Abstract
Gata4, a member of the zinc finger family of GATA transcription factors, is highly expressed in duodenum and jejunum but is nearly undetectable in distal ileum of adult mice. We show here that the caudal reduction of Gata4 is conserved in humans. To test the hypothesis that the regional expression of Gata4 is critical for the maintenance of jejunal-ileal homeostasis in the adult small intestine in vivo, we established an inducible, intestine-specific model that results in the synthesis of a transcriptionally inactive Gata4 mutant. Synthesis of mutant Gata4 in jejuna of 6- to 8-week-old mice resulted in an attenuation of absorptive enterocyte genes normally expressed in jejunum but not in ileum, including those for the anticipated targets liver fatty acid binding protein (Fabp1) and lactase-phlorizin hydrolase (LPH), and a surprising induction of genes normally silent in jejunum but highly expressed in ileum, specifically those involved in bile acid transport. Inactivation of Gata4 resulted in an increase in the goblet cell population and a redistribution of the enteroendocrine subpopulations, all toward an ileal phenotype. The gene encoding Math1, a known activator of the secretory cell fate, was induced ∼75% (P < 0.05). Gata4 is thus an important positional signal required for the maintenance of jejunal-ileal identities in the adult mouse small intestine.
The mammalian small intestine is lined by a highly specialized epithelium that displays a wide-ranging yet tightly regulated functional diversity along its cephalo-caudal axis (13). The functional diversity is linked to a continuous renewal process in which stem cells located near the base of crypts produce transit-amplifying cells that ultimately differentiate into four principal cell types. Absorptive enterocytes, which constitute the majority of intestinal epithelial cells, goblet cells, and enteroendocrine cells, migrate up the villi and are shed into the intestinal lumen every 3 to 5 days, whereas Paneth cells reside at the base of crypts and turn over more slowly. Absorptive enterocyte genes that encode proteins responsible for the terminal digestion and absorption of most nutrients are expressed primarily in duodenum and jejunum, whereas those that mediate absorption of conjugated bile salts and intrinsic factor-bound vitamin B12 are localized to the distal ileum. Goblet and Paneth cells are more numerous in distal small intestine, presumably to provide greater protection against bacterial infiltration, and enteroendocrine subpopulations display a functional diversity characterized by the regional segregation of hormones that activate or repress gastrointestinal processes. Gene knockout and overexpression models have resulted in the discovery that Wnt, Hedgehog, and Notch signaling are critical regulatory pathways for intestinal differentiation and development in vivo (1). However, little is known about the processes involved in the maintenance of regional identities in the mature intestine.
Gata4 is a member of an evolutionarily conserved family of zinc finger transcription factors that are necessary for a variety of developmental processes, including embryonic morphogenesis and cellular differentiation (23). Inactivation of GATA homologues in lower animals, such as elt-2 in Caenorhabditis elegans (11), serpent in Drosophila melanogaster (28), or gata4 in zebrafish (15), results in defects in heart development and gastrointestinal organogenesis, demonstrating a conservation of function. In mice, Gata4 is expressed in visceral endoderm in embryos and in developing and mature heart, ovary, testis, adrenal glands, pancreas, liver, stomach, and small intestine (23). Gata4 inactivation models in mice reveal abnormalities in ventral morphogenesis resulting in a failure to form a primitive heart tube and foregut (20, 24), as well as specific defects in cardiac morphogenesis and proliferation later in embryonic development (27, 46, 48). However, due to the embryonic lethality of these models, the importance of Gata4 for intestinal function in vivo in mature mice is currently unknown.
Gata4 is highly expressed in duodenum and jejunum but is greatly reduced in distal ileum of adult mice (44), implying a regulatory role along the cephalo-caudal axis. It is not known if this regional pattern occurs in humans. Gata4 is expressed in absorptive enterocytes on villi (3, 7, 9, 44), where it is thought to transactivate specific target genes (3, 7, 8, 10, 12, 25, 44). Isolated Gata4−/− cells in the small intestines of chimeric E18.5 mice do not express the intestinal differentiation marker liver fatty acid binding protein (Fabp1 product) (7), providing key in vivo data demonstrating a critical importance for Gata4 in the regulation of intestinal gene expression and a lack of overlapping functions with other intestinal GATA factors. We have shown that Gata4 binds and activates in vitro the promoter of the putative absorptive enterocyte target lactase-phlorizin hydrolase (LPH). We have further shown that Gata4 physically associates with hepatocyte nuclear factor 1α (Hnf1α), a homeodomain transcription factor that is also expressed in the intestinal epithelium (2, 3, 32), to synergistically activate the LPH promoter through an evolutionarily conserved mechanism (44, 45). Finally, we have hypothesized that the known distal decline in LPH gene expression is linked to the caudal reduction in Gata4 expression (44). In the present study, we establish an inducible, intestine-specific Gata4 knockout model to test the hypothesis that Gata4 is required for the maintenance of jejunal-ileal homeostasis in the adult small intestine in vivo.
MATERIALS AND METHODS
Mice.
Mice were housed under standard conditions in the Animal Research at Children's Hospital facility and were provided food and water ad libitum. Gata4flox/flox mice (27) were crossed with transgenic Villin-CreERT2 mice (21) (generous gift of S. Robine, Institut Curie, Paris, France) to generate experimental Gata4flox/flox/Villin-CreERT2 mice. In these mice, Cre is expressed in the epithelial cells of the small and large intestines. The expressed Cre is fused to a mutated estrogen receptor and resides in the cytoplasm until it is translocated into the nucleus upon tamoxifen treatment, where it then excises floxed DNA. Gata4flox/flox mice positive for the Villin-CreERT2 transgene were designated Gata4 mutant mice, whereas Gata4flox/flox/Villin-CreERT2-negative littermates were used as controls.
Genotyping was conducted on tail DNA as described previously (2), using specific primers for Cre and exon 2 of Gata4. Nuclear translocation of Cre was induced in mice at 6 to 8 weeks of age by five daily intraperitoneal injections of tamoxifen (100 μl, 10 mg/ml; Sigma-Aldrich Company, St. Louis, MO) as described previously (14, 21), and study mice were sacrificed 14 days after the last injection. During this 14-day span, mice were weighed and were observed daily for general activity, skin and hair condition, and stool firmness. In selected experiments, mice were treated for 1, 2, or 3 days with tamoxifen and then sacrificed 24 h after the last injection. At the time of sacrifice, mice were anesthetized with avertin (2,2,2-tribromoethanol; Sigma) anesthesia, and the intestine was removed from a midline incision, placed on a glass plate on a bed of ice, and rinsed with ice-cold phosphate-buffered saline (PBS). Intestinal tissue was obtained along the length of the small intestine for isolation of RNA and nuclear extracts and sectioning, as described previously (2, 44). Approval was obtained from the Institutional Animal Care and Use Committee for all mouse experiments described here.
Immunofluorescence.
Intestinal segments were flushed with ice-cold PBS, fixed for 4 h in 4% paraformaldehyde at 4°C, and rinsed with 70% ethanol in PBS. The tissue was embedded in paraffin, sectioned (5 μm), and stained with hemotoxylin and eosin (H&E) in the Department of Pathology at Children's Hospital, Boston, Mass. For selected sections, the periodic acid-Schiff (PAS) reaction to stain for glycoproteins was conducted.
For immunofluorescence, sections were heated for 15 min at 65°C, deparaffinized, and rehydrated. Antigen retrieval was performed by boiling slides for 10 min in 10 mM sodium citrate, pH 6.0. After cooling, slides were washed three times for 5 min each in PBS and incubated in a blocking solution containing 10% donkey serum in PBS for 1 h in a humidified chamber at room temperature. Blocking serum was replaced by the primary antibody in 10% donkey serum-PBS, and the mixture was incubated overnight at 4°C in a humidified chamber. Slides were then washed three times for 5 min each in PBS and incubated with the secondary antibody in 10% donkey serum-PBS for 4 h in a humidified chamber at room temperature. In some experiments, a solution containing DAPI (4′,6-diamino-2-phenylindole dihydrochloride) nucleic acid stain (2 μg/ml) (D1306; Molecular Probes) in PBS was added and incubated for 15 min at room temperature. Slides were washed in PBS, mounted in Mowiol mounting medium (Calbiochem, San Diego, CA), and allowed to dry overnight.
The primary antibodies included mouse anti-Gata4 (1:250) (sc-25310; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), rabbit anti-Gata4 (1:100) (sc-9053; Santa Cruz), goat anti-Gata4 (1:500) (sc-1237; Santa Cruz), goat anti-Gata5 (1:40 or 1:400) (sc-7280; Santa Cruz), rabbit anti-Gata5 (1:40 or 1:400) (sc-9054; Santa Cruz), rabbit anti-Gata6 (1:20) (sc-9055; Santa Cruz), rabbit anti-chromogranin A (1:1,000) (20085; Immunostar, Inc., Hudson, WI), rabbit antilysozyme (1:200) (18-0039; Zymed Laboratories, Inc., San Francisco, CA), rabbit anti-Ki67 (1:100) (RM-9106; Lab Vision), rabbit anti-Fabp1 (1:1,000) (a kind gift of J. Gordon, Washington University), rabbit anti-Asbt (1:500) (a kind gift of P. Dawson, Wake Forest University), and rabbit anti-cleaved caspase-3 (1:100) (9664; Cell Signaling Technology, Inc., Danvers, MA). The secondary antibodies used were Alexa fluor 594 donkey anti-goat antibody, Alexa fluor 488 donkey anti-mouse antibody, and Alexa fluor 488 donkey anti-rabbit antibody (1:500) (Invitrogen Corporation, Carlsbad, CA). In selected experiments, a Gata4-blocking peptide (1:500) (sc-1237P; Santa Cruz) was used to verify specific immunostaining.
