Abstract
Landmark features of imprinted genes are differentially methylated domains (DMDs), in which one parental allele is methylated on CpG dinucleotides and the opposite allele is unmethylated. Genetic experiments in the mouse have shown that DMDs are required for the parent-specific expression of linked clusters of imprinted genes. To understand the mechanism whereby the differential methylation is established and maintained, we analyzed a series of transgenes containing DMD sequences and showed that imperfect tandem repeats from DMDs associated with the Snurf/Snrpn, Kcnq1, and Igf2r gene clusters govern transgene imprinting. For the Igf2r DMD the minimal imprinting signal is two unit copies of the tandem repeat. This imprinted transgene behaves identically to endogenous imprinted genes in Dnmt1o and Dnmt3L mutant mouse backgrounds. The primary function of the imprinting signal within the transgene DMD is to maintain, during embryogenesis and a critical period of genomic reprogramming, parent-specific DNA methylation states established in the germ line. This work advances our understanding of the imprinting mechanism by defining a genomic signal that dependably perpetuates an epigenetic state during postzygotic development.
Imprinted genes are defined by a difference in expression between the two parental alleles (20). For a typical imprinted gene, one parental allele is silenced, and the opposite parental allele is expressed. Genetic studies in humans and mice have identified imprinting centers (ICs) within clusters of imprinted genes that control the differential expression of linked genes (3, 4, 10, 18, 25, 28). Targeted deletion of the Igf2r (insulin-like growth factor type 2 receptor gene) IC in the mouse leads to the loss of imprinting of a cluster of linked imprinted genes on chromosome 17. Similar findings of targeted deletions in the mouse, or spontaneous deletions in the human, led to the identification of Snurf/Snrpn and Kcnq1 ICs. The Snurf/Snrpn IC is linked to the regulation of three imprinted genes, the Kcnq1 IC is linked to the regulation of seven imprinted genes, and the Igf2r IC is important for the monoallelic expression of four genes.
The aforementioned ICs coincide with differentially methylated domains (DMDs) (11, 23, 24). Prominent DMDs associated with the Snurf/Snrpn, Kcnq1 (called KvDMR), and Igf2r (called DMD2) gene clusters in the mouse are highly methylated on the maternal allele and unmethylated on the paternal allele. There is strong evidence that these different epigenetic states are first established in the gametes and then maintained during embryogenesis (9, 14, 26). The DMD sequences of the Snurf/Snrpn, Igf2r, and Kcnq1 genes have several common features including promoter elements, CpG islands, and imperfect tandem repeats (Fig. 1A; CpG islands not shown). These three DMDs contain six to nine tandem repeats, ranging in size from 18 to 170 bp and containing 1 to 14 CpG dinucleotides per repeat copy (Fig. 1A) (20). Within a given DMD the repeated copies are, on average, 50% to 73% similar to one another, and the array of tandem repeats spans 600 bp to 700 bp of genomic sequence (including intervening sequence). There is no obvious sequence similarity among the three mouse DMDs. To determine the roles of these DMD features in genomic imprinting, DMD sequences were analyzed in a mouse model system based on the imprinted RSVIgmyc transgene (Fig. 1B) (5, 6, 21).
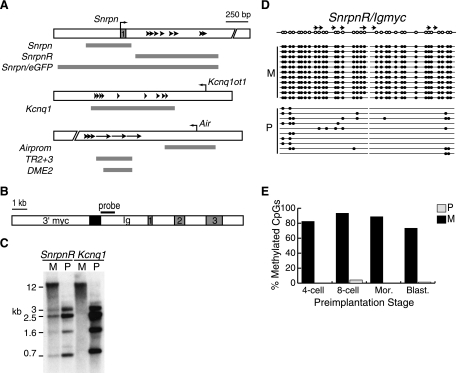
FIG. 1.
DMD sequences containing tandem repeats restore maternal-specific methylation to the Ig/myc transgene. (A) Schematics of the mouse Snurf/Snrpn, Kcnq1, and Igf2r DMDs (white boxes). The Snrpn DMD contains the promoter and exon 1 of the Snurf/Snrpn locus, and the Kcnq1 and Igf2r DMDs contain promoters for untranslated antisense transcripts (bent arrows). Arrowheads or arrows within each DMD indicate tandem repeats (different for each DMD). Gray bars represent the regions analyzed in Ig/myc transgenes in this study and previously (21). The Kcnq1 region analyzed in this study included the conserved repeats examined by Mancini-Dinardo et al. plus additional DMD repeats (15). (B) Design of transgenes analyzed in this study. The Cα and Sα regions of the mouse IgA locus (Ig) and the 3′ untranslated region and c-myc exons (1-3) are shown. A black bar depicts the probe for the Southern blots. The DMD sequences depicted by gray bars in panel A were subcloned into an EcoRI site (flanking the black box). (C) Southern blot analysis of DNAs obtained from maternal and paternal carriers of each transgene. DNAs were digested with HpaII and hybridized with the probe shown in panel B. Band sizes are shown in kilobases (kb). SnrpnR, SnrpnR/Igmyc; Kcnq1, Kcnq1/Igmyc. (D) Summary of bisulfite genomic sequencing analyses performed on DNAs obtained from maternal and paternal carriers of the SnrpnR/Igmyc transgene. The top line shows the location of tandem repeats (arrows) and the CpGs analyzed (open circles). Each line below represents one sequenced allele. Filled circles indicate the positions of methylated CpGs. (E) Summary of bisulfite genomic sequencing data obtained from preimplantation stage embryos following maternal or paternal inheritance of the SnrpnR/Igmyc transgene. Mor, morulae; Blast, blastocysts. For the paternal allele only the eight-cell and blastocyst stages were examined. Each bar represents the percentage of CpGs found to be methylated in all of the subclones analyzed. M, maternal; P, paternal.
We chose the RSVIgmyc transgene model system for a number of reasons. Targeted mutagenesis to remove entire DMDs has identified them as regulatory sequences that physically overlap ICs. However, the use of targeted deletions to more specifically define imprinting elements within a DMD is limited because of potential difficulties in interpreting the effects of small sequence deletions. More importantly, because the three aforementioned DMDs are largely comprised of imperfect tandem repeats, deletions of critical portions of tandem repeats could result in persistence of imprinting in the deleted mice because of a redundancy of function encoded by the repeats.
