Abstract
MAK (male germ cell-associated protein kinase) and MRK/ICK (MAK-related kinase/intestinal cell kinase) are human homologs of Ime2p in Saccharomyces cerevisiae and of Mde3 and Pit1 in Schizosaccharomyces pombe and are similar to human cyclin-dependent kinase 2 (CDK2) and extracellular signal-regulated kinase 2 (ERK2). MAK and MRK require dual phosphorylation in a TDY motif catalyzed by an unidentified human threonine kinase and tyrosine autophosphorylation. Herein, we establish that human CDK-related kinase CCRK (cell cycle-related kinase) is an activating T157 kinase for MRK, whereas active CDK7/cyclin H/MAT1 complexes phosphorylate CDK2 but not MRK. Protein phosphatase 5 (PP5) interacts with MRK in a complex and dephosphorylates MRK at T157 in vitro and in situ. Thus, CCRK and PP5 are yin-yang regulators of T157 phosphorylation. To determine a substrate consensus, we screened a combinatorial peptide library with active MRK. MRK preferentially phosphorylates R-P-X-S/T-P sites, with the preference for arginine at position −3 (P−3) being more stringent than for prolines at P−2 and P+1. Using the consensus, we identified a putative phosphorylation site (RPLT1080S) for MRK in human Scythe, an antiapoptotic protein that interacts with MRK. MRK phosphorylates Scythe at T1080 in vitro as determined by site-directed mutagenesis and mass spectrometry, supporting the consensus and suggesting Scythe as a physiological substrate for MRK.
Human MAK (male germ cell-associated kinase) (38) and MRK/ICK (MAK-related kinase/intestinal cell kinase) (2, 55) are interesting for their similarity to human cyclin-dependent kinase 2 (CDK2) in the catalytic domain and regulation by dual phosphorylation within a mitogen-activated protein kinase (MAPK)-like TDY motif (17). These kinases are also conserved from yeasts to humans. Saccharomyces cerevisiae has one closely related kinase, Ime2p (inducer of meiosis) (reviewed in reference 23). Ime2p is a meiosis-specific homolog of human CDK2 (44) required for timing meiotic S phase (10, 16) and for regulation of exit from pachytene by control of the key meiotic transcription factor Ndt80 (62).
Schizosaccharomyces pombe encodes two Ime2p-like kinases that are related to MAK and MRK, Mde3 (Mei4-dependent expression) and Pit1 (S. pombe Ime two homolog). Mde3 was discovered as a transcriptional target gene of Mei4 (1), a forkhead transcription factor that is essential for meiotic prophase I (24). Cells that lack Mei4+ fail to make double-strand breaks and arrest before the onset of meiosis I (65). Mde3+ is not transcribed in mei4Δ/mei4Δ cells, suggesting that Mde3+ may be directly downstream of Mei4+ targeting for functions in meiotic recombination in meiosis I or exit from pachytene. Cells lacking Mde3+ grow vegetatively and produce no viable progeny (1). Pit1 may have an overlapping function(s) with Mde3 for meiosis, since deletion of Pit1+ causes a similar meiotic phenotype, and the meiotic phenotype after double deletion (pit1Δ mde3Δ) is more severe (1). Pit1+ unlike Mde3+ is also expressed in vegetative cells executing mitotic cell cycles, suggesting that Pit1 evolved to function in mitotic cell cycles in fission yeast. Remarkably, both MAK and MRK are most similar to Pit1 not Mde3 in the T-loop (see Fig. 1A).
FIG. 1.
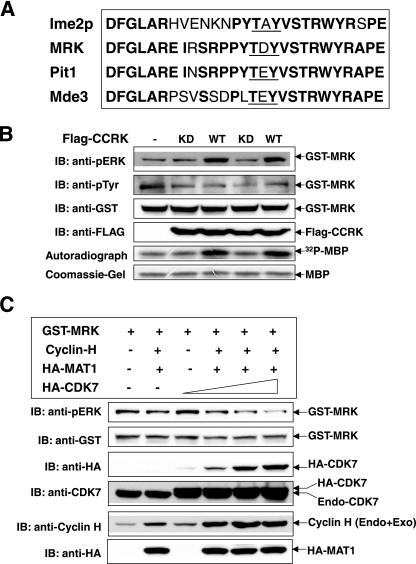
CCRK, but not the Cdk7 complex, increases MRK phosphorylation in the TDY motif in cells. (A) Alignment of the T-loops of MRK, MAK, and their putative homologs in yeasts, Ime2p, Pit1, and Mde3. Identical amino acids are shown in bold type, and the TDY motif is underlined. (B and C) GST-MRK was coexpressed with either WT and KD Flag-CCRK (B) or recombinant Cdk7/cyclin H/MAT1 complex (C) in HEK293T cells. GST fusion proteins were pulled down by glutathione-Sepharose beads and analyzed for MRK phosphorylation in the TDY motif by Western blotting (immunoblotting [IB]) against the anti-phospho-ERK antibody (anti-pERK). The amounts of recombinant proteins on beads and in cell lysate were indicated by Western blotting. After coexpression with either WT or KD Flag-CCRK, GST-MRK bead samples were assayed for kinase activity in vitro using myelin basic protein as the substrate (B). Endo+Exo, endogenous and exogenous.
MAK transcription is induced in prophase I of meiosis of the male germ cells in the rat testis (34, 38), and the MAK protein is expressed in prophase I, peaking in late pachytene (28). This is parallel to the patterns for Mei4 and Mde3 in S. pombe (1). MRK transcription in the gut is confined to the proliferative crypts (55), the same localization as the forkhead transcription factor FOXM1 that is required for mitotic progression (56, 64). (We will refer to the protein herein as MRK, not ICK [55].) MRK can be activated by budding yeast CAK (17), paralleling a genetic requirement of CAK1 for function of IME2 (44). These few points support speculation that MAK and MRK evolved from Mde3 and Pit1 and have important roles in regulation of cell cycle progression in meiosis and/or mitosis.
Functions are yet to be defined for both MAK and MRK. A MAK knockout mouse is viable and fertile (46). Given that MAK and MRK are nearly identical in the catalytic domain, a double knockout of both genes may be required to avoid complementary effects. Meanwhile, very little is known about their physiologic substrates and regulators. Previously, we have shown that MRK is activated by dual phosphorylation of the TDY motif (17). Autophosphorylation on Tyr-159 in the TDY motif confers basal kinase activity only. Full activation of MRK requires additional phosphorylation of Thr-157 in the TDY motif. Furthermore, we showed that yeast Cak1p can serve as a T157 kinase, implying that a mammalian CAK (CDK-activating kinase) may emerge as a physiologic MRK activator (17).
Here, we report new biochemical parallels of MRK to what is known about the yeast homologs. We show that a human kinase CCRK (cell cycle-related kinase)/p42/PNQLARE (35, 59) that is related to yeast Cak1p, but not CDK7, can phosphorylate MRK at the essential Thr-157 in the T-loop. We performed a yeast two-hybrid screen for MRK interactors and identified human protein phosphatase 5 (PP5) and human Scythe (BAT3 [HLA-B-associated transcript 3]). We show that PP5 can deactivate MRK by dephosphorylation of the activating phosphorylation site Thr-157 in the T-loop catalyzed by CCRK, thus establishing CCRK and PP5 as the first human kinase/phosphatase pair to regulate the T-loop phosphorylation of MRK. We also establish in vitro that MRK can phosphorylate Scythe, an antiapoptotic protein required for apoptosis and proliferation during mammalian development (13). Using a novel, rapid method (25) for determining a phosphorylation consensus, we found that MRK prefers sites in an R-P-X-S/T-P, with less stringency for the position +1 (P+1) proline than CDKs or MAP kinases. Using this consensus, we identified an in vivo site T1080 in the BAG domain of Scythe as a major MRK phosphorylation site in vitro. The site is similar to the only identified phosphorylation site for Ime2p in yeast replication protein A (10). Our findings suggest a hypothesis that MRK and MAK are involved in regulation of the cell cycle and cell fate, functions that may have evolved from, and still overlap with, yeast functions.
MATERIALS AND METHODS
Reagents.
The human fetal brain cDNA Matchmaker library (Clontech) was provided by Bert Vogelstein (Howard Hughes Medical Institute, Johns Hopkins University). Sandra Rossie (Department of Biochemistry, Purdue University) provided homogeneous PP5 purified from Escherichia coli and Flag-tagged human PP5(H304Q) plasmid. Full-length PP5, PP5 containing amino acids (aa) 1 to 159 [PP5(1-159)], and PP5(160-499) were PCR amplified and subcloned into pCDNA3-Flag vector at 5′ BamHI and 3′ XhoI sites by Guofei Zhou of Richard Honkanen's lab (Department of Biochemistry and Molecular Biology, University of South Alabama). His6-tagged murine MRK(1-296) [His-MRK(1-296)] was purified from Sf9 cells by Xiaoshan Min of Elizabeth Goldsmith's lab (Department of Biochemistry, University of Texas at Southwestern Medical Center). Andrew Hubberstey (Department of Biological Sciences, University of Windsor, Ontario, Canada) provided the pCI-HA-Scythe plasmid (37). Philipp Kaldis (Mouse Cancer Genetic Program, National Cancer Institute) and David Morgan (Department of Physiology, University of California at San Francisco) provided pCDLSRα296-HA-Cdk7, pCDLSRα296-cyclin H, and pCDNA3.1-HA-MAT1 plasmids. Konstantin Galaktionov (Department of Molecular and Human Genetics, Baylor College of Medicine) provided wild-type (WT) pCDNA3.1-Flag-CCRK and T161A mutant plasmids (35). Didier Trono's laboratory (National Center for Competence in Research, Switzerland) provided the pCMV-dR8.74psPAX2 packaging plasmid and the pMD2G envelope plasmid. Active Cdk7/cyclin H/MAT1 complex and mouse monoclonal antiphosphotyrosine antibody (clone 4G10) were purchased from Upstate Biotechnology. Rabbit polyclonal antibody against cyclin H was purchased from Cell Signaling Technology. Mouse monoclonal antibody against Cdk7 (clone MO1.1) was purchased from Sigma-Aldrich. Rabbit polyclonal antibody against CCRK was a kind gift from Robert Fisher (Memorial Sloan-Kettering Cancer Center) (59). Mouse monoclonal antibody against protein phosphatase 5 was purchased from BD Transduction Laboratories. Arachidonic acid, okadaic acid, anti-FLAG M2 affinity gel, 1× FLAG peptide, MISSION TRC (The RNA: Consortium) short hairpin RNA (shRNA) target set for CCRK, and the nontarget shRNA control vector were all purchased from Sigma-Aldrich. Hydrogen peroxide was purchased from Fisher Scientific.
Yeast two-hybrid screen.
