Abstract
Modular interaction domains that recognize peptide motifs in target proteins can impart selectivity in signaling pathways. Phosphotyrosine binding (PTB) domains are components of cytoplasmic docking proteins that bind cell surface receptors through NPXY motifs. We have employed a library of human proteome-derived NXXY sequences to explore PTB domain specificity and function. SPOTS peptide arrays were used to create a comprehensive matrix of receptor motifs that were probed with a set of 10 diverse PTB domains. This approach confirmed that individual PTB domains have selective and distinct recognition properties and provided a means to explore over 2,500 potential PTB domain-NXXY interactions. The results correlated well with previously known associations between full-length proteins and predicted novel interactions, as well as consensus binding data for specific PTB domains. Using the Ret, MuSK, and ErbB2 receptor tyrosine kinases, we show that interactions of these receptors with PTB domains predicted to bind by the NXXY arrays do occur in cells. Proteome-based peptide arrays can therefore identify networks of receptor interactions with scaffold proteins that may be physiologically relevant.
External signaling molecules typically bind receptors that span the plasma membrane and thereby activate intracellular targets. This is often achieved through the selective recognition of short peptide motifs in receptors by proteins containing modular interaction domains. These binding events form the backbone of cellular signaling, and their regulation is a fundamental process by which cells organize their growth and survival (60, 62).
Posttranslational modifications represent an important mechanism by which cell surface receptors recruit cytoplasmic targets (61, 67). Tyrosine phosphorylation of receptor tyrosine kinases (RTKs), cytokine receptors and adhesion proteins, for example, creates recognition motifs for the Src homology 2 (SH2) or PTB (phosphotyrosine binding) domains of cytoplasmic proteins that control responses such as cytoskeletal organization, metabolism, survival, differentiation, and division.
PTB domains are found in scaffold proteins that often contain additional modular domains and motifs and thereby nucleate the formation of multiprotein complexes (24, 37, 50, 79). They have been grouped into three broad families (insulin receptor substrate 1 [IRS-1]/Dok-like, Shc-like, and Dab-like) based on structural comparisons (75). The minimal PTB domain fold consists of a central β-sandwich comprised of seven antiparallel β-strands that is capped on one end by the C-terminal α-helix and on the other by a variable length α-helix found between strands β1 and β2 (or β2 and β3). Additional helices can be present in members of the Shc and Dab families. The canonical peptide-binding groove is located between the fifth β-strand and the conserved C-terminal α-helix. Peptide ligands typically adopt a β-turn that provides an anchor point for binding and is often initiated by −3 asparagine and −2 proline residues (relative to tyrosine at position 0), contained in an NPXY motif (where X is any amino acid). Although PTB domains can potentially bind a range of peptide and phospholipid ligands, they most frequently recognize NPXY- or NXXY-containing peptides, with further specificity being conferred by tyrosine phosphorylation and by contact with residues N- and C-terminal to this core motif (20, 21, 34, 46, 49, 83).
PTB domains vary in their dependence on phosphorylation of the NPXY tyrosine for high-affinity binding. Members of the Shc and IRS-1/Dok families bind with higher affinity to phosphorylated motifs and therefore serve as scaffolds in both normal and oncogenic RTK signaling. However, the majority of PTB domains, of which Dab and Numb are representative, bind irrespective of ligand phosphorylation or preferentially recognize unphosphorylated ligands. Such phosphorylation-independent PTB domain interactions have been implicated in a wide range of cellular functions, including the signaling and trafficking of reelin receptors (31, 70), the low-density lipoprotein receptors (12, 32), and the amyloid precursor protein (6, 53, 80, 82), as well as asymmetric cell division (13, 84) and integrin-mediated adhesion (9, 10, 22, 74).
The availability of genome sequences and the classification of open reading frames have made it possible to explore the proteome, or subsets thereof, for full sets of short linear motifs conforming to a particular consensus recognition site (44). By probing the full array of ligands that an individual domain might encounter, it is possible to generate a map of potential protein-protein interactions based on physiological motifs. This would provide information that is complementary to that obtained from yeast two-hybrid-, mass spectrometry-, or Lumier-based interaction screens and is distinct from binding data derived from phage display or degenerate peptide libraries. Though the three-dimensional structures of PTB domains are very similar, their relatively low sequence homology makes it difficult to predict the binding properties of uncharacterized domains. Thus, we took an array-based approach, employing a library of NPXY sites from cellular proteins, to examine PTB domain binding.
Here we show that individual PTB domains demonstrate distinct binding specificities towards arrays of NPXY/NXXY peptides derived from human receptors. Data taken from these interactions not only allow us to determine consensus binding preferences for each domain but also provide a network of potential protein-protein interactions relevant to signaling from the cell membrane.
MATERIALS AND METHODS
Plasmid constructs and antibodies.
Human cDNAs encoding full-length Apbb1 (accession no. BC010854), Appl (BC028599), Cten (AK027856), Dok2 (BC032623), Eps8 (BC030010), Frs2 (BC021562), Icap1α (BC012264), NumbL (BC001794), ShcA (BC014158), ShcB (AB001451), ShcC (BC026314), and ShcD (BC033907) were cloned into pDNR-SA (V309) donor vector for use in the modified Creator system (14). Fragments of Apbb1 (residues 345 to 531), Appl (460 to 654), Cten (332 to 480), Dok2 (133 to 267), Eps8 (1 to 223), Frs2 (1 to 130), Icap1α (34 to 200), NumbL (30 to 237), ShcA (35 to 211), ShcB (36 to 238), ShcC (133 to 336), and ShcD (170 to 373) encoding their PTB domains were subcloned into pGEX-4T2 (Amersham Pharmacia Biotech, Piscataway, NJ) for expression as glutathione S-transferase (GST) fusion proteins, in addition to pDNR-SA (V309). A fragment of ShcD representing the SH2 domain (residues 524 to 603) was also cloned into pGEX-4T2 and sequence encoding Apbb1ΔPTB2 (residues 1 to 531) into pDNR-SA (V309). Recombination of the full-length clones or subdomains into the acceptor vectors pLP-EGFP-C1 (V4) for expression of enhanced green fluorescent protein (EGFP) fusion proteins, or pLP-dMycSD (V517) for production of Myc-tagged proteins, was performed as previously described (14). Expression vectors containing the wild-type or K758M versions of human Ret (accession no. NM_020630; ID 5979) were from Carlos Ibanez (Karolinska Institute, Sweden). cDNAs encoding rat ErbB2 (AY116182; ID 24337) or mouse muscle-specific kinase (MuSK) (NM_001037130; ID 18198) were cloned into the pCAGGS vector, putting them under control of the β-actin promoter (58). Sequences encoding red fluorescent protein (RFP) or GFP were subcloned at the C-terminal ends of the receptors. Mutations were generated using the Quikchange II system (Stratagene, La Jolla, CA). Constructs expressing Flag-tagged Dok2 or a Dok2-PTB domain mutant were made in pCMV-3xFLAG (Sigma, Oakville, ON, Canada). All constructs were sequence verified.
Mouse monoclonal antibodies were purchased as follows: anti-FLAG M2 was from Sigma, anti-Myc 9E10 and anti-pTyr PY99/PY20 from Santa Cruz Biotechnology (Santa Cruz, CA), and anti-pTyr 4G10 from Upstate Biotechnology (Lake Placid, NY). Rabbit polyclonal anti-GFP antibodies were purchased from Abcam (Cambridge, MA), and the rabbit polyclonal anti-GST used for SPOTS detection was from Sigal Gelkop (Ben Gurion University, Israel).
Cell culture and immunofluorescence.
Human embryonic kidney 293T (HEK 293T) cells were maintained in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal calf serum and antibiotics. For exogenous expression of proteins, cells were transiently transfected with cDNAs using polyethylenimine as previously described (7). For immunofluorescence, microscopy was carried out on live cells 24 h after transfection, using the Leica DMIRE2 inverted microscope (Heerbrugg, Switzerland) equipped with fluorescence and transmitted light optics and with Openlab software (Quorum Technologies, Guelph, ON, Canada).
Purification of recombinant GST fusion proteins.
GST-PTB or SH2 domains were expressed in Escherichia coli BL21 cells (Amersham Pharmacia Biotech). Cells were lysed in PLC buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1.5 mM MgCl2, 1 mM EDTA, 100 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 10 μg ml−1 aprotinin, 10 μg ml−1 leupeptin, and 1 mM dithiothreitol) and sonicated. Lysate was cleared by centrifugation and incubated with glutathione beads (Amersham Pharmacia Biotech) at 4°C for 1 to 2 h. Bound proteins were eluted using 40 mM glutathione (Sigma) in phosphate-buffered saline over 2 h at 4°C then concentrated.
Immunoprecipitations and Western blotting.
Transfected HEK 293T cells were lysed in TX100 buffer (20 mM Tris-HCl [pH 7.5], 137 mM NaCl, 10% glycerol, 1% Triton X-100, 2 mM EDTA, 1 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride, 10 μg ml−1 aprotinin, and 10 μg ml−1 leupeptin). Proteins were immunoprecipitated for 1 h at 4°C and washed three times (TX100 buffer with or without 0.2% sodium dodecyl sulfate). Bound proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (Schleicher and Schuell Bioscience, Keene, NH). Membranes were blocked in Tris-buffered saline-Tween-20 containing 5% skim milk and then immunoblotted. Primary antibodies were detected with anti-mouse immunoglobulin (Ig) or anti-rabbit Ig antibodies conjugated to horseradish peroxidase (HRP), followed by treatment with enhanced chemiluminescence reagents (Pierce, Rockford, IL).
Peptide SPOTS array synthesis.
Peptide arrays were produced according to the SPOTS-synthesis method (25). Acid-hardened cellulose membranes prederivatized with a polyethylene glycol spacer (Intavis AG, Cologne, Germany) and standard 9-fluorenylmethoxy carbonyl (Fmoc) chemistry were used for synthesis. Fmoc-protected and activated amino acids (Intavis) were spotted in high-density 20 by 30 arrays on 150- by 100-mm membranes using an Intavis MultiPep robot. Membranes were blocked overnight in 10% skim milk, incubated with a 0.25 μM concentration of purified GST fusion proteins in Tris-buffered saline-Tween-20 for 2 h at 4°C, washed three times, and probed with rabbit polyclonal anti-GST antibody. Primary antibodies were detected by HRP-conjugated anti-rabbit Ig followed by enhanced chemiluminescence detection. Labeling of full-length peptides with biotin was done by reacting biotinamidocaproate N-hydroxysuccinimide ester (Sigma) with Fmoc-cleaved, side-chain-protected peptides in dimethylsulfoxide. Incorporation of biotin was assessed using HRP-conjugated streptavidin from Bio-Rad Laboratories (Mississauga, ON, Canada).
