Abstract
AUF1 is an RNA-binding protein that targets mRNAs containing A+U-rich elements (AREs) for rapid cytoplasmic turnover. Alternative pre-mRNA splicing produces five variants of AUF1 mRNA that differ in the composition of their 3′-untranslated regions (3′-UTRs). Previous work suggested that this heterogeneity in 3′-UTR sequence could regulate AUF1 expression by two potential mechanisms. First, AUF1 may regulate its own expression by binding to AREs in 3′-UTR splice variants that retain intron 9. The second potential mechanism, and the focus of this report, is regulation of a subset of 3′-UTR splice variants by the nonsense-mediated mRNA decay (NMD) pathway. Two of the five AUF1 mRNA 3′-UTR variants position the translational termination codon more than 50 nucleotides upstream of an exon-exon junction, creating a potential triggering signal for NMD in mammalian cells. Disruption of cellular NMD pathways by RNA interference-mediated knockdown of Upf1/Rent1 or Upf2/Rent2 or transfection of a dominant-negative Upf1 mutant specifically enhanced expression of these two candidate NMD substrate mRNAs in cells, involving stabilization of each transcript. Ribonucleoprotein immunoprecipitation experiments revealed that both Upf1 and Upf2 can associate with an NMD-sensitive AUF1 mRNA 3′-UTR variant in cells. Finally, quantitation of AUF1 mRNA 3′-UTR splice variants during murine embryonic development showed that the expression of NMD-sensitive AUF1 mRNAs is specifically enhanced as development proceeds, contributing to dynamic changes in AUF1 3′-UTR structures during embryogenesis. Together, these studies provide the first evidence of linkage between the nonsense- and ARE-mediated mRNA decay pathways, which may constitute a new mechanism regulating the expression of ARE-containing mRNAs.
mRNA decay is an important component of regulated gene expression in eukaryotic cells. Together, the rates of transcription, pre-mRNA splicing, nucleocytoplasmic transport, and cytoplasmic mRNA degradation control the steady-state concentrations of cytoplasmic mRNAs, and hence their potential to program protein synthesis at any given time. The mRNAs that encode many cytokines, oncoproteins, growth factors, and signaling components are highly labile, providing a mechanism for rapidly changing mRNA levels in response to extracellular stimuli (50). Many of these mRNAs are targeted for rapid degradation by A+U-rich elements (AREs) within their 3′-untranslated regions (3′-UTRs) (32, 58). AREs range in length from 50 to 150 nt and often possess one or more copies of the AUUUA pentamer or UUAUUUA(U/A)(U/A) nonamer. Destruction of mRNAs via the ARE-mediated mRNA decay (AMD) pathway is initiated by rapid 3′→5′ deadenylation, followed by degradation of the mRNA body (25, 58).
Both the turnover kinetics and translational efficiency of ARE-containing mRNAs may be regulated through the activity of cellular ARE-binding proteins. Over the past 15 years, a variety of these factors have been identified, including AUF1 [ARE- and poly(U)-binding and degradation factor 1], BRF1 (butyrate response factor 1), Hsc/Hsp70, the Hu family of proteins (HuR, HuB, HuC, and HuD), KSRP (KH domain splicing regulatory protein), PM-Scl75 (polymyositis-scleroderma overlap syndrome 75-kDa antigen), TIA-1 (T-cell internal antigen 1), TIAR (TIA-1-related protein), and tristetraprolin (TTP) (2, 8, 58). AUF1, also known as hnRNP D, was first identified by its ability to promote degradation of c-myc mRNA in a cell-free mRNA decay system (7, 56). mRNP immunoprecipitation and microarray analyses indicated that AUF1 binds to ARE-containing mRNAs encoding many cytokines, oncoproteins, cell cycle regulators, and G protein-coupled receptors (34). Small interfering RNA (siRNA)-mediated depletion of AUF1 stabilizes ARE-containing mRNAs, including those for GADD45α, cyclin D1, and the cell cycle inhibitors p21 and p16INK4a, and a reporter mRNA containing the interleukin-3 ARE (33, 34, 49). In addition to its role in AMD, AUF1 participates in other cellular processes, including telomere maintenance and transcriptional activation/repression (12, 14, 16, 18, 20, 26).
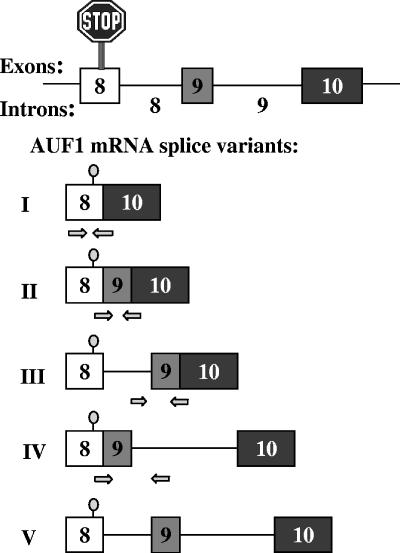
AUF1 is expressed as a family of four protein isoforms generated by alternative splicing of a common pre-mRNA (13, 15, 30, 54). While the AUF1/HNRPD gene consists of 10 exons, the translational termination codon lies in exon 8 rather than the 3′-terminal exon, which is the case for most mRNAs (13, 48, 54). As such, the unusual 3′-end structure of the AUF1/HNRPD gene presents the opportunity for multiple pre-mRNA splicing patterns, potentially creating transcripts with five distinct 3′-UTR structures (Fig. 1) (also see reference 57). In earlier work, we identified four of these AUF1 3′-UTR splice variants (I, II, IV, and V) (Fig. 1) in the human chronic myelogenous leukemia cell line K562 (57). Two observations further suggested that these alternative 3′-UTR structures could play a role in regulating AUF1 expression. First, these transcripts are polysome associated, suggesting that they are not simply inactive nuclear by-products of inefficient pre-mRNA splicing. Second, the 107-nucleotide (nt) exon 9 is 100% conserved between the mouse and human AUF1 loci, while the 3′ 130 nt of intron 8 and the 5′ half of intron 9 retain 99% identity between these species. The extreme conservation of these untranslated mRNA sequences present in variants II, III, and IV suggests that they have critical, albeit unknown, functions in regulating AUF1 expression, which may include control of mRNA translation, stability, or subcellular localization (11). Our earlier work also suggested two mechanisms by which 3′-UTR sequences could regulate levels of specific splice variants (57). First, intron 9, which is present in splice variants IV and V, contains two AUF1-binding sites that could target these transcripts for degradation by AMD. Second, selective excision of intron 9, generating variants II and III, would position an exon-exon junction >50 nt downstream of the stop codon, which may target these transcripts for degradation by the nonsense-mediated mRNA decay (NMD) pathway (48).
FIG. 1.
Potential splicing variants of the AUF1 3′-UTR. Five possible AUF1 3′-UTR splice variants are shown based upon the exon-intron organization of the AUF1 gene. Arrows below the diagrams of variant mRNAs I to IV depict the locations of forward and reverse primers for qRT-PCR amplification of each specific splice variant.
NMD is an RNA surveillance mechanism that detects and degrades transcripts containing a premature termination codon (PTC) (19, 27, 44, 55), thus preventing the synthesis of truncated proteins that could have deleterious effects on cellular physiology. In Saccharomyces cerevisiae, NMD triggers rapid deadenylation-independent mRNA decapping followed by 5′→3′ exonucleolytic decay of the transcript (23). A similar pathway may function in mammalian cells as well, but there is evidence that accelerated deadenylation is also involved (9, 39, 59). A major mechanism by which mammalian cells distinguish a bona fide translational termination codon from a PTC is the position of the stop codon in relation to an exon-exon junction in an mRNA molecule; stop codons located >50 nt upstream of an exon-exon junction normally trigger NMD (27, 44, 48), although T-cell receptor transcripts appear to be exceptions to this rule (41). In the nucleus, pre-mRNA splicing leads to the deposition of the exon junction protein complex 20 to 24 nt upstream of each exon-exon junction. The exon junction protein complex serves as a platform for the binding of Upf3/Upf3X and Upf2 (36-38). Upf2 recruits Upf1, an ATP-dependent 5′-to-3′ RNA helicase and ATPase (4, 46, 52). The nuclear cap-binding proteins CBP20-CBP80 associate with the 5′ cap, and the poly(A)-binding protein PABP2 associates with the 3′ poly(A) tract (28). The mRNP complex then undergoes a pioneer round of translation that establishes whether the mRNA contains a PTC (10, 29). Subsequent remodeling of the protein constellation of the mRNP then permits either mRNA degradation for nonsense-containing transcripts or additional rounds of translation for normal transcripts (10, 21, 29, 40).
