Abstract
Whereas the PML protein has been reported to have both transcriptional coactivator and corepressor potential, the contribution of the PML nuclear body (PML NB) itself to transcriptional regulation is not well understood. Here we demonstrate that plasmid DNA artificially tethered to PML or the PML NB-targeting domain of Sp100 is preferentially localized to PML NBs. Using the tethering technique, we targeted a simian virus 40 promoter-driven luciferase reporter plasmid to PML NBs, resulting in the repression of the transgene transcriptional activity. Conversely, the tethering of a cytomegalovirus promoter-containing reporter plasmid resulted in activation. Targeting a minimal eukaryotic promoter did not affect its activity. The expression of targeted promoters could be modulated by altering the cellular concentration of PML NB components, including Sp100 and isoforms of the PML protein. Finally, we demonstrate that ICP0, the promiscuous herpes simplex virus transactivator, increases the level of transcriptional activation of plasmid DNA tethered to the PML NB. We conclude that when PML NB components are artificially tethered to reporter plasmids, the PML NB contributes to the regulation of the tethered DNA in a promoter-dependent manner. Our findings demonstrate that transient transcription assays are sensitive to the subnuclear localization of the transgene plasmid.
The promyelocytic leukemia (PML) tumor suppressor gene was identified as the translocation partner of the retinoic acid receptor (RARα) in patients with acute promyelocytic leukemia (APL) (12, 20). Antibodies directed towards PML revealed that PML formed discrete foci within the nucleus and that those foci were disrupted in cells derived from APL patients. Later experiments identified PML as the necessary component for the formation of the PML nuclear body (PML NB) (18), a protein-based subnuclear domain whose protein core physically interacts with the surrounding chromatin fibers (4, 14). PML NBs vary in size, number, and biochemical composition depending on cell type, stage of the cell cycle, and environmental conditions (6). PML exists as splice variant isoforms that differ at their C termini. This region of the protein may be responsible for specific interactions with other cellular components and constrain the subcellular localization of PML protein (2, 7). PML I and IV have been the most intensely studied isoforms to date. The overexpression of PML isoforms will alter the size of the PML NB and will also alter the relative levels of PML NB components relative to the nucleoplasmic background (3). The Nuclear Protein Database summarizes over 77 proteins that localize to the PML NB (http://npd.hgu.mrc.ac.uk/) (10). Given the wide range of proteins that localize in PML NBs, it may not be surprising that they have been implicated in many different nuclear processes, including DNA repair, replication, and transcriptional regulation (9). These proteins include well-known coactivators of transcription, such as the acetyltransferase CBP (5, 21) as well as corepressors, such as Sp100 (13) and Daxx (25). Although PML NBs form functional contacts with chromatin (14) and are involved in both aberrant differentiation in APL and early viral gene transcription (reviewed in reference 5), a defined role for PML NBs in transcriptional regulation has remained elusive.
The complement of proteins within PML NBs may reflect the functional heterogeneity of PML NBs at any given time. This is especially true when considering a possible role of the PML NB in transcriptional regulation (39). Some models have postulated that PML NBs can function to sequester transcription factors away from their cognate gene sequences in the soluble nuclear fraction (22). For example, the overexpression of PML protein leads to the recruitment of Sp1 to PML NBs from the nucleoplasm, which might explain the reduced expression of promoter elements of the Sp1-responsive epidermal growth factor receptor (EGFR) (34). Similarly, the sequestration of the Daxx corepressor protein to PML NBs upon PML overexpression may lead to the derepression of the glucocorticoid receptor, as measured by changes in gene expression from hormone-responsive reporter plasmids (23). Other models have implied that the biochemical composition of the PML NB can influence the posttranslational modifications of trafficking transcription factors and consequently modulate their downstream interactions with promoter elements (17, 26).
The transcriptional potential of PML has also been investigated by artificially tethering PML to constitutive viral promoters. This was accomplished by creating an in-frame fusion protein of the yeast DNA binding protein, Gal4, on the N terminus of PML. This ectopically expressed fusion protein is able to physically interact with expression plasmids that contain the Gal4 binding element (upstream activator sequence) 5′ of the constitutive viral promoter, which drives the expression of a reporter gene. The conclusion from these studies is that PML protein functions as a transcriptional corepressor (36, 37). In contrast, other studies have implicated PML as a transcriptional activator. Although transcriptional activation of a promoter artificially tethered to PML has not been observed, the overexpression of PML leads to increased expression of CD18 as well as the major histocompatibility complex (MHC) class I transporter TAP-1 (38). In all of these studies, however, the role of PML NBs in gene regulation was not contemplated, even though a large fraction of PML protein resides in nuclear bodies.
We devised a strategy to target reporter plasmids to PML NBs, which enabled the sampling of the transcriptional environment in the immediate vicinity of these subnuclear domains and is a significant step to aid in identifying the cellular factors that affect transcription at PML NBs. A plasmid containing a Tet operon was targeted to PML NBs in cells expressing a Tet repressor-PML fusion. A simian virus 40 (SV40) promoter element in this plasmid was repressed when targeted to the PML NB. In contrast, we observed the up-regulation of a reporter plasmid containing the cytomegalovirus (CMV) reporter when it was targeted to PML NBs using a similar strategy. The targeting approach provided an opportunity to manipulate the biochemical composition of PML NBs and to measure the outcome with regard to the transcription of a luciferase reporter plasmid targeted to this subnuclear environment. These results are discussed in the broader context of gene regulation at PML NBs. We also discuss the implication that in vivo transcription assays involving transfected reporter plasmids are sensitive to the subnuclear localization of the plasmid and that this must be considered in interpreting the results of these assays.
MATERIALS AND METHODS
Cloning and plasmids.
Firefly luciferase reporter constructs (pGL3; Promega) were driven by either the SV40 promoter derived from the early region of simian virus 40 or the CMV promoter derived from the intermediate-early region of cytomegalovirus. Reporter constructs used for Lac (pGL3-LacO)- or Tet (pGL3-TETO)-based targeting were created by first cloning eight copies of LacO and seven copies of TetO sequences into the multiple cloning site of pBluescript (Stratagene). To create pGL3-LacO and pGL3-TetO, the corresponding arrays were excised from pBluescript using XhoI and BamHI restriction enzymes and cloned downstream of the luciferase gene poly(A) at compatible SalI/BamHI sites. All plasmids containing repeat arrays were grown in Stbl2 Escherichia coli cells to prevent recombination (Invitrogen Corporation). The p21waf promoter was provided by S. Benchimol (University Health Network, Toronto, Canada) and cloned into the pGL3 luciferase construct. Biotin binding peptide (BBP)-PML IV was created by cloning oligonucleotides corresponding to the biotin binding peptide (AGA GGA GAA TTC ACT GGA ACT TAT ATT ACT GCT GTT ACT) and a glycine/serine linker (16a) into pBluescript to create pBBP-GS-BBP, after which the sequences encoding the biotin binding peptide were excised from pBluescript by using BamHI/HindIII and ligated into TetR-PML (for details, see reference 11) between the corresponding restriction sites. Flag-LacI-SpT was cloned into Flag-LacI by using the Sp100 PML NB-targeting domain (amino acids 33 to 139) (11). More details are available upon request.