To identify MRK-interacting proteins, a human fetal brain cDNA library (Clontech) was amplified and screened with human MRKb. The human MRKb variant cDNA encodes 292 amino acid residues, with its residues 1 to 277 being identical to human MRK, overlapping most of the catalytic domain (aa 1 to 284) of MRK (17). MRKb was cloned into bait vector pGBKT7 that encodes an N-terminal Myc tag. The bait plasmid pGBKT7-MRKb was transformed into the yeast host strain AH109. Transformants were selected on plates lacking Trp. The expression of MRKb in AH109 transformants was confirmed by Western blotting against the mouse anti-Myc monoclonal antibody (clone 9E10) (Invitrogen). The yeast MRKb transformants were subsequently transformed with prey cDNA library in a pACT2 vector. The screen for interaction between bait and prey was performed first with medium stringency on plates lacking Leu, Trp, and His. Candidates obtained (501 clones) were further evaluated either for growth under higher stringency on plates lacking Leu, Trp, His, and Ade or growth under medium stringency on plates containing 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (X-Gal). A total of 20 candidates grew on plates at higher stringency. A total of 65 candidates regrew on plates at medium stringency and were visibly blue. Ten candidate clones that were positive on both rescreens were transformed back into yeast AH109 together with either pGBKT7-MRKb or pGBKT7-p53 (control) or prey vector pGBKT7 (control). Four of them were confirmed as real positives in yeast. Blast search at NCBI revealed that clone 3 is a full-length, in-frame cDNA clone encoding protein phosphatase 5, and clone 389 is a partial, in-frame cDNA clone encoding human BAT3, also known as human Scythe or BAG-6 (Bcl2-athanogene 6).
Cell culture and transient transfection.
HEK293T cells were maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium with a high level of glucose supplemented with 10% fetal bovine serum. At about 30% to 40% confluence, HEK293T cells in 10-cm dishes were transfected by using a calcium phosphate protocol (17).
GST pull-down binding assay.
Glutathione S-transferase (GST)-MRK (10 to 12 μg) or control GST (1 to 2 μg) was cotransfected with 6 to 8 μg of either Flag-tagged PP5 (Flag-PP5), or Flag-tagged CCRK (Flag-CCRK), or hemagglutinin (HA)-tagged Scythe (HA-Scythe) in HEK293T cells. Forty-eight hours after transfection, cells were harvested in ice-cold phosphate-buffered saline and lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 2 mM EGTA, and supplemented with complete protease inhibitors [Roche], 1 mM Na3VO4, 1 μM microcystin LR, and 5 mM β-glycerophosphate). The cell lysate was cleared by centrifugation. A portion of the cell lysate was saved for Western blotting to indicate protein signal input. The rest of the cell lysate was incubated with glutathione-Sepharose beads (Amersham Biosciences) for 2 h at 4°C to absorb GST fusion proteins. The beads were washed extensively with lysis buffer followed by phosphate-buffered saline buffer. The beads were boiled in sodium dodecyl sulfate (SDS) sample buffer (0.5 M Tris, ph 6.8, 10% SDS, 10% glycerol, 0.1% bromphenol blue) for 5 min to elute binding proteins. The lysate and bead eluates were subjected to Western blotting against the following antibodies: 0.2 μg/ml anti-GST (Santa Cruz B-14), 0.1 μg/ml anti-Flag M2 (Sigma), 0.1 μg/ml anti-HA (Sigma) and 2.0 μg/ml mouse anti-protein phosphatase 5 (BD Transduction Laboratories).
In vivo CCRK/Cdk7 phosphorylation.
To test CCRK, GST-MRK (10 μg) was cotransfected into HEK293T cells with either WT Flag-CCRK or T161A mutant (6 μg). To test Cdk7 complex, GST-MRK (10 μg) was cotransfected into HEK293T cells with cyclin H (4 μg), HA-MAT1 (6 μg), and HA-Cdk7 (2, 4, or 8 μg). For a control, GST-MRK was either transfected alone, cotransfected with only cyclin H and HA-MAT1, or cotransfected with only HA-Cdk7. Glutathione-Sepharose beads were incubated with cell lysate to pull down GST-MRK fusion proteins. The beads were extensively washed, boiled in SDS sample buffer, and subjected to Western blotting against the anti-phosphorylated extracellular signal-regulated kinase (anti-phospho-ERK) and anti-GST antibodies. To assess the kinase activity of GST-MRK after coexpression with either WT Flag-CCRK or T161 mutant, the bead samples were subjected to in vitro kinase assay using myelin basic protein (MBP) as the substrate (17).
Silencing CCRK gene expression by lentiviral shRNAs.
The MISSION TRC shRNA target set contains four short hairpin sequences targeting human CCRK mRNA: 5′-TCTTGAGGAGTCGCTGTTGAA-3′ (TRCN2215), 5′-AGAACGATATTGAACAGCTTT-3′ (TRCN2216), 5′-CCCAATATAATCCCAGCTAGT-3′ (TRCN2217), and 5′-TGCCGGACTACAACAAGATCT-3′ (TRCN2218). The nontarget shRNA control vector contains a hairpin insert sequence (5′-CAACAAGATGAAGAGCACCAA-3′) that contains 4-base-pair mismatches to any known human or mouse genes.
To generate lentiviral vector particles, HEK293T cells were cotransfected with three plasmids (pLKO.1-puro-shRNA, pCMV-dR8.74psPAX2 packaging plasmid, and pMD2G envelope plasmid) using a calcium phosphate protocol. Cell medium containing viral particles was harvested and used to infect 50% to 60% confluent HEK293T cells. Forty-eight hours after infection, cells were harvested and subjected to either analysis of CCRK gene expression by reverse transcription-PCR and Western blotting or split for transfection with GST-MRK. Forty-eight hours after transfection, cells were harvested and analyzed for CCRK gene expression and TDY motif phosphorylation of GST-MRK.
Purification of Flag-CCRK and in vitro CCRK phosphorylation.
WT Flag-CCRK and T161A mutant were each expressed in 10 10-cm plates of HEK293T cells (10 μg plasmid DNA per plate). Cell lysates pooled from 10 plates were incubated with 0.5 ml of anti-FLAG M2 affinity resin at 4°C for 3 to 4 h. After batch binding, the sample mixture was poured onto a 2-ml affinity column. The column was extensively washed in cold Tris-buffered saline (TBS) until the optical density at 280 nm was almost zero, and then the column was eluted with 0.1 mg/ml 1× FLAG peptides in TBS. The eluates were pooled, dialyzed in TBS, concentrated, and stored frozen.
GST-MRK(1-300) (1 to 2 μg) purified from E. coli was incubated with WT Flag-CCRK or T161A mutant (2 to 3 μg), 5 μCi [γ-32P]ATP (7,000 Ci/mmol), and 50 μM ATP in 30 μl kinase buffer (50 mM HEPES, pH 7.5, and 10 mM MgCl2 supplemented with 5 mM dithiothreitol [DTT], complete protease inhibitors [Roche], 1 mM Na3VO4, 1 μM microcystin LR, and 5 mM β-glycerophosphate) at 30°C for 30 min. The reaction was terminated by the addition of 30 μl of 2× SDS sample buffer.
In vitro phosphatase assay.
GST-MRK was expressed in HEK293T cells and purified as described previously (17) using glutathione-Sepharose beads. The GST-MRK on bead samples was washed in lysis buffer followed by HEPES buffer (50 mM HEPES, pH 7.5, 1 mM EDTA, 1 mM EGTA, and 5 mM DTT supplemented with complete protease inhibitors [Roche]) and resuspended in a final volume of 50 μl HEPES buffer. The reaction was initiated by the addition of 0.2 to 0.4 μM purified Flag-PP5 enzyme (26) in the presence or absence of 50 μM arachidonic acid, and the mixture was incubated at 30°C for 10 to 20 min. The phosphatase reaction was terminated by the addition of 50 μl 2× SDS sample buffer. The sample was boiled for 5 min and subjected to Western blotting against the anti-phospho-ERK and anti-MRK (17) antibodies. To inhibit phosphatase activity of Flag-PP5, 1 μM okadaic acid was added to the reaction mixture.
In vivo PP5 dephosphorylation.
GST-MRK (10 μg) was cotransfected with either full-length Flag-PP5 (2, 4, or 8 μg) or the tetratricopeptide repeat (TPR) domain only Flag-PP5(1-159) (1.5, 3.0, or 6.0 μg) into HEK293T cells. For a control, WT and kinase-defective (KD) GST-MRK were transfected alone into HEK293T cells. The cell lysate was incubated with glutathione-Sepharose beads to pull down GST-MRK. The beads were extensively washed, boiled in SDS sample buffer, and subjected to Western blotting against the anti-phospho-ERK and anti-MRK (17) antibodies. To assess the kinase activity of GST-MRK after coexpression with either full-length Flag-PP5 or TPR domain only [Flag-PP5(1-159)], the bead samples were subjected to in vitro kinase assay using MBP as the substrate (17).
Plasmid construction.
The point mutations S362A, T1080A, and S362A T1080A in full-length Scythe were generated by using a four-primer PCR method (17) and cloned into pCI-HA vector (37) at 5′ XbaI and 3′ NotI sites. All mutations were confirmed by DNA sequencing.
In vitro Scythe phosphorylation.
HA-tagged human WT Scythe or mutants (S362A, T1080A and S362A T1080A) were transfected into HEK293T cells. Forty-eight hours after transfection, cells were harvested and lysed. To pull down HA-Scythe, the cell lysate was cleared by centrifugation and subsequently incubated with anti-HA (2 to 3 μg/ml) monoclonal antibody plus protein A-Sepharose beads at 4°C for 3 to 4 h. The beads were extensively washed in lysis buffer followed by kinase buffer (50 mM HEPES, pH 7.5, and 10 mM MgCl2 supplemented with 5 mM DTT and complete protease inhibitors [Roche], 1 mM Na3VO4, 1 μM microcystin LR, and 5 mM β-glycerophosphate). The bead samples were incubated with 5 μCi [γ-32P]ATP (7,000 Ci/mmol), 100 μM ATP, and 1 to 2 μg of affinity-purified His-MRK(1-296) in 50 μl kinase buffer at 30°C for 15 to 30 min with gentle agitation. The reaction was terminated by the addition of 50 μl of 2× SDS sample buffer. The reaction sample was heated at 95°C for 5 min and separated on a 10% SDS gel. The gel was dried and exposed for autoradiographic and PhosphorImager analyses.
Purification of His-MRK(1-296) from Sf9 cells and GST-MRK(1-300) from E. coli.
MRK(1-296) was PCR amplified and subcloned into pFastBac at 5′ EcoRI and 3′ SpeI sites. Baculovirus generation and His-tagged fusion protein expression and purification were performed with Bac-to-Bac baculovirus expression system (Invitrogen) per the manufacturer's instructions by Xiaoshan Min in the laboratory of Elizabeth Goldsmith (University of Texas at Southwestern Medical Center).
Cloning of MRK(1-300) into pGEX4T-1 and production of GST-MRK(1-300) fusion protein in E. coli were done essentially as described previously (17). GST fusion was affinity purified on a column containing glutathione-Sepharose beads following the manufacturer's (Pierce) instructions.
Determination of MRK phosphorylation specificity.
A combinatorial peptide library method (25) that provides phosphorylation motifs for serine/threonine kinases was used to determine MRK phosphorylation specificity. The library consists of 198 peptides having the general sequence M-A-X-X-X-X-X-S/T-X-X-X-X-A-G-K-K(biotin), where X is an equimolar mixture of all amino acids save Ser, Thr, and Cys, and S/T indicates an even mix of Ser and Thr. In a given peptide, one X position is fixed as one of the 20 unmodified amino acids, phosphothreonine or phosphotyrosine. Peptides (50 μM) were arrayed in microtiter plates and phosphorylated by either His-MRK(1-296) purified from Sf9 cells (at 10 μg/ml) or GST-MRK(1-300) purified from E. coli (at 50 μg/ml) in a solution of 50 mM Tris, pH 7.5, 10 mM MgCl2, 5 mM DTT, 5 mM β-glycerophosphate, 0.1 mM Na3VO4, 0.1% Tween 20 with 50 μM [33P]ATP (0.033 μCi/μl). After incubation for 2 h at 30°C, aliquots were spotted onto a streptavidin membrane, which was washed, dried, and exposed to a phosphor screen as described previously (25).
