Abstract
In the compact Dictyostelium discoideum genome, non-long terminal repeat (non-LTR) retrotransposons known as TREs avoid accidental integration-mediated gene disruption by targeting the vicinity of tRNA genes. In this study we provide the first evidence that proteins of a non-LTR retrotransposon interact with a target-specific transcription factor to direct its integration. We applied an in vivo selection system that allows for the isolation of natural TRE5-A integrations into a known genomic location upstream of tRNA genes. TRE5-A frequently modified the integration site in a way characteristic of other non-LTR retrotransposons by adding nontemplated extra nucleotides and generating small and extended target site deletions. Mutations within the B-box promoter of the targeted tRNA genes interfered with both the in vitro binding of RNA polymerase III transcription factor TFIIIC and the ability of TRE5-A to target these genes. An isolated B box was sufficient to enhance TRE5-A integration in the absence of a surrounding tRNA gene. The RNA polymerase III-transcribed ribosomal 5S gene recruits TFIIIC in a B-box-independent manner, yet it was readily targeted by TRE5-A in our assay. These results suggest a direct role of an RNA polymerase III transcription factor in the targeting process.
Retrotransposons are mobile genetic entities that amplify within a host cell genome by reverse transcription of RNA intermediates (9). Integration of a cDNA copy into a new site of the host cell genome is a default event in the amplification of a retrotransposon. In compact genomes retrotransposon integrations can hardly occur without the risk of deleterious gene disruptions. The yeast Saccharomyces cerevisiae and the social amoeba Dictyostelium discoideum have similarly high gene densities of ca. 70% and 65%, respectively (11, 20, 22). As a consequence, some retrotransposable elements in both S. cerevisiae and D. discoideum have evolved mechanisms to actively avoid integration into genes by targeting to noncoding regions. An intriguing example is integration in the vicinity of genes transcribed by RNA polymerase III (pol III), notably tRNA genes. Mobile elements that specifically target tRNA genes are found in both classes of retrotransposons, namely, the well-characterized long terminal repeat (LTR) retrotransposons Ty1 and Ty3 in yeast (3, 32) and the non-LTR retrotransposon TRE5-A (formerly known as DRE) in D. discoideum (reviewed in reference 39).
All non-LTR retrotransposons in the genome of D. discoideum belong to a monophyletic family known as “tRNA gene-targeted retrotransposable elements” (TREs). There are two subgroups within the TRE family. The TRE5 elements insert exclusively 48 ± 3 bp upstream of the first coding nucleotide of the targeted tRNA gene. All as-yet-analyzed chromosomal TRE5 insertions occurred in an orientation-specific manner, with the 5′ ends of the retrotransposons facing the targeted tRNA genes (2). On the other hand, TRE3 elements are found exclusively in a region ∼100 bp downstream of tRNA genes (37). The tRNA genes in the D. discoideum genome do not share conserved flanking sequences, suggesting that integration site selection is not facilitated by direct binding of TRE-encoded proteins to DNA sequences flanking tRNA genes.
Besides tRNA genes, pol III transcribes other small, untranslated RNAs including the ribosomal 5S RNA and U6 small nuclear RNA (16). Three distinct promoter types of pol III-transcribed genes can be recognized. Type 1 (ribosomal 5S) genes and type 2 (tRNA) genes both have gene-internal control regions recognized by pol III-specific transcription factors. Transcription factor IIIC (TFIIIC) binds to a promoter element of type 2 genes known as the B box. Type 1 genes contain a C box instead of the B box and depend on an additional factor, TFIIIA, for transcription initiation. TFIIIA binds to the C box and then recruits TFIIIC. DNA-anchored TFIIIC mediates the binding of TFIIIB near the transcription start of type 1 and type 2 genes, which in turn recruits pol III to start transcription. Type 3 pol III genes (e.g., U6 snRNA) lack internal promoter elements but have upstream regulatory promoter and enhancer elements such as a TATA box (17, 33).
To date, the in vitro study of tRNA gene-targeted retrotransposition in D. discoideum using PCR-based plasmid assays and cell extracts is impeded by the fact that pol III transcription complexes probably required for targeted integration of TREs are irreversibly disrupted during extract preparation. Furthermore, we have not yet succeeded in cloning an autonomous TRE that would allow for the analysis of retrotransposition of tagged versions of the element, a strategy that is very successfully applied to study retrotransposable elements in mammalian cells (12, 24, 29). Taking advantage of the strong preference of TRE5-A to integrate at tRNA gene loci, we have recently established an in vivo selection system that allows for the isolation of new integrations of naturally active TRE5-A copies from a population of D. discoideum cells (2). The “TRE trap” is a plasmid-borne pyr5-6 gene, which encodes UMP synthase, tagged with an intron carrying a D. discoideum tRNA gene (see Fig. 1). When the TRE trap plasmid is stably inserted into a D. discoideum uracil-auxotrophic strain, the plasmid-borne pyr5-6 gene is transcribed and spliced and the cells are converted to uracil prototrophy. These cells are sensitive to the cytostatic drug 5-fluoroorotic acid (5-FOA). If the pyr5-6 gene is disrupted by mutation, e.g., by targeted integration of a TRE near the “bait” tRNA gene within the TRE trap, then the affected cells gain resistance to 5-FOA and grow out as clones (2).
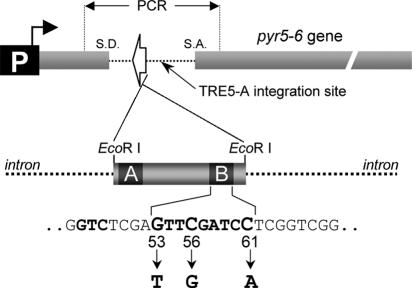
FIG. 1.
Setup of the TRE trap. The D. discoideum gene encoding UMP synthase (pyr5-6) is equipped with an intron derived from the D. discoideum cbfA gene (2). The intron sequence is indicated as a dashed line. The white arrow indicates the transcription orientation of the pol III gene inserted into the intron. All tested pol III genes were inserted into the trap as EcoRI fragments, schematically exemplified by D. discoideum ValUAC. TRE5-A integrations into the trap were isolated by PCR using pyr5-6 exon-specific primers as indicated. The tRNA gene-internal A-box and B-box promoter elements are indicated. Mutations introduced into the consensus GTCnnnnG53TTC56RANYC61 B-box motif of the D. discoideum ValUAC tRNA gene are indicated.
In this study we used the TRE trap assay to determine DNA sequences that support target site recognition by TRE5-A. The data suggest that active pol III genes are landmarks for TRE5-A integration and support a model in which pol III-specific transcription factors are involved in the targeting process by providing direct protein contacts for interaction with TRE5-A-derived proteins.
MATERIALS AND METHODS
Plasmids.
Plasmid pGEM-pyr5-6(cbfA/ValUAC) contains the pyr5-6 gene of D. discoideum including its own promoter and terminator. The pyr5-6 gene is tagged with the intron of the D. discoideum cbfA gene (2). This reporter gene can be equipped with any suitable pol III gene to attract TREs for integration (referred to as the “TRE trap”). The ValUAC tRNA gene used in this and our previous study (2) consists only of the coding sequence (74 bp), flanked by EcoRI sites. B-box mutations were introduced into the ValUAC gene in plasmid pGEM-pyr5-6(cbfA/ValUAC) by oligonucleotide-directed mutagenesis to generate the following mutants: ValUAC(G53T), ValUAC(C56G), and ValUAC(C61A).
A Glusup tRNA gene (10) was amplified by PCR from plasmid pGTET + 1 (40) using primers Glu-01 (5′-GGAATTCTCCTCATTGGTGTAGTCGGTAACAC-3′) and Glu-02 (5′-GGAATTCTAATTTTGGTCGGAATAAAAACCTCC-3′). The resulting PCR fragment was inserted as an EcoRI fragment into the TRE trap. Note that this tRNA gene contains 22 bp of original downstream flanking sequence including a functional pol III terminator sequence. B-box derivatives of the Glusup tRNA gene were generated by site-directed mutagenesis to achieve the following mutants: Glusup(G53T), Glusup(C56G), and Glusup(C61A).