An alternative approach to analyzing small sequence elements for their imprinting role is to move the sequences into a heterologous context. This is a standard, time-honored approach for studying and defining cis-regulatory sequences, and many investigators have applied such an approach to genomic imprinting by generating mouse transgenes. Generally, large bacterial artificial chromosome- or yeast artificial chromosome-sized transgenes centered over DMDs are consistently imprinted with the same parent-specific methylation and transcriptional expression as the endogenous gene (1, 27). Attempts to develop much smaller, consistently imprinted transgenes from the same endogenous imprinted genes that contain DMDs have not succeeded (19, 27). Presumably, a critical non-DMD sequence(s) or the appropriate genomic context is lost in the smaller transgenes. If consistently imprinted, these smaller transgenes would be manageable experimental systems to study the function of putative imprinting sequences from many different genes. For these reasons, we developed a transgene system based on the small RSVIgmyc transgene to identify functionally important imprinting sequences (5, 6, 21).
We previously showed that a portion of the Igf2r DMD2 containing tandem repeats restored imprinting to the Ig/myc transgene, a nonimprinted version of RSVIgmyc in which its DMD sequences have been excised (21). Other DMD sequences, particularly the promoter region of the Snurf/Snrpn gene, were not able to restore transgene imprinting. These findings on the analysis of transgenes comprised solely of mouse genomic sequences suggest that the tandem repeats found within mouse DMDs are cardinal elements of the imprinting mechanism (21). To explore this issue further, we generated additional Igf2r- and Snurf/Snrpn-containing transgenes, as well as Kcnq1-containing transgenes, and studied their parent-specific methylation and expression in mice.
MATERIALS AND METHODS
Transgene constructs.
All transgene constructs were derivatives of the RSVIgmyc transgene and were generated as previously described (21). All transgenes, with the exception of Snrpn/eGFP, were generated by the following method. The Ig/myc portion of the RSVIgmyc transgene was obtained by removal of an EcoRI fragment containing the pBR322 and Rous sarcoma virus (RSV) sequences. The unique EcoRI site was used to insert all DMD sequences. The DMD sequences were amplified by PCR from mouse genomic DNA with oligonucleotide primers containing EcoRI restriction sites, and EcoRI restriction fragments were subcloned into the Ig/myc transgene. The Snrpn/eGFP transgene was generated by removal of the region encompassing pBR322, RSV, Ig, and exon 1 of c-myc. The Snurf/Snrpn promoter, exon 1, and tandem repeats were inserted upstream of c-myc exon 2, and the enhanced green fluorescent protein (eGFP) coding sequence was placed in frame with c-myc. The TR2+3/Igmyc transgene was previously described (21). The following primers were used: for SnrpnRIgmyc, GAGAATTCGTGAGCAATCCTTTGG (forward) and CTGAATTCCTTGAAGCCACATGAAG (reverse); for Kcnq1/Igmyc, GGTGAATTCTGGTCCAGTCAGG (forward) and AAGAATTCCCCTGCTTCTGTAAG (reverse); for DME2/Igmyc, CTCGGAATTCCTCCGAGCCGTCT (forward) and CTCGAATTCTGCTGTGCGATTCTGG (reverse); for DME1.5L/Igmyc, TGAATTCAGAACCCTTCGAATC (forward) and AGGGGAATTCAAAGCTCAGAGGG (reverse); for DME1.5C/ Igmyc, CTCGGAATTCCTCCGAGCCGTCT (forward) and AGGAGGGAATTCGGAGGGTTTTAGAGGG (reverse); for DME1/Igmyc transgene, GCTCACGAATTCCCGCGGAACCCTC (forward) and AGGAGGGAATTCGGAGGGTTTTAGAGGG (reverse); for Snrpn/eGFP, GAGCTGGAATTCCTGGATGGCATT (forward) and GCAACAGAGCTCCCAGAATTCC (reverse).
Transgenic mice.
Transgene fragments for injection were removed from the pKS+ plasmid by NotI digestion and gel isolated using a QiaexII kit (QIAGEN). Fragments for injection were resuspended to a concentration of 5 μg/ml in TE buffer (5 mM Tris-Cl, pH 8, 0.2 mM EDTA, pH 8) and injected into pronuclei of fertilized eggs. All transgenic mouse lines were generated in the inbred FVB/N strain background (Taconic).
Southern blot analysis of DNA methylation.
Genomic DNAs were obtained from tail biopsies performed at the time of weaning (3 to 4 weeks of age) (21). Resulting DNAs were digested with HpaII (New England Biolabs), electrophoresed on a 1% agarose gel, and transferred to Genescreen nylon membrane (NEN, Boston, MA). DNAs were hybridized with a 32P-labeled probe to the Cα region of the transgene (Fig. 1B). Southern blots were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 0.1% sodium dodecyl sulfate at room temperature and with 0.1× SSC with 0.1% sodium dodecyl sulfate at 65°C.
Bisulfite genomic sequencing.
Bisulfite genomic sequencing was performed as previously described (7). Following bisulfite treatment, the DMD sequences were amplified with PCR primers flanking the DMD (within the Ig and c-myc sequences). This design allowed all transgene constructs, with the exception of the Snrpn/eGFP transgene, to be amplified with the same primers. Transgene-specific primers were also designed for the Snrpn/eGFP construct. Primer sequences are available upon request.
Collection of preimplantation embryos.
Preimplantation embryos were collected in M2 medium (Specialty Media) following natural matings. Blastocyst stage embryos were flushed from the uterine horns at 3.5 days postcoitum (dpc). Preimplantation embryos at the two-cell stage were removed from the oviducts at 1.5 dpc; four-cell embryos, eight-cell embryos, and morulae were isolated from the oviducts between 2 to 3 dpc. Meiosis II oocytes were collected from the oviducts of superovulated female mice 24 h after injection with human choriogonadotropin. Hyaluronidase was used to remove cumulus cells. All oocytes and preimplantation embryos were washed in M2 medium followed by 1× phosphate-buffered saline. DNA was collected from pools of 30 to 100 preimplantation embryos by proteinase K digestion.