Mass spectrometry.
On-bead digestion was performed as described previously (17) to identify the phosphorylation state of the TDY motif in MRK. Peptides obtained after trypsin digestion were separated by online nanoflow high-pressure liquid chromatography (HPLC) and analyzed using an LTQ-FT mass spectrometer (Thermo Electron, San Jose, CA). A gradient of 0 to 60% B (70% acetonitrile, 100 mM acetic acid in water) in 40 min was used. A targeted mode, as previously described (17), was used for the identification of the phosphorylation state of the TDY motif in MRK.
To identify in vitro and in vivo phosphorylation sites in Scythe, in-gel digestion with trypsin and chymotrypsin were performed (45). Extracted peptides from Scythe in-gel digestion were analyzed online, using the HPLC conditions described above, in a data-dependent tandem mass spectrometry (MS) mode. One full MS in the Fourier transform ion cyclotron resonance mass spectrometer was followed by tandem MS of the 10 most abundant ions in the ion trap (isolation width of 3 and collision energy of 35%). The relative abundance, reported as a percentage, of each form of a peptide (mass accuracy within 2 ppm) was calculated as a ratio to the total abundance of all forms of that peptide. These percentages were then used to compare the relative amounts of phosphorylation in the samples.
RESULTS
Human CCRK but not CDK7 CAK phosphorylates MRK at T157 in vivo.
Since we have shown previously that yeast Cak1p functions as an activating T157 kinase for MRK (17), we first tested the Cdk7 complex (CDK7/cyclin H/MAT1) that has the highest CAK activity for CDK2 after biochemical fractionation and purification from HeLa cells (30). CDK7/cyclin H/MAT1 (Upstate) was active, as it phosphorylated CDK2/cyclin A as a control, but CDK7/cyclin H/MAT1 (Upstate) did not detectably phosphorylate MRK in vitro (data not shown). Human p42/PNQLARE, which we will refer to herein by the NCBI locus name CCRK, was identified bioinformatically as the human kinase with highest similarity to yeast Cak1p (35). Using SSEARCH (42) (at fasta.bioch.virginia.edu), the most closely related kinases to yeast Cak1p in humans are confirmed to be CCRK first, and then CDK3 followed by CDK7.
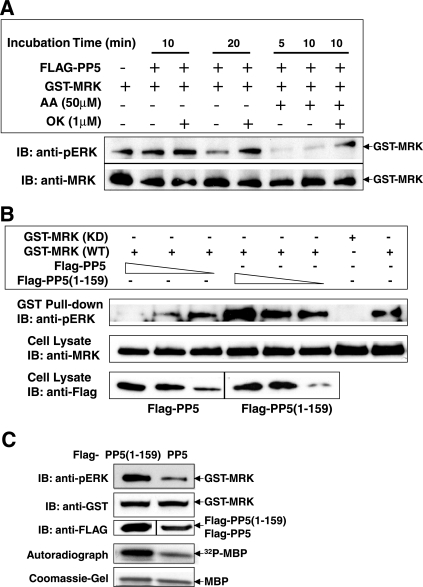
To determine whether human CCRK is an activator of MRK, we coexpressed GST-MRK (17) with wild-type or kinase-defective FLAG-tagged CCRK (35) in cells. There are three alternative transcripts of CCRK, and we used the longer isoform (346-aa isoform [NCBI accession no. NM_001039803]). The two shorter isoforms (338-aa isoform [accession no. NM_178432] and 326-aa isoform [accession no. NM_012119]) lack an alternative in-frame coding exon, thus may lack the protein kinase activity found in the longer isoform. Transfection of wild-type CCRK but not the kinase-defective form increased phosphorylation of GST-MRK in the regulatory TDY motif and the kinase activity of GST-MRK (Fig. 1B), assessed by cross-reactivity of the dually phosphorylated pTDpY motif to anti-phospho-ERK (17) and MBP phosphotransferase activity, respectively. This result establishes that CCRK can phosphorylate and activate MRK in cells. Increased phosphorylation of MRK in the TDY motif required the kinase activity of CCRK, demonstrated in parallel control experiments using kinase-defective CCRK (Fig. 1B). Since CCRK is Ser/Thr specific, CCRK most likely phosphorylates T157 in the TDY motif. The reactivity of GST-MRK to phosphotyrosine antibody was not significantly altered by coexpression with the wild-type and kinase-defective forms of CCRK (Fig. 1B), suggesting that CCRK specifically targets T157 in the TDY motif of MRK. T157 aligns with the activation loop threonine of CDK2 that is phosphorylated by the CDK7/cyclin H/MAT1 complex (17).
We also coexpressed CDK7/cyclin H/MAT1 with GST-MRK in cells (Fig. 1C). Western blotting showed that all three components were successfully overexpressed, but GST-MRK phosphorylation in the TDY motif was not increased; instead phosphorylation was slightly decreased with an increasing amount of overexpressed Cdk7. Given the recent evidence that certain fractions of Cdk7 and CCRK coexist in the same complex (59), it is conceivable that overexpressed HA-Cdk7 may behave in a dominant-negative fashion by sequestering endogenous CCRK from interacting with GST-MRK. This negative result cannot totally exclude CDK7 as an activator, but CCRK phosphorylates MRK in vivo, whereas in experiments with the same design, CDK7/cyclin H/MAT1 does not. Therefore, our data suggest that CCRK is an upstream activating kinase for MRK in vivo.
Downregulation of endogenous CCRK gene expression significantly decreased recombinant MRK phosphorylation in the TDY motif.
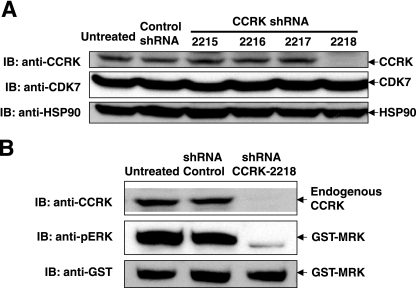
To determine whether endogenous CCRK is an upstream activating kinase for MRK, four different MISSION TRC lentiviral shRNAs designed to specifically target CCRK (Sigma) were tested to downregulate CCRK expression in HEK293T cells. In contrast to the nontarget control shRNA, shRNA-2218 was able to significantly reduce the expression of CCRK at both the message level (reverse transcription-PCR data not shown) and the protein level but had no detectable effect on the expression of its nonspecific targets, such as CDK7 and heat shock protein 90 (Hsp90) (Fig. 2A). Using shRNA-2218, we then tested whether downregulation of CCRK expression would reduce MRK phosphorylation in the TDY motif. Since a phospho-MRK antibody specific for endogenous MRK is not available, we examined the TDY phosphorylation of recombinant MRK in HEK293T cells by Western blotting against the anti-phospho-ERK antibody (17) and by mass spectrometry. Knockdown of endogenous CCRK significantly impaired the TDY motif phosphorylation of GST-MRK, assessed by the anti-phospho-ERK signal (Fig. 2B). By mass spectrometry, we have shown previously that a small fraction of GST-MRK overexpressed in HEK293T cells is either dually phosphorylated or singly tyrosine phosphorylated in the TDY motif (17). Downregulation of CCRK expression resulted in a decrease in the abundance of doubly phosphorylated GST-MRK but essentially no change in the abundance of singly tyrosine-phosphorylated GST-MRK (data not shown), suggesting that CCRK specifically phophorylates T157 in the TDY motif.
FIG. 2.
Downregulation of endogenous CCRK expression impairs phosphorylation of GST-MRK in the TDY motif. (A) HEK293T cells were infected with either four different lentiviral shRNAs targeting the CCRK gene or a nontarget control lentiviral shRNA. Two days after infection, cells were harvested and lysed. Expression of endogenous CCRK in whole-cell lysate was analyzed by Western blotting (immunoblotting [IB]) against the anti-CCRK antibody. Expression of CDK7 and Hsp90 was analyzed as controls. (B) HEK293T cells that were either uninfected or infected with nontarget control lentiviral shRNA or CCRK-specific lentiviral shRNA-2218 were transfected with GST-MRK. Two days after transfection, cells were harvested and lysed. Expression of CCRK and GST-MRK in whole-cell lysate was analyzed by Western blotting against the anti-CCRK and anti-GST antibodies, respectively. Phosphorylation of GST-MRK in the TDY motif was assessed by Western blotting against the anti-phospho-ERK antibody (anti-pERK).
CCRK interacts with MRK and phosphorylates MRK in vitro.
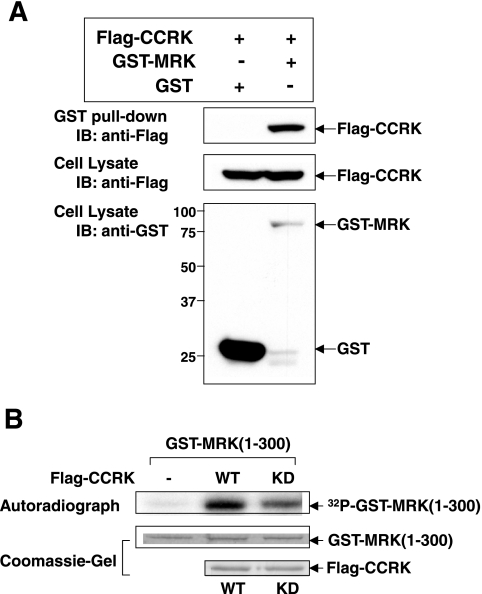
To test whether CCRK has affinity for binding MRK, we examined association of Flag-CCRK with GST-MRK versus control GST after purification from lysates with glutathione beads (Fig. 3A). GST is expressed at a much higher level than GST-MRK is; nevertheless, no Flag-CCRK was detectable in washed beads with bound GST, but a portion of Flag-CCRK was bound by GST-MRK. We conclude that CCRK has a selective affinity for binding MRK.
FIG. 3.
Recombinant CCRK interacts with recombinant MRK in cells and phosphorylates recombinant MRK in vitro. (A) GST-MRK or GST was coexpressed with Flag-CCRK in HEK293T cells. GST fusion proteins were pulled down by glutathione-Sepharose beads and analyzed for association with Flag-CCRK by Western blotting (immunoblotting [IB]) against the anti-Flag antibody. The amounts of GST, GST-MRK, and Flag-CCRK in cell lysate were indicated by Western blotting against the anti-GST and anti-Flag antibodies. The positions of molecular mass markers (in kilodaltons) are shown to the left of the blot. (B) GST-MRK(1-300) purified from E. coli was assayed for in vitro phosphorylation by WT or KD Flag-CCRK purified from HEK293T cells. MRK phosphorylation (autoradiograph) and the amounts of substrates and kinases (Coomassie blue-stained gels) are shown.
To test whether CCRK phosphorylates MRK in vitro, we purified both wild-type and kinase-defective Flag-tagged CCRK proteins from HEK293T cells. Since E. coli-expressed GST-MRK(1-300) is not phosphorylated on Thr-157 in the T-loop (17), we chose it as the in vitro substrate to test for phosphorylation by CCRK. The autoradiograph indicated that wild-type CCRK phosphorylated the MRK substrate to a greater extent than kinase-defective CCRK did, suggesting MRK is an in vitro substrate for CCRK (Fig. 3B). This effect is not due to the difference in the amount of proteins between the wild type and kinase-defective mutant because a Coomassie blue-stained SDS gel indicated equal loading.