Given the unusual 3′-end structure of the AUF1 gene and our prior work demonstrating that alternative pre-mRNA splicing permits the expression of highly conserved 3′-UTR elements (intron 8, exon 9, and intron 9) in AUF1 mRNA, we predicted that NMD might control the expression of selected AUF1 3′-UTR splice variants. In this study, we show that disruption of the NMD pathway, either by reduction of human Upf1 or Upf2 or by expression of a dominant-negative Upf1 mutant, enhances the expression of selected 3′-UTR splice variants of AUF1 mRNA involving transcript stabilization and that Upf1 and Upf2 can associate with an NMD-responsive AUF1 mRNA in cells. Finally, we demonstrate that AUF1 3′-UTR variant mRNAs are differentially expressed during murine embryogenesis, which may impact the spatiotemporal expression of AUF1 during development and ultimately provide a model system for defining the functional and gene regulatory consequences of alternative pre-mRNA splicing events in the AUF1 3′-UTR.
MATERIALS AND METHODS
siRNA synthesis.
siRNAs were chemically synthesized by Dharmacon Research, using previously described sequences for hUpf1/Rent1 and hUpf2/Rent2 (22, 46) and a scrambled control sequence described by Liao et al. (42). The sequence specificity of each siRNA was verified using BLAST searches against the human genome database (1).
Cell culture and transfections.
The human cervical carcinoma cell line HeLa was grown in Dulbecco's modified Eagle medium (Gibco BRL) containing 10% fetal bovine serum (HyClone) supplemented with l-glutamine, penicillin, and streptomycin at 37°C with 5% CO2. For transfections, 1.5 × 105 cells per well were plated in a six-well plate and cultured overnight. The next day, siRNAs were transfected at a final concentration of 100 nM in the presence of serum-free medium (OptiMEM; Invitrogen), using Lipofectamine 2000 (Invitrogen). Analyses of endogenous mRNA and protein expression were performed 72 h after transfection. In two-hit transfection experiments, a second round of siRNA transfection was performed 24 h after the first round. Where indicated, a plasmid carrying either a Upf1 or Upf2 siRNA-resistant cDNA (22, 31) was cotransfected with the second siRNA application. These plasmids contain silent nucleotide exchanges within the sequences to which the respective siRNAs anneal, thus making the encoded mRNAs refractory to RNA interference (RNAi).
The construction of plasmids expressing firefly luciferase-AUF1 3′-UTR chimeric mRNAs downstream of the simian virus 40 core promoter was described previously (57). For experiments utilizing luciferase-AUF1 3′-UTR constructs, HeLa cells were transfected 48 h after siRNA transfection with 1 μg of firefly luciferase reporter plasmid/well and 10 ng/well control Renilla luciferase vector pRL-SV40 (Promega), using Lipofectamine 2000. Where indicated, HeLa cells were cotransfected with 200 ng of either pCI-neo-hUPF1 or pCI-neo-hUPF1(R844C), encoding wild-type (wt) Upf1 or a helicase-defective mutant of Upf1, respectively (52). Preparation of RNA and protein samples for downstream analyses was performed at 48 h posttransfection. Where indicated, firefly luciferase activity was measured using a dual-luciferase kit (Promega), with Renilla luciferase activity as an internal control.
Western blot analyses.
Two million cells per 60-mm dish were resuspended in 0.25 ml of polysome lysis buffer (100 mM KCl, 50 mM MgCl2, 10 mM HEPES [pH 7.9], 0.5% NP-40, 0.1 mM dithiothreitol, and 1× protease inhibitor cocktail [Roche] [7× protease inhibitor cocktail = one tablet dissolved in 1.5 ml deionized water]). The suspension was incubated on ice for 15 min and centrifuged at 4°C for 30 min at 12,000 × g to pellet nuclei and cell debris. The protein concentrations of cytoplasmic supernatants were determined by the Bradford assay, using bovine serum albumin standards.
For Western blot analyses, cytoplasmic proteins (30 to 50 μg) were size fractionated by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Fisher). To detect Upf1 or Upf2, membranes were incubated with polyclonal goat antibodies (1:300; Santa Cruz) followed by rabbit anti-goat immunoglobulin G (IgG)-horseradish peroxidase conjugate (1:5,000; Promega). Immunoreactive bands were detected using a SuperSignal West Pico chemiluminescent substrate kit (Pierce) and exposure to X-ray film. For normalization, blots were reincubated with a mouse monoclonal antibody to α-tubulin (1:10,000; Sigma) followed by goat anti-mouse IgG-horseradish peroxidase conjugate (1:3,000; Promega) and were visualized by chemiluminescence detection. Western blot data were quantified by scanning multiple film exposures using a DC120 Zoom digital image system (Kodak).
qRT-PCR.
Purified total RNA was incubated with RNase-free DNase I (Roche) to remove any contaminating DNA, repurified using an RNeasy kit (QIAGEN), and quantified spectrophotometrically by measuring the A260. One-step quantitative reverse transcription-PCR (qRT-PCR) was performed using SYBR green, a Taqman reverse transcription kit (Applied Biosystems), and an Mx4000 multiplex quantitative PCR system (Stratagene, La Jolla, CA). Primer sets for each mRNA were designed using Primer-3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and were synthesized by Integrated DNA Technologies. The sequences of all qRT-PCR primer sets are displayed in Table S1 in the supplemental material. For quantitation of specific endogenous mRNAs, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as an internal control. Renilla luciferase mRNA was used as a standardization control in luciferase cotransfection experiments.
qRT-PCR mixtures were assembled in triplicate in a final volume of 50 μl containing sense and antisense primers (50 pmol each). The reverse transcription reaction was performed at 48°C for 30 min, and then the reverse transcriptase was inactivated at 95°C for 10 min. The PCR cycles were as follows: 40 cycles of 95°C for 15 s, 52°C for 1 min, and 68°C for 2 min. Single-fragment amplification by each qRT-PCR primer set was verified by thermal denaturation and 3% agarose gel electrophoresis. Reaction mixtures lacking template or reverse transcriptase were used as negative controls. Data were analyzed by the ΔΔCT method, using software provided with the Mx4000 system (Strategene), and are presented as means ± standard deviations (SD) for at least three independent experiments.
mRNP immunoprecipitation.
Twenty million HeLa cells in a 150-mm plate were cotransfected with plasmid pFLAG, pFLAG-Upf1, or pT7-Upf2 (3 μg), together with a luciferase expression vector (1 μg) lacking (pGL3-Promoter) or containing the spliceable AUF1 exon 9-intron 9-exon 10 cassette (pGL3-Ex9:In9:Ex10). Two days after transfection, cells were released with trypsin, pelleted, and washed twice with ice-cold phosphate-buffered saline (PBS). The cell pellet was resuspended in an equal volume of polysome lysis buffer (described above) supplemented with 40 U/ml RNasin and 40 μl of 7× protease inhibitor cocktail (described above). Cells were disrupted by sonication, and debris was removed by centrifugation at 10,000 × g for 30 min at 4°C. The lysates were precleared by incubation with 30 μl of normal mouse serum and protein A-Sepharose beads for 45 min at 4°C. The precleared lysates were then fractionated by mRNP immunoprecipitation as described previously (29), using protein A-Sepharose beads preloaded with either mouse IgG, anti-FLAG antibody (Sigma), or anti-T7-tag antibody (Novagen). Proteins retained in antibody-bead pellets were analyzed for FLAG-Upf1 or T7-Upf2 by Western blotting using antibodies directed at each epitope tag, as well as anti-Upf1 and anti-Upf2 antibodies. RNAs retained in antibody-bead pellets were analyzed for firefly luciferase mRNA by RT-PCR. Amplified DNA products were resolved by electrophoresis through 1.5% agarose gels and visualized by ethidium bromide staining.
mRNA decay assays.
Seventy-two hours after being transfected with control, Upf1, or Upf2 siRNA, HeLa cells were treated with 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) to inhibit polymerase II (Pol II) transcription (47). At selected time points thereafter, total RNA was extracted using Trizol reagent (Invitrogen). Levels of AUF1 mRNA splice variants were quantified at each time point by qRT-PCR and were normalized to GAPDH mRNA levels. First-order decay constants (k) and associated mRNA half-lives were calculated from plots of percent AUF1 variant mRNA remaining versus time of DRB treatment, using Prism 3.03 software (GraphPad).
Mouse embryos.
All animal protocols were approved by the Institutional Animal Care and Use Committee of Robert Wood Johnson Medical School. Matings between C57BL/6 mice were timed, and pregnant females were sacrificed at various days postcoitum. Embryos at selected developmental stages (E5.5, E9.5, E13.5, and E16.5) were dissected from the uterus, and extra-embryonic membranes were removed. Embryos were rinsed in PBS and either fixed in 4% paraformaldehyde in PBS overnight at 4°C or frozen in liquid nitrogen following a PBS wash. Total RNA from each embryo was extracted using an RNeasy kit (QIAGEN). Total RNA was then incubated with RNase-free DNase I (Roche) to remove genomic DNA and repurified using an RNeasy kit. RNA yields were determined by measuring the A260, and integrity was verified by 1% agarose gel electrophoresis.
Statistical analyses.