Semiquantitative and real-time RT-PCR.
Semiquantitative and real-time reverse transcription-PCRs (RT-PCRs) were conducted as previously described (2). Briefly, RNA was isolated using the RNeasy kit (QIAGEN, Inc., Valencia, CA) with DNase treatment (Ambion, Inc., Austin, TX), quantified by optical density at 260 nm, and checked on an agarose gel for intact rRNA bands. cDNA was synthesized using iScript (Bio-Rad Laboratories, Inc, Hercules, CA) according to the manufacturer's instructions. Primer pairs were designed using Beacon Design software (PREMIER Biosoft International, Palo Alto, CA) and optimized as described previously (2). Primer sequences are available upon request. Semiquantitative RT-PCR was terminated in the linear range of amplification, and real-time RT-PCR was carried out using an iCycler and iQ SYBR Green Supermix (Bio-Rad) on four to six animals in each group, corrected for the glyceraldehyde-3-phosphate dehydrogenase gene (Gapdh), and expressed relative to the calibrator, which was either pooled adult jejunal or adult ileal RNA.
Western blotting.
Western blot analysis was performed as previously described (44), using 25 μg of nuclear extracts from the jejuna of control and Gata4 mutant mice and an affinity-purified goat polyclonal antibody directed against the C-terminal domain of Gata4 (sc-1237; Santa Cruz). Nuclear extracts were prepared from isolated epithelial cells as described previously (2).
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were carried out as previously described (44), using nuclear extracts from isolated epithelial cells and a probe that specifically binds GATA proteins (12, 19). For supershift analyses, a goat polyclonal Gata4 antibody (0.1 μg/μl) (sc-1237X; Santa Cruz Biotechnology) was preincubated with the nuclear extracts for 20 min prior to the addition of the probe.
Cloning and construction of a mutant Gata4 expression vector.
During the course of these studies, we found that the mutant mice produced a truncated form of Gata4, termed Gata4Δex2. To clone Gata4Δex2 for further analysis, total RNA was isolated from Gata4wt/Δex2 heart tissue and reverse transcribed using random hexamers and Superscript II (Invitrogen). cDNA was PCR amplified using primers spanning exon 2, and a 450-bp amplicon containing the mutant 5′ end was subcloned into the pCRII-TOPO plasmid (Invitrogen). The mutant 5′ end was then excised by digestion of internal HindIII and MfeI restriction sites and used to replace the 5′ end of the full-length, wild-type Gata4 cDNA. HindIII/NotI digests were used to transfer the 1.3-kb mutant Gata4 cDNA into the pcDNA3.1(+) expression vector. The expression vector was confirmed by sequencing.
Transient cotransfection.
Transient-cotransfection assays were carried out with HeLa cells, using expression vectors for wild-type or mutant Gata4, Gata6, and a 118-bp mouse LPH promoter-human growth hormone reporter construct as described previously (44). Briefly, plasmid DNA (0.4 μg/well in six-well plates) was transfected into preconfluent HeLa cells (∼90%) by using Lipofectamine 2000 (Invitrogen). After 48 h, human growth hormone was quantified from medium by using the human growth hormone DA kit (MP Biomedicals, Orangeburg, NY). Transcriptional activity was expressed relative to that of a metallothionein promoter-human growth hormone reporter transfected in parallel wells. All experiments were conducted in triplicate, and statistically significant differences were determined. HeLa cells were tested before and after experimentation to document an absence of mycoplasma infection, using the MycoTect kit (Invitrogen).
Statistical analyses.
Statistically significant differences were determined by Student's t test or analysis of variance followed by the Tukey-Kramer multiple comparison test. Goblet cell counts were performed by eight independent, masked observers counting a total of 10 villi on PAS-stained slides. Data were expressed as the means from six control and six Gata4 mutant mice.
RESULTS
Gata4 is expressed in absorptive enterocytes on villi and throughout the crypt epithelium.
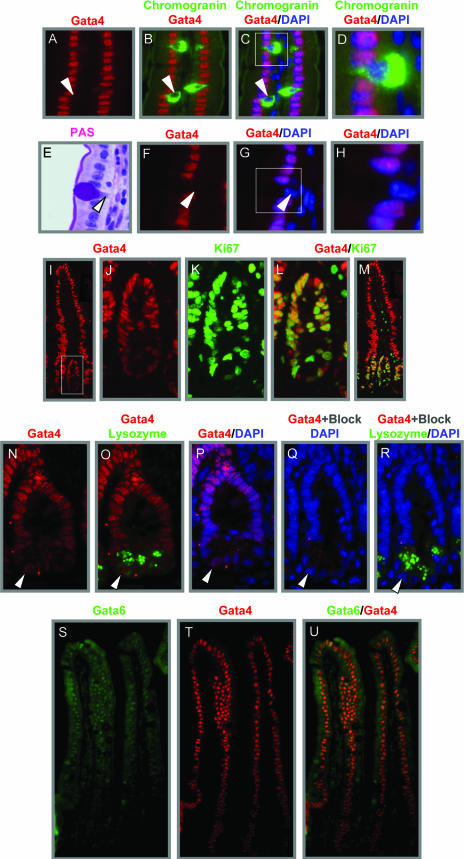
Although it is well recognized that Gata4 is highly expressed in absorptive enterocytes on villi (3, 7, 9, 44), its expression in proliferating crypt cells has not been fully appreciated. We compared three different anti-Gata4 antibodies by immunofluorescence and found that goat anti-Gata4 produced immunostaining with the highest specificity and lowest background. Using this antibody and an antibody for the enteroendocrine marker chromogranin A (Fig. 1A to D), we found no evidence of Gata4 in this lineage, as previously shown (9). Using serial sections that included PAS-stained slides and a criterion that goblet cell nuclei must be outside the plane of absorptive enterocyte nuclei, we also found no evidence of Gata4 in this lineage (Fig. 1E to H), also as previously shown (9). Gata4 immunofluorescence, however, was found throughout the crypt epithelium (Fig. 1I and J), though at a lower intensity than that which occurs on villi. Gata4 was coexpressed with the marker for proliferating cells, Ki67, in the nuclei of epithelial cells of the upper crypt (Fig. 1J to M), as well as lysozyme-positive Paneth cells at the base of the crypt (Fig. 1N to P). Blocking experiments using the specific peptide that was used to raise the goat anti-Gata4 antibody demonstrated that the crypt staining was specific for the Gata4 antibody. Although background immunofluorescence in secretory granules of Paneth cells remained detectable (Fig. 1Q), the blocking peptide completely attenuated Gata4 immunostaining in nuclei of crypts (Fig. 1Q and R) and villi (not shown), demonstrating that the Gata4 immunostaining in crypts was specific. Gata6 was coexpressed with Gata4 in absorptive enterocytes on villi (Fig. 1S to U), in agreement with previous studies (7, 37), but the presence of Gata6 in either enteroendocrine cells, as previously reported (9), or crypts (not shown) could not be documented. Although Gata5 has been shown by immunohistochemistry to be in cells of the secretory lineages (goblet, enteroendocrine, and Paneth cells) (8), we could not verify Gata5 by either immunofluorescence or immunohistochemistry with two different commercial antibodies (not shown). Thus, on villi, Gata4 is localized specifically to the absorptive enterocyte lineage, where it is coexpressed with Gata6. In crypts, Gata4 is expressed throughout the epithelium, including proliferating Ki67-positive cells and nonproliferating Paneth cells.
FIG. 1.
Gata4 is expressed in absorptive enterocytes on villi and throughout the crypt epithelium in the adult mouse jejunum. (A to D) Immunofluorescence showing that Gata4 (red) is not expressed in the nuclei (DAPI, blue) of chromogranin A (green) enteroendocrine cells. Arrowheads indicate the absence of Gata4 in the nucleus of a specific enteroendocrine cell. (E to H) Serial section of a PAS-stained goblet cell, showing absence of Gata4 immunofluorescence (red) in the nucleus (DAPI, blue) of this cell (arrowhead). (I to M) Coimmunofluorescence for Gata4 (red) and Ki67 (green), showing that Gata4 is expressed in proliferating epithelial cells of the upper crypt (yellow). (N to P) Coimmunofluorescence for Gata4 (red) and lysozyme (green), showing that Gata4 is expressed in the nuclei (DAPI, blue) of Paneth cells. (Q and R) Serial section using a blocking peptide, showing that the nuclear Gata4 expression in Paneth cells is not due to nonspecific fluorescence. (S to U) Coimmunofluorescence for Gata6 (green) and Gata4 (red), showing that these Gata factors are coexpressed in the absorptive enterocytes on villi (yellow).
