Abstract
The generalized process of mRNA decay involves deadenylation followed by release from translating polysomes, decapping, and exonuclease decay of the mRNA body. In contrast the mRNA endonuclease PMR1 forms a selective complex with its translating substrate mRNA, where it initiates decay by cleaving within the mRNA body. In stressed cells the phosphorylation of the α subunit of eukaryotic initiation factor 2 causes translating mRNAs to accumulate with stalled 48S subunits in large subcellular structures termed stress granules (SGs), wherein mRNAs undergo sorting for reinitiation, storage, or decay. Given the unique relationship between translation and PMR1-mediated mRNA decay, we examined the impact of stress-induced dissociation of polysomes on this process. Arsenite stress disrupts the polysome binding of PMR1 and its substrate mRNA but has no impact on the critical tyrosine phosphorylation of PMR1, its association with substrate mRNA, or its association with the functional ∼680-kDa mRNP complex in which it normally resides on polysomes. We show that arsenite stress drives PMR1 into an RNase-resistant complex with TIA-1, and we identify a distinct domain in the N terminus of PMR1 that facilitates its interaction with TIA-1. Finally, we show that arsenite promotes the delayed association of PMR1 with SGs under conditions which cause tristetraprolin and butyrate response factor 1, proteins that facilitate exonucleolytic mRNA, to exit SGs.
Translation and mRNA decay are intimately linked processes, and the identification of cytoplasmic foci containing enzymes involved in mRNA decay in yeast (29) suggested that a dynamic process partitions these two key steps in posttranscriptional control into distinct physical complexes. These cytoplasmic foci, termed “processing bodies” (P-bodies) (PBs), were originally thought to function only as sites of mRNA degradation, particularly since related foci (also termed GW bodies or Dcp bodies) (2, 3, 8-11, 17, 34) were identified in mammalian cells and since other studies link these foci to the decay of mRNAs targeted by the RISC complex (24, 28). However, at least in yeast, PBs also serve as sites where mRNA is stored under conditions of stress or arrested initiation, with the stored mRNA returning to translating polysomes after removal of stress conditions (4, 7, 32). It has yet to be determined whether PBs exhibit a similar function in higher eukaryotes or whether this function is served instead by mammalian stress granules (SGs), which are not present in budding yeast.
Stress granules are large cytoplasmic aggregates of poly(A)+ mRNA that accumulate in cells under conditions where translation initiation has been reduced or inhibited (1, 19). This typically occurs in cells exposed to environmental stressors, resulting in the phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF-2α) (1), which in turn blocks formation of the ternary complex of eIF-2, GTP, and methionyl-tRNA (19), causing stalled translation and polysome disassembly. SGs are dynamic subcellular domains where untranslated mRNPs are sorted for reinitiation, degradation, or storage (21) and whose formation is enhanced by drugs that promote termination (e.g., puromycin) and is reversed by agents that freeze elongating ribosomes on translating mRNA (e.g., cycloheximide) (18). Fluorescence recovery after photobleaching analysis of stressed cells shows that SGs undergo continuous exchange of mRNA and proteins, but the block to ternary complex formation resulting from phosphorylation of eIF-2α causes mRNPs from disassembled polysomes to reassemble into SGs until the stress has been resolved (20). SGs and PBs interact in stressed cells (21), and this interaction is enhanced by proteins (e.g., tristetraprolin [TTP] and butyrate response factor 1 [BRF-1]) that accelerate the degradation of unstable mRNAs. Nevertheless, at least some unstable mRNAs are stabilized in stressed cells (23), supporting the notion that SGs in higher eukaryotes may serve the storage function of yeast PBs.
Although vertebrate mRNAs are generally thought to be degraded in PBs or by the exosome (5, 13, 25, 36), a subset are degraded by mRNA endonucleases. To date three protein mRNA endonucleases have been identified, i.e., G3BP-1 (12), IRE1 (16), and PMR1 (6), and there is strong evidence for another endonuclease that is enriched in erythroid cells (35). PMR1, the best characterized of these enzymes, was originally identified as an estrogen-induced endonuclease activity that appeared concomitantly with the estrogen-induced destabilization of serum protein mRNAs in Xenopus liver (26, 27). PMR1-mediated mRNA decay differs fundamentally from other forms of mRNA decay in that PMR1 forms a specific complex with its translating substrate mRNA, and it is in this context that cleavage initiates mRNA decay (38). Interestingly, a similar mechanism was recently reported for IRE1, which is activated by unfolded protein to cleave endoplasmic reticulum-associated mRNAs (16). For PMR1 to target polysomes and substrate mRNA, a tyrosine residue at position 650 within a consensus class I Src homology 2 sequence must be phosphorylated (37). Importantly, the overall process of endonuclease-mediated mRNA decay depends on active translation of substrate mRNA (38), and inserting a stable stem-loop structure in the 5′ untranslated region results in mRNA stabilization.
Because PMR1-mediated mRNA decay typically occurs over hours and cells respond to environmental stressors within minutes, it is difficult to assess whether changes in mRNA steady state represent a direct or indirect response to stress. Given this technical limitation, the current study sought to examine the impact of cell stress on the association of PMR1 with polysomes, substrate mRNP, and SGs, in order to gauge the impact of stress on endonuclease-mediated mRNA decay. Using a catalytically inactive form of PMR1 employed in previous studies on polysome targeting, we show that in arsenite-treated cells PMR1-containing mRNP redistributes from polysomes to SGs. Stress had no impact on the tyrosine phosphorylation of PMR1 or its association with substrate mRNA in a ∼680-kDa mRNP complex. The TIA proteins nucleate SG formation through aggregation of the C-terminal prion-like domain (PRD) (14). We show that arsenite-induced cell stress is required for colocalization of PMR1 with TIA-1 in SGs and for the biochemical recovery of TIA-1 with PMR1 in an RNase-resistant complex. Finally, the region of PMR1 responsible for stress-dependent interaction with TIA-1 mapped to the N terminus of the protein, a domain that is distinct from those involved in targeting to polysomes and association with substrate mRNA.
MATERIALS AND METHODS
Plasmids.