Quantitative comparisons between data sets were performed using the unpaired two-tailed t test (PRISM v3.03), with P values of <0.05 considered significant.
RESULTS
siRNA-directed reduction of Upf1 enhances expression of selected AUF1 3′-UTR splice variants.
The organization of the 3′ region of the AUF1 gene suggested five potential 3′-UTR splice variants (Fig. 1), all of which have been observed in cells (57; this work). The current understanding of NMD indicates that a stop codon located >50 nt upstream of an exon-exon junction should trigger this mRNA decay pathway (48). Since the translational termination codons of AUF1 mRNA 3′-UTR variants II and III are ≥137 nt upstream of the exon 9-exon 10 junction, these mRNA products were predicted to be NMD targets. To test this hypothesis, changes in the expression of AUF1 mRNA splice variants were evaluated following siRNA-directed reduction of Upf1, a central component of the cellular NMD machinery.
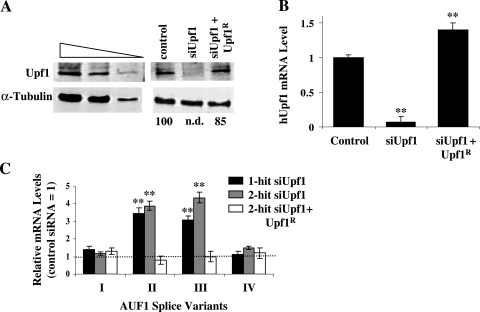
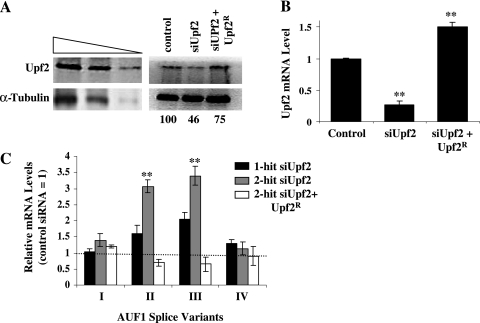
A single-hit Upf1 siRNA transfection protocol yielded a 65 to 70% decrease in Upf1 protein levels over 72 h relative to those in cells transfected with a control siRNA (data not shown), while a two-hit siRNA transfection strategy reduced Upf1 to undetectable levels (Fig. 2A). Extended film exposures and comparisons against an immunoblot containing a titration of lysate (Fig. 2A, left panel) indicate that Upf1 levels were reduced >90% by the two-hit siRNA transfection. A similarly dramatic siRNA-dependent repression of Upf1 mRNA (Fig. 2B) was consistent with the reduction in Upf1 protein levels.
FIG. 2.
Effects of Upf1 reduction on expression of endogenous AUF1 3′-UTR splice variants. (A) Western blot analysis of Upf1 levels. A two-hit strategy was used to transfect control or Upf1-specific siRNA into HeLa cells. A parallel culture was cotransfected with a plasmid encoding an siRNA-resistant Upf1 cDNA (Upf1R) and Upf1 siRNA as described in Materials and Methods. A twofold dilution series of HeLa cytoplasmic lysate was probed with antibodies for Upf1 and α-tubulin to permit estimations of the Upf1 knockdown efficiency (left). Estimates of Upf1 protein levels in the cytoplasm of transfected cells are expressed as percentages of Upf1 in cells transfected with control siRNA (right). n.d., not detectable. (B) qRT-PCR analysis of changes in Upf1 mRNA levels resulting from two-hit transfection of Upf1 siRNA, with or without cotransfected Upf1R, relative to control siRNA transfection, expressed as means ± SD (n = 3). **, P < 0.01 versus control siRNA. (C) Upf1-dependent changes in expression of endogenous AUF1 3′-UTR splice variants. Total RNA was isolated from the transfected cells described in panel A or from cells transfected with a single hit of Upf1 siRNA and analyzed by qRT-PCR, using primer pairs specific for individual AUF1 3′-UTR splice variants (see Fig. 1). The bars indicate the means ± SD (n = 4), where the level of each AUF1 variant mRNA is relative to that measured in cells transfected with the control siRNA (dotted line) **, P < 0.01 versus control siRNA. The same RNA samples were utilized to perform the qRT-PCRs shown in panels B and C.
Using primer sets designed to specifically amplify individual 3′-UTR splice variants of endogenous AUF1 mRNA (Fig. 1, arrows), qRT-PCR analyses revealed that levels of AUF1 mRNA splice variants II and III were elevated >3-fold following Upf1 knockdown (Fig. 2C). Interestingly, increasing the knockdown efficiency of Upf1 from 65% (one-hit) to >90% (two-hit) only modestly influenced these results. By contrast, levels of AUF1 mRNA splice variants I and IV, which are not predicted NMD substrates, were not significantly influenced by diminution of Upf1 expression (Fig. 2C). To control for potential off-target effects of the Upf1 siRNA, cells were subjected to two-hit Upf1 siRNA knockdown but cotransfected with a plasmid encoding an siRNA-resistant form of Upf1 mRNA (Upf1R), which returned total cellular Upf1 protein (Fig. 2A, right panel) and mRNA (Fig. 2B) to near-normal levels. Ectopic restoration with Upf1R abrogated Upf1 siRNA-induced increases in expression of AUF1 splice variant II and III mRNAs, while levels of variant I and IV mRNAs remained unaffected (Fig. 2C). Taken together, these results indicate that Upf1 plays a significant role in repressing the expression of AUF1 mRNA 3′-UTR splice variants in which the stop codon resides >50 nt upstream of an exon-exon junction. However, we also noted that Upf1 knockdown only modestly increased (20%) endogenous AUF1 protein levels in HeLa cells (data not shown). This result was not unexpected, since the most abundant AUF1 mRNA 3′-UTR variant observed in the cultured cell models used in this (data not shown) and previous studies (57) is fully spliced (variant I) and therefore exempt from NMD.
3′-UTR elements mediate control of AUF1 mRNA splice variant expression by Upf1.
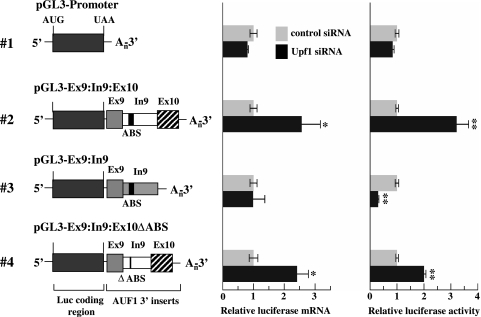
To define elements within AUF1 mRNA required for Upf1-dependent regulatory control, we tested whether selected regions of the AUF1 3′-UTR could confer Upf1-dependent expression on a luciferase reporter gene. Previous work showed that no sequence in either exon 8, 9, or 10 significantly influenced luciferase reporter expression in HeLa cells (57). However, inclusion of intron 9 sequences together with intact 5′ and 3′ splice sites (to permit intron 9 splicing) reduced reporter gene expression at both the protein and mRNA levels. Since the stop codon is >50 nt upstream of the resulting junction between exons 9 and 10, we predicted that this Ex9:In9:Ex10 transcript could be a substrate for the NMD pathway.
siRNA-directed reduction of Upf1 increased levels of the pGL3-Ex9:In9:Ex10 reporter mRNA 2.5-fold (Fig. 3, construct 2, middle panel) and increased luciferase reporter activity 3-fold (Fig. 3, construct 2, right panel) compared to those in cells transfected with the control siRNA. These results are consistent with elevated expression of endogenous AUF1 mRNA splice variants II and III following Upf1 knockdown (Fig. 2C), since both of these mRNAs also contain a splice junction between exons 9 and 10 (Fig. 1). By contrast, expression of the pGL3-Promoter reporter was not influenced at either the mRNA or protein level by transfection of either control or Upf1 siRNA (Fig. 3), verifying that sequence elements in the AUF1 3′-UTR are required for Upf1-dependent regulatory control. The influence of Upf1 on the expression of luciferase-AUF1 3′UTR chimeric mRNAs also required intron 9 splicing, since deletion of the 3′ splice site abrogated Upf1 siRNA-dependent changes in reporter mRNA levels (Fig. 3, construct 3, middle panel). Interestingly, however, Upf1 knockdown reduced expression from pGL3-Ex9:In9 by 80% at the protein level compared to that in the presence of the control siRNA (Fig. 3, construct 3, right panel). This result suggests that, at least under some conditions, Upf1 may contribute to the translational efficiency of some mRNAs (see Discussion).
FIG. 3.