Absence of Gata4 in the ileum is conserved in humans.
Quantification of Gata4 mRNA in five equidistant segments along the length of the adult mouse small intestine by real-time RT-PCR revealed that Gata4 mRNA abundance in the most distal segment (segment 5) was ∼5% of that in all other intestinal segments studied (P < 0.05), demonstrating a sharp decline in Gata4 expression localized to the distal ileum (Fig. 2A) as previously shown (8, 44). This is consistent with our previous Western analysis which showed a greatly reduced abundance of Gata4 protein in distal ileum compared to all other segments of adult mouse small intestine (44). Gata4 was expressed in the epithelial cells of the adult human jejunum (Fig. 2B) but was not detected in human ileum (Fig. 2C), as determined by immunofluorescence. H&E staining (Fig. 2D and E) and positive nuclear staining for Hnf1α (data not shown) confirmed the integrity of the tissue sections. These data demonstrate that the regional expression of Gata4 is conserved in humans and that data obtained from mice on the regulation of anterior-posterior homeostasis by Gata4 is likely relevant to humans.
FIG. 2.
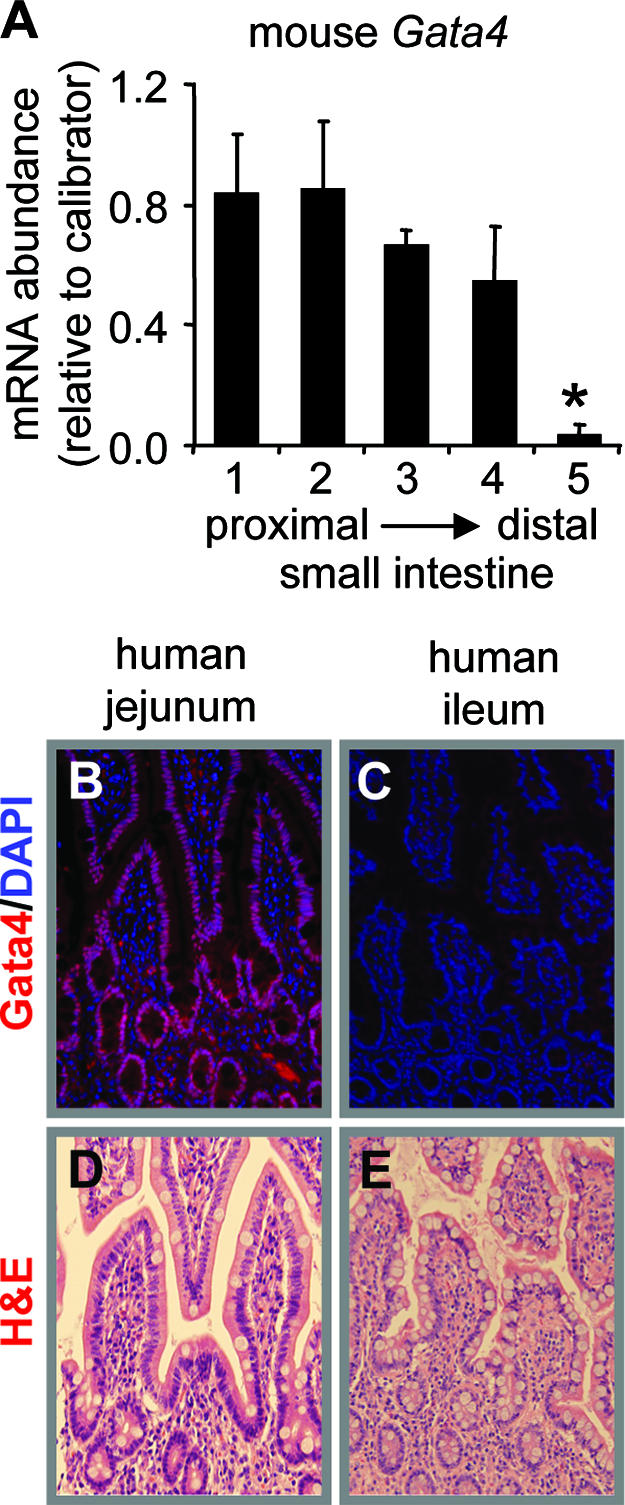
The absence of Gata4 in adult ileum is conserved in humans. (A) Real-time RT-PCR analysis of Gata4 mRNA abundance along the length of the adult mouse small intestine, showing that the level of Gata4 mRNA is significantly lower in the distal ileum than in all other segments ( , P < 0.05; mean ± SEM, n = 3). The calibrator was a pooled sample of jejunal RNA. (B and C) Immunofluorescence of adult human intestinal epithelium for Gata4 (red) and nucleic acid by DAPI (blue), showing the presence of Gata4 in jejunum (B) and the absence of Gata4 in ileum (C). (D and E). H&E staining of adult human jejunum and ileum, showing intact morphology.
, P < 0.05; mean ± SEM, n = 3). The calibrator was a pooled sample of jejunal RNA. (B and C) Immunofluorescence of adult human intestinal epithelium for Gata4 (red) and nucleic acid by DAPI (blue), showing the presence of Gata4 in jejunum (B) and the absence of Gata4 in ileum (C). (D and E). H&E staining of adult human jejunum and ileum, showing intact morphology.
Establishment of an inducible, intestine-specific Gata4 inactivation model.
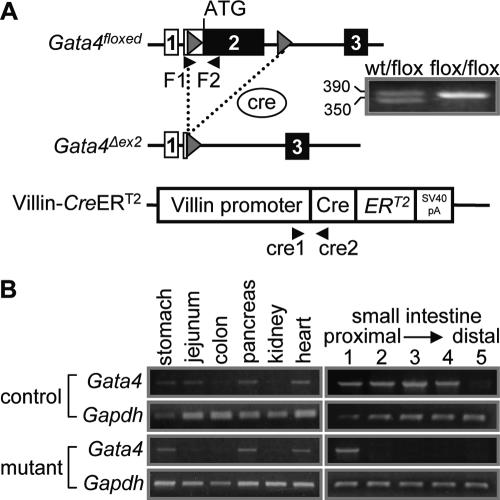
To overcome the limitation of the early lethal phenotype in germ line Gata4 null mice (20, 24), we established an inducible, intestine-specific Gata4 inactivation model (Fig. 3). In this model, a previously validated mouse line was used, in which a portion of exon 2 containing the translational start site and N-terminal activation domains of Gata4 was flanked by loxP sites (floxed) (Fig. 3A) (27). Germ line Cre expression in mice homozygous for the Gata4 floxed allele (Gata4flox/flox) exhibited a phenotype that was indistinguishable from that in published germ line Gata4 knockout models (20, 24) (data not shown), verifying these mice. Gata4flox/flox mice were mated with an established transgenic line (Villin-CreERT2) in which Cre is expressed in the epithelial cells of the small and large intestines under the control of the villin promoter (Fig. 3A) (21). The expressed Cre is fused to a mutated estrogen receptor and resides in the cytoplasm until it is translocated into the nucleus upon tamoxifen treatment, where it then excises floxed DNA. The Villin-CreERT2 transgene is expressed in stem cells, since recombination is stably maintained for up to 60 days after tamoxifen treatment (21).
FIG. 3.
Villin-CreERT2-mediated recombination of the Gata4flox allele in mouse small intestine. (A) Schematic representation of the Gata4 and Villin-CreERT2 alleles. The Gata4flox allele encodes wild-type Gata4, whereas the Gata4Δex2 allele represents the Gata4 locus after Cre-mediated recombination. Arrows indicate locations of primers used for genotyping, and the inset shows the PCR products that distinguish the wild-type (wt) (350 bp) from the floxed (390 bp) Gata4 alleles. (B) Semiquantitative RT-PCR analyses of exon 2 with the F1 and F2 primers reveal normal Gata4 expression in stomach, pancreas, and heart but null expression in the small intestines of the mutant mice, with the exception of duodenum (segment 1), where Gata4 mRNA remains detectable.
As shown in Fig. 3B, Gata4 was expressed normally in stomach, pancreas, and heart but was inactivated in the jejunum of Gata4 mutant mice. Recombination was complete along the length of the small intestine with the exception of the duodenum, where exon 2 was not excised completely (Fig. 3B). This was confirmed by experiments using Rosa26 mice (38), which revealed Cre excision specifically in small and large intestines but not in duodenum (data not shown). Inactivation of Gata4 in the jejunum was complete from 1 day to 4 weeks after the completion of tamoxifen treatment (data not shown).