Peptide synthesis and fluorescence polarization.
Peptides were synthesized on an AbiMed 431 synthesizer using the standard Fastmoc protocol. Authenticity of product was confirmed by matrix-assisted laser desorption ionization mass spectrometry. Equilibrium binding constant determination was carried out on a Beacon fluorescence polarization system (Pan Vera, WI) equipped with a 100-μl sample chamber. Fluorescein-labeled probes were prepared through the reaction of carboxy terminal peptides with 5-(and -6)-carboxyfluorescein succinimidyl ester (Molecular Probes, Eugene, OR). Binding studies were conducted with 5 nM fluorescein-labeled probes dissolved in phosphate-buffered saline containing 100 μg ml−1 bovine serum albumin and 1 mM dithiothreitol. Increasing quantities of protein were added up to 100 μM. Reaction mixtures were allowed to equilibrate for 30 to 60 min at room temperature. All fluorescence polarization measurements were conducted at 22°C.
RESULTS
Assembly and synthesis of an NPXY motif database.
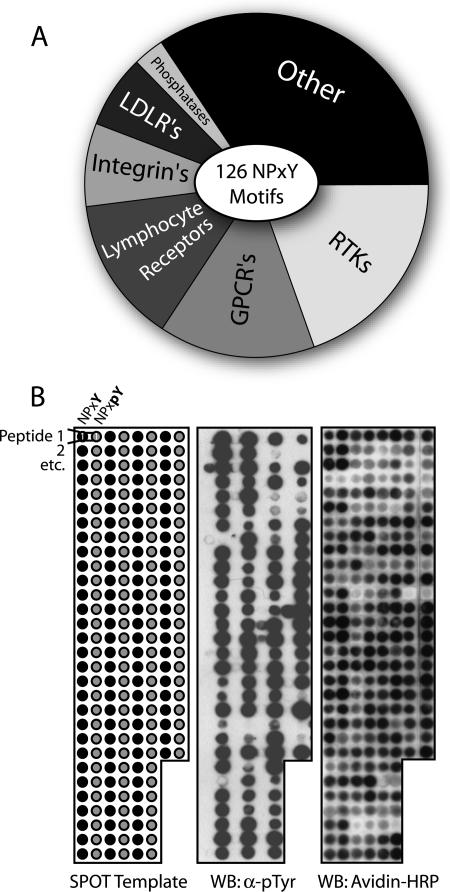
To investigate connections between cytoplasmic scaffolding proteins and cell surface receptors, we used the consensus binding sequence for PTB domains to generate a set of 126 NPXY peptide motifs. These were derived from proteins classified as receptors in a compilation of human proteome databases. The resulting set contains all NPXY motifs found in human receptors, including RTKs, integrins, G-protein-coupled receptors, and phosphatases (Fig. 1A). Several NXXY motifs, missing the −2 proline, were also included. No discrimination was made against sequences contained in the extracellular region of receptors, as we wished to examine a large number of diverse motifs. To study their binding to individual PTB domains, the 126 motifs were synthesized on SPOTS arrays as membrane-bound 12-mer peptides in both tyrosine phosphorylated (pY) and unphosphorylated (Y) forms (Fig. 1B). They encompassed only three flanking residues C-terminal to tyrosine and eight residues toward the N terminus, as additional contacts between PTB domains and their cognate ligands are often made upstream of the NPXY motif. Successful synthesis was confirmed by Western blotting using antibodies against phosphotyrosine (pTyr), as well as the selective coupling of biotin to the extreme N terminus, marking only full-length peptides. The SPOTS assay was chosen as it can be used to study interactions with modified peptides (11) and detect equilibrium binding with affinities down to 1 mM (40). While a subset of the reactions may represent binding events too weak to be biologically relevant, there would be few false negatives.
FIG. 1.
Composition of the NPXY/NXXY motif database and peptide synthesis. (A) Origin of all 126 NXXY motifs identified by surveying human protein sequences classified as receptors. Twenty percent of the sites were derived from RTKs, several of which contain multiple motifs. (B) Peptide arrays comprised of the entire database were synthesized on cellulose membranes. The SPOT template represents a typical synthesis composed of both unphosphorylated (black) and phosphorylated (gray) versions of each 12-mer motif. Western blotting (WB) with anti-pTyr antibodies was able to confirm its presence in most peptides. Also, selective coupling of biotin to only full-length products, with subsequent detection using avidin-HRP, demonstrated that all peptides were adequately synthesized.
Individual PTB domains selectively recognize multiple NPXY-containing peptides.
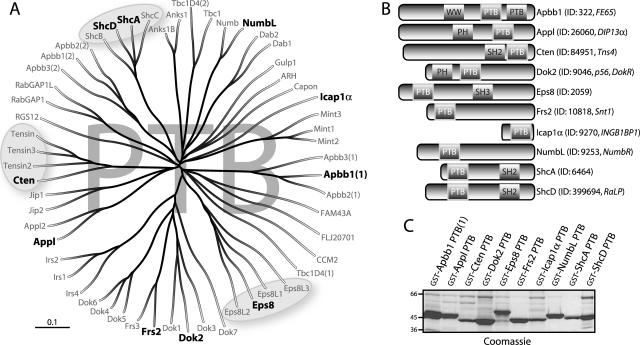
To begin assessing the NPXY binding network, we identified 56 human PTB domains encoded by 52 open reading frames. Protein sequence alignment revealed a number of conserved PTB-containing families, represented by Shc, Dok, IRS, Eps8, and others (Fig. 2A). Ten PTB domains representative of the various families were chosen for further analysis, based on their sequence diversity and predicted domain specificity for motifs containing pTyr (Dok2, Frs2, ShcA, and ShcD) or lacking pTyr (Apbb1, Appl, Cten, Eps8, Icap1α, and NumbL) (50, 75). ShcA was included due to the extensive previous analysis of its structure, specificity, and signaling properties (2, 17, 20, 43, 63, 69, 83). While both the Dok2 and Frs2 PTB domains have also been implicated in a number of signaling pathways, the other seven domains have been poorly characterized and possess few known binding partners. Their source proteins contain varied domain architectures and are predicted to act as scaffolds or adaptors (Fig. 2B).
FIG. 2.
A diverse subset of PTB domains from the human proteome was used to probe the NXXY motifs. (A) Tree representation derived from a protein alignment of the full complement of 56 human PTB domains. Paralogous protein families, such as those represented by Shc, Eps, and tensin (circled), are evident based solely on the similarity of their PTB domain sequences. Ten domains representing various branches of the alignment were selected for binding assays (shown in bold). (B) Domain architecture of the full-length proteins from which the 10 PTB domains were taken. The majority contain associated signaling modules such as WW, PH, SH3, or SH2 domains. Of note, the Apbb (Fe65) family contains two PTB domains, and Icap1α is composed entirely of a single PTB domain. Gene identifiers and aliases (italics) are listed beside the common names. (C) Production of PTB domains for screening of peptide arrays. Individual domains were expressed in E. coli as recombinant proteins containing N-terminal GST tags for their subsequent detection on SPOTS.
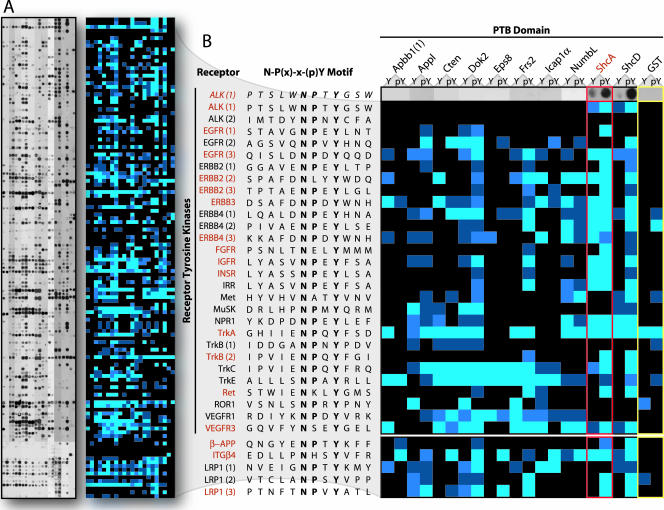
To probe the peptide set, PTB domains were expressed as GST-tagged proteins (Fig. 2C). The SPOTS membranes were incubated with a 0.25 μM concentration of each recombinant domain or 2 μM GST alone as a control. Bound proteins were detected using anti-GST antibodies. Reactions were then subjected to densitometry measurements and classified as strong, intermediate, weak, or undetectable (Fig. 3A; see Fig. S1 in the supplemental material).
FIG. 3.
Scanning of the peptide arrays demonstrates individual PTB domain specificities and evaluates over 2,500 potential interactions. (A) Full array results using the 10 selected PTB domains (see also Fig. S1 in the supplemental material). Peptide SPOTS representing the complete in vitro reaction set are shown on the left. The strength of individual reactions was determined by densitometry and categorized as strong (bright blue), intermediate (blue), weak (dark blue), or undetectable (black). The resulting color-coded array is shown at right of the SPOTS data. (B) A subset of the array showing PTB domain binding to NXXY motifs found in human RTKs. Common names for the receptors and their motif sequences are listed alphabetically down the left side. PTB domains are at top, with binding to both unphosphorylated (Y) and phosphorylated (pY) motifs shown. Both the original reactions, as well as the densitometry measurements, are presented for the anaplastic lymphoma kinase (ALK) receptor (top row). Probing with 2 μM (an eight times excess) of GST alone was used as a control (far right, yellow outline). The ShcA PTB domain (red outline) has previously been shown to recognize several NXXY peptides derived from RTKs (names in red), as well as motifs from additional receptors on the arrays (below RTKs). All of these interactions were scored positive. EGFR, epidermal growth factor receptor; FGFR, fibroblast growth factor receptor; IGFR, insulin-like growth factor receptor; INSR, insulin receptor; IRR, insulin receptor-related receptor; NPR1, natriuretic peptide receptor; ROR1, receptor tyrosine kinase-like orphan receptor; VEGFR, vascular endothelial growth factor receptor; APP, amyloid precursor protein; ITG, integrin; LRP, low-density lipoprotein-related protein.