Identification of AUF1 3′-UTR elements required for regulation by Upf1. Schematics of firefly luciferase-AUF1 3′-UTR chimeric mRNAs are shown on the left and are numbered for text reference. AUF1 exon and intron sequences are labeled, and the ABS within intron 9 are indicated by a black box. In constructs where intron 9 cannot be excised by splicing, the intron sequence is shaded. HeLa cells were first transfected with control or Upf1-specific siRNA, followed 48 h later by cotransfection of luciferase-AUF1 3′-UTR constructs and a Renilla luciferase control vector. Two days following luciferase transfections, firefly and Renilla luciferase mRNA and activity levels were measured as described in Materials and Methods. For each chimeric mRNA, bars represent the means ± SD (n = 3) of firefly luciferase mRNA levels (middle panel) or activities (right panel) relative to those for the control siRNA transfection following normalization to Renilla luciferase mRNA and activity, respectively. *, P < 0.05; **, P < 0.01 versus control siRNA.
Previous work also indicated that intron 9 contains binding sites for the AUF1 protein that can function independently of splicing to reduce reporter gene expression (57). To ensure that the Upf1 siRNA-dependent enhancement of pGL3-Ex9:In9:Ex10 expression was not a secondary effect of Upf1-dependent changes in endogenous AUF1 expression or activity, an additional reporter construct lacking the AUF1-binding sites (ABS) but retaining intron 9 splicing potential was assembled (Fig. 3, pGL3-Ex9:In9:Ex10ΔABS). Similar to the case with the Ex9:In9:Ex10 chimera, Upf1 knockdown significantly enhanced expression of the ΔABS reporter at both the mRNA and protein levels (Fig. 3, cf. constructs 2 and 4), indicating that Upf1-mediated repression of these mRNA substrates does not require the AUF1-binding sites.
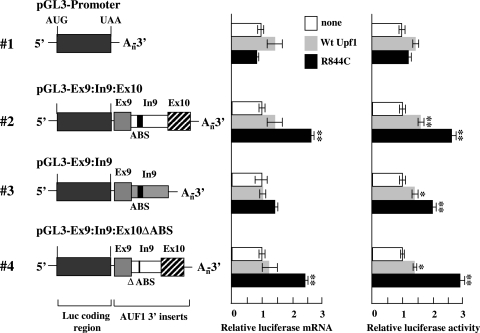
Taken together, these data are consistent with a model whereby removal of intron 9 by pre-mRNA splicing is necessary and sufficient for suppression of AUF1 mRNA splice variants II and III by Upf1. However, subsequent experiments using an independent method for disrupting cellular Upf1 function were performed to validate these findings. In these experiments, the expression of selected luciferase-AUF1 3′-UTR chimeric mRNAs was evaluated in the presence of ectopically expressed wild-type Upf1 or a mutant containing an Arg→Cys substitution in the helicase domain (R844C). The Upf1 R844C mutant displays dominant-negative activity in cells and has been reported to modestly (approximately twofold) increase the levels of mRNAs containing PTCs (52).
As expected, coexpression of wt Upf1 or Upf1(R844C) did not exert any significant effect on expression from the plasmid pGL3-Promoter, which lacks any AUF1 sequences (Fig. 4). Overexpression of Upf1(R844C) increased luciferase mRNA levels approximately 2.5-fold from reporter chimeras containing the spliceable AUF1 intron 9, regardless of the presence (Ex9:In9:Ex10) or absence (Ex9:In9:Ex10ΔABS) of the AUF1-binding sites (Fig. 4, constructs 2 and 4, middle panel, black bars). By contrast, cotransfection of wt Upf1 had no significant effect on expression of the Ex9:In9:Ex10 and Ex9:In9:Ex10ΔABS chimeric mRNAs (Fig. 4, constructs 2 and 4, middle panel, gray bars). Luciferase activities from these reporter constructs were modestly (40 to 60%) but significantly (P < 0.05) enhanced by cotransfection of wt Upf1 (Fig. 4, constructs 2 and 4, right panel, cf. gray and white bars). However, expression of Upf1(R844C) induced much more dramatic (2.5- to 3-fold) increases in luciferase activity from reporter chimeras containing the spliceable intron 9 (Fig. 4, constructs 2 and 4, right panel, black bars), indicating that expression of the putative NMD substrate mRNAs was more profoundly influenced by the dominant-negative than wild-type Upf1. Similar to the Upf1 siRNA data (Fig. 3), cotransfection of either wild-type or dominant-negative Upf1 had no significant effect on expression of the splicing-defective Ex9:In9 reporter mRNA (Fig. 4, construct 3, middle panel). Together, these data demonstrate that intron 9 splicing is required for the regulation of luciferase-AUF1 3′-UTR reporter mRNAs by Upf1. Furthermore, a specific role for Upf1 in restricting the expression of these candidate NMD substrate mRNAs is supported by the enhanced reporter mRNA levels observed concomitant with either siRNA-mediated reduction of endogenous Upf1 (Fig. 3) or coexpression of a dominant-negative Upf1 mutant (Fig. 4).
FIG. 4.
Control of luciferase-AUF1 3′-UTR splice variant expression by a dominant-negative Upf1 mutant. Schematics of firefly luciferase-AUF1 3′-UTR chimeric mRNAs are shown as described in the legend to Fig. 3 (left). HeLa cells were cotransfected with luciferase-AUF1 3′-UTR constructs and a Renilla luciferase control vector in the absence (white bars) or presence of an expression vector encoding wild-type Upf1 (gray bars) or the dominant-negative Upf1 R844C mutant (black bars). At 2 days posttransfection, firefly and Renilla luciferase mRNA and activity levels were analyzed as described in Materials and Methods. For each reporter construct, bars represent the means ± SD (n = 3) of firefly luciferase mRNA levels (middle panel) or activities (right panel) relative to those for cotransfections lacking ectopic Upf1 (or the R844C mutant) following normalization to Renilla luciferase mRNA and activity, respectively. *, P < 0.05; **, P < 0.01 versus control.
Similar to the Upf1 siRNA results shown in Fig. 3, ectopic expression of wild-type or dominant-negative Upf1 appeared to influence translation of the Ex9:In9 reporter mRNA. While overexpression of wild-type Upf1 enhanced luciferase protein activity by 40% compared to the control level, luciferase activity was increased twofold in cells expressing Upf1(R844C) (Fig. 4, construct 3, right panel). Curiously, the influence of Upf1(R844C) expression on luciferase activity from the Ex9:In9 reporter mRNA was opposite to that observed by siRNA-directed Upf1 reduction, since Upf1 siRNA reduced luciferase activity from this reporter construct by 80% (Fig. 3). While these observations are consistent with the possibility that Upf1 may play some role in the maintenance of basal translation of selected mRNAs, they also suggest that this function may be independent of the RNA helicase activity of Upf1. This concept is explored further in Discussion.
Upf2 regulates expression of selected AUF1 3′-UTR splice variant mRNAs.
Recent studies indicated that the stability of some mRNAs can be regulated by Upf1 independently of NMD. For example, the protein Staufen can recruit Upf1 to the 3′-UTR of ADP-ribosylation factor 1 (Arf1) mRNA to promote its degradation in an NMD-independent fashion (31). Thus, to validate a transcript as a bona fide NMD substrate, it is necessary to examine the influence of a second essential effector of NMD, such as Upf2 (31, 47).
To determine whether Upf2 also contributes to the regulated expression of selected AUF1 3′-UTR splice variant mRNAs, HeLa cells were transfected with either control or Upf2-specific siRNA, using a one-hit or two-hit protocol. At 72 hours posttransfection, the two-hit siRNA strategy decreased endogenous Upf2 protein levels by 54% and Upf2 mRNA by 75% relative to those in control siRNA transfections (Fig. 5A and B). A single-hit siRNA transfection reduced Upf2 protein levels <50% (data not shown). AUF1 splice variant-specific qRT-PCR analyses revealed that levels of AUF1 mRNA variants II and III were elevated >3-fold following Upf2 knockdown by two-hit siRNA transfection (Fig. 5C, gray bars), similar to the results observed in Upf1-depleted cells (Fig. 2C). Single-hit siRNA transfections were less effective in enhancing levels of AUF1 mRNA variants II and III (Fig. 5C, black bars), likely owing to the poor suppression of Upf2 expression possible by the one-hit approach. As expected, levels of AUF1 variant I and IV mRNAs, which are not predicted NMD substrates, were not significantly influenced by siRNA-directed knockdown of Upf2 expression. To control for potential off-target effects of the Upf2 siRNA, cells were cotransfected with two-hit Upf2 siRNA and a plasmid encoding siRNA-resistant Upf2 mRNA (Upf2R). Ectopic expression of Upf2R restored total cellular Upf2 protein (Fig. 5A, right panel) and mRNA (Fig. 5B) to near-normal levels and prevented the increased expression of AUF1 variant II and III mRNAs resulting from Upf2 siRNA transfection (Fig. 5C, white bars). Again, the levels of AUF1 variant I and IV mRNAs remained unaffected by manipulation of Upf2 expression. From this experiment, we conclude that suppression of cellular Upf2 levels is singularly responsible for the increased expression of AUF1 variant II and III mRNAs observed following transfection of Upf2 siRNA. Taken together, the data presented to this point indicate that both Upf1 and Upf2 play key roles in repressing the cellular expression of these AUF1 mRNA splice variants. As such, these results indicate that AUF1 3′-UTR variant II and III mRNAs are both bona fide substrates of the NMD pathway (44).
