Abstract
The “classical” nuclear protein import pathway depends on importin α and importin β. Importin α binds nuclear localization signal (NLS)-bearing proteins and functions as an adapter to access the importin β-dependent import pathway. In humans, only one importin β is known to interact with importin α, while six α importins have been described. Various experimental approaches provided evidence that several substrates are transported specifically by particular α importins. Whether the NLS is sufficient to mediate importin α specificity is unclear. To address this question, we exchanged the NLSs of two well-characterized import substrates, the seven-bladed propeller protein RCC1, preferentially transported into the nucleus by importin α3, and the less specifically imported substrate nucleoplasmin. In vitro binding studies and nuclear import assays revealed that both NLS and protein context contribute to the specificity of importin α binding and transport.
Nuclear import substrates possess nuclear localization signals (NLSs) required for recognition by distinct nuclear import factors. The so-called “classical” NLS consists of either one cluster of basic amino acids (monopartite NLS) or two clusters of basic amino acids separated by a linker (bipartite NLS). Nuclear transport of substrates bearing a classical NLS is mediated by the importin α/β heterodimer, also known as karyopherin α/β. Importin α functions as an adapter by binding both the import substrate via the NLS and importin β. Importin β docks the ternary import complex at the nuclear pore complex and facilitates its translocation through the nuclear pore complex into the nucleus (13, 20). In addition, importin α and β are also involved in other processes associated with nuclear functions, ranging from spindle formation to nuclear envelope assembly (17-19, 35, 48, 52).
Importin α is composed of a short basic N-terminal importin β binding domain (14, 50) and a large NLS binding domain comprised of 10 tandem armadillo (ARM) repeats (4, 24). The series of ARM repeats generates a superhelical structure that has a shallow, concave NLS binding groove containing two NLS binding sites consisting of ARM repeats 1 to 4 (major site) and 4 to 8 (minor site) (4, 11). Bipartite NLS sequences span the two binding sites, with each site recognizing one of the basic clusters (3, 10, 11). Monopartite NLS sequences are able to bind both sites, but only the binding at the major site, corresponding to the C-terminal basic cluster of the bipartite NLS, is likely to be physiologically relevant (3, 4, 11, 12). The N-terminal importin β binding domain of importin α serves a dual role. It binds to importin β but also contains an autoinhibitory sequence that mimics an NLS. This autoinhibitory sequence interacts with the NLS binding domain when importin α is not bound to importin β and/or to an NLS cargo (9, 24, 34). Accordingly, the affinity of importin α for import substrates is increased in the presence of importin β (2, 9, 41).
While only one importin β isoform exists for interaction with importin α, six human α importins have been described previously (5, 6, 25, 28, 49). In contrast, the yeast Saccharomyces cerevisiae possesses only one gene for importin α, which is essential (53). Based on the similarity of their primary structures, the α importins are grouped into three subfamilies. The first subfamily consists of importin α1/Rch1. Its most closely related homologue, importin α2 from Xenopus laevis (15), has also been found in other vertebrates, but so far not in mammals. Importin α3/Qip1 and importin α4/hSRP1γ are members of the second subfamily. The third subfamily consists of importin α5/hSRP1, importin α6, and importin α7. Members of different subfamilies have about 50% sequence identity, but within one subfamily, the identity is at least 80% (13, 25, 28). The α importins display differences in their cell- and tissue-specific expression patterns. However, all isoforms except for importin α6 are expressed within the same tissue (21, 25, 26, 28, 47).
Although some import substrates can be transported into the nucleus by various α importins, many experimental approaches provided evidence that several substrates are recognized and transported specifically by particular α importins (8, 27, 28, 38, 39, 45, 51). NLS sequences are both necessary and sufficient for nuclear protein import via the importin α/β-dependent pathway (23, 30, 42). Even though earlier studies indicated that the NLS may also play a role in mediating importin α specificity (33, 36), whether the NLS is indeed sufficient to determine importin α specificity or whether other parts of the protein contribute to this selectivity remains to be elucidated. To address this question, we exchanged the NLSs of two well-characterized import substrates. We found that although the NLS contributes, the NLS alone is not sufficient to determine strong importin α specificity. Only the combination of the NLS and additional structural features within the cargo mediates high selectivity for a particular α importin.
MATERIALS AND METHODS
DNA constructs.
Importin α1 and α3 gene constructs encoding C-terminally glutathione S-transferase (GST)-tagged proteins have been described previously (32). GST fusion plasmids of importins α4, α5, and α7 and importin β were generated by replacing the C-terminal His tags of the previously described constructs (16, 28) by GST tags from pFA6a-GST-His3MX6 (31) via BamHI/HindIII using PCR techniques.
Plasmids encoding full-length, C-terminally His-tagged RCC1 (RCC1/pQE60) and nucleoplasmin (nucleoplasmin/pQE70) have been described earlier (15, 28). The RCC1 gene region encoding amino acids 25 to 77 was amplified by using the primers 5′-CGC CAT GGT CTC ACA CAG GTC CCA C-3′ and 5′-GGT GTG CAT GCC CCC AGC-3′. After NcoI/SphI digestion, the insert was cloned into the equally digested RCC1/pQE60 plasmid, resulting in a truncated RCC1 gene spanning amino acids 25 to 421 (RCC1Δ24). The RCC1 deletion construct RCC1Δ13 was cloned in the same way, but with the forward primer 5′-CGC CAT GGC AGA TGC CAT CCC CAA AAG-3′. N-terminal mutation constructs RCC1mt1, RCC1mt2, and RCC1mt3 were generated by using the QuikChange site-directed mutagenesis kit (Stratagene) and appropriate primers. The creation of the nucleoplasmin NLS [NLS(N)]-propeller (prop) construct was accomplished by PCR amplification of the nucleoplasmin region encoding amino acids 150 to 170 by using the primers 5′-CGC CAT GGC TCC ACC CAA AGC TGT AAA G-3′ and 5′-CGC CAT GGC TTT CTT CTT CTT TGC CTG GCC-3′. The PCR product was digested with NcoI/NcoI and inserted 5′ to the truncated RCC1 gene into the RCC1Δ24 plasmid. The construct NLS(N)-nucleoplasmin core domain (core) was generated by PCR amplification of the same nucleoplasmin region by using the primers 5′-CGG CAT GCC ACC CAA AGC TGT AAA GAG G-3′ and 5′-CGG CAT GCC TTT CTT CTT CTT TGC CTG GCC-3′. The PCR product was ligated as a SphI/SphI fragment 5′ to the nucleoplasmin core domain of the earlier-described core/pQE70 plasmid (15). The creation of the construct RCC1 NLS [NLS(R)]-core was accomplished by PCR amplification of the first 35 amino acids of RCC1 by using the primers 5′-CCG CAT GCA TGT CAC CCA AGC GCA TAG C-3′ and 5′-CCG CAT GCC GGG TTC TGT GCT GTG G-3′. The PCR product was digested with SphI/SphI and inserted 5′ to the nucleoplasmin core domain into the core/pQE70 plasmid. Bases ATGC at the first SphI site were deleted to bring the start codon of the construct into the right position for proper expression. The construct NLS(RΔ12)-core was generated by PCR amplification of the RCC1 gene region encoding amino acids 13 to 35 by using the NLS(R)-core plasmid as the template and the primers 5′-CGG CAT GCC AGC AGA TGC CAT CCC C-3′ and 5′-GAG CAT GCC GGG TTC TGT G-3′ for PCR. The PCR product was ligated as a SphI/SphI fragment 5′ to the nucleoplasmin core domain of the core/pQE70 plasmid. The constructs were verified by sequencing. Dirk Görlich (ZMBH, Heidelberg) kindly provided the expression clone encoding N-terminally zz-tagged RCC1.