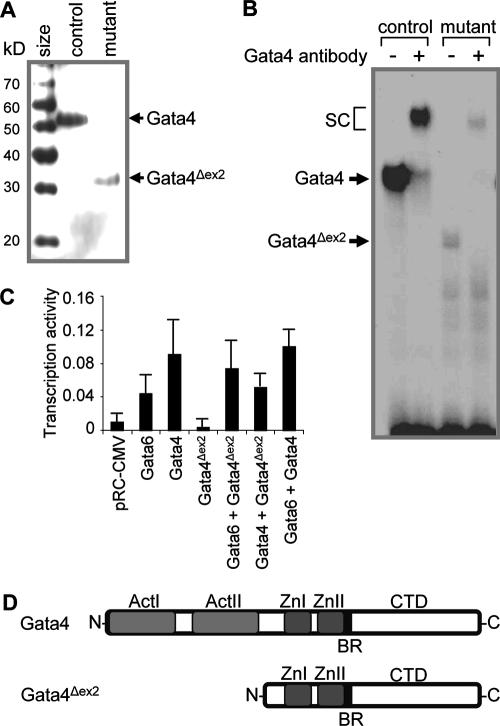
Western analysis using nuclear extracts isolated from the jejuna of control and Gata4 mutant mice and an antibody specific for the C-terminal domain of Gata4 revealed that the mutant mice synthesize a truncated form of Gata4 (∼33 kDa; Gata4Δex2) in the jejunum (Fig. 4A). Quantitative analysis revealed that the abundance of Gata4Δex2 in mutant jejunum was similar to that of Gata4 in control jejunum (n = 4 in each group) (data not shown). Although Gata4Δex2 was capable of binding to DNA as shown by using EMSAs (Fig. 4B), it was unable to activate the LPH promoter in cell culture coexpression experiments (Fig. 4C), similar to the case for a previously engineered deletion of the Gata4 activation domains (44). Gata4 and Gata6 both activated the LPH promoter as anticipated (10, 19), but Gata4Δex2 did not. Cotransfection of Gata6 or Gata4 with Gata4Δex2 revealed evidence of a dominant-negative effect on Gata4 activation but not on Gata6 activation. The predicted structure (Fig. 4D) is based on excision of the native translational start site and activation domains in exon 2, translation of a predicted in-frame ATG in exon 3, the size of the protein on Western blots (Fig. 4A), and recognition by a C-terminally directed antibody. These data demonstrate that treatment of Gata4 mutant mice with tamoxifen results in the synthesis of a truncated, transcriptionally inactive form of Gata4 in the jejunum that is unlikely to have a dominant-negative effect with Gata6.
FIG. 4.
An inactive, truncated form of Gata4 is synthesized in the jejuna of Gata4 mutant mice. (A) Western blot analysis using an antibody specific for the C-terminal domain of Gata4, demonstrating a specific band of ∼54 kDa in the jejuna of control mice (Gata4) and the absence of this band in Gata4 mutant animals. In the Gata4 mutant mice, another specific band of ∼33 kDa reveals the presence of a truncated Gata4 protein (Gata4Δex2). Identical results were obtained from three other mutant animals. (B) EMSA using a standardized GATA binding site as a probe (44) and nuclear extracts from control and Gata4 mutant mice, showing that both Gata4 and Gata4Δex2 bind DNA. Supershift complexes (SC) are formed using an antibody directed against the C-terminal domain of Gata4. (C) Transient-cotransfection assay in HeLa cells, demonstrating that Gata4Δex2 is transcriptionally inactive and does not demonstrate dominant-negative activity with Gata6. A human LPH promoter/human growth hormone reporter plasmid was cotransfected into HeLa cells with the empty expression vector pRC-CMV or individually or in combinations of expression vectors for Gata6, Gata4, and Gata4Δex2. Transcriptional activity is expressed as the ratio of the amount of human growth hormone synthesized from the human LPH promoter/human growth hormone reporter relative to that of the metallothionein promoter fused to human growth hormone. Data are expressed as means ± SEMs (n = 5 assays). (D) Schematic representation of the predicted Gata4 protein synthesized in the jejuna of the Gata4 mutant mice, showing the deletion of the N-terminal activation domains (ActI and ActII) but intact zinc fingers (ZnI and ZnII), basic region (BR), and C-terminal domain (CTD).
Absorptive enterocytes in the jejunum acquire an ileal-like gene expression program in the Gata4 mutant mice.
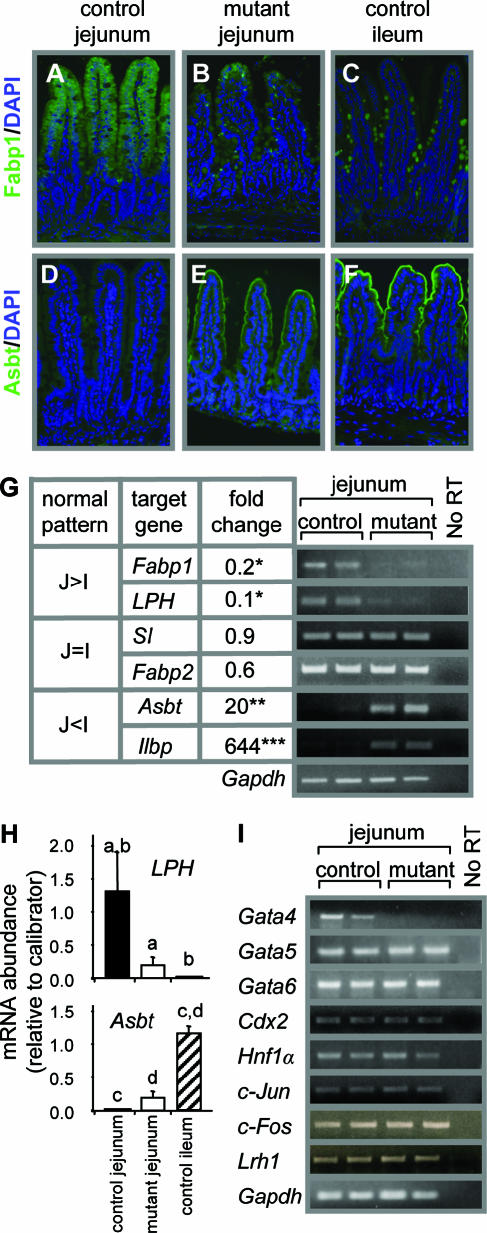
During the 2-week interval between the completion of tamoxifen treatment and the end of the study, it was noted that body weight, general activity, skin and hair condition, and stool firmness of the Gata4 mutant mice were indistinguishable from those of controls. To define the role of Gata4 in the regulation of proximal-distal patterns of absorptive enterocyte gene expression, we immunostained for Fabp1, which is highly expressed in jejunum but nearly undetectable in ileum (36). We also immunostained for the ileal apical sodium-dependent bile acid transporter (Asbt; SLC10A), which is undetectable in mid-jejunum but highly expressed in distal ileum. Fabp1 was expressed normally in the cytoplasm of absorptive enterocytes of control jejunum (Fig. 5A) but was greatly reduced in absorptive enterocytes of Gata4 mutant jejunum (Fig. 5B), approaching levels that are characteristic in distal ileum of control mice (Fig. 5C). Asbt immunofluorescence is normally absent in jejunum (Fig. 5D) but was induced in the jejuna of Gata4 mutant mice (Fig. 5E), though at a lower intensity than in control ileum (Fig. 5F).
FIG. 5.
The absorptive enterocyte gene expression program in jejunum is partially transformed into an ileal-like pattern in Gata4 mutant mice. (A to C) Cytoplasmic Fabp1 immunofluorescence (green) and DAPI nuclear staining (blue) of the absorptive enterocytes of control jejunum (A), mutant jejunum (B), and control ileum (C), showing a reduced expression of Fabp1 in mutant jejunum. (D to F) Microvillus membrane Asbt immunofluorescence (green) and DAPI nuclear staining (blue) of control jejunum (D), mutant jejunum (E), and control ileum (F), showing an induction of Asbt in mutant jejunum. (G) Real-time and semiquantitative RT-PCRs conducted on jejunal RNA from control and Gata4 mutant mice, showing differential effects on absorptive enterocyte gene expression. Real-time RT-PCR (left) is shown as a ratio of mRNA abundance of Gata4 mutant jejunum compared to controls ( , P < 0.05;
, P < 0.05; 
 , P < 0.01;
, P < 0.01; 

 , P < 0.001) of genes normally expressed at higher levels in jejunum than in ileum (J>I), equally in jejunum and ileum (J=I), and at lower levels in jejunum than in ileum (J<I). Semiquantitative RT-PCR (right) is shown for two representative samples each from control and Gata4 mutant mice. A reaction without reverse transcriptase (No RT) served as a control for DNA contamination. (H) Comparison of LPH and Asbt mRNAs in control jejunum, mutant jejunum, and control ileum by real-time RT-PCR, showing that the transformation to an ileal-like phenotype is not complete. Data are means ± SEMs (n = 5). Bars with the same letter are significantly different from each other (P < 0.05). The calibrators were adult jejunal RNA for LPH and adult ileal RNA for Asbt. (I) Semiquantitative RT-PCR analysis for Gata5, Gata6, Cdx2, Hnf1α, c-Jun, c-Fos, Lrh1, and Gapdh on RNA from jejunum, showing that the mRNA abundances in control and Gata4 mutant mice are not different. A reaction without reverse transcriptase (No RT) served as a control for DNA contamination.