The preparation of plasmids expressing catalytically active PMR1, catalytically inactive PMR1, and deletions with or without a tandem affinity (TAP) tag on the C terminus was described previously (38), as was the nomenclature used here. Briefly, full-length PMR1 consisting of the active 60-kDa form of the protein is termed PMR60, and a mutant protein that is catalytically inactive as a consequence of mutating the critical active site histidine residues at 393 and 479 to alanine (38) is termed PMR60o. N- and C-terminal deletions of PMR60o are identified as Δ50, Δ100, or Δ150 to indicate the number of deleted amino acids from each portion of the protein. The central portion of PMR60 was deleted in mutant ΔM, leaving the N-terminal 150 and C-terminal 150 amino acids joined together. The deletion was generated by inducing NheI sites at positions 150 and 571 by using primers 5′-GTGTGTACCCCAGGATCTCCTGCTAGCGTACGGGAGCAAATTAATATAC-3′ and 5′-ATATGCAAAGAGGCCGGGAAGCTAGCCATGGGCTGCCAGGATACAATG-3′ with the GeneEditor system (Promega). The product was digested with NheI to remove the fragment containing amino acids 151 to 570 and religated to generate the middle-domain deletion. All of the PMR1 constructs have an N-terminal myc epitope tag. The preparation of pcDNA3-GFP and pcDNA3-100N-GFP was described previously (38). pcDNA3-50N-GFP was cloned by PCR amplification with forward primer 5′-AGTCGCTAGCAGGCAGAGGAAAGCCATTGATG-3′ and reverse primer 5′-GACTGCGGCCGCTGTTGACCTTGCAGGTGTC-3′ and inserted between the EcoRI and NheI sites of pcDNA3-GFP. The preparation of pMT2-HA-TIA-1 and pMT2-HA-TIA-PRD was described previously (14). pcDNA3-FLAG-TIA-1 was made by inserting the PCR-amplified coding region of human TIA-1 into pcDNA3-FLAG in frame with the FLAG tag by using the BamHI and EcoRI sites. GFP-TIA-1 was described previously (20). GFP-G3BP1 was a kind gift from Jamal Tazi (University of Montpellier). All plasmid constructs used in this study were verified by sequencing before use in transfection experiments.
Antibodies and reagents.
Mouse monoclonal antibodies to the c-myc epitope tag (9E10), eIF-3b, eIF-4G, TIA-1, and GFP (B2) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Horseradish peroxidase-coupled rabbit anti-mouse immunoglobulin G (IgG) and IgG-Sepharose were purchased from Amersham Biosciences. Monoclonal antibodies to eIF-4E and phosphotyrosine (PY20) were purchased from BD Biosciences. Sodium arsenite was purchased from Sigma, and the hemagglutinin (HA) monoclonal antibody was purchased from Covance (Princeton, NJ). The 3E6 antibody to TIA-1/TIAR was described previously (31). For immunofluorescence experiments, PMR60o was visualized using a mouse anti-myc antibody (Affinity BioReagents) and donkey anti-mouse Cy3-IgG (Jackson Immunoresearch Laboratories, West Grove, PA). All species-specific secondary antibodies conjugated to Cy2, Cy3, or Cy5 were donkey ML grade and were obtained from Jackson Immunoresearch Laboratories (West Grove, PA). DAPI (4′,6-diamidine-2′-phenylindole dihydrochloride) was purchased from Roche.
Cell culture and transient transfection.
Cos-1 cells were cultured in Dulbecco modified Eagle medium plus 10% fetal bovine serum and 2 mM glutamine, and 2.5 × 106 cells in log-phase growth were transfected with 10 μg (total) of plasmid DNA by using Lipofectamine Plus (Invitrogen) according to the manufacturer's protocol. Unless otherwise indicated, cells were collected 40 h after transfection. To activate the stress response, cells were treated with 0.5 mM sodium arsenite for 30 min, followed by a 30 min recovery prior to fixation or harvest. In experiments using cycloheximide to dissociate SGs, cells were treated with 0.5 mM sodium arsenite for 30 min, washed, and incubated for 1 h with 200 μg of cycloheximide. In transient transfection for microscopy, 5 × 105 Cos-7 cells were plated per well in six-well dishes 5 h prior to transfection. Each well was transfected with 2.0 μg total plasmid DNA complexed with 10 μl SuperFect (QIAGEN) in serum-free medium for 15 min prior to application on cells. Cells were exposed to the DNA complexes for 12 h and then trypsinized, replated onto 12-mm coverslips (Fisher), and cultured for an additional 24 h prior to drug treatments, fixation, and staining.
Immunofluorescence microscopy.
Cells grown on coverslips were rinsed with phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde in PBS for 15 min at 25°C, and then permeabilized by 5 min of treatment in −20°C methanol. Cells were incubated in blocking buffer (5% horse serum in PBS) for 1 h at 25°C. Cells were exposed to primary antibody combinations (as indicated in the figure legends) for 1 h at room temperature, followed by three 5-min washes in PBS. Cells were then incubated in secondary antibody cocktails (e.g., donkey anti-goat Cy2, donkey anti-mouse Cy3, and donkey anti-rabbit Cy5) containing DAPI made in blocking buffer and incubated for 1 h. Cells were then washed three times in PBS for 5 min, mounted in a homemade polyvinyl alcohol-based mounting medium, and viewed using a Nikon E800 microscope. Images were obtained using a National Diagnostics CCD-SPOT RT digital camera and compiled using Adobe Photoshop CS.
Preparation of cell extracts, polysome profile analysis, and glycerol gradients.