Recombinant protein expression and purification.
C-terminally GST-tagged fusion proteins were expressed in Escherichia coli BL21/Rep4 (importins α1- and α7-GST) or in E. coli JM101 (importins α3-, α4-, α5-, and β-GST). After induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 15°C, cells were lysed by sonication in phosphate-buffered saline (PBS)-KMT (PBS supplemented with 1 mM MgCl2, 3 mM KCl and 0.1% Tween 20) containing the Complete protease inhibitor mixture (added according to the manufacturer's instructions) (Roche). The lysate was cleared by centrifugation for 1 h at 20,000 rpm in an SS 34 rotor and incubated for 2 h at 4°C with glutathione-Sepharose (Amersham Biosciences) preequilibrated in PBS-KMT. The Sepharose was washed with PBS-KM (PBS-KMT without Tween 20), and the protein was eluted with elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM reduced glutathione). Samples were dialyzed against dialysis buffer (50 mM HEPES-KOH, pH 7.5, 200 mM NaCl, 5% glycerol) and concentrated in a Centriprep centrifugal filter device (Millipore). Integrity of the protein N termini was confirmed by protein digestion with trypsin (Promega), peptide separation by high-pressure liquid chromatography (UltiMate 3000 high-pressure liquid chromatography system; Dionex), and detection of the N-terminal peptides via mass spectrometry analysis (4000 Q TRAP mass spectrometer; Applied Biosystems/MDS Sciex).
The expression of the His-tagged proteins was induced by 1 mM IPTG in E. coli BL21/Rep4 at 37°C for 2 h (RCC1Δ24) or at 22°C for 3.5 h (other proteins). Cells were lysed by sonication in sonication buffer [50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 5 mM Mg(OAc)2 and 5% glycerol] containing protease inhibitors (Roche). The lysate was cleared by ultracentrifugation in a Ti 70 rotor at 70,000 rpm for 1 h and incubated for 2 h at 4°C with Talon metal affinity resin (BD Biosciences) preequilibrated in sonication buffer. Proteins were eluted with an imidazole gradient, and peak fractions were pooled. Dialysis and protein concentration was performed as described previously for GST fusion proteins.
In vitro nuclear import assay.
Import assays were performed as described earlier (28) based on the method described by Adam et al. (1). Briefly, HeLa cells were grown on three-well microscopy slides (Roth) to 40 to 80% confluence, washed twice in ice-cold PBS, and permeabilized for 8 min in ice-cold import buffer [20 mM HEPES-KOH, pH 7.5, 100 mM KOAc, 0.5 mM EGTA, 5 mM Mg(OAc)2, 250 mM sucrose] containing 30 μg/ml digitonin. After extensive washing with import buffer, permeabilized cells were incubated with 20 μl of import mixture containing the fluorescence-labeled import substrate(s) for 8 min at room temperature. After washing with import buffer and PBS, cells were fixed with 3.7% formaldehyde in PBS for 15 min at room temperature. Hoechst staining in a 1-μg/ml solution was performed for 3 min at room temperature. After extensive washing, cells were mounted and slides were analyzed by fluorescence microscopy by using a Zeiss microscope (Axioplan 2) with a 40× objective lens.
The import reaction mixtures consisted of an energy-regenerating system (0.5 mM ATP, 0.5 mM GTP, 10 mM creatine phosphate, 50 μg/ml creatine kinase), core buffer (nucleoplasmin core at 2 mg/ml in import buffer), 10% reticulocyte lysate, 0.5 mM EGTA, 3 μM RanGDP, 0.2 μM Rna1p, 0.3 μM RanBP1, 0.4 μM NTF2, 1 μM importin β, and one of the His-tagged importin α proteins at 0.4 μM.
Fluorescence labeling of purified import substrates was performed with fluorescein (Fl) 5′-maleimide or Texas red (TR) for 2 h on ice as described earlier (29).
Importin α binding assay.
GST fusion proteins (approximately 100 pmol) were immobilized on glutathione-Sepharose (Amersham Biosciences) preequilibrated in import buffer (specified above) for 1 h at 4°C. Beads were washed twice with import buffer, and His-tagged proteins were added to the beads in 0.8 ml import buffer containing 2 mg/ml bovine serum albumin and protease inhibitors (Roche). The substrates were used in an approximately threefold molar excess in comparison to the GST fusion proteins, apart from RCC1 and RCC1Δ24 in the control binding assay. In the latter case, both proteins were used with only slight excess in order to visualize the diminished amount of RCC1 in the supernatant in contrast to the unchanged RCC1Δ24 amount. When His-tagged importin β was used as a supplementary component, approximately 100 pmol was added into each binding reaction except for the control sample containing immobilized importin β-GST. After the binding reaction (1 h at 4°C), beads were washed three times with import buffer. Bound proteins were eluted with sodium dodecyl sulfate (SDS) gel-loading buffer, separated by SDS-polyacrylamide gel electrophoresis (PAGE), and stained with Coomassie brilliant blue R 250.
RESULTS
Importin α specificity is displayed in in vitro binding assays.
To analyze the impact of the NLS on importin α preference, we selected two proteins with a well-established difference in importin α specificity: RCC1, the guanine nucleotide exchange factor for the GTPase Ran, and nucleoplasmin, an acidic chaperone. Both proteins are composed of a terminally located NLS and a main part that can fold stably and independently (7, 40). The main part of RCC1 forms a seven-bladed propeller (40), whereas the primarily unstructured N terminus harbors two clusters of basic amino acids (Fig. 1A) and functions as an NLS (37, 43, 46). The well-characterized NLS of nucleoplasmin has already been crystallized in complex with importin α (3, 11) and resides C terminally to the main part of nucleoplasmin, the core domain (Fig. 1B).
FIG. 1.