To evaluate the assay, we studied the results obtained by probing with the ShcA PTB domain. Figure 3B shows the complete set of RTK NXXY motifs from the analysis, many of which have been shown to recruit ShcA in cells or to interact with its PTB domain in vitro (Fig. 3B, red names). Considering binding to the phosphorylated peptides and eliminating all sequences recognized by the GST control, it is evident that every previously described ShcA interaction was scored as positive. Only the first NPXY motif from ErbB2 was identified as a possible false positive, as this site reportedly does not associate with ShcA in vivo or in vitro (16, 17) though it does conform to the ShcA consensus.
While binding to the phosphorylated peptides gave the expected results, a fraction of the interactions thought to be phospho dependent were not identified as such on the arrays. This is likely due to the sensitivity of the assay and PTB domain recognition of residues other than pTyr. Indeed, PTB domains show extensive contacts along the entire length of their peptide ligands (68), exemplified by the fact that a majority do not require pTyr for binding. Despite the apparent lack of phospho specificity in a small subset of the data, the results correlate well with previously described findings for ShcA.
The full binding matrix revealed a complex pattern of domain-peptide recognition, suggesting that individual PTB domains have distinct specificities. Because the array was composed entirely of peptides containing the consensus NPXY/NXXY motif, a significant amount of the selectivity must depend on residues outside of this core.
Verification of PTB domain binding data using full-length receptors.
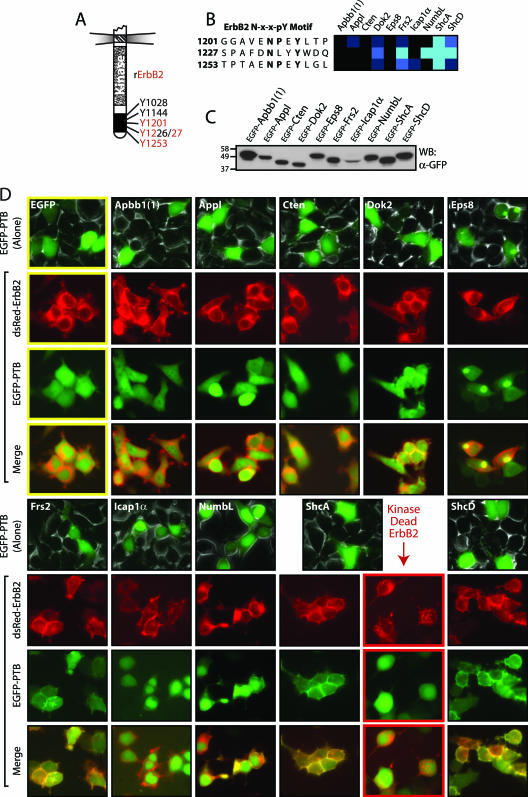
To pursue the possible relevance of the peptide binding data, we focused on two RTKs for which there is already a significant body of knowledge: ErbB2 (Neu/Her2) and Ret, both of which possess NXXY sequences recognized by some PTB domains used in the screen. ErbB2 belongs to the ErbB or epidermal growth factor family of RTKs and possesses at least four phosphorylated Tyr residues in its C-terminal tail that mediate its transforming potential (18, 39). Three of these ErbB2 tyrosine sites are in NXXY motifs present on the arrays (Fig. 4A). The SPOTS data indicated that the PTB domains from ShcA, ShcD, Frs2, and NumbL could interact strongly with motifs in ErbB2 (Fig. 4B). Dok2 recognized one peptide with intermediate affinity, and there were weak interactions between Appl and Icap1α with single sites. Neither Apbb1 nor Cten nor Eps8 positively reacted with any ErbB2 motifs. To determine if the data obtained from the array analysis could predict which of the studied PTB domains can colocalize with ErbB2 in cells, we expressed EGFP-tagged PTB domains in HEK 293T cells (Fig. 4C). Individual domains displayed no discrete localization when expressed alone, except Eps8, which resides in a large punctate structure outside the nucleus in ∼50% of the cells. Upon cotransfection with an RFP-tagged, activated ErbB2 (NeuNT/V664E) (3), all of the domains that bound with intermediate to strong affinity to ErbB2-derived peptides colocalized with the receptor at the plasma membrane (Fig. 4D, Dok2, Frs2, NumbL, ShcA, and ShcD). None of the domains that either failed to recognize an ErbB2 motif on the peptide array, or bound only weakly, had an altered localization (similar to the EGFP control: Apbb1, Appl, Cten, Eps8, and Icap1α). Additionally, the ShcA PTB domain did not colocalize with a kinase-dead form of ErbB2 (K757M), verifying a dependence for this interaction on Tyr phosphorylation. These results correlate with previously described data and the peptide binding assays. Significantly, three new PTB domains capable of recognizing NXXY motifs in ErbB2 were identified: Frs2, NumbL, and ShcD.
FIG. 4.
Colocalization of PTB domains with full-length ErbB2 verifies data from the interaction matrix. (A) Representation of the intracellular region of ErbB2. Common numbering of tyrosines is from Rattus norvegicus protein. Tyrosine residues contained in NXXY motifs present on the arrays are shown in red. (B) Array results for PTB domain binding to three tyrosine-phosphorylated motifs of ErbB2. Frs2, NumbL, ShcA, and ShcD all recognized at least one motif with high affinity. An intermediate interaction with Dok2, as well as weak associations between Appl and Icap1α with a single motif were observed. Amino acid numbers are for corresponding tyrosines in R. norvegicus ErbB2. (C) Individual PTB domains were expressed with fluorescent tags to examine their localization. Recombinant proteins N-terminally labeled with EGFP were expressed in HEK 293T cells. Production of PTB domains was monitored by Western blotting (WB), using antibodies against GFP. (C) Domains able to bind peptide motifs from ErbB2 colocalize with the receptor at the plasma membrane. EGFP-PTB domains show no obvious localization when expressed alone (top rows), save for Eps8 which localizes in a large punctate area outside of the nucleus in ∼50% of the cells. PTB domains were coexpressed with an activated, RFP-tagged ErbB2. Shown are the localizations of ErbB2 (red, second row), the EGFP-PTB domains (green, third row), and their merged images (fourth row). An EGFP control (top left, yellow outline) showed no colocalization with the receptor. The ShcA PTB domain was also coexpressed with a kinase-dead ErbB2 (lower right, red outline), to determine the requirement for kinase activity.
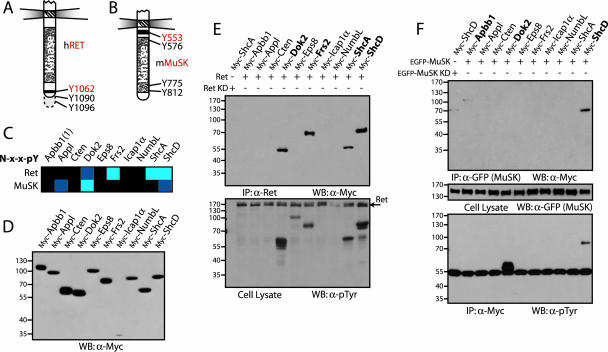
We also pursued interactions involving Ret, a receptor for ligands of the glial cell-line derived neurotrophic factor family (1, 51). Autophosphorylation of an NXXY motif in the C-terminal tail of the receptor (Y1062) is able to activate the Ras/mitogen-activated protein kinase (MAPK), phosphatidylinositol 3-kinase/Akt, and Jun N-terminal protein kinase pathways (Fig. 5A) (42, 51). This correlates with signaling through the PTB domains of adaptor proteins like ShcA, Frs2, and Dok2, all of which bound the phosphorylated Y1062 motif on our arrays (Fig. 5C), as did the ShcD PTB domain. Apbb1, Appl, Cten, Eps8, Icap1α, and NumbL did not positively react with the Ret peptide. To test these data, we transfected Myc-tagged forms of the full-length PTB domain adaptor proteins in HEK 293T cells (Fig. 5D) together with Ret, which is constitutively phosphorylated due to overexpression. All of the adaptors predicted to associate with the receptor by in vitro peptide binding, including ShcD, coprecipitated with Ret from cell lysates (Fig. 5E, upper panel). Western blotting of whole-cell lysates with anti-pTyr antibodies showed that Frs2, Dok2, ShcA, and ShcD were also phosphorylated in cells expressing activated Ret (Fig. 5E, lower panel). As a control, ShcA was not phosphorylated or coprecipitated with a kinase-dead version of the receptor (K758M). None of the other six PTB domain-containing proteins we examined interacted with Ret, as assessed by this method. The coprecipitations provide further evidence that the peptide-binding matrix is representative of interactions that can occur between full-length proteins.
FIG. 5.
Peptide arrays identify interactions between full-length receptors and PTB domain-containing adaptor proteins. (A and B) Representation of the intracellular regions from Ret and MuSK. Common numbering of tyrosines from the Ret receptor is from Homo sapiens and from Mus musculus for MuSK. Tyrosine residues contained in NXXY motifs present on the arrays are shown in red. (C) Interaction data for PTB domains with tyrosine-phosphorylated NXXY motifs from the Ret and MuSK receptors. Domains from Dok2, Frs2, ShcA, and ShcD bound the Ret peptide. Interactions between the MuSK motif and the PTB domains of Appl, Dok2, and ShcD represented novel and potentially interesting associations. (D) Full-length PTB domain adaptor proteins were used to assess the interactions. Proteins were cloned and expressed in HEK 293T cells with N-terminal Myc tags. Production was monitored by Western blotting, using antibodies against Myc. (E) PTB domain-containing proteins were coexpressed with activated Ret. Immunoprecipitation using antibodies against the receptor, and immunoblotting with anti-Myc, revealed associated proteins (upper panel). Only those adaptors containing PTB domains able to recognize the NXXY motif of the Ret receptor in vitro were coprecipitated (shown in bold). To determine if these proteins were also phosphorylated by Ret, a Western blot using antibodies against pTyr was performed (lower panel). As a control, ShcA was also coexpressed with a kinase-dead Ret (lane 1). (F) Adaptor protein precipitation or phosphorylation by MuSK. The Myc-tagged PTB proteins were coexpressed with recombinant, EGFP-tagged MuSK in HEK 293T cells. Immunoprecipitating the receptor with anti-GFP antibodies and a Western blot using anti-Myc revealed associated proteins (upper panel). Both Apbb1 and ShcD were precipitated (in bold). Immunoblotting anti-Myc precipitations using antibodies against pTyr determined whether potential interaction partners were phosphorylated (lower panel). Dok2 and ShcD were highly phosphorylated by MuSK (in bold). The middle panel shows a Western blot on cell lysates using anti-GFP antibodies to confirm expression of the receptor. As a control, ShcD was also coexpressed with a kinase-dead MuSK (lane 1). WB, Western blotting; IP, immunoprecipitation; KD, kinase dead.