, P < 0.001) of genes normally expressed at higher levels in jejunum than in ileum (J>I), equally in jejunum and ileum (J=I), and at lower levels in jejunum than in ileum (J<I). Semiquantitative RT-PCR (right) is shown for two representative samples each from control and Gata4 mutant mice. A reaction without reverse transcriptase (No RT) served as a control for DNA contamination. (H) Comparison of LPH and Asbt mRNAs in control jejunum, mutant jejunum, and control ileum by real-time RT-PCR, showing that the transformation to an ileal-like phenotype is not complete. Data are means ± SEMs (n = 5). Bars with the same letter are significantly different from each other (P < 0.05). The calibrators were adult jejunal RNA for LPH and adult ileal RNA for Asbt. (I) Semiquantitative RT-PCR analysis for Gata5, Gata6, Cdx2, Hnf1α, c-Jun, c-Fos, Lrh1, and Gapdh on RNA from jejunum, showing that the mRNA abundances in control and Gata4 mutant mice are not different. A reaction without reverse transcriptase (No RT) served as a control for DNA contamination.
Quantitative analysis revealed that jejunal Fabp1 mRNA in the Gata4 mutant mice was ∼20% (P < 0.05) of that in control mice (Fig. 5G), confirming the reduced immunofluorescence signal for Fabp1 in Gata4 mutant mice. Other hypothesized targets of Gata4 include the genes that encode the disaccharidases LPH (44) and sucrase-isomaltase (SI) (3, 19), as well as intestinal fatty acid binding protein (Fabp2) (12). In mice, LPH is normally highly expressed in jejunum but is low or undetectable in distal ileum (18), whereas SI and Fabp2 are normally expressed in both jejunum and ileum (22, 42). In Gata4 mutant mice, LPH mRNA abundance was significantly attenuated (∼10% of control levels; P < 0.05), whereas the mRNAs for SI and Fabp2 were only marginally reduced by the inactivation of Gata4 (Fig. 5G).
The unexpected induction of Asbt due to the inactivation of Gata4 was confirmed by real-time RT-PCR, which revealed a 20-fold increase (P < 0.01) in Asbt mRNA in Gata4 mutant mice (Fig. 5G). The expression of the ileal lipid binding protein (Ilbp; Fabp6), an ileal absorptive enterocyte marker (6) thought to be important for intracellular bile acid transport, was also markedly induced (644-fold; P < 0.001) in the jejuna of Gata4 mutant mice (Fig. 5G), demonstrating that the induction of ileal genes is not exclusive to Asbt.
To define the extent of the transformation, the mRNA abundances of LPH and Asbt in mutant mice were quantitatively compared to those in control ileum. As shown in Fig. 5H, LPH and Asbt mRNAs changed in the direction of the ileum but did not attain ileal levels, indicating that the transformation to an ileal gene expression program in absorptive enterocytes is not complete.
Gata5, Gata6, and Cdx2 have been implicated as transcriptional activators of Fabp1 and/or LPH in vitro (7, 43), and Hnf1α is indispensable for the expression of these two genes in vivo (2). We found no difference in Gata5, Gata6, Cdx2, or Hnf1α mRNA levels between control and Gata4 mutant mice, indicating that the attenuation of Fabp1 and LPH is not secondary to changes in the expression of these regulators (Fig. 5I). In cardiac tissue, Gata6 is up-regulated in the absence of Gata4 (20, 24), but this does not appear to be the case in the small intestine.
The molecular control of Asbt gene expression in the mouse ileum has been studied in detail, and Hnf1α (33), the AP-1 proteins c-Jun and c-Fos (4), and liver receptor homologue-1 (Lrh1) (5) have been implicated as activators of Asbt gene expression. In the jejuna of Gata4 mutant mice, the mRNA abundances of Hnf1α, c-Jun, c-Fos, and Lrh1 were indistinguishable from those in control jejunum (Fig. 5I) and ileum (data not shown), indicating that the induction of Asbt is not secondary to an increase in these activators.
Inactivation of Gata4 results in a jejunal-to-ileal shift in secretory cell composition.
Although differential expression of specific absorptive enterocyte genes between jejunum and ileum is readily detectable, differences in composition and gene expression of the secretory lineages (Paneth, goblet, and enteroendocrine cells) between jejunum and ileum are more difficult to quantify due to the subtlety of the regional differences. This is particularly true for Paneth cells, in which we have been unable to document differences in cryptdin-1, cryptdin-4, and lysozyme mRNA abundances between control jejunum and ileum (data not shown). As a consequence, we found no difference in Paneth cell-specific expression of these genes in the Gata4 mutant mice (data not shown).
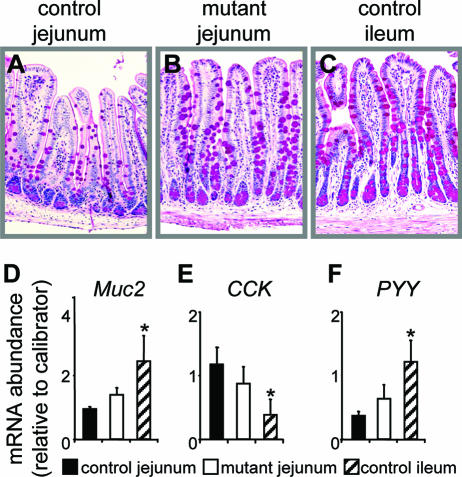
Perhaps the most distinguishable change in the secretory cells is the volume density of goblet cells, which normally increases from duodenum to distal ileum, and this is reflected by an increase in goblet cell number on villi and an increase in abundance of small intestinal goblet cell markers such as mucins (39). In the jejuna of Gata4 mutant mice, the number of PAS-positive goblet cells on villi (Fig. 6A to C) was significantly increased (∼40%) from 11.3 ± 1.4 goblet cells/villus (mean ± standard error of the mean [SEM]) in control jejunum to 15.8 ± 1.1 goblet cells/villus in Gata4 mutant jejunum (P < 0.05, n = 6), as determined by unbiased counting. Quantification of mucin-2 (Muc2) mRNA, the principal small intestinal goblet marker and indicator of goblet cell volume, revealed an increase toward ileal levels (Fig. 6D) but, like for the absorptive enterocyte genes, did not demonstrate a complete transformation to ileal levels. These data show that inactivation of Gata4 in the adult jejunum results in an increase in the goblet cell population, supporting a partial jejunal-to-ileal transformation in cell fate allocation.
FIG. 6.
Secretory lineages are redistributed towards an ileal-like composition. (A to C) PAS staining of control jejunum (A), mutant jejunum (B), and control ileum (C), showing expansion of the goblet cell population in Gata4 mutant jejunum. (D to F) Real-time RT-PCR of Muc2 (D), CCK (E), and PYY (F) mRNA abundances in control jejunum, mutant jejunum, and control ileum, showing a redistribution toward an ideal pattern in Gata4 mutant jejunum.  , P < 0.05 compared to control jejunum; data are means ± SEMs.
, P < 0.05 compared to control jejunum; data are means ± SEMs.
Enteroendocrine cells are equally distributed throughout the small intestine, but subpopulations are localized to distinct regions based on function. For example, hormones that stimulate pancreatic secretions, such as cholecystokinin (CCK), are synthesized and secreted from enteroendocrine cells in the duodenum and jejunum, whereas hormones that inhibit pancreatic secretions, including peptide YY (PYY), are more abundant distally. To test the hypothesis that the absence of Gata4 causes a jejunal-to-ileal transformation in enteroendocrine subpopulations, real-time RT-PCR for CCK and PYY mRNAs was undertaken. In the jejuna of Gata4 mutant mice, CCK mRNA was decreased, whereas PYY mRNA was increased, with both shifting towards ileal levels (Fig. 6E and F). These data suggest that the inactivation of Gata4 in the adult jejunum results in a partial redistribution of enteroendocrine subpopulations towards an ileal-like composition.
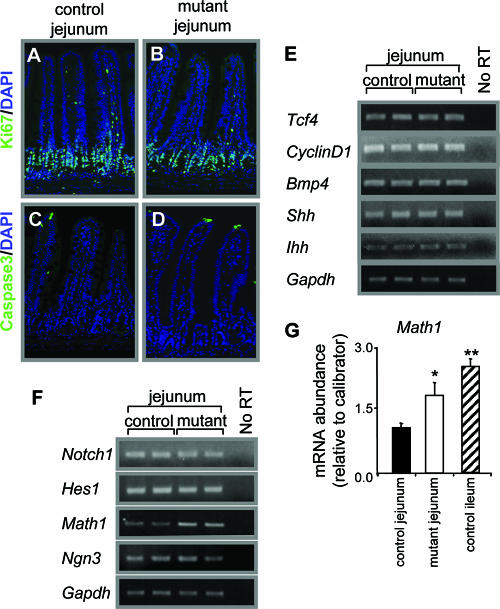
There was no evidence of abnormal proliferation or apoptosis identified by anti-Ki67 and anti-cleaved caspase-3 staining in the jejuna of Gata4 mutant mice compared to controls (Fig. 7A to D). This was true not only 2 weeks after the completion of tamoxifen treatment but also 1, 2, and 3 days after the beginning of tamoxifen treatment (data not shown). We also found no difference in the abundances in the mRNAs for Wnt and Hedgehog signaling proteins, including those for T-cell factor 4 (Tcf4), cyclin D1, bone morphogenetic protein 4 (Bmp4), sonic hedgehog (Shh), and Indian hedgehog (Ihh), between control and Gata4 mutant mice (Fig. 7E). Together, these data indicate that inactivation of Gata4 in the small intestine does not appreciably alter proliferation or the normal apoptotic fate of differentiated cells and likely does not involve Wnt and Hedgehog signaling pathways.
