Abstract
Proteins containing the DM domain, a zinc finger-like DNA binding motif, have been implicated in sexual differentiation in diverse metazoan organisms. Of seven mammalian DM domain genes, only Dmrt1 and Dmrt2 have been functionally analyzed. Here, we report expression analysis and targeted disruption of Dmrt4 (also called DmrtA1) in the mouse. Dmrt4 is widely expressed during embryonic and postnatal development. However, we find that mice homozygous for a putative null mutation in Dmrt4 develop essentially normally, undergo full sexual differentiation in both sexes, and are fertile. We observed two potential mutant phenotypes in Dmrt4 mutant mice. First, ovaries of most mutant females have polyovular follicles, suggesting a role in folliculogenesis. Second, 25% of mutant males consistently exhibited copulatory behavior toward other males. We also tested potential redundancy between Dmrt4 and two other gonadally expressed DM domain genes, Dmrt1 and Dmrt7. We observed no enhancement of gonadal phenotypes in the double mutants, suggesting that these genes function independently in gonadal development.
Regulators of sex determination and sexual differentiation are generally not conserved between phyla. An apparent exception that has emerged in the past few years involves genes related to the Drosophila sexual regulator Doublesex (Dsx) (28). Genes sharing a novel DNA binding motif identified in Dsx, the DM domain, have been shown to regulate sexual differentiation in insects and nematodes (1, 4, 12, 19, 22). In particular, DSX and the nematode protein MAB-3 have been shown to control several analogous aspects of sexual differentiation (1, 26) and regulate some analogous genes (2, 27). Moreover, the male isoform DSX-M can functionally replace MAB-3 in Caenorhabditis elegans (19). These results have raised the possibility that this gene family has an ancient and conserved function in sexual development.
Mammals have seven DM domain-encoding genes, but functional analysis has been reported for only two. The first, Dmrt1, is expressed only in the gonad and is required in mice for several aspects of testicular differentiation (18). Dmrt1 is expressed testis specifically in a variety of vertebrates with different primary sex determination mechanisms, and a recently duplicated copy of Dmrt1 (DMY/Dmrt1bY) has become the Y-linked testis-determining gene in the Medaka fish (14, 16). By contrast, Dmrt2 does not appear to play a role in gonadal development or sexual differentiation. Dmrt2 null mutant mice have defects in segmentation and die perinatally of lung defects but do not have obvious defects in gonadal development or signs of incomplete sexual differentiation at the time of death (21).
Based on the few examples studied so far, it is unclear whether DM domain genes comprise a family of general developmental regulators (e.g., Dmrt2) or are primarily involved in sexual differentiation (e.g., Dsx, mab-3, and Dmrt1). To better define the functional repertoire of these genes in mammals, we conducted a mutational analysis of the murine Dmrt genes. Here we report the targeted disruption of Dmrt4. Dmrt4 is widely expressed in embryos and adults. Dmrt4 mutants are viable and fertile, with no obvious anatomical defects, but ovaries of mutant females have elevated numbers of polyovular follicles. Thus, Dmrt4 appears to play a role in folliculogenesis. In addition, 25% of mutant males attempted to copulate with other males, suggesting a possible behavioral abnormality.
MATERIALS AND METHODS
Animals.
Mice were maintained under controlled temperature and a 12-h dark/12-h light cycle; protocols were approved by the University of Minnesota Institutional Animal Care and Use Committee. Animals were of a mixed C57BL/6J and 129/S1 background, unless indicated otherwise. Pups were weaned at 21 to 28 days and group housed by sex except as noted. Behavioral tests were conducted during hours 4 to 8 of the light cycle.
Generation of Dmrt4 mutant animals.
Dmrt4 was disrupted by homologous recombination in CJ7 embryonic stem (ES) cells using the targeting vector pJB15; details of the targeting vector are available on request. Southern blotting probes for identification of targeted ES cells were made by PCR using the following DNA primers: for the 5′ probe (SP1), 5′-GAGTTTCTGTGTCACCAGCA-3′ (forward) and 5′-TGATGCTCTACTTTCCTGAA-3′ (reverse); for the 3′ probe (SP2), 5′-TTATGATGCGTTATGTAGTC (forward) and 5′-GATAAGTAATTCCATCCCAA-3′ (reverse).
A correctly targeted ES cell clone was injected into C57BL/6J blastocysts, and the resulting chimeras were bred to generate animals heterozygous for the floxed allele Dmrt4loxP. Subsequently, the DM domain was deleted by breeding to β-actin Cre recombinase mice (11) (gift of Mark Lewandoski) to generate animals carrying the deleted putative null allele Dmrt4Δ.
Genotyping.
Genotyping of the Dmrt4 wild-type and deleted alleles was performed by PCR using a mixture of four primers: P1, 5′-GAGAAAGATTCATCCTCCCT-3′; P2, 5′-AGATCTGCAGTTTTGACAAC-3′; P3, 5′-GAGCCGGTCAGTCCCAACTT; and P4, 5′-CCGGTTTCCTGTGCAAGAAC-3′. PCR conditions were 94°C for 5 min and 35 cycles of 94°C for 45 s, 52°C for 45 s, and 72°C for 1 min, with a final extension step at 72°C for 10 min.