MRK interacts with protein phosphatase 5.
We performed a yeast two-hybrid screen of a human fetal brain cDNA library using as bait C-terminally truncated MRK, encoded by an alternatively spliced cDNA (see Materials and Methods for details). This cDNA encodes almost the entire catalytic domain of murine MRK. We identified a full-length clone for human PP5 in this screen.
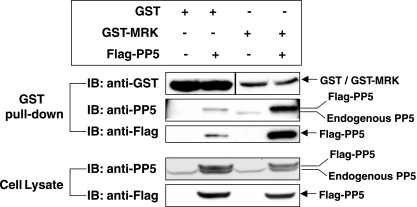
To test for interaction in cells, we either expressed GST-MRK alone or coexpressed GST-MRK with Flag-PP5 in cells and then searched for the presence of both Flag-PP5 and endogenous PP5 copurifying with GST-MRK compared to GST on glutathione beads. Both Flag-PP5 and endogenous PP5 interacted, directly or indirectly, with MRK (Fig. 4).
FIG. 4.
Recombinant MRK interacts with both recombinant and endogenous PP5 in cells. GST-MRK or GST was expressed alone or coexpressed with Flag-PP5 in HEK293T cells. GST fusion proteins were purified on glutathione-Sepharose beads and analyzed for association with recombinant and/or endogenous PP5 by Western blotting (immunoblotting [IB]) against the anti-Flag and anti-PP5 antibodies, respectively. The amounts of GST fusion proteins on beads and the amounts of Flag-PP5 and endogenous PP5 in cell lysate were indicated by Western blotting.
Interaction of MRK and PP5 requires the TPR domain.
PP5 has an N-terminal regulatory tetratricopeptide repeat domain and a C-terminal catalytic domain (63). TPR domains mediate protein-protein interactions. For example, Rac1-GTP interacts with the TPR domain of PP5 and was proposed as a physiologic regulator of PP5 (18). Therefore, we examined whether the TPR domain is required for interaction of MRK with PP5.
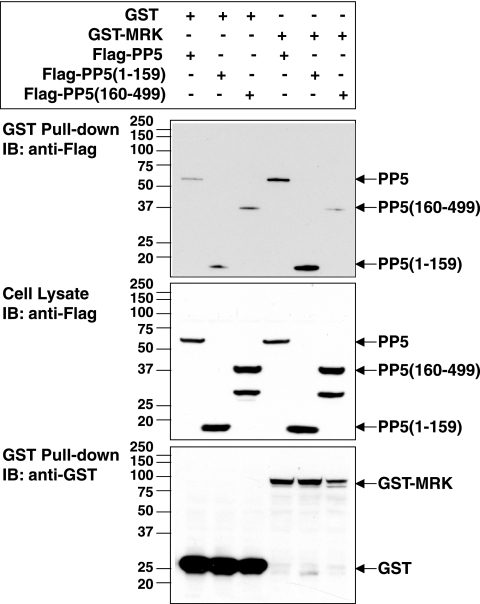
We compared the abilities of Flag-PP5, Flag-PP5(1-159) possessing the TPR domain, and Flag-PP5(160-499) lacking the TPR domain to interact with GST-MRK in cells using a glutathione bead pull-down assay (Fig. 5). GST was expressed more robustly than GST-MRK was, even with the much smaller amount of GST plasmid than of GST-MRK plasmid used for transfection. Flag-PP5, Flag-PP5(1-159), and Flag-PP5(160-499) were present in detectable amounts with GST, due to nonspecific binding to the large amount of GST on beads. In contrast to Flag-PP5(160-499) lacking the TPR domain, Flag-PP5 and Flag-PP5(1-159) were reproducibly copurified in significantly greater amounts with GST-MRK than with GST. We conclude that the TPR domain is sufficient for binding with GST-MRK complex. The binding may be direct or indirect through chaperones because MRK and MAK are known to interact with Hsp90 and Cdc37 (39) and PP5 is present in complexes with Hsp90 (11, 66).
FIG. 5.
The TPR domain of PP5 is required for the interaction between PP5 and MRK. GST-MRK or GST was coexpressed with Flag-tagged full-length PP5 or PP5(1-159), consisting of only the N-terminal TPR domain, or PP5(160-499), lacking the TPR domain, in HEK293T cells. GST fusion proteins were purified on glutathione-Sepharose beads and analyzed for association with Flag-PP5 as described in the legend to Fig. 4. Note that the anti-Flag antibody also recognized a 30-kDa band in addition to the 37-kDa band of Flag-PP5(160-499) (middle gel), which is probably a product of proteolysis or truncation in translation. IB, immunoblotting.
The TPR domain and the C terminus constrain PP5 catalytic activity in its unliganded state by forming the TPR-phosphatase domain interface (51, 63). Both Hsp90 and fatty acids, including arachidonic acid, interact with the TPR domain and stimulate phosphatase activity by either disrupting or destabilizing the autoinhibitory TPR-phosphatase domain interface (63). Thus, proteins that are acceptors for the PP5 TPR domain or in the same complex are potential regulators and/or substrates. Therefore, we tested whether MRK is a substrate for PP5.
PP5 dephosphorylates MRK at T157 in vitro and in vivo.
A portion of HEK293T-expressed GST-MRK recovered on glutathione beads is dually phosphorylated on the TDY motif (17). Dual phosphorylation of the pTDpY form of MRK can be detected by cross-reactivity to anti-phospho-ERK (17). To test whether PP5 is a MRK phosphatase in vitro, we incubated GST-MRK on beads with homogeneous Flag-PP5 purified from E. coli (26), after washing and equilibrating the beads in phosphatase assay buffer to remove phosphatase inhibitors (Fig. 6A). PP5 significantly decreased the Western blot signal of MRK against the anti-phospho-ERK after 20 min of incubation. This indicates dephosphorylation of MRK at T157, since PP5 is specific for phosphoserine and phosphothreonine (9). Activation of PP5 by the addition of arachidonic acid (8, 48) caused nearly complete loss of the anti-phospho-ERK signal after a shorter incubation period (5 min) under the same assay conditions. Furthermore, the dephosphorylation of MRK in the TDY motif by activated PP5 can be prevented by the PP5 inhibitor okadaic acid.
FIG. 6.
PP5 dephosphorylates MRK in the TDY motif in vitro and in vivo. (A) GST-MRK purified on glutathione-Sepharose beads from HEK293T cells was incubated with purified Flag-PP5 protein in the absence (−) or presence (+) of 50 μM arachidonic acid (AA) in vitro. For a control, 1 μM okadaic acid (OK) was added to inhibit PP5 phosphatase activity. After the reaction, the GST-MRK bead samples were analyzed for MRK phosphorylation in the TDY motif by Western blotting (immunoblotting [IB]) against the anti-phospho-ERK antibody (anti-pERK). The amount of GST-MRK on beads was indicated by Western blotting against the anti-MRK antibody. (B) GST-MRK was coexpressed with either the full-length Flag-PP5 or the catalytic inactive form Flag-PP5(1-159) in HEK293T cells. GST fusion proteins were purified on glutathione-Sepharose beads and analyzed for MRK phosphorylation in the TDY motif as described above for panel A. The amounts of GST-MRK, Flag-PP5, and Flag-PP5(1-159) in cell lysate were indicated by Western blotting against the anti-MRK and anti-Flag antibodies. KD GST-MRK was used as a negative control for TDY motif phosphorylation. (C) GST-MRK was coexpressed with either the full-length Flag-PP5 or the catalytic inactive form Flag-PP5(1-159) in HEK293T cells. GST fusion proteins were analyzed for MRK phosphorylation in the TDY motif as described above for panel B and for MRK kinase activity in vitro using MBP as the substrate.
To test whether PP5 is a phosphatase for MRK in vivo, we coexpressed GST-MRK with either full-length Flag-PP5 or the catalytically inactive form Flag-PP5(1-159) in HEK293T cells (Fig. 6B). Increasing the expression level of the full-length PP5, but not the catalytically inactive form of PP5, caused a marked decrease in the TDY motif phosphorylation of MRK detected by the anti-phospho-ERK antibody. This decrease in the anti-phospho-ERK signal is not due to a decrease in the amount of GST-MRK because the anti-MRK blotting indicated a constant expression level of MRK. Similar to the TPR domain-only form of PP5 [Flag-PP5(1 to 159)], the full-length PP5 containing a catalytic inactive mutation (H308Q) also failed to alter the TDY motif phosphorylation of GST-MRK compared to wild-type PP5 (data not shown). It is also worth pointing out that overexpression of an increasing amount of PP5(1-159) seems to cause an increase in the reactivity of GST-MRK against the anti-phospho-ERK. This effect may be explained by the dominant-negative effect of the TPR domain-only form of PP5 because the TPR domain is sufficient to interact with GST-MRK (Fig. 5), thus capable of preventing GST-MRK from interacting with and being dephosphorylated by endogenous PP5. Dephosphorylation of MRK in the TDY motif by PP5, but not the catalytically inactive form of PP5, significantly decreased the kinase activity of MRK, assessed by MBP phosphotransferase activity in vitro (Fig. 6C). We have both in vitro and in vivo evidence that PP5 can dephosphorylate MRK in the essential phosphothreonine residue within the TDY motif, thus downregulating MRK kinase activity.
Hydrogen peroxide activates PP5 and induces MRK dephosphorylation.
Hydrogen peroxide treatment is a cellular stress that induces activation of ASK1 (apoptosis signal-regulating kinase 1), a MAPK kinase kinase that functions upstream of MEK4 and MEK6 to activate Jun N-terminal protein kinase and p38 MAPKs (19, 40). Concurrently, a competing negative-feedback pathway that specifically affects the MEK4-Jun kinase module and that is mediated by PP5-dependent dephosphorylation of ASK1 is activated (40, 68). Hydrogen peroxide is also a DNA-damaging agent that activates ATM (ataxia-telangiectasia mutated)- or ATR (ATM- and Rad3-related)-dependent checkpoints (5).
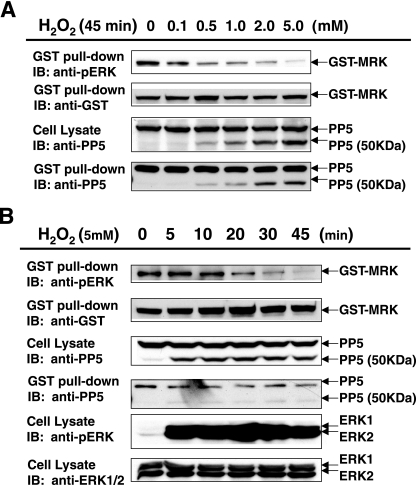
With this motivation, we examined the effect of hydrogen peroxide treatment of cells expressing GST-MRK (Fig. 7). The lowest concentration tested (0.1 mM final concentration) sufficed to induce significant dephosphorylation of GST-MRK within the TDY motif (Fig. 7A). The hydrogen peroxide effect was concentration dependent, with 5.0 mM near maximal. At higher concentrations, the abundance of a 50-kDa form of PP5 increased. This 50-kDa form results from a C-terminal cleavage that also activates PP5 enzymatic activity (47, 66). The time course of dephosphorylation was studied after treatment of cells with a final concentration of 5 mM hydrogen peroxide (Fig. 7B). Generation of the 50-kDa form was a rapid event, as rapid as activation of endogenous ERK1 and ERK2 MAP kinases. The loss of Western blot signal against the anti-phospho-ERK for GST-MRK was due to dephosphorylation because GST-MRK protein remained constant over this time course. We conclude that hydrogen peroxide induces dephosphorylation of GST-MRK and rapid cleavage of a portion of endogenous PP5 to an active form. It is worth noting that, unlike ASK1 interaction with PP5, which was inducible (40, 68), hydrogen peroxide treatment does not enhance PP5 binding to MRK complexes, instead it induces generation of the active form(s) of PP5 that is capable of dephosphorylating MRK. The correlation of a steady decrease in the anti-phospho-ERK signal for MRK with a steady increase in the amount of the active form of PP5 bound to MRK provided a strong argument that endogenous PP5 is a MRK phosphatase (Fig. 7A), although we cannot exclude the possibility that another phosphatase would bind to MRK in response to hydrogen peroxide treatment and contributes to MRK dephosphorylation in the TDY motif.