To determine the impact of stress on the sedimentation properties of PMR1, cells were treated with 0.5 mM sodium arsenite for 30 min and then allowed to recover for 30 min. They were washed twice with ice-cold PBS and then suspended in cell lysis buffer (10 mM HEPES-KOH [pH 7.5], 10 mM KCl, 5 mM MgCl2, 50 mM NaF, 0.5% [vol/vol] NP-40, 2 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 25 μl/ml of protease inhibitor cocktail [Sigma], 10 μl/ml of phosphatase inhibitor [Sigma], and 10 μl/ml of RNaseOUT [Invitrogen]). After incubation for 15 min on ice, the cells were homogenized with 30 strokes of a Dounce homogenizer (pestle A), and the homogenate was centrifuged for 10 min at 15,000 × g. To disassociate polysomes, the postmitochondrial supernatant was adjusted to 50 mM of EDTA. Linear gradients of 15% to 40% (wt/vol) sucrose were prepared in 10 mM HEPES-KOH (pH 7.5)-2 mM dithiothreitol (DTT) containing 5 mM MgCl2. Postmitochondrial extracts were gently layered onto gradients and centrifuged for 3.5 h at 228,000 × g in a Sorvall TH641 rotor. Fractions of 0.5 ml were collected from the bottom of the tube. Linear gradients of 10% to 40% (vol/vol) glycerol were prepared in buffer containing 10 mM HEPES-KOH (pH 7.5), 5 mM MgCl2, and 2 mM DTT. Postmitochondrial extracts were separated by centrifugation for 20 h at 83,000 × g in a Sorvall TH641 rotor. Molecular size markers containing a mixture of thyroglobulin (molecular weight, 669,000), ferritin (440,000), catalase (232,000), lactate dehydrogenase (140,000), and bovine serum albumin (67,000) were fractionated on a parallel gradient.
IgG-Sepharose selection of PMR1-TAP complexes.
To map the stress domain Cos-1, cells were transiently transfected with 6 μg of vector (pcDNA3) or plasmid myc-PMR60° or its various N-terminal, C-terminal or middle deletions fused to TAP, plus 4 μg of pcDNA3-FLAG-TIA-1 and 2 μg of pcDNA-GFP-TAP. A 250-μl amount of postmitochondrial extract in lysis buffer containing 10 mM EDTA was mixed with 50 μl of drained IgG-Sepharose 6 beads (Amersham Pharmacia Biotech) that were preblocked with lysate from nontransfected cells. This was incubated by end-over-end rotation for 1 h at 4°C. The beads were washed three times with 0.5 ml of washing buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% NP-40) and once with Tev buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5 mM EDTA, 0.1% NP-40, and 1 mM DTT) at 4°C. Bound complexes were eluted by cleavage with 50 U of Tev protease (Invitrogen) in 100 μl of Tev buffer at 4°C for 2 h. Protein released from the resin was analyzed by Western blotting using the monoclonal antibody against the myc epitope tag for PMR1 and antibody to the FLAG epitope tag for TIA-1. To determine the impact of arsenite on tyrosine phosphorylation of PMR1, 2 × 106 Cos-1 cells were transfected as described above with 10 μg of myc-PMR60o-TAP or GFP-TAP, and protein recovered following Tev protease cleavage of complexes bound to IgG-Sepharose was analyzed by Western blotting using antibodies to the myc epitope tag, anti-GFP, and the phosphotyrosine-specific monoclonal antibody PY20 as described previously (37). To determine the impact of arsenite-induced stress on the interaction of PMR1 with albumin mRNA, 2 × 106 Cos-1 cells were transfected with 6 μg of myc-PMR60o-TAP or GFP-TAP plus 2 μg of plasmid expressing albumin mRNA and 2 μg of plasmid expressing firefly luciferase. Postmitochondrial extracts were applied to IgG-Sepharose and recovered by Tev protease cleavage as described above, except that RNA was extracted from 200 μl of recovered material and analyzed by semiquantitative reverse transcription-PCR (RT-PCR) as described previously (38).
RT-PCR analysis of albumin and luciferase mRNAs.
Albumin and luciferase mRNAs recovered with IgG-Sepharose beads were analyzed by RT-PCR as described previously (38). Reverse transcription was performed with oligo(dT)15 as the primer and Superscript II reverse transcriptase (Invitrogen). Albumin mRNA was amplified using 5′-CAAAGACCAGCCTTCAAACTC-3′ (sense) and 5′-CACAGTTGAATGCTCTAAGCA-3′ (antisense). Luciferase mRNA was amplified using 5′-AGATCCACAACCTTCGCTTC-3′ (sense) and 5′-CTGAGGAGCCTTCAGGATTAC-3′ (antisense). In each set the sense primers was 5′-32P labeled. PCR amplification was performed for 18 cycles of 95°C for 45 s, 61°C for 45 s, and 72°C for 60 s, conditions that were empirically determined to lie in the linear range. Serial dilutions of albumin and luciferase mRNAs were analyzed in parallel, the products from each reaction were separated on a denaturing 6% polyacrylamide-urea gel, and the results were quantified by phosphorimager analysis.
Recovery of TIA-1 with PMR1-TAP on IgG-Sepharose.
Cells (2.0 × 106) were transiently transfected with 5.0 μg of myc-PMR60o-TAP plus 5.0 μg of pMT2 (vector), pMT2-HA-TIA-1, or pMT2-HA-TIA-1-PRD. Control cells were transfected with 5.0 μg of GFP-TAP plus 5.0 μg of pMT2-HA-TIA-1. At 40 hours after transfection, cells were treated with or without 0.5 mM sodium arsenite for 30 min, followed by 30 min of recovery. Postmitochondrial extract (250 μl) was incubated on ice for 10 min with or without 50 ng/μl RNase A, followed by addition of lysis buffer containing 10 mM EDTA and 50 μl of drained IgG-Sepharose 6 beads (Amersham Pharmacia Biotech) that were preblocked with lysate from nontransfected cells. The beads were incubated by end-over-end rotation for 2 h at 4°C and then washed three times with 0.5 ml of washing buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% NP-40) and once with Tev buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5 mM EDTA, 0.1% NP-40, and 1 mM DTT) at 4°C. Bound complexes were eluted by cleavage with 50 U of Tev protease (Invitrogen) in 100 μl of Tev buffer at 4°C for 2 h. Proteins released from the resin were precipitated using trichloroacetic acid (TCA), PMR60o was analyzed by Western blotting using the monoclonal antibody against the myc epitope tag, and TIA-1 was analyzed by using the monoclonal antibody against the HA epitope tag. To map the TIA-1-interacting domain, 2 × 106 cells were transfected with 6.0 μg of myc-PMR60o-TAP plasmid or the indicated deletions fused to TAP, plus 4.0 μg of pcDNA3-FLAG-TIA-1 and 2 μg of GFP-TAP as controls for efficiency of recovery. Forty hours later cells were treated with 0.5 mM of arsenite for 30 min, followed by 30 min of recovery. Postmitochondrial extract was recovered as described above by binding to IgG-Sepharose and elution with Tev protease cleavage. Recovered protein was TCA precipitated and analyzed by Western blotting. In each sample the recovery of GFP was monitored by Western blotting with a GFP antibody.