Recombinant proteins used in the study. (A) Schematic diagrams of RCC1-derived substrates. RCC1 is shown with indicated positions of the propeller domain and the NLS. The sequence of the NLS-containing N terminus is listed. Basic residues are in bold. N-terminal mutation and deletion constructs are listed below. Underlining indicates amino acid mutations. zzRCC1 consists of RCC1 and an N-terminal fused zz-tag (immunoglobulin G binding domains from protein A). NLS(N)-prop was obtained by fusing the nucleoplasmin (NPL) region listed in panel B N terminal to the RCC1Δ24 construct. (B) Schematic diagrams of nucleoplasm in-derived substrates. Nucleoplasmin is shown with indicated positions of the core domain and the NLS. The sequence of the NLS-containing region that was used for further constructs is listed. The crystallographic analysis-confirmed bipartite NLS sequence (11) is underlined. Basic clusters involved in importin α binding are in bold. The NLS-containing nucleoplasmin region was cloned N terminal to the core domain to achieve NLS(N)-core. NLS(R)-core was generated by fusing the RCC1 N terminus listed in panel A N terminal to the core domain. Amino acids 13 to 35 of RCC1 were cloned N terminal to the core domain to obtain NLS(RΔ12)-core. (C) Coomassie staining of bead-associated importins. Equal amounts of the indicated importins were immobilized via their C-terminal GST tag on glutathione-Sepharose beads and separated by SDS-PAGE. Mass spectrometry analysis of tryptic cleavage peptides confirmed the integrity of the N termini and revealed that contaminations of the importin α4-GST preparation (enclosed by the circle) are GST fragments. α, importin α; β, importin β.
Earlier studies demonstrated that import substrates and NLS sequences can bind to importin α in vitro in the absence of any other import factor (12, 22, 37, 38, 44, 49). This binding could reproduce importin α specificities observed in in vitro nuclear import assays (46, 51), indicating that importin α transport specificity is linked to the binding strength of substrates to individual α importins. We therefore established an in vitro pull-down assay and determined the binding patterns of our model substrates (RCC1 and nucleoplasmin) to the ubiquitously expressed α importins, importin α1, α3, α4, α5, and α7. For this purpose, all α importins were equipped with a C-terminal GST tag, recombinantly expressed, and purified (Fig. 1C). We found that RCC1 bound very strongly to importin α3, less efficiently to importin α4, and only very weakly to the other isoforms (Fig. 2A). This finding is in perfect agreement with the published importin α specificity of RCC1 observed in different in vitro transport assays (28, 39, 46). No specific binding was observed to importin β. Approximately equimolar amounts of RCC1 bound to importin α3 (Fig. 2B, lanes 1, 3, and 5). Binding was dependent on the presence of the proposed NLS, since a construct lacking the main part of the RCC1 N terminus (RCC1Δ24) (Fig. 1A) did not bind to importin α3 in either the absence (data not shown) or the presence (Fig. 2B, lanes 2, 4, and 6) of importin. In contrast to RCC1 and in line with earlier studies (28), nucleoplasmin displayed binding to various α importins. Nucleoplasmin bound strongly to importins α1 and α5, to a lesser extent to importins α3 and α7, but only weakly to importin α4 (Fig. 2C). After the removal of the NLS-bearing C terminus, the remaining nucleoplasmin core domain (Fig. 1B) showed no binding to any α importin, even in the presence of importin β (Fig. 2D). The results of our binding assays confirm the previously reported different importin α preferences of RCC1 and nucleoplasmin and the dependency of importin α binding on the presence of an NLS.
FIG. 2.
Importin α binding specificity of RCC1 and nucleoplasmin is represented in in vitro binding assays. (A) Coomassie staining of bead-associated proteins derived from binding reactions of RCC1 with the various α importins. Equal amounts of the indicated C-terminally GST-tagged importins were immobilized on glutathione-Sepharose beads and incubated with purified recombinant RCC1. Bound proteins were separated by SDS-PAGE. The asterisk marks bovine serum albumin that was used in the binding reactions at a concentration of 2 mg/ml and remains to some extent in the beads despite the washing procedure. (B) Binding assays of RCC1 and RCC1Δ24 with immobilized importin α3-GST in the presence of importin β. Bound and unbound fractions were subjected to SDS-PAGE and analyzed by Coomassie staining (bound proteins) or Western blotting using an RCC1-specific antibody (unbound proteins and input samples). + or −, presence or absence of RCC1 and RCC1Δ24, respectively. (C) Binding of nucleoplasmin to the various α importins, as described for panel A. (D) Similar to panel A, with the exception that binding reactions were carried out with the nucleoplasmin core domain in the presence of importin β. Bound proteins and an input sample were separated by SDS-PAGE and analyzed by Coomassie staining. α, importin α; β, importin β; NPL, nucleoplasmin.
The NLS is not the only determinant for importin α binding specificity.
If the NLS was the only determinant for the observed importin α specificity of RCC1, specificity would be lost upon exchanging the RCC1 NLS with the nucleoplasmin NLS. Furthermore, specificity should be transferable by fusing the RCC1 NLS to another protein. Therefore, we set out to exchange the NLSs of the two model substrates. For a better comparison with RCC1, the protein with a pronounced importin α specificity, we constructed all proteins with the NLS fused N terminally. Consequently, we first transferred the nucleoplasmin NLS from the C terminus to the N terminus of the core domain (Fig. 1B). Similar to wild-type nucleoplasmin, NLS(N)-core displayed binding to importins α1, α3, α5, and α7, whereas binding to importin α4 was less pronounced (Fig. 3A), indicating that the position of the NLS may be not important for importin α selectivity.
FIG. 3.
The NLS is not sufficient to mediate importin α binding specificity. Equal amounts of the indicated C-terminally GST-tagged importins were used in binding experiments with single substrates. The importin-GSTs were immobilized on glutathione-Sepharose beads and incubated with the purified recombinant His-tagged proteins. Bead-associated proteins were separated by SDS-PAGE and visualized by Coomassie staining. Binding experiments were carried out with the following substrates: (A) NLS(N)-core, (B) NLS(N)-prop, and (C) NLS(R)-core. α, importin α; β, importin β.
Thereafter, we created two different fusion proteins: NLS(N)-prop, consisting of the nucleoplasmin NLS and the propeller domain of RCC1 (Fig. 1A), and NLS(R)-core, a fusion between the RCC1 N terminus and the nucleoplasmin core domain (Fig. 1B). In contrast to RCC1, NLS(N)-prop was bound by all α importins (Fig. 3B), showing that the binding specificity of RCC1 is dependent on its NLS. Surprisingly, NLS(R)-core associated with importins α1, α3, α5, and α7, but only weakly with importin α4 (Fig. 3C). This binding pattern showed no significant difference to the binding pattern of NLS(N)-core. Thus, the RCC1 NLS was not able to transfer the robust importin α specificity of RCC1 to the nucleoplasmin core domain. These data demonstrate that the RCC1 NLS is required but not sufficient to mediate strong importin α specificity.
Both NLS and whole-protein context determine importin α binding specificity.
In living cells, many substrates coexist in the cytoplasm and compete for their transport into the nucleus by a particular α importin. Assays reflecting this situation have been shown to be more sensitive with regard to importin α specificity (28). Therefore, we again analyzed the contribution of the NLS to importin α specificity by performing competition experiments. For this purpose, two substrates were added simultaneously to the binding reactions. In analogy to the situation in living cells, binding was performed in the presence of importin β to avoid additional competition with the autoinhibitory domain of importin α.