The D. discoideum ribosomal 5S gene (r5S) was amplified from vector pUd5S (26) by PCR using primers r5S-01 (5′-GGGAATTCGTATACGGCCATACTAGGTTG-3′) and r5S-02 (5′-GGGAATTCAAAAAATAAATAAAGTATACAGCACCC-3′). A human MetCAU tRNA gene was amplified by PCR using plasmid pXlt1met (kindly provided by W. Meißner, Universität Marburg) (38) as template and primers hMet-01 (5′-GGGGAATTCAGCAGAGTGGCGCAGCGG-3′) and hMet-02 (5′-GGGGAATTCAAAAAAAAAAAGGACCTAGC-3′). Both the human MetCAU tRNA gene and the D. discoideum r5S gene were inserted as EcoRI fragments into the TRE trap plasmid.
D. discoideum cell culture and TRE trap assay.
The detailed protocol of the selection procedure to isolate 5-FOA-resistant cells is given in the work of Beck et al. (2). Briefly, TRE trap plasmids carrying the individual pol III genes were transformed into D. discoideum DH1 cells by electroporation. Ura+ strains were recovered after selection of transformed cells in FM medium in the absence of uracil. In order to isolate new TRE5 insertions, 1 × 106 to 1 × 107 Ura+ cells transformed with the individual TRE trap plasmids were cultured in 5-FOA (150 μg/ml for 3 days, followed by 100 μg/ml) and uracil (20 μg/ml) until clones became visible. Clones from up to 15 petri dishes were counted, and the values presented normalized for 1 × 106 cells ± standard deviation (SD) (or percentage of control ± SD with control being the respective wild-type pol III genes). Distances of TRE5-A elements integrated upstream of the bait pol III genes were determined by direct DNA sequencing of PCR products or sequencing of subcloned PCR fragments as previously described for the D. discoideum ValUAC tRNA gene (2).
Suppressor tRNA gene activity.
D. discoideum vector pA15-Gal (kindly provided by J. Williams, University of Dundee) allows for the expression of Escherichia coli β-galactosidase under the control of the constitutive D. discoideum act15 promoter (23). The β-galactosidase-encoding gene lacZ was modified by oligonucleotide-directed mutagenesis with an amber (TAG) translation stop codon at amino acid position 18 of the β-galactosidase protein. The resulting plasmid, pA15-Gal(amber), was transformed into D. discoideum AX2 cells together with the plasmids carrying the TRE trap equipped with the individual Glusup tRNA gene mutants. Stable transformants were selected in the presence of 10 μg/ml G418. Of these clones 2 × 107 logarithmically growing cells were pelleted and stored at −80°C for further use. β-Galactosidase activity of 107 cells was measured using chlorophenol red-β-d-galactoside (CPRG) as described previously (34). One katal is defined as the enzyme activity that hydrolyzes 1 mol of CPRG per second at 22°C.
Nuclear extracts and EMSAs.
Nuclei of D. discoideum cells were isolated and extracted with 600 mM KCl as described previously (5). The extracts are referred to as NE600. Electrophoretic mobility shift assays (EMSAs) were performed as detailed elsewhere (5). The Glusup derivatives were prepared from the pGEM-pyr5-6(cbfA/Glusup) derivatives described above. EcoRI fragments were labeled by fill-in with Klenow polymerase in the presence of [α-32P]dATP. Each incubation mixture contained 1 μg of each of the nonspecific competitors poly(dAdT) · poly(dAdT) (Sigma no. 0883) and pGEM7Zf(−) vector. About 0.5 μg NE600 proteins was preincubated for 30 min at ambient temperature with ∼10,000 cpm of 32P-labeled tRNA genes. Free 32P-labeled probe was separated from DNA-protein complexes by polyacrylamide gel electrophoresis (5).
RESULTS
A tRNA gene embedded in the TRE trap is actively transcribed.
The TRE trap is illustrated in Fig. 1. In a previous study, we used the D. discoideum ValUAC tRNA gene as a bait to isolate de novo integrations of active TRE5-A elements from a population of D. discoideum cells (2). In this configuration the bait tRNA gene is in an unusual environment since a pol III gene was embedded in a pol II gene. Our hypothesis is that TRE5-A-encoded proteins determine integration sites by directly interacting with unique transcription factors required to initiate tRNA gene transcription. If this model holds true, only actively transcribed tRNA genes should be targets for TRE5-A. It has been noted that proper transcription of adjacent pol II and pol III genes may require a minimum distance to avoid unfavorable interference of the transcription machineries (4). Thus, it was desirable to test whether a bait tRNA gene within the TRE trap would be bound by pol III transcription factors and actively transcribed. A method to test for tRNA gene activity in D. discoideum cells in vivo is the use of translation stop codon suppression (10). We modified a lacZ reporter gene with an amber (TAG) translation stop codon at amino acid position 18 of β-galactosidase. The resulting lacZ(Am) gene was expressed under the control of a strong D. discoideum promoter and placed on a plasmid that conferred G418 resistance to D. discoideum cells. We cloned a suppressor tRNA gene (Glusup) (10) into the TRE trap and cotransformed vectors containing either the wild-type lacZ gene or the lacZ(Am) gene together with the TRE trap plasmid into D. discoideum cells. Stably transformed D. discoideum cells were isolated by selection in G418, and expression of β-galactosidase activity from the lacZ(Am) gene was taken as a measure of the activity of the suppressor tRNA gene expressed in the same cells from the TRE trap. Cells transformed with the wild-type lacZ gene expressed 1,323.5 ± 464.2 pkat/108 cells (n = 10). Cells transformed with the lacZ(Am) reporter gene alone had background β-galactosidase activity (0.7 ± 0.1 pkat/108 cells), whereas cells cotransformed with the lacZ(Am) reporter gene and the Glusup TRE trap plasmid showed a β-galactosidase activity of 16.2 ± 5.9 pkat/108 cells. This observed suppression rate of 1.2% was in good agreement with previous data obtained with a slightly different lacZ(Am) reporter gene (10) and suggested that the tRNA gene within the TRE trap was accessible for the pol III transcription machinery.
The TRE trap assay picks up authentic TRE5-A integrations.
We use the term “empty trap” if the intron within the pyr5-6 gene does not contain a bait tRNA gene, and we interpret clones obtained with the empty trap as being due to natural mutations (data not shown). Specific targeting of a tRNA gene embedded in the reporter gene is indicated by increased clone numbers obtained after selection (Fig. 2A). In typical experiments no clones were recovered from the empty trap when 1.0 × 106 to 1.5 × 106 D. discoideum cells were used to carry out a 5-FOA selection. By contrast, >50 clones were usually obtained when the trap was “loaded” with the D. discoideum ValUAC tRNA gene (Fig. 2A). Integrated retrotransposons were studied after performing PCRs with primers specific for exons 1 and 2 of the pyr5-6 gene (Fig. 1).
FIG. 2.