Recovery of TIA-1 with PMR1-GFP fusion.
Cells (2 × 106) were transiently transfected with 5.0 μg of pcDNA3-GFP, pcDNA3-50N-GFP, or pcDNA3-100N-GFP, plus 5.0 μg of plasmid pMT2-HA-TIA-1. At 40 hours after transfection, cells were treated with 0.5 mM of arsenite for 30 min, followed by 30 min of recovery. A 250-μl amount of postmitochondrial extract in lysis buffer containing 10 mM EDTA was mixed with 20 μl of drained GFP antibody-conjugated beads (Santa Cruz Biotech) that were preblocked with lysate from nontransfected cells. This was incubated by end-over-end rotation for 2 h at 4°C. The beads were washed three times with 0.5 ml of washing buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% NP-40), and bound complexes were eluted with 100 μl of 0.1 M glycine (pH 3.5) at 4°C. Proteins released from the resin were precipitated using TCA, PMR1-GFP fusions were analyzed by Western blotting using the monoclonal antibody against GFP, and TIA-1 was analyzed by using the monoclonal antibody against the HA epitope tag.
RESULTS
Arsenite-induced stress disrupts polysome binding of PMR60o and its substrate mRNA.
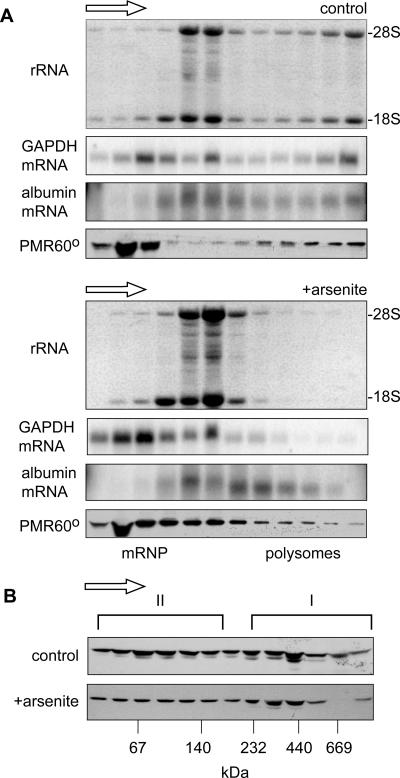
Under normal conditions, PMR60 is found in two complexes that can be physically distinguished on both sucrose and glycerol gradients (38). The functional complex containing PMR60 and its substrate mRNA sediments with polysomes on sucrose gradients, and enforced disassembly of polysomes using EDTA or puromycin releases both PMR60 and substrate mRNA as a ∼680-kDa complex (termed complex I (37). PMR60 is also present in a smaller complex (complex II) that sediments toward the top of sucrose gradients and at ∼140 kDa on glycerol gradients. The latter is thought to be the precursor to complex I and the likely site where tyrosine phosphorylation occurs (37). The stress-induced phosphorylation of eIF-2α results in bulk dissociation of polysomes due to the block in reinitiation (22), although some mRNAs are resistant to this. Given the unique relationship between translation and PMR60-mediated mRNA decay, we first examined the impact of arsenite treatment on the sedimentation of polysome-bound PMR1 and compared this to the sedimentations of its substrate (albumin) mRNA and an endogenous control, GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA. In the experiment in Fig. 1A, Cos-1 cells expressing catalytically inactive PMR60o (38) and albumin mRNA were cultured without arsenite (upper panels) or treated with arsenite for 30 min followed by 30 min of recovery (lower panels). Cytoplasmic extracts of these cells were fractionated on linear 15 to 40% sucrose gradients, and RNA extracted from odd-numbered fractions was denatured with glyoxal and electrophoresed on agarose gels. The first panel of each set shows the membrane stained with Coomassie blue to visualize rRNA. The middle panels show the distribution of albumin mRNA and endogenous GAPDH mRNA, and the bottom panels show Western blots for PMR60o using a monoclonal antibody to the N-terminal myc tag.
FIG. 1.
Impact of cell stress on the gradient distribution of PMR60o. A. Cytoplasmic extracts of control cells or cells treated for 30 min with arsenite followed by 30 min of recovery were separated on linear 15 to 40% sucrose gradients. RNA isolated from even-numbered fractions was denatured with glyoxal, separated on a 1% agarose gel, blotted onto a nylon membrane that was stained with Coomassie blue to visualize the gradient distribution of 18S and 28S rRNAs (upper panels), and then probed for the distribution of GAPDH and albumin mRNAs (middle panels). Protein present in odd-numbered fractions was TCA precipitated and analyzed by Western blotting with antibody to the myc tag on PMR60o (lower panels). The arrow above each set of panels indicates the direction of sedimentation. B. Cytoplasmic extracts prepared as for panel A were treated with EDTA to dissociate polysomes and separated on linear 10 to 40% glycerol gradients. Protein present in odd-numbered fractions was assayed by Western blotting as for panel A to determine the gradient distribution of PMR60o. Note that the second-to-the-bottom fraction was lost from the gradient from arsenite-treated cells.
The effectiveness of arsenite stress in disrupting polysomes is seen in the redistribution of rRNA from the polysome fractions to 80S and above, and the shift in sedimentation of GAPDH mRNA from polysomes to mRNPs confirms that arsenite stress caused a general dissociation of ribosomes from translating mRNA. In control cells, albumin mRNA sediments with 80S monosomes and throughout the polysome fractions. Like GAPDH, it is lost from the heaviest polysome fractions following arsenite stress; however, much of this mRNA still sediments more rapidly than 80S, suggesting either that the overall impact on ribosome binding to this mRNA is less pronounced or that albumin mRNA becomes associated with another large complex.