FIG. 5.
Effects of Upf2 reduction on expression of endogenous AUF1 3′-UTR splice variants. (A) Western blot analysis of Upf2 levels. A two-hit strategy was used to transfect control or Upf2-specific siRNA into HeLa cells or to cotransfect a combination of Upf2 siRNA and siRNA-resistant Upf2 cDNA (Upf2R) as described in Materials and Methods. Estimation of the Upf2 knockdown efficiency was performed by probing immunoblots of cytoplasm from each transfected cell population with antibodies for Upf2 and α-tubulin (right panel) and by comparison to a twofold dilution series of cytoplasmic lysate from nontransfected HeLa cells (left panel). Estimates of Upf2 protein levels in each cell population are expressed as percentages of the Upf2 in cells transfected with control siRNA. (B) Changes in Upf2 mRNA levels resulting from two-hit transfection of Upf2 siRNA, with or without cotransfected Upf2R, relative to those with control siRNA, measured by qRT-PCR and expressed as means ± SD (n = 3). **, P < 0.01 versus control siRNA. (C) Upf2-dependent changes in expression of endogenous AUF1 3′-UTR splice variants. Total RNA was isolated from the transfected cells described in panel A or from cells transfected with a single hit of Upf2 siRNA and analyzed for individual AUF1 3′-UTR variant mRNAs by qRT-PCR. The bars indicate the means ± SD (n = 4), where the level of each mRNA variant is shown relative to that measured in cells transfected with the control siRNA (dotted line). **, P < 0.01 versus control siRNA. The same RNA samples were utilized to perform the qRT-PCRs shown in panels B and C.
Upf1 and Upf2 can associate with AUF1 mRNA 3′-UTR sequences.
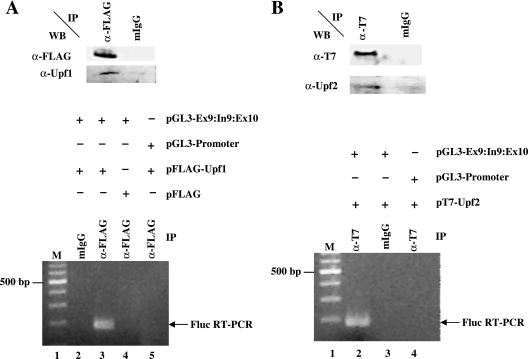
The destruction of most substrate mRNAs by NMD requires an association of both Upf1 and Upf2 with the mRNP complexes (28). To determine whether Upf1 and/or Upf2 can interact with the 3′-UTR of an NMD-sensitive AUF1 splice variant mRNA in cells, mRNP immunoprecipitation assays were performed. Epitope-tagged variants of Upf1 or Upf2 were coexpressed with a luciferase reporter mRNA lacking (pGL3-Promoter) or containing (Ex9:In9:Ex10) NMD-targeted AUF1 3′-UTR sequences. Western blots revealed that FLAG-Upf1 and T7-Upf2 were immunopurified using the appropriate antibodies but not by control IgG (Fig. 6A and B, top panels). The identities of immunoprecipitated proteins were further verified by immunoreactivity to anti-Upf1 and anti-Upf2 antibodies. RT-PCR analyses of copurified RNAs revealed that only mRNA encoded by plasmid pGL3-Ex9:In9:Ex10 coimmunopurified with FLAG-Upf1 (Fig. 6A, lower panel, lane 3) or T7-Upf2 (Fig. 6B, lower panel, lane 2). Luciferase mRNA lacking AUF1 3′-end sequences was not immunoprecipitated by antibodies targeting either epitope (Fig. 6A and B, lower panels, lanes 5 and 4, respectively), and neither reporter mRNA was detected in samples immunoprecipitated with control IgG. These results demonstrate that FLAG-Upf1 and T7-Upf2 can associate with 3′-UTR sequences of an AUF1 mRNA splice variant subject to NMD.
FIG. 6.
Association of Upf1 and Upf2 with AUF1 3′-UTR sequences. (A) Analyses of Upf1 association. HeLa cells were transiently transfected with plasmid pFLAG-Upf1 or a control vector lacking Upf1 sequences (pFLAG), together with luciferase reporter vectors lacking (pGL3-Promoter) or containing (pGL3-Ex9:In9:Ex10) the AUF1 exon 9-intron 9-exon 10 sequence downstream of the luciferase coding region. At 2 days posttransfection, whole-cell lysates were prepared and fractionated by ribonucleoprotein immunoprecipitation (IP), using anti-FLAG antibodies or control mouse IgG (mIgG) as described in Materials and Methods. Immunoprecipitated material was analyzed by Western blotting to validate anti-FLAG-dependent recovery of FLAG-Upf1 (top), while RT-PCR was used to identify copurifying firefly luciferase mRNA (bottom). Lane 1 is a 100-bp ladder, and the position of the 500-bp marker is noted to the left of the panel. (B) Analyses of Upf2 association. HeLa cells were cotransfected with plasmid pT7-Upf2 and either pGL3-Promoter or pGL3-Ex9:In9:Ex10. Cell lysates were prepared and immunoprecipitated, using control IgG or anti-T7-tag antibody. Immunoprecipitated material was analyzed by Western blotting for Upf2 and by RT-PCR for luciferase mRNA, essentially as described for panel A.
NMD-sensitive AUF1 mRNA variants are stabilized by reduction of cellular Upf1 or Upf2.
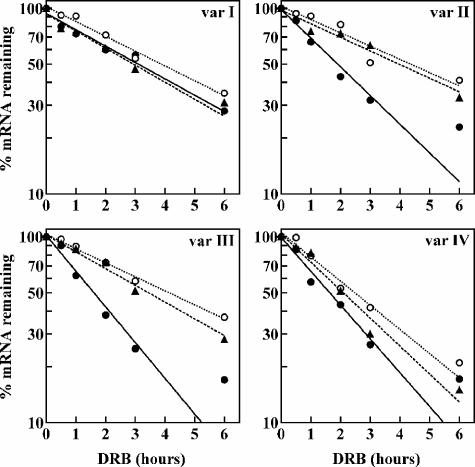
If Upf1 and Upf2 regulate AUF1 3′-UTR variant II and III mRNAs by NMD, then it follows that the enhanced expression of these mRNAs accompanying a reduction of cellular Upf1 or Upf2 (Fig. 2 and 5) should include stabilization of variant II and III mRNAs. To test this hypothesis, the stabilities of AUF1 3′-UTR splice variants I to IV were compared between HeLa cells transfected with control, Upf1, or Upf2 siRNA, using DRB time course assays (Fig. 7).
FIG. 7.
Decay kinetics of endogenous AUF1 3′-UTR splice variants following reduction of Upf1 or Upf2. The turnover rates of endogenous AUF1 3′-UTR variant I, II, III, and IV mRNAs were measured using DRB time course assays following two-hit transfections of control (solid circles, solid lines), Upf1 (open circles, dotted lines), or Upf2 (triangles, dashed lines) siRNA into HeLa cells. Levels of each AUF1 3′-UTR mRNA variant were normalized to that of GAPDH mRNA and plotted as the percent AUF1 variant mRNA remaining as a function of time following DRB treatment. Nonlinear regression analysis yielded first-order decay constants (k) and associated cellular mRNA half-lives, which are listed in Table 1.
All AUF1 3′-UTR splice variant mRNAs were relatively unstable (Table 1), but in the presence of control siRNA, AUF1 mRNA variants II, III, and IV were degraded approximately twofold faster than variant I (P < 0.005 for each variant versus variant I). This observation is consistent with the model that additional sequences retained in the longer AUF1 3′-UTR variants repress mRNA levels by enhancing mRNA decay kinetics through NMD (variants II and III) and/or AMD (variant IV). The reduction of Upf1 stabilized variant II mRNA threefold relative to that in control samples (cf. the half-lives (t1/2) [4.6 h versus 1.5 h]), while Upf2 knockdown was only slightly less effective in stabilizing this mRNA (t1/2 = 4.0 h). Similarly, the stability of AUF1 variant III mRNA was significantly increased by reduction of either Upf1 (t1/2 = 4.1 h) or Upf2 (t1/2 = 3.5 h) relative to that of the control (t1/2 = 1.6 h). A two- to threefold stabilization of NMD substrate mRNAs is consistent with observations reported by others following siRNA-directed suppression of cellular Upf1 or Upf2 (47). By contrast, reduction of either Upf1 or Upf2 had no significant effect on the half-lives of AUF1 mRNA 3′-UTR variants I and IV. Based upon these experiments, we deduce that AUF1 3′-UTR variant II and III mRNAs are selectively targeted for degradation by the Upf1/Upf2-dependent NMD pathway.
TABLE 1.
