Abstract
Virus-induced activation of the beta interferon (IFN-β) gene requires orderly recruitment of chromatin-remodeling complexes and time-regulated acetylation of histone residues K8H4 and K14H3 on the promoter region. We have previously shown that transcription factor Yin Yang 1 (YY1) binds the murine IFN-β promoter at two sites (−122 and −90) regulating promoter transcriptional capacity with a dual activator/repressor role. In this work we demonstrate that both YY1 −122 and −90 sites are required for CBP recruitment and K8H4/K14H3 acetylation to take place on the IFN-β promoter region after virus infection. A single point mutation introduced at either one of these two sites inhibiting YY1 binding completely disrupted CBP recruitment and K8H4/K14H3 acetylation independently of HMGI or IRF3 binding to the promoter. We have previously demonstrated that YY1 represses the transcriptional capacity of the IFN-β promoter through its −90 site via histone deacetylation. Here we demonstrate that, in vivo, the binding of YY1 to the −90 site is constant all through virus infection whereas the binding of YY1 to the −122 site is activated after infection. We discuss here the capacity of YY1 to either repress (through histone deacetylase recruitment) or activate (through CBP recruitment) IFN-β gene expression according to the occupancy of either only its −90 site or both its −122 and −90 sites.
Beta interferon (IFN-β) plays an essential role during the establishment of an antiviral state (8, 30). The transcriptional capacity of the IFN-β promoter is constitutively repressed in an adult normal cell and remains inactivated until an external stimulus such as virus infection triggers its activation. Activation of the transcriptional capacity of the IFN-β promoter is transient. It is turned on 4 to 6 h after virus infection, and it is turned off 10 to 12 h after it (10, 36). Regulation of the promoter transcriptional capacity requires specific binding of transcription factors as well as the orderly recruitment of chromatin-remodeling complexes on the promoter region (1, 25, 33).
In the absence of virus infection, histone deacetylase activity maintains deacetylated lysine residues of histones H3 and H4 positioned on the IFN-β promoter region (22, 27). Shortly after virus infection, transcription factors and protein HMGI bind to the promoter on the nucleosome-free virus-responsive element region. The subsequent recruitment of histone acetyltransferases (HATs) CBP/p300 and GCN5/PCAF leads to the specific acetylation of certain lysine residues of histone H4 and H3, specially K8H4 and K14H3, which are essential for the recruitment of the RNA polymerase II holoenzyme complex and the SWI/SNF nucleosome-remodeling complex (2). Finally, nucleosome remodeling allows the binding of TFIID to the TATA box, triggering initiation of transcription of the IFN-β gene.
We have recently published data indicating that transcription factor Yin Yang 1 (YY1) binds to the murine IFN-β (muIFN-β) promoter at two different sites and regulates promoter transcriptional capacity with a dual activator/repressor role (35). The repressor role of YY1 appeared linked to its capacity to interact and recruit a histone deacetylase (HDAC) on the promoter region through its −90 site, while the exact molecular mechanisms governing the capacity of YY1 to activate the muIFN-β promoter remained to be elucidated.
YY1 is a ubiquitous, highly conserved zinc finger transcription factor (3, 9, 32) that activates or represses several different eukaryotic genes, among which are the c-Myc, c-Fos, β-casein, α-actin, interleukin 3 and 5, and IFN-γ genes, as well as some viral promoters (15, 16, 29, 32, 38). It binds to DNA through the recognition of a specific sequence containing a consensus (C/t/aCAT [uppercase letters represent preferred nucleotides; lowercase letters represent nucleotides tolerated to a lesser extent]) core motif (12). Promoter context (20), intracellular concentration (6), and posttranslational modifications (37) as well as the capacity of YY1 to interact with transcription factors and cofactors can influence the capacity of YY1 to act either as an activator or a repressor (7, 26, 32). Particularly, the interaction of YY1 with HATs or HDACs can orient YY1 towards either an activator or a repressor role.
Using gel retardation and recombinant glutathione S-transferase (GST)-YY1 protein we demonstrate in this work that the sequences surrounding the consensus YY1 DNA-binding core motifs present on the muIFN-β promoter strongly influence the binding of YY1 to these sites so that YY1 effectively binds only to the core motifs present at positions −122 and −90. Using the DNase I footprinting technique we have analyzed the capacity of YY1 for binding to its respective sites in the context of the entire promoter. We have observed that YY1 is able to bind both sites simultaneously protecting a region that extends beyond the −122 and −90 sites. Disruption of YY1 binding to either the −90 or the −122 site did not disrupt the binding of YY1 to the remaining intact site. In order to decipher the molecular mechanism governing the role of YY1 as activator of the transcriptional capacity of the murine IFN-β promoter, we have carried out chromatin immunoprecipitation (ChIP) assays on murine L929 cells and established cell lines containing either the wild-type muIFN-β promoter or the corresponding promoters mutated at either the −122 or the −90 YY1 site fused to a chloramphenicol acetyltransferase (CAT) reporter gene integrated in their genomes. The in vivo binding of CBP, AcK8H4, AcK14H3, IRF3, and YY1 to either the wild-type promoter or to the promoters mutated at the −122 (mut122) or −90 (mut90) site was analyzed before and at different times after virus infection. The results we have obtained here clearly indicated that the simultaneous presence of intact −90 and −122 sites was required to allow virus-induced CBP recruitment and K8H4/K14H3 acetylation on the muIFN-β promoter region as well as to reach virus-induced promoter transcriptional activation. The binding of YY1 to only one intact site, either −122 or −90, was not sufficient to allow CBP recruitment and K8H4/K14H3 acetylation. The binding of YY1 to the −90 site appeared constant, visible before as well as after infection, whereas the binding of YY1 to the −122 site was induced after infection so that simultaneous occupancy of both sites seemed possible only after infection. Contrary to the role of YY1 as activator of the muIFN-β promoter that, as demonstrated here, required that both the −122 and −90 sites be intact, only an intact −90 site has been previously described as required for the YY1-dependent repression of the promoter (35).
We analyze here the essential role of YY1 during virus-induced CBP recruitment, K8H4/K14H3 acetylation, and muIFN-β promoter transcriptional activation and show that its role is predominant with respect to the main role generally attributed to IRF3 as a regulator of virus-induced CBP recruitment and IFN-β promoter activation. We also discuss here the possibility to regulate the orientation of YY1 either as a repressor or an activator by regulating the degree of occupancy of the respective YY1 binding sites present in a same promoter region.
MATERIALS AND METHODS
Expression and purification of recombinant GST-YY1 and HMGI protein.
The plasmid encoding GST-YY1 was obtained from Martin Montecino (Chile). The GST fusion protein was isolated by transformation of the plasmid into Escherichia coli BL21 strain followed by batch purification using glutathione agarose (Sigma) and the procedure recommended by the manufacturer except that phosphate-buffered saline (PBS) was used as washing buffer instead of PBS-Tween 20. The purity of the protein was evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Coomassie blue staining and Western blot analysis using anti-YY1-specific antibodies (Santa Cruz). The protein concentration was determined by Bradford assay. Purification of recombinant HMGI protein was carried out as previously described (5).
Gel retardation assays.
Purified GST-YY1 or nuclear extracts of murine L929 cells prepared by microextraction were incubated with the corresponding 5′ 32P-labeled probes as previously indicated (35) in 20 μl (final volume) of 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM EDTA, 10% glycerol, and 5 mM dithiothreitol in the presence of an excess of either unlabeled poly(dI/dC) or unlabeled sonicated salmon sperm DNA as indicated in the figure legends. In each gel, equivalent amounts of radioactive probes (in cpm) were used for each labeled probe tested. During competition experiments, the corresponding unlabeled DNA probes were incubated with the protein for 10 min at room temperature before the labeled probe was added. When indicated (see Fig. 1B and 8C), 2 μg of either monoclonal H-10 anti-YY1 (sc-7341X), polyclonal PL-425 anti-IRF3 (sc-9082X), or polyclonal A-22 anti-CBP (sc-369X) antibodies or normal rabbit immunoglobulin G (IgG) (sc-2027) was added to the protein and incubated 1 h at 4°C prior to the addition of the labeled probe.