FIG. 7.
Activation of endogenous PP5 by H2O2-induced oxidative stress causes dephosphorylation in the TDY motif of MRK. HEK293T cells transfected with GST-MRK were treated either with an increasing concentration of H2O2 (0 to 5 mM) for 45 min (A) or with 5 mM H2O2 for an increasing length of time (0 to 45 min) (B) before harvesting. GST fusion proteins were pulled down by glutathione-Sepharose beads and analyzed for MRK phosphorylation in the TDY motif by Western blotting (immunoblotting [IB]) against the anti-phospho-ERK antibody (pERK). The amounts of GST-MRK on beads were indicated by Western blotting against the anti-GST antibody. The amounts of endogenous full-length PP5 (55 kDa) and the C-terminally truncated form of PP5 (50 kDa) in cell lysate and on beads were indicated by the anti-PP5 antibody. For comparison, the TEY motif phosphorylation of ERK1 and ERK2 under oxidative stress was also analyzed by the anti-phospho-ERK antibody (B).
Taking all these data together, we have demonstrated that CCRK phosphorylates MRK at T157 and PP5 dephosphorylates phospho-T157, making CCRK and PP5 the first human kinase/phosphatase pair targeting the T-loop of MRK.
Identification of a MRK phosphorylation consensus.
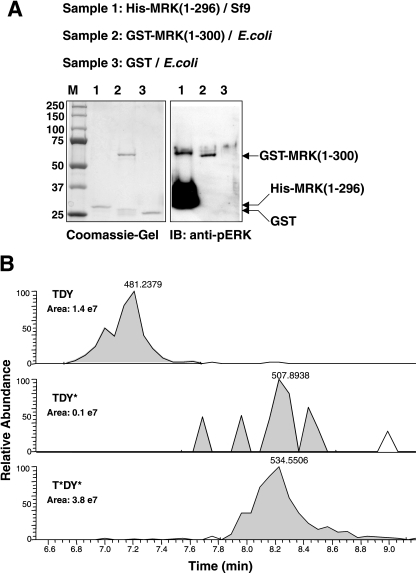
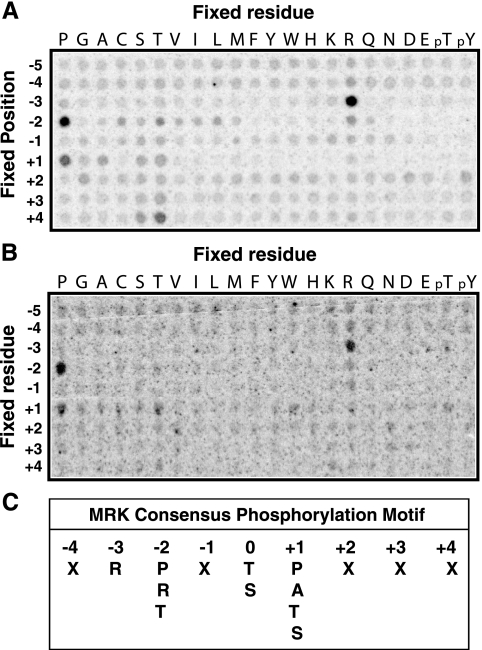
So far, no physiological substrate for MRK has been identified. Protein kinases recognize specific amino acid sequences surrounding the site of phosphorylation (50), and knowledge of this sequence specificity can be useful for identifying protein substrates (41). To define a phosphorylation consensus for MRK, we used a recently described positional scanning peptide array method (25). For the active kinase, we used purified His-tagged MRK(1-296) produced in Sf9 cells, as well as purified GST-MRK(1-300) produced from E. coli (Fig. 8A). Sf9-purified His-MRK(1-296) reacted very strongly to the anti-phospho-ERK compared to E. coli-purified GST-MRK(1-300), suggesting that His-MRK(1-296) expressed in Sf9 cells is much more active. Mass spectrometry studies (Fig. 8B) revealed that a high proportion (about 70%) of Sf9 cell-generated MRK(1-296) was dually phosphorylated at the activating TDY site, compared to E. coli-generated MRK(1-300) that is only singly tyrosine phosphorylated and not detectably dually phosphorylated at the TDY site (17). Note that a higher-mobility band reactive to the anti-phospho-ERK antibody was also present in the His-MRK(1-296) sample (Fig. 8A, right gel, lane 1), but its corresponding protein band was not readily detected on the Coomassie blue-stained gel (Fig. 8A, left gel, lane 1). We speculate that this band may be an endogenous protein from Sf9 cells that copurified with His-MRK(1-296) fusion protein. The reactivity of this higher-mobility band to the anti-phospho-ERK antibody was much lower than that of His-MRK(1-296), suggesting that it is much less active than His-MRK(1-296) and most likely has no significant contribution to the kinase activity of the purified His-MRK(1-296) sample.
FIG. 8.
His-MRK(1-296) purified from Sf9 cells is highly active and is predominantly doubly phosphorylated in the TDY motif. His-MRK(1-296) was expressed in Sf9 cells and affinity purified. The purity was indicated on a Coomassie blue-stained SDS gel (A), and the state of the TDY motif phosphorylation was analyzed by Western blotting (immunoblotting [IB]) against the anti-phospho-ERK antibody (pERK) (A) and by mass spectrometry (B). The positions of molecular mass markers (M) are indicated (in kilodaltons) to the left of the gel. (B) Comparison of selected ion chromatograms from MS targeted analysis of tryptic peptides containing the TDY motif is shown. The peak area of each form of the TDY motif phosphorylation was calculated to indicate relative abundance. The control is GST-MRK(1-300) purified from E. coli that is weakly reactive to the anti-phospho-ERK antibody (A) and not detectably doubly phosphorylated in the TDY motif by mass spectrometry (17).
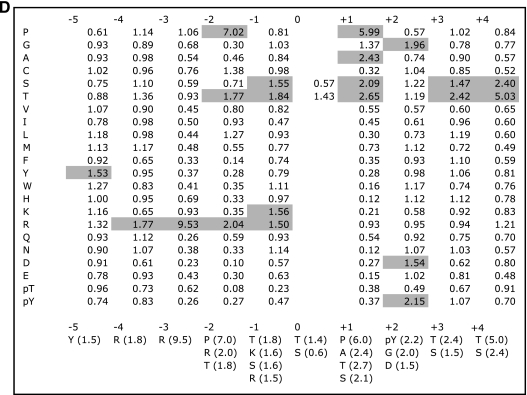
His-MRK(1-296) from insect cells phosphorylated peptides in an R-P-X-S/T-P consensus (Fig. 9A). A preference for proline at P+1 was observed and was expected, given the similarity of MRK to CDK2 and ERK2 (17). The strongest selection was for arginine at P−3 and proline at P−2. Alanine, threonine, and serine were also selected at P+1 but less well. These data suggest that MRK may not always require the P+1 proline. Human Dyrk1A, which is also related to MAPKs and CDKs but is only 31% identical to MRK in its catalytic domain, has been reported to phosphorylate a similar consensus sequence (21). Because other kinases could copurify with MRK from insect cells, we confirmed this result using GST-MRK(1-300) obtained by expression in E. coli (17). Though E. coli-expressed MRK as anticipated had much less activity on the peptide substrates, the essential features of the motif observed with the baculovirus-expressed protein were clearly reproducible (Fig. 9B).
FIG. 9.
Determination of MRK phosphorylation specificity. Using a combinatorial peptide library method (25), we analyzed MRK phosphorylation specificity using purified His-MRK(1-296) protein from Sf9 cells (A) and purified GST-MRK(1-300) protein from E. coli (B). Note that the apparent preferences for peptides with fixed Ser and Thr residues may reflect phosphorylation at the fixed residue rather than at the central position and should be interpreted with caution. (C) The substrate phosphorylation consensus for MRK is R-P-X-S/T-P, with the preference for arginine at P−3 being more stringent than for prolines at P−2 and P+1. (D) Sequence preferences for phosphorylation by MRK. The amount of radioactivity incorporated into each peptide mixture was measured by a phosphorimager using ImageQuant software (Molecular Dynamics). Data are normalized so that the average value for the 20 natural amino acids within a given position is 1; thus, values greater than 1 indicate positive selection. Selectivity values are therefore given relative to the other residues at the same position. Note that cysteine selections should be treated more qualitatively because oxidation is likely to make quantitation of cysteine unreliable. Also note that the apparent preferences for peptides with fixed Ser and Thr residues may reflect phosphorylation at the fixed residue rather than at the central position and should be interpreted with caution. Values for preferred residues are shaded.
MRK interacts with and phosphorylates the antiapoptotic protein Scythe.
From the yeast two-hybrid screen, we also isolated the cDNA encoding a large fragment of human Scythe (accession no. NM_004639) that includes the nuclear localization sequence and the BAG domain of Scythe, but not the N-terminal ubiquitin-like (UBL) domain. The BAG domain was named for a region at the C terminus of Bcl2-athanogene 1 (BAG-1) that interacts with Hsc70 (reviewed in reference 14). Scythe is a nuclear protein (37), is prominent in developing germ cells in the testis (57), and is essential for development (13). Gene deletion of Scythe in mice causes embryonic lethality, and the tissues have disturbed proliferation and apoptosis, particularly the lung and kidney tissues (13).
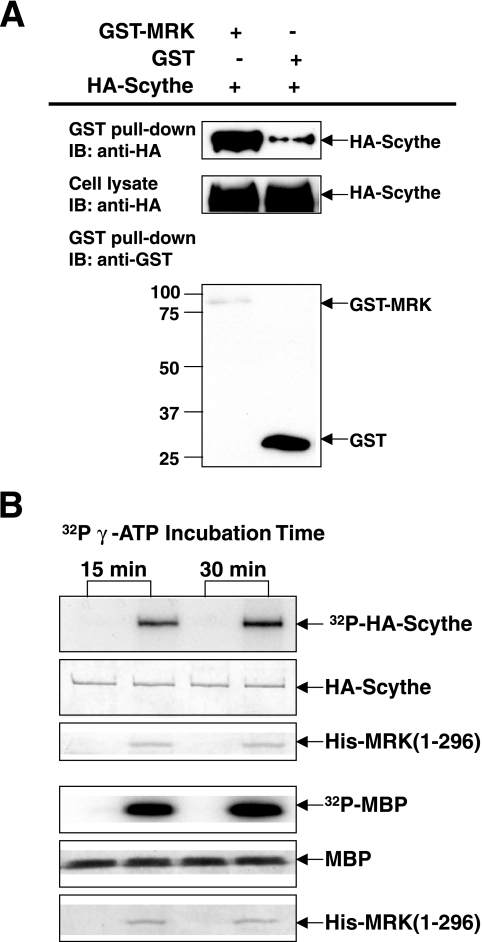
We attempted to confirm this interaction in mammalian cells by cotransfecting HA-Scythe with either GST-MRK or control GST in HEK293T cells. GST-MRK, although less well expressed than GST, was able to pull down a significantly greater amount of HA-Scythe (Fig. 10A), suggesting that GST-MRK selectively interacts with HA-Scythe.