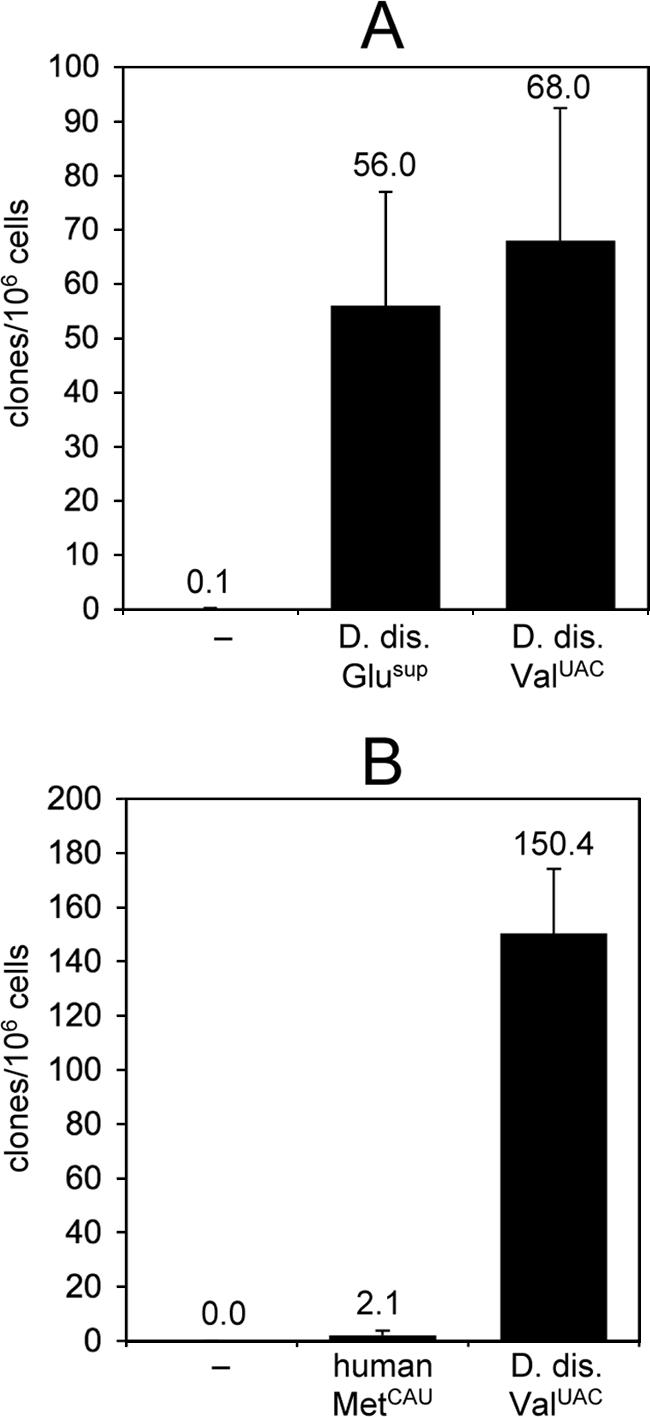
Results from TRE trap assays. (A) In this experiment 4 × 106 D. discoideum cells carrying the empty trap (−), a D. discoideum Glusup gene, or a D. discoideum ValUAC gene were subjected to selection in 5-FOA and uracil. (B) Comparison of targeting frequencies at a human MetCAU gene and a D. discoideum ValUAC gene. D. discoideum cells (5 × 106 of each strain) were used for 5-FOA selection. Mean clone numbers from 10 petri dishes are shown ± SD.
To further demonstrate the specificity and the sensitivity of the assay, we tested two other bait tRNA genes, a D. discoideum Glusup tRNA gene and a human MetCAU tRNA gene. The former was used to show that any D. discoideum tRNA gene may serve as a landmark for integration of TREs in our assay, whereas the latter was used to test whether the TRE trap would allow for the isolation of specific targeting events near a heterologous tRNA gene. Integration frequencies upstream of the D. discoideum Glusup bait were slightly lower than those obtained with the D. discoideum ValUAC tRNA gene (Fig. 2A). As shown in Fig. 2B, the human MetCAU tRNA gene was targeted at very low frequency, but clone numbers were significantly above background (empty trap) and most 5-FOA-resistant clones had bona fide integrations of TRE5-A in the 5′-flanking region of the MetCAU bait (Fig. 3A; discussed below). Most TRE5-A copies integrated upstream of the Glusup and MetCAU tRNA gene showed extended 5′ deletions, a known characteristic property of non-LTR retrotransposons.
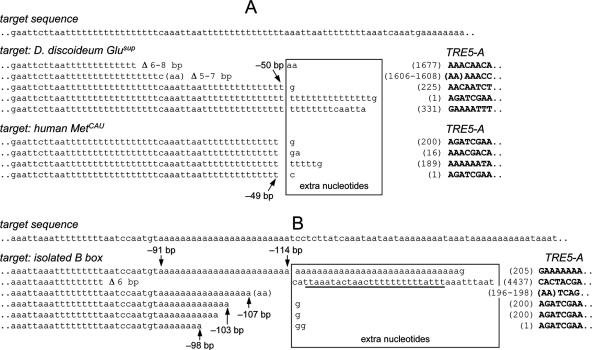
FIG. 3.
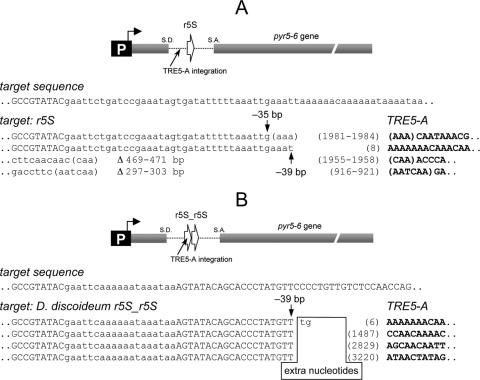
5′ junctions of de novo integrations of TRE5-A into the TRE trap. (A) Targeting of the D. discoideum Glusup tRNA gene and the human MetCAU gene. The target sequence is written in the first line in lowercase letters. Vertical arrows point to the integration sites of TRE5-A elements; the numbers indicate the distance of the TRE5-A to the first nucleotide of the targeted tRNA gene. The numbers in parentheses indicate the first nucleotides of the inserted TRE5-As, which are written in bold uppercase letters. Extra nucleotides are boxed, and target site deletions are indicated as “Δ.” (B) Integrations of TRE5-A upstream of an isolated B box. The target sequence is shown in lowercase letters. The underlined sequence of 26 bp represents duplication of the intron sequence located 120 bp downstream of the integration site. Note that all integrations shown were isolated from different selection plates and represent independent integration events by definition, even if the integrated elements and integration sites look very similar. Note that the 5′ end of a full-length TRE5-A consists of a 271-bp A module which is composed of a 199-bp core sequence and 72 bp identical with the 5′ end of the core. Thus, elements whose 5′ ends are designated “(1)” have full-length A modules (199 + 72 bp), whereas elements labeled “(200)” contain only the 72-bp repeat.
TRE5-A integrations cause characteristic alterations at the target site.
Several examples of TRE5-A integrations upstream of the D. discoideum Glusup and the human MetCAU tRNA gene are depicted in Fig. 3A. All integrated TRE5-A elements had the expected orientation relative to the targeted tRNA genes, and integrations occurred at positions 49 to 50 bp upstream of the bait tRNA genes. In most cases, integrations of the TRE5-A elements caused the addition of nontemplated extra nucleotides of 1 to 40 bp in length. Noteworthy is the frequent expansion of homopolymeric thymidine or adenine stretches into which the element had integrated. This characteristic property of TRE5-A cannot be attributed to the unusual genomic sequence of the integration target in the TRE trap, since the D. discoideum genome is generally very enriched in A/T nucleotides (11) and long homopolymeric nucleotide runs can occur at any natural target site as well. Several de novo integrations of TRE5-A were analyzed for the presence of target site duplications (TSDs). TRE5-As were flanked by TSDs of 11 to 19 bp in length (data not shown) in all analyzed cases except for those integration events that apparently occurred at shorter distances from the tRNA gene target than average. These integration events have caused small deletions of target sequence (indicated as “Δ” in Fig. 3) that seemed to be guided by microhomologies between the upstream target sequence and the 5′-truncated TRE5-A copies (Fig. 3). Due to the fact that target site deletions also eliminate the 5′ TSD sequence, we could not assign exact integration sites in these cases, but assuming a ca. 15-bp TSD, integration of these TRE5-A elements was in the range of 50 bp upstream of the tRNA gene target. It should be noted that templated and nontemplated extra nucleotides as well as microhomology-mediated target site deletions are known integration characteristics of other non LTR-retrotransposons including human L1 (18, 19, 36).
TRE5-A targets an isolated B box.