FIG. 1.
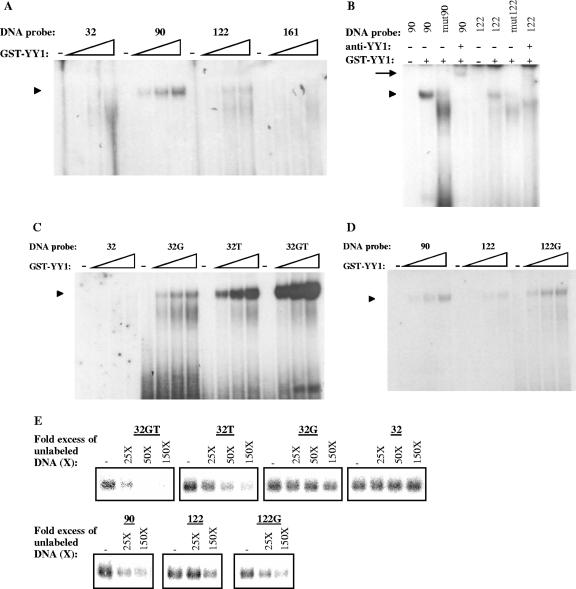
Bases surrounding the YY1 core motif strongly influence YY1 DNA-binding affinity. A) Equal amounts of GST-YY1 (0, 12.5, 25, and 37.5 ng/μl) were incubated with labeled probes 32, 90, 122, and 161 (in the presence of 500 ng of unlabeled poly[dI/dC] and 125 ng of unlabeled sonicated salmon sperm DNA) and subjected to a gel retardation assay. The arrowhead (also in panels B, C, and D) indicates protein-DNA complexes containing GST-YY1. B) GST-YY1 (37.5 ng/μl) was incubated with labeled probes 90, mut90, 122, and mut122 (in the presence of 500 ng of unlabeled poly[dI/dC] and 125 ng of unlabeled sonicated salmon sperm DNA) in the presence or absence of 2 μg of monoclonal anti-YY1 antibody H-10 (sc-7341) raised against the full-length YY1 protein. The arrow indicates supershifted complexes. C) Equal amounts of GST-YY1 protein (0, 12.5, 25, and 37.5 ng/μl) were incubated with labeled probes 32, 32G, 32T, and 32GT (in the presence of 500 ng of unlabeled poly[dI/dC] and 125 ng of unlabeled sonicated salmon sperm DNA) and subjected to a gel retardation assay. D) Equal amounts of GST-YY1 protein (0, 12.5, 25, and 37.5 ng/μl) were incubated with labeled probes 90, 122, and 122G (in the presence of 500 ng of unlabeled poly[dI/dC] and 125 ng of unlabeled sonicated salmon sperm DNA) and subjected to a gel retardation assay. E) GST-YY1 (37.5 ng/μl) was incubated with labeled probe 32GT in the absence (−) or presence of a 25-, 50-, or 150-fold excess of the indicated unlabeled probes. Protein-DNA complexes were subjected to a gel retardation assay.
FIG. 8.
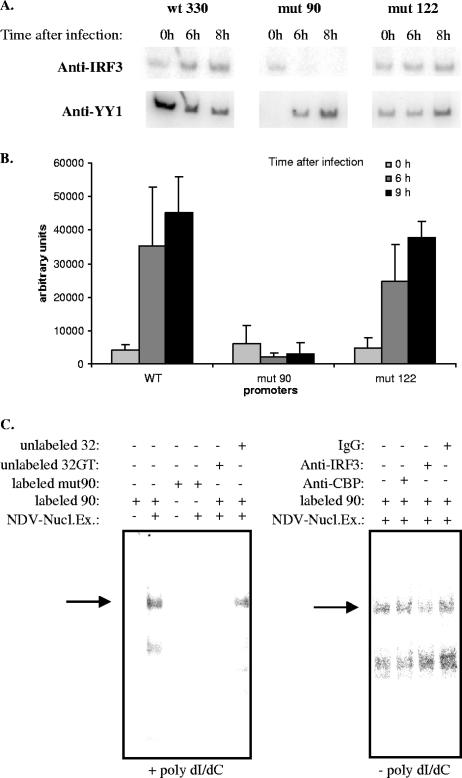
CBP recruitment is independent of IRF3 binding to muIFN-β. A) Equal amounts of genomic DNA from L929wt330, L929mut90, and L929mut122 noninfected (0 h) or NDV-infected (collected at 6 and 8 after infection) cells were immunoprecipitated with antibodies directed against IRF-3 and YY1. The corresponding immunoprecipitated DNA was amplified with primers specific for the integrated IFN-β promoter. B) Intensity of the band amplified from anti-IRF3-immunoprecipitated DNA corresponding to the integrated wt330, mut90, and mut122 promoters, respectively, 0, 6, and 8 h after NDV infection was quantified using a PhosphorImager. Results correspond to the averages of two independent amplification reactions. C) (Left panel) Zero (−) or 5 μg (+) of total nuclear extracts prepared from NDV-infected L929 cells was incubated with labeled probe 90 or mut90 in the presence of 1.5 μg of unlabeled sonicated poly(dI/dC) and subjected to gel retardation. When indicated, a 50-fold excess of unlabeled probe 32GT or 32 was added to the reaction mixture. (Right panel) Five micrograms (+) of total nuclear extracts prepared from NDV-infected L929 cells was incubated with labeled probe 90, in the absence of poly(dI/dC), and subjected to gel retardation. When indicated, 2 μg of anti-CBP, anti-IRF3, or total rabbit IgG was added to the reaction mixture. Arrows indicate the most retarded complex, behaving as a YY1-DNA binding complex.
DNase I footprint.
For the footprinting experiments the noncoding strand of the muIFN-β promoter fragment (from position −330 to +20), either wild type (wt330) or mutated in the corresponding YY1 binding site (mut90 and mut122), was 5′ end labeled at position +20. The fragments were prepared from 20 μg of the respective plasmids pBLCAT3-muIFNβwt330, pBLCAT3-muIFNβmut90, and pBLCAT3-muIFNβmut122 (35). The plasmids were digested with BamHI, dephosphorylated with alkaline phosphatase, 5′ 32P end labeled with T4 polynucleotide kinase, and further digested with PstI. Before DNase I digestion, the 350-bp fragments, 5′ 32P end labeled at the BamHI site (+20) were incubated with various amounts of recombinant GST-YY1 protein in 100 μl (final volume) of YY1 binding buffer containing 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM EDTA, and 5 mM dithiothreitol in the presence of an excess of unlabeled poly(dI/dC). The digested DNA was precipitated and resuspended in formamide-Tris-borate-EDTA (TBE). The samples were loaded onto an 8 M urea-6% acrylamide gel buffered in TBE. The migration was carried out at 40 W.
Cell line and transfection.
L929wt330, L929mut90, and L929mut122 cell lines have been described previously (35). These cell lines carry integrated into their genomes the muIFN-β promoter (from position −330 to +20), either wild type (wt330) or mutated in the corresponding YY1 binding sites (mut90 and mut122), fused to the CAT reporter gene. Cell culture, Newcastle disease virus (NDV) infection, and CAT assays were as previously described (5).
Chromatin immunoprecipitation.
