Abstract
The inclusion of exons 2 and 3 of α-tropomyosin is governed through tissue-specific alternative splicing. These exons are mutually exclusive, with exon 2 included in smooth muscle cells and exon 3 included in nearly all other cell types. Several cis-acting sequences contribute to this splicing decision: the branchpoints and pyrimidine tracts upstream of both exons, UGC-repeat elements flanking exon 3, and a series of purine-rich enhancers in exon 2. Previous work showed that proteins rich in serine-arginine (SR) dipeptides act through the exon 2 enhancers, but the specific proteins responsible for such activation remained unknown. Here we show that a 35-kDa member of the SR protein family, 9G8, can activate the splicing of α-tropomyosin exon 2. Using RNA affinity chromatography and cross-linking competition assays, we also demonstrate that the heterogeneous nuclear ribonucleoproteins (hnRNPs) H and F bind to and compete for the same elements. Overexpression of hnRNPs H and F blocked 9G8-mediated splicing both in vivo and in vitro, and small interfering RNA-directed depletion of H and F led to an increase in exon 2 splicing. These data suggest that the activation of exon 2 is dependent on the antagonistic activities of 9G8 and hnRNPs H and F.
The mechanism of pre-mRNA splicing has proven quite complex with the discovery that ca. 74% of human genes are subject to alternative splicing (42). Cell-, tissue- or development-specific splicing allows for the production of functionally distinct protein isoforms, enabling a large proteome yet limiting the genome size (30, 54). As in stepwise, constitutive pre-mRNA splicing, the sequence signals that mediate control of alternative splicing include the 5′ splice site, the 3′ splice site, branchpoint, and polypyrimidine tract (2). However, the strength of these signals is often weak in alternatively spliced genes, requiring the assistance of additional factors and sequences to accurately define exons and introns. The best characterized of these sequence elements are splicing enhancers that function to facilitate exon and intron definition through the formation of bridge complexes (9, 48, 64, 78).
A variety of RNA-binding proteins have been identified that interact with intronic and exonic splicing enhancers to assist in splice site recognition. For example, TIA-1 acts on U-rich intronic sequences to activate multiple splicing events (17, 24), as does Nova-1, which interacts with the intronic sequence (UCAUY)3 to regulate splicing of the glycine receptor α2 and GABAAγ2L genes (39). Proteins that bind to exonic splicing enhancers include Tra and Tra-2 (47, 52, 70), the CELF (for CUG-BP1- and ETR-3-like factor) family of splicing regulators (10, 45), and the SR protein family (25, 31). SR proteins contain carboxy-terminal domains rich in serine-arginine dipeptides with either one or two amino-terminal RNA recognition motifs. Core SR proteins are characterized by their ability to precipitate in high magnesium concentrations and by the presence of a conserved phosphoepitope. Although they can interact with pre-mRNA sequences to promote skipping of exons (40), the majority of examples point toward SR proteins as positive regulators of splicing by binding to splicing enhancer elements (23, 31, 72).
In contrast to enhancers, splicing silencers regulate exon inclusion by recruiting RNA-binding proteins that block splice site recognition (73). These proteins are most commonly members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family, with hnRNPs A1 and I being the most extensively studied (6, 15, 18, 19, 22, 34, 43, 56, 69, 71, 81, 83). hnRNP I, or polypyrimidine tract-binding protein (PTB), acts with hnRNP A1 to repress the alternative exon N1 in c-src (65). This repression is relieved in part by SR proteins binding to enhancer sequences. PTB has been implicated in the repression of splicing of a variety of substrates, including its own pre-mRNA (3, 68, 69, 71, 77). However, hnRNPs are not the only RNA-binding proteins known to interact with and negatively influence splice site selection. For example, the “muscleblind” (MBNL) proteins bind to CUG repeats to both positively and negatively regulate alternative exons (16, 35). Likewise, Fox-1 can act both positively and negatively to regulate splice site choice (41). Thus, overall splicing regulation is controlled by the combinatorial action of multiple proteins and RNA sequence elements.
α-Tropomyosin (α-TM) is a well-studied alternative splicing system with multiple tissue-specific isoforms (14, 29, 67, 75). Exons 2 and 3 of α-TM are mutually exclusive due to the proximity of the strong exon 3 branchpoint/polypyrimidine tract adjacent to the upstream 5′ splice site (67). Inclusion of exon 3 is the default pattern due to the competitive strength of its branchpoint/polypyrimidine tract, whereas exon 2 is selected primarily in smooth muscle cells (21, 58). Previous work has identified several cis-acting regulatory elements flanking exon 3 that control tissue-specific splicing. These include pyrimidine-rich regions upstream and downstream of exon 3 that bind PTB and flanking groups of UGC repeats that bind an as-yet-undiscovered protein (27, 28, 33, 62) (see Fig. 1). In addition, there are four purine-rich enhancers located in exon 2 that interact with one or more SR proteins to activate its inclusion in smooth muscle cells concomitant with the repression of exon 3 (5, 20, 49).
FIG. 1.
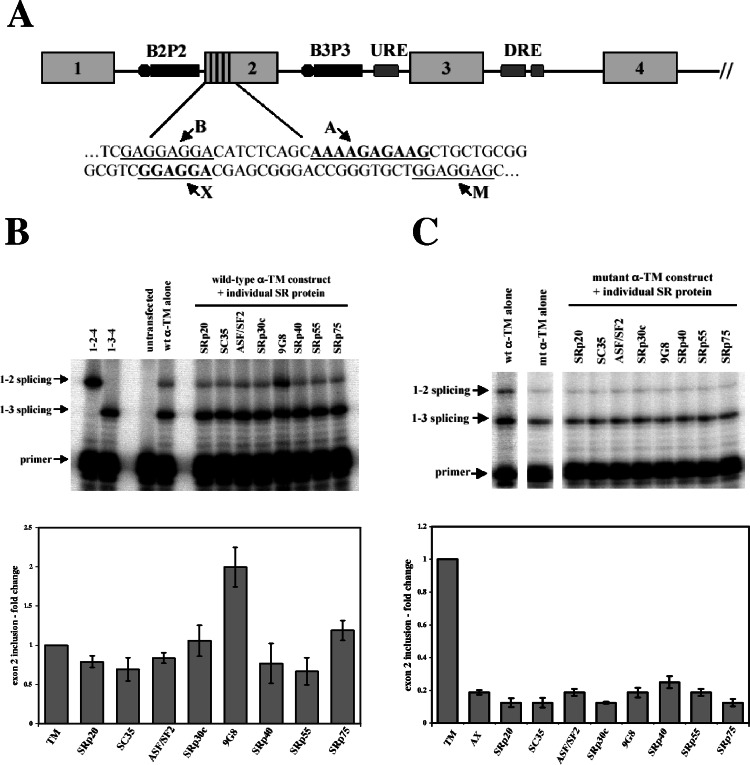
9G8 activates α-TM exon 2 inclusion. (A) Exons 1 to 4 of α-TM and splicing regulatory elements are shown, encompassing the branchpoint/pyrimidine tracts of exons 2 (B2P2) and 3 (B3P3), upstream and downstream regulatory elements (URE and DRE), and purine-rich enhancers in exon 2 (denoted by vertical lines). A portion of the exon 2 sequence is shown below, with the four enhancers underlined, denoted B, A, X, and M (20). Previous work showed that only the two central enhancers are needed for activity (5, 20). (B) A minigene (400 ng) encompassing the first four exons of α-TM (pSVpA α-TM 1-4) were cotransfected with 800 ng of eight individual SR protein expression vectors. Transfections were carried out in PAC1 cells, and RT-PCR products were analyzed by dideoxy termination primer extension. The graph below shows averages and standard errors derived from at least three independent transfections. (C) Mutation of the A and X elements (AAAAGAGAAG to AAAACTTAAG, GGAGGAC to GGAGCTT) abolished activation by 9G8. Samples were analyzed and quantitated as described above.
To further understand the alternative splicing of α-TM, we sought to identify specific SR proteins that activate exon 2 splicing. Through in vivo and in vitro studies, a 35-kDa SR protein, 9G8, was identified that interacts with purine-rich enhancers to activate exon 2. Enhancer-driven activation could be blocked by overexpression of hnRNP H and hnRNP F, whereas small interfering RNA (siRNA) knockdown of both proteins reversed such repression. UV cross-linking assays showed that hnRNPs H and F directly compete with 9G8 for binding to the exon 2 enhancer sequences. These findings are consistent with a model of antagonism between 9G8 and hnRNPs H and F for the activation or repression of exon 2 splicing.
MATERIALS AND METHODS
RT-PCR and primer extension.
Reverse transcription-PCR (RT-PCR) and primer extension assays were performed as described previously (62, 63), except 200 fmol of γ-32P-labeled TM1PE was used in the primer extensions, and reactions were visualized on 15% polyacrylamide-8 M urea gels.
RNA affinity chromatography.
Chromatography columns were prepared as previously described (1). A total of 200 nmol of modified RNAs (pur-AX, 5′-N5-AAAGAGAAGGGAGGAG-3′; mut-AX, 5′-N5-AAACUUAAGGGAGCUU-3′) was deprotected and incubated in 4.3 mM sulfo-SIAB and 190 mM sodium phosphate (pH 8.0) for 6 h in the dark at room temperature. The RNAs were then precipitated and resuspended in 220 mM sodium phosphate (pH 8.0) and 22 mM NaCl. After denaturing at 90°C for 3 min and cooling on ice for 15 min, the RNAs were loaded onto the modified columns and incubated overnight in the dark at room temperature. Uncoupled RNAs were removed with 200 mM Tris-HCl (pH 7.5)-20 mM NaCl, and unreacted thiol groups were blocked with 20 mM iodoacetamide in 50 mM sodium phosphate (pH 8.0).