Decay kinetics of AUF1 3′-UTR variant mRNAs in control versus Upf1/Upf2-depleted HeLa cells
| Variant | Control siRNA
|
Upf1 siRNA
|
Upf2 siRNA
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| k (h−1)a | t1/2 (h)b | n | k (h−1)a | t1/2 (h)b | n | k (h−1)a | t1/2 (h)b | n | |
| I | 0.22 ± 0.04 | 3.1 | 4 | 0.23 ± 0.04 | 3.0 | 3 | 0.23 ± 0.02 | 3.0 | 3 |
| II | 0.46 ± 0.07 | 1.5 | 4 | 0.15 ± 0.04* | 4.6 | 3 | 0.17 ± 0.04* | 4.0 | 3 |
| III | 0.43 ± 0.06 | 1.6 | 5 | 0.17 ± 0.04* | 4.1 | 3 | 0.20 ± 0.03* | 3.5 | 3 |
| IV | 0.42 ± 0.07 | 1.7 | 4 | 0.30 ± 0.05 | 2.3 | 3 | 0.36 ± 0.01 | 1.9 | 3 |
First-order decay constants derived from plots of percent AUF1 3′-UTR variant mRNA remaining versus time following DRB treatment, as described in Materials and Methods. Values represent the means ± SD for n independent experiments. *, P < 0.005 versus control siRNA.
Average mRNA half-lives were calculated as follows: t1/2 = ln2/k.
AUF1 3′-end splice variant mRNAs are differentially expressed during murine embryogenesis.
At present, the functional significance of alternative pre-mRNA splicing within the AUF1 3′-UTR is unclear. However, two observations suggest that the role of NMD in regulating the expression of selected AUF1 3′-UTR variant mRNAs is to control the abundance of AUF1 mRNAs containing exon 9 sequences. First, the AUF1 variant mRNAs sensitive to NMD (II and III) are generated by a pre-mRNA splicing event that excises intron 9 but retains exon 9 (Fig. 1). Second, AUF1 sequences downstream of the stop codon are highly conserved. In particular, the 107-nt sequence of exon 9 is identical in humans and mice (57), suggesting that this region of AUF1 mRNA may contribute some essential but currently undefined regulatory role.
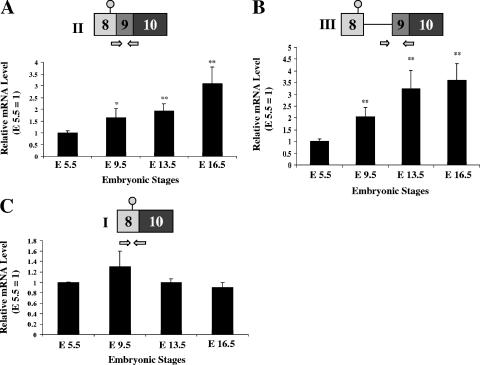
One situation where dynamic spatiotemporal patterns of AUF1 expression have been noted occurs during embryonic development. Gouble and Morello have reported variations in AUF1 protein expression between different tissues and as development progresses from stage E8.5 to adulthood (24). For example, at E10.5, AUF1 is expressed predominantly in the branchial arches, neural tube, and limb buds. By contrast, AUF1 levels in the brain are high between E10.5 and E16.5 but decline sharply thereafter. To test whether regulated expression of AUF1 3′-UTR variants might contribute to the dynamic changes in AUF1 levels observed during embryonic development, total RNAs were prepared from mouse embryos at E5.5, E9.5, E13.5, and E16.5 and analyzed for the expression of AUF1 3′-UTR variant mRNAs. Levels of both variant II and III mRNAs gradually increased as embryonic development proceeded from E5.5 to E16.5 (Fig. 8A and B). By contrast, the expression of AUF1 variant I mRNA did not change significantly between E5.5 and E16.5 (Fig. 8C). Enhanced levels of AUF1 variant II and III mRNAs in late murine development indicate that selected AUF1 3′-UTR variant mRNAs are expressed dynamically during embryogenesis. The potential significance of this novel regulatory event is considered below.
FIG. 8.
Dynamic expression of AUF1 3′-UTR splice variants during murine embryogenesis. Total RNA was purified from C57BL/6 embryos at developmental stages E5.5, E9.5, E13.5, and E16.5 as described in Materials and Methods. qRT-PCR was performed using RNAs from each developmental stage to quantify mRNA levels of AUF1 splice variant II (A), variant III (B), and variant I (C). Bars represent the means ± SD for levels of each AUF1 mRNA variant measured from four separate embryos at each stage, normalized to GAPDH mRNA and shown relative to the expression of each AUF1 variant mRNA at stage E5.5. *, P < 0.05; **, P < 0.01.
DISCUSSION
The unusual organization of the 3′ region of the AUF1 locus permits up to five different 3′-UTR structures. Earlier work demonstrated that at least two distinct mechanisms could repress the expression of specific AUF1 3′-UTR splice variants (57). The first involves high-affinity AUF1-binding sites within intron 9, permitting AMD to repress the expression of splice variants that retain intron 9 (variants IV and V) (Fig. 1). The second mechanism occurs concomitant with or results from selective excision of intron 9, which is required for the maturation of AUF1 variant II and III mRNAs. This processing event creates an exon-exon junction >50 nt downstream of the translation termination codon, which led to the hypothesis that expression of AUF1 3′-UTR variant II and III mRNAs could be regulated by the NMD pathway. In this work, RNAi-directed reduction of Upf1, a key regulator of NMD, specifically enhanced the expression of endogenous AUF1 variant II and III mRNAs (Fig. 2), which included significant stabilization of each mRNA (Fig. 7). Importantly, neither the steady-state levels nor the turnover kinetics of AUF1 variant mRNAs lacking an exon-exon junction >50 nt downstream of the stop codon (variants I and IV) were significantly influenced by Upf1 suppression. Expression of luciferase-AUF1 3′-UTR chimeric mRNAs demonstrated that Upf1-dependent changes in mRNA abundance required not only AUF1 3′-UTR sequences but also the splicing potential of intron 9 (Fig. 3). These data were further supported by parallel experiments where endogenous Upf1 activity was inhibited by ectopic expression of the dominant-negative Upf1 R844C mutant (Fig. 4). The magnitudes of AUF1 variant II and III mRNA induction following disruption of Upf1 function by siRNA or the dominant-negative mutant (two- to fivefold) are consistent with those observed for other endogenous Upf1-targeted mRNAs in comparable experiments (47, 52). Similar to the results of Upf1 experiments, RNAi-directed reduction of Upf2, a second key mediator of NMD, also led to an increase in AUF1 variant II and III mRNA levels (Fig. 5) involving stabilization of these mRNA variants (Fig. 7). Thus, we conclude that AUF1 3′-UTR variant II and III mRNAs are regulated by the NMD pathway (43, 44).
In the AUF1 3′-UTR, elements contained within intron 8, exon 9, and intron 9 are responsible for the control of AUF1 3′-UTR splice variant expression by NMD or AMD. Interestingly, within this domain there are sequences with extraordinarily strong conservation between the human and murine loci. Specifically, a 195-nt region roughly centered around exon 9 is 100% conserved between these species (57). Recent genomic analyses have identified 481 regions of at least 200 bp that are absolutely conserved (100% identity, with no insertions or deletions) between orthologous regions of the human and rodent genomes, which have been designated “ultraconserved elements” (3, 5, 6, 51). Moreover, the human and rodent genomes share 5,000 sequences with at least 100 bp of perfectly conserved sequence. Many of these ultraconserved elements overlap exons within genes that encode RNA processing factors or reside within introns or adjacent to genes that are involved in transcriptional control and development. While sequence conservation among genetic elements is a well-characterized indicator of functional significance, the absolute identity of ultraconserved elements has prompted speculation that these DNA sequences encode multiple critical functions (3). This principle raises the question of whether the ultraconserved region of the AUF1 gene containing exon 9 may be multifunctional. Supporting this model are observations that this region of the AUF1 gene is involved in the control of AUF1 splice variant expression by NMD and AMD (57; this paper). Additionally, bioinformatic analyses suggest that exon 9 may be a target for selected microRNAs (L. Banihashemi, S. Adusumalli, and G. Brewer, unpublished observations), presenting the possibility that microRNAs might act in concert with AMD and/or NMD, thereby contributing to processes dictating temporal, hormonal, or developmental expression patterns of AUF1 3′-UTR splice variants.