Aliquots of 30 μg of genomic DNA from L929, L929wt330, L929mut90, and L929mut122 cells were immunoprecipitated as previously described (35) using anti-YY1 H-10 (sc-4703; Santa Cruz) monoclonal antibody, anti-CBP A-22 (sc-369; Santa Cruz) polyclonal antibody, anti-acetyl-histoneH4(Lys8) (06-760; Upstate) polyclonal antibody, anti-acetyl-histoneH3(Lys14) (06-911; Upstate) polyclonal antibody, anti-IRF3 polyclonal antibody from Michael David (San Diego, CA), and anti-NSs polyclonal antibody from Michèle Bouloy (Institut Pasteur, Paris, France). The same amount of genomic DNA (30 μg) was used for input amplifications. PCR analysis of inputs or immunoprecipitated DNA was performed in 25-μl final volume. The previously described (35) F-40 and CAT oligonucleotides, specific for the integrated muIFN-β promoters, were used as primers to amplify the integrated wt330, mut90, and mut122 IFN-β promoters using the following PCR conditions: 1 cycle of 94°C for 5 min; 20 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 30 s; and 1 cycle of 72°C for 10 min. A first “cold” PCR was carried out in the presence of 25 pmol of each primer; 2 μl of the product of the first PCR was subjected to a second “hot” PCR carried out in the presence of 0.1 μl of [α-32P]dATP (6,000 Ci/mmol) and 10 pmol of each primer. For the amplification of the endogenous wild-type IFN-β promoter present in the L929 strain we used as primers oligonucleotides 5.233 (5′-CCTTTGCTCCAGCAATTGGTGA-3′) and 3.27 (5′-CCGGATCCTGGCAGTGAGAATGAT-3′) under the following PCR conditions: 1 cycle of 94°C for 5 min; 20 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min; and 1 cycle of 72°C for 10 min. A first “cold” PCR was carried out in the presence of 25 pmol of each primer; 2 μl of the product of the first PCR was subjected to a second “hot” PCR carried out in the presence of 0.1 μl of [α-32P]dATP (6,000 Ci/mmol) and 10 pmol of each primer. For amplification of the endogenous wild-type IFN-β promoter present in L929mut122 strains we used as primers oligonucleotides 5.334 (5′-AGCTACTCTGCCTGGCTT-3′) and 3.+125 (5′-GGA GAA GCA CAG CAG GAA-3′), which specifically amplify the endogenous IFN-β promoter without amplifying the integrated mut122 IFN-β promoter. With these primers, PCR conditions were as follows: 1 cycle of 94°C for 5 min; 20 cycles of 94°C for 30 s, 63°C for 1 min, and 72°C for 1.5 min; and 1 cycle of 72°C for 10 min. A first “cold” PCR was carried out in the presence of 25 pmol of each primer; 2 μl of the product of the first PCR was subjected to a second “hot” PCR carried out in the presence of 0.1 μl of [α-32P]dATP (6,000 Ci/mmol) and 10 pmol of each primer. For amplification of the β-actin gene we used as primers oligonucleotides actineBC (5′-TGACGGGGTCACCCACACTGT-3′) and actineBNC (5′-CTAGAAGCATTTGCGGTGGAC-3′) with the following PCR conditions: 1 cycle of 94°C for 5 min; 20 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 2 min; and 1 cycle of 72°C for 10 min. A first “cold” PCR was carried out in the presence of 25 pmol of each primer; 2 μl of the product of the first PCR was subjected to a second “hot” PCR carried out in the presence of 0.1 μl of [α-32P]dATP (6,000 Ci/mmol) and 10 pmol of each primer.
RESULTS
Bases surrounding the YY1 DNA-binding core motif strongly influence the binding of YY1 to the muIFN-β promoter.
Four potential YY1 binding sites are present in the muIFN-β promoter at positions −32, −90, −122, and −161 (Table 1). Using total murine L929 nuclear extracts we have previously shown that protein YY1 could bind only to the sites present at positions −122 and −90, with a stronger affinity to the −90 site (35). In order to determine if the differential binding affinities of YY1 for the −32, −90, −122, and −161 sites observed with nuclear extracts were due to the sequences surrounding the corresponding YY1 core motifs present at these positions rather than to a factor present in the nuclear extracts susceptible of influencing YY1 binding to the different DNA sequences, we carried out gel retardation experiments using recombinant GST-YY1 protein (instead of total nuclear extracts) and double-stranded DNA probes corresponding to the −32, −90, −122, and −161 sites (Table 1). As we had previously observed with nuclear extracts, we observed the binding of GST-YY1 only to probes 90 and 122, with an apparent stronger affinity for probe 90. No binding to probes 32 and 161 was observed (Fig. 1A). As expected for a YY1-DNA complex, the complexes formed by GST-YY1 with probes 90 and 122 were disrupted after the introduction of a mutation on the corresponding core motifs (oligonucleotides mut90 and mut122 in Table 1) as well as in the presence of anti-YY1 antibodies (Fig. 1B).
TABLE 1.
Sequences of wild-type or mutated YY1 DNA-binding sites
| Name | Sequencea (5′-3′) | muIFN-β promoter position |
|---|---|---|
| Consensus | GA(C/g/a)(G/t)(C/a/t)CATN(T/a)(T/g/c) | |
| 32 | GCAGAAAGGACCATCCCTTATA | −32 coding strand |
| 32G | GCAGAAAGGgCCATCCCTTATA | −32 coding strand |
| 32T | GCAGAAAGGACCATCttTTATA | −32 coding strand |
| 32GT | GCAGAAAGGgCCATCttTTATA | −32 coding strand |
| 90 | TTTTCCTCTGTCATTTTCTCTT | −90 noncoding strand |
| mut90 | TTTTCCTCTGTaATTTTCTCTT | −90 noncoding strand |
| 122 | CTTCTAATATTCATTTTATTCA | −122 noncoding strand |
| mut122 | CTTCTAATATTgATTTTATTCA | −122 noncoding strand |
| 122G | CTTCTAATAgTCATTTTATTCA | −122 noncoding strand |
| 161 | TTAACCCAGTACATAGCATATA | −161 coding strand |
In the consensus sequence as defined by Hyde-DeRuyscher et al. (12) uppercase letters represent the preferred nucleotides and lowercase letters represent nucleotides tolerated to a lesser extent. Boldface letters indicate the nucleotides corresponding to the YY1 core motif. Underlining indicates the nucleotides present outside the core motif characterized as important for binding affinity and specificity. Lightface lowercase letters in the muIFN-β sites indicate mutations introduced in the sequence of the potential YY1-binding sites.
The results we show here with recombinant GST-YY1 protein are equivalent to those we have previously obtained with YY1 protein present in total L929 nuclear extracts (35). Therefore, the inability of YY1 to bind to the sites present at positions −161 and −32 and the weaker affinity of YY1 for the −122 site compared to the −90 site cannot be assigned to an eventual inhibitory factor present in the nuclear extracts.
The presence of thymidines 3′ and a guanine 5′ of the YY1 core motif (underlined in Table 1) has been shown to be important for binding affinity and specificity during YY1-DNA complex formation (12). The inability of YY1 to bind to the site present at position −32 (GACCATCCC), which carries a canonical CCAT core motif, could be related to the absence of two thymidines 3′ and of a guanine 5′ of the core motif. In order to verify this hypothesis, we synthesized oligonucleotides corresponding to probe 32 carrying modifications 5′ and/or 3′ of the core motif. Oligonucleotide 32G carries a G 5′ (GGCCATCCC), oligonucleotide 32T carries two TTs 3′ (GACCATCTT), and oligonucleotide 32GT carries both a G 5′ and two TTs 3′ (GGCCATCTT) of the core motif. In Fig. 1C we compared, using gel retardation, the binding capacities of GST-YY1 to probes 32, 32G, 32T, and 32GT. As previously shown, no GST-YY1-DNA complex is observed with probe 32 whereas GST-YY1-DNA complexes are formed with probes 32G, 32T, and 32GT, with the apparent corresponding affinities increasing from 32G to 32GT as follows 32G < 32T < 32GT. A similar experiment was carried out with probe 122, which carries two Ts 3′, but lacks a G 5′, of the core motif. In Fig. 1D we compared, using gel retardation, the binding capacities of GST-YY1 to probes 90 (TGTCATTTT), 122 (ATTCATTTT), and 122G (AGTCATTTT). As in the case of probes 32 and 32G, the addition of a G 5′ of the core motif of probe 122 enhanced the DNA-binding affinity of YY1 for this probe rendering it equivalent to probe 90.