HeLa nuclear extracts were applied to the columns and allowed to incubate at room temperature for 20 min. Extracts were then washed through with buffer D (20 mM Tris-HCl [pH 8.0], 100 mM KCl, 0.5 mM dithiothreitol [DTT], 0.2 mM EDTA, 5% glycerol) and reapplied for 10 min. After being washed in buffer D, bound proteins were eluted in buffer D with increasing KCl concentrations (0.1 to 1.0 M) with a final elution in 8 M urea-20 mM Tris-HCl (pH 8.0). Fractions were analyzed on 10% sodium dodecyl sulfate (SDS) gels and either Coomassie blue or silver stained.
Identification of hnRNP H and hnRNP F by mass spectrometry and database analysis.
Individual proteins were excised and subjected to in-gel trypsin digestion, and peptide mixtures were analyzed by matrix-assisted laser desorption ionization-time of flight (TOF) and TOF/TOF tandem mass spectrometry using a Voyager 4700 mass spectrometer (Applied Biosystems, Framingham, MA). Mass spectral data, in the form of peptide mass maps/fingerprints (PMF) of the intact molecular peptide ions (M+H), as well as fragmentation data derived from individual peptide ions, were used to interrogate the Swiss-Prot and NCBInr protein databases for statistically significant protein matches using GPS Explorer software (Applied Biosystems) running the MASCOT search engine (Matrix Science).
Protein expression and purification.
pET-15b-hnRNP H and pET-15b-hnRNP F were generous gifts from Doug Black. Bacterial protein preparations from these vectors were as described previously (12), except that proteins were eluted prior to renaturing. Dialysis was performed by a linear urea gradient from 6 to 0.25 M, with a final wash in buffer D. p-AcHLT-C SRp20, p-AcHLT-A ASF/SF2, pAcHLT-A 9G8, and p-AcHLT-A SRp55 were cotransfected into Sf9 cells with linearized BaculoGold, and recombinant viruses were isolated 4 days later. Sf9 cells were retransfected with the viruses for two successive passages. Hi5 cells were then infected for recombinant protein expression and incubated for 3 days. Cells were harvested and resuspended in lysis buffer (10 mM Tris-HCl [pH 7.5], 130 mM NaCl, 1% Triton X-100, 10 mM NaF, 10 mM NaPi, 10 mM NaPPi, 0.1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin/ml, 1 μg of aprotinin/ml) on ice for 30 min and sonicated twice for 30 s. Lysed cells were then incubated with Ni-nitrilotriacetic acid beads for 1 h at 4°C; washed three times with buffer containing 50 mM NaPO4 (pH 7.5), 300 mM NaCl, 10% glycerol, and 10 mM imidazole; and eluted in the same buffer with increasing imidazole concentrations from 50 mM to 500 mM. Peak fractions were dialyzed against buffer D. Purified SR proteins from calf thymus were prepared as described previously (82), except proteins were resuspended in buffer D.
DNA constructs.
Three micrograms of oligonucleotides 5′-CGAACAAAAGAGAAGCTGCTGCGGGCGTCGGAGGACGTT-3′ and 5′-CGAACGTCCTCCGACGCCCGCAGCAGCTTCTCTTTTGTT-3′ was annealed together in 100 mM potassium acetate, 30 mM HEPES-KOH (pH 7.4), and 2 mM magnesium acetate; heated to 95°C for 5 min; placed in a 70°C heat block for 10 min; allowed to cool to room temperature; and then incubated on ice for 30 min. A total of 300 ng of annealed oligonucleotides was phosphorylated in reactions containing 70 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 5 mM DTT, 10 U of T4 polynucleotide kinase (NEB), and 1 mM ATP. Phosphorylated oligonucleotides were ligated into the BstBI site of dsx-ΔE (kindly provided by Brenton Graveley) to make dsx-TM. The primer pairs 5′-CGAACAAAACTTAAGCTGCTGCGGGCGTCGGAGGACGTT-3′ and 5′-CGAACAAGCTCCGACGCCCGCAGCAGCTTAAGTTTTGTT-3′, 5′-CGAACAAAAGAGAAGTACGTGCGGGCGTCGGAGGACGTT-3′ and 5′-CGAACGTCCTCCGACGCCCGCACGTACTTCTCTTTTGTT-3′, 5′-CGAACAAAAGAGAAGCTGCACGTGGCGTCGGAGGACGTT-3′ and 5′-CGAACGTCCTCCGACGCCACGTGCAGCTTCTCTTTTGTT-3′, and 5′-CGAACAAAAGAGAAGCTGCTGCGACGTTCGGAGGACGTT-3′ and 5′-CGAACGTCCTCCGAACGTCGCAGCAGCTTCTCTTTTGTT-3′ were annealed and phosphorylated as described above and inserted into the BstBI site of dsx-ΔE to make dsx-AX, dsx-M1, dsx-M2, and dsx-M3, respectively. The primer pairs 5′-AATTCAAAAGAGAAGCTGCTGCGGGCGTCGGAGGACG-3′ and 5′-AATTCGTCCTCCGACGCCCGCAGCAGCTTCTCTTTTG-3′, 5′-AATTCACTTCTTAAGCTGCTGCGGGCGTCGCTTCTTG-3′ and 5′-AATTCAAGAAGCGACGCCCGCAGCAGCTTAAGAAGTG-3′, 5′-AATTCACTTCTTAAGCTGCTGCGGGCGTCGGAGGACG-3′ and 5′-AATTCGTCCTCCGACGCCCGCAGCAGCTTAAGAAGTG, 5′-AATTCAAAAGAGAAGCTGCTGCGGGCGTCGCTTCTTG-3′ and 5′-AATTCAAGAAGCGACGCCCGCAGCAGCTTCTCTTTTG, and 5′-AATTCAAAAGAGAAGTCATCATAAATACTGGAGGACG-3′ and 5′-AATTCGTCCTCCAGTATTTATGATGACTTCTCTTTTG-3′ were annealed together as described above and ligated into an EcoRI-digested pGEM-T Easy Vector (Promega) to make pGEM TM, AX, A, X, and INSIDE, respectively. Sequences between the transcription site and insert were removed by reverse PCR. pCGT7-SRp30c and pSG5-SRp55 were provided by Adrian Krainer and Robert Lafyatis, respectively. hnRNP H and F cDNAs were digested with BamHI and XhoI and cloned into the same sites in pCS2.
Western blots.
Protein samples were loaded onto 10% SDS gels and transferred to Nitropure nitrocellulose membranes (Osmonics) at 150 mA for 1.5 h. Membranes were then blocked for 30 min in 5% nonfat dry milk in 10 mM Tris-HCl (pH 8.0), 0.9% NaCl, and 0.1% Tween 20. Primary antibodies were diluted in milk and incubated with the membranes for 1 h, washed, and incubated with secondary antibody for 1 h. 9G8 was detected by monoclonal N-terminal 2B12 (a generous gift from James Stevenin) diluted 1:3,333, and the monoclonal T7-tag antibody (Novagen) was diluted 1:10,000. The secondary antibody for 9G8 and T7-tag was peroxidase-conjugated goat anti-mouse immunoglobulin G at 1:5,000. hnRNP H and hnRNP F were detected by polyclonal N-terminal antibodies (from Doug Black) at 1:5,000 and 1:3,333 concentrations, respectively. The primary antibody for α-tubulin (Abcam) was diluted 1:2,000. The secondary antibody for hnRNPs H and F and α-tubulin was peroxidase-conjugated donkey anti-rabbit immunoglobulin G at 1:5,000. Protein bands were visualized by ECL (Perkin-Elmer) and analyzed with NIH-Image (version 1.63; http://rsb.info.nih.gov/nih-image/).
Cell culture, transfections, and RNA extraction.
HeLa and PAC1 cells (66) were incubated at 37°C in Dulbecco modified Eagle medium supplemented with 1% penicillin-streptomyocin and either 10 or 20% fetal bovine serum, respectively. At 16 to 24 h before transfection, cells were washed with phosphate-buffered saline-EDTA, removed from the plate with pancreatin, and replated in 5 or 3 ml of antibiotic-free medium on a 60-mm plate or six-well dish. Transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Cells were harvested 48 h after transfection by using Tri-Reagent (Molecular Research Center). Precipitated RNAs were used for RT-PCR and primer extension (62).
In vitro transcription and splicing.
Transcription and splicing of substrate RNAs was performed as previously described (59, 60). All dsx constructs were linearized with MluI. An adenovirus-derived substrate was linearized with BamHI. pGEM A, X, and INSIDE constructs were linearized with PstI. These constructs were transcribed with T7 RNA polymerase (Promega). pGEM TM and AX constructs were linearized with SacII and transcribed with Sp6 RNA polymerase (NEB). For splicing reactions, 10 ng of substrate was spliced in either 40% HeLa nuclear extract or 40% S100 extract supplemented with the indicated proteins. Proteins were incubated with RNA for 5 min on ice before the addition of extracts or reaction mixtures. Adenovirus splicing was performed at 30°C for 1 h and visualized on 15% polyacrylamide 8 M urea gels by phosphorimager analysis. All other reactions were performed at 30°C for 2 h, and products were resolved on 8% gels.
