increased levels of O-GlcNAc.
The association of OGT with nuclear hormone corepressors in vivo (Fig. 6A) contributes to its ability to specifically repress nuclear hormone signaling pathways. Surprisingly, we observed that increased levels of O-GlcNAc fostered an increase in nuclear hormone receptor corepressors, along with a decrease in coactivators as well as nuclear hormone receptors themselves, at the mRNA level. We demonstrate that transcriptional regulation of the transcriptional activators Sp1 and AIB1 is more sensitive to changes in O-GlcNAc levels than that of SRC1 (Fig. 6). This observation suggests that an additional level of specificity may exist by which some coactivator molecules are more strictly regulated by O-GlcNAc. Thus, increased O-GlcNAc levels foster a state of transcriptional inhibition by increasing transcription of corepressors and decreasing transcription of coactivators. We previously showed that OGT may associate directly with corepressors NCoR and SMRT in a pull-down assay and in immunoprecipitation in which overexpressed tagged proteins were employed (29). Herein, we suggest that this association occurs in vivo and creates a corepressor complex at nuclear hormone receptors (Fig. 6A). By increasing the levels of transcriptional corepressors with which it interacts, OGT enhances its targeting to sites of transcriptional repression. Consistently, estrogen-responsive promoters are associated with O-GlcNAc-modified transcription factors in the absence of hormone (32).
O-GlcNAc regulation of the proteolytic protein degradation by the proteasome also plays a role in regulating complex formation and thus nuclear hormone transactivation. We previously reported that O-GlcNAc modification of the RPT2 ATPase subunit of the proteasome 19S cap acts as an endogenous inhibitor of chymotryptic protein degradation (33). The proteasome is indirectly linked to transcriptional activation. Proteasome-dependent turnover of nuclear hormone receptors and transcription factors is a critical step in transcriptional activation (17). Receptors cycle on and off the chromatin in both the presence and the absence of ligand (19). In the presence of ligand, activation is accomplished by recruitment of a host of coactivator complexes that bind to the liganded receptor. In addition, there is a concomitant disassociation of corepressor complexes which are degraded locally by the proteasome (14, 19). In the case of ERα, the receptor is degraded after binding to a consensus estrogen response element in the presence or absence of hormone (19). During proteasome inhibition, this degradation of ERα cannot occur and the ERα that is present is immobilized on the nuclear envelope (19). ERα is degraded by the proteasome when transcription is blocked by HDAC inhibition by TSA. But, the increased level of O-GlcNAc in MCF10AT cells blocks this proteasomal degradation (Fig. 5). These data imply that O-GlcNAc regulation of the proteasome plays some role in maintaining transcriptional repression. Besides regulating the proteasomal degradation of the corepressor complexes, the increased O-GlcNAc level could do other things to transcription. For example, O-GlcNAc could regulate ERα turnover both directly and indirectly. The coactivator, AIB1, was previously shown to play a role in ERα stability in the presence of hormone (22). We demonstrate that AIB1 expression is impaired at the mRNA level by increased O-GlcNAc levels (Fig. 5). The known direct effect of O-GlcNAc on the proteasome and the effect of this modification on AIB1 suggest that O-GlcNAc regulates ERα proteolytic turnover and therefore activity by more than one mechanism.
We observed that an increase in O-GlcNAc has a specific effect on nuclear hormone receptor signaling, versus other signaling pathways, in vivo. While many proteins are O-GlcNAc modified in vivo, the current work provides evidence that the effects of O-GlcNAc are highly specific in the complex developmental context of organ development. This specificity suggests at least two potential scenarios for O-GlcNAc regulation. Our evidence suggests that OGT is targeted to sites of transcriptional repression by its interaction with other proteins, such as mSin3A, NCoR, and SMRT (1), and that OGT promiscuously associates with other proteins and is activated by a posttranslational modification at distinct intracellular sites (2). Thus, we propose a model in which O-GlcNAc is able to regulate global hormone receptor signaling (Fig. 6B). By inhibition of the proteasome as well as by the association of OGT with corepressor molecules which target O-GlcNAc modification to essential components of the transcriptome, O-GlcNAc maintains a state of transcriptional repression. Specificity for this state of transcriptional repression is hypothesized to be a function of the association of O-GlcNAc with specific corepressor molecules. In the developing mouse mammary gland, this includes inhibition of nuclear hormone receptor signaling pathways. The removal of O-GlcNAc residues at both transcription factors and the proteasome by NCOAT is essential for activation to occur. Proteolytic activity is necessary for the breakdown/removal of corepressor molecules from the system as well as for turnover of coactivator molecules, including ERα. By enabling recycling of transcriptional activator proteins, the removal of O-GlcNAc fosters a system that is primed for activation. In addition, the removal of O-GlcNAc moieties from essential elements of the transcriptome, including Sp1 and RNA polymerase II, plays a critical role in activation. Thus, O-GlcNAc appears to govern the switch from transcriptional repression to activation by integrating proteasome function with direct regulation of transcriptional cofactors. We conclude that O-GlcNAc may operate as a broad regulator of nuclear hormone receptor signaling in diverse processes ranging from development to disease.