In this work, we observed dynamic expression of AUF1 3′-UTR splice variants during murine embryogenesis (Fig. 8), since levels of both NMD-sensitive mRNA variants (II and III) increased as embryogenesis proceeded. By contrast, levels of the NMD-insensitive splice variant I were unchanged across the developmental profile. These observations suggest that combinations of alternative pre-mRNA splicing, AMD, and NMD may be modulated during development and may contribute to stage-specific changes in the distribution of AUF1 3′-UTR variant mRNAs. Future experiments will explore the cause-and-effect relationships between NMD and dynamic expression of AUF1 3′-UTR splice variants during embryogenesis. Currently, these studies are hampered by observations that Upf1/Rent1−/− mice are embryonically lethal (45). However, it might be informative to examine the effects of targeted AUF1 3′-UTR knockouts on embryogenesis and the ability of embryonic stem (ES) cells to commit to different cell lineages following administration of appropriate stimuli. A related issue is whether specific 3′-UTR structures might be coupled to an open reading frame encoding a particular protein isoform, since the selective inclusion or exclusion of exons 2 and 7 dictates which of the four AUF1 protein isoforms are synthesized (13, 30, 54). Resolution of this problem will be challenging, however, given that the four potential splicing variants of the coding region, each linked to any of five potential 3′-UTR variants, may yield as many as 20 different mRNA structures.
The work of Dietz and colleagues indicates that NMD controls the expression of several hundred native transcripts in mammalian cells, encoding proteins responsible for a broad array of biochemical functions (47). To our knowledge, however, the current study demonstrating the regulation of AUF1 3′-UTR splice variant expression by Upf1 and Upf2 provides the first mechanistic link between the NMD and AMD pathways. By regulating AUF1 expression, the population of genes and biochemical pathways subject to control by NMD would be dramatically enlarged through a combination of direct and indirect effects (Fig. 9). For example, as an AMD factor, AUF1 promotes the decay of mRNAs encoding proteins involved in cell growth, apoptosis, and signaling (34, 35). Moreover, the AUF1 protein isoforms are multifunctional and are known to regulate the transcription of several genes (12, 14, 20, 26, 53) as well as to contribute to telomere maintenance through interaction with telomere repeats (18). Interestingly, a link between NMD and telomere maintenance via regulation of RNA-binding proteins has already been reported for simple eukaryotes. In S. cerevisiae, EST1 encodes an RNA-binding protein with structural homology to AUF1 (18). The yeast NMD pathway regulates several genes involved in modulating telomerase activity, including EST1, EST2, EST3, and STN1 (17). Moreover, the human homologue of Est1p, termed hEST1A, is similar to SMG5 and SMG7a, which are Caenorhabditis elegans gene products involved in NMD (17).
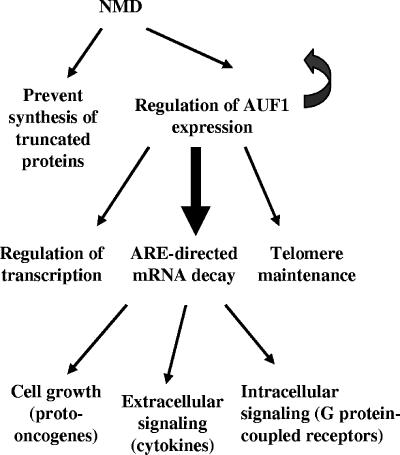
FIG. 9.
Model for regulation of AUF1 expression and possible downstream pathways by NMD. NMD was first described as a mechanism to prevent the synthesis of truncated proteins by degrading mRNAs containing PTCs. Emerging models indicate that NMD may also regulate a variety of naturally occurring mRNAs, including AUF1 mRNA, as demonstrated in the current work. By controlling the production of cellular AUF1, NMD may thus indirectly influence a broad variety of biological pathways regulated through the various AUF1 isoforms, including telomere maintenance, cell growth, extracellular and intracellular signaling, and possibly many others. This model is discussed further in the text.
Finally, the data presented in this report raise the possibility that Upf1 may directly or indirectly influence the translational efficiency of selected mRNAs. Levels of the luciferase-AUF1 Ex9:In9 chimeric mRNA, which is incapable of splicing intron 9, were unaffected by Upf1 reduction (Fig. 3). However, luciferase activity from this reporter was reduced by 80% in cells transfected with Upf1 siRNA. Neither the mRNA level nor luciferase activity from the pGL3-Promoter vector was influenced by the reduction of Upf1. Together, these observations suggest that the translational efficiency of the luciferase-AUF1 Ex9:In9 mRNA may be enhanced in the presence of Upf1 and that this regulation requires sequences within the AUF1 Ex8:In9 domain. This effect is likely not related to the reduction of 5′→3′ helicase activity following Upf1 knockdown, since ectopic expression of the Upf1 R844C mutant, which lacks helicase activity (52), resulted in twofold more luciferase expression from the Ex9:In9 reporter without significantly changing the mRNA level (Fig. 4). Accordingly, enhancement of luciferase-AUF1 Ex8:In9 translation through Upf1 is more likely mediated by some other function of the protein, such as direct RNA binding or RNA/DNA-dependent ATPase activity. Alternatively, the effect on translation of the Ex9:In9 reporter might be indirect. For example, Upf1 reduction could influence the expression of an unknown factor that is required to maintain translation of the Ex9:In9 reporter at normal levels. Future studies will address these possibilities and determine whether specific sequence elements within the AUF1 3′-UTR are required for Upf1-dependent changes in Ex9:In9 reporter translation.
In conclusion, we have identified the AUF1/HNRPD gene as a native target of NMD, thus providing a mechanistic link between the AMD and NMD pathways. The plethora of potential mRNA variants derived from the combination of five possible 3′-UTR structures linked to four alternate open reading frames provides abundant regulatory possibilities for developmental and tissue-specific expression of AUF1 protein isoforms. This, in turn, could have profound consequences for a broad range of biochemical processes, including transcriptional and posttranscriptional control of gene expression, both intracellular and extracellular signaling, and telomere maintenance.
Supplementary Material
Acknowledgments
We thank Anwar Hossein and Stuart Peltz for the Western blotting protocol and Kiran Chada and Anna Azarova for their help with the mice. We also thank Lynne Maquat, Josh Mendell, Hal Dietz, Niels Gehring, and Andreas Kulozik for their generous gifts of plasmids.
This work was supported by Public Health Service grants CA052443 (to G.B.) and CA102428 (to G.M.W.) from the National Institutes of Health.
Footnotes
Published ahead of print on 25 September 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barreau, C., L. Paillard, and H. B. Osborne. 2005. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33:7138-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bejerano, G., M. Pheasant, I. Makunin, S. Stephen, W. J. Kent, J. S. Mattick, and D. Haussler. 2004. Ultraconserved elements in the human genome. Science 304:1321-1325. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya, A., K. Czaplinski, P. Trifillis, F. He, A. Jacobson, and S. W. Peltz. 2000. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6:1226-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boffelli, D., M. A. Nobrega, and E. M. Rubin. 2004. Comparative genomics at the vertebrate extremes. Nat. Rev. Genet. 5:456-465. [DOI] [PubMed] [Google Scholar]
- 6.Boffelli, D., C. V. Weer, L. Weng, K. D. Lewis, M. I. Shoukry, L. Pachter, D. N. Keys, and E. M. Rubin. 2004. Intraspecies sequence comparisons for annotating genomes. Genome Res. 14:2406-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brewer, G. 1991. An A+U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol. 11:2460-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brewer, G. 2002. Messenger RNA decay during aging and development. Ageing Res. Rev. 1:607-625. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C. Y., and A. B. Shyu. 2003. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol. Cell. Biol. 23:4805-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu, S. Y., F. Lejeune, A. C. Ranganathan, and L. E. Maquat. 2004. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 18:745-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decker, C. J., and R. Parker. 1995. Diversity of cytoplasmic functions for the 3′ untranslated region of eukaryotic transcripts. Curr. Opin. Cell Biol. 7:386-392. [DOI] [PubMed] [Google Scholar]
- 12.Dempsey, L. A., L. A. Hanakahi, and N. Maizels. 1998. A specific isoform of hnRNP D interacts with DNA in the LR1 heterodimer: canonical RNA binding motifs in a sequence-specific duplex DNA binding protein. J. Biol. Chem. 273:29224-29229. [DOI] [PubMed] [Google Scholar]
- 13.Dempsey, L. A., M. J. Li, A. DePace, P. Bray-Ward, and N. Maizels. 1998. The human HNRPD locus maps to 4q21 and encodes a highly conserved protein. Genomics 49:378-384. [DOI] [PubMed] [Google Scholar]