UV cross-linking.
UV cross-linking reactions were carried out in a final volume of 15 μl in the presence of 0.5 mM DTT, 2.5 mM MgCl2, 16.67 mM creatine phosphate, 25 nM radiolabeled pGEM TM transcript, and the indicated proteins and/or cold competitors. Briefly, reactions were set up and left on ice for 20 min, incubated for 20 min in a 30°C water bath, and subjected to UV irradiation (254 nm at a distance of 6 cm) for 8 min. Then, 30 μg of RNase A was added, followed by incubation at 37°C for 30 min, after which SDS buffer was added and the samples were loaded onto 12% SDS gels. Cross-linking was visualized by phosphorimager analysis.
siRNA transfections and analysis.
Double-stranded, preannealed siRNA oligonucleotides against hnRNPs H and F (target sequences: hnRNP H, 5′-AAAGCAGUUGAAUUAUGUUAA-3′; hnRNP F, 5′-AAUGAGUAAACUAAAACUAUU-3′ [sequences from Paul Boutz in the Doug Black lab]) and 9G8 (si-1, 5′-AAAGGGACAUUAUGCUUAUUU-3′; si-2, 5′-GAGGAGAAACCAAGGUGUAUU-3′; si-3, 5′-CGACGUCCCUUUGAUCCAAUU-3′; si-4, 5′-CAGUUAUUAUGGUCCUUUAUU-3′) were purchased from Dharmacon. Lyophilized siRNA pellets were resuspended in buffer containing 20 mM KCl, 6 mM HEPES-KOH (pH 7.5), and 0.2 mM MgCl2 to a concentration of 20 μM. siRNA transfections were performed in PAC1 and HeLa cells with TransIt-TKO (Mirus). Final siRNA concentrations for transfections were 10 nM (PAC1) or 20 nM (HeLa). At 24 h after the initial siRNA transfections, the indicated expression constructs were transfected using TransIT-LT1 and siRNA transfections were repeated. At 24 h later, siRNA transfections were performed a third time. At 72 h after initial transfections, cells were harvested and assayed for protein or mRNA expression.
RESULTS
9G8 activates exon 2 splicing in vivo.
We previously showed that splicing of α-TM exon 2 can be activated by the addition of SR proteins purified from calf thymus (20, 49). To identify individual SR proteins that are responsible for splicing activation, a candidate gene approach was adopted. Vectors expressing individual SR proteins were cotransfected into rat pulmonary artery (PAC1) cells (66), along with a construct containing the first four exons of α-TM (58). PAC1 cells dedifferentiate with increasing passage number meaning the levels of exon 2 inclusion decrease and the exon 3 levels increase over time. Nevertheless, except for primary cultures of smooth muscle cells, PAC1 cells are the best cell type to examine exon 2 inclusion. As shown in Fig. 1B, overexpression of the SR protein 9G8 (sixfold compared to endogenous 9G8 levels; see Fig. S1 in the supplemental material) with the reporter construct caused a twofold increase in exon 2 inclusion. In contrast, none of the other tested SR proteins were able to activate exon 2 inclusion. To ensure that the effect was not restricted to the minigene reporter, we also examined the splicing of endogenous α-TM transcripts and observed a similar activation of exon 2 splicing by 9G8 (data not shown).
Exon 2 contains four purine-rich enhancer elements (Fig. 1A), with the two central enhancers responsible for most exon 2 inclusion (20). To ensure that the effect of 9G8 was a result of SR activation through the exon 2 enhancers, PAC1 transfections were repeated using an α-TM construct with mutations in the two central enhancer elements. As shown in Fig. 1C, loss of the enhancers caused a fivefold decrease in exon 2 inclusion, and cotransfection of 9G8 was unable to activate splicing. Taken together, these results indicate that the 9G8 acts through the exon 2 enhancer elements to activate splicing.
9G8 activates splicing of a heterologous splicing construct containing α-TM exon 2 enhancers.
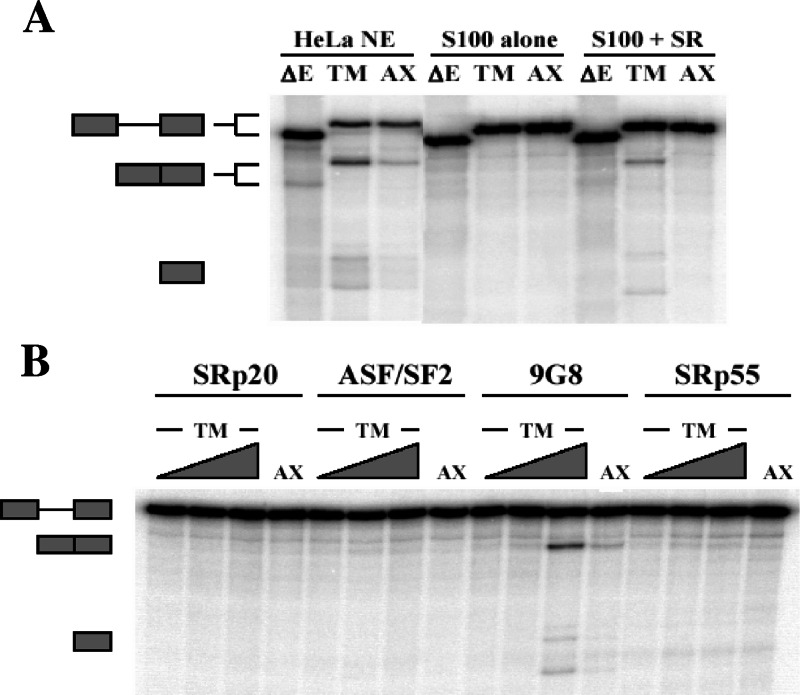
To validate the in vivo activation of splicing by 9G8, an enhancer-dependent splicing substrate derived from the Drosophila melanogaster gene doublesex was utilized. The DNA sequence encompassing the central exon 2 enhancers was inserted into this construct (dsx-ΔE) to create the wild-type construct dsx-TM. A control construct that included the same enhancer mutations from Fig. 1C was also created (dsx-AX). In vitro splicing was then performed in HeLa nuclear extracts, in splicing defective cytoplasmic extracts (S100), or in S100 extracts supplemented with calf thymus-purified SR proteins. Figure 2A shows that, in the absence of enhancer elements, little spliced product was observed (ΔE). In contrast, spliced product formation was readily detectable with the dsx substrate containing the exon 2 enhancers, either in HeLa nuclear extracts or in S100 extracts supplemented with purified SR proteins. As described above, mutation of the enhancer elements substantially inhibited splicing, a finding consistent with sequence-specific activation of splicing. Thus, the α-TM purine-rich elements function as splicing enhancers, and SR proteins can activate splicing in the presence of these sequences in both wild-type and heterologous settings.
FIG. 2.
9G8 activates in vitro splicing of a heterologous construct containing the exon 2 enhancers. (A) In vitro splicing reactions were performed with the indicated substrates in either 40% HeLa nuclear extract, 40% cytoplasmic S100 extracts or 40% S100 extract supplemented with purified SR proteins. The two central exon 2 enhancers elements were inserted into an enhancerless doublesex construct (dsx-ΔE) to make wild-type dsx-TM, whereas enhancer mutations identical to those in Fig. 1 were used to create dsx-AX. After splicing, radiolabeled RNAs were separated on 8% gels with the identity of individual bands as indicated. (B) Individual recombinant SR proteins were added to S100 splicing reactions, and spliced products were analyzed as in panel A. Reactions included increasing amounts of SRp20, ASF/SF2, 9G8, and SRp55 (250 nM, 750 nM, and 1.5 μM). The lanes utilizing substrates with mutated enhancer elements (AX) used 1.5 μM concentrations of each SR protein.
To determine whether 9G8 can activate exon 2 in vitro, splicing was carried out with the dsx constructs in S100 extracts supplemented with individual SR proteins. Each SR protein was purified from baculovirus-infected insect cells, and the activity was verified in S100 rescue experiments using an adenovirus-derived splicing substrate (data not shown) and tested for exon 2 activation across a range of concentrations (Fig. 2B). Consistent with the PAC1 transfections, only 9G8 was able to activate splicing in the presence of the wild-type α-TM enhancer elements. Mutation of the enhancer elements inhibited the rescue, a finding consistent with the in vivo results described above. Thus, 9G8 can activate splicing through the exon 2 enhancer elements both in vivo and in vitro.
9G8, hnRNP H, and hnRNP F bind the exon 2 enhancer elements.
Splicing of α-TM in HeLa cells or HeLa nuclear extracts results in predominant selection of exon 3 with small but detectable levels of exon 2 inclusion. We initially tested whether overexpression of 9G8 might be able to activate exon 2 splicing in HeLa cells, but little activation was detected (data not shown). Given the fact that most alternative splicing decisions are combinatorially controlled, this is perhaps not too surprising. However, it does suggest the possibility that other factors could be present in HeLa cells that could block the ability of 9G8 to activate splicing. As a means to identify such factors as well as to determine whether 9G8 binds to the exon 2 enhancers, RNA affinity chromatography was used. RNA oligonucleotides consisting of the wild-type or mutant enhancer sequences were covalently linked to modified-Sepharose resin (1), HeLa nuclear extracts were passed over the columns, and interacting proteins eluted with increasing salt concentrations. Fractions were separated on SDS gels, and Western blots were performed with antibodies to the amino terminus of 9G8 (Fig. 3A). A band corresponding to 9G8 was detected in the low-salt washes from the wild-type column but was not detectable in any of the fractions derived from the mutated enhancer sequences. Thus, 9G8 binds to the exon 2 enhancer elements, a finding consistent with the functional splicing assays above.