FIG. 7.
Proliferation, apoptosis, and signaling pathways in Gata4 mutant mice. (A and B) Immunofluorescence for the proliferation marker Ki67 (green) and DAPI (blue), showing that the proliferative compartment in control (A) and mutant (B) jejuna were not different. (C and D) Immunofluorescence for cleaved caspase-3 (green) and DAPI (blue), showing that cell death in control (C) and mutant (D) jejuna were not different. (E) Semiquantitative RT-PCR analysis of Tcf4, cyclin D1, Bmp4, Shh, Ihh, and Gapdh, showing that the mRNA abundances in control and Gata4 mutant mice are not different. (F) Semiquantitative RT-PCR analysis of Notch1, Hes1, Math1, neurogenin3 (Ngn3), and Gapdh, showing that jejunal mRNA abundances in control and Gata4 mutant mice are not different, with the exception of Math1, which is significantly greater by real-time RT-PCR (G) in both mutant jejunum (P < 0.05) and control ileum (P < 0.01) compared to control jejunum (data are means ± SEMs). The calibrator was adult jejunal RNA.
Cell lineage allocation is determined in large part by the Notch signaling pathway, whereby Notch activates Hes1 in intestinal progenitor cells, which in turn represses Math1, resulting in the specification of the absorptive enterocyte cell fate (30). In occasional cells that do not express Hes1, Math1 is expressed, resulting in a commitment to the secretory lineages. Inactivation of Hes1 (17) results in low numbers of absorptive enterocytes and an expansion of the secretory lineages, whereas ectopic activation of Notch (40) or deletion of Math1 (47) results in a decrease or elimination, respectively, of secretory cells. Expression of the neurogenin-3 gene (Ngn3) in Math1-positive cells specifies the enteroendocrine lineage. Notch1, Hes1, and Ngn3 mRNA abundances were unaffected by the inactivation of Gata4 as shown by semiquantitative (Fig. 7F) and real-time (data not shown) RT-PCR, but Math1 mRNA in the jejuna of the Gata4 mutant mice was significantly increased (P < 0.05) and was comparable to the higher levels of Math1 mRNA in the control ileum (Fig. 7G), which is consistent with an expansion of the goblet cell lineage.
DISCUSSION
Gata4, a member of an evolutionarily conserved transcription factor family, is expressed in multiple tissues throughout development and into adulthood, but due to the embryonic lethality of published Gata4 knockout models (20, 24, 27, 46, 48), its function in vivo in adults is unknown. In the mature small intestine of mice, Gata4 is expressed in the absorptive enterocytes on villi and in all epithelial cells of the crypt (Fig. 1). In both mice and humans, Gata4 is highly expressed in jejunum but is undetectable in distal ileum (Fig. 2), suggesting a role in the regulation of proximal-distal homeostasis within the intestine. In the present study, we established an inducible, intestine-specific inactivation model (Fig. 3) in which a truncated, transcriptionally inactive Gata4 mutant is synthesized (Fig. 4). Adult mice expressing mutant Gata4 in the jejunum display a partial jejunal-to-ileal transformation characterized by an attenuation of absorptive enterocyte genes normally expressed in jejunum but not in ileum, an induction of genes not normally expressed in jejunum but highly expressed in ileum (Fig. 5), and a redistribution of secretory cells toward an ileal phenotype (Fig. 6). Taken together, these data indicate that Gata4 plays a role in maintaining the regional differences in intestinal function between the jejunum and ileum.
The synthesis of a truncated form of Gata4 in the mutant mice was unexpected. This protein, designated Gata4Δex2, is transcriptionally inactive but is able to bind to DNA and thus potentially act as a dominant-negative Gata factor (Fig. 4). Since Gata4 is coexpressed with Gata6 in absorptive enterocytes on villi (Fig. 1), and possibly in crypts, the putative dominant-negative activity of Gata4Δex2 could conceivably mask underlying compensation by Gata6. However, we could not document a dominant-negative effect on Gata6 either by competition EMSAs (data not shown) or by transient-cotransfection assays (Fig. 4C). There was, however, evidence of a dominant-negative effect on Gata4, suggesting that the Gata4 and Gata6 targets are independent. Furthermore, the same competition for DNA elements that might occur between Gata4Δex2 and Gata6 in mutant mice would also occur between Gata4 and Gata6 in control mice, especially since Gata4Δex2 and Gata4 have identical DNA binding domains and are synthesized in similar amounts. We believe that under these conditions, Gata4Δex2 would occupy the identical cadre of binding sites in mutant mice as Gata4 would in control mice. Thus, although we cannot unequivocally rule out compensation by Gata6 in vivo, our data are highly indicative that the phenotype observed in the present study in the Gata4 mutant mice is reflective of Gata4 function.
In this study, we have uncovered three distinct features of Gata4 regulation in the adult jejunum in vivo. These features are (i) activation of a jejunal gene expression program in absorptive enterocytes (44), (ii) repression of an ileal gene expression program in absorptive enterocytes, and (iii) alteration in cell fate specification (Fig. 8). It is possible that Gata4 acts as a global regulator at an early step in cellular differentiation, whereby cells of all lineages are fated toward a jejunal phenotype in the presence of Gata4 and toward an ileal phenotype in the absence of Gata4, but this mechanism does not take into account a function for Gata4 in absorptive enterocytes on villi where it is highly expressed (Fig. 1). A more plausible hypothesis, which is supported by promoter binding and activation studies of enterocytes genes and is more consistent with Gata4 expression patterns, is that Gata4 regulates absorptive enterocyte genes by direct interactions with target promoters within these cells and modulates cell fate in the cells of the progenitor crypt compartment.
FIG. 8.
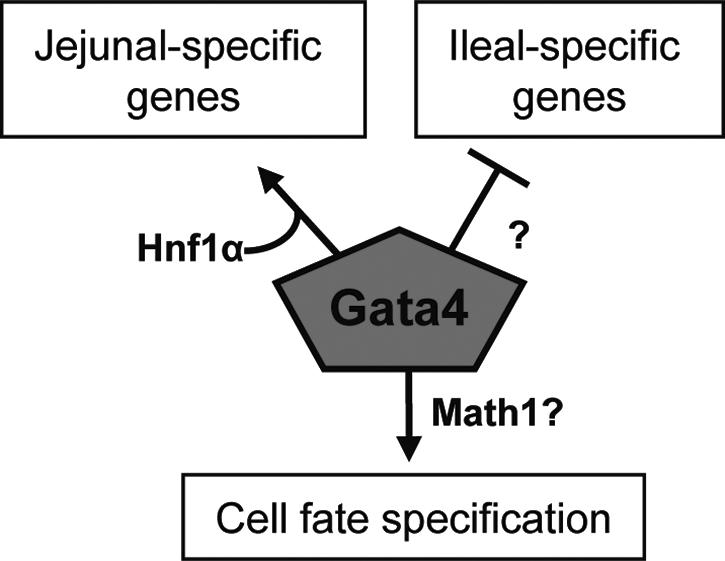
Model showing that Gata4 maintains a jejunal phenotype through (i) activation of a jejunal gene expression program in absorptive enterocytes, possibly through interactions with Hnf1α; (ii) repression of an ileal gene expression program in absorptive enterocytes through an as-yet-unknown mechanism; and (iii) alteration of cell fate specification, possibly through a Math1-dependent pathway.
Activation of absorptive enterocyte genes in the jejunum by Gata4 has been hypothesized in our previous work to occur in cooperation with Hnf1α through an evolutionarily conserved mechanism requiring physical interactions between these two transcription factors (44, 45). Cooperative activation by Gata4 and Hnf1α has been demonstrated in cell culture overexpression experiments not only for LPH (19, 44, 45) but also for Fabp1 (7) and SI (3, 19, 44, 45). In the present study, the expression of LPH and Fabp1 was significantly reduced in the mutant mice, but SI was unaffected (Fig. 5). Previously, we found that in the jejuna of adult Hnf1α−/− mice, gene expression of both Fabp1 and LPH was attenuated, whereas that of SI was similar to that in wild-type mice (2). Given the abundant in vitro data implicating Gata4 and Hnf1α as activators of SI gene expression (3, 19), it was surprising to find that neither regulate SI in vivo in adult intestine. However, the identical effects in both the adult Gata4 mutant and Hnf1α null mice is supportive of coregulation by Gata4 and Hnf1α and is further consistent with our hypothesis that these two transcription factors act in concert to activate a subset of absorptive enterocyte target genes.