Gel shift competition experiments were carried out in order to further confirm the different affinities of YY1 for binding to the 32GT, 32T, 32G, 32, 90, 122, and 122G YY1-binding sites. In the experiment for which results are presented in Fig. 1E, 32P-labeled probe 32GT was incubated with GST-YY1 in the presence of 25-, 50-, and 150-fold excesses of unlabeled probes 32GT, 32T, 32G, and 32 or in the presence of 25- and 150-fold excesses of unlabeled probes 90, 122, and 122G. A clear competition was observed in the presence of a 25-fold excess of unlabeled probe 32GT, the competition being total in the presence of a 50-fold excess. In the case of probe 32T, a clear competition was observed in the presence of a 50-fold excess of unlabeled probe 32T, with competition being total in the presence of a 150-fold excess. No competition was observed with probe 32, and only a faint competition was observed in the presence of a 150-fold excess of probe 32G. Unlabeled probes 90 and 122G equally competed labeled probe 32GT, in a way similar to probe 32G, whereas competition observed with unlabeled probe 122 was weaker than that obtained with unlabeled probes 90 and 122G. Overall, the results obtained during gel shift competition experiments fully agreed and confirmed the different DNA binding affinities of YY1 for probes 32GT, 32T, 32G, 32, 90, 122, and 122G observed in Fig. 1C and D, with the apparent corresponding affinities increasing as follows 32 < 32G < 122 < 90 ≈ 122 ≈ 32G < 32GT.
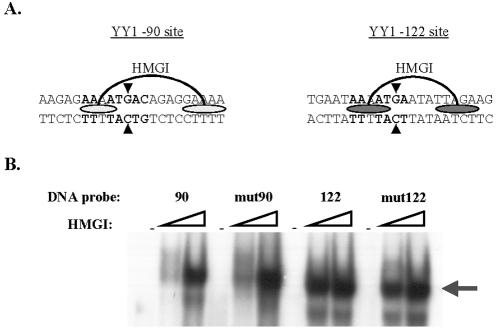
The DNA regions containing either the −90 or the −122 YY1 binding sites also contain previously described HMGI binding sites (Fig. 2A). The HMGI binding site overlapping the YY1 −90 site has been described as a weak HMGI binding site whereas the HMGI binding site overlapping the YY1 −122 site is a strong HMGI binding site (5). Single point mutations mut90 and mut122, introduced in the YY1 −90 and YY1 −122 sites (indicated by arrowheads in Fig. 2A), respectively, were chosen in order to modify the corresponding YY1 core binding motifs and disrupt YY1 binding without affecting the bases previously described as necessary for HMGI binding (5). Results obtained during gel retardation experiments carried out with recombinant HMGI protein and radioactively labeled 90, mut90, 122, and mut122 sequences (Fig. 2B) demonstrated that, as expected, mutations mut90 and mut122 did not affect the DNA binding of HMGI to its respective binding sites present in these two regions.
FIG. 2.
Mutations introduced in YY1 binding sites −90 (mut90) and −122 (mut122) do not affect HMGI binding. A) Schematic representation of HMGI binding sites present in the −90 (weak HMGI binding site) and −122 (strong HMGI binding site) regions as described previously (5). Ovals indicate the bases directly interacting with HMGI, and boldface letters indicate the bases directly interacting with YY1. Arrowheads indicate the bases mutated to give rise to mut90 and mut122 sequences. B) Equal amounts of recombinant HMGI protein (0, 1, and 3 ng/μl) were incubated with labeled probes 90, mut90, 122, and mut122 (in the presence of 125 ng of unlabeled sonicated salmon sperm DNA) and subjected to a gel retardation assay. The arrow indicates protein-DNA complexes containing HMGI.
In vitro, YY1 can bind the −90 and −122 sites either simultaneously or independently of one another.
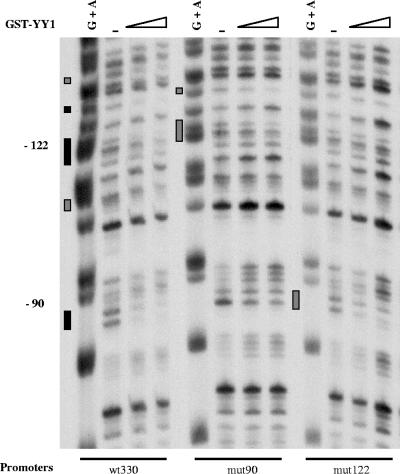
We have carried out DNase I footprinting experiments in order to analyze the binding of GST-YY1 to the −90 and −122 sites in the context of the entire promoter, either wild type (wt300) or mutated at the −90 site (mut90) or the −122 site (mut122). Results are shown in Fig. 3. Protections translating YY1 binding are visible at both the −90 and −122 regions of the wild-type (wt330) promoter, indicating that YY1 is capable of binding both wild-type sites simultaneously. Regions situated near and between the −90 and −122 sites were protected, and protection extended beyond the −122 site, up to the −140 region. Some regions were completely protected (indicated by black rectangles), and some of them were only partially protected (indicated by gray rectangles). Introduction of a mutation at either the −90 site (mut90) or the −122 site (mut122) disrupted the binding of YY1 to the corresponding mutated site without preventing, even though slightly diminishing, the binding of YY1 to the remaining wild-type site.
FIG. 3.
YY1 can bind the −122 and −90 sites simultaneously as well as independently of one another. DNase I footprinting was carried out on the noncoding strand of the wild-type muIFN-β promoter (−330 to +20) (wt330) or an muIFN-β promoter mutated in the −90 site (mut90) or −122 site (mut122). The corresponding 5′-end-labeled promoter fragments were incubated with 0, 80, 160, and 240 ng/μl of GST-YY1 in the presence of unlabeled poly(dI/dC), digested with DNase I, and analyzed on an 8 M urea-6% polyacrylamide gel. A specific G+A DNA sequencing reaction of the corresponding labeled DNA fragments is shown. Positions of the −90 and −122 sites are indicated. Regions completely protected (black rectangles) or partially protected (gray rectangles) by YY1 are indicated.
In vivo, YY1 binds to the murine IFN-β promoter before as well as after virus infection.