First, we used substrates differing in their NLSs only. Both RCC1 and NLS(N)-prop were able to bind to importins α3 and α4 when they were added as single substrates. Due to the fact that the two proteins are too similar in size to be separated by SDS-PAGE, we made use of a RCC1 construct with a N-terminal zz-tag (immunoglobulin G binding domains from protein A) (Fig. 1A). zzRCC1 has an increased size (∼60 kDa) compared to that of RCC1 (∼45 kDa) but displayed the same specificity to α importins (Fig. 4A). In competition experiments with NLS(N)-prop, only zzRCC1 was bound by importins α3 and α4, whereas NLS(N)-prop was bound by importins α1, α5, and α7 (Fig. 4B). Thus, the RCC1 NLS shows a higher affinity to importins α3 and α4 in comparison to that of the nucleoplasmin NLS.
FIG. 4.
The NLS contributes to importin α binding specificity. Competition binding assays were performed using substrates differing only in their NLS. Equal amounts of the indicated C-terminally GST-tagged importins were immobilized on glutathione-Sepharose beads and incubated with one single substrate (A) or with two competing substrates in equimolar amounts in the presence of importin β (B and C). Bound proteins were separated by SDS-PAGE and visualized by Coomassie staining. The following substrates were used: (A) zzRCC1, (B) zzRCC1 and NLS(N)-prop, and (C) NLS(R)-core and NLS(N)-core. α, importin α; β, importin β.
Both NLS were also tested for competition in the context of the nucleoplasmin core domain. The constructs NLS(R)-core and NLS(N)-core displayed similar binding patterns to the α importins when they were added as single substrates (Fig. 3A and C). However, in a competing situation, mainly NLS(R)-core was bound by importins α3 and α4, whereas NLS(N)-core was the major substrate detected with importin α1. Both substrates bound to importins α5 and α7 with similar efficiencies (Fig. 4C). Thus, even if the RCC1 NLS is torn out of its wild-type protein context and fused to another protein core domain, it shows a higher affinity to importins α3 and α4 in comparison to the nucleoplasmin NLS.
We next carried out competition assays with substrates bearing the same NLS but differing in their main protein part. Both substrates carrying the RCC1 NLS, RCC1 and NLS(R)-core, were able to bind to importin α3 when they were added as single substrates into the binding reactions (Fig. 2A and 3C). However, in competition experiments, only RCC1 was bound by importins α3 and α4. In contrast, NLS(R)-core was efficiently bound by importins α1 and α5 and, to a minor extent, by importin α7, but no binding was observed to importins α3 and α4 (Fig. 5A). Thus, the affinity of the RCC1 NLS for importins α3 and α4 is enhanced in the context of the RCC1 propeller.
FIG. 5.
The RCC1 propeller increases substrate affinity for importins α3 and α4. Competition binding assays were performed using substrates containing the same NLS fused to different protein core domains. Equal amounts of the indicated C-terminally GST-tagged importins were immobilized on glutathione-Sepharose beads and incubated with two competing substrates in equimolar amounts and importin β. Bead-associated proteins were separated by SDS-PAGE and visualized by Coomassie staining. The following substrates were used in the competition experiments: (A) RCC1 and NLS(R)-core, (B) NLS(N)-prop and NLS(N)-core, and (C) NLS(N)-prop and nucleoplasmin. α, importin α; β, importin β; NPL, nucleoplasmin.
Likewise, both substrates bearing the nucleoplasmin NLS, NLS(N)-prop and NLS(N)-core, bound to importin α3 when they were added as single substrates into the binding assays (Fig. 3A and B). During competition, NLS(N)-prop was able to maintain a strong association with all α importins, whereas NLS(N)-core could be detected with only importins α1 and α5 and a very faint binding to importin α7 was observed (Fig. 5B). To exclude the possibility that the last-mentioned result was affected by poor accessibility of the nucleoplasmin NLS in the NLS(N)-core construct, we repeated the competition experiment with wild-type nucleoplasmin. Again, NLS(N)-prop bound efficiently to all α importins. Nucleoplasmin associated with importins α1 and α5 in an intensity similar to that observed for NLS(N)-core. Additionally, weak binding of nucleoplasmin was detected to importins α4 and α7, but no binding to importin α3 was observed (Fig. 5C). Thus, the relative affinity of the nucleoplasmin NLS to α importins is also influenced by the presence of either the RCC1 propeller or the nucleoplasmin core domain. Taken together, these data demonstrate that both NLS and protein context contribute to specificity in importin α binding.
The protein context also contributes to importin α specificity in in vitro nuclear import.
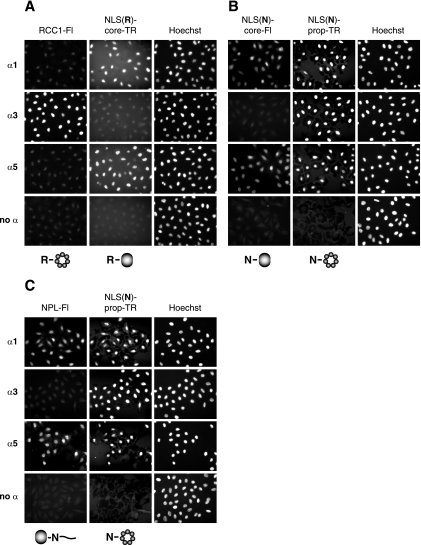
Due to the possibility that binding assays may display artificial interactions that do not involve cargo import, we further analyzed the different constructs using in vitro nuclear import assays with semipermeabilized HeLa cells. We selected one importin α of each subfamily, namely, importins α1, α3, and α5, since importin α members of the same subfamily (α3/α4 α5/α7, respectively) had shown very similar behavior in our competition binding assays. The substrates were labeled either with fluorescein [for labeling of RCC1, nucleoplasmin, and NLS(N)-core] or with Texas red [for labeling of NLS(N)-prop and NLS(R)-core]. The labeled proteins were first added into the import assay reaction mixtures as single substrates. RCC1-Fl was imported into the nuclei by importin α3 only (Fig. 6A). Nucleoplasmin-Fl was transported by importins α1, α3, and α5, although importin α3 displayed a weaker effect on the nuclear import than importins α1 and α5 did (Fig. 6B). The same transport pattern was observed for NLS(N)-core-Fl (Fig. 6C), which represents a slight difference from the binding assays where NLS(N)-core was bound to importins α1, α3, α5, and α7 with similar efficiencies (Fig. 3A). NLS(N)-prop-TR and NLS(R)-core-TR were imported efficiently into the nuclei by all α importins (Fig. 6D and E). Thus, these import assays confirmed the results of the single-substrate binding assays. The robust importin α specificity of RCC1 was lost upon exchanging the NLS, but it was also not transferable by fusing the RCC1 NLS to the nucleoplasmin core domain.
FIG. 6.