RT-PCR.
Tissues were harvested and stored in liquid nitrogen prior to RNA extraction. Total RNA was extracted from adult mouse tissues using Trizol reagent according to the manufacturer's protocol (Invitrogen Corporation). cDNA was synthesized using a Superscript II polymerase kit (Invitrogen). Reverse transcription-PCR (RT-PCR) primers for genes tested were designed to span an intron. Primers were as follows: 5′-GGAGCCGGTCAGTCCCAACT-3′ (forward) and 5′-AATGTAGTCTCTGGCCCAC-3′ (reverse) for Dmrt4; 5′-CATTCAGAGAGAAAGATCGC-3′ (forward) and 5′-GAGAAAGACCTGGGACTGTC-3′ (reverse) for the alternative Dmrt4 RNA initiated from upstream (altDmrt4). PCRs were performed using the following conditions: 94°C for 5 min and 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s, with a final extension step at 72°C for 10 min. Primers and PCR conditions for Hprt cDNA were as previously described (10).
Histology.
Organs and tissues from adult animals were fixed in 10% neutral buffered formalin and were paraffin embedded. Sectioning and hematoxylin-eosin staining were performed by the University of Minnesota Cancer Center Histopathology Core using standard protocols. Whole-mount immunohistochemistry was performed on embryonic day 11.5 (E11.5) embryos fixed with 4% paraformaldehyde (5) using anti-neural cell adhesion molecule (NCAM) antibody (AB5032; Chemicon International) at a dilution of 1:500. Necropsy was performed by the University of Minnesota Cancer Center Histopathology Core, which sectioned and examined major organs.
Skeletons of wild-type and Dmrt4 mutant pups at postnatal day 1 were stained with alizarin red and alcian blue as previously described (5).
Fertility test.
We established long-term breeding pairs for two homozygous mutant and two wild-type females with CD1 males as well as two mutant and two wild-type males with CD1 females, starting at 8 to 10 weeks of age. Numbers and sizes of litters were recorded for 1 year.
Rotarod test.
Motor coordination and balance were assessed by performance on a rotarod. After an initial 5-min trial on a rotarod rotating at a constant speed of 5 rpm, a 5-min test session was performed with acceleration from 4 rpm to 40 rpm. The trial and the first test were conducted on the same day at least 1 h apart. A second test was performed on the following day.
Buried food retrieval test.
General olfactory function was investigated based on the ability to find hidden food. Food was withheld from male and female test animals for 16 h prior to the test to increase motivation. Asteroid-shaped Cheetos (Frito-Lay) were randomly buried 3 cm under clean bedding, and the test animal was placed into the cage. Time required to retrieve the Cheeto was recorded.
Intermale behavior test.
This test was based on a “resident-intruder” paradigm. Sexually naive 8- to 12-week-old males (wild type, heterozygous, or Dmrt4 homozygous mutant) were used as “residents.” Resident males were individually housed for 1 week prior to the test session, with bedding unchanged for 3 to 4 days to allow establishment of territory. Eight-week old males of the passive A/J strain were then introduced as “intruders.” The intruder was placed into the resident's cage for a maximum of 10 min, and the session was filmed. The intruder was immediately removed if an aggressive attack occurred. Recordings were coded prior to analysis of behaviors. We scored the latency to the first aggressive attack and the number and duration of all nonaggressive mounts. A mount was scored as strongly sexual if a resident approached an intruder from behind, grasped it with its front paws, and exhibited a strong spinal curvature with rapid pelvic thrust.
Male olfactory preference test.
The bedding preference test was essentially as previously described (3). The test was performed in a large Plexiglas cage (45 by 25 cm by 21 cm deep) containing three color-coded bowls (7.5-cm diameter) placed 15 cm apart. Sexually naïve 8- to 12-week-old males of each genotype were tested. On two consecutive days prior to the test, the animals were habituated for 10 min in the test cage containing bowls filled with clean bedding. For testing, bowls were filled with clean bedding or soiled bedding was collected immediately prior to testing from group-housed adults of each sex whose cages had not been changed for 1 week. Test sessions were video recorded, and time spent investigating each bedding type was assessed by an investigator blind to the content of each bowl.
Maternal aggression and pup retrieval tests.
Sexually mature females were paired with wild-type males for 10 days and then housed singly. Litters were culled to six pups, and maternal aggression was tested on postpartum days 6 to 8. To avoid injuries, pups were removed from the cage just before the test. A wild-type “intruder” male was placed into the cage for 10 min. The intruder was a group-housed male of a mixed background. The test sessions were video recorded. The number of females that exhibited aggression toward the intruder was recorded. Immediately after the maternal aggression test, pups were returned to the home cage in a scattered manner. The time to retrieve the pups to the nest was recorded.
Statistical analysis.