Arsenite causes a change in the sedimentation pattern of PMR60o that parallels that of albumin mRNA, with that portion of cellular PMR60o that sedimented with the heavy polysome fractions redistributing to 80S and somewhat larger complexes. This result is consistent with the stress-induced formation of a macromolecular complex containing PMR60o and albumin mRNA. As noted above, when polysomes are dissociated with EDTA or puromycin, the mRNP that contains PMR60o and albumin mRNA (complex I) sediments at ∼680 kDa on glycerol gradients, with the remaining protein sedimenting primarily in a ∼140-kDa complex (complex II). To determine if arsenite stress significantly altered the distribution of PMR60o between these complexes, extracts from control and arsenite-treated cells were treated with EDTA and separated on glycerol gradients (Fig. 2B). Although one fraction from the gradient of arsenite-treated cells was lost, overall the distributions of PMR60o are similar for the two samples. Thus, each of the PMR60o-containing complexes is intact in stressed cells.
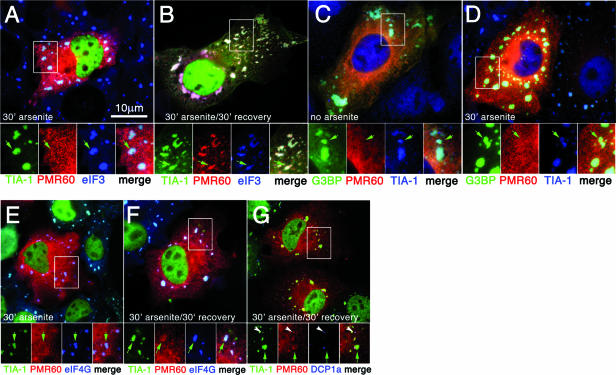
FIG. 2.
Stress causes delayed recruitment of PMR60o into stress granules. Cos-7 cells were transfected with plasmids encoding the indicated proteins, treated as indicated in each panel, and processed for immunofluorescence. A and B. Cells expressing GFP-TIA-1 (green) and PMR60o (red), counterstained for endogenous eIF-3b (blue). C and D. Cells expressing GFP-G3BP1 (green) and PMR60o (red), counterstained for endogenous TIA-1. E and F. Cells expressing PMR60o (red), counterstained for endogenous TIA-1 (green) and eIF-4G (blue). G. Cells expressing PMR60o (red), counterstained for endogenous TIA-1 (green) and DCP1a (blue). Bar, 10 μm. Boxed areas are shown enlarged below the corresponding panels, with separate color channels and a merged view. Green arrows indicate positions of SGs; white arrowheads indicate PBs.
Arsenite-induced cell stress results in the delayed accumulation of PMR60o in stress granules.
PMR60o is seen to distribute uniformly throughout the cytoplasm when cells expressing this protein or a GFP-PMR60o fusion are examined by fluorescence microscopy (38). To examine whether PMR1 can be recruited to SGs, Cos cells were transfected with a catalytically inactive form of the mature protein bearing an N-terminal myc tag (myc-PRM60o) and GFP-TIA-1, a protein marker of SGs and a facilitator of SG assembly. Following 30 min of treatment with 500 mM sodium arsenite, virtually 100% of the transfected cells display SGs containing GFP-TIA-1 (Fig. 2A). Counterstaining these cells with another marker of SGs (eIF-3) confirms that these large TIA-1-positive foci are bona fide SGs. Despite the robust recruitment of both GFP-TIA-1 and eIF-3 to SGs, PMR60° was not recruited to SGs during this initial period of SG assembly (Fig. 2A, inset). However, clear recruitment of PMR60o to SGs was seen (Fig. 2B, inset) in 30 to 50% of the transfected cells when cells were treated with arsenite for 30 min, washed, and subsequently allowed to recover for 30 min in arsenite-free medium (Fig. 2B). This observation suggests that SG assembly per se is not sufficient to induce recruitment of PMR60o to SGs but that some later event triggered by arsenite is involved in recruiting PMR60° to SGs. To test this hypothesis, cells were transfected with plasmids expressing PMR60° and GFP-G3BP1, a potent inducer of SG assembly whose overexpression is able to induce SGs without arsenite treatment (21, 33). As expected, cells expressing GFP-G3BP1 (Fig. 2C) exhibit large SGs which also contain eIF-3; however, no recruitment of PMR60o (Fig. 2C) to SGs is observed in these cells. Arsenite treatment of GFP-G3BP1/PMR60o transfectants for 30 min (Fig. 2D) did not result in the recruitment of PMR60o into SGs, nor did we observe PMR60o recruitment to SGs in these cells following washout and recovery (not shown), suggesting that recruitment of PMR60o to SGs involves a direct stoichiometric interaction between PMR60o and TIA-1. This was confirmed by experiments in which PMR60o was cotransfected with other SG-inducing proteins, such as FAST, TTP, and SMN, which also induced the formation of SGs lacking PMR60o (data not shown). Consistent with this idea, in cells transfected with PRM60o and empty vector, arsenite treatment (Fig. 2E) was sufficient to induce SG assembly (assessed by endogenous TIA-1 and confirmed by counterstaining for eIF-4G) but failed to promote PRM60o recruitment to SGs. In cells treated with arsenite followed by 30-min recovery, a fraction of cells (2 to 5%) showed weak recruitment of PMR60o to SGs (Fig. 2F).
Previous work showed that little if any PMR60o is associated with PBs in Cos-1 cells that were cotransfected with enhanced GFP-Dcp1a (38). To test whether this was changed by stress, cells expressing myc-PMR60o (Fig. 1G) were treated with arsenite for 30 min, followed by 30 min of recovery, and then were fixed and stained for myc-PMR60o (Fig. 1G), endogenous TIA-1 (SGs), and the endogenous PB marker protein DCP1a. A slight enrichment of PMR60o at PBs is seen, but expression of PMR60o does not promote PB-SG interactions, as is seen upon overexpression of the mRNA decay proteins TTP and BRF-1 (22). Taken together, our data suggest that arsenite activates an interaction between TIA-1 and PMR1 that occurs independently of SG formation. Thus, the interaction of PMR60o with TIA-1 facilitates its recruitment to SGs, accounting for the higher frequency of SG-associated PMR60o seen in cells also overexpressing TIA-1.