PTB domain-mediated interactions with MuSK.
The MuSK RTK is involved in clustering acetylcholine receptors at the developing neuromuscular junction. MuSK signaling depends on an autophosphorylated tyrosine residue in its juxtamembrane region (Y553) contained within an NPXY motif for which Dok7 is the only known binding partner and target (Fig. 5B) (8, 30, 59, 71). This motif was recognized, to various affinities, by the Appl, Dok2, and ShcD PTB domains in the array-based screen (Fig. 5C). To verify these interactions, EGFP-tagged MuSK was coexpressed with the Myc-tagged adaptor proteins in HEK 293T cells. The receptor, which is constitutively phosphorylated, was then precipitated with an anti-GFP antibody, and bound proteins were identified using anti-Myc (Fig. 5F, upper panel). Of the three proteins predicted to bind MuSK, only ShcD was consistently detected by this approach. Dok2 association was not detectable by immunoprecipitating MuSK, but it became highly tyrosine phosphorylated in MuSK-expressing cells, as did ShcD (Fig. 5F, lower panel). Unexpectedly, we also observed a weak interaction with Apbb1 in cells, which was not detected in the array analysis.
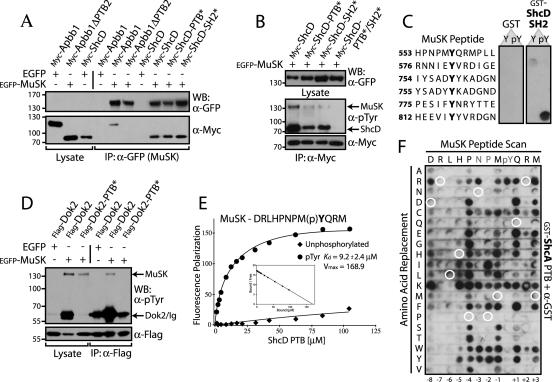
Apbb1, Dok2, and ShcD can bind MuSK through their PTB domains.
We have pursued the interactions between MuSK and the ShcD, Dok2, and Apbb1 proteins, as they might contribute to intracellular signaling from this RTK, in addition to Dok7. These three adaptors contain multiple interaction domains (Fig. 2B). Apbb1 has a second PTB domain at the C-terminal end of the protein, and ShcD has an SH2 domain toward its C terminus, both with the potential for pTyr recognition. To explore how these proteins interact with MuSK, we employed inactivating mutations of their PTB or SH2 domains and coexpressed wild-type or mutant proteins with MuSK in HEK 293T cells (Fig. 6A). A truncation mutant of Apbb1 that removed the second PTB domain (ΔPTB2, with a deletion of residues 532 to 710) was not coprecipitated with activated MuSK, indicating that the interaction is likely mediated by the C-terminal Apbb1 PTB domain and consequently would not have been detected by the peptide arrays as it was not included in the screen. In contrast, single amino acid substitutions in either the PTB (R315Q) or SH2 domains (R549K) of ShcD, which ablate pTyr recognition, did not eliminate binding to MuSK. Such mutants were able to coprecipitate with and serve as substrates for MuSK, albeit at lower levels than wild type (Fig. 6B). ShcD tyrosine phosphorylation was only entirely eliminated following mutation of both the SH2 and PTB domains.
FIG. 6.
Apbb1, ShcD, and Dok2 associate with the MuSK receptor via PTB domain interactions, whereas ShcA does not. (A) The C-terminal PTB domain of Apbb1 and both the PTB and SH2 domains of ShcD bind to MuSK. EGFP-tagged MuSK or EGFP alone as a control was coexpressed with Myc-Apbb1 or a truncated version lacking the second PTB domain (ΔPTB2). Wild-type ShcD or individual PTB/SH2 domain mutants of ShcD (PTB* or SH2*) were also coexpressed. Immunoprecipitation of MuSK using anti-GFP antibodies and immunoblotting with anti-Myc revealed bound proteins (bottom panel). Whole-cell lysates (lanes 1 to 3) and binding to EGFP alone (lanes 4 and 7) served as controls. An anti-GFP Western blot confirmed expression of the receptor (top panel). While both ShcD mutants were able to bind, Apbb1ΔPTB2 was not. (B) Mutation of both the SH2 and PTB domains of ShcD is necessary to relieve its phosphorylation and abolish binding to the receptor. Myc-ShcD or ShcD carrying point mutations in the SH2 and/or PTB domains (PTB*, SH2*, or PTB*/SH2*) was coexpressed with EGFP-MuSK. Immunoprecipitation with anti-Myc and anti-pTyr immunoblots determined the amount of bound receptor, as well as the phosphorylation level of the ShcD proteins (middle panel). Expression of ShcD was verified by Western blotting with anti-Myc (bottom panel) and blotting of whole-cell lysates using anti-GFP confirmed MuSK expression (top panel). (C) The ShcA SH2 domain binds Y812 of MuSK in vitro. SPOTS peptides representing pTyr residues from the intracellular region of MuSK were synthesized. Membranes were incubated with recombinant ShcA SH2 domain tagged with GST or GST alone as a control. Bound proteins were identified using anti-GST antibodies. Peptide sequences, synthesized as both phosphorylated (pY) and unphosphorylated (Y) motifs are listed down the left. (D) MuSK interaction with, and phosphorylation of Dok2 requires its PTB domain. Flag-tagged Dok2, wild-type or with a PTB domain point mutation (PTB*), was coexpressed with EGFP-MuSK. Precipitation of Dok2 using anti-Flag antibodies and Western blotting with anti-pTyr determined the amount of associated MuSK (top panel). Whole-cell lysate (lanes 1 to 3) served as a control and revealed the extent of Dok2 phosphorylation. An anti-Flag immunoblot confirmed expression of Dok2 (bottom panel). (E) The ShcD PTB domain binds specifically to phosphorylated MuSK, as determined by fluorescence polarization. Purified, untagged ShcD PTB domain was incubated with fluorescently labeled peptides derived from the MuSK NPXY motif (sequence at top). Both tyrosine-phosphorylated (circle) and -unphosphorylated (diamond) versions were used. Displayed are the Michaelis-Menten and Scatchard (inset) plots. Only the phosphorylated peptide bound with significant affinity, and the Kd and Vmax for this interaction are shown. (F) The ShcA PTB domain binding specificity differs from that of ShcD, and it is unable to recognize MuSK ligand. All 20 natural amino acids were substituted at each position of the MuSK NPXY motif. The 12-mer peptide sequence is indicated along the top, with amino acid substitutions listed down the left side. The membrane was incubated with GST-ShcA PTB domain, and bound protein was revealed with anti-GST antibodies. Wild-type peptide sequence did not bind (circled in white), though a number of single residue substitutions conferred a positive interaction. IP, immunoprecipitation.
To determine where the ShcD SH2 domain might recognize the activated receptor, we synthesized SPOTS peptides corresponding to the six intracellular tyrosines known to be phosphorylated in MuSK (78). The membrane was incubated with 0.25 μM GST-ShcD SH2 domain, and binding was revealed using anti-GST antibodies (Fig. 6C). A single, phospho-specific interaction with the MuSK Y812 site was observed. This argues that both SH2 and PTB domain-mediated interactions could couple ShcD to MuSK, resulting in its tyrosine phosphorylation and subsequent ability to transduce signals downstream of the receptor, and suggests that these domains may act synergistically to associate with MuSK.
In the case of Dok2, substitution of two PTB domain residues involved in pTyr recognition (R202/217E) (35) resulted in a loss of Dok2 association with MuSK and an attenuation of its phosphorylation on tyrosine (Fig. 6D). These data confirm that the PTB domain of Dok2 can also mediate an interaction with MuSK.
The ShcD and ShcA PTB domains differ in their ability to recognize MuSK.
Due to the high sequence similarity between the ShcA and ShcD PTB domains (91%), we anticipated that they would show like binding profiles. Indeed, their overall pattern of peptide recognition was extremely similar; however, only ShcD detectably recognized MuSK. We therefore measured the affinity with which the ShcD PTB domain binds the NPXY peptide motif from MuSK using fluorescence polarization. Purified PTB domain was incubated with fluorescently labeled MuSK peptide and determined to bind with a Kd of 9.2 μM (Fig. 6E), in the range of previously described PTB domain-NPXpY peptide interactions. We observed no binding to the unphosphorylated MuSK motif, and the ShcA PTB domain was unable to detectably associate with either peptide using the same assay. This indicated that the two PTB domains do possess distinct binding properties, despite having a general capacity to recognize very similar motifs.
To test how closely the MuSK NPXY sequence resembles a ShcA PTB domain substrate, SPOTS peptides were synthesized that replaced each position of this motif with all 20 natural amino acids. The membrane was then probed with GST-ShcA PTB domain (Fig. 6F). None of the control (wild-type) peptides positively interacted with the ShcA PTB domain, but several peptides containing single residue changes were recognized. This suggested that the PTB domain binding site in MuSK is not far from being a ShcA ligand. However, single substitutions introduced into full-length MuSK (P549L or Q554D) only induced a weak ability to precipitate ShcA and did not result in ShcA phosphorylation (data not shown). It is therefore likely that multiple amino acid substitutions would be required to endow upon the receptor a capacity to bind ShcA. Phosphorylated NPXY in the context of the MuSK motif is therefore a poor ShcA ligand in comparison to ShcD.