FIG. 10.
Recombinant MRK interacts with recombinant Scythe in cells and phosphorylates recombinant Scythe in vitro. (A) GST-MRK or GST was coexpressed with Flag-Scythe in HEK293T cells. GST fusion proteins were pulled down by glutathione-Sepharose beads and analyzed for association with Flag-Scythe by Western blotting (immunoblotting [IB]) against the anti-Flag antibody. The amounts of GST, GST-MRK and Flag-Scythe in cell lysate were indicated by Western blotting. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the blot. (B) Flag-Scythe immunoprecipitated from HEK293T cells was assayed for phosphorylation by MRK in vitro using purified His-MRK(1-296) (top panel). MBP served as a positive-control substrate for MRK kinase activity (bottom panel).
To determine whether Scythe is a substrate for MRK, we tested the ability of active His-MRK(1-296) to phosphorylate HA-Scythe immunoprecipitated from cells (Fig. 10B). MRK phosphorylated HA-Scythe and myelin basic protein in vitro. The phosphorylation of Scythe by MRK at 15 min is robust in comparison to phosphorylation of myelin basic protein, considering the amount and concentration of Scythe are much lower than that of myelin basic protein in these assays. 32P incorporation of HA-Scythe was increased minimally by doubling the time of incubation, suggesting that phosphorylation by MRK rapidly plateaus. We conclude that Scythe is an interactor and in vitro substrate for MRK.
Scan for putative phosphorylation sites on Scythe by MRK consensus.
HA-tagged human Scythe is a relatively large protein. Transcript variant 1 (accession no. NM_004639) used as a substrate is a 1,132-residue protein, containing 158 total Ser/Thr residues, including 20 occurring as Ser/Thr-Pro motifs. We used the working MRK consensus to scan the Scythe sequence for good matches to the consensus. Since RPXS/T was most strongly selected (Fig. 9), we focused on these motifs present in Scythe. There are just two: RPXS362HYTT and RPXT1080SPES, with T1080 being a stronger candidate than S362 because Ser appears to be more preferable than His at the P+1 position (Fig. 9D).
T1080 is an in vitro MRK phosphorylation site.
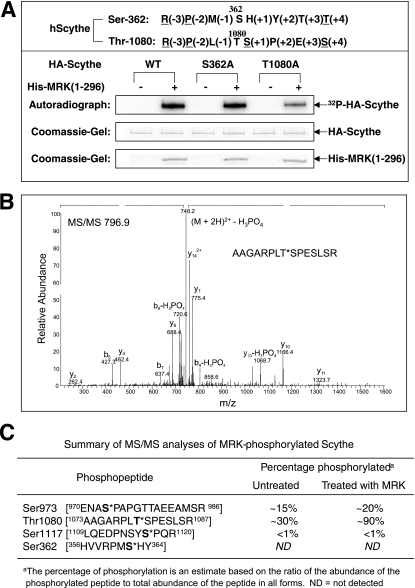
S362 and T1080 in HA-Scythe were singly and doubly mutated to alanine for comparison to wild-type Scythe as kinase assay substrates (Fig. 11A). Coomassie blue-stained SDS gel confirmed that approximately equal amounts of each substrate were recovered on beads. Mutant S362A was not appreciably less phosphorylated by MRK than the wild-type. T1080A mutation caused a reproducible, significant decrease in 32P incorporation (about 80% decrease analyzed by phosphorimager [quantitation data not shown]). The T1080A S362A double mutation did not cause a further decrease in 32P incorporation compared to the effect of a single T1080A mutation (data not shown). We conclude that T1080 is a MRK phosphorylation site, supporting the phosphorylation consensus used to select T1080 as the strongest candidate.
FIG. 11.
Thr-1080 is an in vivo phosphorylation site located within the BAG domain of Scythe that can be phosphorylated by MRK in vitro. (A) In vitro kinase assay of two point mutations (S362A and T1080A) on Scythe by MRK. hScythe, human Scythe. (B) Tandem mass spectrometry (MS/MS) of the phosphorylated form of the RPLT1080SP tryptic peptide that is derived from in-gel digestion of affinity-purified Flag-Scythe (see Materials and Methods). (C) Summary of mass spectrometry analyses of the relative phosphorylation level of Scythe on several sites (shown in bold type with an asterisk) with and without MRK treatment in vitro. The in vitro kinase assay was done as described above for panel A, except that only nonradiolabeled ATP was used.
T1080 is an in vivo phosphorylation site.
We examined mass spectra for evidence of sites in HA-Scythe that become phosphorylated in HEK293T cells. After an in-gel trypsin digestion and analyses, we found two major sites (Ser973 and Thr1080) and one minor site (Ser1117) of phosphorylation (mass accuracy within 2 ppm). Ser973 in peptide ENAS*PAPGTTAEEAMSR was ∼15% phosphorylated (indicated by an asterisk). Thr1080 in peptide AAGARPLT*SPESLSR was ∼30% phosphorylated (Fig. 11B). Ser1117 in peptide LQEDPNYS*PQR was minimally phosphorylated (<1%). Theoretical digestion predicts that Ser362 exists in a proline-rich 89-residue peptide sequence. We did not observe this peptide in our in-gel trypsin analyses; thus, we pursued in-gel chymotrypsin digestion as an alternative. In the reverse-phase HPLC analyses, phosphorylation of S362 in peptide HVVRPMSHY, was not detected, suggesting that the phosphorylation, if present, is <1%.
T1080 phosphorylation in vitro by MRK is stoichiometric.
To determine whether phosphorylation of HA-Scythe in vitro by His-MRK(1-296) caused a significant change in the stoichiometry of T1080 phosphorylation, we performed site analyses by mass spectrometry (Fig. 11C). The stoichiometry of phosphorylation of T1080 was estimated from ion currents as described in Materials and Methods. T1080 was ∼30% phosphorylated omitting His-MRK(1-296) in the kinase assay with ATP/Mg. Incubation with His-MRK(1-296) and ATP/Mg increased stoichiometry at T1080 to ∼90%. In comparison to T1080, the stoichiometry at S973 is merely increased by about 5%, and no change was detected at Ser1117 and Ser362 sites. We conclude that His-MRK(1-296) phosphorylates T1080 in vitro with specificity and selectivity. These experiments also establish that MRK can phosphorylate sites that lack a P+1 proline in an otherwise favorable sequence context. Further studies are required to test whether T1080 is an in vivo phosphorylation site for MRK.
T1080 is within the BAG domain of Scythe that interacts with Hsc70 (32, 53). Six human proteins, including Scythe/BAG-6, have C-terminal BAG domains, function as adapter molecules, and are generally antiapoptotic (reviewed in reference 14). The T1080 motif is conserved in rodent and human Scythe, but this segment is not highly conserved in Xenopus laevis or Danio rerio. Whether T1080 plays a key role in BAG domain-mediated interaction with Hsc70 and apoptosis awaits further studies.
DISCUSSION
In summary, we have identified CCRK and PP5 as a human kinase and phosphatase pair for the critical activation site, T157, in the T-loop of MRK. The phosphorylation consensus determined for MRK is RPX-S/T-P with the stringency for proline at P+1 less striking than for the proline at P−2. Using this consensus, we searched for phosphorylation sites in an in vitro substrate for MRK, the antiapoptotic protein Scythe. MRK phosphorylates Scythe in vitro stoichiometrically at Thr1080, within GARPLT1080SPES. This MRK phosphorylation site is similar to the only identified Ime2 phosphorylation site Ser27 (shown underlined) in a RPGSGER motif in yeast replication protein A2 (10).
CCRK is the leading candidate for the in vivo T157 kinase for MRK. The cellular localization of CCRK is consistent with an activator of MRK and MAK. CCRK is predominantly nuclear (35), like MRK (17) and MAK (61). CCRK is also widely expressed (35, 59) and very abundant in the testis (59) where MAK is most abundant. CCRK may support a role for MRK and MAK in regulation of proliferation and/or apoptosis. CCRK was identified in a large-scale small interfering RNA (siRNA) screen for suppressors of apoptosis (36). A specific chemical inhibitor of CDK family members (RGB-286147) inhibits CCRK and promotes apoptosis in the absence of cell cycle progression (6). This is intriguing because it supports structural similarity of CCRK to other CDKs (35), but the target(s) for RGB-286147 that promotes apoptosis is unknown. Clones of HeLa cells that stably express a CCRK siRNA establish a positive correlation between clonal growth and residual CCRK expression (35). Recently, the role of CCRK both as a CAK and as a cell cycle regulator was investigated in HCT116 and U2OS cells transfected with two different siRNAs (59). Both CCRK siRNAs significantly inhibit proliferation (59). Fluorescence-activated cell sorting analyses of DNA content showed no prominent arrest in G1, S, or G2/M fraction but did show an increase in the sub-G1 fraction, which is usually indicative of apoptosis (59). Thus, we hypothesize that some of the phenotypic effects of silencing CCRK may channel through MRK and/or MAK. Since MRK and MAK have nearly identical catalytic domains, defining the functions of MRK and MAK will probably require knocking out or silencing both genes in animals and cultured cells.
CCRK is most closely related to yeast Cak1p. CAK1 regulates both mitotic and meiotic cell cycles by control of two different downstream effectors. In the mitotic cycle of the budding yeast S. cerevisiae, Cak1p is an activating kinase phosphorylating Cdc28 on threonine 169 (15, 31, 54). Cak1p is also required to activate Ime2p during meiosis through a mechanism that requires threonine 242 and tyrosine 244 in Ime2p's activation loop (43, 44). Thus, an intriguing question arises as to whether CCRK has a similar dual role in vertebrates. There are two divergent reports about the intrinsic CAK activity of CCRK with CDK2 (35, 59). The reasons are unclear, requiring resolution as to whether CCRK is an activating kinase for CDKs. Our data implicate both MRK and MAK as physiologic downstream targets of CCRK, implying that CCRK is capable of multiple functions through different downstream effectors.
The physiologic regulators of PP5 and its substrates are incompletely defined, but reports implicate PP5 in cell cycle checkpoints, DNA damage response, and proliferation. Inhibition of PP5 expression causes a marked antiproliferative effect by activation of the p53-dependent G1 checkpoint (69). PP5 is required for both ATM and ATR checkpoint signaling (3, 67), operant in S and G2/M. Inhibition of PP5 also causes premature mitosis after hydroxyurea treatment (67). Hydrogen peroxide treatment is a stress that can produce apoptosis (12) or G2/M arrest (7). Hydrogen peroxide treatment induces activation of PP5 to negatively regulate ASK1 and inhibit apoptosis (40). PP5 interacts with DNA-dependent protein kinase catalytic subunit (DNA-PKc) and dephosphorylates two functional sites in DNA-PKc for DNA repair of double-strand breaks (58). Similar to ASK1 and DNA-PKc, PP5 can deactivate MRK by dephosphorylation of an essential phosphothreonine residue within the T-loop. We have also shown that hydrogen peroxide treatment induces activation of endogenous PP5 to negatively regulate MRK phosphorylation in the T-loop, although via a different mechanism than that proposed for ASK1. Finding MRK as a new downstream target of PP5 allows us to speculate that PP5 may inactivate MRK to mediate some branch of checkpoint signaling in response to stress and DNA damage.