Arsenite-induced stress does not alter the interaction of PMR60 with its substrate-containing mRNP.
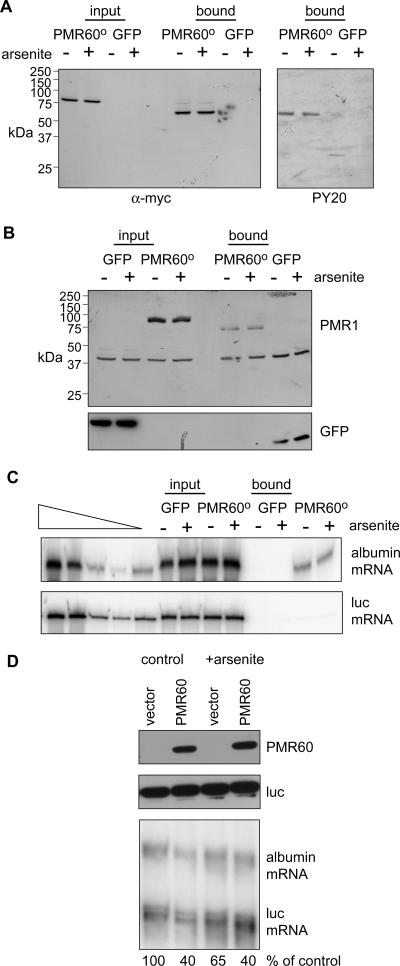
The binding of PMR60o to complex I requires phosphorylation of the tyrosine residue at position 650 (37). To determine whether stress alters PRM60o tyrosine phosphorylation, cytoplasmic extracts from control and arsenite-treated cells expressing myc-PMR60o-TAP or GFP-TAP were absorbed with IgG-Sepharose, washed, and then eluted using Tev protease cleavage (Fig. 3A) (38). In agreement with the glycerol gradient profile in Fig. 1B, Western blots probed with anti-myc or antiphosphotyrosine show that arsenite has no impact on PMR60o tyrosine phosphorylation.
FIG. 3.
Impact of cell stress on the tyrosine phosphorylation of PMR60o, its association with substrate mRNA, and mRNA decay. A. Cells expressing myc-PMR60o bearing a C-terminal TAP tag (myc-PMR60o-TAP) or GFP-TAP were treated without (−) or with (+) arsenite for 30 min, followed by a 30-min recovery period. Cytoplasmic extracts were either analyzed directly or bound to IgG-Sepharose and eluted with Tev protease cleavage. Western blots of input and recovered protein were probed with antibody to the myc tag to visualize PMR60o and with PY20 to visualize tyrosine phosphorylation. B. Cells expressing PMR60o-TAP or GFP-TAP, albumin, and luciferase mRNAs were treated as for panel A, and input protein and protein recovered from IgG-Sepharose after Tev protease cleavage were analyzed by Western blotting using antibody to the myc tag on PMR60o-TAP or GFP. C. Albumin and luciferase mRNAs in the input and bound fractions from panel B were analyzed by semiquantitative RT-PCR (38). The samples under the triangle are twofold dilutions of mRNA standards. D. Cells transfected with empty vector or plasmid expressing catalytically active PMR60 plus plasmids expressing albumin and luciferase mRNAs were treated as described above prior to isolating RNA and protein. Protein samples were analyzed by Western blotting for PMR60 and luciferase (upper panels) and by RNase protection assay for albumin and luciferase mRNAs (lower panel). Phosphorimager analysis was used to normalize the amount of albumin mRNA to luciferase mRNA in each sample, and the quantified results are presented underneath the autoradiogram.
The same approach was employed to determine whether the arsenite-induced cell stress affects the association of PMR1 with its substrate mRNA in complex I (Fig. 3B and C). Cos-1 cells were transfected with PMR60o-TAP or GFP-TAP together with plasmid vectors expressing albumin and firefly luciferase mRNAs. Western blots of input material and TAP-purified complexes (Fig. 3B) confirm that arsenite has no impact on the overall level of either PMR60o-TAP or GFP-TAP in these extracts (input) or on their recovery (bound). Semiquantitative RT-PCR analysis of input RNA shows that arsenite has no effect on the overall amount of albumin or luciferase mRNA expressed in these cells. Importantly, arsenite stress also does not alter the recovery of albumin mRNA with PMR60o-TAP. As shown previously (37, 38), luciferase mRNA is absent from complexes recovered with PMR60o-TAP, and neither albumin nor luciferase mRNA is recovered with GFP-TAP. Taken together, these results indicate that stress does not alter the association of PMR60o with its substrate-containing mRNP.
Given that the association of PMR60o with albumin mRNA remained unchanged, we next asked whether changes in PMR1-mediated mRNA decay could be seen during this brief period of oxidative stress. In the experiment in Fig. 3D, Cos-1 cells were transfected with empty vector or a plasmid vector expressing catalytically active PMR60, luciferase, and albumin mRNAs as described previously (38). Twenty-four hours later they were treated with arsenite, and protein and RNA were recovered for Western blotting and RNase protection assay. As seen above, arsenite treatment has no impact on the amount of PMR60. When normalized to the cotransfected luciferase control, PMR60 caused a 60% reduction in albumin mRNA under the conditions used here (Fig. 3D, lane 2). A similar result was seen after arsenite treatment, indicating that this period of stress neither increased nor significantly inhibited PMR1-mediated mRNA decay.
Stress recruits PMR60o into an RNase-resistant complex with TIA-1.