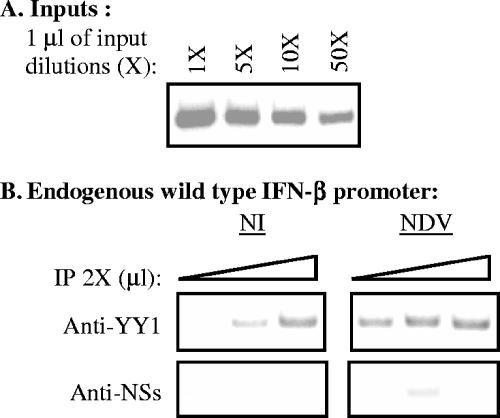
Using chromatin immunoprecipitation, we have analyzed the in vivo binding of YY1 to the wild-type endogenous promoter present in murine L929 cells before as well as after virus infection. In order to do this, equal amounts of genomic DNA collected from noninfected as well as from NDV-infected cells were immunoprecipitated with anti-YY1 monoclonal antibody H-10 or with irrelevant anti-NSs polyclonal antibody directed against the nonstructural protein NSs of Rift Valley fever virus. Equal amounts of immunoprecipitated DNA were amplified with primers specific for the endogenous wild-type murine IFN-β promoter. Under the PCR conditions used in these experiments the intensity of the radioactively labeled amplified band corresponding to the IFN-β promoter appeared proportional to the amount of DNA present in the reaction mixture (Fig. 4A). As shown in Fig. 4B, the endogenous IFN-β promoter was immunoprecipitated with anti-YY1 antibody in noninfected as well as in infected cells. The immunoprecipitation obtained with anti-YY1 antibody appeared specific since under the same conditions the IFN-β promoter was not immunoprecipitated with an irrelevant anti-NSs antibody.
FIG. 4.
In vivo, YY1 binds to the endogenous IFN-β promoter before as well as after virus infection. Input (1 μl of 1-, 5-, 10-, and 50-fold dilutions) (A) and anti-YY1 and anti-NSs immunoprecipitated (IP; 1, 2, and 3 μl of a 2-fold dilution) (B) DNA from L929 cells noninfected (NI) or NDV infected (NDV) collected 6 h after infection was amplified with primers specific for the endogenous wild-type murine IFN-β promoter.
Intact −122 and −90 YY1 binding sites are required for CBP recruitment on the IFN-β promoter after virus infection.
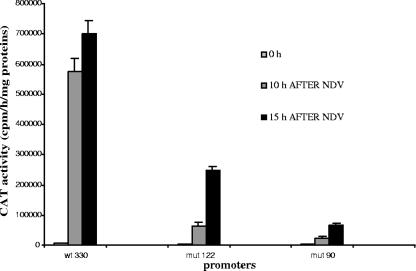
The IFN-β promoter reaches its highest transcriptional capacity around 10 h after infection, followed by a posttranscriptional turnoff that is established between 10 and 12 h after infection. In Fig. 5, we show the virus-induced transcriptional capacities of the stably integrated wild-type (wt330) and mutated (mut122 and mut90) muIFN-β promoter CAT reporter constructs measured 10 and 15 h after infection. Ten hours after infection, both the mut122 and mut90 promoters displayed weak virus-induced transcriptional activities compared to the wild-type wt330 promoter, corresponding to 10% and 5%, respectively, of the activity displayed by the wild-type promoter at the same time. Between 10 and 15 h after infection, the activity of the wt330 promoter remained almost constant, with a 1.2-fold increase, whereas the activity of the mut122 and mut90 promoters continued to progress, with 4.0-fold and 2.7-fold increases, respectively. Therefore, under the conditions used in these experiments, both the mut122 and mut90 promoters displayed similar phenotypes corresponding to (i) weak virus-induced transcriptional capacities and (ii) a retarded posttranscriptional turnoff.
FIG. 5.
Mutated promoters mut122 and mut90 display abnormal virus-induced activities. Cells from the wild-type L929wt330 strain or the mutated L929mut122 and L929mut90 strains were either mock infected (0 h) or infected with NDV and collected 10 and 15 h after infection. The corresponding CAT activities were measured. The results correspond to the averages of two independent experiments with each point in duplicate. The CAT activities for mock infection were as follows: L929wt330, 6,926 ± 132.5 cpm/h/mg; L929mut90, 2,427.5 ± 104.5 cpm/h/mg; L929mut122, 2,774.5 ± 132.5 cpm/h/mg.
Cofactor CBP is recruited on the IFN-β promoter starting 4 h and peaking 8 h after infection (1). CBP recruitment on the IFN-β promoter has been described as being required for correct promoter transcriptional activation as well as for the establishment of promoter transcriptional turnoff (19). Since transcription factor YY1 directly interacts with cofactor CBP, we analyzed the eventual role of YY1 during the recruitment of CBP on the muIFN-β promoter.
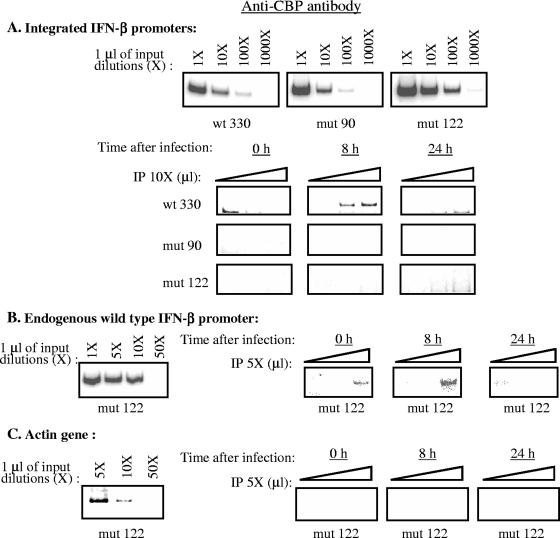
Using chromatin immunoprecipitation we have compared the recruitment of CBP on the wild-type (wt330) promoter to its recruitment on the mut90 and mut122 promoters, which displayed a weak transcriptional capacity as well as a retarded transcriptional turnoff. Genomic DNA from wt330, mut90, and mut122 strains was collected before (0 h) as well as 8 and 24 h after infection. Equal amounts of DNA collected at these times were immunoprecipitated with an anti-CBP antibody and amplified with primers specific for either the wt330, mut90, and mut122 integrated promoters (Fig. 6A); the endogenous IFN-β promoter (Fig. 6B); or the endogenous β-actin gene (Fig. 6C). Under the PCR conditions used in these experiments, the intensity of the radioactively labeled amplified band appeared proportional to the amount of DNA present in the reaction mixture (see inputs in Fig. 6A, B, and C). In agreement with the results previously described for the human endogenous IFN-β promoter (1), CBP recruitment on the wild-type integrated wt330 IFN-β promoter was induced after virus infection (Fig. 6A). Compared to the wild-type integrated promoter, neither the mut90 nor the mut122 integrated promoter was able to recruit CBP after virus infection (Fig. 6A).
FIG. 6.
Intact −122 and −90 YY1 binding sites are required to allow virus-induced CBP recruitment on the muIFN-β promoter. A) Input (1 μl of 1, 10, 100, and 1,000-fold dilutions) and anti-CBP-immunoprecipitated (IP) (1, 2, and 3 μl of a 10-fold dilution) DNA from L929wt330, L929mut90, and L929mut122 strains collected 0, 8, or 24 h after infection was amplified with primers specific for the integrated wt330, mut90, and mut122 IFN-β promoters. B) Input (1 μl of 1, 5, 10, and 50-fold dilutions) and anti-CBP-immunoprecipitated (1, 2, and 3 μl of a 5-fold dilution) DNA from the L929mut122 strain collected 0, 8, and 24 h after infection was amplified with primers specific for the wild-type endogenous IFN-β promoter. C) Input (1 μl of 5, 10, and 50-fold dilutions) and anti-CBP-immunoprecipitated (1, 2, and 3 μl of a 5-fold dilution) DNA from the L929mut122 strain collected 0, 8, and 24 h after infection was amplified with primers specific for the endogenous β-actin gene.
The absence of immunoprecipitation with anti-CBP antibody of either the mut90 or the mut122 promoter was specific since under the same conditions the endogenous wild-type IFN-β promoter present in the L929mut122 strain was immunoprecipitated with anti-CBP antibody (Fig. 6B), displaying a phenotype similar to the one obtained for the wt330 promoter on the L929wt330 strain and similar to what has been described in the literature for the endogenous human IFN-β promoter (1). As expected, no amplification of the murine β-actin gene was obtained from the genomic DNA of the L929mut122 strain immunoprecipitated with anti-CBP antibody either in noninfected or in infected cells. We therefore demonstrate here that CBP recruitment on the murine IFN-β promoter required that both YY1 binding sites be intact. The presence of only one intact site, either −90 or −122, was not sufficient to allow CBP recruitment on the promoter region after virus infection.