We emphasize that despite demonstrated success in using the consensus data to identify a site in Scythe, this is a working consensus. Although T1080 proved to be a major phosphorylation site for MRK in vitro, mutating T1080 alone did not completely abolish in vitro phosphorylation of Scythe by MRK, indicating that additional minor phosphorylation sites exist that may not conform strictly to the working consensus. Due to limited coverage of Ser/Thr sites by mass spectrometry, we were unable to define additional phosphorylation sites without ambiguity. With that caveat, the consensus we obtained for MRK has similarities to ERK1 (49) and ERK2 (25) and CDK2/cyclin A (22, 49). ERK2 and other MAP kinases have a P-X-S/T-P consensus sequence that is found in many physiologic sites of phosphorylation, such as T188 in C/EBPβ (20), but only the P+1 proline residue appears to be absolutely required in substrates. MAP kinase specificity for substrates is also determined by highly specific docking interactions (52). The strongest preferences defined for CDK2/cyclin A in detailed studies are for P+1 proline and for a basic residue (K/R) at P+3 (22, 49). We did not observe a preference for basic residues at the P+3 position for MRK, under the conditions of our studies. The similarity of the MRK consensus to Dyrk1a (21) was surprising, since MRK is phylogenetically closer to CDK2 and ERK2 (29) within the CMGC subgroup. Though MRK has clear preferences at three positions near the phosphorylation site, only arginine at P−3 position appears to be very stringent and neither of the proline residues at P−2 and P+1 positions appears to be an absolute requirement for phosphorylation, as suggested by a lack of P+1 proline in the major MRK phosphorylation site in Scythe. We obtained the same consensus with E. coli-purified MRK protein as well as the Sf9-purified His-MRK protein, indicating that active MRK and not a contaminating kinase phosphorylated R-P-X-S/T-P in the array.
Given that MRK and MAK are essentially identical in the catalytic domain, we expect human Scythe to be a MAK substrate in vitro as well. Scythe is an alias for BAT3 (4). BAT3 is especially enriched in testis and abundant in the male germ cells (57). This pattern is strikingly similar to MAK (28, 34, 38). Both Scythe and MAK mRNAs increase dramatically in the mouse testis ∼14 to 20 days after birth. These correlations suggest that MAK and Scythe monitor or function in a general process that is in great demand in spermatogenesis, such as DNA repair, which is a normal part of meiosis.
Scythe is considered an antiapoptotic protein (13, 33), but the evidence for Scythe as a direct player in apoptotic signaling has caveats (37). Of several apoptotic stimuli tested, thapsigargin- and menadione-induced cell death was enhanced by Scythe depletion and rescued by Scythe transfection (13). Thapsigargin treatment negates the ability of caffeine (an ATM inhibitor) to release the G2 delay caused by γ radiation in tumor cells (27). Menadione is a model quinone compound used to study the effects of oxidative stress because it generates superoxide in mammalian cells that causes strand breaks in DNA (60). In addition to the link of PP5 to DNA damage control, the link of Scythe to apoptosis induced by DNA damage further motivated us to speculate that MRK and/or MAK may play a key role in cell cycle checkpoint control and apoptosis in response to stress conditions, such as DNA damage.
Acknowledgments
We are especially grateful to all the generous colleagues who have made our studies possible by provision of their reagents and advice: Richard Honkanen, Sandra Rossie, Elizabeth Goldsmith, Xiaoshan Min, Andrew Hubberstey, Bert Vogelstein, Robert Fisher, and Sally Kornbluth. We are indebted to Katrina Clines for her support and technical assistance with the yeast two-hybrid screen. We thank members of Sturgill Lab (Carol Chrestensen and Darlene Bruce) for discussions and support and members of John Lawrence Lab (Dale Choi and Thurl Harris) for advice on lentivirus infection.
This work was supported by NIH grants GM62890 (to T.W.S.), GM37537 (to D.F.H.) and DK064751 (to S.M.C.).
Footnotes
Published ahead of print on 5 September 2006.
REFERENCES
- 1.Abe, H., and C. Shimoda. 2000. Autoregulated expression of Schizosaccharomyces pombe meiosis-specific transcription factor Mei4 and a genome-wide search for its target genes. Genetics 154:1497-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abe, S., T. Yagi, S. Ishiyama, M. Hiroe, F. Marumo, and Y. Ikawa. 1995. Molecular cloning of a novel serine/threonine kinase, MRK, possibly involved in cardiac development. Oncogene 11:2187-2195. [PubMed] [Google Scholar]
- 3.Ali, A., J. Zhang, S. Bao, I. Liu, D. Otterness, N. M. Dean, R. T. Abraham, and X. F. Wang. 2004. Requirement of protein phosphatase 5 in DNA-damage-induced ATM activation. Genes Dev. 18:249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerji, J., J. Sands, J. L. Strominger, and T. Spies. 1990. A gene pair from the human major histocompatibility complex encodes large proline-rich proteins with multiple repeated motifs and a single ubiquitin-like domain. Proc. Natl. Acad. Sci. USA 87:2374-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barzilai, A., and K. Yamamoto. 2004. DNA damage responses to oxidative stress. DNA Repair 3:1109-1115. [DOI] [PubMed] [Google Scholar]
- 6.Caligiuri, M., F. Becker, K. Murthi, F. Kaplan, S. Dedier, C. Kaufmann, A. Machl, G. Zybarth, J. Richard, N. Bockovich, A. Kluge, and N. Kley. 2005. A proteome-wide CDK/CRK-specific kinase inhibitor promotes tumor cell death in the absence of cell cycle progression. Chem. Biol. 12:1103-1115. [DOI] [PubMed] [Google Scholar]
- 7.Chang, D. K., A. Goel, L. Ricciardiello, D. H. Lee, C. L. Chang, J. M. Carethers, and C. R. Boland. 2003. Effect of H2O2 on cell cycle and survival in DNA mismatch repair-deficient and -proficient cell lines. Cancer Lett. 195:243-251. [DOI] [PubMed] [Google Scholar]
- 8.Chen, M. X., and P. T. Cohen. 1997. Activation of protein phosphatase 5 by limited proteolysis or the binding of polyunsaturated fatty acids to the TPR domain. FEBS Lett. 400:136-140. [DOI] [PubMed] [Google Scholar]
- 9.Chen, M. X., A. E. McPartlin, L. Brown, Y. H. Chen, H. M. Barker, and P. T. Cohen. 1994. A novel human protein serine/threonine phosphatase, which possesses four tetratricopeptide repeat motifs and localizes to the nucleus. EMBO J. 13:4278-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clifford, D. M., K. E. Stark, K. E. Gardner, S. Hoffmann-Benning, and G. S. Brush. 2005. Mechanistic insight into the Cdc28-related protein kinase Ime2 through analysis of replication protein A phosphorylation. Cell Cycle 4:1826-1833. [DOI] [PubMed] [Google Scholar]
- 11.Conde, R., J. Xavier, C. McLoughlin, M. Chinkers, and N. C. Ovsenek. 2005. Protein phosphatase 5 is a negative modulator of heat shock factor 1. J. Biol. Chem. 280:28989-28996. [DOI] [PubMed] [Google Scholar]
- 12.Daroui, P., S. D. Desai, T. K. Li, A. A. Liu, and L. F. Liu. 2004. Hydrogen peroxide induces topoisomerase I-mediated DNA damage and cell death. J. Biol. Chem. 279:14587-14594. [DOI] [PubMed] [Google Scholar]
- 13.Desmots, F., H. R. Russell, Y. Lee, K. Boyd, and P. J. McKinnon. 2005. The Reaper-binding protein Scythe modulates apoptosis and proliferation during mammalian development. Mol. Cell. Biol. 25:10329-10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doong, H., A. Vrailas, and E. C. Kohn. 2002. What's in the “BAG”?—a functional domain analysis of the BAG-family proteins. Cancer Lett. 188:25-32. [DOI] [PubMed] [Google Scholar]
- 15.Espinoza, F. H., A. Farrell, H. Erdjument-Bromage, P. Tempst, and D. O. Morgan. 1996. A cyclin-dependent kinase-activating kinase (CAK) in budding yeast unrelated to vertebrate CAK. Science 273:1714-1717. [DOI] [PubMed] [Google Scholar]
- 16.Foiani, M., E. Nadjar-Boger, R. Capone, S. Sagee, T. Hashimshoni, and Y. Kassir. 1996. A meiosis-specific protein kinase, Ime2, is required for the correct timing of DNA replication and for spore formation in yeast meiosis. Mol. Gen. Genet. 253:278-288. [DOI] [PubMed] [Google Scholar]
- 17.Fu, Z., M. J. Schroeder, J. Shabanowitz, P. Kaldis, K. Togawa, A. K. Rustgi, D. F. Hunt, and T. W. Sturgill. 2005. Activation of a nuclear Cdc2-related kinase within a mitogen-activated protein kinase-like TDY motif by autophosphorylation and cyclin-dependent protein kinase-activating kinase. Mol. Cell. Biol. 25:6047-6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentile, S., T. Darden, C. Erxleben, C. Romeo, A. Russo, N. Martin, S. Rossie, and D. L. Armstrong. 2006. Rac GTPase signaling through the PP5 protein phosphatase. Proc. Natl. Acad. Sci. USA 103:5202-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldman, E. H., L. Chen, and H. Fu. 2004. Activation of apoptosis signal-regulating kinase 1 by reactive oxygen species through dephosphorylation at serine 967 and 14-3-3 dissociation. J. Biol. Chem. 279:10442-10449. [DOI] [PubMed] [Google Scholar]
- 20.Hanlon, M., T. W. Sturgill, and L. Sealy. 2001. ERK2- and p90Rsk2-dependent pathways regulate the CCAAT/enhancer-binding protein-beta interaction with serum response factor. J. Biol. Chem. 276:38449-38456. [DOI] [PubMed] [Google Scholar]
- 21.Himpel, S., W. Tegge, R. Frank, S. Leder, H. G. Joost, and W. Becker. 2000. Specificity determinants of substrate recognition by the protein kinase DYRK1A. J. Biol. Chem. 275:2431-2438. [DOI] [PubMed] [Google Scholar]
- 22.Holmes, J. K., and M. J. Solomon. 1996. A predictive scale for evaluating cyclin-dependent kinase substrates. A comparison of p34cdc2 and p33cdk2. J. Biol. Chem. 271:25240-25246. [DOI] [PubMed] [Google Scholar]
- 23.Honigberg, S. M. 2004. Ime2p and Cdc28p: co-pilots driving meiotic development. J. Cell. Biochem. 92:1025-1033. [DOI] [PubMed] [Google Scholar]
- 24.Horie, S., Y. Watanabe, K. Tanaka, S. Nishiwaki, H. Fujioka, H. Abe, M. Yamamoto, and C. Shimoda. 1998. The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol. Cell. Biol. 18:2118-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutti, J. E., E. T. Jarrell, J. D. Chang, D. W. Abbott, P. Storz, A. Toker, L. C. Cantley, and B. E. Turk. 2004. A rapid method for determining protein kinase specificity. Nat. Methods 1:27-29. [DOI] [PubMed] [Google Scholar]