Determining PTB domain specificity using interaction array data.
Phage display or degenerate peptide libraries have been used to determine consensus binding motifs for modular domains. While these are important approaches, the resulting consensus sites are not necessarily prevalent in potential interacting proteins. The arrays described here consist entirely of motifs found in the human proteome but can still be used to derive consensus binding data. To identify residues preferred by individual PTB domains, other than those in the NPXY core, we clustered the peptides interacting with high intensity and searched for amino acid similarities at each position. Figure 7A shows results using the ShcD PTB domain as a probe (see also Fig. S2 in the supplemental material). There is a strong partiality to aliphatic residues at the −5 position, as well as hydrophobic residues at the +1 and +3 positions and hydrophilic amino acids at −7. This is similar to results obtained for the ShcA PTB domain using degenerate peptides (69) and phage display (20), reinforcing the similarity in binding specificity of the two domains.
FIG. 7.
Individual PTB domain specificities can be derived from clustering motifs with sets of defined binding properties. (A) The ShcD PTB domain prefers aliphatic residues at the −5 position, hydrophobic amino acids at the +1 and +3 positions, and hydrophilic residues at position −7 (relative to pTyr). Peptides recognized by the ShcD PTB domain were clustered according to signal intensity, with higher affinity interactions grouped at top (along left side). It is clear that ShcD prefers peptides containing pTyr (right side of cluster). By studying the favored sequences (receptor name and motif listed), we derived information on residue selection at specific positions in the peptide. Conserved amino acid classes are boxed and shown in bold (symbols in panels A, B, and E are by convention: ζ, hydrophilic; Φ, hydrophobic; Ω, aromatic; Ψ, aliphatic; π, small side chain; [−], acidic; [+], basic). (B) The Eps8 PTB domain shows preference for hydrophobic amino acids at the −5 position and basic residues C-terminal to tyrosine. Clustering the high-affinity Eps8 binding peptides confirmed the domain has no requirement for pTyr (along left side). The favored ligands typically contain arginine, lysine, or histidine at the +1 to +3 positions. (C) Examining the full complement of peptides recognized by Eps8 corroborates a predisposition to basic residues. A frequency plot containing information from all of the Eps8 PTB domain binding motifs shows a bias towards basic residues C-terminal to tyrosine (black), as well as hydrophobic residues (black outline) at the −5 position. Arginine at +1 is particularly prevalent (asterisk). (D) Confirmation of Arg requirement for Eps8 PTB domain interactions. All 20 natural amino acids were substituted at each position of a synthetic NPXY motif conforming to a consensus Eps8 binding site. The 12-mer peptide sequence is indicated along the top, with residue substitutions listed down the left side. The membrane was incubated with GST-Eps8 PTB domain, and bound protein was revealed using anti-GST antibodies. A requirement for arginine at the +1 position can be observed. (E) Clustering motifs recognized by ShcD but not ShcA. Five phosphorylated NXXY peptides binding to the ShcD PTB domain (with low to high affinity), and not to ShcA, show similar amino acid properties (boxed and shown in bold), including an aromatic residue at −5 and a basic residue at +2. (F) Arginine at the +2 position of Ret9 impedes ShcA binding but not ShcD, ShcB, or ShcC.12-mer peptides representing the NXXY motif from the long and short isoforms of Ret were synthesized as SPOTS in phosphorylated or unphosphorylated forms and incubated with GST-PTB domains from all four Shc proteins. Sequence differences C-terminal to the NXXY tyrosine are shown in italics. Only ShcA preferred the Ret51 motif, lacking arginine at +2, over the Ret9 motif. ALK, anaplastic lymphoma kinase; EGFR, epidermal growth factor receptor; GPCR, G-protein-coupled receptor; OR1A, odorant receptor 1A; OX2R, orexin receptor type 2; CD200R, cell surface glycoprotein receptor CD200; LOXC, lysyl oxidase-like 4 receptor; LRPDIT, low density lipoprotein receptor-related protein deleted in tumor; TU12B1-TY, nucleotidase domain-containing receptor; VIPR, vasoactive intestinal peptide receptor; GP330, low-density lipoprotein receptor-related protein 2; P2RX2, purinergic receptor; PACAPR, adenylate cyclase activating polypeptide receptor; TAS1R2, G protein-coupled taste receptor.
We also examined the peptides recognized by Eps8, as it has no previously known ligands (Fig. 7B). Contrary to the ShcD binding pattern and agreeing with previous predictions (75), it was evident that Eps8 is not a phospho-dependent binding module. The Eps8 PTB domain is slightly predisposed to hydrophobic amino acids at the −5 position but most strikingly binds peptides with basic residues C-terminal to the NPXY tyrosine. A frequency plot containing information from every interacting peptide (weak or strong) confirmed a preference for arginine at the +1 position (Fig. 7C). To measure the relevance of this arginine, an artificial peptide corresponding to the determined PTB consensus binding sequence was synthesized on SPOTS membranes in which each position was replaced with all 20 amino acids. By probing with GST-Eps8 PTB domain, it was evident that the +1 arginine could not be replaced by any other amino acid (Fig. 7D). This verified that the Eps8 PTB domain selects basic residues C-terminal to the NPXY tyrosine and supports the utility of this approach as a tool for determining consensus binding data.
Using a similar clustering method, we also sought to identify the properties of peptide ligands that were discretely recognized by separate PTB domains. Figure 7E lists five phosphorylated motifs, including MuSK, that positively reacted with the ShcD PTB domain but were not bound by ShcA. As four of these peptides possess basic residues at the +2 position, it seemed possible that this may account for their slight differences in specificity. One example where this may have biological significance is binding to the Ret receptor. There are two alternatively spliced isoforms of Ret that differ immediately downstream of the Y1062 NXXY motif, one containing an arginine at the +2 position (short isoform, Ret9), and the other, a methionine (long isoform, Ret51) (34, 45, 48). To test their binding to Shc, we synthesized the motifs as peptide SPOTS and probed with GST-PTB domains from all four Shc adaptor proteins (Fig. 7F). Ret51, which was included in the original NXXY arrays, interacted strongly with both ShcA and ShcD as before and not with ShcB or ShcC. Ret9 was bound with higher affinity by ShcB, ShcC, and ShcD but only weakly by ShcA. This confirms that the +2 arginine present in Ret9 does reduce ShcA PTB binding and that clustering motifs binding in a mutually exclusive manner can help explain variance in domain specificities. These data suggest that the distinct sequences immediately C-terminal to the NXXY motif in alternatively spliced Ret isoforms may be biologically important. Coupled with the Eps8 consensus data and MuSK interactions, they imply that basic residues C-terminal to NPXY tyrosines may also be a more general means of regulating PTB domain binding.
DISCUSSION
Peptide arrays based on physiological motifs.
We have examined PTB domain binding properties using a comprehensive array of all 117 NPXY motifs found in membrane-associated receptors, and an additional 9 NXXY sites. These data indicate binding preferences for 10 of the 56 human PTB domains, based on peptides they are likely to encounter within the cell, and therefore reveal potential interactions between adaptor proteins and NXXY-containing receptors. In addition, they present information on consensus ligand-binding properties of individual domains and provide a basis for dissecting the contribution of different modules within a multidomain protein to receptor binding.
The systematic screen evaluated over 2,500 possible receptor-adaptor interactions. To test the effectiveness of the system, the arrays were probed with the ShcA PTB domain. Interactions were observed with motifs corresponding to previously identified binding sites on RTKs of the ErbB (15, 16, 66, 76) and Trk families (54), as well as the insulin receptor (23, 27) and a number of other signaling and adhesion proteins (amyloid precursor protein [65, 73], interleukin receptors [5, 64], integrins [19, 52, 77], and low-density lipoprotein receptors [4]. The positive identification of established ShcA interactions indicated that the assay provides a useful prediction for associations between full-length proteins.
In addition to their high sensitivity, the arrays also provided a semiquantitative approach where interactions between equally represented substrates could be classified based on signal intensity. Moreover, cluster analysis provided a data set from which preferred binding motifs could be extracted, with the advantage of being derived solely from sequences in the human proteome. There are some limitations to using peptide arrays. For example, not all of the NXXY peptides may be available for binding to PTB domains in the context of the intact protein (i.e., buried in the three-dimensional structure or present in the extracellular region), and the relevant binding partners may not necessarily be coexpressed.
PTB domain interactions with RTK motifs.
We have extended the data from the interaction matrix by characterizing the binding of PTB domain proteins to Ret and ErbB2, both of which are involved in mammalian development and the etiology of some cancers (1, 17, 18, 39, 51, 72). By observing PTB domain association with these RTKs in cells, we verified known and several novel interactions detected on the arrays. The recently identified ShcD (or RaLP) protein bears high sequence identity with ShcA and potentially contributes to signals downstream of ErbB2 and Ret. NumbL was unique in having the only PTB domain that likely does not require tyrosine phosphorylation for strong binding but which nonetheless colocalizes with ErbB2, albeit more weakly than the other binding partners. The second NXXY motif in the C-terminal tail of the receptor was a favored target of NumbL, independent of its phosphorylation state, and if expressed in similar tissues and developmental stages, this might represent an interaction with biological implications.
Several of the array motifs represent receptors for which there is little information about downstream signaling. The presence of consensus NPXY sites in these proteins suggests that PTB domain-containing adaptors may be an important component of intracellular signaling complexes. The juxtamembrane region of MuSK, which is necessary and sufficient for the clustering of acetylcholine receptors at the developing neuromuscular junction, contains a canonical NPXY motif (29, 30, 71). The PTB domain of the scaffold protein Dok7 has recently been shown to signal from this site and to exert effects on neuromuscular synaptogenesis (59). We find that the ShcD and Dok2 proteins can also interact with MuSK Y553 via their PTB domains. While inactivating mutations in the PTB domain of Dok2 completely abrogated its association with the receptor, we found synergistic interactions with the SH2 and PTB domains of ShcD. Like MuSK, both Dok2 (36) and ShcD (unpublished results) are expressed in skeletal muscle, though Dok2 is mostly found in spleen and lungs and ShcD in neuronal tissues. We identified a third interaction with the C-terminal PTB domain of Apbb1. Apbb1 and its closely related family members Apbb2 (Fe65L), and Apbb3 (Fe65L2), are primarily neuronal proteins (38). Of relevance, MuSK is also expressed in the brain where it is involved in memory retention (26). It will be interesting to explore whether these three candidates play a role in transducing MuSK signals in vivo, whether in skeletal muscle or the nervous system.