The data described above supported the hypothesis that a DNA sequence within a tRNA gene mediates target site recognition by TRE5-A. In support of this view, the human MetCAU tRNA gene, which is readily targeted by TRE5-A, has no similarities with D. discoideum tRNA genes except for the highly conserved gene-internal A box and B box. These sequences are part of the tRNA gene promoter and contribute to the binding site of pol III transcription factor TFIIIC. We developed a consensus sequence of a D. discoideum tRNA gene-internal B box based on the DNA sequences of 390 tRNA genes of the D. discoideum genome (11). The derived B-box consensus GTCNNNNG53TTCRANYC closely matches B-box sequences from the tRNA genes of other eukaryotes. The presence of “extra” B (exB) boxes located downstream of D. discoideum tRNA genes has been previously noted (25). We inspected the complete list of D. discoideum tRNA genes and confirmed the presence of tRNA gene-associated exB boxes that are 41 bp on average (ranging from 33 to 60 bp; n = 261) downstream of the gene-internal B boxes of 66% of all D. discoideum tRNA genes.
We tested whether an isolated B box can mediate TRE5-A integration in the absence of a surrounding tRNA gene sequence. For this purpose, we inserted the B-box sequence GTCGCAGGTTCGAATC either into the empty trap or downstream of the ValUAC tRNA gene. The number of clones obtained with the ValUAC/exB target was similar to that of the ValUAC tRNA gene alone (Fig. 4, columns 1 and 2), suggesting that a downstream B box does not influence targeting of an intact tRNA gene.
FIG. 4.
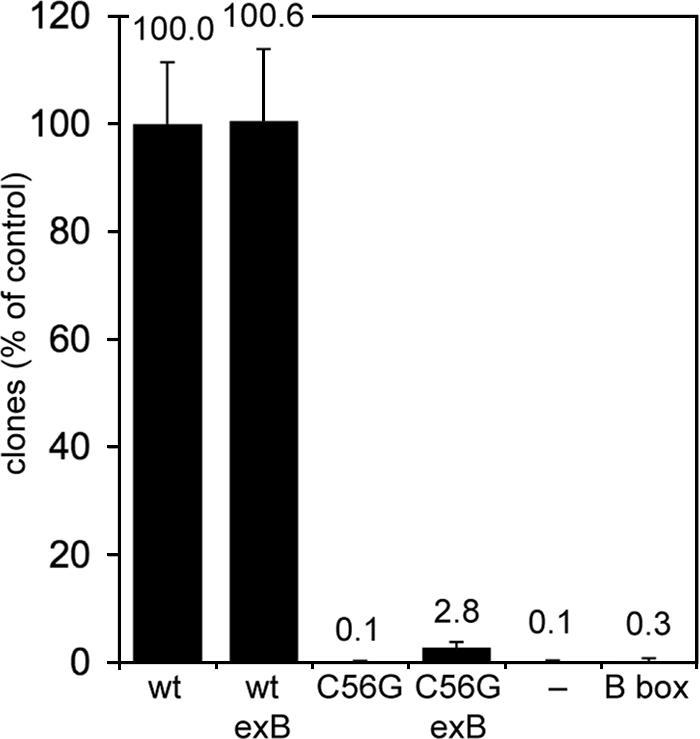
Frequencies of TRE integrations upstream of an isolated B box. The TRE trap assay was performed with the ValUAC gene (wt), the mutant ValUAC(C56G) gene, and the empty trap (−). A B box was inserted 34 bp downstream of the wild-type and mutant tRNA gene (referred to as the exB box) and at the corresponding position in the intron of the empty trap. Clone numbers from 10 petri dishes of two independent clones were counted after 5-FOA selection and normalized for the wild-type ValUAC gene without the exB box and presented ± SDs.
When the B box was used as bait in the absence of a tRNA gene, clone numbers dropped to background levels, which at first glance indicated that an isolated B box could not mediate TRE5-A integration (Fig. 4, columns 5 and 6). The difference in clone numbers of 0.3 (B-box target) versus 0.1 (empty trap) clones per 106 cells seemed not to be significant (Fig. 4). Hence, it was surprising to find that some clones obtained after 5-FOA selection of cells containing the isolated B box were in fact due to targeted integration of TRE5-A into the trap (Fig. 3B). However, these clones were hidden in the background of clones that obtained 5-FOA resistance due to natural mutations (25 of 70 analyzed clones contained TRE5-A insertions). It should be noted at this point that PCR and Southern blot analyses of clones isolated with the empty trap did not show any sign of TRE insertions (40 clones analyzed; data not shown), which further demonstrates the efficiency of the TRE trap in picking up TRE integration if a proper target is provided. B-box targeting by TRE5-A was finally proven by analyzing the 5′ junctions of some inserted TRE5-A copies (Fig. 3B). All integrations occurred into a 24-mer polyadenine run in the TRE trap. Since the 3′ end of TRE5-A is also a poly(A) tail, we could not exactly determine the integration sites. Occasionally integration sites could not be accurately determined due to extension of a poly(A) tract at the target site. In one example shown in Fig. 3B the poly(A) run was extended by 30 adenines and one guanine, the integration site being located somewhere between 91 and 114 bp upstream of the B box (Fig. 3B). Four other de novo integrations were more informative due to the characteristic insertion of single guanine nucleotides that separate the target sequence from the 5′ ends of integrated TRE5-A elements. These elements had integrated between 98 and 107 bp upstream of the B-box target (Fig. 3B). One additional integration resulted in a 6-bp target site deletion (Fig. 3B). Although we did not determine the TSD for this element, we can calculate that this element integrated between 97 and 106 bp upstream of the B box (86 bp plus TSD range of 11 of 19 bp; Fig. 3B). Interestingly, the average distance of the favorite TRE5-A integration site 48 bp upstream from a canonical tRNA gene reflects integration of TRE5-A at about 100 bp upstream of a tRNA gene-internal B box. Thus, it is evident that targeting of tRNA gene-internal B boxes and that of isolated B boxes occur at very similar distances by the same targeting mechanism.
B-box mutations in tRNA genes affect targeting by TREs.
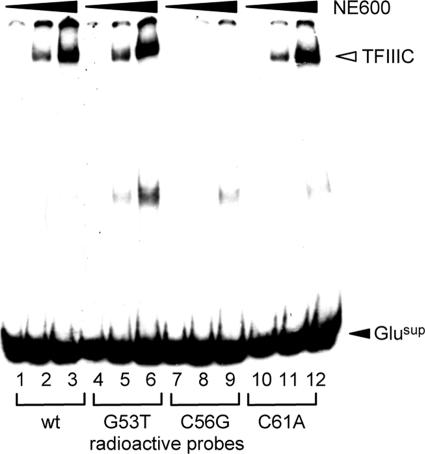
Based on the literature (15, 16, 30) and on the consensus sequence of D. discoideum tRNA gene B boxes, we generated two B-box mutations in the D. discoideum ValUAC and Glusup tRNA genes (G53T and C56G) predicted to interfere with binding to TFIIIC in vitro and in vivo and one control mutation that was predicted to have only a limited effect (C61A). We first tested whether the mutant tRNA genes were bound by TFIIIC in vitro. For this purpose we applied an EMSA that was previously used to characterize D. discoideum TFIIIC in highly purified fractions (5). The Glusup tRNA gene was isolated from the TRE trap plasmid, and binding of TFIIIC was analyzed in EMSAs (Fig. 5). TFIIIC recognized the wild-type Glusup gene as well as the Glusup(G53T) and Glusup(C61A) mutant tRNA genes (Fig. 5, lanes 4 to 6 and 10 to 12). In contrast, no complexes of TFIIIC with the Glusup(C56G) tRNA gene could be detected (Fig. 5, lanes 7 to 9). Similar results were obtained with the ValUAC tRNA gene mutants (data not shown). Mutant analyses of the two D. discoideum tRNA genes, taken together, showed that position C56 of D. discoideum tRNA genes was most critical for the interaction with TFIIIC in vitro. The data predicted that C56G tRNA gene mutants should no longer be targets of TRE integrations if TFIIIC was in fact involved in target site selection.
FIG. 5.