RNA isolation and RT-PCR.
Brains were dissected from adult mice and stored in RNALater stabilization reagent (QIAGEN). Brain tissue was homogenized using a Minibeadbeater (BioSpec Products) and zirconia/silica beads (BioSpec Products). Total RNA was extracted using an RNeasy MiniKit (QIAGEN). Reverse transcription was performed on 1 μg of total brain RNA, using oligo(T) hexamers and 200 U of Moloney murine leukemia virus reverse transcriptase (Promega) at 37°C for 1 h. The eGFP/cMyc transcript was PCR amplified from brain cDNA with Taq Polymerase (Invitrogen). The primers were Snex1a (GATGCCAGACGCTTGGTTCTG) and Mex2b (CGTAGCGACCGCAACATAGG). The hypoxanthine guanine phosphoribosyl transferase (Hprt) transcript was PCR amplified as a positive control with the primers HprtF (TCTCATGCCGACCCGCAGTCC) and HprtR (ATTCAACTTGCGCTCATCTTA).
RESULTS
Tandem repeats from other DMDs restore Ig/myc imprinting.
Because tandem repeats of the Igf2r DMD restored Ig/myc imprinting and because the promoter sequences of the Snurf/Snrpn DMD were unable to restore Ig/myc imprinting, we examined the effects of introducing the Igf2r DMD (Air) promoter and the Snurf/Snrpn DMD tandem repeats on Ig/myc imprinting. We constructed the SnrpnR/Igmyc transgene in the same manner as the previously analyzed Igf2r-containing transgenes (Fig. 1A and B) (21). When analyzed by Southern blotting of transgenic mouse DNA digested with the methylation-sensitive restriction enzyme HpaII, the maternal SnrpnR/Igmyc alleles were always methylated, and the paternal alleles were always undermethylated (Fig. 1C). These results were seen in each of three transgenic lines. We conclude from this that the SnrpnR/Igmyc transgene is consistently imprinted in the mouse.
To demonstrate that the SnrpnR/Igmyc parental allele methylation differences evident on Southern blots (Fig. 1C) reflect methylation differences within the SnrpnR/Igmyc tandem repeats, we analyzed the methylation of the tandem repeats using the bisulfite genomic sequencing method. Bisulfite genomic sequencing primers were designed within the transgene sequences flanking the repeats to distinguish the transgene DMD sequences from endogenous Snurf/Snrpn sequence. One of the three SnrpnR/Igmyc lines was analyzed in this way, and its DMD-derived repeat sequences were differentially methylated (Fig. 1D). In adult tissue the maternal alleles of the transgene were methylated at 99.7% of CpG dinucleotides analyzed. In contrast, only 10.5% of CpGs on paternally inherited transgene alleles were methylated.
The parent-specific methylation observed on the SnrpnR/Igmyc transgene DMD in adult tissues was also seen in preimplantation embryos. The DMD of the maternally inherited transgene was methylated in all stages examined, namely, four-cell embryos, eight-cell embryos, morulae, and blastocysts (Fig. 1E). Because endogenous Snurf/Snrpn and RSVIgmyc DMD methylation is established in oocytes, we interpret these results to indicate that the imprinted SnrpnR/Igmyc transgene derives its maternal allele methylation from the gamete and then maintains it throughout preimplantation development. In contrast, the DMD of the paternally inherited SnrpnR/Igmyc transgene was unmethylated in all stages examined, namely, the eight-cell and blastocyst stages. Methylation on the paternal transgene allele was not examined in other preimplantation stages or in sperm. These data are consistent with the notion that gamete-derived methylation is actively maintained during preimplantation development, a developmental period in which much genomic, but not DMD, methylation is lost (6, 16).
To further test the role of DMD-associated tandem repeats in genomic imprinting, the repeat region from the Kcnq1 DMD was used to generate the Kcnq1/Igmyc transgene. Maternal Kcnq1/Igmyc alleles were methylated, and paternal alleles were undermethylated when analyzed by Southern blotting (Fig. 1C). This pattern of transgene imprinting was also found in a separate transgenic mouse line generated with the same transgene construct (data not shown). Imprinting of the Kcnq1/Igmyc transgene was not affected by the orientation of the tandem repeat array within the Ig/myc transgene, as the opposite orientation of the Kcnq1 tandem repeats produced consistently imprinted transgenes (data not shown). We conclude from the imprinting of Snrpn- and Kcnq1-containing transgenes that tandem repeats are likely important for parent-specific methylation of DMD sequences.
Snurf/Snrpn tandem repeats control expression of a linker reporter gene.
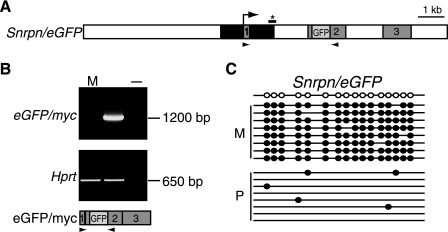
To determine if methylation on the Snurf/Snrpn repeat sequences is able to silence a linked promoter, a genomic fragment containing the Snurf/Snrpn first exon, 800 bp of upstream sequence, and the tandem repeats was coupled to a genomic c-myc reporter construct. GFP coding sequence was inserted in frame into c-myc coding sequence to distinguish the transgene-specific transcript from endogenous c-myc expression (Fig. 2A). One Snrpn/eGFP transgenic line was created and analyzed for expression and methylation. The transgene-specific transcript predicted by splicing of Snurf/Snrpn and c-myc exons is shown in Fig. 2B. A paternal-specific transcript was detected in adult brain of transgenic mice, indicating that the transgene was imprinted (Fig. 2B). We used bisulfite genomic sequencing to determine the methylation of transgene Snurf/Snrpn repeats adjacent to c-myc sequences; only the silenced maternal allele of the Snrpn/eGFP transgene was methylated in this region (Fig. 2C). Thus, the tandem repeats and promoter of a DMD can establish an imprinted transcriptional unit when moved to a heterologous position 5′ of a c-myc reporter gene. We conclude that the Snurf/Snrpn intronic tandem repeats are a key element for imprinting the reporter construct for a number of reasons: the endogenous c-myc gene is not imprinted (8); deletion of the endogenous Snurf/Snrpn promoter had no effect on imprinting within the gene cluster (4); the Snurf/Snrpn promoter alone in the Ig/myc transgene was not imprinted (21); and the Snurf/Snrpn tandem repeats in the Ig/myc transgene were consistently imprinted (Fig. 1).