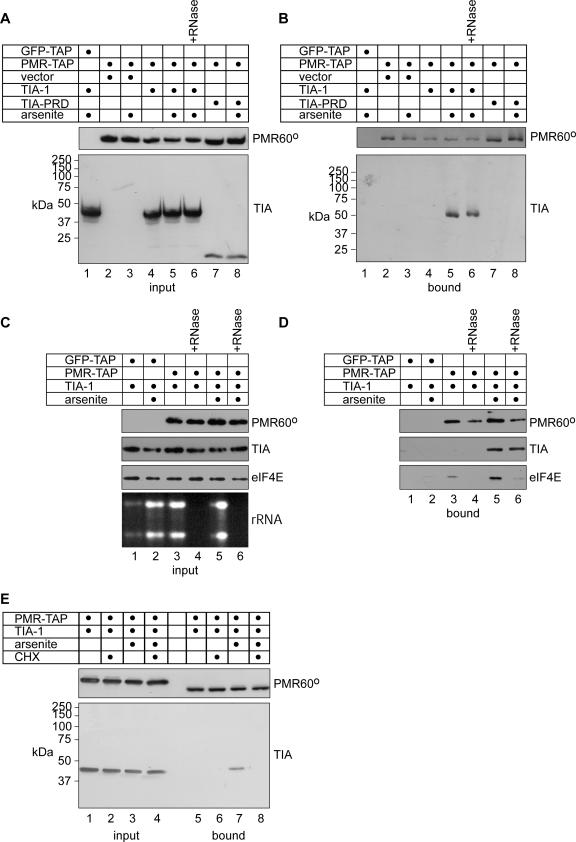
Next we asked whether the accumulation of PMR60o in SGs was a manifestation of the generalized accumulation of nontranslating mRNPs in stressed cells or the product of a specific interaction between PMR60o and one or more TIA proteins. In the experiment shown in Fig. 4, Cos-1 cells were transfected with plasmid vectors expressing HA-TIA-1 plus GFP-TAP or PMR60o-TAP and treated with or without arsenite, followed by 30 min of recovery prior to lysis. Arsenite treatment has no impact on the protein levels of HA-TIA-1 and PMR60o-TAP (Fig. 4A, lanes 4 to 6), and HA-TIA-1 is recovered with PMR60o only following arsenite treatment (Fig. 4B, lane 5), indicating that cell stress is required for the interaction of PMR60o with TIA-1. Treating cytoplasmic extracts with RNase A prior to application to IgG-Sepharose had no impact on the recovery of HA-TIA-1 with PMR60o in arsenite-treated cells (Fig. 4B, lane 6), suggesting either that the stress-induced interaction of these two proteins represents a direct interaction between PMR1 and TIA-1 or that PMR1 and TIA-1 are brought together in SGs as part of a larger protein complex.
FIG. 4.
Stress-induced association of TIA-1 with PMR60o. Cells transfected with the indicated combinations of plasmids expressing GFP-TAP, PMR60o-TAP (PMR-TAP), HA-TIA-1, the TIA-PRD, or vector were treated without or with arsenite for 30 min then allowed to recover for 30 min. A. Western blots of input samples for PMR60o (upper panels), HA-TIA-1, or HA-TIA-PRD. B. The samples in panel A were bound to IgG-Sepharose, recovered by Tev protease cleavage, and assayed as for panel. In lane 6 of panels A and B, cytoplasmic extract was treated with RNase A before binding onto IgG-Sepharose. C. Extracts of cells transfected with GFP-TAP and PMR60o-TAP plus HA-TIA-1 as for panel A were treated without or with RNase A and analyzed by Western blotting for PMR60o, HA-TIA-1, and eIF-4E. rRNA present before and after RNase digestion was detected by ethidium bromide staining of a nondenaturing agarose gel. D. The samples in panel C were recovered on IgG-Sepharose as for panel B and assayed by Western blotting for PMR60o, TIA-1, and eIF-4E. E. Cells coexpressing PMR60o-TAP and HA-TIA-1 were treated with arsenite for 30 min (lanes 3, 4, 7, 8), with cycloheximide (CHX) for 1 h (lanes 2 and 6) or with arsenite for 30 min followed by CHX for 1 h (lanes 4 and 8). Input protein (lanes 1 to 4) and protein recovered from IgG-Sepharose by Tev protease cleavage (lanes 5 to 8) were analyzed as for panel A.
To confirm that this was indeed a direct interaction between these proteins, we looked at the impact of RNase A on the recovery of eIF-4E, a protein recruited to stress granules through its binding to the mRNA cap (Fig. 4C and D). In Fig. 4C, cytoplasmic extracts from cells transfected with the same plasmid vectors as in Fig. 4A and B were treated without (lanes 1, 2, 3, and 5) or with (lanes 4 and 6) RNase A. Western blotting showed that neither arsenite treatment of the cells nor RNase A treatment of extracts had any effect on the overall amounts of PMR60o, TIA-1, and eIF-4E. Native gel electrophoresis of RNA recovered from each of these samples confirmed that RNase A effectively removed RNA present in these samples. Each of these samples was then applied to IgG-Sepharose, and recovered complexes were again analyzed by Western blotting (Fig. 4D). In agreement with the results in Fig. 4B, arsenite treatment resulted in formation of an RNase-resistant complex between PMR60o and TIA-1. In extracts that were not treated with RNase A, eIF-4E was recovered with PMR60o-TAP from control and arsenite-treated cells (lanes 3 and 5); however, this association was lost following digestion with RNase A (lanes 4 and 6). Thus, the stress-induced association of PMR1 with TIA-1 is likely the result of direct interaction between these proteins.
The TIA-1 C terminus has a glutamine-rich prion-like domain that facilitates SG formation through self-aggregation (14). Expression of the TIA-PRD alone effects a dominant-negative inhibition of SG formation (22). In the experiment shown in lanes 7 and 8 of Fig. 4A and B, TIA-PRD was substituted for full-length HA-TIA-1. As one might anticipate from the insoluble properties of this form of TIA-1, the TIA-PRD is not recovered with PMR60o-TAP regardless of arsenite treatment (Fig. 4B, lanes 7 and 8). However, PMR60o-TAP remains soluble, suggesting that it fails to interact with the isolated TIA-1-PRD, as it does with HSP27 and HSP70 (14). This suggests that either SG formation is required for the interaction of PMR60o with TIA-1 or PMR60o binds to a domain of TIA-1 other than the PRD, although we cannot rule out the possibility that the isolated TIA-1-PRD is inaccessible to PMR60o-TAP.