In vitro binding of TFIIIC to mutant tRNA genes. The figure shows results of EMSAs with the D. discoideum Glusup tDNA derivatives as radiolabeled probe. Radiolabeled probes were wild-type Glusup (lanes 1 to 3), Glusup(G53T) (lanes 4 to 6), Glusup(C56G) (lanes 7 to 9), and Glusup(C61A) (lanes 10 to 12). One microgram of plasmid carrying the empty trap was used as competitor. DNA-TFIIIC complexes are indicated by the white arrowhead. Three increasing amounts (0.1 μg, 0.5 μg, and 1 μg) of NE600 fraction were used as a source of TFIIIC.
We next tested whether the B-box mutations affected the targeting of the mutant tRNA genes by TREs in vivo. ValUAC and Glusup tRNA mutant genes were placed into the TRE trap, and cells were selected in the presence of 5-FOA and uracil. As shown in Fig. 6, the C56G mutation of both the ValUAC and Glusup tRNA genes completely abolished targeting by TREs, as the clone numbers obtained with this tRNA gene mutant dropped to background and PCR analysis of 26 5-FOA-resistant clones derived from the Glusup(C56G) and ValUAC(C56G) tRNA gene targets did not reveal any signs of TRE integrations (data not shown). When we performed 5-FOA selections of cells transformed with the TRE trap containing the G53T and C61A mutations of the bait tRNA genes, clone numbers were well above background, albeit drastically reduced compared to the wild-type tRNA genes (Fig. 6).
FIG. 6.
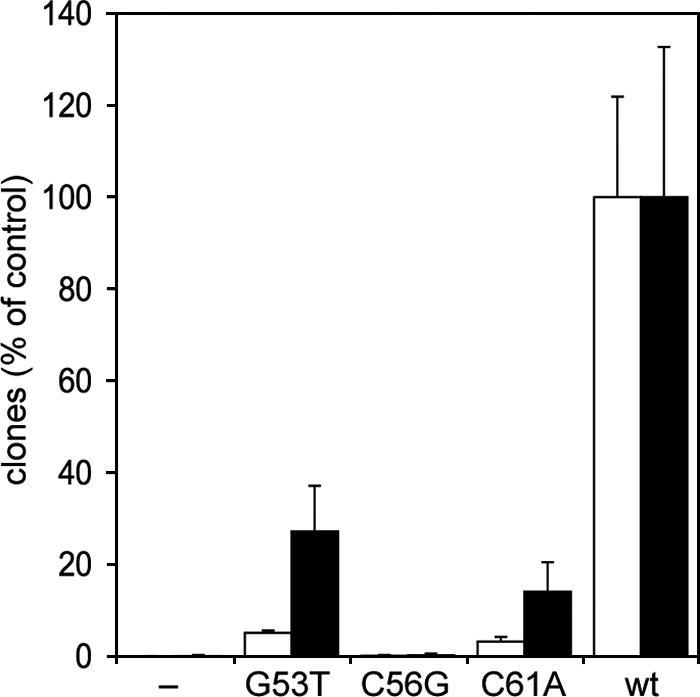
TRE integration frequencies at mutant tRNA genes. The figure shows the results of the TRE trap assay with ValUAC (white bars) and Glusup (black bars) as bait tRNA genes. Mutant tRNA genes (as indicated), empty TRE trap (−), and wild-type tRNA genes were tested. Clone numbers from 10 petri dishes of two independent clones of each strain are normalized for the wild-type (wt) tRNA genes and expressed ± SDs.
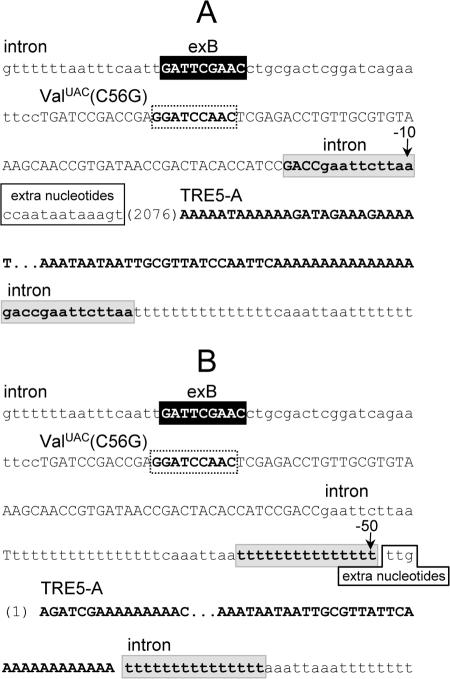
As mentioned above, we have not been able to show integrations of TREs near the putatively inactive Glusup(C56G) or ValUAC(C56G) tRNA genes. Since an isolated B box was a target for TRE integrations, we next tested whether a B box located downstream of an inactive ValUAC(C56G) tRNA gene would restore targeting activity by TRE5-A. In the experiments presented in Fig. 4 and Fig. 7, a B box was placed 34 bp downstream of the internal B boxes of the wild-type ValUAC gene and the mutant ValUAC(C56G) tRNA gene, respectively. We refer to this tRNA gene-external B box as the “exB box” in Fig. 7. D. discoideum cells transformed with a TRE trap equipped with the ValUAC(C56G)/exB derivative were cultured in 5-FOA and uracil until clones became visible. As expected from previous experiments, the C56G mutation reduced targeting of the ValUAC tRNA gene to background level (Fig. 4, column 3). Combination of the targeting-defective tRNA gene with a downstream B box increased clone numbers moderately (Fig. 4, column 4), suggesting that an exB box could rescue the “dead” tRNA gene with low efficiency. Some clones analyzed by PCR revealed integration of TRE5-A into the ValUAC(C56G)/exB target. Interestingly, however, DNA sequencing of the obtained PCR fragments yielded two different results. Firstly, TRE5-A integrated into the trap by targeting the downstream B box, while ignoring the inactive tRNA gene. In this case, shown in Fig. 7A, TRE5-A was found inserted in a position 10 bp upstream of the ValUAC(C56G) gene, which corresponds to 106 bp upstream of the exB box. Thus, the inserted TRE5-A ignored the presence of the inactive tRNA gene and specifically targeted the functional exB box. Secondly, TRE5-A integrated exactly at position −50 relative to the ValUAC(C56G) gene (Fig. 7B), suggesting that the presence of the downstream B box somehow restored the function of the ValUAC(C56G) gene well enough to allow integration of TRE5-A in the normal position.
FIG. 7.
Examples of TRE5-A integrations upstream of an inactivated tRNA gene equipped with a downstream extra B box. DNA sequences of representative TRE5-A insertions at positions 10 bp (A) and 50 bp (B) upstream of an inactive ValUAC(C56G) are shown. The intron sequence is shown in lowercase letters. The exB box is presented in uppercase letters inside the black box; the mutated B box of the tRNA gene is boxed with a dashed line. The tRNA gene sequence is shown in uppercase letters. The insertion site is indicated by the vertical arrow. The DNA sequences of the inserted TRE5-As are shown in bold uppercase letters. The first nucleotides of the 5′-deleted TRE5-As are shown in parentheses and TSDs in gray boxes.
TRE5-A integrates upstream of a ribosomal 5S gene.