Transfections.
A total of 2.5 × 105 cells were plated 24 h prior to transfection. A total of 2 μg of DNA was used to transfect cells by using Lipofectamine 2000 according to the protocol of the manufacturer (Invitrogen). Cells were prepared for immunofluorescence or luciferase assays 24 h following transfection.
Cell lines.
SK-N-SH cells and HeLa cells were obtained from the American Type Culture Collection and maintained as recommended on the ATCC website (http://www.atcc.org).
Immunofluorescence.
Cells were grown on coverslips and fixed using 2% paraformaldehyde-phosphate-buffered saline (PBS) for 10 min, washed three times for 5 min, permeabilized in 0.5% Triton X-100 for 5 min, and thereafter washed three times for 5 min in PBS. Cells were incubated in diluted primary antibody for 1 h at room temperature or overnight at 4°C. The primary antibodies (with concentrations and sources) that were used were as follows: PML (1/1,000; Chemicon), Sp100 (1/200; Chemicon), Daxx (1/500; Santa Cruz), CBP (1/500; Santa Cruz), Flag tag (M5, 1/2,000; Sigma), and hemagglutinin (HA) tag (anti-HA, 1/500; Sigma). Coverslips were washed three times for 5 min prior to incubation with secondary antibody for 1 h. Coverslips were mounted on glass slides using PBS-90% glycerol containing 1 mg/ml paraphenylenediamine and 1 μg/ml of the DNA-specific stain DAPI (4′,6′-diamidino-2-phenylindole).
Luciferase assays.
Using pBluescript as a carrier, 2 μg plasmid DNA was used to transfect 2.5 × 105 SK-N-SH cells that were plated 24 h prior to the experiment. Cells were harvested according to the Promega luciferase assay kit protocol 24 h after transfection. Briefly, cells were washed with PBS, incubated in reporter lysis buffer, freeze-thawed, and scraped into microfuge tubes. The supernatant was transferred to a 96-well plate, and bioluminescence was measured using an automated luminometer (Fisher Scientific). For experiments in which Renilla luciferase was used as a control, cells were lysed using Renilla lysis buffer and immediately transferred to a 96-well plate for analysis. To minimize variation of cell number per well, freshly harvested cells were resuspended into a 10-ml pipette and evenly distributed into a six-well dish (2 ml/well). Statistical analysis of data to determine significance employed a t test (a one-sample test of mean or two-sample test of mean) using Smith's Statistical Package.
Plasmid labeling.
Plasmid DNA was labeled using the Label-IT Track-It biotin or CY3 kit (Mirus Biotechnology) at a ratio of 0.5 μl total labeling reagent (Cy3 or biotin) to 1 μg DNA according to the manufacturer's protocol. Labeled DNA was purified by ethanol precipitation.
RESULTS
Plasmid DNA can be targeted to PML NBs.
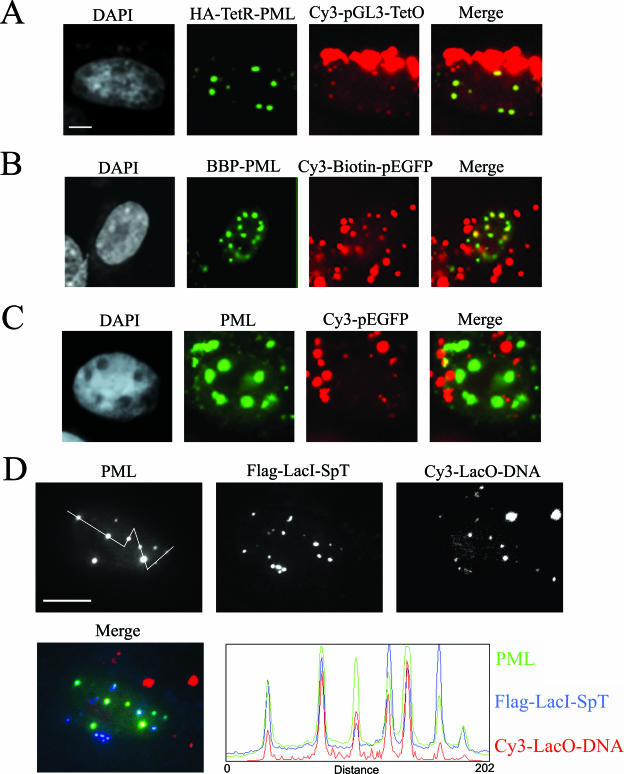
The up-regulation of PML isoform IV expression leads to an increase in the size of the existing PML NBs (14, 15). Thus, we predicted that the overexpression of a PML fusion protein having the ability to interact with an ectopic plasmid DNA would accumulate in PML NBs and thereby target the plasmid DNA to the PML NB. One approach was to fuse Tet repressor (TetR) protein to the N terminus of the most intensely studied PML IV isoform (TetR-PMLIV). A Tet operator array (TetO) specific for TetR was inserted downstream of the luciferase gene in the plasmid pGL3 (pGL3-TETO). pGL3-TetO was covalently labeled with the fluorescent tag Cy3 and cotransfected into SK-N-SH cells along with the TetR-PMLIV construct. Cells expressing TetR-PMLIV showed enrichment of Cy3-labeled plasmid at PML NBs 24 h after transfection (TetR-PML) (Fig. 1A). We obtained a similar result when we fused a 13-amino-acid biotin binding peptide in frame with PML IV (BBP-PML). The plasmid DNA encoding enhanced green fluorescent protein (EGFP) was targeted to PML NBs when labeled with biotin and Cy3 before being transfected into cells (Cy3-biotin-peGFP). Cy3 labeling was used for detection of the plasmid by fluorescence microscopy. GFP-expressing cells displayed an enrichment of labeled plasmid at PML NBs (Fig. 1B). As a control, cells transfected with DNA that was not biotinylated showed no enrichment of the plasmid at PML NBs in cells expressing BBP-PML IV (Fig. 1C).
FIG. 1.
Plasmid DNA can be targeted to PML NBs. (A) Immunofluorescence micrographs of SK-N-SH cells transfected with Cy3-labeled pGL3-TetO and TetR-PMLIV constructs and detected with antibodies directed against an N-terminal HA tag. Chromatin was counterstained with DAPI. PML was detected by immunofluorescence. (B) SK-N-SH cells transfected with BBP-PMLIV and Cy3- and biotin-labeled pEGFP. Transfected cells were identified by GFP expression. (C) SK-N-SH cells transfected with BBP-PMLIV and Cy3-labeled, but not biotinylated, p-EGFP. (D) SK-N-SH cells transfected with Flag-LacI-SpT and Cy3-labeled plasmid DNA containing the LacO sequence. A line scan through several PML bodies (from left to right) reveals the relative distributions of PML protein, SpT protein, and the targeted DNA. Bars, 5 μm in panels A to C and 10 μm in panel D.