FIG. 2.
A transgene containing the Snrpn promoter and tandem repeats shows maternal-specific methylation and paternal-specific expression of an eGFP/myc fusion transcript. (A) The transgene is a derivative of the transgene shown in Fig. 1B. The black box indicates Snrpn sequence (exon 1 of Snrpn is shown). Exon 2 of c-myc contains an in-frame fusion of the entire eGFP protein-coding region (GFP). Arrowheads represent primers used for RT-PCR. (B) RT-PCR of cDNAs obtained from adult brain of transgenic mice. M, maternal inheritance; P, paternal inheritance; −, negative control. Paternal-specific eGFP/myc expression (top) and control amplification of the Hprt gene (bottom) are shown. The transgene-specific spliced transcript is shown below. (C) Bisulfite genomic sequencing of maternal (M) and paternal (P) alleles of the transgene (as described in the legend of Fig. 1). Position of sequence amplified by PCR with modified, semi-nested primers is shown in panel A (*).
The Igf2r tandem repeats and not the Air promoter are required for transgene imprinting.
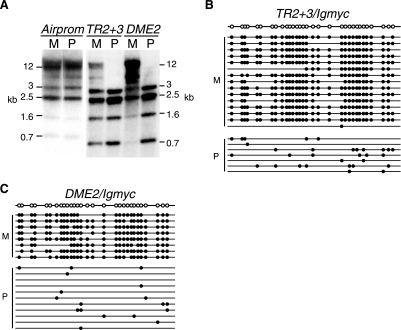
An Airprom/Igmyc transgene was created that contained a 550-bp region of the Igf2r DMD2 encompassing the Air transcription start site and promoter (Fig. 1A). One Airprom/Igmyc transgenic line was generated. Maternal and paternal transgene alleles were analyzed by Southern blotting for methylation differences (Fig. 3A). Both parental alleles of the Airprom/Igmyc transgene were equally methylated. Thus, the Air promoter does not confer imprinting to the Ig/myc transgene.
FIG. 3.
The TR2+3 repeat from the Igf2r DMD, but not the Air promoter, is able to create an imprinted transgene locus. (A) Southern blot analysis of DNAs obtained from maternal and paternal carriers of each transgene construct. DNAs were digested with HpaII and hybridized with the probe depicted in Fig. 1B. Band sizes are shown in kilobases (kb). Airprom, Airprom/Igmyc; TR2+3, TR2+3/Igmyc; DME2, DME2/Igmyc. Positions within the DMD are shown in Fig. 1A. (B and C) Summary of bisulfite genomic sequencing analyses performed on DNAs obtained from maternal and paternal carriers of the TR2+3/Igmyc (B) and DME2/Igmyc (C) transgenes (as described in the legend of Fig. 1). M, maternal; P, paternal.
We had previously shown that 2.5 unit copies of the TR2+3 repeat within the Igf2r DMD2 was differentially methylated in one transgenic line (17, 21). To confirm the imprinting function of the TR2+3 repeats, two additional TR2+3/Igmyc lines were generated and tested for parental allele-specific differences in methylation (Fig. 1A). Both new lines were imprinted when maternal and paternal allele methylation patterns were compared using HpaII digests or bisulfite genomic sequencing (Fig. 3A and B). These data demonstrate that we have identified sequences in the Igf2r DMD2 that are able to generate a maternally methylated, imprinted transgene locus. Together, these observations indicate that the Igf2r tandem repeats, and not the promoter, are capable of creating a maternally methylated transgene locus. The ability of tandem repeat sequences from the Igf2r, Snurf/Snrpn, and Kcnq1 DMDs to restore allelic methylation differences to the Ig/myc and eGFP/myc transgenes is the first demonstration of an essential imprinting function for a feature present in multiple DMDs.
Identification of a minimal Igf2r DMD imprinting element.
We have designated the 459-bp region of Igf2r that possesses the ability to establish an imprinted transgene locus the differential methylation element (DME) of the Igf2r DMD2. The DME was used as a starting point to define the minimal Igf2r DMD sequence needed to generate an imprint. Five transgenes were generated that eliminated portions of the DME sequence (Fig. 1A and Fig. 4A). At the 5′ end of the DME, an 83-bp segment that contained two CpG dinucleotides was removed, leaving a sequence equivalent to two unit copies of the TR2+3 repeat. In each of three independently generated DME2/Igmyc transgenic lines, maternal alleles were heavily methylated, and paternal alleles were undermethylated (Fig. 3A and C). Thus, the DME2/Igmyc transgene retained the imprinting function of the DME.
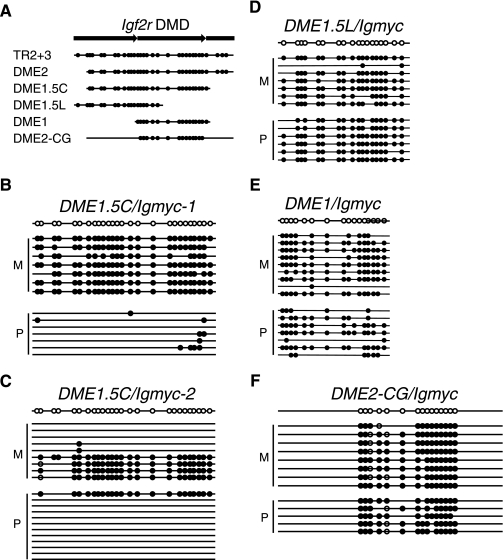
FIG. 4.
Igf2r DMD sequences smaller than two unit copies of the TR2+3 repeat are not capable of creating an imprinted transgene locus. (A) Arrows depict the TR2+3 repeats of the Igf2r DMD2. Lines below indicate the DNA fragments used to create several hybrid transgenes. Filled circles represent the number and approximate location of CpGs (not to scale). All transgenes were constructed as described in the legend of Fig. 1. (B to F) Bisulfite genomic sequencing analysis of the DMD region of each transgene. For each panel bisulfite genomic sequencing was performed as described in the legend of Fig. 1D.