FIG. 3.
9G8, hnRNP H, and hnRNP F bind to the exon 2 enhancer elements. (A) RNA oligonucleotides containing wild-type or mutant exon 2 enhancers were covalently linked to modified Sepharose. HeLa nuclear extracts were passed over the columns, and associated proteins were eluted with increasing salt, with a final elution in urea. Fractions were separated on 10% SDS gels, and Western blots performed with antibodies to 9G8. (B) Fractions from both columns were silver stained. The indicated bands were excised and identified by mass spectrometry.
To identify other proteins that interact with the exon 2 enhancer elements, fractions were separated on SDS gels and silver stained (Fig. 3B). On both mutant and wild-type columns, most proteins eluted between 100 and 500 mM KCl. However, a doublet of proteins was observed in the higher-salt washes with the wild-type column but not with the mutant column. These ∼55-kDa proteins were gel purified, analyzed by mass spectrometry, and found to be hnRNPs H and F.
hnRNPs H and F repress 9G8-activated splicing in vitro and in vivo.
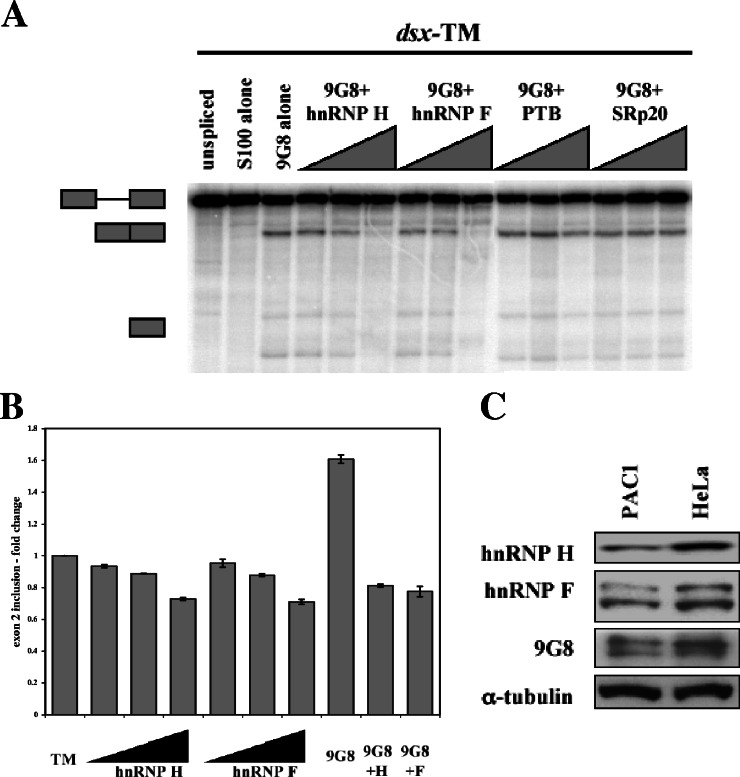
To determine the roles, if any, that hnRNP H and hnRNP F play in exon 2 splicing, we used the dsx constructs to perform in vitro splicing assays supplemented with combinations of 9G8, hnRNP H, and/or hnRNP F. As shown in Fig. 4A, supplementing HeLa S100 extracts with recombinant 9G8 activated splicing, but such activation could be blocked by the addition of increasing amounts of either hnRNP H or F. As controls, we added recombinant PTB or SRp20, neither of which was able to block the effect of 9G8. This suggests that hnRNPs H and F interact with the enhancer elements to block 9G8-mediated splicing.
FIG. 4.
hnRNP H and hnRNP F antagonize 9G8-mediated activation. (A) In vitro splicing reactions were performed with the enhancer-containing dsx-TM in the presence of 1.5 μM 9G8 and increasing amounts of hnRNPs H and F (750 nM, 1.5 μM, and 3 μM). Similar amounts of PTB and SRp20 were added as controls. (B) The wild-type α-TM construct (pSVpA α-TM 1-4; 400 ng) was cotransfected into PAC1 cells with increasing amounts of hnRNP H or hnRNP F (100 ng, 800 ng, and 3.2 μg), and splicing patterns were analyzed as in Fig. 1. At the highest transfection levels, hnRNP H and hnRNP F fractions were increased 3.1- and 3.4-fold, respectively (see Fig. S2 in the supplemental material). (C) Western blots of whole-cell lysates from PAC1 and HeLa cells were performed with antibodies to hnRNP H, hnRNP F, 9G8, and α-tubulin.
To test whether hnRNPs H and F maintain their repressor activity in PAC1 cells, we cotransfected plasmids expressing these proteins together with the wild-type α-TM reporter. Upon transfection of increasing levels of either hnRNP H or F, a modest but reproducible decrease (ca. 25%) in exon 2 inclusion was noted (Fig. 4B). However, when cotransfected with 9G8, both hnRNP H and F were able to block activation of exon 2 splicing. Thus, both in vitro and in vivo, hnRNPs H and F can block 9G8-driven activation of exon 2.
A prediction from the findings presented above is that PAC1 cells should contain lower levels of hnRNPs H and F and, conversely, increased levels of 9G8. Keeping in mind that potential changes in concentration could be subtle and that posttranslational modifications could also affect activity, we nevertheless performed Western blots with antibodies to 9G8 and hnRNPs H and F (Fig. 4C). When normalized to α-tubulin levels, clear decreases were detected for hnRNPs H (50%) and F (42%) in PAC1 versus HeLa cells. In contrast, 9G8 levels did not correlate with splicing activity. This could be due to differential regulation of phosphorylation of 9G8 between the two cell types, a finding consistent with the observation that the hyperphosphorylated form was predominant in HeLa cells (Fig. 4C). Since 9G8 is dephosphorylated during splicing (38), one possibility is that hnRNPs H and F could modulate 9G8 activity by altering phosphorylation or dephosphorylation.
hnRNPs H and F directly compete with 9G8 for binding to the exon 2 enhancers.
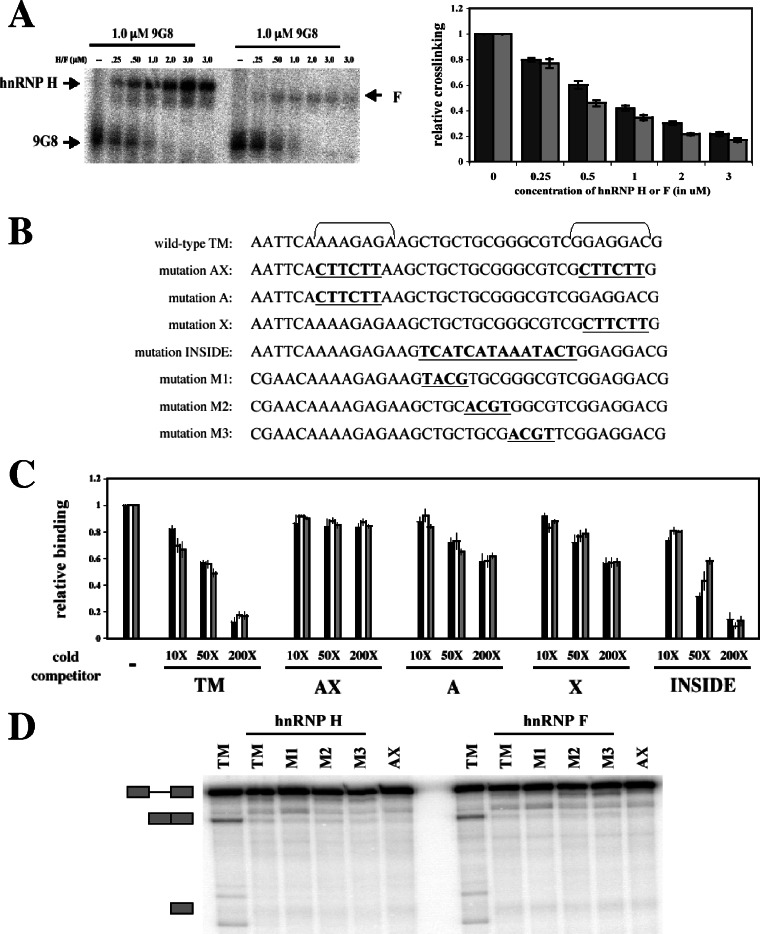
To examine the mechanism of antagonism between 9G8 and hnRNPs H and F, we performed UV cross-linking competition assays. Radiolabeled transcripts containing the intact exon 2 enhancers were subjected to UV irradiation in the presence of recombinant 9G8 and increasing amounts of either hnRNP H or hnRNP F. Consistent with the antagonism seen in Fig. 4, hnRNPs H and F were able to outcompete 9G8 for binding to the enhancer elements in a dose-dependent manner (Fig. 5A). To localize the binding sites for these proteins, wild-type and mutant RNA sequences were used as cold competitors in UV cross-linking assays (Fig. 5B and C). The mutant RNA sequences included base substitutions designed to abolish the enhancer elements, either individually or in combination (A, X, and AX). As shown in Fig. 5C, the wild-type TM sequence was an effective cross-linking self-competitor, as expected. In contrast, mutation of both enhancer elements completely blocked the ability of such RNAs to compete for cross-linking. Competitor RNAs containing mutations in either the A or X enhancer were still able to compete for cross-linking but at levels consistent with the fact that each contained only a single enhancer element. These experiments suggest that hnRNPs H and F compete with 9G8 for binding to the same sequences within exon 2.