Importin α-dependent nuclear import of single substrates. In vitro nuclear import assays with fluorescein- and Texas red-labeled proteins were performed with digitonin-permeabilized HeLa cells by using the indicated importin α isoforms. Sections on the left display nuclear import of the following substrates: (A) RCC1-Fl, (B) NPL-Fl, (C) NLS(N)-core-Fl, (D) NLS(N)-prop-TR, and (E) NLS(R)-core-TR. Right panels show Hoechst nuclear DNA staining. α, importin α; NPL, nucleoplasmin.
Next, we performed competition import assays in equivalence to the binding studies by adding two substrates simultaneously in equimolar amounts into the import assay reaction mixtures. Again, we started with substrates differing only in their NLS. The competition of RCC1-Fl and NLS(N)-prop-TR revealed that RCC1-Fl was mainly imported by importin α3, whereas NLS(N)-prop-TR was imported by importins α1 and α5. Thus, the RCC1 NLS results in a higher import efficiency with importin α3 in comparison to the nucleoplasmin NLS (Fig. 7A). The competition experiment with NLS(N)-core-Fl and NLS(R)-core-TR showed that only NLS(N)-core-Fl was imported by importin α1, whereas only NLS(R)-core-TR was transported into the nuclei by importin α3 (Fig. 7B). Both substrates were imported by importin α5, but their transport was less efficient, as if they were added as single substrates (Fig. 6C and E). These data confirm the results of the competition binding assays by pointing out the contribution of the NLS to importin α specificity.
FIG. 7.
The NLS contributes to importin α transport specificity. In vitro nuclear import competition assays were carried out with substrates differing in their NLSs only. The competition reactions were performed on digitonin-permeabilized HeLa cells. Indicated importin α isoforms and equimolar amounts of the following competing substrates were added into the import assay reaction mixtures: (A) RCC1-Fl and NLS(N)-prop-TR, and (B) NLS(N)-core-Fl and NLS(R)-core-TR. Left sections display nuclear import of fluorescein-labeled substrates; middle panels show import of Texas red-labeled proteins. Nuclei were visualized by Hoechst staining (right panels). α, importin α.
Furthermore, we carried out competition import assays with substrates containing the same NLS fused to different protein core domains. Both substrates bearing the RCC1 NLS, RCC1-Fl and NLS(R)-core-TR, were imported into HeLa cell nuclei by importin α3 when they were used as single substrates (Fig. 6A and E). However, during competition, only RCC1-Fl was imported by importin α3, whereas only NLS(R)-core-TR was transported by importins α1 and α5 (Fig. 8A). Hence, the RCC1 NLS leads to greater import efficiency with importin α3 in the context of the RCC1 propeller. The competition of the substrates containing the nucleoplasmin NLS, NLS(N)-core-Fl and NLS(N)-prop-TR, revealed that only NLS(N)-prop-TR was transported into the nuclei by importin α3, whereas both substrates were imported by importins α1 and α5 (Fig. 8B). The transport of both substrates by importins α1 and α5 was less efficient, as if they were used as single substrates (Fig. 6C and D). The same result was obtained when we used fluorescein-labeled wild-type nucleoplasmin instead of NLS(N)-core-Fl in the competition experiment with NLS(N)-prop-TR (Fig. 8C). According to this, the nucleoplasmin NLS also results in a higher import efficiency with importin α3 in the context of the RCC1 propeller. Taken together, the nuclear import assays confirm the results of the importin α binding assays insofar as the NLS of a substrate as well as the whole-protein context turns out to be important for importin α specificity.
FIG. 8.
The RCC1 propeller increases nuclear transport efficiency of importin α3. In vitro nuclear import competition assays were carried out with substrates containing the same NLS fused to different protein core domains. Competition reactions were performed using digitonin-permeabilized HeLa cells. Indicated importin α isoforms and equimolar amounts of the following competing substrates were added into the import assay reaction mixtures: (A) RCC1-Fl and NLS(R)-core-TR, (B) NLS(N)-core-Fl and NLS(N)-prop-TR, and (C) NPL-Fl and NLS(N)-prop-TR. Left sections display nuclear import of fluorescein-labeled substrates; middle panels show import of Texas red-labeled proteins. Nuclei were visualized by Hoechst staining (right panels). α, importin α; NPL, nucleoplasmin.
Both basic clusters of the RCC1 N terminus contribute to importin α binding.
The sequence of the RCC1 N terminus resembles a bipartite NLS bearing two basic clusters. Each of these clusters contains five basic residues separated by a linker of eight nonbasic residues (Fig. 1A). To analyze the contribution of these basic stretches to importin α binding, we expressed RCC1 mutants having individual clusters altered or deleted (Fig. 1A). We determined the binding of these mutants to the various α importins in the absence or presence of importin β using the pull-down assay described above. As aforementioned, wild-type RCC1 showed a strong binding preference to importins α3 and α4 in the absence of importin β (Fig. 2A). As importin β increases the affinity of substrates to importin α by preventing any competition with the autoinhibitory domain, the addition of importin β into the binding reactions of single-substrate binding assays, leaving the molarities of the other components unaltered, almost completely impeded the detection of importin α binding specificity (Fig. 9A and data not shown). However, if the first (N-terminal) basic cluster of the RCC1 N terminus was modified by exchanging lysine 4 and arginine 5 (RCC1mt1) or lysine 8 and arginine 9 (RCC1mt2) with alanines (Fig. 1A), these mutants displayed specificity for importins α3 and α4 both in the absence and in the presence of importin β (Fig. 9B and data not shown). This effect was even more pronounced if the first 13 residues of the RCC1 N terminus were deleted, and hence, the complete first basic cluster was absent (RCC1Δ13) (Fig. 1A). RCC1Δ13 was exclusively bound by importins α3 and α4 in the presence of importin β (Fig. 9C, right part), albeit with reduced affinity compared to that of the wild-type RCC1 and to the mutants RCC1mt1 and RCC1mt2. In the absence of importin β, even the interaction of RCC1Δ13 with importin α4 was almost lost (Fig. 9C, left part). When lysines 21 and 22 in the second (C-terminal) basic cluster of the RCC1 N terminus were mutated to alanines (RCC1mt3) (Fig. 1A), no significant binding to any α importin was observed in either the absence or the presence of importin β (Fig. 9D).
FIG. 9.
Both basic clusters of the RCC1 N terminus contribute to importin α binding. Equal amounts of the indicated C-terminally GST-tagged importins were used in binding experiments with single substrates. The importin-GSTs were immobilized on glutathione-Sepharose beads and incubated with substrates in the presence (A) or in both the presence (+) and absence (B, C, and D) of importin β. Bound proteins were separated by SDS-PAGE and visualized by Coomassie staining. The following substrates were used: (A) RCC1, (B) RCC1mt2 (K8A, R9A), (C) RCC1Δ13, and (D) RCC1mt3 (K21A, K22A). α, importin α; β, importin β; Imp., importin.