We wished to determine whether a PML NB component other than PML protein was able to target and retain DNA in the vicinity of the body. We utilized a domain consisting of amino acids 33 to 139 from Sp100. This domain (SpT), responsible for Sp100 localization in PML NBs, was originally described as the HSR domain by Stemsdorf and colleagues (33). When SpT was fused in frame at its N terminus with a Lac repressor (LacI) protein (Flag-LacI-SpT), plasmid DNA containing the Lac operator sequences (pGL3-LacO) was also targeted to PML NBs. SpT localized to PML NBs at very low levels of transfection of Flag-LacI-SpT (10 ng of transfected DNA). At higher levels of the targeting protein expression vector (e.g., 72 ng of transfected DNA), SpT was found at PML NBs, but it was also at other structures that contained no detectable PML (data not shown). Twenty-four hours following transfection of 10 ng of DNA, the plasmid was enriched in the nucleus at sites enriched with Flag-LacI-SpT (Fig. 1D). The line scans reveal that the amount of SpT at PML NBs is not proportional to the PML signal (i.e., the size of the bodies). Moreover, the amount of DNA targeted at a body is not directly proportional to the amount of targeting protein (LacI-SpT). The accessibility of the plasmid DNA molecules to the PML NB domains may be affected by other properties of the bodies. The line scans also reveal that the nucleoplasmic plasmid DNA signal is very low relative to that found at the bodies. Cells showing no expression of SpT showed no enrichment of plasmid DNA at PML NBs (data not shown).
Targeting of transgenes to PML NBs alters their expression.
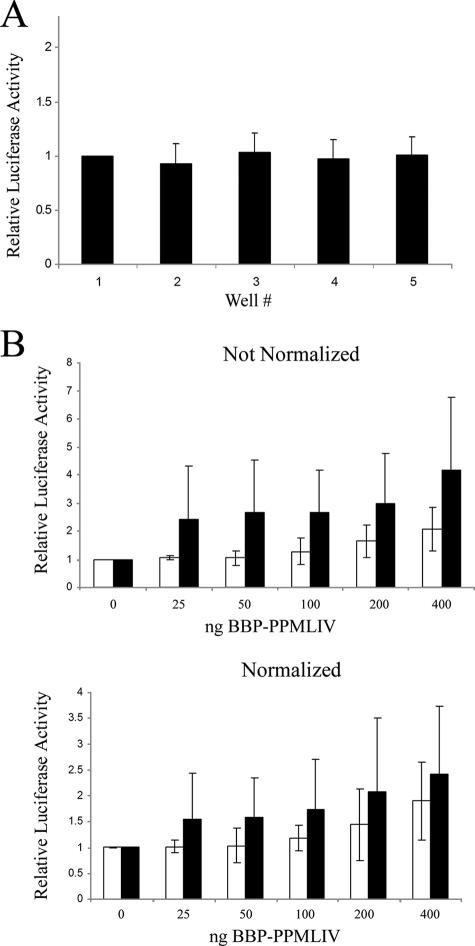
Our first concern in testing whether the PML NB environment is conducive to transcriptional regulation was to verify that transfection reporter assays were quantitative and that the results between experiments were comparable. Controls for transfection and expression efficiency using a firefly luciferase assay typically employ a second independent reporter gene, such as Renilla luciferase. Unfortunately, with some of the experiments, we were concerned that proteins being expressed could also affect this gene's reporter or that the Renilla luciferase expression plasmid might negatively affect expression from the reporter construct through effects in trans, such as promoter quenching (19). We also wanted to limit the number of plasmids being transfected to the minimum required. Hence, we devised a strategy to measure reporter gene activity using a single reporter plasmid, eliminating the need for the Renilla luciferase reporter. The first criterion of the technique is that transfections within a single experiment are comparable. By plating a very precise volume of cells from a single suspension into each well of a single six-well plate and delivering the same amount of DNA into each well by lipofection, we demonstrated that the variation in the detected luminescence was within an acceptable degree of variation (Fig. 2A). Thus, pipetting errors, transfection efficiency, and cell number could be controlled. For control experiments using different six-well plates or experiments performed on different days, we first normalized the values obtained for each well to that of the first well of each plate. Thus, by setting the first well as a baseline control for each experiment, we could then compare the results from one six-well plate to those of another. The efficacy of this approach is demonstrated in an experiment where we compare results with and without the use of Renilla luciferase in an experiment where promoter quenching is not a likely concern. In this experiment, we transfected SK-N-SH cells with constant amounts of the reporter pGL3-p21waf promoter (Fig. 2B), increasing amounts of the targeting protein plasmid expressing BBP-PMLIV and the Bluescript carrier into each well of a six-well dish. The effect of targeting the expression promoter to PML NBs was tested by first biotinylating the pGL3-p21waf plasmid before transfection (Fig. 2B, upper panel). A comparison of expression levels from targeted and nontargeted vectors was possible by normalizing to the first well of two six-well dishes used. Although increasing levels of the targeting protein vector had a small positive effect on the expression of this minimal promoter, the differences between targeted and nontargeted promoters were not significant (P > 0.1). The above experiment was repeated with an additional Renilla luciferase promoter used for normalizing expression levels. Similar results were obtained with this additional normalization step (Fig. 2B, lower panel) compared to that with no Renilla luciferase normalization (upper panel). We conclude that our approach does not require normalizing with a Renilla luciferase plasmid, and in the process of demonstrating this, we conclude that the p21waf minimal promoter element is unaffected by targeting to PML NBs.
FIG. 2.
Quantification of expression from reporter plasmids does not require normalization with a Renilla luciferase construct. (A) A total of 100 ng pGL3 and 1.5 μg pBluescript carrier was transfected into each well of a six-well plate. Luciferase expression from each well was measured and normalized to the expression of the first well. Variation was tested using a single sample t test (H0 = 1) to determine whether any given well varied statistically from the first. No statistically significant variation was observed (P ≫ 0.05). (B) Cells were transfected with 100 ng of biotinylated (white bars) or unbiotinylated (black bars) pGL3-p21waf (reporter), 50 ng SV40 promoter-driven Renilla luciferase (normalization protein), and increasing amounts of the BBP-PML IV plasmid (targeting protein). The top and bottom graphs were derived from the same data, whereas the data for the bottom graph were normalized to that of Renilla luciferase. Error bars in panels A and B represent the standard deviations of three separate measurements made from each well.