The removal of an additional 73 bp of sequence that contained three CpG dinucleotides from the 3′ end of DME2 disrupted transgene imprinting (Fig. 4B and C). This DME1.5C/Igmyc transgene contained just over 1.5 copies of the TR2+3 repeat in the center of the DME. In a single transgenic line, paternal alleles were largely unmethylated, and methylation on maternal alleles was variable; some maternal alleles were completely methylated, and the remaining alleles were completely unmethylated. In a second DME1.5C/Igmyc line, maternal alleles were methylated, and paternal alleles were unmethylated. These results suggested that the DME1.5C/Igmyc transgene retained some aspects of the imprinting function of the DME, but it was not consistently imprinted.
Two additional transgenes clearly lost the imprinting function of the DME. The DME1.5L/Igmyc transgene contained just under 1.5 unit copies of the TR2+3 repeat at the 5′ end of the DME, and the DME1/Igmyc transgene contained a single complete unit copy of the TR2+3 repeat at the center of the DME. Two transgenic lines were generated for each construct. In all four transgenic lines, the maternal and paternal transgene alleles acquired comparable levels of methylation (Fig. 4D and E). Therefore, we conclude that there is no identifiable sequence element smaller than the 376-bp DME2 that completely restores Ig/myc imprinting. Different sequences were removed from DME2 in generating the DME1/Igmyc, the DME1.5L/gmyc, and the DME1.5C/Igmyc transgenes, yet these transgenes were either not imprinted or only partially imprinted.
DME2 CpG dinucleotides are required for transgene imprinting.
The imprinting function of the DME2/Igmyc transgene was unambiguously lost when it was reduced to one unit copy (DME1/Igmyc). This result could be due to a loss of one unit copy of the repeat or to a loss of CpG dinucleotides commensurate with the reduction in size. Based on this reasoning, we constructed a version of the DME2 transgene that eliminated a portion of its CpG dinucleotides but retained its size. The CpG dinucleotides within the central DME1 sequence were retained, and the remaining 15 CpGs were changed to TpGs by site-directed mutagenesis to generate the DME2-CG/Igmyc transgene. This left a DME2-CG sequence that contained 15 fewer CpG dinucleotides but was otherwise identical to the original DME2 (95% of the sequence remained unchanged). One DME2-CG/Igmyc transgenic line was created and analyzed for allele-specific methylation by bisulfite genomic sequencing (Fig. 4F). Both maternal and paternal transgene alleles showed comparable levels of methylation. These results are consistent with a specific CpG dinucleotide requirement for DME2/Igmyc imprinting and suggest that a DME2 transgene that keeps all CpG dinucleotides, but eliminates tandem repetitiveness, would remain imprinted.
Dnmt1o and Dnmt3L are required for DME2/Igmyc imprinting.
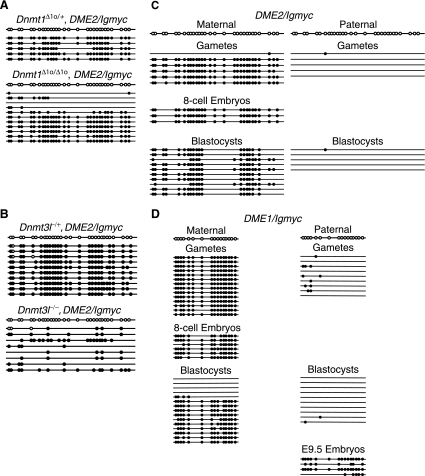
To determine if the same molecular machinery controlling the imprinting of endogenous genes governs DME2/Igmyc imprinting, we studied the effect of Dnmt1o methyltransferase deficiency and Dnmt3L protein deficiency on DME2/Igmyc imprinting (2, 12, 13). Homozygous Dnmt1Δ1o/Δ1o female mice or heterozygous Dnmt1Δ1o/+ female mice carrying the DME2/Igmyc transgene were mated to wild-type FVB/N males, and methylation of the transgene was analyzed in embryonic day 9.5 (E9.5) embryos. In an embryo obtained from a Dnmt1Δ1o/+ heterozygous female, the maternal DME2/Igmyc allele was methylated on every allele examined (Fig. 5A, top). In contrast, in an embryo obtained from a Dnmt1Δ1o/Δ1o homozygous mutant female, there were two types of maternal alleles (Fig. 5A, bottom). Seven of 11 sequenced alleles had the high level of methylation normally seen on a wild-type Dnmt1 background. No methylation, or a very low level of methylation, was seen on the remaining maternal DME2/Igmyc alleles. Such a distribution of methylated and unmethylated maternal alleles was also seen at endogenous imprinted loci in embryos that developed in the absence of Dnmt1o protein. We conclude from this analysis that DME2/Igmyc imprinting requires Dnmt1o protein from the oocyte, much as oocyte-derived Dnmt1o is required for the imprinting of endogenous genes.
FIG. 5.
Transgene methylation in mutant mouse backgrounds and during preimplantation development. For each panel bisulfite genomic sequencing was performed as described in the legend of Fig. 1D. (A) The DME2/Igmyc transgene was placed into the Dnmt1Δ1o mutant mouse background, and methylation of the DME2/Igmyc DMD was analyzed in E9.5 transgenic embryos derived from heterozygous Dnmt1Δ1o/+ (top) or homozygous Dnmt1Δ1o/Δ1o (bottom) transgenic females. (B) The DME2/Igmyc transgene was placed into the Dnmt3l mutant background, and methylation of the DME2/Igmyc DMD was analyzed in E9.5 transgenic embryos derived from heterozygous Dnmt3l−/+ (top) or homozygous Dnmt3l−/− (bottom) transgenic females. (C and D) Bisulfite genomic sequencing of transgene DMD methylation for DME2/Igmyc (C) and DME1/Igmyc (D). Methylation of maternal transgene alleles was compared to paternal transgene alleles. Not all stages were analyzed for the paternal allele.