Acknowledgments
D. B. Bowe was supported by institutional predoctoral training grant DAMD17-00-1-0119 and is currently supported by a fellowship from the Pharmaceutical Research and Manufacturers of America (PhRMA) Foundation. J. E. Kudlow is supported by an endowment from the Ruth Lawson Foundation. This work was supported by RO-1 grant 2RO1DK43652-14A1 from the NIDDK.
Footnotes
Published ahead of print on 11 September 2006.
REFERENCES
- 1.Bence, N. F., R. M. Sampat, and R. R. Kopito. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292:1552-1555. [DOI] [PubMed] [Google Scholar]
- 2.Bowe, D. B., N. J. Kenney, Y. Adereth, and I. G. Maroulakou. 2002. Suppression of Neu-induced mammary tumor growth in cyclin D1 deficient mice is compensated for by cyclin E. Oncogene 21:291-298. [DOI] [PubMed] [Google Scholar]
- 3.Brisken, C., S. Park, T. Vass, J. P. Lydon, B. W. O'Malley, and R. A. Weinberg. 1998. A paracrine role for the epithelial progesterone receptor in mammary gland development. Proc. Natl. Acad. Sci. USA 95:5076-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 5.Brzozowski, A. M., A. C. Pike, Z. Dauter, R. E. Hubbard, T. Bonn, O. Engstrom, L. Ohman, G. L. Greene, J. A. Gustafsson, and M. Carlquist. 1997. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 389:753-758. [DOI] [PubMed] [Google Scholar]
- 6.Gunther, E. J., G. K. Belka, G. B. Wertheim, J. Wang, J. L. Hartman, R. B. Boxer, and L. A. Chodosh. 2002. A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology. FASEB J. 16:283-292. [DOI] [PubMed] [Google Scholar]
- 7.Humphreys, R. C., J. Lydon, B. W. O'Malley, and J. M. Rosen. 1997. Mammary gland development is mediated by both stromal and epithelial progesterone receptors. Mol. Endocrinol. 11:801-811. [DOI] [PubMed] [Google Scholar]
- 8.Kelly, W. G., M. E. Dahmus, and G. W. Hart. 1993. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J. Biol. Chem. 268:10416-10424. [PubMed] [Google Scholar]
- 9.Li, X., D. M. Lonard, S. Y. Jung, A. Malovannaya, Q. Feng, J. Qin, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 2006. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by the REGgamma proteasome. Cell 124:381-392. [DOI] [PubMed] [Google Scholar]
- 10.Liu, K., A. J. Paterson, F. Zhang, J. McAndrew, K.-I. Fukuchi, J. M. Wyss, L. Peng, Y. Hu, and J. E. Kudlow. 2004. Accumulation of protein O-GlcNAc modification inhibits proteasomes in the brain and coincides with neuronal apoptosis in brain areas with high O-GlcNAc metabolism. J. Neurochem. 89:1044-1055. [DOI] [PubMed] [Google Scholar]
- 11.Lydon, J. P., F. J. DeMayo, C. R. Funk, S. K. Mani, A. R. Hughes, C. A. Montgomery, Jr., G. Shyamala, O. M. Conneely, and B. W. O'Malley. 1995. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 9:2266-2278. [DOI] [PubMed] [Google Scholar]
- 12.Mallepell, S., A. Krust, P. Chambon, and C. Brisken. 2006. Paracrine signaling through the epithelial estrogen receptor alpha is required for proliferation and morphogenesis in the mammary gland. Proc. Natl. Acad. Sci. USA 103:2196-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naar, A. M., B. D. Lemon, and R. Tjian. 2001. Transcriptional coactivator complexes. Annu. Rev. Biochem. 70:475-501. [DOI] [PubMed] [Google Scholar]
- 14.Nagaich, A. K., D. A. Walker, R. Wolford, and G. L. Hager. 2004. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol. Cell 14:163-174. [DOI] [PubMed] [Google Scholar]
- 15.Nandi, S. 1958. Endocrine control of mammary gland development and function in the C3H/He Crgl mouse. J. Natl. Cancer Inst. 21:1039-1063. [PubMed] [Google Scholar]
- 16.Perissi, V., A. Aggarwal, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2004. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116:511-526. [DOI] [PubMed] [Google Scholar]
- 17.Perissi, V., and M. G. Rosenfeld. 2005. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat. Rev. Mol. Cell Biol. 6:542-554. [DOI] [PubMed] [Google Scholar]