It was of interest to determine whether the r5S gene, which recruits TFIIIC by a B-box-independent mechanism involving TFIIIA, may be a target for TRE integrations. The D. discoideum r5S gene is located in two copies on an extrachromosomal ribosomal DNA (rDNA) palindrome that accumulates to about 100 copies per cell (35). Despite the large number of putative r5S targets, in silico analysis of the D. discoideum genome has not revealed integrations of TRE5-A or other TREs at natural r5S loci. To test whether an r5S gene mediates targeted integration of TRE5-A under the experimental conditions of our TRE trap assay, we PCR amplified an r5S gene and cloned it into the TRE trap. After 5-FOA selection of cells transformed with the r5S target, we readily obtained resistant clones at frequencies of about 10% of the ValUAC tRNA gene target (Fig. 8). PCR analysis indicated that integrations of TREs near the r5S gene had occurred in most cases (data not shown). DNA sequencing of some of these clones confirmed integrations of TRE5-A elements upstream of the r5S gene in the same orientation as if targeting tRNA genes (Fig. 9A). In one example shown in Fig. 9A a TRE5-A was inserted at position 35 or 38 upstream of the 5S gene; the exact position could not be determined due to microhomology-guided 5′ truncation of the element. A second example of a TRE5-A integration 39 bp upstream of the 5S target is also shown in Fig. 9A. In two other cases, microhomologies of 3 and 6 bp between genomic target sequence and the retrotransposon prompted large target site deletions of 297 to 471, thus leaving the inserted elements without TSDs (Fig. 9B). Worth mentioning is that we also used a TRE trap construct in which we accidentally cloned an r5S gene tandem (Fig. 9B). We analyzed four TRE5-A integrations into this target in detail. All de novo integrated elements had canonical TSDs (data not shown), and integration had occurred exactly at the same position 39 bp upstream of the second r5S target of the tandem, i.e., the elements inserted within the first r5S copy of the tandem. Yet the individual integrations could be distinguished by the presence of extra nucleotide additions and different extents of 5′ deletions of the inserted retroelements (Fig. 9B).
FIG. 8.
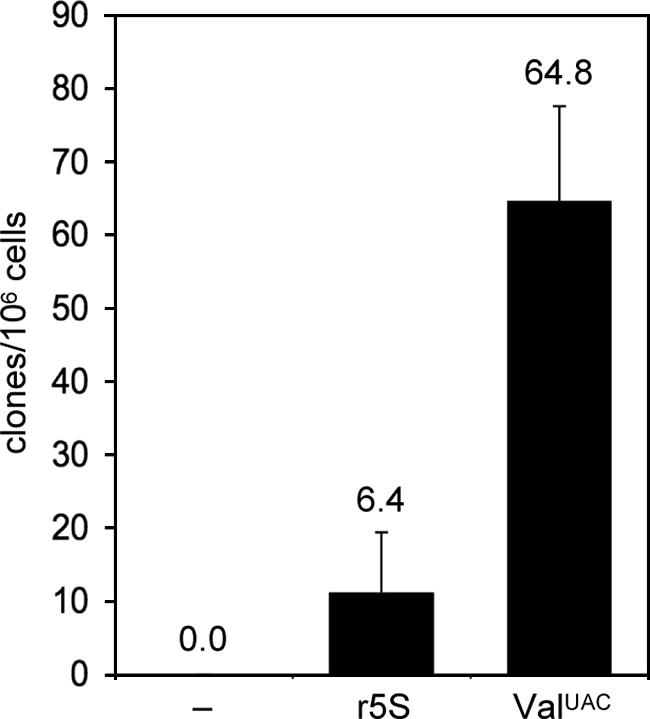
Results from a TRE trap assay using the D. discoideum ribosomal 5S gene as bait. The targeting frequency at the r5S gene is compared to the empty trap (−) and the trap loaded with a D. discoideum ValUAC gene. Mean clone numbers from 10 petri dishes are shown ± SDs.
FIG. 9.
5′ junctions of new TRE5-A integrations. (A) Targeting of the D. discoideum r5S gene. The target sequence is written in the first line in lowercase letters. The 5′ end of the r5S gene (reverse orientation) is shown in uppercase letters. The vertical arrow points at the integration site of a TRE5-A element; the numbers indicate the distance of the TRE5-A to the first nucleotide of the targeted r5S gene. The numbers in parentheses indicate the first nucleotides of the inserted TRE5-A, which are written in bold uppercase letters. Target site deletions are indicated as “Δ.” (B) Integrations of TRE5-A upstream of an r5S tandem. The first line indicates the target sequence, which consists of two r5S genes (uppercase) separated by an EcoRI restriction site and the transcription terminator sequence of the first r5S gene (lowercase). The first nucleotides of the inserted TRE5-As (bold uppercase) are written in parentheses; extra nucleotides are boxed.
DISCUSSION
Targeted integration of TRE5-A requires TFIIIC.
We are confident that TRE5-A integrations picked up with the TRE trap assay mimic authentic retrotransposition events that continuously shape the D. discoideum genome. Accumulative data using three different tRNA genes have shown that (i) TRE5-A integrations occur upstream of the bait tRNA genes at distances of about 48 bp, (ii) the TRE5-As insert without exceptions in the “correct” orientation relative to the targeted tRNA gene and the insertion sites of TRE5-As change according to the orientation of the bait tRNA gene in the trap (data not shown), (iii) integration produced TSDs or in some cases target site deletions of variable lengths, (iv) most integrated TRE5-As were 5′ deleted, (v) integrations were accompanied by the generation of templated and nontemplated extra nucleotides, and (vi) single extra guanine nucleotides at the 5′ junctions of full-length TRE5-As suggest the use of the 7-methylguanine cap as the last RNA template nucleotide. These properties (except for tRNA gene recognition) of the newly integrated TRE5-A elements and target site alterations are known properties of other non-LTR retrotransposons like L1 as well (18, 19, 36, 42), suggesting that TRE5-A is a bona fide non-LTR retrotransposon that amplifies via target-primed reverse transcription.
It has been debated whether TRE5-A recognizes tRNA genes by DNA-specific binding of a TRE5-A-derived protein to the tRNA gene itself or to conserved motifs in transfer tRNA-flanking regions. This possibility is ruled out by two considerations. Firstly, the ribosomal 5S gene lacks a B-box motif and recruits TFIIIC in a B-box-independent manner. Secondly, if target-printed reverse transcription is the underlying mechanism of TRE5-A integration, then initiation of reverse transcription is primed at the site of first-strand DNA cleavage. D. discoideum TRE5-A belongs to the group of non-LTR retrotransposons that contain apurinic/apyrimidinic endonucleases. These endonucleases are thought to bind to target DNA directly at the cleavage site (6, 7, 13, 14). Since we have observed that single nucleotide substitutions within the B box strongly interfere with TRE5-A integration, it seems inconceivable that binding of TRE5-A-derived endonuclease protein at the B box and subsequent cleavage of the genomic DNA there will lead to the integration of the retrotransposon at a site 100 bp upstream.
The TRE5-As isolated in this study targeted B boxes of canonical tRNA genes at about 100 bp upstream; very similar distances to isolated B boxes in the absence of a tRNA gene have been observed. Thus, B boxes provide important cis-acting sequences at the target sites of TRE5-A integrations. We calculated the distances of TRE5-A to the targeted tRNA genes by counting nucleotides between the retrotransposon's 5′ end and the first coding nucleotide of the targeted tRNA gene (48 ± 3 bp). However, we noticed that the distances of the tRNA gene-internal B boxes to the 5′ ends of the tRNA genes vary between 40 and 65 bp (O. Siol, unpublished observation). Hence, the quite precise integration upstream of tRNA genes is best explained by an active involvement of the A box, which is located at positions 8 to 19 bp of D. discoideum tRNA genes (T. Winckler and G. Glöckner, unpublished analysis of genome sequencing data). The putative involvement of the A box is in fact another strong argument to support the interpretation that tRNA genes occupied by TFIIIC are landmarks for integration.
Model of target site selection by TRE5-A.
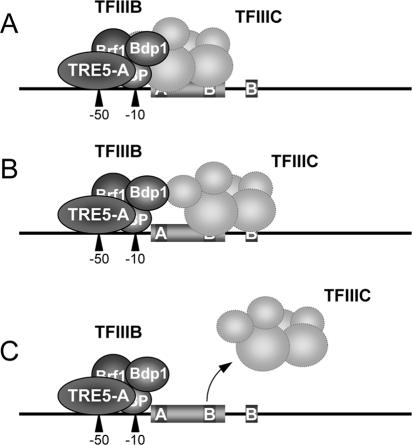
The question remains what function tRNA gene-external B boxes might have in vivo. We have consistently observed that an exB box is not sufficient to fully restore integration frequencies at a mutant tRNA gene whose internal B box has been inactivated (Fig. 4). Nevertheless, we have observed TRE5-A integrations into this target at low rates but at two alternative positions 50 and 10 bp upstream of the inactivated tRNA gene. We inspected many natural TRE5-A integrations in the D. discoideum genome, and we never found TRE5-A integrations in the −10 position relative to the tRNA gene that would indicate targeting of downstream exB boxes of intact tRNA genes. Thus, under natural conditions, the tRNA gene-internal B box and not the exB box is the primary target for integration of TRE5-A. In this respect TRE5-A differs from TRE3 elements, which favor two alternative insertion sites downstream of tRNA genes that can be attributed to the targeting of either the tRNA gene-internal B box or the exB box (37). This observation suggests that an exB box can be occupied by TFIIIC in vivo and that targeting of tRNA genes by TRE3s occurs via direct protein interactions with TFIIIC.