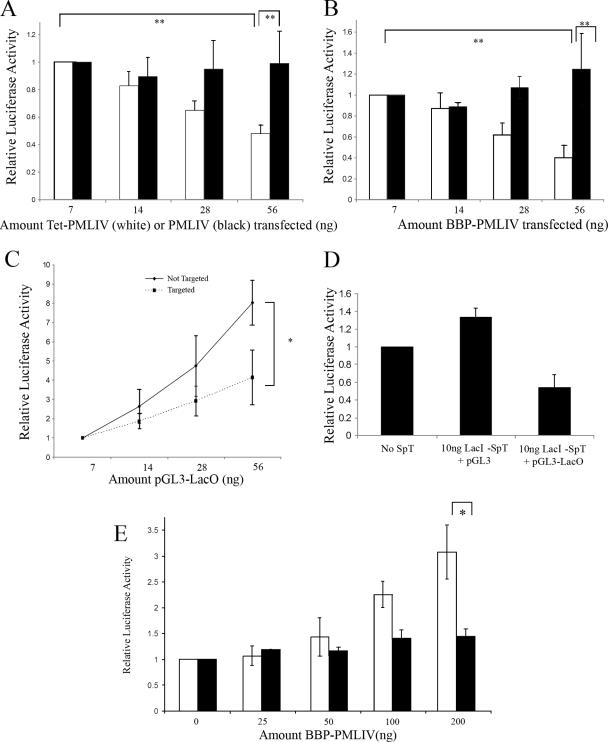
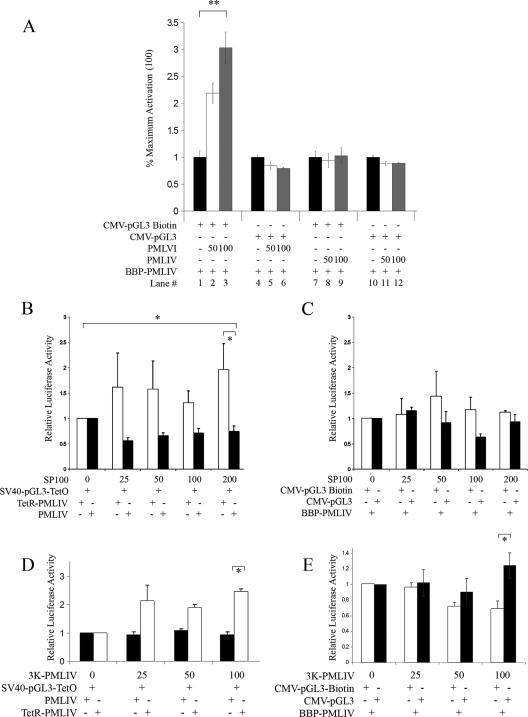
We then asked whether a more robust promoter, such as the SV40 promoter, is affected by targeting to PML NBs in SK-N-SH cells. The targeting of pGL3-TetO to PML-NBs via TetR-PMLIV resulted in the repression of the SV40 promoter (Fig. 3A). In these experiments, constant amounts of pGL3-TetO were transfected into SK-N-SH cells along with increasing amounts of TetR-PMLIV. When PMLIV was used as a nontargeting control, no repression of pGL3-TetO was observed; thus, increasing amounts of the targeting form of PML IV (TetR-PML IV), not PML IV itself, was responsible for targeting and hence the observed repression. To account for the possible repressive effect of using bacterial DNA binding proteins in our targeting scheme, we also tested the transcriptional effect of targeting using biotinylated reporter DNA and the biotin binding peptide fused in frame with PML (Fig. 3B). Here, constant amounts of biotinylated or unbiotinylated pGL3 were transfected, along with increasing amounts of BBP-PMLIV (Fig. 3B). Using the biotin-targeting strategy, we observed a similar repression of the SV40 promoter. There was no repression of the SV40 promoter in the unbiotinylated control. As expected, increasing amounts of the untargeted reporter plasmid produced a linear increase in reporter gene expression (see Fig. S1a in the supplemental material). The slope of the linear expression profile was decreased with increasing amounts of cotransfected expression plasmid coding for the TetR-PML IV or BBP-PMLIV targeting protein. The repression at high levels of reporter DNA (200 ng) relative to where no or low levels of targeting protein are present indicates that the targeting protein is not limiting.
FIG. 3.
Targeting luciferase expressing reporter constructs to PML NBs alters expression in a promoter-dependent fashion. (A) SK-N-SH cells were transfected with 100 ng pGL3-TetO and increasing amounts of TetR-PML (white bars) or PMLIV (black bars). (B) SK-N-SH cells were transfected with 100 ng biotinylated (white bars) or unbiotinylated (black bars) pGL3 plasmid and increasing amounts of BBP-PMLIV plasmid as indicated. (C) SK-N-SH cells were transfected with increasing amounts of pGL3-LacO and either 10 ng LacI-SpT DNA (dotted line) or no targeting vector as a control (solid line). (D) To ensure that LacI-SpT did not induce repression alone, cells were transfected with 100 ng pGL3-LacO with no SpT, 100 ng pGL3 with 10 ng LacI-SpT, or 100 ng pGL3-LacO with 10 ng LacI-SpT. (E) SK-N-SH cells were transfected with 100 ng biotinylated (white bars) or unbiotinylated (black bars) CMV-pGL3 with increasing amounts of BBP-PMLIV as indicated. *, P was <0.05; **, P was <0.01. Error bars represent the standard deviations of three separate measurements.
To rule out a role of the PML protein itself in these effects on the expression of targeted promoters, we used a targeting strategy that did not rely on PML protein. Instead, DNA was targeted to PML NBs with the domain of Sp100 (SpT) that is responsible for its accumulation in PML NBs (Fig. 3C). The repression of the SV40 promoter was observed when the targeting protein was expressed relative to the levels observed without the targeting protein. Only reporter DNA containing the Lac operator sequence was repressed when transfected into cells expressing the targeting protein (Flag-LacI-SpT) (Fig. 3D). These results were not cell line dependent, as we saw a similar repression by artificial tethering to PML NBs in other cell lines, including murine embryonic fibroblasts, human U2OS, and HeLa cells (data not shown).
We then asked whether repression was a general property of promoters targeted to PML NBs. To address this question, we targeted a reporter construct driven by the CMV promoter rather than the SV40 promoter (Fig. 3E). We used the biotin-targeting strategy and carried out the experiment in SK-N-SH cells. The levels of transcription from the targeted DNA (biotinylated) increased proportionally with the amount of targeting protein, whereas increasing amounts of the targeting protein had no effect on expression from untargeted DNA (not biotinylated). Similar results were obtained with HeLa cells (data not shown). Considering the responses of the p21waf, SV40, and CMV promoters, we conclude that promoters are differentially regulated when brought into the vicinity of PML NBs.