Intact −122 and −90 YY1 binding sites are required for virus-induced acetylation of K8H4 and K14H3 on the IFN-β promoter.
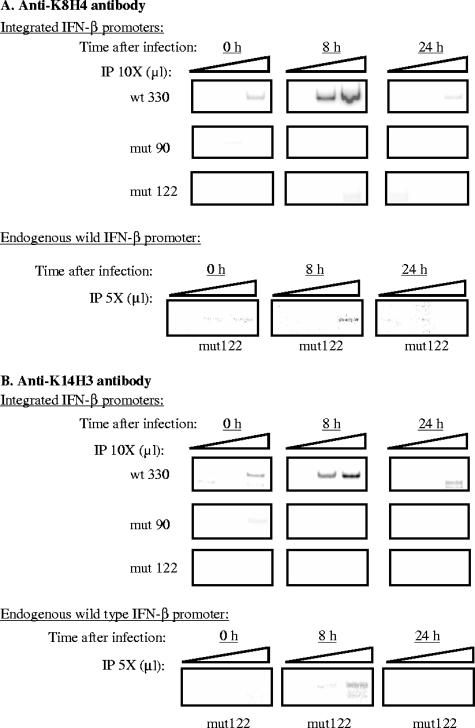
Acetylation of K8H4 and K14H3 has been shown to be essential for the induction of the transcriptional capacity of the IFN-β promoter (2). The capacity of YY1 to directly interact and regulate CBP recruitment on the IFN-β promoter, which itself interacts with PCAF, the HAT responsible of K8H4 and K14H3 acetylation on the IFN-β promoter, made us consider YY1 as a potential regulator of the rate of AcK8H4 and AcK14H3 on the IFN-β promoter after virus infection. We tested this hypothesis using ChIP assays. The same fractions of genomic DNA from L929 strains containing the stably integrated wt330, mut90, and mut122 promoters immunoprecipitated with anti-CBP antibodies in Fig. 6 were also immunoprecipitated with anti-AcK8H4 (Fig. 7A) and anti-AcK14H3 (Fig. 7B) antibodies. In agreement with the results previously described for the endogenous human IFN-β promoter (2), acetylation of K8H4 and K14H3 was transiently induced on the integrated wild-type murine IFN-β wt330 promoter 8 h after infection, whereas acetylation of K8H4 and K14H3 was completely inhibited on the mut90 and mut122 promoters. As in the case of the immunoprecipitates obtained with anti-CBP antibodies shown in Fig. 6, the absence of immunoprecipitations with anti-K8H4 and anti-K14H3 antibodies of either the mut90 or the mut122 promoter was specific since under the same conditions the endogenous wild-type IFN-β promoter present in the L929mut122 strain was immunoprecipitated with anti-K8H4 and anti-K14H3 antibodies, displaying a phenotype similar to the one obtained for the wt330 promoter on the L929wt330 strain and similar to what has been described in the literature for the endogenous human IFN-β promoter (1). Therefore the presence of two intact YY1 binding sites appeared to be required not only for CBP recruitment but also for K8H4 and K14H3 acetylation on the IFN-β promoter after virus infection.
FIG. 7.
Intact −122 and −90 YY1 binding sites are required to allow virus-induced K8H4 and K14H3 acetylation on the muIFN-β promoter. For integrated IFN-β promoters, anti-K8H4 (A)- and anti-K14H3 (B)-immunoprecipitated (IP) DNA from L929wt330, L929mut90, and L929mut122 strains collected 0, 8, or 24 h after infection (1, 2, and 3 μl of a 10-fold dilution) was amplified with primers specific for the integrated wt330, mut90, and mut122 IFN-β promoter. For the endogenous wild-type IFN-β promoter, anti-K8H4 (A)- and anti-K14H3 (B)-immunoprecipitated DNA from the L929mut122 strain collected 0, 8, or 24 h after infection (1, 2, and 3 μl of a fivefold dilution) was amplified with primers specific for the wild-type endogenous IFN-β promoter.
The lack of CBP recruitment and K8H4/K14H3 acetylation on the mut122 promoter is independent of IRF3 binding to the promoter.
Virus-induced transcriptional activation of the IFN-β gene requires IRF3 binding to the IFN-β promoter. After virus infection, transcription factor IRF3 is activated and translocated to the nucleus, where it binds to the PRDI-III region of the IFN-β promoter. Cofactor CBP and IRF3 can directly interact with each other, and it has been proposed that the binding of IRF3 to the IFN-β promoter occurs concomitantly with CBP recruitment on the promoter region (11, 17, 34, 39). We carried out ChIP assays in order to analyze if IRF3 binding to the IFN-β promoter was affected in the context of the mut90 and mut122 promoters, where no recruitment of CBP was observed. Genomic DNA collected at 0, 6, and 8 h after virus infection from the wt330, mut90, and mut122 strains was immunoprecipitated with anti-IRF3 and anti-YY1 antibodies. Results are shown in Fig. 8A. As expected, IRF3 binding to the wild-type wt330 promoter was enhanced after infection. Quantification of the amount of IFN-β promoter DNA amplified from wt330, mut90, and mut122 anti-IRF3 immunoprecipitates (Fig. 8B) showed that the profile for the binding of IRF3 to the mut122 promoter was very similar to the one observed for the wild-type wt330 promoter, demonstrating that mutation mut122 did not dramatically affect IRF3 binding. Therefore, the absence of CBP recruitment and K8H4/K14H3 acetylation observed on the mut122 promoter could not be assigned to default of IRF3 binding that occurred here independently of CBP. In contrast to what was found for the mut122 promoter, no binding of IRF3 to the mut90 promoter was observed after virus infection. The weaker transcriptional capacity displayed by the mut90 promoter compared to the mut122 promoter (Fig. 5) could be related to the absence of IRF3 on promoter mut90 compared to promoter mut122.
In order to analyze the eventual role of the YY1 binding site present at position −90 on the formation of a protein-DNA complex containing IRF3 and probe 90, we carried out in vitro gel retardation experiments. In a first set of experiments (Fig. 8C, left panel), nuclear extracts prepared from NDV-infected L929 cells were incubated with labeled probe 90 in the presence of an excess of sonicated poly(dI/dC) as nonspecific competitor DNA. The main most retarded complex formed, indicated by an arrow, behaved as a YY1-containing protein-DNA complex since it disappeared when the same nuclear extracts were incubated with labeled probe mut90 (carrying a mutation on the YY1 DNA-binding core motif present at position −90) instead of probe 90. Also, the formation of this complex was inhibited after addition of an excess of unlabeled probe 32GT (containing a good YY1-binding site, used here as a specific competitor DNA) but not in the presence of an excess of unlabeled probe 32 (containing a very weak YY1-binding site). In a second set of experiments (Fig. 8C, right panel), the same nuclear extracts prepared from NDV-infected L929 cells were incubated in the absence of poly(dI/dC) but in the presence of 2 μg of either anti-CBP, anti-IRF3 polyclonal antibodies, or total normal rabbit IgGs. Under these conditions, the formation of the main most retarded complex, indicated again by an arrow, was specifically inhibited after incubation of the nuclear extracts with anti-IRF3 antibody but not by the anti-CBP antibody or rabbit IgGs. In agreement with the results obtained in vivo during ChIP assays (shown in Fig. 8A, upper line), the experiments shown in Fig. 8C indicate that IRF3 can form a protein-DNA complex in vitro with probe 90 in the absence of CBP provided that the YY1 −90 site is intact.