The Dnmt3L protein is required for the establishment of maternal-specific methylation in oocytes, specifically at imprinted gene DMDs (2, 12). Homozygous Dnmt3l−/− mice are viable, but males and females are infertile. Embryos from homozygous Dnmt3l−/− mothers died by E10.5 of gestation and showed nearly a complete loss of methylation at maternally methylated imprinted gene DMDs. Homozygous Dnmt3l−/− female mice carrying the DME2/Igmyc transgene were mated to wild-type males, and embryos were collected at E9.5 of gestation. Heterozygous Dnmt3l−/+female mice carrying the DME2/Igmyc transgene were mated to wild-type males, and E9.5 embryos were used as controls. DNA was collected from individual embryos and analyzed by bisulfite genomic sequencing. In a control embryo, the transgene was nearly completely methylated on all alleles examined (Fig. 5B, top). In contrast, methylation on the transgene in an embryo obtained from the homozygous Dnmt3l−/− female was almost completely lost (Fig. 5B, bottom). A few methylated CpGs remained on each allele, and only one allele contained a high level of methylation. These data indicate that the imprinted DME2/Igmyc transgene requires the Dnmt3L protein for establishment of methylation, as seen at endogenous imprinted loci.
Developmental function of the minimal DME2 element.
We have shown that the DME2/Igmyc transgene requires Dnmt3L and Dnmt1o proteins, present in oocytes and in preimplantation embryos, for the differential methylation on its DME2 element. These observations suggest that the transgene's DME2 sequences are needed for the germ line establishment and/or early embryonic maintenance of DME2/Igmyc imprinting. We analyzed the methylation of maternal and paternal alleles of DME1/Igmyc and DME2/Igmyc transgenes in the gametes and in preimplantation embryos. The DME1/Igmyc transgene is a nonimprinted mutant version of the imprinted DME2/Igmyc transgene. Abnormalities in the establishment and/or inheritance of methylation of the nonimprinted DME1/Igmyc transgene, in comparison to methylation on the imprinted DME2/Igmyc transgene, should identify the developmental times at which DME2 sequences are required for genomic imprinting.
The maternal and paternal differences in DME2/Igmyc transgene methylation were evident in the gametes (Fig. 5C). Moreover, these gametic methylation states were maintained in eight-cell embryos and preimplantation blastocysts (Fig. 5C). At the blastocyst stage, each maternal allele was methylated at over 60% of CpG dinucleotides. By comparison, paternal DME2/Igmyc alleles were methylated in blastocysts at only 0.7% of CpG dinucleotides. These data demonstrate that the two distinct epigenetic states of the DME2/Igmyc parental alleles are established and maintained in the same manner as the imprinted SnrpnR/Igmyc transgene (Fig. 1E) and many endogenous imprinted genes (14, 26).
The maternal and paternal alleles of the nonimprinted DME1/Igmyc transgene were partially methylated to approximately equal levels in adult tissue (Fig. 2F). Specifically, 68% of CpG dinucleotides on maternal alleles were methylated, and 55.5% of CpG dinucleotides of paternal alleles were methylated in adult transgene carriers. Paternal transgene alleles were unmethylated in sperm, indicating that the partially methylated paternal state developed entirely during postzygotic development (Fig. 5D). Because only 1.25% of CpGs were methylated on paternal alleles in blastocysts collected at 3.5 dpc, the paternal allele methylation was established de novo at a later blastocyst stage or following implantation. Although the precise time of acquisition of this methylation is unknown, the level of 65.5% methylation of paternal DME1/Igmyc alleles at E9.5 (Fig. 5D) is approximately the same as the level in adult paternal carriers, indicating that the acquisition occurred between E3.5 and E9.5 of development.
The time course of the acquisition of the adult methylation of the maternal DME1/Igmyc was different from that of the paternal allele. Maternal alleles were highly methylated in oocytes. Notably, the obvious epigenetic differences between DME1/Igmyc alleles in sperm (unmethylated) and oocytes (methylated) indicate that imprinting is established on the DME1/Igmyc transgene. The perpetuation of the methylation on oocyte-derived alleles was examined by measuring the methylation of maternal DME1/Igmyc alleles at different preimplantation stages (Fig. 5D). The maternal allele was highly methylated in eight-cell embryos to a level comparable to the level seen in oocytes. However, in blastocysts two epigenetic forms of DME1/Igmyc alleles were found. Five of 15 maternal DME1/Igmyc alleles examined were unmethylated at all 16 CpG dinucleotides. All other maternal alleles were nearly completely methylated. Interestingly, the unmethylated alleles acquired significant methylation after the blastocyst stage, as almost all maternal DME1/Igmyc alleles were partially methylated in the adult.
DISCUSSION
Developmental function of the DMD epigenetic maintenance signal.
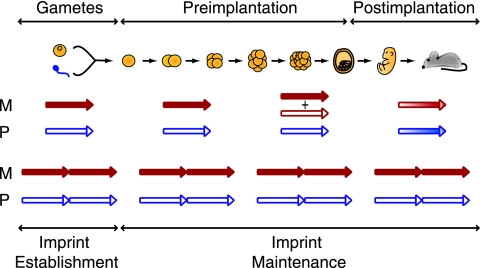
We have identified a bifunctional genomic element (DME2) that is essential for the maintenance of differential methylation in the DME2/Igmyc imprinted transgene. The DME2 sequence is a component of the Igf2r DMD2, an element central to the epigenetic regulation of the Igf2r cluster of imprinted genes (28). A model of DME2 function is presented in Fig. 6. The imprinted DME2/Igmyc transgene maintains DNA methylation on alleles inherited from the oocyte and keeps the alleles inherited from the sperm unmethylated (Fig. 6, bottom). These two epigenetic maintenance functions may be controlled by the same sequence, as both functions were eliminated in the nonimprinted DME1/Igmyc transgene (Fig. 6, top). Our data suggest that similar elements exist in the Snurf/Snrpn and Kcnq1 DMDs, and it is these bifunctional elements that likely distinguish imprinted genes from nonimprinted genes.
FIG. 6.
Model for the establishment and maintenance of transgene DMD methylation during development. The DME1/Igmyc transgene (single arrow) is compared to the DME2/Igmyc transgene (double arrows). The methylation state of the maternal (M) allele is on the top and the methylation state of the paternal allele (P) is on the bottom. The DME2/Igmyc transgene represents the “wild-type” methylation pattern on each allele throughout development, while the “mutant” DME1/Igmyc transgene pattern depicts when and to what extent methylation levels change during development.