TFIIIC binds to the tRNA gene via the gene-internal B box and recruits TFIIIB to the 5′ flanking region of the tRNA gene to establish a stable preinitiation complex. Roberts et al. (31) have suggested that pol III may displace TFIIIC from its binding site during transcription elongation but leave TFIIIB in place. Reinitiation after the first round of transcription may not require TFIIIC. During tRNA gene transcription, D. discoideum TFIIIC may remain in a standby position by binding to the exB box in close proximity to the transcribed tRNA gene, whereas TFIIIB remains bound to the tRNA gene. We propose that TFIIIB and not TFIIIC is targeted by TRE5-A proteins. This model is favored by the principal agreement of distances of integration sites to the tRNA genes (∼48 bp) and the r5S gene (∼40 bp) and the extents of TFIIIB footprints on these genes (8, 27). In our model (Fig. 10), TFIIIC is required only to put TFIIIB in place, and integration would be feasible as long as TFIIIB stays bound to the DNA and pol III allows the TRE5-A preintegration complex to enter the tRNA gene. This model explains why we do not find natural integrations of TRE5-A in the −10 position, since this position would be permanently blocked by TFIIIB.
FIG. 10.
Model of tRNA gene recognition by TRE5-A proteins. A tRNA gene is indicated with an A box and a B box and a downstream exB box. D. discoideum TFIIIB likely consists of three subunits: TATA binding protein, Brf1, and Bdp1 (T. Winckler, unpublished observation). The exact subunit composition of D. discoideum TFIIIC is unknown. The TRE5-A preintegration complex, consisting of ORF1 and/or ORF2 proteins and TRE5-A RNA, is indicated as a single sphere. (A) TFIIIC binds to the tRNA gene-internal B box and recruits TFIIIB to the 5′ end of the tRNA gene. Integration of TRE5-A occurs via interaction with TFIIIB, which leaves the −50 position unprotected. (B) If TFIIIC slides to the exB box during transcription of the tRNA gene, TFIIIB stays at its position, still supporting integration of TRE5-A in the −50 position, while the −10 position is blocked by DNA-bound TFIIIB. (C) If TFIIIC dissociates from the tRNA gene, TFIIIB may stay and further support TRE5-A integration in the −50 position.
Targeting of pol III genes is a successful evolutionary strategy to avoid mutagenesis of compact genomes.
In this study, we have for the first time provided evidence that a non-LTR retrotransposable element uses protein interactions with chromatin components to select integration sites. In the D. discoideum genome, the targeting of tRNA genes has been invented several times by (i) the TRE5 elements, including TRE5-A discussed in this study; (ii) the TRE3s, close relatives of TRE5-A, which integrate approximately 100 bp downstream of the tRNA gene-internal B box or extra B boxes (37); and (iii) the poorly characterized putative LTR retrotransposon DGLT-A, which integrates at positions ∼30 bp upstream of tRNA genes (21).
Selection of integration sites by TRE5-A shows striking similarity with the pol III gene-targeted integration of yeast LTR retrotransposons. Ty3 elements integrate precisely at the transcription start of pol III genes, whereas Ty1 shows a preference to integrate within a window of about 700 bp upstream of tRNA genes (1, 3, 28, 32). For Ty3 it is known that targeting of pol III genes by Ty3 is mediated by specific protein interactions of the Ty3 preintegration complex with the pol III-specific transcription factor TFIIIB subunit(s) TATA binding protein and/or Brf (41). With reference to the model of tRNA gene targeting by TRE5-A described above, it is tempting to speculate that the strong selection pressure to avoid devastating insertional mutagenesis of compact genomes has led to convergent evolution of pol III gene targeting by diverse mobile genetic elements via interactions with protein factors that specifically tag chromosomal sites of pol III transcription.
Acknowledgments
This work was supported by grant WI 1142/5-4 from the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print on 18 September 2006.
REFERENCES
- 1.Bachmann, N., M. E. Gelbart, T. Tsukiyama, and J. D. Boeke. 2005. TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes Dev. 19:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck, P., T. Dingermann, and T. Winckler. 2002. Transfer RNA gene-targeted retrotransposition of Dictyostelium TRE5-A into a chromosomal UMP synthase gene trap. J. Mol. Biol. 318:273-285. [DOI] [PubMed] [Google Scholar]
- 3.Boeke, J. D., and S. E. Devine. 1998. Yeast retrotransposons: finding a nice quiet neighborhood. Cell 93:1087-1089. [DOI] [PubMed] [Google Scholar]
- 4.Bolton, E. C., and J. D. Boeke. 2003. Transcriptional interactions between yeast tRNA genes, flanking genes and Ty elements: a genomic point of view. Genome Res. 13:254-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukenberger, M., T. Dingermann, W. Meissner, K. H. Seifart, and T. Winckler. 1994. Isolation of transcription factor IIIC from Dictyostelium discoideum. Eur. J. Biochem. 220:839-846. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, S. M., and T. H. Eickbush. 2005. R2 target-primed reverse transcription: ordered cleavage and polymerization steps by protein subunits asymmetrically bound to the target DNA. Mol. Cell. Biol. 25:6617-6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cost, G. J., and J. D. Boeke. 1998. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry 37:18081-18093. [DOI] [PubMed] [Google Scholar]
- 8.Costanzo, G., S. Camier, P. Carlucci, L. Burderi, and R. Negri. 2001. RNA polymerase III transcription complexes on chromosomal 5S rRNA genes in vivo: TFIIIB occupancy and promoter opening. Mol. Cell. Biol. 21:3166-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craig, N. L., R. Craigie, M. Gellert, and A. M. Lambowitz (ed.). 2002. Mobile DNA II. ASM Press, Washington, D.C.