One-way analysis of variance (ANOVA) was used to analyze significance of differences between the means of three genotypic groups for the majority of the measurements (rotarod, food retrieval, bedding preference test, etc). A chi-square test was used to calculate P value for differences in incidence of male-male mounting among the three genotypic groups.
RESULTS
Dmrt4 mRNA is widely expressed in adult tissues.
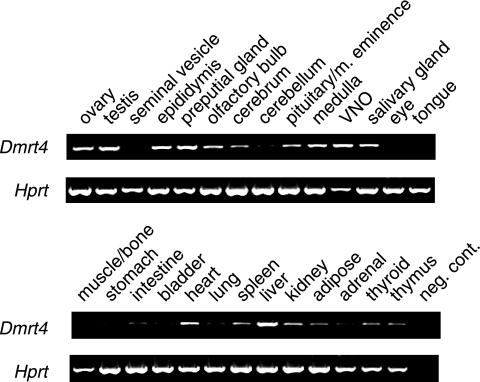
During embryogenesis murine Dmrt4 mRNA is expressed in a variety of tissues including testis, ovary, brain, heart, and kidney (10). Postnatal expression of murine Dmrt4 has not been described, so we assayed expression in adult tissues by RT-PCR (Fig. 1). As in the embryo, Dmrt4 mRNA was expressed in most adult tissues tested. The highest mRNA expression was observed in ovary, testis, epididymis, preputial gland, vomeronasal organ (VNO), liver, salivary glands, and heart. Dmrt4 also is expressed throughout the brain, with higher expression in the olfactory bulbs and medulla.
FIG. 1.
Dmrt4 adult mRNA expression. RT-PCR analysis of cDNA from the adult tissues indicated, performed as described in Materials Methods. Equal amounts of cDNA were amplified with primers directed against Dmrt4 and Hprt. m. eminence, median eminence; neg. cont., negative control (no cDNA template).
Conditional targeting of Dmrt4.
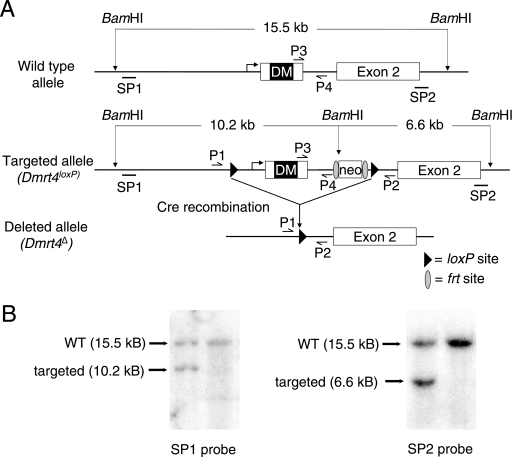
To test the function of Dmrt4, we disrupted the gene by homologous recombination in ES cells, as diagrammed in Fig. 2. The DM domain has been shown to be essential for all functions of several genes containing this motif (4, 18, 19). We therefore deleted exon 1, which contains the DM domain, as well as 1.2 kb upstream of the translational start site (including the 5′ untranslated region, transcriptional start, and proximal promoter) and 450 bp of intron 1. The resulting mutant allele is designated Dmrt4Δ.
FIG. 2.
Disruption of Dmrt4 by homologous recombination. (A) Targeting strategy. Homologous recombination of targeting vector pJB15 inserts a loxP site (recognition site for Cre DNA recombinase; black triangles) upstream of the Dmrt4 transcriptional start site and a second loxP site into intron 1. A neomycin positive selection cassette is inserted into intron 1 and is flanked by frt sites (gray ovals), which are recognized by Flpe DNA recombinase. Homologous recombination generates the targeted allele Dmrt4loxP and introduces a new BamHI site, as indicated. Breeding to Flpe-expressing mice can delete the neomycin cassette (not shown). Breeding to Cre-expressing mice deletes sequences between the loxP sites, generating the putative null allele Dmrt4Δ. SP1 and SP2 denote probes used for Southern analysis of neomycin resistant clones. P1, P2, P3, and P4 are PCR primers used to genotype animals. (B) Homologously targeted ES cell clone. Southern blots of ES cell DNA digested with BamH1 and probed with SP1 (left panel) and SP2 (right panel) probes. Both probes detect the 15.5-kb wild-type BamHI fragment from the untargeted Dmrt4 allele. In each panel the first lane contains DNA from a correctly targeted clone, as indicated by the presence of a novel 10.2-kb BamHI fragment detected by SP1 and a novel 6.2-kb BamHI fragment detected by SP2. The second lane contains DNA from a control ES cell clone. WT, wild type.