The two functions of DME2 operate at different developmental stages (Fig. 6). Because approximately half of the maternal alleles of the imprinted DME2/Igmyc transgene lose methylation in the absence of preimplantation Dnmt1o, normal DME2/Igmyc imprinting depends critically on a Dnmt1o maintenance methyltransferase activity during preimplantation development (Fig. 5A). This view is strengthened by the observation that similar proportions of methylated and unmethylated maternal alleles of the nonimprinted DME1/Igmyc transgene are found in preimplantation blastocysts soon after Dnmt1o's maintenance function (Fig. 5D). In contrast, paternal alleles of the DME1/Igmyc transgene do not lose their unmethylated state until embryogenesis, suggesting that it is at this time that the DME2 element is required to block de novo methylation of an imprinted locus (Fig. 5D).
Role of the genomic context of the epigenetic maintenance signal in genomic imprinting.
Small DMDs of endogenous imprinted gene clusters function within a large genomic context of different chromatin conformations and different levels of transcriptional activity of several genes. These characteristics complicate the analysis of function for any small sequence element residing within that genomic context. The significant influence that genomic context has on imprinting can best be appreciated by the collection of transgenes built around the Igf2r DMD2. Consistent imprinting was achieved with a 300-kb transgene construct whose imprinting was dependent upon DMD2 (27). However, smaller Igf2r transgenes were often not imprinted or were inconsistently imprinted. A transgene containing 44 kb of sequence surrounding the Igf2r DMD2 was imprinted in one of three transgenic lines, and smaller transgenes of 3 kb, 9 kb, and 14 kb were not imprinted (22, 27). These results indicate that the ability of DMD2 to attain the correct parent-specific methylation can be found in greater than 40 kb but less than 300 kb of surrounding sequence. From the analysis of our Igf2r-containing transgenes, we conclude that the DME element is required for imprint maintenance and not for imprint establishment. The much larger genomic context surrounding a DMD would provide the signal to establish gametic methylation differences on that DMD (21). Loss of all or a significant portion of this genomic context would therefore preclude the establishment of gametic methylation differences.
In moving the DMD sequences from endogenous genes into the Ig/myc model transgene, we have significantly reduced the size of the genomic context required for transgene imprinting. The genomic environment provided by this transgene results in consistent imprinting upon addition of endogenous DMD sequences. Therefore, we are able to address a specific function of the DMD sequences, namely their ability to maintain parent-specific DNA methylation patterns. This provides a decided advantage over deleting the sequences from their endogenous genomic context and monitoring the effect, particularly when the goal is to monitor small changes in methylation at early preimplantation stages.
Roles of DMD tandem repeats and CpG dinucleotides in genomic imprinting.
The Snurf/Snrpn, Kcnq1, and Igf2r DMDs all contain unique tandem repeats. In all three cases the imperfect tandem repeats are important for creating a differentially methylated transgene locus, and for Snurf/Snrpn the tandem repeats are important for the imprinted expression of a heterologous reporter gene from the adjacent Snurf/Snrpn promoter. In defining the minimal sequence required to create a differentially methylated transgene containing Igf2r DMD sequences, we established that no sequence containing less than two unit copies of the repeat was capable of reproducibly creating an imprinted locus. This result suggests that the repeated nature of the DMD sequence is critical.
Importantly, the tandem repeats of the Igf2r DMD2 are within a region rich in CpG dinucleotides. Because the positions of many CpGs are conserved among adjacent unit copies of the TR2+3 repeats (21), an ordered arrangement of CpGs is obtained in the DME tandem repeats. Moreover, a similar ordered arrangement of CpG dinucleotides is imposed by tandem repeats of the Snurf/Snrpn and Kcnq1 DMDs (20). It is interesting to speculate that, once differential DMD methylation is established in the gametes, the maintenance of the methylated state on the maternal allele would be governed by the regular pattern of methylated CpGs, and the maintenance of the unmethylated state on the paternal allele is governed by the similar arrangement of unmethylated CpGs. In this way the two distinct epigenetic states of the DME2 element direct their own perpetuation during embryogenesis.
The CpG dinucleotide content of the tandem repeats changes across the 560-bp repeated region of the Igf2r DMD2; TR1 repeats have a low CpG content compared to the TR2+3 repeats (21). We can speculate that the low CpG content of the TR1 repeats is related to the inability of the TR1 repeats to imprint the Ig/myc transgene, and the high CpG content of the TR2+3 repeats is related to their ability to imprint Ig/myc. In this regard, it is interesting that the CpG content varies across the repeated regions of both the Snurf/Snrpn and the Kcnq1 DMDs (20). Certain tandem repeats lie within CpG-rich DMD regions, whereas other repeats lie in relatively CpG-poor regions. We would predict from this observed variation that only particular subsets of Snurf/Snrpn and Kcnq1 tandem repeats govern the maintenance of the parent-specific DMD methylation.
If CpG dinucleotide content and/or position within an epigenetic maintenance element is indeed essential for imprinting, then these CpGs and their direct interaction with maintenance methyltransferases may alone determine the maintenance of parental imprinted methylation states. Such a model would require an innate ability of the Dnmt1 maintenance methyltransferase to distinguish sequence or structural features of genomic DNA. Although no such distinguishing ability of Dnmt1 has been defined, this possibility has not been directly addressed in vivo. It may be important to address this particular issue during preimplantation development, a period in which the maintenance of imprinted methylation patterns may be most important.
Acknowledgments
This work was funded by a grant from the NIH.
We thank Leonardo D'Aiuto for helpful comments on the manuscript.
Footnotes
Published ahead of print on 5 September 2006.