We next fused amino acids 13 to 35 of RCC1 containing the second basic cluster N terminally to the nucleoplasmin core domain [NLS(RΔ12)-core] (Fig. 1B). Binding assays revealed that the interaction of this construct with α importins was considerably weaker than that of NLS(R)-core which contains the whole RCC1 N terminus, since binding of NLS(RΔ12)-core to α importins could be detected only in the presence of importin β (compare Fig. 10 and 3C). Interestingly, the observed binding of NLS(RΔ12)-core to α importins was most intense to importins α1 and α5 (Fig. 10, right part). This binding pattern is similar to the binding preferences observed for wild-type nucleoplasmin in the single-substrate binding assay (Fig. 2C). In combination with the results obtained for RCC1Δ13, this indicates that the missing N-terminal half of the RCC1 NLS contains no information essential for binding to any of the α importins. Moreover, this result again demonstrates that the protein context of NLS-bearing substrates is crucial for importin α binding specificity.
FIG. 10.
Importin α binding specificity of the second basic cluster of the RCC1 N terminus is mediated by the protein context. Equal amounts of the indicated C-terminally GST-tagged importins were immobilized on glutathione-Sepharose beads and incubated with NLS(RΔ12)-core either in the absence or in the presence (+) of importin β. Bead-associated proteins were separated by SDS-PAGE and visualized by Coomassie staining. α, importin α; β, importin β; Imp., importin.
Taken together, these data show that the second basic cluster of the RCC1 N terminus is essential for importin α binding, whereas the first basic cluster strengthens this interaction. Furthermore, the C-terminal half of the NLS is sufficient for the maintenance of importin α specificity in the context of the RCC1 propeller.
DISCUSSION
We aimed to elucidate the determinants for specific interaction between an import substrate and a particular α importin. For our analysis, we used two well-characterized substrates, RCC1, which displays a significant importin α preference, and nucleoplasmin that can be imported by various α importins. The NLS of nucleoplasmin is one of the best- characterized examples of a bipartite NLS (3, 11, 42). Our data support the previous suggestion (46) that the NLS of RCC1 also belongs to the class of bipartite NLS, since both basic clusters contribute to importin α binding. The N-terminal cluster of the RCC1 NLS displayed a weaker contribution to the high-affinity importin α binding compared to that of the C-terminal basic cluster, as the deletion of the N-terminal cluster strongly reduced importin α binding, whereas the mutation of the C-terminal cluster abolished any significant binding. The essential nature of the lysine doublet (K21, K22) of the C-terminal basic stretch suggests that it interacts with the major binding site of importin α. This assumption is supported by a striking similarity of the C-terminal cluster (KSKKVK) with the sequence of the C-terminal basic stretch of the bipartite N1N2 NLS (KAKKSK), which has been shown to contact the major NLS binding site of importin α (10). We are not able to predict the binding of particular basic residues within the first basic stretch of the RCC1 NLS to importin α. Individual mutations of both KR doublets had the same effect on importin α binding, making it impossible to predict which residues might bind to the binding pockets P1′ and P2′ (3, 11) of the minor NLS binding site.
The importin α specificity of RCC1 measured in our single-substrate binding assays was identical to that found in competition binding assays in the presence of importin β or in in vitro nuclear import assays. This finding indicates that the importin α specificity of RCC1 is independent of potentially different affinities of the autoinhibitory domains to their particular importin α binding sites. Similar results were also obtained for the other substrates. Possibly, the different autoinhibitory domains exhibit quite similar affinities to their corresponding α importins. Interestingly, all human α importins contain the same autoinhibitory core sequence, KRRNV, which has been shown to bind to the major NLS binding site of importin α (24).
We exchanged the NLS of RCC1 with the nucleoplasmin NLS, and we fused the N terminus of RCC1 N terminally to the nucleoplasmin core domain. We found that the RCC1 NLS was able to transfer the importin α specificity of its native protein to some extent to an artificial protein containing the nucleoplasmin core domain. However, this finding was detectable only by means of the more sensitive competition assays, indicating strong influence of the protein context on importin α specificity. Even more compelling, by using identical NLSs either in the context of the RCC1 propeller or in the context of the nucleoplasmin core domain, we found that the presence of the propeller domain always resulted in stronger interactions with importin α3. This result was obtained in both competition binding studies as well as competition import assays. Interestingly, this important influence of the main protein part was observed even if two artificial fusion proteins were competing for importin α binding (Fig. 5B and 8B), excluding the possibility that sequences of the main part adjacent to the cognate NLS in wild-type proteins are responsible for this effect.
Thus far, we cannot distinguish precisely whether the main part of RCC1 improves binding to importins α3 and α4 or whether the nucleoplasmin core domain somehow impairs this interaction. Our data do not exclude the possibility that both mechanisms may be operative. On the one hand, the absence of the RCC1 propeller domain weakens the binding preference of the RCC1 NLS to importins α3 and α4; this fact points towards a positive influence of the propeller. On the other hand, poor binding efficiency of NLS(R)-core to importin α4 in the single-substrate binding assay could be interpreted as a negative influence of the nucleoplasmin core domain. However, such a model would not explain why the position of the core domain either upstream or downstream of the nucleoplasmin NLS causes a similar importin α binding pattern (Fig. 2C and 3A).
How could the RCC1 propeller mediate a preferred binding to importins α3 and α4? We believe that weak interactions between the propeller and importin α3/α4 are the basis for this effect. This possibility is not excluded by the fact that the propeller does not bind to any α importin in the absence of an NLS. We speculate that additional interactions between the propeller and importin α are likely to occur outside the NLS binding groove. The long groove created by the entire ARM domain of importin α accommodates NLS peptides and flanking sequences but leaves no extensive space for further interactions with more-distal parts of an import substrate (3, 11, 12). However, the possibility that small conformational changes triggered by weak interactions of the RCC1 propeller and importin α3/α4 could strengthen the interaction between NLS and importin α is not excluded.
How could the RCC1 NLS mediate preferred binding to importins α3 and α4? Our results suggest that the N-terminal basic cluster of the RCC1 N terminus increases the affinity to any importin α. Therefore, an impact on the importin α3/α4 preference of the RCC1 NLS seems unlikely. The C-terminal part of the RCC1 N terminus displayed the same protein context-dependent binding behavior as that of the complete RCC1 NLS since it showed the most prominent binding to importin α3 in the context of the RCC1 propeller but not in the context of the nucleoplasmin core domain. Possibly, the amino acids of the C-terminal basic cluster, which most likely bind to the major binding site of importin α, and/or the residues preceding this cluster somehow participate in mediating specificity. The last-mentioned residues have been shown to be important for the binding of both monopartite and bipartite NLS (10, 12, 33). If we compare the RCC1 NLS and the nucleoplasmin NLS, remarkable differences are found within the linker region preceding the C-terminal basic cluster. For instance, nucleoplasmin harbors additional basic amino acids in this region, whereas RCC1 accommodates an acidic residue in the corresponding position. Such differences could modulate the affinity for distinct α importins.