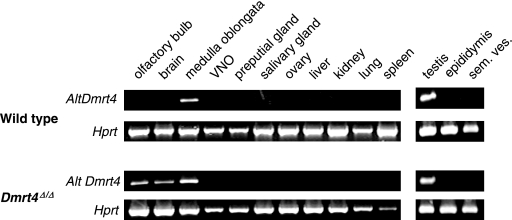
RT-PCR confirmed that Dmrt4Δ/Δ homozygotes do not express the major DM domain-encoding Dmrt4 mRNA (data not shown). However, a Dmrt4 cDNA lacking the DM domain (from E14.5 retina) has been described (BU924300), and the genomic sequences encoding this RNA were not removed by our deletion. This alternative RNA initiates ∼9 kb upstream of Dmrt4 exon 1, presumably under control of an alternative upstream promoter, and splices into Dmrt4 exon 2, bypassing the DM domain. We investigated the expression of this RNA in tissues of wild-type and Dmrt4Δ/Δ adults by RT-PCR (Fig. 3). In wild-type animals, the DM domain-less RNA was detectable only in testis and medulla oblongata (brain stem). However, in Dmrt4Δ/Δ mutants the alternative RNA was ectopically expressed in all brain regions tested, including the olfactory bulbs and medulla. It is unknown whether the alternative RNA is translated, but in principle it could encode a 39-amino-acid peptide containing amino acids 251 to 290 of Dmrt4. Based on these results we can conclude that the Dmrt4 mutation we generated should eliminate all DM domain-dependent functions of the gene. However, we cannot exclude the possibility that the mutant allele may retain DM domain-independent activity or have neomorphic activity.
FIG. 3.
Expression of Dmrt4 RNA from an alternative upstream promoter. RT-PCR of cDNA from indicated wild-type and Dmrt4Δ/Δ adult tissues, amplified with primers specific for the alternative Dmrt4 transcript (altDmrt4), as described in Materials and Methods, and Hprt primers as a positive control. sem. ves., seminal vesicle.
Dmrt4 mutants are viable and fertile.
Interbreeding of Dmrt4Δ/+ heterozygotes produced homozygous progeny at the expected frequency, and these animals were of normal size and had no obvious physical abnormalities. As a test of fertility we bred two wild-type and two mutant animals of each sex for 12 months. There was no significant difference in the numbers of litters, average litter sizes, or the lengths of the fertile period between mutant and wild-type males. For females, the average number of litters was 6.5 ± 0.7 for wild type and 9.5 ± 0.7 for Dmrt4Δ/Δ, with the mutants continuing to produce litters longer than the wild-type females. There was no difference in average numbers of pups per litter between wild-type (7.8 ± 2.2) and mutant females (8.4 ± 0.9). These results indicate that fertility of Dmrt4 mutants of both sexes is not significantly compromised.
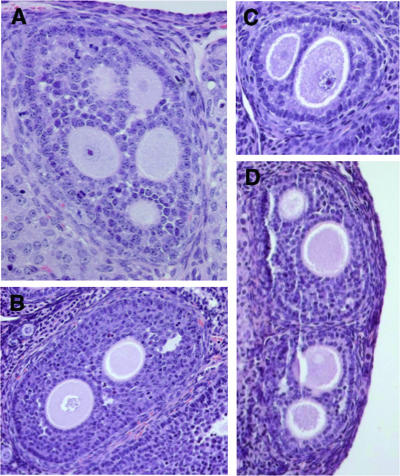
Polyovular follicles in Dmrt4 mutant ovaries.
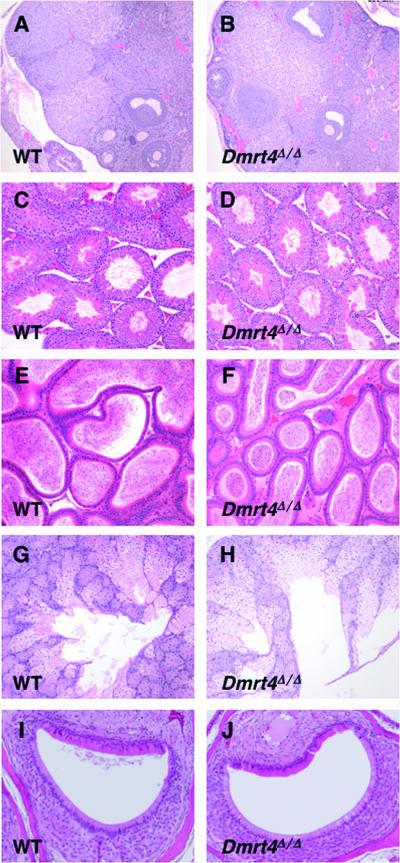
To look for more subtle phenotypes, we performed full necropsy on an adult wild-type and Dmrt4 mutant littermate of each sex, including histological examination of all major organs. For tissues and organs with high Dmrt4 mRNA expression, we examined a larger number of animals. No histopathological abnormalities were evident in most adult tissues examined, including testis, epididymis, salivary gland, and VNO (Fig. 4). However, most Dmrt4 mutant ovaries sectioned (seven of eight) contained at least one polyovular follicle (Fig. 5) and, in some cases, more than one (Fig. 5D). Most of these were biovular (Fig. 5B to D), but we observed one with five oocytes (Fig. 5A). We also observed polyovular follicles in two of six heterozygous ovaries sectioned, whereas we did not observe polyovular follicles in the ovaries of six wild-type females of the same mixed genetic background. These results suggest that our Dmrt4 mutant allele is haploinsufficient or semidominant for this phenotype.