FIG. 5.
hnRNPs H and F directly compete with 9G8 for binding to the exon 2 enhancers. (A) Radiolabeled wild-type α-TM transcripts (25 nM) were incubated with 1 μM 9G8 in the presence or absence of increasing amounts of hnRNPs H and F. Reactions were subjected to UV cross-linking, and labeled proteins were analyzed on 10% SDS gels. At least three independent competitions were performed, and averages and standard errors are shown at right (hnRNP H, ▪; hnRNP F,  ). (B) RNA sequences used for cross-linking (TM, AX, A, X, and INSIDE; panels A and C) and in vitro splicing (M1, M2, and M3; panel D) assays. The wild-type sequence consists of the two central exon 2 enhancers (bracketed above) and the sequence between them (shown in Fig. 1A). Nucleotides underlined and in boldface denote mutated nucleotides. (C) UV cross-linking competition. Radiolabeled wild-type TM RNAs (25 nM) were incubated with 9G8 (▪), hnRNP H (□), or hnRNP F (
). (B) RNA sequences used for cross-linking (TM, AX, A, X, and INSIDE; panels A and C) and in vitro splicing (M1, M2, and M3; panel D) assays. The wild-type sequence consists of the two central exon 2 enhancers (bracketed above) and the sequence between them (shown in Fig. 1A). Nucleotides underlined and in boldface denote mutated nucleotides. (C) UV cross-linking competition. Radiolabeled wild-type TM RNAs (25 nM) were incubated with 9G8 (▪), hnRNP H (□), or hnRNP F ( ) in the presence of increasing amounts of cold competitor RNAs. Samples were subjected to UV cross-linking and analyzed as described above. Competitor RNAs were incubated at the indicated molar excess to radiolabeled RNA. RNAs are as indicated in panel B (TM, AX, A, X, and INSIDE). At least three independent competitions were performed, and averages and standard errors are as shown. (D) In vitro splicing reactions were performed with the enhancer-containing dsx-TM and the dsx-M1, M2, M3, and AX mutants in the presence of 9G8 alone (1.5 μM) or when combined with hnRNPs H or F (3 μM).
) in the presence of increasing amounts of cold competitor RNAs. Samples were subjected to UV cross-linking and analyzed as described above. Competitor RNAs were incubated at the indicated molar excess to radiolabeled RNA. RNAs are as indicated in panel B (TM, AX, A, X, and INSIDE). At least three independent competitions were performed, and averages and standard errors are as shown. (D) In vitro splicing reactions were performed with the enhancer-containing dsx-TM and the dsx-M1, M2, M3, and AX mutants in the presence of 9G8 alone (1.5 μM) or when combined with hnRNPs H or F (3 μM).
hnRNPs H and F have previously been shown to interact with G-rich sequences (4, 7, 8, 55), including a G-rich silencer element that serves to block a nearby exonic enhancer in the β-tropomyosin gene (11). Since there is a stretch of G residues in the sequence between the exon 2 enhancers (Fig. 5B), we performed UV cross-linking and splicing assays with a series of RNAs to determine whether hnRNPs H and F bind to the G-rich element as opposed to the exon 2 enhancer elements. The mutants included changes to alter the entire G-rich stretch (INSIDE), as well as three smaller substitutions across the region (M1, M2, and M3). When used in UV cross-linking competition assays, the INSIDE mutant RNA was an effective competitor on par with the wild-type TM RNA, suggesting that hnRNPs H and F do not bind to the internal G-rich stretch (Fig. 5C). The M1, M2, and M3 mutants were incorporated into splicing substrates and tested for the ability of hnRNPs H and F to repress the activation of splicing by 9G8 (Fig. 5D). As shown, splicing of the dsx-TM construct could be activated by the addition of 9G8 protein, but this activation could be abolished by the addition of hnRNPs H or F or by mutation of the enhancer elements (AX). Importantly, none of the three mutations altering the G-rich stretch between the exon 2 enhancers affected the ability of either hnRNP H or F to repress splicing. If either protein recognized these elements, decreased repression should have been observed upon mutation. Since none of the mutants altered repression, it appears that the G-rich sequence between the two enhancer elements within exon 2 does not interact with either hnRNP H or F. Together with the UV cross-linking data, these results suggest that hnRNPs H and F antagonize 9G8-mediated activation of exon 2 inclusion by directly competing for the enhancer elements within exon 2.
siRNA depletion of hnRNP H, hnRNP F, and 9G8.
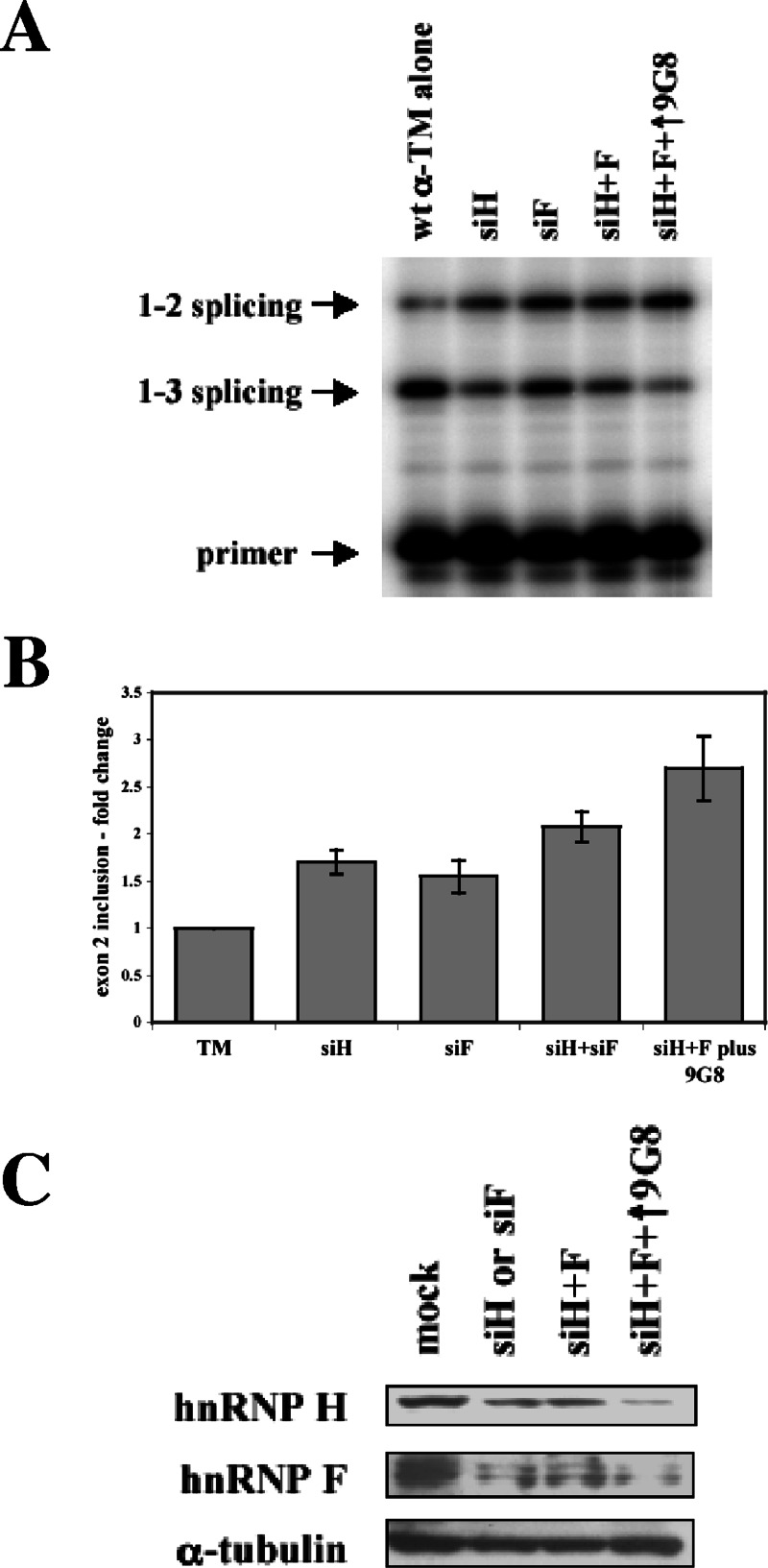
To further test the role that these proteins play in regulating splicing, we used RNA interference to assay splice site choice after knockdown of each factor. If hnRNPs H and F act as repressors, exon 2 inclusion should increase upon their depletion. In contrast, siRNAs against 9G8 should have the opposite effect. Double-stranded siRNAs were cotransfected into PAC1 cells with the wild-type α-TM reporter construct, and spliced products were analyzed. As shown in Fig. 6A and B, siRNA-directed knockdown of hnRNP H and/or F led to increased exon 2 inclusion. When hnRNP H and F levels were decreased in cells in which 9G8 was concurrently overexpressed, a much more dramatic increase in exon 2 inclusion was detected.