How could variations in NLS fit into the known structure of importin α? Most of the mouse importin α1 (PTAC58/pendulin) residues that have been shown to be important for binding to the C-terminal basic cluster or the linker region of bipartite NLS (10, 11) are also found in all human α importins. Only a few mouse importin α1 residues like E266 (a major binding determinant of pocket P3), R106 and E107 (both important residues of pocket P4), or T311 (which interacts with the linker region of the N1N2 NLS) display slight differences between human importin α subfamilies and could contribute to importin α specificity. Nevertheless, other variations in the amino acid composition of the different importin α subfamilies could account for importin α specificity by influencing the position or flexibility of the conserved residues within the binding groove. For example, considerable variations are found in ARM repeat 4 (32), close to residues of mouse importin α1 which are involved in NLS interaction (10, 11).
A thorough understanding of the mechanisms underlying importin α binding specificity will require structural analyses of the complete substrate-importin α-importin β complex. Nevertheless, even at the current state of knowledge, our findings have clinical consequences. The possibility that interactions between the main part of the import substrate and the respective α importin may be relevant for an efficient nuclear import offers new prospects for the design of drugs that are intended to interfere with the function of nuclear key regulatory proteins.
Acknowledgments
We thank Dirk Görlich for the kind gifts of recombinant proteins (RanGDP, Rna1p, RanBP1, and NTF2) and expression clones. We are thankful to Albrecht Otto and Eva-Christina Müller for performing mass spectrometry analysis. We also thank Brigitte Nentwig for excellent technical help and the Sommer group for helpful comments on the project. We are grateful to Friedrich C. Luft for critical reading of the manuscript.
The Deutsche Forschungsgemeinschaft supported Matthias Köhler, Enno Hartmann, and Thomas Sommer. Matthias Köhler was the recipient of a Helmholtz Fellowship.
Footnotes
Published ahead of print on 25 September 2006.
REFERENCES
- 1.Adam, S. A., R. S. Marr, and L. Gerace. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Catimel, B., T. Teh, M. R. Fontes, I. G. Jennings, D. A. Jans, G. J. Howlett, E. C. Nice, and B. Kobe. 2001. Biophysical characterization of interactions involving importin-alpha during nuclear import. J. Biol. Chem. 276:34189-34198. [DOI] [PubMed] [Google Scholar]
- 3.Conti, E., and J. Kuriyan. 2000. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure 8:329-338. [DOI] [PubMed] [Google Scholar]
- 4.Conti, E., M. Uy, L. Leighton, G. Blobel, and J. Kuriyan. 1998. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94:193-204. [DOI] [PubMed] [Google Scholar]
- 5.Cortes, P., Z. S. Ye, and D. Baltimore. 1994. RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc. Natl. Acad. Sci. USA 91:7633-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuomo, C. A., S. A. Kirch, J. Gyuris, R. Brent, and M. A. Oettinger. 1994. Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc. Natl. Acad. Sci. USA 91:6156-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutta, S., I. V. Akey, C. Dingwall, K. L. Hartman, T. Laue, R. T. Nolte, J. F. Head, and C. W. Akey. 2001. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol. Cell 8:841-853. [DOI] [PubMed] [Google Scholar]
- 8.Fagerlund, R., L. Kinnunen, M. Köhler, I. Julkunen, and K. Melen. 2005. NF-κB is transported into the nucleus by importin α3 and importin α4. J. Biol. Chem. 280:15942-15951. [DOI] [PubMed] [Google Scholar]
- 9.Fanara, P., M. R. Hodel, A. H. Corbett, and A. E. Hodel. 2000. Quantitative analysis of nuclear localization signal (NLS)-importin alpha interaction through fluorescence depolarization. Evidence for auto-inhibitory regulation of NLS binding. J. Biol. Chem. 275:21218-21223. [DOI] [PubMed] [Google Scholar]
- 10.Fontes, M. R., T. Teh, D. Jans, R. I. Brinkworth, and B. Kobe. 2003. Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-alpha. J. Biol. Chem. 278:27981-27987. [DOI] [PubMed] [Google Scholar]
- 11.Fontes, M. R., T. Teh, and B. Kobe. 2000. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J. Mol. Biol. 297:1183-1194. [DOI] [PubMed] [Google Scholar]
- 12.Fontes, M. R., T. Teh, G. Toth, A. John, I. Pavo, D. A. Jans, and B. Kobe. 2003. Role of flanking sequences and phosphorylation in the recognition of the simian-virus-40 large T-antigen nuclear localization sequences by importin-alpha. Biochem. J. 375:339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldfarb, D. S., A. H. Corbett, D. A. Mason, M. T. Harreman, and S. A. Adam. 2004. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 14:505-514. [DOI] [PubMed] [Google Scholar]
- 14.Görlich, D., P. Henklein, R. A. Laskey, and E. Hartmann. 1996. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 15:1810-1817. [PMC free article] [PubMed] [Google Scholar]
- 15.Görlich, D., S. Prehn, R. A. Laskey, and E. Hartmann. 1994. Isolation of a protein that is essential for the first step of nuclear protein import. Cell 79:767-778. [DOI] [PubMed] [Google Scholar]
- 16.Görlich, D., F. Vogel, A. D. Mills, E. Hartmann, and R. A. Laskey. 1995. Distinct functions for the two importin subunits in nuclear protein import. Nature 377:246-248. [DOI] [PubMed] [Google Scholar]
- 17.Gruss, O. J., R. E. Carazo-Salas, C. A. Schatz, G. Guarguaglini, J. Kast, M. Wilm, N. Le Bot, I. Vernos, E. Karsenti, and I. W. Mattaj. 2001. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104:83-93. [DOI] [PubMed] [Google Scholar]
- 18.Hachet, V., T. Kocher, M. Wilm, and I. W. Mattaj. 2004. Importin alpha associates with membranes and participates in nuclear envelope assembly in vitro. EMBO J. 23:1526-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harel, A., R. C. Chan, A. Lachish-Zalait, E. Zimmerman, M. Elbaum, and D. J. Forbes. 2003. Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol. Biol. Cell 14:4387-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harel, A., and D. J. Forbes. 2004. Importin beta: conducting a much larger cellular symphony. Mol. Cell 16:319-330. [DOI] [PubMed] [Google Scholar]
- 21.Hogarth, C. A., S. Calanni, D. A. Jans, and K. L. Loveland. 2006. Importin alpha mRNAs have distinct expression profiles during spermatogenesis. Dev. Dyn. 235:253-262. [DOI] [PubMed] [Google Scholar]
- 22.Hu, W., and D. A. Jans. 1999. Efficiency of importin alpha/beta-mediated nuclear localization sequence recognition and nuclear import. Differential role of NTF2. J. Biol. Chem. 274:15820-15827. [DOI] [PubMed] [Google Scholar]