FIG. 4.
Histology of wild-type and Dmrt4Δ/Δ tissues. Left column shows hematoxylin-eosin-stained sections of wild-type (WT) organs, and right column shows Dmrt4Δ/Δ tissues. Tissues are ovary (A and B), testis (C and D), epididymis (E and F), salivary gland (G and H), and VNO (I and J).
FIG. 5.
Polyovular follicles in Dmrt4 mutant ovaries. Shown are examples of polyovular follicles from Dmrt4 mutant adult female ovaries. (A) Penta-ovular follicle. (B and C) Biovular follicles. (D) Pair of biovular follicles.
Skeletal and motoneural development and function.
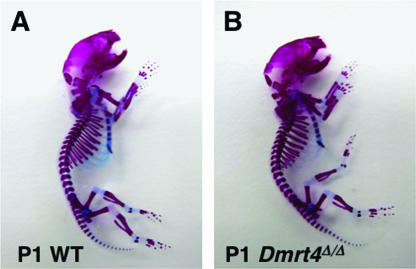
Because Dmrt2 is required for skeletal patterning, we evaluated skeletal development of Dmrt4 mutants, comparing skeletal preparations from wild-type and homozygous littermates at postnatal day 1 (Fig. 6). There were no apparent defects in bone or cartilage development. To assess function of the musculoskeletal system and balance, we performed a rotarod test. This revealed no significant difference between wild-type (n = 18), heterozygous (n = 15), or homozygous mutant (n = 23) animals in average latency to falling off the rotarod (111.8 s, 127.3 s, and 103.9 s, respectively; P = 0.2, one-way ANOVA test).
FIG. 6.
Skeletal development of Dmrt4 mutant embryos. Skeletal preparations of 1 day postnatal (P1) wild-type (WT) and Dmrt4 mutants are shown.
Normal olfactory epithelium formation and general olfaction.
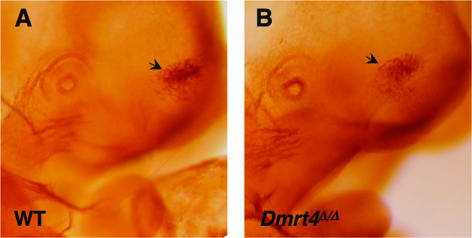
Dmrt4 has been reported to regulate neurogenesis in the olfactory placode during Xenopus development, based on morpholino depletion experiments in embryos (6). The olfactory placode and pit form normally in Dmrt4 mutant mice, and histological examination of adult olfactory tissues revealed no obvious defects. To visualize sensory neurons, we stained embryos at the 35-somite stage with an anti-NCAM antibody. Clustered neurons were present in the sensory epithelium in both wild-type and mutant (Fig. 7), and we did not observe the severe deficit in neurogenesis found in Xenopus. To assay general olfactory function, we tested for the ability to locate hidden food. There was no significant difference in average time required to retrieve buried Cheetos between wild type (163 s; n = 21), heterozygotes (279 s; n = 20), and homozygotes (209 s; n = 36) (P = 0.15, one-way ANOVA test). These results indicate that Dmrt4 is not essential for neurogenesis of olfactory pit epithelium or general olfaction in the mouse, though we cannot exclude mild or transient effects.
FIG. 7.
Olfactory neurons in Dmrt4 mutant embryos. Whole-mount immunohistochemistry with NCAM antibody staining of E11.5 embryos is shown. An arrow indicates the position of nasal placode olfactory epithelium. Both embryos are at the ∼35-somite developmental stage.
Male-male sexual interaction.
During husbandry of Dmrt4Δ/Δ animals, we noted several episodes of sexual mounting by mutant males on other males with which they were housed. We therefore examined intermale behavior further by conducting a resident-intruder assay (Materials and Methods). We tested 17 wild-type, 27 heterozygous, and 16 homozygous mutant males for interaction with wild-type intruder males of the passive A/J strain. There was no significant difference in latency of investigation by residents of intruders (P = 0.24, one-way ANOVA test), with average latencies of 17 s, 18 s, and 31s for wild type, heterozygotes and homozygotes, respectively. Likewise, the incidence of aggression by the residents did not differ significantly between the three genotypes (59%, 59%, and 44% for wild type, heterozygotes, and homozygotes, respectively). Latency to attack also was similar (310 s, 260 s, and 299 s, respectively; P = 0.6, one-way ANOVA test).
The total frequency of mounting was similar between genotypes, but we observed qualitative differences. Instances of a resident mounting an intruder in any manner were observed in 24% of wild-type, 7% of heterozygous, and 31% of homozygous animals (P = 0.2, chi-square test). The average duration of mounts was 0.027 s/min, 0.016 s/min, and 0.055 s/min for wild type, heterozygotes, and homozygotes, respectively (P = 0.2, one-way ANOVA test). Homozygous mutants differed in the occurrence of mounts scored as clearly sexual (involving clasping with both forelimbs, strong spinal curvature, and rapid pelvic thrusting). None of the wild-type animals exhibited mounting of this type, whereas 2/27 of heterozygotes and 4/16 of homozygotes did (P = 0.05, chi-square test). Although the proportion of animals exhibiting same-sex sexual mounting was low, individuals exhibiting this behavior did so consistently upon repeated testing.