Expression of targeted promoters is affected by the biochemical composition of PML NBs.
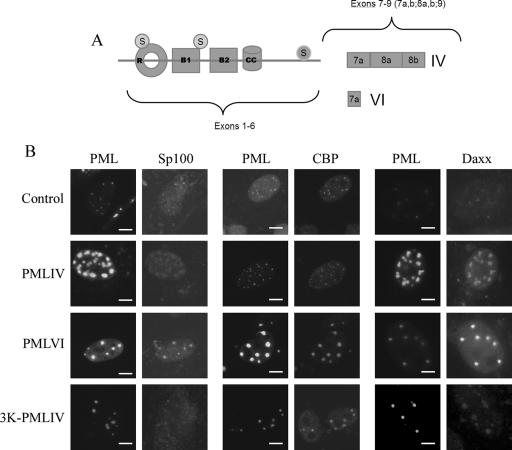
To understand the contribution of the PML NB to the regulation of targeted reporter constructs, we sought to alter the relative levels of PML NB components and measure concomitant changes in the expression of targeted promoters. The overexpression of some isoforms of PML can alter the relative levels of PML NB components within the NB relative to the nucleoplasmic background (Fig. 4A). The degree of the accumulation of these components could be based on differential interactions, direct or indirect, with the variable C termini of the various PML alternate splice isoforms (19). Three PML NB components that we monitored by immunofluorescence detection were Sp100, CBP, and Daxx. The accumulation of these proteins relative to the nucleoplasmic background was qualitatively described in cells after the overexpression of PMLIV, PMLVI, and a PML mutant lacking the three major sumoylation sites (3K-PMLIV) (27). Although the size of PML NBs increases in cells overexpressing PMLIV, the bodies still contain Sp100, Daxx, and CBP (Fig. 4B). We did not determine in this experiment whether slight or even moderate changes in the accumulation of these proteins relative to that in the nucleoplasmic background occurred in cells overexpressing PML IV compared to cells with endogenous levels of PML (Fig. 4B). In contrast, when PML VI was overexpressed, we observed that Daxx accumulation was dramatically increased in the NBs relative to that in the nucleoplasmic background. In addition and in agreement with previous studies (45), the overexpression of 3K-PML IV lead to a significant loss of both Sp100 and Daxx from PML NBs, relative to the nucleoplasmic background. Having established that these two forms of PML affect the biochemical composition of PML NBs, we asked whether the overexpression of these proteins alters the expression of promoters that are targeted to PML NBs.
FIG. 4.
Overexpression of different isoforms of PML as well as a PML mutant alters the biochemical composition of PML NBs. (A) Schematic diagram displaying the variable C-terminal domain of the various PML isoforms as well as the location of the three sumoylation mutation sites of the 3K-PMLIV protein (circled S). (B) Immunofluorescence microscopy of cells transfected with either no plasmid, PMLIV, PMLVI, or 3K-PMLIV. Lanes 1, 3, and 5, cells were labeled using an anti-PML antibody. Lanes 2, 4, and 6, cells were labeled with anti-Sp100, CBP, and Daxx antibodies, respectively. Exposure times were chosen so that the fluorescence signal did not saturate the detector. No contrast enhancement was performed in any of these images, so that the relationship of signal within the PML NBs relative to the nucleoplasmic background could be assessed qualitatively. Bar = 5 μm.
We observed that the overexpression of PMLVI accentuated the activation of the biotinylated CMV promoter-driven plasmid when targeted with BBP-PMLIV (Fig. 5A). PML VI overexpression had no effect on the nontargeted promoter, and the overexpression of PML IV under the same conditions had no effect on either the targeted or the nontargeted promoter. Since Sp100 accumulation in PML NBs was affected by the overexpression of 3K-PML IV, we wished to determine if directly altering Sp100 levels affected the transcription of a targeted transgene. We observed that increased levels of Sp100 did alleviate the repression of the targeted SV40 promoter, leading to approximately threefold more activity from the targeted promoter than from the untargeted promoter (Fig. 5B). Whereas Sp100 overexpression derepressed the targeted SV40 promoter, the overexpression of Sp100 abolished the activation of the expression of the targeted CMV promoter (Fig. 5C). The overexpression of 3K-PMLIV had no significant effect on the repression of the targeted SV40 promoter but significantly impaired the activation of the targeted CMV promoter (Fig. 5D and E, respectively). The overexpression of 3K-PMLIV had no effect on the expression of the untargeted CMV reporter (Fig. 5E). Our conclusion from these results is that the regulation of the transcription of promoters targeted to PML NBs may be dependent on the biochemical composition of the bodies.
FIG. 5.
Alteration of the distribution of PML NB components changes expression from targeted transgene. (A) A total of 100 ng biotinylated CMV-pGL3 (lanes 1 to 3 and 7 to 9) or unbiotinylated CMV-pGL3 (lanes 4 to 6 and 10 to 12) was cotransfected with BBP-PMLIV to achieve targeting of the biotinylated vector but not the unbiotinylated control. Cells were also cotransfected with increasing amounts of PMLVI (lanes 1 to 6) or PMLIV (lanes 7 to 12) as indicated. Data are shown normalized to that of Renilla luciferase. (B) A total of 100 ng SV40-pGL3-TetO was cotransfected with 100 ng TetR-PMLIV (white bars) or PMLIV as a nontargeted control (black bars). Increasing amounts of Sp100 were cotransfected with the targeting constructs as indicated. Luciferase activity was normalized relative to the well containing no Sp100. (C) A total of 100 ng biotinylated (white bars) or unbiotinylated (black bars) CMV-pGL3 was cotransfected with 100 ng BBP-PMLIV. Increasing amounts of Sp100 were cotransfected with the targeting constructs as indicated. Luciferase activity was normalized relative to the well containing no Sp100. (D) A total of 100 ng SV40-pGL3-TetO was cotransfected with 100 ng TetR-PMLIV (white bars) or PMLIV as a nontargeted control (black bars). Increasing amounts of 3K-PMLIV were cotransfected with the targeting constructs as indicated. Luciferase activity was normalized relative to the well containing no 3K-PMLIV. (E) A total of 100 ng biotinylated (black bars) or unbiotinylated (white bars) CMV-pGL3 was cotransfected with 100 ng BBP-PMLIV. Increasing amounts of 3K-PMLIV were cotransfected with the targeting constructs as indicated. Luciferase activity was normalized relative to the well containing no 3K-PMLIV. *, P was <0.05; **, P was <0.01. Error bars represent standard deviations of three separate measurements. −, absence of; +, presence of.
The HSV immediate-early gene product ICP0, but not the adenovirus protein E4orf3, affects the regulatory potential of the PML NB environment.