Consensus binding motifs for PTB domains.
By probing a large set of NXXY peptides, we were able to obtain consensus binding motifs for PTB domains, reflecting their selectivity for residues outside of this core. Clustering the higher-affinity ligands of ShcD, for example, gave an extended consensus sequence of ζxΨxNPxpYΦxΦ (ζ indicates hydrophilic, Ψ indicates aliphatic, and Φ indicates hydrophobic residues), whereas the peptides recognized by ShcA provided a consensus of ΦΦ[-]NPxpYΦ ([−] representing acidic residues). Therefore, despite their high overall identity and propensity to bind similar ligands, there is a slight difference in the substrate binding preferences of the ShcA and ShcD PTB domains, as exemplified by their contrasting interactions with MuSK. The MuSK peptide is not a perfect match to the ShcD consensus, consistent with its moderate binding affinity (Kd = 9.2 μM) but may make a favorable interaction due to the hydrophobic at the +3 position. It is these extended contacts that may permit the ShcD PTB domain to recognize even the unphosphorylated MuSK peptide on the arrays. Such selective interactions between modular domains and sequences flanking their core recognition motifs can contribute to selectivity in signaling (62).
Some of the NXXY sites show rather broad specificities, which may permit signal transduction through multiple targets. For instance, the Ret receptor possesses a single NXXY site that is recognized by PTB domains in the Shc, IRS, Frs, and Dok family proteins to variably signal through the Ras/MAPK, phosphatidylinositol 3-kinase/Akt, Jun N-terminal protein kinase, p38MAPK and ERK5 pathways (28, 41, 55-57). Conversely, proteins containing two PTB domains with potentially divergent binding properties, like the Apbb1 family, represent platforms upon which physiologically important pathways might converge, as has been shown for scaffolds containing multiple WW domains (33). The differential expression and localization patterns of these proteins will also play a key role in determining when and where they undergo productive interactions.
PTB domain-NXXY peptide interactions in signaling networks.
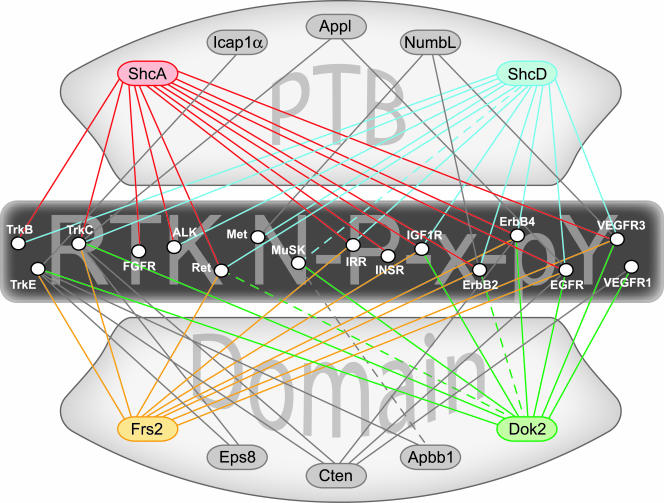
Peptide array data allow the connection of adaptor proteins to the receptors from which they may be signaling to build interaction networks. Figure 8 shows an example of such a system using only high-affinity PTB domain binding to phospho-NXXY peptides. From such a plot we can derive both specific information on possible binding partners, as well as data on which NPXY-mediated interactions play significant roles in signaling from RTKs. The Shc proteins, along with Frs2 and Dok2, are highly connected, suggesting that they are important initiators of intracellular pathways. Some receptors, such as those in the ErbB family, are potentially associated with a variety of adaptors, providing them with the flexibility to signal through a diverse set of pathways. This same analysis using integrins shows that many of their NXXY motifs (from β4, β5, β6, and β7, in particular) are highly connected to PTB domain-containing adaptor proteins as well (see Fig. S3 in the supplemental material). The phospho-independent PTB domains from Cten and Icap1α become more highly integrated in this network, contrary to their lack of participation with phospho-RTKs. This agrees with previous data demonstrating that Icap1α (9, 81) and the tensin family proteins (47, 74) are components of integrin-mediated adhesion. These data could be overlapped with orthogonal data sets from approaches such as mass spectrometry, Lumier, or yeast two-hybrid-based screens to build more extensive networks, which would then contain noncanonical interactions such as the Frs2 PTB domain recognition of an extended peptide sequence in the juxtamembrane of the fibroblast growth factor receptor (21).
FIG. 8.
Protein interaction maps can be assembled using data obtained from the peptide arrays. Exploiting the in vitro binding data to predict interactions between full-length proteins allows us to connect membrane-bound receptors and putative cytoplasmic adaptors. PTB domain-containing adaptor proteins are indicated at top and bottom, and RTKs bearing one or more NXXY motifs in their cytoplasmic domain are listed across the center. Only the higher-affinity binding reactions with phosphorylated motifs are shown (solid lines). Adaptors showing high connectivity in this network are highlighted with color (ShcA, ShcD, Frs2, and Dok2). The interaction map can be further supplemented with data verifying associations that fell below the affinity threshold, such as the interactions described for ShcD-MuSK, Dok2-ErbB2, and Apbb1-MuSK (dashed lines). ALK, anaplastic lymphoma kinase; EGFR, epidermal growth factor receptor; FGFR, fibroblast growth factor receptor; IGFR, insulin-like growth factor receptor.
Here we described an in vitro screen for PTB domain ligands using NPXY sequences derived from receptors in the human proteome. It will be interesting to further exploit these data to determine the significance of these interactions on a biological scale.
Supplementary Material
Acknowledgments
We thank Carlos Ibanez for the wild-type and kinase-dead Ret constructs and Chris Stark for help with bioinformatics.
This work was supported by grants from the Canadian Institutes for Health Research (CIHR), the National Cancer Institute of Canada (NCIC) with funds from the Terry Fox Run, and Genome Canada through the Ontario Genomics Institute. M.J.S. was a recipient of an NSERC Postgraduate Scholarship. W.R.H. was supported by a Terry Fox Ph.D. studentship from the NCIC. J.M.M. is a recipient of a C. J. Martin Fellowship from the National Health and Medical Research Council of Australia. N.J. was supported by fellowships from the CIHR and the NCIC.
Footnotes
Published ahead of print on 18 September 2006.
Supplemental data for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Asai, N., M. Jijiwa, A. Enomoto, K. Kawai, K. Maeda, M. Ichiahara, Y. Murakumo, and M. Takahashi. 2006. RET receptor signaling: Dysfunction in thyroid cancer and Hirschsprung's disease. Pathol. Int. 56:164-172. [DOI] [PubMed] [Google Scholar]
- 2.Barberis, L., K. K. Wary, G. Fiucci, F. Liu, E. Hirsch, M. Brancaccio, F. Altruda, G. Tarone, and F. G. Giancotti. 2000. Distinct roles of the adaptor protein Shc and focal adhesion kinase in Integrin signaling to ERK. J. Biol. Chem. 275:36532-36540. [DOI] [PubMed] [Google Scholar]
- 3.Bargmann, C. I., and R. A. Weinberg. 1988. Increased tyrosine kinase activity associated with the protein encoded by the activated neu oncogene. Proc. Natl. Acad. Sci. USA 85:5394-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes, H., B. Larsen, M. Tyers, and P. van Der Geerqq. 2001. Tyrosine-phosphorylated low density lipoprotein receptor-related protein 1 (Lrp1) associates with the adaptor protein SHC in SRC-transformed cells. J. Biol. Chem. 276:19119-19125. [DOI] [PubMed] [Google Scholar]
- 5.Bates, M. E., W. W. Busse, and P. J. Bertics. 1998. Interleukin 5 signals through Shc and Grb2 in human eosinophils. Am. J. Respir. Cell. Mol. Biol. 18:75-83. [DOI] [PubMed] [Google Scholar]
- 6.Borg, J. P., J. Ooi, E. Levy, and B. Margolis. 1996. The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol. Cell. Biol. 16:6229-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boussif, O., F. Lezoualc'h, M. A. Zanta, M. D. Mergny, D. Scherman, B. Demeneix, and J. P. Behr. 1995. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. USA 92:7297-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowen, D. C., J. S. Park, S. Bodine, J. L. Stark, D. M. Valenzuela, T. N. Stitt, G. D. Yancopoulos, R. M. Lindsay, D. J. Glass, and P. S. DiStefano. 1998. Localization and regulation of MuSK at the neuromuscular junction. Dev. Biol. 199:309-319. [DOI] [PubMed] [Google Scholar]
- 9.Chang, D. D., C. Wong, H. Smith, and J. Liu. 1997. ICAP-1, a novel β1 integrin cytoplasmic domain-associated protein, binds to a conserved and functionally important NPXY sequence motif of β1 integrin. J. Cell Biol. 138:1149-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, H., Z. Zou, K. L. Sarratt, D. Zhou, M. Zhang, E. Sebzda, D. A. Hammer, and M. L. Kahn. 2006. In vivo β1 integrin function requires phosphorylation-independent regulation by cytoplasmic tyrosines. Genes Dev. 20:927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, H. I., A. Einbond, S. J. Kwak, H. Linn, E. Koepf, S. Peterson, J. W. Kelly, and M. Sudol. 1997. Characterization of the WW domain of human yes-associated protein and its polyproline-containing ligands. J. Biol. Chem. 272:17070-17077. [DOI] [PubMed] [Google Scholar]