Olfactory function and sexual preference.
Olfaction and sexual behavior are strongly linked in mice (9, 20). As described earlier, we observed high Dmrt4 expression in olfactory tissues including the VNO, which is implicated in male sexual behavior (9, 20, 24). Because we also observed a possible abnormality in sexual behavior, we further investigated olfactory function and sexual preference.
Although the main and accessory olfactory epithelia appeared histologically normal, we considered the possibility that Dmrt4 might mediate pheromone response by regulating genes required for vomeronasal function. We used RT-PCR to assess mRNA expression of several VNO receptor mRNAs (V1ra1, V1Ra2, V1rC25, and V2r3) (24) and two G proteins (Goα and Gαi2) (13, 17) involved in olfaction. All of the tested mRNAs were expressed at comparable levels in wild-type and Dmrt4 mutant VNOs (not shown).
As a test of sexual preference, we assayed the selectivity of males for fresh bedding versus female- or male-soiled bedding. We tested eight wild-type, eight heterozygous, and four Dmrt4 homozygous mutant animals. As shown in Table 1, males of all genotypes showed a similar preference for female-soiled bedding over fresh or male-soiled bedding. From these data we conclude that Dmrt4 mutants have relatively normal olfactory detection of pheromones as well as food (described above).
TABLE 1.
Results of test of sexual preference
| Genotype | Avg time (s) investigating beddinga
|
||
|---|---|---|---|
| Fresh bedding | Male-soiled | Female-soiled | |
| WTb | 30 | 86 | 126 |
| Dmrt4Δ/+ | 35 | 89 | 105 |
| Dmrt4Δ/Δ | 43 | 95 | 134 |
P values by one-way ANOVA are 0.29, 0.93, and 0.26 for fresh, male-soiled, and female-soiled bedding, respectively.
WT, wild type.
Maternal aggression.
In addition to male-specific behavior, we also assayed two aspects of female-specific behavior: maternal aggression and pup retrieval. The percentage of mothers displaying aggression toward a male intruder was similar for wild type (43%; n = 7) versus mutants (40%; n = 10). Likewise, we observed no differences in time required to retrieve four pups removed from the nest between wild-type (45 s; n = 7) and Dmrt4Δ/Δ (49 s; n = 10) mothers (P = 0.7, one-way ANOVA test).
Testing redundancy with other Dmrt genes.
Dmrt4 expression overlaps that of several other DM domain genes, including two whose expression is gonad-specific, Dmrt1 and Dmrt7. We tested whether Dmrt4 might function redundantly with either gene by analyzing the gonadal phenotypes of double mutants. Both Dmrt1 and Dmrt7 have defects in testicular differentiation (18; also unpublished results), and we looked for more severe phenotypes in double mutants. Doubly heterozygous Dmrt4; Dmrt1 and Dmrt4; Dmrt7 breeding pairs produced pups of all genotypes at the expected ratios, and litter sizes were normal. Males homozygous for Dmrt4 and either Dmrt1 or Dmrt7 mutations had testicular histology indistinguishable from that of the Dmrt1 or Dmrt7 single mutants (data not shown). From the lack of additive phenotypes, we conclude that Dmrt4 is unlikely to function redundantly with Dmrt1 or Dmrt7 in the gonad.
DISCUSSION
We have generated a targeted mouse mutation in Dmrt4, removing the DM domain as well as promoter and intronic sequences. This mutation does not abolish all transcription from the Dmrt4 locus but eliminates all protein isoforms containing the DM domain. Despite its widespread embryonic and postnatal expression, we find that Dmrt4 is not required for viability or fertility, at least on a mixed genetic background.
Our analysis revealed two potential phenotypes in Dmrt4 mutants. The first was the presence of polyovular follicles in most mutant ovaries. The origin of polyovular follicles is poorly understood, but it is likely that they arise during folliculogenesis by the incorporation of multiple oocytes into primordial follicles. The defect in Dmrt4 mutants may therefore reflect inappropriate germ cell-granulosa cell interaction. In this regard, we speculate that Dmrt4 might play a role in the perinatal ovary roughly analogous to that of Dmrt1 in the perinatal testis, where Dmrt1 is required for proper intercalation of gonocytes among Sertoli cells (18). Polyovular follicles occur infrequently in mice and other rodents (25); but they can be induced in mice by forced expression of inhibin-α (15) or by administration of exogenous estrogen (8), and their abundance can be modulated by gonadotropins (7). Because estrogen can affect polyovular follicle formation, we assayed mRNA expression of the estrogen receptors ERα and ERβ but did not observe a difference in Dmrt4 mutant ovaries (not shown). The conditional Dmrt4 mutation we generated might allow a more detailed dissection of where and when Dmrt4 expression is required for normal follicle development.