REFERENCES
- 1.Ainscough, J. F., T. Koide, M. Tada, S. Barton, and M. A. Surani. 1997. Imprinting of Igf2 and H19 from a 130 kb YAC transgene. Development 124:3621-3632. [DOI] [PubMed] [Google Scholar]
- 2.Arnaud, P., K. Hata, M. Kaneda, E. Li, H. Sasaki, R. Feil, and G. Kelsey. 2006. Stochastic imprinting in the progeny of Dnmt3L−/− females. Hum. Mol. Genet. 15:589-598. [DOI] [PubMed] [Google Scholar]
- 3.Bielinska, B., S. M. Blaydes, K. Buiting, T. Yang, M. Krajewska-Walasek, B. Horsthemke, and C. I. Brannan. 2000. De novo deletions of SNRPN exon 1 in early human and mouse embryos result in a paternal to maternal imprint switch. Nat. Genet. 25:74-78. [DOI] [PubMed] [Google Scholar]
- 4.Bressler, J., T. F. Tsai, M. Y. Wu, S. F. Tsai, M. A. Ramirez, D. Armstrong, and A. L. Beaudet. 2001. The SNRPN promoter is not required for genomic imprinting of the Prader-Willi/Angelman domain in mice. Nat. Genet. 28:232-240. [DOI] [PubMed] [Google Scholar]
- 5.Chaillet, J. R., D. S. Bader, and P. Leder. 1995. Regulation of genomic imprinting by gametic and embryonic processes. Genes Dev. 9:1177-1187. [DOI] [PubMed] [Google Scholar]
- 6.Chaillet, J. R., T. F. Vogt, D. R. Beier, and P. Leder. 1991. Parental-specific methylation of an imprinted transgene is established during gametogenesis and progressively changes during embryogenesis. Cell 66:77-83. [DOI] [PubMed] [Google Scholar]
- 7.Clark, S. J., J. Harrison, C. L. Paul, and M. Frommer. 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22:2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, A. C., M. Wims, G. D. Spotts, S. R. Hann, and A. Bradley. 1993. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 7:671-682. [DOI] [PubMed] [Google Scholar]
- 9.Engemann, S., M. Strodicke, M. Paulsen, O. Franck, R. Reinhardt, N. Lane, W. Reik, and J. Walter. 2000. Sequence and functional comparison in the Beckwith-Wiedemann region: implications for a novel imprinting centre and extended imprinting. Hum. Mol. Genet. 9:2691-2706. [DOI] [PubMed] [Google Scholar]
- 10.Fitzpatrick, G. V., P. D. Soloway, and M. J. Higgins. 2002. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 32:426-431. [DOI] [PubMed] [Google Scholar]
- 11.Glenn, C. C., S. Saitoh, M. T. Jong, M. M. Filbrandt, U. Surti, D. J. Driscoll, and R. D. Nicholls. 1996. Gene structure, DNA methylation, and imprinted expression of the human SNRPN gene. Am. J. Hum. Genet. 58:335-346. [PMC free article] [PubMed] [Google Scholar]
- 12.Hata, K., M. Okano, H. Lei, and E. Li. 2002. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129:1983-1993. [DOI] [PubMed] [Google Scholar]
- 13.Howell, C. Y., T. H. Bestor, F. Ding, K. E. Latham, C. Mertineit, J. M. Trasler, and J. R. Chaillet. 2001. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 104:829-838. [DOI] [PubMed] [Google Scholar]
- 14.Lucifero, D., C. Mertineit, H. J. Clarke, T. H. Bestor, and J. M. Trasler. 2002. Methylation dynamics of imprinted genes in mouse germ cells. Genomics 79:530-538. [DOI] [PubMed] [Google Scholar]
- 15.Mancini-Dinardo, D., S. J. Steele, J. M. Levorse, R. S. Ingram, and S. M. Tilghman. 2006. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 20:1268-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monk, M. 1990. Changes in DNA methylation during mouse embryonic development in relation to X-chromosome activity and imprinting. Philos. Trans. R. Soc. Lond. B 326:299-312. [DOI] [PubMed] [Google Scholar]
- 17.Neumann, B., P. Kubicka, and D. P. Barlow. 1995. Characteristics of imprinted genes. Nat. Genet. 9:12-13. [DOI] [PubMed] [Google Scholar]
- 18.Nicholls, R. D., and J. L. Knepper. 2001. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu. Rev. Genomics Hum. Genet. 2:153-175. [DOI] [PubMed] [Google Scholar]
- 19.Pfeifer, K., P. A. Leighton, and S. M. Tilghman. 1996. The structural H19 gene is required for transgene imprinting. Proc. Natl. Acad. Sci. USA 93:13876-13883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reinhart, B., and J. R. Chaillet. 2005. Genomic imprinting: cis-acting sequences and regional control. Int. Rev. Cytol. 243:173-213. [DOI] [PubMed] [Google Scholar]
- 21.Reinhart, B., M. Eljanne, and J. R. Chaillet. 2002. Shared role for differentially methylated domains of imprinted genes. Mol. Cell. Biol. 22:2089-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sleutels, F., and D. P. Barlow. 2001. Investigation of elements sufficient to imprint the mouse Air promoter. Mol. Cell. Biol. 21:5008-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smilinich, N. J., C. D. Day, G. V. Fitzpatrick, G. M. Caldwell, A. C. Lossie, P. R. Cooper, A. C. Smallwood, J. A. Joyce, P. N. Schofield, W. Reik, R. D. Nicholls, R. Weksberg, D. J. Driscoll, E. R. Maher, T. B. Shows, and M. J. Higgins. 1999. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc. Natl. Acad. Sci. USA 96:8064-8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoger, R., P. Kubicka, C. G. Liu, T. Kafri, A. Razin, H. Cedar, and D. P. Barlow. 1993. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell 73:61-71. [DOI] [PubMed] [Google Scholar]
- 25.Thorvaldsen, J. L., K. L. Duran, and M. S. Bartolomei. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 12:3693-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warnecke, P. M., J. R. Mann, M. Frommer, and S. J. Clark. 1998. Bisulfite sequencing in preimplantation embryos: DNA methylation profile of the upstream region of the mouse imprinted H19 gene. Genomics 51:182-190. [DOI] [PubMed] [Google Scholar]
- 27.Wutz, A., O. W. Smrzka, N. Schweifer, K. Schellander, E. F. Wagner, and D. P. Barlow. 1997. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 389:745-749. [DOI] [PubMed] [Google Scholar]
- 28.Zwart, R., F. Sleutels, A. Wutz, A. H. Schinkel, and D. P. Barlow. 2001. Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes. Genes Dev. 15:2361-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]