Lastly, results obtained with the anti-YY1 monoclonal antibody, shown in the bottom line of Fig. 8A, indicated that YY1 remained associated to the integrated wild-type wt330 promoter before as well as after virus infection. This is in agreement with results obtained during ChIP assays with the endogenous wild-type IFN-β promoter shown in Fig. 4. Results obtained here with the integrated mut90 and mut122 promoters indicate that the binding of YY1 to the −122 site was enhanced after virus infection whereas, in noninfected cells, YY1 appeared predominantly bound to the −90 site.
DISCUSSION
The presence of more than one YY1 binding site on the same promoter region as a means to functionally switch YY1 from a repressor to an activator.
Two functional YY1 binding sites are present on the proximal region of the muIFN-β promoter present at positions −122 and −90, respectively. We have shown here that the introduction of single point mutations on either the −122 or the −90 site completely disrupted CBP recruitment and K8H4/K14H3 acetylation on the mutated promoters, strongly affecting the transcriptional capacity of the corresponding promoters.
The presence of more than one YY1 DNA-binding site on several promoters regulated by YY1 has been described (13, 24, 38), as has their presence on YY1-bound pericentromeric γ-satellite DNA sequences (28). Can a functional meaning be ascribed to the presence of more than one YY1 binding site on the same promoter region, considering that binding of YY1 to more than one site did not appear strongly cooperative?
A protein called YY2, closely related to YY1, has been recently isolated and described as able to bind to some, although not all, YY1 binding sites (21). Therefore some of the sites considered until now as YY1 binding sites might actually be YY2 binding sites. The binding of YY2 to the IFN-β promoter has been recently considered by Klar and Bode (14) as a possible means to antagonize the negative effect of YY1 on promoter transcriptional capacity. Nevertheless, notwithstanding evidence indicating probable binding of YY2 to far-upstream control elements present in the human IFN-β promoter at kb −3 and −2, there is for the moment no strong evidence of YY2 binding to the proximal region of the murine IFN-β promoter. Gel retardation experiments with recombinant YY2 protein carried out by Klar and Bode showed that in vitro YY2 could form a protein-DNA complex with a probe carrying the −90 site but that, as stated by the authors themselves, the intensity of the complex formed was particularly weak (14).
Nguyen et al. (21) have analyzed the capacity of YY2 to bind 10 sequences previously characterized as YY1 binding sites. Their results indicated that YY2 was able to bind 5 out of the 10 tested sequences. All the sequences that were not bound by YY2 contained a T (underlined) 3′ of the core motif [C/t/aCAT(N)T], and a T at this position was absent on all the sequences, except one, bound by YY2. In the case of the muIFN-β promoter, both the −122 and −90 sites carry a T 3′ of the core motif. Even though we cannot completely exclude the possibility that the −122 and −90 YY1 binding sites could be YY2 binding sites, there is for the moment no strong argument supporting this hypothesis.
During the YY1 ChIP assays we carried out in this work, we used the monoclonal anti-YY1 antibody (H-10) that according to the manufacturer recognizes only YY1 protein. The results obtained for the endogenous as well as the wild-type wt330 integrated promoter showed that YY1 interacted with the proximal region of the muIFN-β promoter before as well as after virus infection. In noninfected cells YY1 appeared to be predominantly bound to its −90 site and remained bound to this site at least until 8 h after infection. The situation was different in the case of the −122 site. As observed for the mut90 promoter, the binding of YY1 to the −122 site, which was not visible in noninfected cells, was induced only after infection. In a previous work we demonstrated that YY1-dependent repression of the muIFN-β promoter via HDAC recruitment relied mainly on the presence of an intact −90 site (35). The results obtained in this work suggest that the simultaneous double occupancy of both the −122 and −90 sites is necessary to allow CBP recruitment and K8H4/K14H3 acetylation, which are essential for promoter transcriptional activation. The transition from single-site occupancy (−90 site, before virus infection) to double-site occupancy (−90 and −122 sites, after virus infection) appears therefore as a possible means to promote a functional switch of YY1 from a repressor (via HDAC recruitment) to an activator (via CBP recruitment). It is possible to speculate that the interaction of CBP with only one YY1 molecule would not be sufficient to disrupt the YY1-HDAC interaction taking place in noninfected cells, this being only possible if CBP interacts with two YY1 molecules. Qin et al. (23) have suggested that coactivator CBP assumes different conformational changes according to the number of transcription factors bound to it. In the case of IFN-β transcriptional activation, it is possible that the “functionally correct” conformation of CBP could be reached only when interaction is with at least two YY1 molecules.
Of the two YY1 binding sites, the −122 site is the weaker one and therefore the one most likely to be modulated. Several factors, such as acetylation, phosphorylation, and protein-protein interactions, are capable of modifying YY1 DNA-binding affinity. Even though these factors could affect the binding of YY1 to either the −90 or the −122 site, the effect would be expected to be more pronounced in the case of the −122 site, which is the weaker one. A strong HMGI binding site overlaps the YY1 −122 binding site, whereas only a weak HMGI site overlaps the YY1 −90 site. In future work, it would be interesting to analyze the effect of protein HMGI on YY1 binding to the muIFN-β promoter.
Going against the IRF3-CBP dogma during the regulation of the IFN-β promoter.
In this work we have demonstrated that YY1 plays a determinant role during regulation of IFN-β gene expression. Intact YY1 binding sites present at positions −122 and −90 are essential for CBP recruitment and K8H4/K14H3 acetylation on the proximal region of the muIFN-β promoter.
Until now an essential role for virus-induced IFN-β promoter activation has been ascribed to factor IRF3. It has been often suggested not only that IRF3 is the main factor responsible of CBP recruitment on the IFN-β promoter but also that IRF3 and CBP associate prior to binding to the IFN-β promoter (31).
The results we present here clearly demonstrate that IRF3 is not the main factor responsible for CBP recruitment on the IFN-β promoter. Promoter mut122, carrying a single point mutation on the core motif of the YY1 −122 binding site, was not able to allow CBP recruitment notwithstanding IRF3 binding to this promoter, indicating not only that IRF3 was not the main factor responsible of CBP recruitment on the muIFN-β promoter but also that IRF3 could bind to the promoter in the absence of CBP.
Results obtained during ChIP assays with anit-IRF3 antibody and mut90 promoter showing the inability of IRF3 to interact with the promoter carrying a mutation on the YY1 −90 binding site were confirmed during gel retardation experiments. In vitro binding of IRF3 present in nuclear extracts prepared from NDV-infected cells to probe 90, which contains a single IRF3 binding site at its 3′ end, also appeared to require that the YY1-binding site present at position −90 be intact. Overall these results open up the possibility of a “cross talk” between YY1 and IRF3 during binding to the −90 region of the murine IFN-β promoter. Nevertheless, we believe that no definite conclusion can for the moment be reached concerning this potentially interesting point before carrying out further experiments.
Viruses have developed different strategies to counteract the IFN-β-dependent antiviral response. Some viruses have developed strategies to stop IFN-β mRNA synthesis by functionally inhibiting IRF3 and IRF7 factors, which are the main activators of the IFN-α genes (18) whereas other viruses, like the Rift Valley fever virus (a phlebovirus of the family Bunyaviridae transmitted by mosquitoes) specifically blocks IFN-β transcription by a yet-unknown mechanism without functionally counteracting or inhibiting virus-induced activation of virus-responsive element factors such as IRF3, NF-κB, and AP-1 (4). By identifying YY1 as one of the main factors regulating the transcriptional capacity of the IFN-β promoter, our results open up new axis of investigation that could lead us to decipher strategies developed by some viruses to specifically block IFN-β transcription in order to counteract the host antiviral response.