A second potential phenotype we observed was sexual mounting of males by some mutant males. Bedding preference tests indicated that Dmrt4 mutant males retain a preference for female soiled bedding and that mutant males do mate with females, so the behavior we observed is more likely to represent lower selectivity rather than reversed mate preference. We examined Dmrt4 mutants for defects in structures and functions linked to sexual behavior, in particular, in the olfactory system, but did not find any. The nonselective mounting behavior we observed resembles that of males mutant in the VNO putative ion channel TRP2 (24). However, TRP2 mutants lack male-male aggression, which was not significantly effected in Dmrt4 mutants. Only a minority of mutant males exhibited clearly sexual mounting of other males, but the behavior when present was reproducible and was never observed in wild-type siblings. The mice we tested were on a mixed genetic background, which may affect the penetrance of the phenotype. It will be important to retest Dmrt4 mutants for this behavior after extensive breeding onto defined backgrounds. If the male-on-male sexual behavior is robust on an inbred background, it may be possible to identify the anatomical focus of the defect by cell-type-specific gene targeting with the conditional Dmrt4 allele.
In Xenopus, morpholino oligonucleotide depletion of Dmrt4 or overexpression of a C-terminally truncated Dmrt4 protein reduces proliferation of NCAM-expressing neurons in the olfactory placode during embryonic development (6). Moreover, forced expression of Dmrt4 was sufficient to induce expression of neurogenic markers in cultured Xenopus explants. In the mouse, however, the reduction in olfactory placode neurons, if any, was modest during the equivalent period (Fig. 7), and the adult olfactory epithelium of mutants was histologically normal. We also found that a number of markers of olfactory neurons are expressed normally and that mutants have normal olfactory detection of food and pheromones.
There are several possible explanations for this discrepancy. First, the effects of Dmrt4 depletion in Xenopus on adult olfactory placode-derived tissues have not been reported, so it is possible that the reduced embryonic neurogenesis observed in the frog is transient. Second, Dmrt4 expression differs between frog and mouse (6, 10), and the Xenopus Dmrt4 expression pattern more closely resembles that of murine Dmrt3 (23). It may be, therefore, that the functions of these genes have shifted along with their expression patterns during vertebrate evolution. Third, because Dmrt4 expression overlaps that of Dmrt3 in the olfactory placode, it is possible that the two genes function redundantly in mice. It will be important to test the phenotype of Dmrt3; Dmrt4 double mutants, and we are currently pursuing those experiments. The extensive expression of Dmrt4 makes genetic redundancy a possibility in a number of tissues, and it will be important to test this as mutations in other Dmrt genes become available.
In summary, we have used ES cell homologous recombination to create a conditional allele and a deleted allele of the murine Dmrt4 gene and have characterized the phenotype of the deleted allele. We observed defects in the ovary and possibly in male sexual behavior, and these merit more in-depth study in the future, particularly on better-defined genetic backgrounds. However, we can conclude from these studies of mice of mixed background that, despite its widespread expression, Dmrt4 is not essential for normal viability and fertility.
Acknowledgments
We thank members of the Zarkower and Bardwell laboratories and Emily Rissman, Roger Gosden, and Jurrien Dean for helpful discussions; Shinseog Kim for substantial technical advice; and Nicole Kirchhof, Ilze Matin, and Aric Frantz of the University of Minnesota Cancer Center Histopathology Core for technical assistance and expert analysis.
This work was supported by a grant from the NIH (GM59152).
Footnotes
Published ahead of print on 18 September 2006.
REFERENCES
- 1.Baker, B. S., and K. A. Ridge. 1980. Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics 94:383-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coschigano, K. T., and P. C. Wensink. 1993. Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes Dev. 7:42-54. [DOI] [PubMed] [Google Scholar]
- 3.Dominguez-Salazar, E., H. L. Bateman, and E. F. Rissman. 2004. Background matters: the effects of estrogen receptor alpha gene disruption on male sexual behavior are modified by background strain. Horm. Behav. 46:482-490. [DOI] [PubMed] [Google Scholar]