Although the induction of ileal genes such as Asbt and Ilbp in the jejuna of Gata4 mutant mice is consistent with our hypothesis that Gata4 maintains a jejunal identity, this finding was nevertheless surprising because it indicates that Gata4 is responsible for the repression in the jejunum of ileal-specific genes. Gata4 is generally thought of as an activator of gene transcription, although it may recruit friend-of-GATA cofactors and repress the expression of certain Gata4 target genes (41). Gata1 demonstrates multiple functions in hematopoeisis, where it is required for the activation of erythroid genes as well as repression of early hematopoietic genes (29). Using a biotinylated tagging/proteomics approach with mouse erythroleukemic cells, Gata1 was found to coexist in multiple complexes, and divergent complexes were found to associate in vivo with discrete Gata1 target genes, thereby linking specific Gata1 partners to distinct aspects of its functions (29). We propose an analogy with Gata1 in that Gata4 can function as both an activator and repressor depending on the context of the promoter and the transcription factors and cofactors with which Gata4 interacts within the cell.
The induction of Asbt and Ilbp gene expression in the adult jejunum may be clinically relevant for patients who have undergone resection of the terminal ileum or who have suffered disease-related changes in the ileal mucosa, such as in Crohn's disease. These patients develop severe bile acid malabsorption characterized by diarrhea and eventually gallstone and kidney stone formation (34). Thus, further characterization of the Gata4-dependent inhibitory pathway that blocks ileal gene expression in the jejunum may reveal specific targets that can be used for therapeutic intervention in order to induce bile acid absorption in the jejuna of patients with impaired ileal function.
The partial redistribution of the number of goblet cells and type of enteroendocrine cells in jejunum to an ileal-like pattern in the absence of Gata4 indicates that Gata4 plays a role in defining cell fate, and it likely explains the function of Gata4 in crypt progenitor cells where this process occurs. Math1 is expressed in secretory cells on villi as well as isolated progenitor cells in crypts and is responsible for commitment of the secretory lineages (26, 47). Thus, the higher levels of Math1 mRNA in the jejuna of the Gata4 mutant mice as well as in control ileum could be a reflection of the increase in goblet cell number or could represent a novel regulatory pathway whereby Gata4 controls the composition of the secretory cell lineages in jejunum versus ileum through a Math1-dependent process.
Inactivation of jejunal Gata4 resulted in a ∼40% decrease in CCK mRNA and a ∼40% increase in PYY mRNA, all within the same set of RNA samples. It is well known that in addition to the specification of enteroendocrine cells by Math1/Ngn3, other factors, including Pax4, Pax6, BETA2/NeuroD, and pancreatic-duodenal homeobox-1 (Pdx1), regulate the specific subsets of enteroendocrine genes along the cephalo-caudal axis (31). Our data suggest that Gata4 is yet another factor involved in distinguishing the regional fate of enteroendocrine subpopulations, specifically between jejunum and ileum. Since Gata4 is not expressed in mature enteroendocrine cells (Fig. 1) (9), it is likely that this process occurs within the progenitor crypt compartment.
Although there are no other known in vivo examples of a regional shift in phenotype within the small intestinal system, there are examples of homeotic shifts in other organs of the gastrointestinal tract. These include Cdx2, which is normally expressed in the small and large intestines, and Pdx1, which is normally expressed in the pancreas and duodenum. Ectopic expression of Cdx2 in the stomachs of transgenic mice gives rise to patches of intestinal-like tissue (35), whereas ectopic expression of Pdx1 in Xenopus transforms part of the liver into pancreas (16). These data demonstrate that specific transcription factors expressed in restricted patterns along the anterior-posterior axis are responsible for the maintenance of regional identities along the gastrointestinal tract and that Gata4 is one of these factors.
The adult intestinal epithelium is a rapidly renewing tissue with differential functions along the cephalo-caudal axis, and therefore constant positional information to newly emerging cells is essential for the maintenance of local functions. Despite significant advances in our understanding of the molecular control governing intestinal differentiation and development, little is known about the molecular signals required for maintenance of proximal-distal identities in adult intestinal homeostasis. Insight into these molecular signals may be of clinical value in situations where local functions are lost due to disease or resection. In the present study, we have uncovered a member of the well-studied GATA family, Gata4, as a major and essential positional signal that is required for the maintenance of differential functions between jejunum and ileum in vivo.
Acknowledgments
We thank L. Liu and S. White for assistance with histology; J. Wagner of the Harvard Digestive Disease Imaging Core for help with immunofluorescence; J. Glickman for the sections of adult human intestine; S. Robine for the Villin-CreERT2 mice; J. I. Gordon for the Fabp1 antibody; P. A. Dawson for the Asbt antibody; M. R. Neutra, A. B. Leiter, R. A. Shivdasani, Y. Fujiwara, and S. H. Orkin for advice and guidance; and E. Beuling, R. K. Montgomery, and R. J. Grand for valuable insight and suggestions.
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases grant R01-DK-061382 (to S.D.K.) and by grants from the Harvard Digestive Disease Center (5P30-DK-34854) and the Nutricia Research Foundation (to T.B.).
Footnotes
Published ahead of print on 28 August 2006.
REFERENCES
- 1.Bjerknes, M., and H. Chang. 2005. Gastrointestinal stem cells. II. Intestinal stem cells. Am. J. Physiol. 289:G381-G387. [DOI] [PubMed] [Google Scholar]
- 2.Bosse, T., H. M. van Wering, M. Gielen, L. N. Dowling, J. J. Fialkovich, C. M. Piaseckyj, F. J. Gonzalez, T. E. Akiyama, R. K. Montgomery, R. J. Grand, and S. D. Krasinski. 2006. Hepatocyte nuclear factor-1α is required for expression, but dispensable for histone acetylation of the lactase-phlorizin hydrolase gene in vivo. Am. J. Physiol. 290:1016-1024. [DOI] [PubMed] [Google Scholar]
- 3.Boudreau, F., E. H. Rings, H. M. van Wering, R. K. Kim, G. P. Swain, S. D. Krasinski, J. Moffett, R. J. Grand, E. R. Suh, and P. G. Traber. 2002. Hepatocyte nuclear factor-1α, GATA-4, and caudal related homeodomain protein Cdx2 interact funtionally to modulate intestinal gene transcription. Implications for the developmental regulation of the sucrase-isomaltase gene. J. Biol. Chem. 277:31909-31917. [DOI] [PubMed] [Google Scholar]
- 4.Chen, F., L. Ma, N. Al-Ansari, and B. L. Shneider. 2001. The role of AP-1 in the transcriptional regulation of the rat apical sodium-dependent bile acid transporter. J. Biol. Chem. 276:38703-38714. [DOI] [PubMed] [Google Scholar]
- 5.Chen, F., L. Ma, P. A. Dawson, C. J. Sinal, E. Sehayek, F. J. Gonzalez, J. L. Breslow, M. Ananthanarayanan, and B. L. Shneider. 2003. Liver receptor homologue-1 mediates species- and cell line-specific bile acid-dependent negative feedback regulation of the apical sodium-dependent bile acid transporter. J. Biol. Chem. 278:19909-19916. [DOI] [PubMed] [Google Scholar]
- 6.Crossman, M. W., S. M. Hauft, and J. I. Gordon. 1994. The mouse ileal lipid-binding protein gene: a model for studying axial patterning during gut morphogenesis. J. Cell Biol. 126:1547-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Divine, J. K., L. J. Staloch, H. Haveri, C. M. Jacobsen, D. B. Wilson, M. Heikinheimo, and T. C. Simon. 2004. GATA-4, GATA-5, and GATA-6 activate the rat liver fatty acid binding protein gene in concert with HNF-1α. Am. J. Physiol. 287:G1086-G1099. [DOI] [PubMed] [Google Scholar]
- 8.Dusing, M. R., E. A. Florence, and D. A. Wiginton. 2003. High-level activation by a duodenum specific enhancer requires functional GATA binding sites. Am. J. Physiol. 284:G1053-1065. [DOI] [PubMed] [Google Scholar]
- 9.Dusing, M. R., and D. A. Wiginton. 2005. Epithelial lineages of the small intestine have unique patterns of GATA expression. J. Mol. Hist. 36:15-24. [DOI] [PubMed] [Google Scholar]