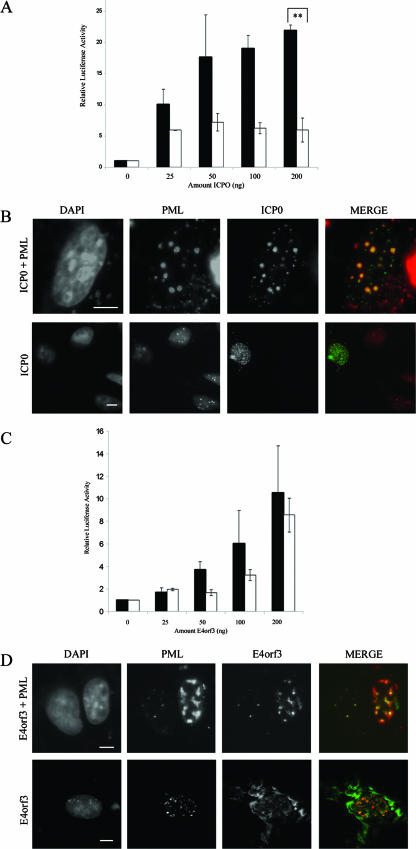
The herpes simplex virus type 1 (HSV-1) immediate-early protein ICP0 is an E3-ubiquitin ligase, which localizes to PML NBs, desumoylates PML, and targets it and other PML NB components for degradation by the 26S proteasome. The overexpression of PML protein, however, prevents PML NB breakdown in the presence of ICP0 (24). In this situation, ICP0 colocalizes to PML NBs as well as to other uncharacterized structures in the nucleus. Aside from its potential to alter PML NBs, ICP0 is also a promiscuous transactivator, although the mechanism of transactivation remains unknown (16). The cotransfection of ICP0 with a plasmid expressing any PML isoform results in the transactivation of the PML gene and thus may further impede the ability of ICP0 to break down NBs completely (28). To test the effect of ICP0 on targeted transgenes, we expressed equal amounts of the targeting protein (BBP-PML IV) and the reporter transgene (biotin-SV40-pGL3) with increasing amounts of ICP0 (Fig. 6A). We observed that the transactivation was 20-fold more efficient when SV40-pGL3 was targeted to PML NBs at the highest level of ICP0 tested. The colocalization of ICP0 with PML NBs was confirmed by immunofluorescence microscopy (Fig. 6B). Although ICP0 was able to increase the expression of the untargeted reporter plasmid, this effect was saturated at relatively low levels of ICP0 expression and the targeted promoter continued to increase significantly above the levels of the untargeted DNA. We conclude that ICP0's ability to affect transcription in the PML NB environment may be due to biochemical changes that the ICP0 protein induces that are independent from its transactivation function observed on untargeted promoters. In contrast to these results with ICP0, we observed that the overexpression of the adenoviral early protein E4orf3 did not significantly affect the expression of the targeted SV40 promoter relative to that of the untargeted promoter (Fig. 6C). Although E4orf3 localizes to PML NBs and alters their morphologies (Fig. 6D), it did not significantly affect the accumulation of Sp100, CBP, or Daxx in the bodies (data not shown). Taken together, these data indicate that PML NB biochemical composition, when manipulated by viral gene products or by changes brought about by PML mutant or specific isoforms, can regulate gene activity in the immediate vicinity of PML NBs.
FIG. 6.
Targeting plasmid DNA to PML NBs enhances the transcriptional potential of ICP0. (A) Cells were transfected with 100 ng pGL3-TetO and TetR-PMLIV (black bars) or PMLIV (white bars) as well as increasing amounts of ICP0 as indicated. Luciferase activity was normalized to the well containing no ICP0. **, P was <0.01. (B) Immunofluorescence images of double labeling using anti-PML and anti-ICP0 antibodies. Top panels show an image for visualization of the presence of PML NBs (imaged with a 100× objective). Bottom panels images show both transfected and untransfected cells. (imaged with a 40× objective). (C) Cells were transfected with 100 ng pGL3-TetO and TetR-PMLIV (black bars) or PMLIV (white bars) as well as increasing amounts of E4orf3 as indicated. Luciferase activity was normalized to the well containing no E4orf3. (D) Cells transfected with E4orf3 and PML (top panels) relative to cells transfected with only E4orf3. Error bars represent standard deviations. Bar = 5 μm.
DISCUSSION
It has been reported that PML NBs are associated with nuclear subregions of transcription activity (6). This is consistent with our observations with correlative fluorescence microscopy and electron spectroscopic imaging, which show that PML NBs are frequently surrounded by chromatin fibers involved in transcription (4, 8). Unfortunately, these data do not provide a direct functional link between transcribing chromatin and PML NBs per se. PML NBs, for example, may fortuitously be located in regions of the nucleus where transcription occurs. Moreover, the discovery that the PML protein can act as both a transcriptional coactivator (43) and a corepressor (35) does not establish whether that function occurs at PML NBs or throughout the nucleoplasm. To test this directly, we developed a novel strategy to target reporter gene plasmids to PML NBs. This technique allows us to determine whether the PML NB “microenvironment” is competent for transcriptional regulation. Three approaches were used for targeting plasmids. Two of these involved bacterial repressors fused in frame with PML or Sp100 (TetR-PML or LacI-SpT, respectively). Low levels of expression of the targeting protein were used so that we did not severely alter the biochemical composition, number, or morphology of the PML NBs. The targeted plasmids contained the operator that is bound by the bacterial repressor. A second approach avoided the use of bacterial repressors. Instead, we constructed an in-frame fusion of a 14-amino-acid biotin binding peptide with PML (BBP-PML). The plasmid DNA was targeted to the bodies when labeled with biotin before transfection into cells expressing the targeting PML fusion protein.
Regardless of the targeting scheme, we observed that when the luciferase reporter was driven by the SV40 promoter, transcription was repressed when the reporter plasmid was targeted to PML NBs. In contrast, the CMV reporter was up-regulated when targeted to PML NBs. (We wish to emphasize that the behavior of the two viral promoters, when targeted as transgenes, does not necessarily reflect their activities in intact viral genomes in virally infected cells. Although the viral genomes may be positioned at PML NBs, viral tegument proteins and early virus-expressed proteins can affect the functional relationships between PML NBs and the viral genomes, including their promoters [reviewed in reference 6]). Compared to those from using a nontargeted reporter, transcription levels from a minimal promoter were not affected by targeting to PML NBs. Whether the immediate vicinity of the PML NB favors activation or repression, therefore, is promoter dependent.
PML protein is classed as a transcriptional corepressor because it represses transcription when tethered as a GAL4 fusion to an upstream activator sequence site near a promoter (36, 37). In such experiments, PML could be functioning as a regulatory factor in the nucleoplasm or at PML NBs and, indeed, in these studies, the possibility that PML could be tethering the reporter plasmid to a subnuclear domain was not considered. We have demonstrated that if PML is artificially tethered to a plasmid, DNA will be targeted to PML NBs. In fact, our results indicate that the conventional transient expression assay may frequently be testing the role of subnuclear localization rather than simply the potential of interactions between factors and DNA promoter elements.