- 14.Dobi, A., M. Szemes, C. Lee, M. Palkovits, F. Lim, A. B. Gyorgy, M. A. Mahan, and D. V. Agoston. 2006. AUF1 is expressed in the developing brain, binds to AT-rich double-stranded DNA, and regulates enkephalin gene expression. J. Biol. Chem. 281:28889-28900. [DOI] [PubMed] [Google Scholar]
- 15.Ehrenman, K., L. Long, B. J. Wagner, and G. Brewer. 1994. Characterization of cDNAs encoding the murine A+U-rich RNA-binding protein AUF1. Gene 149:315-319. [DOI] [PubMed] [Google Scholar]
- 16.Enokizono, Y., Y. Konishi, K. Nagata, K. Ouhashi, S. Uesugi, F. Ishikawa, and M. Katahira. 2005. Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J. Biol. Chem. 280:18862-18870. [DOI] [PubMed] [Google Scholar]
- 17.Enomoto, S., L. Glowczewski, J. Lew-Smith, and J. G. Berman. 2004. Telomere cap components influence the rate of senescence in telomerase-deficient yeast cells. Mol. Cell. Biol. 24:837-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eversole, A., and N. Maizels. 2000. In vitro properties of the conserved mammalian protein hnRNP D suggest a role in telomere maintenance. Mol. Cell. Biol. 20:5425-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frischmeyer, P. A., and H. C. Dietz. 1999. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 8:1893-1900. [DOI] [PubMed] [Google Scholar]
- 20.Fuentes-Panana, E. M., R. Peng, G. Brewer, J. Tan, and P. D. Ling. 2000. Regulation of the Epstein-Barr virus C promoter by AUF1 and the cyclic AMP/protein kinase A signaling pathway. J. Virol. 74:8166-8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao, Q., B. Das, F. Sherman, and L. E. Maquat. 2005. Cap-binding protein 1-mediated and eukaryotic translation initiation factor 4E-mediated pioneer rounds of translation in yeast. Proc. Natl. Acad. Sci. USA 102:4258-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gehring, N. H., J. B. Kunz, G. Neu-Yilik, S. Breit, M. H. Viegas, M. W. Hentze, and A. E. Kulozik. 2005. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell 20:65-75. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez, C. I., W. Wang, and S. W. Peltz. 2001. Nonsense-mediated mRNA decay in Saccharomyces cerevisiae: a quality control mechanism that degrades transcripts harboring premature termination codons. Cold Spring Harb. Symp. Quant. Biol. 66:321-328. [DOI] [PubMed] [Google Scholar]
- 24.Gouble, A., and D. Morello. 2000. Synchronous and regulated expression of two AU-binding proteins, AUF1 and HuR, throughout murine development. Oncogene 19:5377-5384. [DOI] [PubMed] [Google Scholar]
- 25.Guhaniyogi, J., and G. Brewer. 2001. Regulation of mRNA stability in mammalian cells. Gene 265:11-23. [DOI] [PubMed] [Google Scholar]
- 26.Hanakahi, L. A., and N. Maizels. 2000. Transcriptional activation by LR1 at the Emu enhancer and switch region sites. Nucleic Acids Res. 28:2651-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hentze, M. W., and A. E. Kulozik. 1999. A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96:307-310. [DOI] [PubMed] [Google Scholar]
- 28.Hosoda, N., Y. K. Kim, F. Lejeune, and L. E. Maquat. 2005. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 12:893-901. [DOI] [PubMed] [Google Scholar]
- 29.Ishigaki, Y., X. Li, G. Serin, and L. E. Maquat. 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106:607-617. [DOI] [PubMed] [Google Scholar]
- 30.Kajita, Y., J. Nakayama, M. Aizawa, and F. Ishikawa. 1995. The UUAG-specific RNA binding protein, heterogeneous nuclear ribonucleoprotein D0. Common modular structure and binding properties of the 2xRBD-Gly family. J. Biol. Chem. 270:22167-22175. [DOI] [PubMed] [Google Scholar]
- 31.Kim, Y. K., L. Furic, L. Desgroseillers, and L. E. Maquat. 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120:195-208. [DOI] [PubMed] [Google Scholar]
- 32.Knapinska, A. M., P. Irizarry-Barreto, S. Adusumalli, I. Androulakis, and G. Brewer. 2005. Molecular mechanisms regulating mRNA stability: physiological and pathological significance. Curr. Genomics 6:471-486. [Google Scholar]
- 33.Lal, A., K. Abdelmohsen, R. Pullmann, T. Kawai, S. Galban, X. Yang, G. Brewer, and M. Gorospe. 2006. Posttranscriptional derepression of GADD45alpha by genotoxic stress. Mol. Cell 22:117-128. [DOI] [PubMed] [Google Scholar]
- 34.Lal, A., K. Mazan-Mamczarz, T. Kawai, X. Yang, J. L. Martindale, and M. Gorospe. 2004. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 23:3092-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lapucci, A., M. Donnini, L. Papucci, E. Witort, A. Tempestini, A. Bevilacqua, A. Nicolin, G. Brewer, N. Schiavone, and S. Capaccioli. 2002. AUF1 is a bcl-2 A+U-rich element-binding protein involved in bcl-2 mRNA destabilization during apoptosis. J. Biol. Chem. 277:16139-16146. [DOI] [PubMed] [Google Scholar]
- 36.Le Hir, H., D. Gatfield, E. Izaurralde, and M. J. Moore. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20:4987-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Hir, H., E. Izaurralde, L. E. Maquat, and M. J. Moore. 2000. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19:6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Hir, H., M. J. Moore, and L. E. Maquat. 2000. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 14:1098-1108. [PMC free article] [PubMed] [Google Scholar]
- 39.Lejeune, F., X. Li, and L. E. Maquat. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12:675-687. [DOI] [PubMed] [Google Scholar]
- 40.Lejeune, F., A. C. Ranganathan, and L. E. Maquat. 2004. eIF4G is required for the pioneer round of translation in mammalian cells. Nat. Struct. Mol. Biol. 11:992-1000. [DOI] [PubMed] [Google Scholar]
- 41.Li, S., and M. F. Wilkinson. 1998. Nonsense surveillance in lymphocytes? Immunity 8:135-141. [DOI] [PubMed] [Google Scholar]
- 42.Liao, B., Y. Hu, D. J. Herrick, and G. Brewer. 2005. The RNA-binding protein IMP-3 is a translational activator of insulin-like growth factor II leader-3 mRNA during proliferation of human K562 leukemia cells. J. Biol. Chem. 280:18517-18524. [DOI] [PubMed] [Google Scholar]
- 43.Maquat, L. E. 2005. Nonsense-mediated mRNA decay in mammals. J. Cell Sci. 118:1773-1776. [DOI] [PubMed] [Google Scholar]
- 44.Maquat, L. E. 2004. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 5:89-99. [DOI] [PubMed] [Google Scholar]
- 45.Medghalchi, S. M., P. A. Frischmeyer, J. T. Mendell, A. G. Kelly, A. M. Lawler, and H. C. Dietz. 2001. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet. 10:99-105. [DOI] [PubMed] [Google Scholar]
- 46.Mendell, J. T., C. M. ap Rhys, and H. C. Dietz. 2002. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298:419-422. [DOI] [PubMed] [Google Scholar]
- 47.Mendell, J. T., N. A. Sharifi, J. L. Meyers, F. Martinez-Murillo, and H. C. Dietz. 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36:1073-1078. [DOI] [PubMed] [Google Scholar]
- 48.Nagy, E., and L. E. Maquat. 1998. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 23:198-199. [DOI] [PubMed] [Google Scholar]
- 49.Raineri, I., D. Wegmueller, B. Gross, U. Certa, and C. Moroni. 2004. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 32:1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ross, J. 1995. mRNA stability in mammalian cells. Microbiol. Rev. 59:423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siepel, A., G. Bejerano, J. S. Pedersen, A. S. Hinrichs, M. Hou, K. Rosenbloom, H. Clawson, J. Spieth, L. W. Hillier, S. Richards, G. M. Weinstock, R. K. Wilson, R. A. Gibbs, W. J. Kent, W. Miller, and D. Haussler. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15:1034-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun, X., H. A. Perlick, H. C. Dietz, and L. E. Maquat. 1998. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 95:10009-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tolnay, M., J. D. Lambris, and G. C. Tsokos. 1997. Transcriptional regulation of the complement receptor 2 gene: role of a heterogeneous nuclear ribonucleoprotein. J. Immunol. 159:5492-5501. [PubMed] [Google Scholar]
- 54.Wagner, B. J., C. T. DeMaria, Y. Sun, G. M. Wilson, and G. Brewer. 1998. Structure and genomic organization of the human AUF1 gene: Alternative pre-mRNA splicing generates four protein isoforms. Genomics 48:195-202. [DOI] [PubMed] [Google Scholar]
- 55.Wilkinson, M. F. 2003. The cycle of nonsense. Mol. Cell 12:1059-1061. [DOI] [PubMed] [Google Scholar]
- 56.Wilson, G. M., and G. Brewer. 1999. The search for trans-acting factors controlling messenger RNA decay. Prog. Nucleic Acid Res. Mol. Biol. 62:257-291. [DOI] [PubMed] [Google Scholar]
- 57.Wilson, G. M., Y. Sun, J. Sellers, H. Lu, N. Penkar, G. Dillard, and G. Brewer. 1999. Regulation of AUF1 expression via conserved alternatively spliced elements in the 3′ untranslated region. Mol. Cell. Biol. 19:4056-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilusz, C. J., M. Wormington, and S. W. Peltz. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2:237-246. [DOI] [PubMed] [Google Scholar]
- 59.Yamashita, A., T. C. Chang, Y. Yamashita, W. Zhu, Z. Zhong, C. Y. Chen, and A. B. Shyu. 2005. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 12:1054-1063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.