- 4.Erdman, S. E., and K. C. Burtis. 1993. The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. EMBO J. 12:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hogan, B., R. Beddington, F. Constantini, and E. Laci. 1994. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- 6.Huang, X., C. S. Hong, M. O'Donnell, and J. P. Saint-Jeannet. 2005. The doublesex-related gene, XDmrt4, is required for neurogenesis in the olfactory system. Proc. Natl. Acad. Sci. USA 102:11349-11354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iguchi, T., Y. Fukazawa, Y. Uesugi, and N. Takasugi. 1990. Polyovular follicles in mouse ovaries exposed neonatally to diethylstilbestrol in vivo and in vitro. Biol. Reprod. 43:478-484. [DOI] [PubMed] [Google Scholar]
- 8.Iguchi, T., N. Takasugi, H. A. Bern, and K. T. Mills. 1986. Frequent occurrence of polyovular follicles in ovaries of mice exposed neonatally to diethylstilbestrol. Teratology 34:29-35. [DOI] [PubMed] [Google Scholar]
- 9.Keverne, E. B. 2004. Importance of olfactory and vomeronasal systems for male sexual function. Physiol. Behav. 83:177-187. [DOI] [PubMed] [Google Scholar]
- 10.Kim, S., J. R. Kettlewell, R. C. Anderson, V. J. Bardwell, and D. Zarkower. 2003. Sexually dimorphic expression of multiple doublesex-related genes in the embryonic mouse gonad. Gene Expr. Patterns 3:77-82. [DOI] [PubMed] [Google Scholar]
- 11.Lewandoski, M., E. N. Meyers, and G. R. Martin. 1997. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harbor Symp. Quant. Biol. 62:159-168. [PubMed] [Google Scholar]
- 12.Lints, R., and S. W. Emmons. 2002. Regulation of sex-specific differentiation and mating behavior in C. elegans by a new member of the DM domain transcription factor family. Genes Dev. 16:2390-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo, A. H., E. H. Cannon, K. S. Wekesa, R. F. Lyman, J. G. Vandenbergh, and R. R. Anholt. 2002. Impaired olfactory behavior in mice deficient in the alpha subunit of G(o). Brain Res. 941:62-71. [DOI] [PubMed] [Google Scholar]
- 14.Matsuda, M., Y. Nagahama, A. Shinomiya, T. Sato, C. Matsuda, T. Kobayashi, C. E. Morrey, N. Shibata, S. Asakawa, N. Shimizu, H. Hori, S. Hamaguchi, and M. Sakaizumi. 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417:559-563. [DOI] [PubMed] [Google Scholar]
- 15.McMullen, M. L., B. N. Cho, C. J. Yates, and K. E. Mayo. 2001. Gonadal pathologies in transgenic mice expressing the rat inhibin alpha-subunit. Endocrinology 142:5005-5014. [DOI] [PubMed] [Google Scholar]
- 16.Nanda, I., M. Kondo, U. Hornung, S. Asakawa, C. Winkler, A. Shimizu, Z. Shan, T. Haaf, N. Shimizu, A. Shima, M. Schmid, and M. Schartl. 2002. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. USA 99:11778-11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norlin, E. M., F. Gussing, and A. Berghard. 2003. Vomeronasal phenotype and behavioral alterations in G alpha i2 mutant mice. Curr. Biol. 13:1214-1219. [DOI] [PubMed] [Google Scholar]
- 18.Raymond, C. S., M. W. Murphy, M. G. O'Sullivan, V. J. Bardwell, and D. Zarkower. 2000. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 14:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raymond, C. S., C. E. Shamu, M. M. Shen, K. J. Seifert, B. Hirsch, J. Hodgkin, and D. Zarkower. 1998. Evidence for evolutionary conservation of sex-determining genes. Nature 391:691-695. [DOI] [PubMed] [Google Scholar]
- 20.Restrepo, D., J. Arellano, A. M. Oliva, M. L. Schaefer, and W. Lin. 2004. Emerging views on the distinct but related roles of the main and accessory olfactory systems in responsiveness to chemosensory signals in mice. Horm. Behav. 46:247-256. [DOI] [PubMed] [Google Scholar]
- 21.Seo, K. W., Y. Wang, H. Kokubo, J. R. Kettlewell, D. A. Zarkower, and R. L. Johnson. 2006. Targeted disruption of the DM domain containing transcription factor Dmrt2 reveals an essential role in somite patterning. Dev. Biol. 290:200-210. [DOI] [PubMed] [Google Scholar]
- 22.Shen, M. M., and J. Hodgkin. 1988. mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans. Cell 54:1019-1031. [DOI] [PubMed] [Google Scholar]
- 23.Smith, C. A., T. M. Hurley, P. J. McClive, and A. H. Sinclair. 2002. Restricted expression of DMRT3 in chicken and mouse embryos. Mech. Dev. 119(Suppl. 1):S73-S76. [DOI] [PubMed] [Google Scholar]
- 24.Stowers, L., T. E. Holy, M. Meister, C. Dulac, and G. Koentges. 2002. Loss of sex discrimination and male-male aggression in mice deficient for TRP2. Science 295:1493-1500. [DOI] [PubMed] [Google Scholar]
- 25.Telfer, E., and R. G. Gosden. 1987. A quantitative cytological study of polyovular follicles in mammalian ovaries with particular reference to the domestic bitch (Canis familiaris). J. Reprod. Fertil. 81:137-147. [DOI] [PubMed] [Google Scholar]
- 26.Yi, W., J. M. Ross, and D. Zarkower. 2000. Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development 127:4469-4480. [DOI] [PubMed] [Google Scholar]
- 27.Yi, W., and D. Zarkower. 1999. Similarity of DNA binding and transcriptional regulation by Caenorhabditis elegans MAB-3 and Drosophila melanogaster DSX suggests conservation of sex determining mechanisms. Development 126:873-881. [DOI] [PubMed] [Google Scholar]
- 28.Zarkower, D. 2001. Establishing sexual dimorphism: conservation amidst diversity? Nat. Rev. Genet. 2:175-185. [DOI] [PubMed] [Google Scholar]