- 26.Jeong, J. Y., J. Johns, C. Sinclair, J. M. Park, and S. Rossie. 2003. Characterization of Saccharomyces cerevisiae protein Ser/Thr phosphatase T1 and comparison to its mammalian homolog PP5. BMC Cell Biol. 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jha, M. N., J. R. Bamburg, B. W. Bernstein, and J. S. Bedford. 2002. Caffeine eliminates gamma-ray-induced G2-phase delay in human tumor cells but not in normal cells. Radiat. Res. 157:26-31. [DOI] [PubMed] [Google Scholar]
- 28.Jinno, A., K. Tanaka, H. Matsushime, T. Haneji, and M. Shibuya. 1993. Testis-specific Mak protein kinase is expressed specifically in the meiotic phase in spermatogenesis and is associated with a 210-kilodalton cellular phosphoprotein. Mol. Cell. Biol. 13:4146-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, S. A., and T. Hunter. 2005. Kinomics: methods for deciphering the kinome. Nat. Methods 2:17-25. [DOI] [PubMed] [Google Scholar]
- 30.Kaldis, P., and M. J. Solomon. 2000. Analysis of CAK activities from human cells. Eur. J. Biochem. 267:4213-4221. [DOI] [PubMed] [Google Scholar]
- 31.Kaldis, P., A. Sutton, and M. J. Solomon. 1996. The Cdk-activating kinase (CAK) from budding yeast. Cell 86:553-564. [DOI] [PubMed] [Google Scholar]
- 32.Kaye, F. J., S. Modi, I. Ivanovska, E. V. Koonin, K. Thress, A. Kubo, S. Kornbluth, and M. D. Rose. 2000. A family of ubiquitin-like proteins binds the ATPase domain of Hsp70-like Stch. FEBS Lett. 467:348-355. [DOI] [PubMed] [Google Scholar]
- 33.Kikukawa, Y., R. Minami, M. Shimada, M. Kobayashi, K. Tanaka, H. Yokosawa, and H. Kawahara. 2005. Unique proteasome subunit Xrpn10c is a specific receptor for the antiapoptotic ubiquitin-like protein Scythe. FEBS J. 272:6373-6386. [DOI] [PubMed] [Google Scholar]
- 34.Koji, T., A. Jinno, H. Matsushime, M. Shibuya, and P. K. Nakane. 1992. In situ localization of male germ cell-associated kinase (mak) mRNA in adult mouse testis: specific expression in germ cells at stages around meiotic cell division. Cell Biochem. Funct. 10:273-279. [DOI] [PubMed] [Google Scholar]
- 35.Liu, Y., C. Wu, and K. Galaktionov. 2004. p42, a novel cyclin-dependent kinase-activating kinase in mammalian cells. J. Biol. Chem. 279:4507-4514. [DOI] [PubMed] [Google Scholar]
- 36.MacKeigan, J. P., L. O. Murphy, and J. Blenis. 2005. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat. Cell Biol. 7:591-600. [DOI] [PubMed] [Google Scholar]
- 37.Manchen, S. T., and A. V. Hubberstey. 2001. Human Scythe contains a functional nuclear localization sequence and remains in the nucleus during staurosporine-induced apoptosis. Biochem. Biophys. Res. Commun. 287:1075-1082. [DOI] [PubMed] [Google Scholar]
- 38.Matsushime, H., A. Jinno, N. Takagi, and M. Shibuya. 1990. A novel mammalian protein kinase gene (mak) is highly expressed in testicular germ cells at and after meiosis. Mol. Cell. Biol. 10:2261-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyata, Y., and E. Nishida. 2004. CK2 controls multiple protein kinases by phosphorylating a kinase-targeting molecular chaperone, Cdc37. Mol. Cell. Biol. 24:4065-4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morita, K., M. Saitoh, K. Tobiume, H. Matsuura, S. Enomoto, H. Nishitoh, and H. Ichijo. 2001. Negative feedback regulation of ASK1 by protein phosphatase 5 (PP5) in response to oxidative stress. EMBO J. 20:6028-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Obenauer, J. C., L. C. Cantley, and M. B. Yaffe. 2003. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31:3635-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pearson, W. R. 1991. Searching protein sequence libraries: comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Genomics 11:635-650. [DOI] [PubMed] [Google Scholar]
- 43.Schaber, M., A. Lindgren, K. Schindler, D. Bungard, P. Kaldis, and E. Winter. 2002. CAK1 promotes meiosis and spore formation in Saccharomyces cerevisiae in a CDC28-independent fashion. Mol. Cell. Biol. 22:57-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schindler, K., K. R. Benjamin, A. Martin, A. Boglioli, I. Herskowitz, and E. Winter. 2003. The Cdk-activating kinase Cak1p promotes meiotic S phase through Ime2p. Mol. Cell. Biol. 23:8718-8728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 46.Shinkai, Y., H. Satoh, N. Takeda, M. Fukuda, E. Chiba, T. Kato, T. Kuramochi, and Y. Araki. 2002. A testicular germ cell-associated serine-threonine kinase, MAK, is dispensable for sperm formation. Mol. Cell. Biol. 22:3276-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sinclair, C., C. Borchers, C. Parker, K. Tomer, H. Charbonneau, and S. Rossie. 1999. The tetratricopeptide repeat domain and a C-terminal region control the activity of Ser/Thr protein phosphatase 5. J. Biol. Chem. 274:23666-23672. [DOI] [PubMed] [Google Scholar]
- 48.Skinner, J., C. Sinclair, C. Romeo, D. Armstrong, H. Charbonneau, and S. Rossie. 1997. Purification of a fatty acid-stimulated protein-serine/threonine phosphatase from bovine brain and its identification as a homolog of protein phosphatase 5. J. Biol. Chem. 272:22464-22471. [DOI] [PubMed] [Google Scholar]
- 49.Songyang, Z., S. Blechner, N. Hoagland, M. F. Hoekstra, H. Piwnica-Worms, and L. C. Cantley. 1994. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol. 4:973-982. [DOI] [PubMed] [Google Scholar]
- 50.Songyang, Z., K. P. Lu, Y. T. Kwon, L. H. Tsai, O. Filhol, C. Cochet, D. A. Brickey, T. R. Soderling, C. Bartleson, D. J. Graves, A. J. DeMaggio, M. F. Hoekstra, J. Blenis, T. Hunter, and L. C. Cantley. 1996. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol. 16:6486-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swingle, M. R., R. E. Honkanen, and E. M. Ciszak. 2004. Structural basis for the catalytic activity of human serine/threonine protein phosphatase-5. J. Biol. Chem. 279:33992-33999. [DOI] [PubMed] [Google Scholar]
- 52.Tanoue, T., R. Maeda, M. Adachi, and E. Nishida. 2001. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 20:466-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thress, K., J. Song, R. I. Morimoto, and S. Kornbluth. 2001. Reversible inhibition of Hsp70 chaperone function by Scythe and Reaper. EMBO J. 20:1033-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thuret, J. Y., J. G. Valay, G. Faye, and C. Mann. 1996. Civ1 (CAK in vivo), a novel Cdk-activating kinase. Cell 86:565-576. [DOI] [PubMed] [Google Scholar]
- 55.Togawa, K., Y. X. Yan, T. Inomoto, S. Slaugenhaupt, and A. K. Rustgi. 2000. Intestinal cell kinase (ICK) localizes to the crypt region and requires a dual phosphorylation site found in map kinases. J. Cell. Physiol. 183:129-139. [DOI] [PubMed] [Google Scholar]
- 56.Wang, I. C., Y. J. Chen, D. Hughes, V. Petrovic, M. L. Major, H. J. Park, Y. Tan, T. Ackerson, and R. H. Costa. 2005. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol. Cell. Biol. 25:10875-10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, R., and C. C. Liew. 1994. The human BAT3 ortholog in rodents is predominantly and developmentally expressed in testis. Mol. Cell. Biochem. 136:49-57. [DOI] [PubMed] [Google Scholar]
- 58.Wechsler, T., B. P. Chen, R. Harper, K. Morotomi-Yano, B. C. Huang, K. Meek, J. E. Cleaver, D. J. Chen, and M. Wabl. 2004. DNA-PKcs function regulated specifically by protein phosphatase 5. Proc. Natl. Acad. Sci. USA 101:1247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wohlbold, L., S. Larochelle, J. C. Liao, G. Livshits, J. Singer, K. M. Shokat, and R. P. Fisher. 2006. The cyclin-dependent kinase (CDK) family member PNQALRE/CCRK supports cell proliferation but has no intrinsic CDK-activating kinase (CAK) activity. Cell Cycle 5:546-554. [DOI] [PubMed] [Google Scholar]
- 60.Woods, J. A., A. J. Young, I. T. Gilmore, A. Morris, and R. F. Bilton. 1997. Measurement of menadione-mediated DNA damage in human lymphocytes using the comet assay. Free Radic. Res. 26:113-124. [DOI] [PubMed] [Google Scholar]
- 61.Xia, L., D. Robinson, A. H. Ma, H. C. Chen, F. Wu, Y. Qiu, and H. J. Kung. 2002. Identification of human male germ cell-associated kinase, a kinase transcriptionally activated by androgen in prostate cancer cells. J. Biol. Chem. 277:35422-35433. [DOI] [PubMed] [Google Scholar]
- 62.Xu, L., M. Ajimura, R. Padmore, C. Klein, and N. Kleckner. 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6572-6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang, J., S. M. Roe, M. J. Cliff, M. A. Williams, J. E. Ladbury, P. T. Cohen, and D. Barford. 2005. Molecular basis for TPR domain-mediated regulation of protein phosphatase 5. EMBO J. 24:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ye, H., T. F. Kelly, U. Samadani, L. Lim, S. Rubio, D. G. Overdier, K. A. Roebuck, and R. H. Costa. 1997. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol. Cell. Biol. 17:1626-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Young, J. A., R. W. Hyppa, and G. R. Smith. 2004. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics 167:593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeke, T., N. Morrice, C. Vazquez-Martin, and P. T. Cohen. 2005. Human protein phosphatase 5 dissociates from heat-shock proteins and is proteolytically activated in response to arachidonic acid and the microtubule-depolymerizing drug nocodazole. Biochem. J. 385:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, J., S. Bao, R. Furumai, K. S. Kucera, A. Ali, N. M. Dean, and X. F. Wang. 2005. Protein phosphatase 5 is required for ATR-mediated checkpoint activation. Mol. Cell. Biol. 25:9910-9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou, G., T. Golden, I. V. Aragon, and R. E. Honkanen. 2004. Ser/Thr protein phosphatase 5 inactivates hypoxia-induced activation of an apoptosis signal-regulating kinase 1/MKK-4/JNK signaling cascade. J. Biol. Chem. 279:46595-46605. [DOI] [PubMed] [Google Scholar]
- 69.Zuo, Z., N. M. Dean, and R. E. Honkanen. 1998. Serine/threonine protein phosphatase type 5 acts upstream of p53 to regulate the induction of p21WAF1/Cip1 and mediate growth arrest. J. Biol. Chem. 273:12250-12258. [DOI] [PubMed] [Google Scholar]