- 12.Chen, W. J., J. L. Goldstein, and M. S. Brown. 1990. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J. Biol. Chem. 265:3116-3123. [PubMed] [Google Scholar]
- 13.Chien, C. T., S. Wang, M. Rothenberg, L. Y. Jan, and Y. N. Jan. 1998. Numb-associated kinase interacts with the phosphotyrosine binding domain of Numb and antagonizes the function of Numb in vivo. Mol. Cell. Biol. 18:598-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colwill, K., C. D. Wells, K. Elder, M. Goudreault, K. Hersi, S. Kulkarni, W. R. Hardy, T. Pawson, and G. B. Morin. 2006. Modification of the Creator recombination system for proteomics applications-improved expression by addition of splice sites. BMC Biotechnol. 6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Culouscou, J. M., G. W. Carlton, and A. Aruffo. 1995. HER4 receptor activation and phosphorylation of Shc proteins by recombinant heregulin-Fc fusion proteins. J. Biol. Chem. 270:12857-12863. [DOI] [PubMed] [Google Scholar]
- 16.Dankort, D., N. Jeyabalan, N. Jones, D. J. Dumont, and W. J. Muller. 2001. Multiple ErbB-2/Neu phosphorylation sites mediate transformation through distinct effector proteins. J. Biol. Chem. 276:38921-38928. [DOI] [PubMed] [Google Scholar]
- 17.Dankort, D., B. Maslikowski, N. Warner, N. Kanno, H. Kim, Z. Wang, M. F. Moran, R. G. Oshima, R. D. Cardiff, and W. J. Muller. 2001. Grb2 and Shc adapter proteins play distinct roles in Neu (ErbB2)-induced mammary tumorigenesis: implications for human breast cancer. Mol. Cell. Biol. 21:1540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dankort, D. L., Z. Wang, V. Blackmore, M. F. Moran, and W. J. Muller. 1997. Distinct tyrosine autophosphorylation sites negatively and positively modulate neu-mediated transformation. Mol. Cell. Biol. 17:5410-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dans, M., L. Gagnoux-Palacios, P. Blaikie, S. Klein, A. Mariotti, and F. G. Giancotti. 2001. Tyrosine phosphorylation of the β4 integrin cytoplasmic domain mediates Shc signaling to extracellular signal-regulated kinase and antagonizes formation of hemidesmosomes. J. Biol. Chem. 276:1494-1502. [DOI] [PubMed] [Google Scholar]
- 20.Dente, L., C. Vetriani, A. Zucconi, G. Pelicci, L. Lanfrancone, P. G. Pelicci, and G. Cesareni. 1997. Modified phage peptide libraries as a tool to study specificity of phosphorylation and recognition of tyrosine containing peptides. J. Mol. Biol. 269:694-703. [DOI] [PubMed] [Google Scholar]
- 21.Dhalluin, C., K. S. Yan, O. Plotnikova, K. W. Lee, L. Zeng, M. Kuti, S. Mujtaba, M. P. Goldfarb, and M. M. Zhou. 2000. Structural basis of SNT PTB domain interactions with distinct neurotrophic receptors. Mol. Cell 6:921-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filardo, E. J., P. C. Brooks, S. L. Deming, C. Damsky, and D. A. Cheresh. 1995. Requirement of the NPXY motif in the integrin β3 subunit cytoplasmic tail for melanoma cell migration in vitro and in vivo. J. Cell Biol. 130:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finlayson, C. A., J. Chappell, J. W. Leitner, M. L. Goalstone, M. Garrity, S. Nawaz, T. P. Ciaraldi, and B. Draznin. 2003. Enhanced insulin signaling via Shc in human breast cancer. Metabolism 52:1606-1611. [DOI] [PubMed] [Google Scholar]
- 24.Forman-Kay, J. D., and T. Pawson. 1999. Diversity in protein recognition by PTB domains. Curr. Opin. Struct. Biol. 9:690-695. [DOI] [PubMed] [Google Scholar]
- 25.Frank, R. 1992. SPOT-synthesis: an easy technique for positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 48:9217-9232. [Google Scholar]
- 26.Garcia-Osta, A., P. Tsokas, G. Pollonini, E. M. Landau, R. Blitzer, and C. M. Alberini. 2006. MuSK expressed in the brain mediates cholinergic responses, synaptic plasticity, and memory formation. J. Neurosci. 26:7919-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giorgetti, S., P. G. Pelicci, G. Pelicci, and E. Van Obberghen. 1994. Involvement of Src-homology/collagen (SHC) proteins in signaling through the insulin receptor and the insulin-like growth factor-I receptor. Eur. J. Biochem. 223:195-202. [DOI] [PubMed] [Google Scholar]
- 28.Grimm, J., M. Sachs, S. Britsch, S. Di Cesare, T. Schwarz-Romond, K. Alitalo, and W. Birchmeier. 2001. Novel p62Dok family members, Dok-4 and Dok-5, are substrates of the c-Ret receptor tyrosine kinase and mediate neuronal differentiation. J. Cell Biol. 154:345-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herbst, R., E. Avetisova, and S. J. Burden. 2002. Restoration of synapse formation in MuSK mutant mice expressing a MuSK/Trk chimeric receptor. Development 129:5449-5460. [DOI] [PubMed] [Google Scholar]
- 30.Herbst, R., and S. J. Burden. 2000. The juxtamembrane region of MuSK has a critical role in agrin-mediated signaling. EMBO J. 19:67-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herrick, T. M., and J. A. Cooper. 2004. High affinity binding of Dab1 to Reelin receptors promotes normal positioning of upper layer cortical plate neurons. Brain Res. Mol. Brain Res. 126:121-128. [DOI] [PubMed] [Google Scholar]
- 32.Howell, B. W., L. M. Lanier, R. Frank, F. B. Gertler, and J. A. Cooper. 1999. The Disabled 1 phosphotyrosine-binding domain binds to the internalization signals of transmembrane glycoproteins and to phospholipids. Mol. Cell. Biol. 19:5179-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingham, R. J., K. Colwill, C. Howard, S. Dettwiler, C. S. Lim, J. Yu, K. Hersi, J. Raaijmakers, G. Gish, G. Mbamalu, L. Taylor, B. Yeung, G. Vassilovski, M. Amin, F. Chen, L. Matskova, G. Winberg, I. Ernberg, R. Linding, P. O'Donnell, A. Starostine, W. Keller, P. Metalnikov, C. Stark, and T. Pawson. 2005. WW domains provide a platform for the assembly of multiprotein networks. Mol. Cell. Biol. 25:7092-7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishiguro, Y., T. Iwashita, H. Murakami, N. Asai, K. Iida, H. Goto, T. Hayakawa, and M. Takahashi. 1999. The role of amino acids surrounding tyrosine 1062 in Ret in specific binding of the Shc phosphotyrosine-binding domain. Endocrinology 140:3992-3998. [DOI] [PubMed] [Google Scholar]
- 35.Jones, N., and D. J. Dumont. 1999. Recruitment of Dok-R to the EGF receptor through its PTB domain is required for attenuation of Erk MAP kinase activation. Curr. Biol. 9:1057-1060. [DOI] [PubMed] [Google Scholar]
- 36.Jones, N., and D. J. Dumont. 1998. The Tek/Tie2 receptor signals through a novel Dok-related docking protein, Dok-R. Oncogene 17:1097-1108. [DOI] [PubMed] [Google Scholar]
- 37.Kavanaugh, W. M., and L. T. Williams. 1994. An alternative to SH2 domains for binding tyrosine-phosphorylated proteins. Science 266:1862-1865. [DOI] [PubMed] [Google Scholar]
- 38.Kesavapany, S., S. J. Banner, K. F. Lau, C. E. Shaw, C. C. Miller, J. D. Cooper, and D. M. McLoughlin. 2002. Expression of the Fe65 adapter protein in adult and developing mouse brain. Neuroscience 115:951-960. [DOI] [PubMed] [Google Scholar]
- 39.Khoury, H., D. L. Dankort, S. Sadekova, M. A. Naujokas, W. J. Muller, and M. Park. 2001. Distinct tyrosine autophosphorylation sites mediate induction of epithelial mesenchymal like transition by an activated ErbB2/Neu receptor. Oncogene 20:788-799. [DOI] [PubMed] [Google Scholar]
- 40.Kramer, A., U. Reineke, L. Dong, B. Hoffmann, U. Hoffmuller, D. Winkler, R. Volkmer-Engert, and J. Schneider-Mergener. 1999. SPOT synthesis: observations and optimizations. J. Pept. Res. 54:319-327. [DOI] [PubMed] [Google Scholar]
- 41.Kurokawa, K., T. Iwashita, H. Murakami, H. Hayashi, K. Kawai, and M. Takahashi. 2001. Identification of SNT/FRS2 docking site on RET receptor tyrosine kinase and its role for signal transduction. Oncogene 20:1929-1938. [DOI] [PubMed] [Google Scholar]
- 42.Kurokawa, K., K. Kawai, M. Hashimoto, Y. Ito, and M. Takahashi. 2003. Cell signalling and gene expression mediated by RET tyrosine kinase. J. Intern. Med. 253:627-633. [DOI] [PubMed] [Google Scholar]
- 43.Lai, K. M., and T. Pawson. 2000. The ShcA phosphotyrosine docking protein sensitizes cardiovascular signaling in the mouse embryo. Genes Dev. 14:1132-1145. [PMC free article] [PubMed] [Google Scholar]
- 44.Landgraf, C., S. Panni, L. Montecchi-Palazzi, L. Castagnoli, J. Schneider-Mergener, R. Volkmer-Engert, and G. Cesareni. 2004. Protein interaction networks by proteome peptide scanning. PLOS Biol. 2:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee, K. Y., E. T. Samy, M. H. Sham, P. K. Tam, and V. C. Lui. 2003. 3′ Splicing variants of Ret receptor tyrosine kinase are differentially expressed in mouse embryos and in adult mice. Biochim. Biophys. Acta 1627:26-38. [DOI] [PubMed] [Google Scholar]