FIG. 6.
siRNA depletion of hnRNP H and hnRNP F. (A) Wild-type pSVpA α-TM 1-4 minigene (400 ng) was cotransfected into PAC1 cells with 10 nM concentrations of siRNAs directed against hnRNP H and/or hnRNP F, either alone, together, or in combination with 800 ng of 9G8. Splicing patterns were analyzed as described above. (B) At least three independent transfections as in panel A were performed, and averages and standard errors for exon 2 inclusion are shown. (C) Western blots of normal cells (mock) or cells treated with siRNAs against hnRNPs H and F were performed with antibodies to hnRNP H, F, or α-tubulin as a loading control.
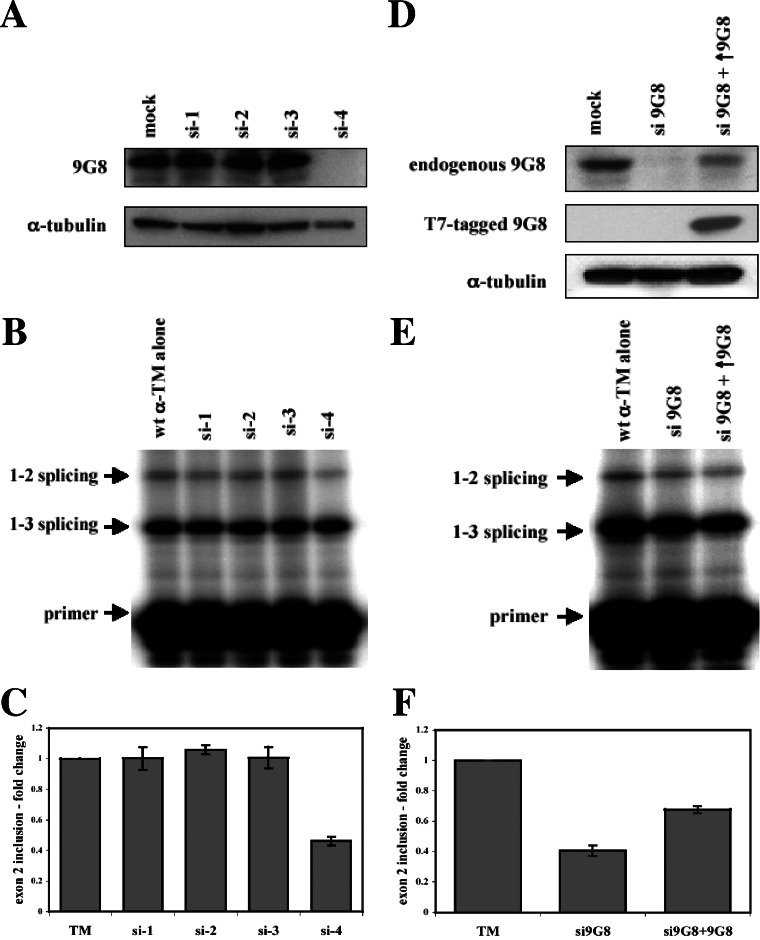
Attempts to reduce 9G8 levels in PAC1 cells using RNAi were unfortunately ineffective, necessitating a change to another cell line. We decided to use HeLa cells since small, but detectable levels of exon 2 inclusion can be visualized upon transfection of α-TM reporter constructs. If such levels of exon 2 inclusion are due in part to the action of 9G8, depletion of 9G8 should result in decreased exon 2 inclusion. Therefore, siRNAs against 9G8 were cotransfected with the wild-type α-TM reporter construct into HeLa cells, and the splicing patterns were analyzed. Four different siRNA sequences were tested. As shown in Fig. 7A, only one of these siRNAs (si-4) was able to decrease the levels of 9G8 protein in HeLa cells. Consistent with a role for 9G8 in activating exon 2, depletion of 9G8 led to an approximate 50% reduction in the level of exon 2 inclusion (Fig. 7B and C). None of the other siRNAs were able to decrease 9G8 levels and, in turn, no changes in splicing were observed. As a further test of specificity, we cotransfected an epitope-tagged version of 9G8 along with si-4 to test whether we could complement the depletion. Partial rescue of exon 2 inclusion was observed (Fig. 7E and F). Thus, both in vitro and in vivo, and under both overexpression and knockdown experiments, 9G8 activates α-TM exon 2 splicing, and such activation is antagonized by hnRNPs H and F.
FIG. 7.
siRNA depletion of 9G8. (A) Wild-type pSVpA α-TM 1-4 minigene (400 ng) was cotransfected into HeLa cells with 20 nM concentrations of each of four individual siRNAs (si-1, si-2. si-3, and si-4) directed against 9G8. Western blots were performed on normal cells (mock) or cells treated with siRNAs against 9G8 using antibodies to 9G8 or α-tubulin. (B) Splicing patterns were analyzed as in Fig. 1, and the average and standard errors from at least three independent transfections are shown below (C). (D) Wild-type pSVpA α-TM 1-4 minigene (400 ng) was transfected into HeLa cells with either 20 nM of the si-4 siRNA directed against 9G8 or the si-4 siRNA plus cotransfection of an epitope-tagged (T7) version of 9G8. Western blots were performed on normal cells (mock) or cells treated with siRNAs against 9G8 using antibodies to 9G8, Τ7, or α-tubulin. (E) Splicing patterns were analyzed as in Fig. 1, and the average and standard errors from at least three independent transfections are shown at bottom (F).
DISCUSSION
To identify individual SR proteins that interact with and activate exon 2, we undertook a candidate gene approach in vivo. Transfection studies were performed in smooth muscle cells with an α-TM reporter construct and core SR proteins. We found that the SR protein 9G8 was able to activate exon 2 splicing in a sequence-dependent manner. In vitro splicing studies using a heterologous construct verified that 9G8 interacts with two enhancer elements within exon 2 for splicing activation. RNA affinity chromatography confirmed the binding of 9G8 to these enhancer sequences but also identified hnRNP H and hnRNP F as associating with the same sequence elements. In vitro and in vivo overexpression studies showed that hnRNPs H and F antagonize the 9G8-mediated activation of splicing, acting as splicing repressors of exon 2. Through UV cross-linking experiments, we found that hnRNPs H and F compete with 9G8 for binding to the exon 2 enhancer sequences. Lastly, siRNA-directed depletion of hnRNP H and F led to an increase in exon 2 inclusion, whereas the loss of 9G8 decreased exon 2 splicing. Together, these results show that 9G8 enhances the inclusion of α-TM exon 2 through its purine-rich sequences, an action that is antagonized by hnRNPs H and F.
Combinatorial control of α-TM splicing.
The α-TM gene has served as an excellent model system to dissect regulatory mechanisms controlling alternative splicing (68). Mutually exclusive splicing of exons 2 and 3 is regulated by steric hindrance due to the unique upstream positioning of the branchpoint upstream of exon 3 (67). With the genomic architecture thereby precluding the joining of exons 2 and 3, regulatory elements and factors have been identified that determine exon inclusion (20, 27, 28, 33, 58, 62). As shown in Fig. 1, the RNA sequence elements include the branchpoint/pyrimidine tracts upstream of each exon, regulatory elements flanking exon 3 (URE and DRE), and enhancer elements within exon 2. PTB and an interacting protein, raver1, have been shown to mediate repression of exon 3 by binding to pyrimidine-rich regulatory sites flanking exon 3 (32, 68). How widely expressed proteins act to mediate tissue-specific splicing remains unclear but may be influenced, in part, by an as-yet-unknown protein(s) that binds to the UGC repeats found within the URE and DRE or could perhaps be due to differential activity of PTB paralogs and alternatively spliced isoforms (53, 61, 76, 80).
As for elements and factors that activate exon 2, initial studies argued against the need for any such enhancers since deletion of exon 3 allows inclusion of exon 2 in all cell types tested (58). Later experiments with heterologous constructs that contained either exon 2 or 3 suggested that exon 2 might contain elements facilitating its inclusion (27). When α-TM minigene substrates were tested, it was found that SR proteins could increase exon 2 inclusion and that purine-rich elements within exon 2 were necessary for such activation (20, 49). Here, we show that increased exon 2 inclusion is dependent upon 9G8 binding to these purine enhancers. Further biochemical analysis led to the discovery that hnRNPs H and F also interact with these enhancer sequences.
hnRNP H, hnRNP F, and splicing regulation.
hnRNPs bind nascent RNA transcripts not only to package pre-mRNA but also to assist and/or regulate specific posttranscriptional events, nucleocytoplasmic transport, and RNA stability (13, 44, 74). In general, hnRNPs are thought to act as negative factors repressing specific exons or splice sites. However, this is not always case, especially for hnRNPs H and F, where activation has been detected in the regulation of the N1 exon of c-src (12, 57), exon 6D of human immunodeficiency virus type 1 (HIV-1) (8), and the apoptotic mediator Bcl-xs (26), and repression has been observed at 5′ splice sites in the NF-1 and TSHβ genes (4). c-src uses a complex of proteins, including hnRNPs H and F, to activate the inclusion of a neural-specific, 18-nucleotide exon (12, 57, 65). In HIV-1, hnRNP H is required for the association of U1 snRNP to the tev-specific exon 6D, enhancing exon inclusion (8). For Bcl-x, hnRNP H and F influence the production of the proapoptotic Bcl-xs isoform by directing 5′-splice site usage (26). Similarly, hnRNP H can bind to G-rich 5′ splice sites and block U1 snRNP binding, leading to exon skipping, especially when coupled to additional mutations that alter U1 snRNP pairing (4).