Acknowledgments
We are grateful to Thibault Mesplede for preparation of nuclear extracts from NDV-infected L929 cells as well as for fruitful discussions and to Martin Montecino and Roberto Paredes (University of Concepcion, Chile) for the gift of plasmid pGEX-YY1.
This work was supported by the Centre National de la Recherche Scientifique and by grants from the Association pour la Recherche sur le Cancer (ARC 3236), Ligue National contre le Cancer (580054), and Agence Nationale pour la Recherche (ANR-05-MIIM-033-02).
Footnotes
Published ahead of print on 5 September 2006.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381-392. [DOI] [PubMed] [Google Scholar]
- 3.Austen, M., B. Luscher, and J. M. Luscher-Firzlaff. 1997. Characterization of the transcriptional regulator YY1. J. Biol. Chem. 272:1709-1717. [DOI] [PubMed] [Google Scholar]
- 4.Billecocq, A., M. Spiegel, P. Vialat, A. Kohl, F. Weber, M. Bouloy, and O. Haller. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 78:9798-9806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnefoy, E., M.-T. Bandu, and J. Doly. 1999. Specific binding of high-mobility-group I (HMGI) protein and histone H1 to the upstream AT-rich region of the murine beta interferon promoter: HMGI protein acts as a potential antirepressor of the promoter. Mol. Cell. Biol. 19:2803-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bushmeyer, S., K. Park, and M. L. Atchison. 1995. Characterization of functional domains within the multifunctional transcription factor YY1. J. Biol. Chem. 270:30213-30220. [DOI] [PubMed] [Google Scholar]
- 7.Coull, J. J., F. Romerio, J.-M. Sun, J. L. Volker, K. M. Galvin, J. R. Davie, Y. Shi, U. Hansen, and D. M. Margolis. 2000. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J. Virol. 74:6790-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeMayer, E., and J. DeMayer-Guignard. 1988. Interferons and other regulatory cytokines. John Wiley & Sons, New York, N.Y.
- 9.Galvin, K. M., and Y. Shi. 1997. Multiple mechanisms of transcriptional repression by YY1. Mol. Cell. Biol. 17:3723-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higashi, Y. 1985. Changes of chromatin conformation around mouse interferon-β gene associated with induction of interferon synthesis. Nucleic Acids Res. 13:5157-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiscott, J., P. Pitha, P. Genin, H. Nguyen, C. Heylbroeck, Y. Mamane, M. Algarte, and R. Lin. 1999. Triggering the interferon response: the role of IRF-3 transcription factor. J. Interferon Cytokine Res. 19:1-13. [DOI] [PubMed] [Google Scholar]
- 12.Hyde-DeRuyscher, R. P., E. Jennings, and T. Shenk. 1995. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 23:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobsen, B. M., and D. G. Skalnik. 1999. YY1 binds five cis-elements and trans-activates the myeloid cell-restricted gp91phox promoter. J. Biol. Chem. 274:29984-29993. [DOI] [PubMed] [Google Scholar]
- 14.Klar, M., and J. Bode. 2005. Enhanceosome formation over the beta interferon promoter underlies a remote-control mechanism mediated by YY1 and YY2. Mol. Cell. Biol. 25:10159-10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, T.-C., Y. Zhang, and R. J. Schwartz. 1994. Bifunctional transcriptional properties of YY1 in regulating muscle actin and c-myc gene expression during myogenesis. Oncogene 9:1047-1052. [PubMed] [Google Scholar]
- 16.Lewis, B. L., G. Tullis, E. Seto, N. Horikoshi, R. Weinmann, and T. Shenk. 1995. Adenovirus E1A proteins interact with the cellular YY1 transcription factor. J. Virol. 69:1628-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mesplede, T., S. Navarro, P. Genin, P. Morin, M.-L. Island, E. Bonnefoy, and A. Civas. 2003. Positive and negative control of virus-induced interferon-A gene expression. Autoimmunity 36:447-455. [DOI] [PubMed] [Google Scholar]
- 19.Munshi, N., M. Merika, J. Ye, K. Senger, G. Chen, and D. Thanos. 1998. Acetylation of HMGI(Y) by CBP turns off IFN-β expression by disrupting the enhanceosome. Mol. Cell 2:457-467. [DOI] [PubMed] [Google Scholar]
- 20.Natesan, S., and M. Z. Gilman. 1993. DNA bending and orientation-dependent function of YY1 in the c-fos promoter. Genes Dev. 7:2497-2509. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen, N., X. Zhang, N. Olashaw, and E. Seto. 2004. Molecular cloning and functional characterization of the transcription factor YY2. J. Biol. Chem. 279:25927-25934. [DOI] [PubMed] [Google Scholar]
- 22.Parekh, B. S., and T. Maniatis. 1999. Virus infection leads to localized hyperacetylation of histone H3 and H4 at the IFN-β promoter. Mol. Cell 3:125-129. [DOI] [PubMed] [Google Scholar]
- 23.Qin, B. Y., C. Liu, H. Srinath, S. S. Lam, J. J. Correia, R. Derynck, and K. Lin. 2005. Crystal structure of IRF-3 in complex with CBP. Structure 13:1269-1277. [DOI] [PubMed] [Google Scholar]
- 24.Riggs, K. J., S. Saleque, K.-K. Wong, K. T. Merell, J.-S. Lee, Y. Shi, and K. Calame. 1993. Yin-Yang 1 activates the c-myc promoter. Mol. Cell. Biol. 13:7487-7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, M. Nakaya, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 26.Seto, E., B. Lewis, and T. Shenk. 1993. Interaction between transcription factors Sp1 and YY1. Nature 365:462-464. [DOI] [PubMed] [Google Scholar]
- 27.Shestakova, E., M.-T. Bandu, J. Doly, and E. Bonnefoy. 2001. Inhibition of histone deacetylation induces constitutive derepression of the beta interferon promoter and confers antiviral activity. J. Virol. 75:3444-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shestakova, E., Z. Mansuroglu, H. Mokrani, N. Ghinea, and E. Bonnefoy. 2004. Transcription factor YY1 associates with pericentromeric γ-satellite DNA in cycling but not in quiescent (G0) cells. Nucleic Acids Res. 32:4390-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shrivastava, A., and L. Calame. 1994. An analysis of genes regulated by multi-functional transcriptional regulator Yin Yang-1. Nucleic Acids Res. 22:5151-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stark, G. R., I. M. Kerr, B. R. G. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 31.Suhara, W., M. Yoneyama, I. Kitabayashi, and T. Fujita. 2002. Direct involvement of CREB-binding protein/p300 in sequence-specific DNA binding of virus-activated interferon regulatory factor-3 holocomplex. J. Biol. Chem. 277:22304-22313. [DOI] [PubMed] [Google Scholar]
- 32.Thomas, M. J., and E. Seto. 1999. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene 236:197-208. [DOI] [PubMed] [Google Scholar]
- 33.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 34.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulated factor 3 and CREB-binding protein /p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weill, L., E. Shestakova, and E. Bonnefoy. 2003. Transcription factor YY1 binds to the murine interferon-beta promoter and regulates its transcriptional capacity with a dual activator/repressor role. J. Virol. 77:2903-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whittemore, L. A., and Maniatis, T. 1990. Postinduction repression of the β-interferon gene is mediated through two positive regulatory domains. Proc. Natl. Acad. Sci. USA. 87:7799-7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao, Y.-L., W.-M. Yang, and E. Seto. 2001. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 21:5979-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye, J., M. Cippitelli, L. Dorman, J. R. Ortaldo, and H. A. Young. 1996. The nuclear factor YY1 suppresses the human gamma interferon promoter through two mechanisms: inhibition of AP1 binding and activation of a silencer element. Mol. Cell. Biol. 16:4744-4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]