- 46.Li, S. C., C. Zwahlen, S. J. Vincent, C. J. McGlade, L. E. Kay, T. Pawson, and J. D. Forman-Kay. 1998. Structure of a Numb PTB domain-peptide complex suggests a basis for diverse binding specificity. Nat. Struct. Biol. 5:1075-1083. [DOI] [PubMed] [Google Scholar]
- 47.Lo, S. H. 2004. Tensin. Int. J. Biochem. Cell. Biol. 36:31-34. [DOI] [PubMed] [Google Scholar]
- 48.Lorenzo, M. J., C. Eng, L. M. Mulligan, T. J. Stonehouse, C. S. Healey, B. A. Ponder, and D. P. Smith. 1995. Multiple mRNA isoforms of the human RET proto-oncogene generated by alternate splicing. Oncogene 10:1377-1383. [PubMed] [Google Scholar]
- 49.Lundgren, T. K., R. P. Scott, M. Smith, T. Pawson, and P. Ernfors. 17 July 2006, posting date. Engineering the recruitment of PTB adaptor proteins reveals distinct roles for Ret receptor-mediated cell survival. J. Biol. Chem. [Online.] doi: 10.1074/jbc.M600473200. [DOI] [PubMed]
- 50.Margolis, B., J. P. Borg, S. Straight, and D. Meyer. 1999. The function of PTB domain proteins. Kidney Int. 56:1230-1237. [DOI] [PubMed] [Google Scholar]
- 51.Mason, I. 2000. The RET receptor tyrosine kinase: activation, signalling and significance in neural development and disease. Pharm. Acta Helv. 74:261-264. [DOI] [PubMed] [Google Scholar]
- 52.Mauro, L., D. Sisci, M. Bartucci, M. Salerno, J. Kim, T. Tam, M. A. Guvakova, S. Ando, and E. Surmacz. 1999. SHC-α5β1 integrin interactions regulate breast cancer cell adhesion and motility. Exp. Cell Res. 252:439-448. [DOI] [PubMed] [Google Scholar]
- 53.McLoughlin, D. M., and C. C. Miller. 1996. The intracellular cytoplasmic domain of the Alzheimer's disease amyloid precursor protein interacts with phosphotyrosine-binding domain proteins in the yeast two-hybrid system. FEBS Lett. 397:197-200. [DOI] [PubMed] [Google Scholar]
- 54.Meakin, S. O., J. I. MacDonald, E. A. Gryz, C. J. Kubu, and J. M. Verdi. 1999. The signaling adapter FRS-2 competes with Shc for binding to the nerve growth factor receptor TrkA. A model for discriminating proliferation and differentiation. J. Biol. Chem. 274:9861-9870. [DOI] [PubMed] [Google Scholar]
- 55.Melillo, R. M., F. Carlomagno, G. De Vita, P. Formisano, G. Vecchio, A. Fusco, M. Billaud, and M. Santoro. 2001. The insulin receptor substrate (IRS)-1 recruits phosphatidylinositol 3-kinase to Ret: evidence for a competition between Shc and IRS-1 for the binding to Ret. Oncogene 20:209-218. [DOI] [PubMed] [Google Scholar]
- 56.Mercalli, E., S. Ghizzoni, E. Arighi, L. Alberti, R. Sangregorio, M. T. Radice, M. L. Gishizky, M. A. Pierotti, and M. G. Borrello. 2001. Key role of Shc signaling in the transforming pathway triggered by Ret/Ptc2 oncoprotein. Oncogene 20:3475-3485. [DOI] [PubMed] [Google Scholar]
- 57.Murakami, H., Y. Yamamura, Y. Shimono, K. Kawai, K. Kurokawa, and M. Takahashi. 2002. Role of Dok1 in cell signaling mediated by RET tyrosine kinase. J. Biol. Chem. 277:32781-32790. [DOI] [PubMed] [Google Scholar]
- 58.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 59.Okada, K., A. Inoue, M. Okada, Y. Murata, S. Kakuta, T. Jigami, S. Kubo, H. Shiraishi, K. Eguchi, M. Motomura, T. Akiyama, Y. Iwakura, O. Higuchi, and Y. Yamanashi. 2006. The muscle protein Dok-7 is essential for neuromuscular synaptogenesis. Science 312:1802-1805. [DOI] [PubMed] [Google Scholar]
- 60.Pawson, T. 2003. Organization of cell-regulatory systems through modular-protein-interaction domains. Philos. Transact. A 361:1251-1262. [DOI] [PubMed] [Google Scholar]
- 61.Pawson, T. 2002. Regulation and targets of receptor tyrosine kinases. Eur. J. Cancer 38(Suppl. 5):S3-S10. [DOI] [PubMed] [Google Scholar]
- 62.Pawson, T., and P. Nash. 2003. Assembly of cell regulatory systems through protein interaction domains. Science 300:445-452. [DOI] [PubMed] [Google Scholar]
- 63.Ravichandran, K. S. 2001. Signaling via Shc family adapter proteins. Oncogene 20:6322-6330. [DOI] [PubMed] [Google Scholar]
- 64.Ravichandran, K. S., and S. J. Burakoff. 1994. The adapter protein Shc interacts with the interleukin-2 (IL-2) receptor upon IL-2 stimulation. J. Biol. Chem. 269:1599-1602. [PubMed] [Google Scholar]
- 65.Russo, C., V. Dolcini, S. Salis, V. Venezia, N. Zambrano, T. Russo, and G. Schettini. 2002. Signal transduction through tyrosine-phosphorylated C-terminal fragments of amyloid precursor protein via an enhanced interaction with Shc/Grb2 adaptor proteins in reactive astrocytes of Alzheimer's disease brain. J. Biol. Chem. 277:35282-35288. [DOI] [PubMed] [Google Scholar]
- 66.Schulze, W. X., L. Deng, and M. Mann. 2005. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol. Syst. Biol. 1:E1-E13. [Online.] doi: 10.1038/msb4100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seet, B. T., I. Dikic, M. M. Zhou, and T. Pawson. 2006. Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell. Biol. 7:473-483. [DOI] [PubMed] [Google Scholar]
- 68.Shi, N., S. Ye, M. Bartlam, M. Yang, J. Wu, Y. Liu, F. Sun, X. Han, X. Peng, B. Qiang, J. Yuan, and Z. Rao. 2004. Structural basis for the specific recognition of RET by the Dok1 phosphotyrosine binding domain. J. Biol. Chem. 279:4962-4969. [DOI] [PubMed] [Google Scholar]
- 69.Songyang, Z., B. Margolis, M. Chaudhuri, S. E. Shoelson, and L. C. Cantley. 1995. The phosphotyrosine interaction domain of SHC recognizes tyrosine-phosphorylated NPXY motif. J. Biol. Chem. 270:14863-14866. [DOI] [PubMed] [Google Scholar]
- 70.Stolt, P. C., and H. H. Bock. 2006. Modulation of lipoprotein receptor functions by intracellular adaptor proteins. Cell. Signal. 18:1560-1571. (First published 24 May 2006; doi: 1010.1016/j.cellsig.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 71.Strochlic, L., A. Cartaud, and J. Cartaud. 2005. The synaptic muscle-specific kinase (MuSK) complex: new partners, new functions. Bioessays 27:1129-1135. [DOI] [PubMed] [Google Scholar]
- 72.Takahashi, M. 2001. The GDNF/RET signaling pathway and human diseases. Cytokine Growth Factor Rev. 12:361-373. [DOI] [PubMed] [Google Scholar]
- 73.Tarr, P. E., R. Roncarati, G. Pelicci, P. G. Pelicci, and L. D'Adamio. 2002. Tyrosine phosphorylation of the β-amyloid precursor protein cytoplasmic tail promotes interaction with Shc. J. Biol. Chem. 277:16798-16804. [DOI] [PubMed] [Google Scholar]
- 74.Torgler, C. N., M. Narasimha, A. L. Knox, C. G. Zervas, M. C. Vernon, and N. H. Brown. 2004. Tensin stabilizes integrin adhesive contacts in Drosophila. Dev. Cell 6:357-369. [DOI] [PubMed] [Google Scholar]
- 75.Uhlik, M. T., B. Temple, S. Bencharit, A. J. Kimple, D. P. Siderovski, and G. L. Johnson. 2005. Structural and evolutionary division of phosphotyrosine binding (PTB) domains. J. Mol. Biol. 345:1-20. [DOI] [PubMed] [Google Scholar]
- 76.Vijapurkar, U., K. Cheng, and J. G. Koland. 1998. Mutation of a Shc binding site tyrosine residue in ErbB3/HER3 blocks heregulin-dependent activation of mitogen-activated protein kinase. J. Biol. Chem. 273:20996-21002. [DOI] [PubMed] [Google Scholar]
- 77.Wary, K. K., F. Mainiero, S. J. Isakoff, E. E. Marcantonio, and F. G. Giancotti. 1996. The adaptor protein Shc couples a class of integrins to the control of cell cycle progression. Cell. 87:733-743. [DOI] [PubMed] [Google Scholar]
- 78.Watty, A., G. Neubauer, M. Dreger, M. Zimmer, M. Wilm, and S. J. Burden. 2000. The in vitro and in vivo phosphotyrosine map of activated MuSK. Proc. Natl. Acad. Sci. USA 97:4585-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yan, K. S., M. Kuti, and M. M. Zhou. 2002. PTB or not PTB—that is the question. FEBS Lett. 513:67-70. [DOI] [PubMed] [Google Scholar]
- 80.Zambrano, N., J. D. Buxbaum, G. Minopoli, F. Fiore, P. De Candia, S. De Renzis, R. Faraonio, S. Sabo, J. Cheetham, M. Sudol, and T. Russo. 1997. Interaction of the phosphotyrosine interaction/phosphotyrosine binding-related domains of Fe65 with wild-type and mutant Alzheimer's β-amyloid precursor proteins. J. Biol. Chem. 272:6399-6405. [DOI] [PubMed] [Google Scholar]
- 81.Zhang, X. A., and M. E. Hemler. 1999. Interaction of the integrin β1 cytoplasmic domain with ICAP-1 protein. J. Biol. Chem. 274:11-19. [DOI] [PubMed] [Google Scholar]
- 82.Zhang, Z., C. H. Lee, V. Mandiyan, J. P. Borg, B. Margolis, J. Schlessinger, and J. Kuriyan. 1997. Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain. EMBO J. 16:6141-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou, M. M., K. S. Ravichandran, E. F. Olejniczak, A. M. Petros, R. P. Meadows, M. Sattler, J. E. Harlan, W. S. Wade, S. J. Burakoff, and S. W. Fesik. 1995. Structure and ligand recognition of the phosphotyrosine binding domain of Shc. Nature 378:584-592. [DOI] [PubMed] [Google Scholar]
- 84.Zwahlen, C., S. C. Li, L. E. Kay, T. Pawson, and J. D. Forman-Kay. 2000. Multiple modes of peptide recognition by the PTB domain of the cell fate determinant Numb. EMBO J. 19:1505-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.