- 10.Fang, R., L. C. Olds, N. A. Santiago, and E. Sibley. 2001. GATA family transcription factors activate the lactase gene promoter in intestinal Caco-2 cells. Am. J. Physiol. 260:G58-G67. [DOI] [PubMed] [Google Scholar]
- 11.Fukushige, T., M. G. Hawkins, and J. D. McGhee. 1998. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol. 198:286-302. [PubMed] [Google Scholar]
- 12.Gao, X., T. Sedgwick, Y. B. Shi, and T. Evans. 1998. Distinct functions are implicated for the GATA-4, -5, and -6 transcription factors in the regulation of intestine epithelial cell differentiation. Mol. Cell. Biol. 18:2901-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon, J. I., G. H. Schmidt, and K. A. Roth. 1992. Studies of intestinal stem cells using normal, chimeric, and transgenic mice. FASEB J. 6:3039-3050. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi, S., and A. P. McMahon. 2002. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 244:305-318. [DOI] [PubMed] [Google Scholar]
- 15.Holtzinger, A., and T. Evans. 2005. Gata4 regulates the formation of multiple organs. Development 132:4005-4014. [DOI] [PubMed] [Google Scholar]
- 16.Horb, M. E., C. N. Shen, D. Tosh, and J. M. Slack. 2003. Experimental conversion of liver to pancreas. Curr. Biol. 13:105-115. [DOI] [PubMed] [Google Scholar]
- 17.Jensen, J., E. E. Pedersen, P. Galante, J. Hald, R. S. Heller, M. Ishibashi, R. Kageyama, F. Guilemot, P. Serup, and O. D. Madsen. 2000. Control of endodermal endocrine development by Hes-1. Nat. Genet. 24:36-44. [DOI] [PubMed] [Google Scholar]
- 18.Krasinski, S. D., B. H. Upchurch, S. J. Irons, R. M. June, K. Mishra, R. J. Grand, and M. Verhave. 1997. Rat lactase-phlorizin hydrolase/human growth hormone transgene is expressed on small intestinal villi in transgenic mice. Gastroenterology 113:844-855. [DOI] [PubMed] [Google Scholar]
- 19.Krasinski, S. D., H. M. Van Wering, M. R. Tannemaat, and R. J. Grand. 2001. Differential activation of intestinal gene promoters: functional interactions between GATA-5 and HNF-1 alpha. Am. J. Physiol. 281:G69-G84. [DOI] [PubMed] [Google Scholar]
- 20.Kuo, C. T., E. E. Morrisey, R. Anandappa, K. Sigrist, M. M. Lu, M. S. Parmacek, C. Soudais, and J. M. Leiden. 1997. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 11:1048-1060. [DOI] [PubMed] [Google Scholar]
- 21.Marjou, F. E., K.-P. Janssen, B. H.-J. Chang, M. Li, V. Hindie, L. Chan, D. Louvard, P. Chambon, D. Metzger, and S. Robine. 2004. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39:186-193. [DOI] [PubMed] [Google Scholar]
- 22.Markowitz, A. J., G. D. Wu, E. H. Birkenmeier, and P. G. Traber. 1993. The human sucrase-isomaltase gene directs complex patterns of gene expression in transgenic mice. Am. J. Physiol. 265:G526-G39. [DOI] [PubMed] [Google Scholar]
- 23.Molkentin, J. D. 2000. The zinc finger-containing transcription factors GATA-4, -5, and -6. J. Biol. Chem. 275:38949-38952. [DOI] [PubMed] [Google Scholar]
- 24.Molkentin, J. D., Q. Lin, S. A. Duncan, and E. N. Olson. 1997. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 11:1061-1072. [DOI] [PubMed] [Google Scholar]
- 25.Oesterreicher, T. J., and S. J. Henning. 2004. Rapid induction of GATA transcription factors in developing mouse intestine following glucocorticoid administration. Am. J. Physiol. 286:G947-G953. [DOI] [PubMed] [Google Scholar]
- 26.Pinto, D., A. Gregorieff, H. PBegthel, and H. Clevers. 2003. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 17:1709-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pu, W. T., T. Ishiwata, A. L. Juraszek, and S. Izumo. 2004. GATA4 is a dosage sensitive regulator of cardiac morphogenesis. Dev. Biol. 275:235-244. [DOI] [PubMed] [Google Scholar]
- 28.Rehorn, K.-P., H. Thelen, A. M. Michelson, and R. Reuter. 1996. A molecular aspect of hematopoiesis and endoderm development common to vertebrates and Drosophila. Development 122:4023-4031. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez, P., E. Bonte, J. Krijgsveld, K. E. Kolodziej, B. Guyot, A. J. R. Heck, P. Vyas, E. de Boer, F. Grosveld, and J. Strouboulis. 2005. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 24:2354-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sancho, E., E. Batlle, and H. Clevers. 2003. Live and let die in the intestinal epithelium. Curr. Opin. Cell Biol. 15:763-770. [DOI] [PubMed] [Google Scholar]
- 31.Schonhoff, S. E., M. Giel-Moloney, and A. B. Leiter. 2004. Development and differentiation of gut endocrine cells. Endocrinology 145:2639-2644. [DOI] [PubMed] [Google Scholar]
- 32.Serfas, M. S., and A. L. Tyner. 1993. HNF-1 alpha and HNF-1 beta expression in mouse intestinal crypts. Am. J. Physiol. 265:G506-G513. [DOI] [PubMed] [Google Scholar]
- 33.Shih, D. Q., M. Bussen, E. Sehayek, M. Ananthanarayanan, B. L. Shneider, F. J. Suchy, S. Shefer, J. S. Bollileni, F. J. Gonzalez, J. L. Breslow, and M. Stoffel. 2001. Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat. Genet. 27:375-382. [DOI] [PubMed] [Google Scholar]
- 34.Shneider, B. L. 2001. Intestinal bile acid transport: biology, physiology and pathophysiology. J. Pediatr. Gastroenterol. Nutr. 32:407-417. [DOI] [PubMed] [Google Scholar]
- 35.Silberg, D. G., J. Sullivan, E. Kang, G. P. Swain, J. Moffett, N. J. Sund, S. D. Sackett, and K. H. Kaestner. 2002. Cdx2 ectopic expression induces gastric intestinal metaplasia in transgenic mice. Gastroenterology 122:689-696. [DOI] [PubMed] [Google Scholar]
- 36.Simon, T. C., K. A. Roth, and J. I. Gordon. 1993. Use of transgenic mice to map cis-acting elements in the liver fatty acid-binding protein gene (Fabpl) that regulate its cell lineage-specific, differentiation-dependent, and spatial patterns of expression in the gut epithelium and in the liver acinus. J. Biol. Chem. 268:18345-18358. [PubMed] [Google Scholar]
- 37.Sodhi, C. P., J. Li, and S. A. Duncan. 2006. Generation of mice harbouring a conditional loss-of-function allele of Gata6. BMC Dev. Biol. 6:1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soriano, P. 1999. Generalized lacZ expression with ROSA26 Cre reporter strain. Nat. Genet. 21:70-71. [DOI] [PubMed] [Google Scholar]
- 39.Specian, R. D., and M. G. Oliver. 1991. Functional biology of intestinal goblet cells. Am. J. Physiol. 260:C183-C193. [DOI] [PubMed] [Google Scholar]
- 40.Stanger, B. Z., R. Datar, L. C. Murtaugh, and D. A. Melton. 2005. Direct regulation of intestinal fate by Notch. Proc. Natl. Acad. Sci. USA 102:12443-12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svensson, E. C., R. L. Tufts, C. E. Polk, and J. M. Leiden. 1999. Molecular cloning of FOG-2: a modulator of transcription factor GATA-4 in cardiomyocytes. Proc. Natl. Acad. Sci. USA 96:956-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sweetser, D. A., E. H. Birkenmeier, I. J. Klisak, S. Zollman, R. S. Sparkes, T. Mohandas, A. J. Lusis, and J. I. Gordon. 1987. The human and rodent intestinal fatty acid binding protein genes. A comparative analysis of their structure, expression, and linkage relationships. J. Biol. Chem. 262:16060-16071. [PubMed] [Google Scholar]
- 43.Troelsen, J. T. 2005. Adult-type hypolactasia and regulation of lactase expression. Biochim. Biophys. Acta 1723:19-32. [DOI] [PubMed] [Google Scholar]
- 44.van Wering, H. M., T. Bosse, A. Musters, E. de Jong, N. de Jong, C. E. Hogen Esch, F. Boudreau, G. P. Swain, L. N. Dowling, R. K. Montgomery, R. J. Grand, and S. D. Krasinski. 2004. Complex regulation of the lactase-phlorizin hydrolase promoter by GATA-4. Am. J. Physiol. 287:G899-G909. [DOI] [PubMed] [Google Scholar]
- 45.van Wering, H. M., I. L. Huibregtse, S. M. van der Zwan, M. S. de Bie, L. N. Dowling, F. Boudreau, E. H. H. M. Rings, R. J. Grand, and S. D. Krasinski. 2002. Physical interaction between GATA-5 and HNF-1α results in synergistic activation of the human lactase-phlorizin hydrolase promoter. J. Biol. Chem. 277:27659-27667. [DOI] [PubMed] [Google Scholar]
- 46.Watt, A. J., M. A. Battle, J. Li, and S. A. Duncan. 2004. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc. Natl. Acad. Sci. USA 101:12573-12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, Q., N. A. Bermingham, M. J. Finegold, and H. Y. Zoghbi. 2001. Requirement of math1 for secretory cell lineage commitment in the mouse intestine. Science 294:2155-2158. [DOI] [PubMed] [Google Scholar]
- 48.Zeisberg, E. M., Q. Ma, A. L. Juraszek, K. Moses, R. J. Schwartz, S. Izumo, and W. T. Pu. 2005. Morphogenesis of the right ventricle requires myocardial expression of Gata4. J. Clin. Investig. 115:1522-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]