This novel targeting strategy permits experiments to test the role of the PML NB itself in regulating transcription. For example, it is possible to manipulate the PML NB composition and measure the outcome on a targeted promoter's function. When we changed the biochemical composition of PML NBs by overexpressing the PML VI isoform, we observed a substantial increase of transcription from the targeted CMV promoter. This PML isoform had a dramatic effect on the accumulation of Daxx at PML NBs. Moreover, the expression of the 3K-PML IV mutant of PML, which is not SUMO-1 modified, also changed the levels of the accumulation of Daxx and Sp100 in the PML NBs (45). Although we did not directly test the ability of Daxx depletion or overexpression to affect the activity of the PML NB-targeted reporter, we did demonstrate that the overexpression of Sp100 alleviated the repression of a PML NB-targeted SV40 promoter. At this point, we do not know the basis for the response of targeted reporters to levels of specific factors accumulating in PML NBs. We have demonstrated, however, that PML NBs are not transcriptionally inert subnuclear compartments, in that changes in PML NB biochemical composition can affect both the repression and the activation of gene expression at these bodies. Further studies are under way to dissect the roles of specific nuclear body components in regulating transcription from targeted transgenes.
We observed that the activation of an SV40 promoter by the HSV immediate-early gene product, ICP0, was accentuated when the plasmid was targeted to the PML NB. This was not seen with the adenoviral gene product E4orf3. Although the effect on gene expression associated with the PML NB environment could be a reflection of the increased concentration of the transcriptional activator ICP0 at the nuclear body, the increased transcriptional activity could also be based on the ability of ICP0 to recruit SENP1, the protease specific for SUMO-1. Recruiting SENP1 to PML NBs would lead to the rapid de-SUMOylation of PML and presumably other PML NB proteins (1, 28). (Under our conditions, where PML is up-regulated or overexpressed, de-SUMOylation occurs but PML NBs remain intact [24]). Although we do not know the precise mechanism by which ICP0 is modulating gene expression in the vicinity of PML NBs, it may involve the loss of PML NB components that require SUMOylation for targeting to PML NBs, such as Sp100 or Daxx (39). It has also been shown that PML can repress the EGFR promoter when tethered to the reporter via GAL4 in a transient assay. Further, this repression was dependent on the Sp1 binding site in this promoter (34, 36, 39). From our experience with PML fusions with bacterial repressors and other targeting strategies, we now know that the EGFR promoter was most likely targeted to PML NBs in these experiments and that such targeting is the basis for the observed repression. Interestingly, it has also been reported that SUMO-1 represses the activity of the Sp1 family of transcriptional activators (30, 32). The SV40 promoter used in our study contains several Sp1 binding sites. Therefore, the recruitment of SENP1 by ICP0 could result in the de-SUMOylation and activation of Sp1, which might explain the observed derepression of the PML NB-targeted SV40 promoter when ICP0 is overexpressed. We are currently testing this hypothesis.
We hypothesize that PML NBs play a role in regulating cellular gene promoters that are found on their immediate periphery. The MHC gene cluster has been reported to be associated with PML NBs (31), and PML may play a role in regulating members of these genes (29, 38). Therefore, it is important to test whether MHC gene cluster genes are subject to transcription regulation when tethered to PML NBs. The targeting approach will also have broad applications for studying other genes that are regulated by factors that are modulated by PML or PML NB components, such as Sp3, p53, p63, p73, pRb, myc, CBP, Sp100, and Daxx. Finally, we suggest that our targeting strategy could provide a means of testing the transcription regulatory environment of subchromosomal domains, such as heterochromatin, or other subnuclear compartments, such as splicing speckles or the nuclear envelope.
Supplementary Material
Acknowledgments
We gratefully acknowledge P. Cheung for providing us with the construct containing ICP0 as well as the corresponding antibody and S. Benchimol for providing the p21waf promoter. We also thank T. Sternsdorf for providing the construct containing the Sp100 targeting domain.
This research was funded by an operating grant from the Canadian Institutes of Health Research. G.D. is a senior research fellow of the Canadian Institutes of Health Research. D.P.B.-J. holds a Canada Research Chair in Molecular and Cellular Imaging.
Footnotes
Published ahead of print on 11 September 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bailey, D., and P. O'Hare. 2002. Herpes simplex virus 1 ICP0 co-localizes with a SUMO-specific protease. J. Gen. Virol. 83:2951-2964. [DOI] [PubMed] [Google Scholar]
- 2.Beech, S. J., K. J. Lethbridge, N. Killick, N. McGlincy, and K. N. Leppard. 2005. Isoforms of the promyelocytic leukemia protein differ in their effects on ND10 organization. Exp. Cell Res. 307:109-117. [DOI] [PubMed] [Google Scholar]
- 3.Bischof, O., O. Kirsh, M. Pearson, K. Itahana, P. G. Pelicci, and A. Dejean. 2002. Deconstructing PML-induced premature senescence. EMBO J. 21:3358-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boisvert, F. M., M. J. Hendzel, and D. P. Bazett-Jones. 2000. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol. 148:283-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boisvert, F. M., M. J. Kruhlak, A. K. Box, M. J. Hendzel, and D. P. Bazett-Jones. 2001. The transcription coactivator CBP is a dynamic component of the promyelocytic leukemia nuclear body. J. Cell Biol. 152:1099-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ching, R. W., G. Dellaire, C. H. Eskiw, and D. P. Bazett-Jones. 2005. PML bodies: a meeting place for genomic loci? J. Cell Sci. 118:847-854. [DOI] [PubMed] [Google Scholar]
- 7.Condemine, W., Y. Takahashi, J. Zhu, F. Puvion-Dutilleul, S. Guegan, A. Janin, and H. de The. 2006. Characterization of endogenous human promyelocytic leukemia isoforms. Cancer Res. 66:6192-6198. [DOI] [PubMed] [Google Scholar]
- 8.Dehghani, H., G. Dellaire, and D. P. Bazett-Jones. 2005. Organization of chromatin in the interphase mammalian cell. Micron 36:95-108. [DOI] [PubMed] [Google Scholar]
- 9.Dellaire, G., and D. P. Bazett-Jones. 2004. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays 26:963-977. [DOI] [PubMed] [Google Scholar]