The question raised by these examples is how can these proteins act as both activators and repressors of splicing? They both bind to G-rich sequences: GGGA in HIV-1 to activate splicing and GGGGU in NF-1 to repress splicing (4, 7, 8, 55). This argues that not only are the binding sites important but the context and/or position of the sites likely influences activity. In the present study, we demonstrate that hnRNP H and hnRNP F interact with purine-rich sequences and not with a nearby G-rich element, bolstering the argument for contextual and positional importance. Besides sequence context, differences in concentration (36), subcellular localization, or tissue-specific posttranslational modifications could also affect activity. For SR proteins, regulated phosphorylation/dephosphorylation affects splicing activity and nucleocytoplasmic shuttling (37, 38, 46, 50). Hyperphosphorylated ASF/SF2 and 9G8 associate with pre-mRNA to initiate splicing and then become hypophosphorylated as splicing proceeds, followed by export to the cytoplasm. Similarly, the nucleocytoplasmic localization of PTB is also affected by phosphorylation (79). Regulated phosphorylation of hnRNP H and hnRNP F has not been detected, but the examples presented above suggest that such modification could account for the ability to exhibit both positive and negative effects on splicing.
Decreased hnRNP H levels cause an upregulation in smooth muscle-specific gene expression and smooth muscle development (51). Consistent with that finding, we showed here that decreased hnRNP H and F levels caused increased expression of the smooth muscle-specific α-TM splicing pattern. Likewise, examination of α-TM exon 2 splicing using a transgenic mouse model showed a broad correlation between smooth muscle splicing patterns and tissues expressing decreased levels of hnRNP H and F (21). This suggests that hnRNPs H and F could play important roles in regulating not only α-TM splicing but also perhaps other smooth muscle or tissue-specific splicing events.
Supplementary Material
Acknowledgments
We thank Doug Black, Brenton Graveley, James Stévenin, Adrian Krainer, Robert Lafyatis, and Tracey Rouault for kindly providing expression constructs, antibodies, cDNA clones, and protocols. We also thank David Friedman and the Vanderbilt Proteomics Laboratory.
This study was supported by an NIH grant to J.G.P. (GM 62487) and the Vanderbilt Mechanisms of Vascular Disease Training Grant (NIH 5 T32 HL07751).
Footnotes
Published ahead of print on 25 September 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Allerson, C. R., A. Martinez, E. Yikilmaz, and T. A. Rouault. 2003. A high capacity RNA affinity column for the purification of human IRP1 and IRP2 overexpressed in Pichia pastoris. RNA 9:364-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 3.Black, D. L., and P. J. Grabowski. 2003. Alternative pre-mRNA splicing and neuronal function. Prog. Mol. Subcell. Biol. 31:187-216. [DOI] [PubMed] [Google Scholar]
- 4.Buratti, E., M. Baralle, L. De Conti, D. Baralle, M. Romano, Y. M. Ayala, and F. E. Baralle. 2004. hnRNP H binding at the 5′ splice site correlates with the pathological effect of two intronic mutations in the NF-1 and TSH-β genes. Nucleic Acids Res. 32:4224-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buvoli, M., S. A. Mayer, and J. G. Patton. 1997. Functional crosstalk between exon enhancers, polypyrimidine tracts, and branchpoint sequences. EMBO J. 16:7174-7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caputi, M., A. Mayeda, A. R. Krainer, and A. M. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caputi, M., and A. M. Zahler. 2001. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J. Biol. Chem. 276:43850-43859. [DOI] [PubMed] [Google Scholar]
- 8.Caputi, M., and A. M. Zahler. 2002. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 21:845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cartegni, L., S. Chew, and A. Krainer. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Gen. 3:285-298. [DOI] [PubMed] [Google Scholar]
- 10.Charlet-B, N., P. Logan, G. Singh, and T. A. Cooper. 2002. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell 9:649-658. [DOI] [PubMed] [Google Scholar]
- 11.Chen, C. D., R. Kobayashi, and D. M. Helfman. 1999. Binding of hnRNP H to an exonic splicing enhancer is involved in the regulation of alternative splicing of the rat β-tropomyosin gene. Genes Dev. 13:593-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou, M.-Y., N. Rooke, C. W. Turck, and D. L. Black. 1999. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 19:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conway, G., J. Wooley, T. Bibring, and W. LeStourgeon. 1988. Ribonucleoproteins package 700 nucleotides of pre-mRNA into a repeating array of regular particles. Mol. Cell. Biol. 8:2884-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooley, B. C., and G. Bergtrom. 2001. Multiple combinations of alternatively spliced exons in rat tropomyosin-alpha gene mRNA: evidence for 20 new isoforms in adult tissues and cultured cells. Arch. Biochem. Biophys. 390:71-77. [DOI] [PubMed] [Google Scholar]
- 15.Cáceres, J. F., S. Stamm, D. M. Helfman, and A. R. Krainer. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265:1706-1708. [DOI] [PubMed] [Google Scholar]
- 16.Dansithong, W., S. Paul, L. Comai, and S. Reddy. 2005. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J. Biol. Chem. 280:5773-5780. [DOI] [PubMed] [Google Scholar]
- 17.Del Gatto-Konczak, F., C. F. Bourgeois, C. Le Guiner, L. Kister, M. C. Gesnel, J. Stevenin, and R. Breathnach. 2000. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 20:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Del Gatto-Konczak, F., M. Olive, M.-C. Gesnel, and R. Breathnach. 1999. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol. 19:251-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domsic, J. K., Y. Wang, A. Mayeda, A. R. Krainer, and C. M. Stoltzfus. 2003. Human immunodeficiency virus type 1 hnRNP A/B-dependent exonic splicing silencer ESSV antagonizes binding of U2AF65 to viral polypyrimidine tracts. Mol. Cell. Biol. 23:8762-8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dye, B. T., M. Buvoli, S. A. Mayer, C.-H. Lin, and J. G. Patton. 1998. Enhancer elements activate the weak 3′ splice site of α-tropomyosin exon 2. RNA 4:1523-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellis, P. D., C. W. Smith, and P. Kemp. 2004. Regulated tissue-specific alternative splicing of enhanced green fluorescent protein transgenes conferred by α-tropomyosin regulatory elements in transgenic mice. J. Biol. Chem. 279:36660-36669. [DOI] [PubMed] [Google Scholar]
- 22.Eperon, I. C., O. V. Makarova, A. Mayeda, S. H. Munroe, J. F. Caceres, D. G. Hayward, and A. R. Krainer. 2000. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol. 20:8303-8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fairbrother, W. G., R. F. Yeh, P. A. Sharp, and C. B. Burge. 2002. Predictive identification of exonic splicing enhancers in human genes. Science 297:1007-1013. [DOI] [PubMed] [Google Scholar]
- 24.Forch, P., O. Puig, N. Kedersha, C. Martinez, S. Granneman, B. Seraphin, P. Anderson, and J. Valcarcel. 2000. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell 6:1089-1098. [DOI] [PubMed] [Google Scholar]
- 25.Fu, X.-D. 1995. The superfamily of arginine/serine-rich splicing factors. RNA 1:663-680. [PMC free article] [PubMed] [Google Scholar]
- 26.Garneau, D., T. Revil, J. F. Fisette, and B. Chabot. 2005. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem. 280:22641-22650. [DOI] [PubMed] [Google Scholar]
- 27.Gooding, C., G. C. Roberts, G. Moreau, B. Nadal-Ginard, and C. W. J. Smith. 1994. Smooth muscle specific switching of α-tropomyosin mutually exclusive exon selection by specific inhibition of the strong default exon. EMBO J. 13:3861-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gooding, C., G. C. Roberts, and C. W. J. Smith. 1998. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated α-tropomyosin exon. RNA 4:85-100. [PMC free article] [PubMed] [Google Scholar]
- 29.Goodwin, L. O., J. P. Lees-Miller, M. A. Leonard, S. B. Cheley, and D. M. Helfman. 1991. Four fibroblast tropomyosin isoforms are expressed from the rat α-tropomyosin gene via alternative RNA splicing and the use of two promoters. J. Biol. Chem. 266:8408-8415. [PubMed] [Google Scholar]
- 30.Graveley, B. R. 2001. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17:100-107. [DOI] [PubMed] [Google Scholar]
- 31.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gromak, N., A. Rideau, J. Southby, A. Scadden, C. Gooding, S. Huttelmaier, R. Singer, and C. Smith. 2003. The PTB interacting protein raver1 regulates α-tropomyosin alternative splicing. EMBO J. 22:6356-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gromak, N., and C. W. Smith. 2002. A splicing silencer that regulates smooth muscle specific alternative splicing is active in multiple cell types. Nucleic Acids Res. 30:3548-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamon, S., C. Le Sommer, A. Mereau, M. R. Allo, and S. Hardy. 2004. Polypyrimidine tract-binding protein is involved in vivo in repression of a composite internal/3′-terminal exon of the Xenopus α-tropomyosin Pre-mRNA. J. Biol. Chem. 279:22166-22175. [DOI] [PubMed] [Google Scholar]