- 23.Kalderon, D., B. L. Roberts, W. D. Richardson, and A. E. Smith. 1984. A short amino acid sequence able to specify nuclear location. Cell 39:499-509. [DOI] [PubMed] [Google Scholar]
- 24.Kobe, B. 1999. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat. Struct. Biol. 6:388-397. [DOI] [PubMed] [Google Scholar]
- 25.Köhler, M., S. Ansieau, S. Prehn, A. Leutz, H. Haller, and E. Hartmann. 1997. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 417:104-108. [DOI] [PubMed] [Google Scholar]
- 26.Köhler, M., A. Fiebeler, M. Hartwig, S. Thiel, S. Prehn, R. Kettritz, F. C. Luft, and E. Hartmann. 2002. Differential expression of classical nuclear transport factors during cellular proliferation and differentiation. Cell. Physiol. Biochem. 12:335-344. [DOI] [PubMed] [Google Scholar]
- 27.Köhler, M., D. Görlich, E. Hartmann, and J. Franke. 2001. Adenoviral E1A protein nuclear import is preferentially mediated by importin α3 in vitro. Virology 289:186-191. [DOI] [PubMed] [Google Scholar]
- 28.Köhler, M., C. Speck, M. Christiansen, F. R. Bischoff, S. Prehn, H. Haller, D. Gorlich, and E. Hartmann. 1999. Evidence for distinct substrate specificities of importin α family members in nuclear protein import. Mol. Cell. Biol. 19:7782-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kutay, U., F. R. Bischoff, S. Kostka, R. Kraft, and D. Görlich. 1997. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell 90:1061-1071. [DOI] [PubMed] [Google Scholar]
- 30.Lanford, R. E., P. Kanda, and R. C. Kennedy. 1986. Induction of nuclear transport with a synthetic peptide homologous to the SV40 T antigen transport signal. Cell 46:575-582. [DOI] [PubMed] [Google Scholar]
- 31.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 32.Melen, K., R. Fagerlund, J. Franke, M. Köhler, L. Kinnunen, and I. Julkunen. 2003. Importin alpha nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J. Biol. Chem. 278:28193-28200. [DOI] [PubMed] [Google Scholar]
- 33.Miyamoto, Y., N. Imamoto, T. Sekimoto, T. Tachibana, T. Seki, S. Tada, T. Enomoto, and Y. Yoneda. 1997. Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J. Biol. Chem. 272:26375-26381. [DOI] [PubMed] [Google Scholar]
- 34.Moroianu, J., G. Blobel, and A. Radu. 1996. The binding site of karyopherin alpha for karyopherin beta overlaps with a nuclear localization sequence. Proc. Natl. Acad. Sci. USA 93:6572-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nachury, M. V., T. J. Maresca, W. C. Salmon, C. M. Waterman-Storer, R. Heald, and K. Weis. 2001. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell 104:95-106. [DOI] [PubMed] [Google Scholar]
- 36.Nadler, S. G., D. Tritschler, O. K. Haffar, J. Blake, A. G. Bruce, and J. S. Cleaveland. 1997. Differential expression and sequence-specific interaction of karyopherin alpha with nuclear localization sequences. J. Biol. Chem. 272:4310-4315. [DOI] [PubMed] [Google Scholar]
- 37.Nemergut, M. E., and I. G. Macara. 2000. Nuclear import of the ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J. Cell Biol. 149:835-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishinaka, Y., H. Masutani, S. Oka, Y. Matsuo, Y. Yamaguchi, K. Nishio, Y. Ishii, and J. Yodoi. 2004. Importin α1 (Rch1) mediates nuclear translocation of thioredoxin binding protein-2/vitamin D3-up-regulated protein 1. J. Biol. Chem. 279:37559-37565. [DOI] [PubMed] [Google Scholar]
- 39.Quensel, C., B. Friedrich, T. Sommer, E. Hartmann, and M. Köhler. 2004. In vivo analysis of importin α proteins reveals cellular proliferation inhibition and substrate specificity. Mol. Cell. Biol. 24:10246-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renault, L., N. Nassar, I. Vetter, J. Becker, C. Klebe, M. Roth, and A. Wittinghofer. 1998. The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature 392:97-101. [DOI] [PubMed] [Google Scholar]
- 41.Rexach, M., and G. Blobel. 1995. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83:683-692. [DOI] [PubMed] [Google Scholar]
- 42.Robbins, J., S. M. Dilworth, R. A. Laskey, and C. Dingwall. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64:615-623. [DOI] [PubMed] [Google Scholar]
- 43.Seino, H., N. Hisamoto, S. Uzawa, T. Sekiguchi, and T. Nishimoto. 1992. DNA binding domain of RCC1 protein is not essential for coupling mitosis with DNA replication. J. Cell Sci. 102:393-400. [DOI] [PubMed] [Google Scholar]
- 44.Seki, T., S. Tada, T. Katada, and T. Enomoto. 1997. Cloning of a cDNA encoding a novel importin-alpha homologue, Qip1: discrimination of Qip1 and Rch1 from hSrp1 by their ability to interact with DNA helicase Q1/RecQL. Biochem. Biophys. Res. Commun. 234:48-53. [DOI] [PubMed] [Google Scholar]
- 45.Sekimoto, T., N. Imamoto, K. Nakajima, T. Hirano, and Y. Yoneda. 1997. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 16:7067-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Talcott, B., and M. S. Moore. 2000. The nuclear import of RCC1 requires a specific nuclear localization sequence receptor, karyopherin alpha3/Qip. J. Biol. Chem. 275:10099-10104. [DOI] [PubMed] [Google Scholar]
- 47.Tsuji, L., T. Takumi, N. Imamoto, and Y. Yoneda. 1997. Identification of novel homologues of mouse importin alpha, the alpha subunit of the nuclear pore-targeting complex, and their tissue-specific expression. FEBS Lett. 416:30-34. [DOI] [PubMed] [Google Scholar]
- 48.Walther, T. C., P. Askjaer, M. Gentzel, A. Habermann, G. Griffiths, M. Wilm, I. W. Mattaj, and M. Hetzer. 2003. RanGTP mediates nuclear pore complex assembly. Nature 424:689-694. [DOI] [PubMed] [Google Scholar]
- 49.Weis, K., I. W. Mattaj, and A. I. Lamond. 1995. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science 268:1049-1053. [DOI] [PubMed] [Google Scholar]
- 50.Weis, K., U. Ryder, and A. I. Lamond. 1996. The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. EMBO J. 15:1818-1825. [PMC free article] [PubMed] [Google Scholar]
- 51.Welch, K., J. Franke, M. Köhler, and I. G. Macara. 1999. RanBP3 contains an unusual nuclear localization signal that is imported preferentially by importin-α3. Mol. Cell. Biol. 19:8400-8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiese, C., A. Wilde, M. S. Moore, S. A. Adam, A. Merdes, and Y. Zheng. 2001. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science 291:653-656. [DOI] [PubMed] [Google Scholar]
- 53.Yano, R., M. Oakes, M. Yamaghishi, J. A. Dodd, and M. Nomura. 1992. Cloning and characterization of SRP1, a suppressor of temperature-sensitive RNA polymerase I mutations, in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:5640-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]