- 10.Dellaire, G., R. Farrall, and W. A. Bickmore. 2003. The Nuclear Protein Database (NPD): sub-nuclear localisation and functional annotation of the nuclear proteome. Nucleic Acids Res. 31:328-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dellaire, G., R. Nisman, C. H. Eskiw, and D. P. Bazett-Jones. 2004. In situ imaging and isolation of proteins using dsDNA oligonucleotides. Nucleic Acids Res. 32:e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de The, H., C. Lavau, A. Marchio, C. Chomienne, L. Degos, and A. Dejean. 1991. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell 66:675-684. [DOI] [PubMed] [Google Scholar]
- 13.Dyck, J. A., G. G. Maul, W. H. Miller, Jr., J. D. Chen, A. Kakizuka, and R. M. Evans. 1994. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell 76:333-343. [DOI] [PubMed] [Google Scholar]
- 14.Eskiw, C. H., G. Dellaire, and D. P. Bazett-Jones. 2004. Chromatin contributes to structural integrity of promyelocytic leukemia bodies through a SUMO-1-independent mechanism. J. Biol. Chem. 279:9577-9585. [DOI] [PubMed] [Google Scholar]
- 15.Eskiw, C. H., G. Dellaire, J. S. Mymryk, and D. P. Bazett-Jones. 2003. Size, position and dynamic behavior of PML nuclear bodies following cell stress as a paradigm for supramolecular trafficking and assembly. J. Cell Sci. 116:4455-4466. [DOI] [PubMed] [Google Scholar]
- 16.Hagglund, R., and B. Roizman. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78:2169-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Hiller, Y., E. A. Bayer, and M. Wilchek. 1991. Studies on the biotin-binding site of avidin. Minimized fragments that bind biotin. Biochem J. 278:573-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann, T. G., A. Moller, H. Sirma, H. Zentgraf, Y. Taya, W. Droge, H. Will, and M. L. Schmitz. 2002. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 4:1-10. [DOI] [PubMed] [Google Scholar]
- 18.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen, K., C. Shiels, and P. S. Freemont. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene 20:7223-7233. [DOI] [PubMed] [Google Scholar]
- 20.Kastner, P., A. Perez, Y. Lutz, C. Rochette-Egly, M. P. Gaub, and P. Chambon. 1991. [Fusion proteins between PML and alpha-RAR in acute promyelocytic leukemia]. C. R. Seances Soc. Biol. Fil. 185:391-401. (In French.) [PubMed] [Google Scholar]
- 21.LaMorte, V. J., J. A. Dyck, R. L. Ochs, and R. M. Evans. 1998. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc. Natl. Acad. Sci. USA 95:4991-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, H., C. Leo, J. Zhu, X. Wu, J. O'Neil, E. J. Park, and J. D. Chen. 2000. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol. Cell. Biol. 20:1784-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, D. Y., M. Z. Lai, D. K. Ann, and H. M. Shih. 2003. Promyelocytic leukemia protein (PML) functions as a glucocorticoid receptor co-activator by sequestering Daxx to the PML oncogenic domains (PODs) to enhance its transactivation potential. J. Biol. Chem. 278:15958-15965. [DOI] [PubMed] [Google Scholar]
- 24.Lopez, P., R. J. Jacob, and B. Roizman. 2002. Overexpression of promyelocytic leukemia protein precludes the dispersal of ND10 structures and has no effect on accumulation of infectious herpes simplex virus 1 or its proteins. J. Virol. 76:9355-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melnick, A., and J. D. Licht. 1999. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood 93:3167-3215. [PubMed] [Google Scholar]
- 26.Moller, A., H. Sirma, T. G. Hofmann, S. Rueffer, E. Klimczak, W. Droge, H. Will, and M. L. Schmitz. 2003. PML is required for homeodomain-interacting protein kinase 2 (HIPK2)-mediated p53 phosphorylation and cell cycle arrest but is dispensable for the formation of HIPK domains. Cancer Res. 63:4310-4314. [PubMed] [Google Scholar]
- 27.Muller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nefkens, I., D. G. Negorev, A. M. Ishov, J. S. Michaelson, E. T. Yeh, R. M.Tanguay, W. E. Muller, and G. G. Maul. 2003. Heat shock and Cd2+ exposure regulate PML and Daxx release from ND10 by independent mechanisms that modify the induction of heat-shock proteins 70 and 25 differently. J. Cell Sci. 116:513-524. [DOI] [PubMed] [Google Scholar]
- 29.Rotem-Yehudar, R., M. Groettrup, A. Soza, P. M. Kloetzel, and R. Ehrlich. 1996. LMP-associated proteolytic activities and TAP-dependent peptide transport for class 1 MHC molecules are suppressed in cell lines transformed by the highly oncogenic adenovirus 12. J. Exp. Med. 183:499-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sapetschnig, A., G. Rischitor, H. Braun, A. Doll, M. Schergaut, F. Melchior, and G. Suske. 2002. Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J. 21:5206-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiels, C., S. A. Islam, R. Vatcheva, P. Sasieni, M. J. Sternberg, P. S. Freemont, and D. Sheer. 2001. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J. Cell Sci. 114:3705-3716. [DOI] [PubMed] [Google Scholar]
- 32.Spengler, M. L., and M. G. Brattain. 2006. Sumoylation inhibits cleavage of Sp1 N-terminal negative regulatory domain and inhibits Sp1-dependent transcription. J. Biol. Chem. 281:5567-5574. [DOI] [PubMed] [Google Scholar]
- 33.Sternsdorf, T., K. Jensen, B. Reich, and H. Will. 1999. The nuclear dot protein Sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J. Biol. Chem. 274:12555-12566. [DOI] [PubMed] [Google Scholar]
- 34.Vallian, S., K. V. Chin, and K. S. Chang. 1998. The promyelocytic leukemia protein interacts with Sp1 and inhibits its transactivation of the epidermal growth factor receptor promoter. Mol. Cell. Biol. 18:7147-7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vallian, S., J. A. Gaken, E. B. Gingold, T. Kouzarides, K. S. Chang, and F. Farzaneh. 1998. Modulation of Fos-mediated AP-1 transcription by the promyelocytic leukemia protein. Oncogene 16:2843-2853. [DOI] [PubMed] [Google Scholar]
- 36.Vallian, S., J. A. Gaken, I. D. Trayner, E. B. Gingold, T. Kouzarides, K. S. Chang, and F. Farzaneh. 1997. Transcriptional repression by the promyelocytic leukemia protein, PML. Exp. Cell Res. 237:371-382. [DOI] [PubMed] [Google Scholar]
- 37.Wu, W. S., S. Vallian, E. Seto, W. M. Yang, D. Edmondson, S. Roth, and K. S. Chang. 2001. The growth suppressor PML represses transcription by functionally and physically interacting with histone deacetylases. Mol. Cell. Biol. 21:2259-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng, P., Y. Guo, Q. Niu, D. E. Levy, J. A. Dyck, S. Lu, L. A. Sheiman, and Y. Liu. 1998. Proto-oncogene PML controls genes devoted to MHC class I antigen presentation. Nature 396:373-376. [DOI] [PubMed] [Google Scholar]
- 39.Zhong, S., P. Salomoni, and P. P. Pandolfi. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2:E85-E90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.