- 18.Petz, L. N., Y. S. Ziegler, J. R. Schultz, H. Kim, J. K. Kemper, and A. M. Nardulli. 2004. Differential regulation of the human progesterone receptor gene through an estrogen response element half site and Sp1 sites. J. Steroid Biochem. Mol. Biol. 88:113-122. [DOI] [PubMed] [Google Scholar]
- 19.Reid, G., M. R. Hubner, R. Metivier, H. Brand, S. Denger, D. Manu, J. Beaudouin, J. Ellenberg, and F. Gannon. 2003. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol. Cell 11:695-707. [DOI] [PubMed] [Google Scholar]
- 20.Richert, M. M., K. L. Schwertfeger, J. W. Ryder, and S. M. Anderson. 2000. An atlas of mouse mammary gland development. J. Mammary Gland Biol. Neoplasia 5:227-241. [DOI] [PubMed] [Google Scholar]
- 21.Sadlonova, A., S. Mukherjee, D. B. Bowe, S. R. Gault, N. A. Dumas, B. Van Tine, G. P. Page, D. R. Welch, L. Novak, and A. R. Frost. Human breast fibroblasts inhibit growth of the MCF10AT xenograft model of proliferative breast disease. Am. J. Pathol., in press. [DOI] [PMC free article] [PubMed]
- 22.Shao, W., E. K. Keeton, D. P. McDonnell, and M. Brown. 2004. Coactivator AIB1 links estrogen receptor transcriptional activity and stability. Proc. Natl. Acad. Sci. USA 101:11599-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shekhar, P. V., M. L. Chen, J. Werdell, G. H. Heppner, F. R. Miller, and J. K. Christman. 1998. Transcriptional activation of functional endogenous estrogen receptor gene expression in MCF10AT cells: a model for early breast cancer. Int. J. Oncol. 13:907-915. [DOI] [PubMed] [Google Scholar]
- 24.Slawson, C., N. E. Zachara, K. Vosseller, W. D. Cheung, M. D. Lane, and G. W. Hart. 2005. Perturbations in O-linked beta-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J. Biol. Chem. 280:32944-32956. [DOI] [PubMed] [Google Scholar]
- 25.Toleman, C., A. J. Paterson, and J. E. Kudlow. 2006. Location and characterization of the O-GlcNAcase active site. Biochim. Biophys. Acta 1760:829-839. [DOI] [PubMed] [Google Scholar]
- 26.Toleman, C., A. J. Paterson, T. R. Whisenhunt, and J. E. Kudlow. 2004. Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J. Biol. Chem. 279:53665-53673. [DOI] [PubMed] [Google Scholar]
- 27.Van Tine, B. A., A. J. Patterson, and J. E. Kudlow. 2003. Assignment of N-acetyl-d-glucosaminidase (Mgea5) to rat chromosome 1q5 by tyramide fluorescence in situ hybridization (T-FISH): synteny between rat, mouse and human with insulin degradation enzyme (IDE). Cytogenet. Genome Res. 103:202-204. [DOI] [PubMed] [Google Scholar]
- 28.Wells, L., and G. W. Hart. 2003. O-GlcNAc turns twenty: functional implications for post-translational modification of nuclear and cytosolic proteins with a sugar. FEBS Lett. 546:154-158. [DOI] [PubMed] [Google Scholar]
- 29.Whisenhunt, T. R., X. Yang, D. B. Bowe, A. J. Paterson, B. A. Van Tine, and J. E. Kudlow. 2006. Disrupting the enzyme complex regulating O-GlcNAcylation blocks signaling and development. Glycobiology 16:551-563. [DOI] [PubMed] [Google Scholar]
- 30.Xie, W., A. J. Paterson, E. Chin, L. M. Nabell, and J. E. Kudlow. 1997. Targeted expression of a dominant negative epidermal growth factor receptor in the mammary gland of transgenic mice inhibits pubertal mammary duct development. Mol. Endocrinol. 11:1766-1781. [DOI] [PubMed] [Google Scholar]
- 31.Yang, X., K. Su, M. D. Roos, Q. Chang, A. J. Paterson, and J. E. Kudlow. 2001. O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc. Natl. Acad. Sci. USA 98:6611-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, X., F. Zhang, and J. E. Kudlow. 2002. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell 110:69-80. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, F., K. Su, X. Yang, D. B. Bowe, A. J. Paterson, and J. E. Kudlow. 2003. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell 115:715-725. [DOI] [PubMed] [Google Scholar]