- 10.Dingermann, T., N. Reindl, T. Brechner, H. Werner, and K. Nerke. 1990. Nonsense suppression in Dictyostelium discoideum. Dev. Genet. 11:410-417. [DOI] [PubMed] [Google Scholar]
- 11.Eichinger, L., J. A. Pachebat, G. Glöckner, M.-A. Rajandream, R. Sucgang, M. Berriman, J. Song, R. Olsen, K. Szafranski, Q. Xu, B. Tunggal, S. Kummerfeld, M. Madera, B. A. Konfortov, F. Rivero, A. T. Bankier, R. Lehmann, N. Hamlin, R. Davies, P. Gaudet, P. Fey, K. Pilcher, G. Chen, D. Saunders, E. Sodergren, P. Davis, A. Kerhornou, X. Nie, N. Hall, C. Anjard, L. Hemphill, N. Bason, P. Farbrother, B. Desany, E. Just, T. Morio, R. Rost, C. Churcher, J. Cooper, S. Haydock, N. van Driessche, A. Cronin, I. Goodhead, D. Muzny, T. Mourier, A. Pain, M. Lu, D. Harper, R. Lindsay, H. Hauser, K. James, M. Quiles, M. Madan Babu, T. Saito, C. Buchrieser, A. Wardroper, M. Felder, M. Thangavelu, D. Johnson, A. Knights, H. Loulseged, K. Mungall, K. Oliver, C. Price, M. A. Quail, H. Urushihara, J. Hernandez, E. Rabbinowitsch, D. Steffen, M. Sanders, J. Ma, Y. Kohara, S. Sharp, M. Simmonds, S. Spiegler, A. Tivey, S. Sugano, B. White, D. Walker, J. Woodward, T. Winckler, Y. Tanaka, G. Shaulsky, M. Schleicher, G. Weinstock, A. Rosenthal, E. C. Cox, R. L. Chisholm, R. Gibbs, W. F. Loomis, M. Platzer, R. R. Kay, J. Williams, P. H. Dear, A. A. Noegel, B. Barrell, and A. Kuspa. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435:43-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esnault, C., J. Maestre, and T. Heidmann. 2000. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24:363-367. [DOI] [PubMed] [Google Scholar]
- 13.Feng, Q. H., J. V. Moran, H. H. Kazazian, and J. D. Boeke. 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87:905-916. [DOI] [PubMed] [Google Scholar]
- 14.Feng, Q. H., G. Schumann, and J. D. Boeke. 1998. Retrotransposon R1Bm endonuclease cleaves the target sequence. Proc. Natl. Acad. Sci. USA 95:2083-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaëta, B. A., S. J. Sharp, and T. S. Stewart. 1990. Saturation mutagenesis of the Drosophila tRNAArg gene B-box intragenic promoter element: requirements for transcription activation and stable complex formation. Nucleic Acids Res. 18:1541-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geiduschek, E., and G. Tocchini-Valentini. 1988. Transcription by RNA polymerase III. Annu. Rev. Biochem. 57:873-914. [DOI] [PubMed] [Google Scholar]
- 17.Geiduschek, E. P., and G. A. Kassavetis. 2001. The RNA polymerase III transcription apparatus. J. Mol. Biol. 310:1-26. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert, N., S. Lutz, T. A. Morrish, and J. V. Moran. 2005. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 25:7780-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert, N., S. Lutz-Prigge, and J. V. Moran. 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell 110:315-325. [DOI] [PubMed] [Google Scholar]
- 20.Glöckner, G., L. Eichinger, K. Szafranski, J. Pachebat, P. Dear, R. Lehmann, C. Baumgart, J. F. Abril, G. Parra, R. Guigó, K. Kumpf, B. Tunggal, E. Cox, M. A. Quail, M. Platzer, A. Rosenthal, A. A. Noegel, and the Dictyostelium Genome Sequencing Consortium. 2002. Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature 418:79-85. [DOI] [PubMed] [Google Scholar]
- 21.Glöckner, G., K. Szafranski, T. Winckler, T. Dingermann, M. Quail, E. Cox, L. Eichinger, A. A. Noegel, and A. Rosenthal. 2001. The complex repeats of Dictyostelium discoideum. Genome Res. 11:585-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldmann, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6000 genes. Science 274:546-567. [DOI] [PubMed] [Google Scholar]
- 23.Harwood, A. J., and L. Drury. 1990. New vectors for expression of the E. coli lacZ gene in Dictyostelium. Nucleic Acids Res. 18:4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heidmann, O., and T. Heidmann. 1991. Retrotransposition of a mouse IAP sequence tagged with an indicator gene. Cell 64:159-170. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann, J., G. Schumann, G. Borschet, R. Gosseringer, M. Bach, W. M. Bertling, R. Marschalek, and T. Dingermann. 1991. Transfer RNA genes from Dictyostelium discoideum are frequently associated with repetitive elements and contain consensus boxes in their 5′-flanking and 3′-flanking regions. J. Mol. Biol. 222:537-552. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann, J., T. Winckler, A. Hanenkamp, M. Bukenberger, G. Schumann, R. Marschalek, and T. Dingermann. 1993. The Dictyostelium discoideum 5S rDNA is organized in the same transcriptional orientation as the other rDNAs. Biochem. Biophys. Res. Commun. 191:558-564. [DOI] [PubMed] [Google Scholar]
- 27.Kassavetis, G. A., D. L. Riggs, R. Negri, L. H. Nguyen, and E. P. Geiduschek. 1989. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol. Cell. Biol. 9:2551-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, J. M., S. Vanguri, J. D. Boeke, A. Gabriel, and D. F. Voytas. 1998. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8:464-478. [DOI] [PubMed] [Google Scholar]
- 29.Moran, J. V., S. E. Holmes, T. P. Naas, R. J. DeBerardinis, J. D. Boeke, and H. H. Kazazian. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87:917-927. [DOI] [PubMed] [Google Scholar]
- 30.Nichols, M., J. Bell, M. S. Klekamp, A. Weil, and D. Söll. 1989. Multiple mutations of the first gene of a dimeric tRNA gene abolish in vitro tRNA gene transcription. J. Biol. Chem. 264:17084-17090. [PubMed] [Google Scholar]
- 31.Roberts, D. N., A. J. Stewart, J. T. Huff, and B. R. Cairns. 2003. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc. Natl. Acad. Sci. USA 100:14695-14700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandmeyer, S. B. 1998. Targeting retrotransposition: at home in the genome. Genome Res. 8:416-418. [DOI] [PubMed] [Google Scholar]
- 33.Schramm, L., and N. Hernandez. 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 16:2593-2620. [DOI] [PubMed] [Google Scholar]
- 34.Schumann, G., I. Zündorf, J. Hofmann, R. Marschalek, and T. Dingermann. 1994. Internally located and oppositely oriented polymerase II promoters direct convergent transcription of a LINE-like retroelement, the Dictyostelium repetitive element, from Dictyostelium discoideum. Mol. Cell. Biol. 14:3074-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sucgang, R., G. K. Chen, W. Liu, R. Lindsay, J. Lu, D. Muzny, G. Shaulsky, W. F. Loomis, R. Gibbs, and A. Kuspa. 2003. Sequence and structure of the extrachromosomal palindrome encoding the ribosomal RNA genes in Dictyostelium. Nucleic Acids Res. 31:2361-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Symer, D. E., C. Connelly, S. T. Szak, E. M. Caputo, G. J. Cost, G. Parmiggiani, and J. D. Boeke. 2002. Human L1 retrotransposition is associated with genetic instability in vivo. Cell 110:327-338. [DOI] [PubMed] [Google Scholar]
- 37.Szafranski, K., G. Glöckner, T. Dingermann, K. Dannat, A. A. Noegel, L. Eichinger, A. Rosenthal, and T. Winckler. 1999. Non-LTR retrotransposons with unique integration preferences downstream of Dictyostelium discoideum transfer RNA genes. Mol. Gen. Genet. 262:772-780. [DOI] [PubMed] [Google Scholar]
- 38.Waldschmidt, R., D. Jahn, and K. H. Seifart. 1988. Purification of transcription factor IIIB from HeLa cells. J. Biol. Chem. 263:13350-13356. [PubMed] [Google Scholar]
- 39.Winckler, T., K. Szafranski, and G. Glöckner. 2005. Transfer RNA gene-targeted integration: an adaptation of retrotransposable elements to survive in the compact Dictyostelium discoideum genome. Cytogenet. Genome Res. 110:288-298. [DOI] [PubMed] [Google Scholar]
- 40.Winckler, T., C. Trautwein, C. Tschepke, C. Neuhäuser, I. Zündorf, P. Beck, G. Vogel, and T. Dingermann. 2001. Gene function analysis by amber stop codon suppression: CMBF is a nuclear protein that supports growth and development of Dictyostelium amoebae. J. Mol. Biol. 305:703-714. [DOI] [PubMed] [Google Scholar]
- 41.Yieh, L., G. Kassavetis, E. P. Geiduschek, and S. B. Sandmeyer. 2000. The Brf and TBP subunits of the RNA polymerase III transcription factor IIIB mediate position-specific integration of the gypsy-like element Ty3. J. Biol. Chem. 275:29800-29807. [DOI] [PubMed] [Google Scholar]
- 42.Zingler, N., U. Willhoeft, H.-P. Brose, V. Schoder, T. Jahns, K. M. O. Hanschmann, T. A. Morrish, J. Löwer, and G. Schumann. 2005. Analysis of 5′ junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5′ end attachment requiring microhomology-mediated end-joining. Genome Res. 15:780-789. [DOI] [PMC free article] [PubMed] [Google Scholar]