- 35.Ho, T. H., B. N. Charlet, M. G. Poulos, G. Singh, M. S. Swanson, and T. A. Cooper. 2004. Muscleblind proteins regulate alternative splicing. EMBO J. 23:3103-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Honore, B., U. Baandrup, and H. Vorum. 2004. Heterogeneous nuclear ribonucleoproteins F and H/H′ show differential expression in normal and selected cancer tissues. Exp. Cell Res. 294:199-209. [DOI] [PubMed] [Google Scholar]
- 37.Huang, Y., and J. A. Steitz. 2005. SRprises along a messenger's journey. Mol. Cell 17:613-615. [DOI] [PubMed] [Google Scholar]
- 38.Huang, Y., T. A. Yario, and J. A. Steitz. 2004. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. USA 101:9666-9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jensen, K. B., B. K. Dredge, G. Stefani, R. Zhong, R. J. Buckanovich, H. J. Okano, Y. Y. L. Yang, and R. B. Darnell. 2000. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 25:359-371. [DOI] [PubMed] [Google Scholar]
- 40.Jiang, Z.-H., W.-J. Zhang, Y. Rao, and J. Y. Wu. 1998. Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc. Natl. Acad. Sci. USA 95:9155-9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin, Y., H. Suzuki, S. Maegawa, H. Endo, S. Sugano, K. Hashimoto, K. Yasuda, and K. Inoue. 2003. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 22:905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson, J., J. Castle, P. Garrett-Engele, Z. Kan, P. Loerch, C. Armour, R. Santos, E. Schadt, R. Stoughton, and D. Shoemaker. 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302:2141-2144. [DOI] [PubMed] [Google Scholar]
- 43.Kashima, T., and J. L. Manley. 2003. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 34:460-463. [DOI] [PubMed] [Google Scholar]
- 44.Krecic, A. M., and M. S. Swanson. 1999. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11:363-371. [DOI] [PubMed] [Google Scholar]
- 45.Ladd, A. N., N. Charlet, and T. A. Cooper. 2001. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 21:1285-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai, M. C., and W. Y. Tarn. 2004. Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J. Biol. Chem. 279:31745-31749. [DOI] [PubMed] [Google Scholar]
- 47.Lam, B. J., A. Bakshi, F. Y. Ekinci, J. Webb, B. R. Graveley, and K. J. Hertel. 2003. Enhancer-dependent 5′-splice site control of fruitless pre-mRNA splicing. J. Biol. Chem. 278:22740-22747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lam, B. J., and K. J. Hertel. 2002. A general role for splicing enhancers in exon definition. RNA 8:1233-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin, C.-H., and J. G. Patton. 1995. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA 1:234-245. [PMC free article] [PubMed] [Google Scholar]
- 50.Lin, S., R. Xiao, P. Sun, X. Xu, and X. D. Fu. 2005. Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Mol. Cell 20:413-425. [DOI] [PubMed] [Google Scholar]
- 51.Liu, J., S. Beqaj, Y. Yang, B. Honore, and L. Schuger. 2001. Heterogeneous nuclear ribonucleoprotein-H plays a suppressive role in visceral myogenesis. Mech. Dev. 104:79-87. [DOI] [PubMed] [Google Scholar]
- 52.Lynch, K. W., and T. Maniatis. 1996. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 10:2089-2101. [DOI] [PubMed] [Google Scholar]
- 53.Markovtsov, V., J. M. Nikolic, J. A. Goldman, C. W. Turck, M.-Y. Chou, and D. L. Black. 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 20:7463-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matlin, A., F. Clark, and C. Smith. 2005. Understanding alternative splicing: toward a cellular code. Nat. Rev. Mol. Cell. Biol. 6:386-398. [DOI] [PubMed] [Google Scholar]
- 55.Matunis, M. J., J. Xing, and G. Dreyfuss. 1994. The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 22:1059-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mayeda, A., and A. R. Krainer. 1992. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68:365-375. [DOI] [PubMed] [Google Scholar]
- 57.Min, H., R. C. Chan, and D. L. Black. 1995. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 9:2659-2671. [DOI] [PubMed] [Google Scholar]
- 58.Mullen, M. P., C. W. J. Smith, J. G. Patton, and B. Nadal-Ginard. 1991. α-Tropomyosin mutually exclusive exon selection: competition between branch point/polypyrimidine tracts determines exon choice. Genes Dev. 5:642-655. [DOI] [PubMed] [Google Scholar]
- 59.Patton, J. G., S. A. Mayer, P. Tempst, and B. Nadal-Ginard. 1991. Characterization and molecular cloning of polypyrimidine tract binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 5:1237-1251. [DOI] [PubMed] [Google Scholar]
- 60.Patton, J. G., E. B. Porro, J. Galceran, P. Tempst, and B. Nadal-Ginard. 1993. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 7:393-406. [DOI] [PubMed] [Google Scholar]
- 61.Polydorides, A. D., H. J. Okano, Y. Y. L. Yang, G. Stefani, and R. B. Darnell. 2000. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc. Natl. Acad. Sci. USA 97:6350-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pérez, I., C.-H. Lin, J. G. McAfee, and J. G. Patton. 1997. Mutation of PTB binding sites causes misregulation of α-tropomyosin alternative 3′ splice site selection in vivo. RNA 3:764-778. [PMC free article] [PubMed] [Google Scholar]
- 63.Reuter, S. M., C. M. Burns, S. A. Coode, P. Mookherjee, and R. B. Emeson. 1995. Glutamate receptor RNA editing by enzymatic conversion of adenosine to inosine. Science 267:1491-1494. [DOI] [PubMed] [Google Scholar]
- 64.Robberson, B. L., G. Cote, and S. M. Berget. 1990. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell. Biol. 10:84-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rooke, N., V. Markovtsov, E. Cagavi, and D. L. Black. 2003. Roles for SR proteins and hnRNP A1 in the regulation of c-src exon N1. Mol. Cell. Biol. 23:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rothman, A., T. J. Kulik, M. B. Taubman, B. C. Berk, C. W. J. Smith, and B. Nadal-Ginard. 1992. Characterization of a newly established rat pulmonary arterial smooth muscle cell line. Circulation 86:1977-1986. [DOI] [PubMed] [Google Scholar]
- 67.Smith, C. W. J., and B. Nadal-Ginard. 1989. Mutually exclusive splicing of α-tropomyosin exons enforced by an unusual lariat branch point location; implications for constitutive splicing. Cell 56:749-758. [DOI] [PubMed] [Google Scholar]
- 68.Spellman, R., A. Rideau, A. Matlin, C. Gooding, F. Robinson, N. McGlincy, S. N. Grellscheid, J. Southby, M. Wollerton, and C. W. Smith. 2005. Regulation of alternative splicing by PTB and associated factors. Biochem. Soc. Trans. 33:457-460. [DOI] [PubMed] [Google Scholar]
- 69.Spellman, R., and C. W. Smith. 2006. Novel modes of splicing repression by PTB. Trends Biochem. Sci. 31:73-76. [DOI] [PubMed] [Google Scholar]
- 70.Tian, M., and T. Maniatis. 1992. Positive control of pre-mRNA splicing in vitro. Science 256:237-240. [DOI] [PubMed] [Google Scholar]
- 71.Wagner, E. J., and M. A. Garcia-Blanco. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21:3281-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, J., P. J. Smith, A. R. Krainer, and M. Q. Zhang. 2005. Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic Acids Res. 33:5053-5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang, Z., M. E. Rolish, G. Yeo, V. Tung, M. Mawson, and C. B. Burge. 2004. Systematic identification and analysis of exonic splicing silencers. Cell 119:831-845. [DOI] [PubMed] [Google Scholar]
- 74.Weighardt, F., G. Biamonti, and S. Riva. 1996. The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays 18:747-756. [DOI] [PubMed] [Google Scholar]
- 75.Wieczorek, D., C. W. J. Smith, and B. Nadal-Ginard. 1988. The rat α-tropomyosin gene generates a minimum of six different mRNAs coding for striated, smooth, and non-muscle isoforms by alternative splicing. Mol. Cell. Biol. 8:679-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wollerton, M. C., C. Gooding, F. Robinson, E. C. Brown, R. J. Jackson, and C. W. J. Smith. 2001. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB). RNA 7:819-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wollerton, M. C., C. Gooding, E. J. Wagner, M. A. Garcia-Blanco, and C. W. Smith. 2004. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 13:91-100. [DOI] [PubMed] [Google Scholar]
- 78.Wu, J. Y., and T. Maniatis. 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75:1061-1070. [DOI] [PubMed] [Google Scholar]
- 79.Xie, J., J. A. Lee, T. L. Kress, K. L. Mowry, and D. L. Black. 2003. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 100:8776-8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yamamoto, H., K. Tsukahara, Y. Kanaoka, S. Jinno, and H. Okayama. 1999. Isolation of a mammalian homologue of a fission yeast differentiation regulator. Mol. Cell. Biol. 19:3829-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zahler, A. M., C. K. Damgaard, J. Kjems, and M. Caputi. 2004. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J. Biol. Chem. 279:10077-10084. [DOI] [PubMed] [Google Scholar]
- 82.Zahler, A. M., W. S. Lane, J. A. Stolk, and M. B. Roth. 1992. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 6:837-847. [DOI] [PubMed] [Google Scholar]
- 83.Zhu, J., A. Mayeda, and A. R. Krainer. 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8:1351-1361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.