As noted previously, translation elongation inhibitors such as emetine or cycloheximide cause SGs to disappear by shifting the equilibrium of mRNPs to ribosome-bound complexes (21). In the experiment shown in Fig. 4E, cells expressing PMR60o-TAP and HA-TIA-1 were treated with arsenite (lanes 3 and 7) or with arsenite followed by cycloheximide (lanes 4 and 8) and examined for recovery of TIA-1 with PMR60o-TAP. Cycloheximide treatment caused the dissolution of SGs as seen by fluorescence microscopy (not shown) and, concurrently, reversed the arsenite-induced association of PMR60o with HA-TIA-1. Taken together, the results in Fig. 4 indicate that the appearance of PMR60o in SGs represents a specific stress-induced recruitment of this mRNA endonuclease and that PMR60o may interact directly with TIA-1, in a portion of TIA-1 distinct from its PRD.
The N terminus of PMR60o contains a stress-responsive domain.
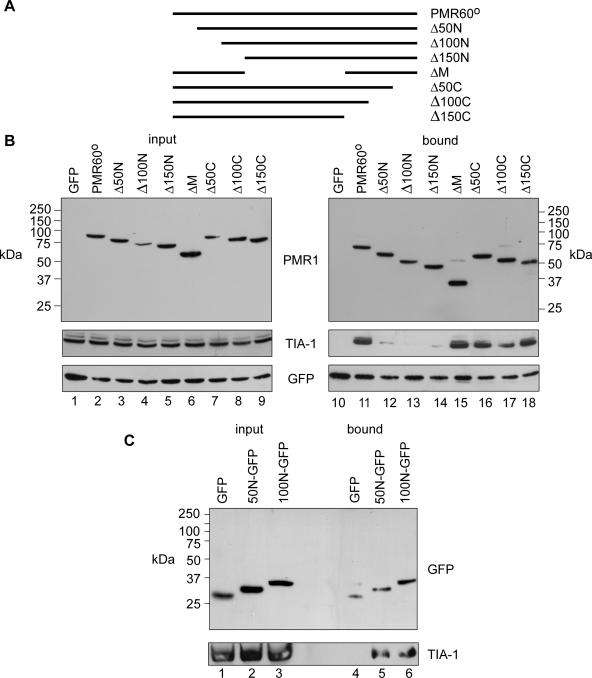
The domain of PMR60° responsible for its arsenite-induced accumulation in SGs was mapped using a series of PMR60° truncations used previously to map the polysome-targeting domains (38). Cos-1 cells were transfected with TAP-tagged fragments of the PMR60° deletion constructs shown in Fig. 5A together with a plasmid expressing TIA-1. As indicated for some samples, GFP-TAP was used as a control to normalize recovery on IgG-Sepharose.
FIG. 5.
Mapping a stress-responsive domain to the N terminus of PMR1. Cells expressing the forms of myc-PMR60o-TAP shown in panel A, GFP-TAP, and HA-TIA-1 were treated with arsenite for 30 min, followed by 30 min recovery prior to harvest. B. Western blotting was performed on input cytoplasmic extracts (left panel) and protein recovered by Tev protease cleavage from IgG-Sepharose (right panel). C. Extracts from cells expressing HA-TIA-1 plus GFP or N-terminal fusions of GFP bearing amino acids 1 to 50 (50N) or 1 to 100 (100N) of PMR60° were analyzed by Western blotting with the indicated antibodies before and after immunoprecipitation with anti-GFP.
Cytoplasmic extracts from arsenite-treated cells were then analyzed for each form of PMR60° and for TIA-1 in complexes recovered by Tev protease cleavage (Fig. 5B). As seen in Fig. 4, TIA-1 is recovered with full-length PMR60o-TAP but not GFP-TAP. The interaction of PMR-TAP with TIA-1 is lost when the N-terminal 50 amino acids of PMR60° are deleted, and recovery is not restored with further deletions of 100 and 150 amino acids from the N terminus. Deleting the central 150 amino acids (ΔM) or up to 150 amino acids from the C terminus from PMR60o has no impact on TIA-1 recovery, supporting the notion that the N terminus has a specific stress-responsive TIA-1-interacting domain. To determine if this domain acts independently of the rest of PMR60°, the N-terminal 50 and 100 amino acids were fused to the N terminus of GFP and transfected together with a TIA-1-expressing plasmid. When cytoplasmic extracts from arsenite-treated cells were immunoprecipitated with antibody to GFP and analyzed by Western blotting, TIA-1 was recovered with both 50N-GFP (Fig. 5C, lane 5) and 100N-GFP (lane 6) but not with GFP (lane 4). Taken together, these data indicate that the N-terminal region of PMR60° is responsible for its stress-induced interaction with TIA-1.
DISCUSSION
Environmental stress disrupts the reciprocal distribution of mRNAs between translating polysomes and untranslated mRNAs, which are routed to specific sites within the cytoplasm. In yeast, nontranslating mRNAs accumulate in PBs (4, 32); however, the situation in mammalian cells is more complex. Mammalian cells display similar PBs containing a cohort of related proteins, but stressors that activate phosphorylation of eIF-2α induce the assembly of larger structures (SGs) not seen in yeast. Unlike PBs, SGs contain poly(A)+ mRNA, poly(A)-binding protein, eIF-4G, eIF-3 and 40S subunits (19, 21). SGs are in equilibrium with polysomes, and SG contents are in rapid flux, leading to the proposal that SGs are subcellular sorting structures, wherein mRNPs are targeted for storage, recycling back to the translating pool, or decay (1). Furthermore, it is likely that subvisible SGs are present in normal cells, where they undergo rapid assembly and disassembly, and their accumulation in stressed cells is a manifestation of the large-scale inhibition of translation and enforced polysome disassembly resulting from the block to ternary complex formation. SGs lack Dcp1, Dcp2, and Xrn1, but the enhanced interaction of SGs with PBs in cells overexpressing TTP or BRF-1 suggests that these proteins accelerate mRNA decay by facilitating the handoff of mRNPs from SGs to PBs (21).
The process of endonuclease-mediated mRNA decay differs fundamentally from that of mRNA decay in PBs. Whereas decay enzymes in PBs act on nontranslating mRNA, PMR1 acts in the context of a selective complex that it forms with its translating substrate mRNA, in which it initiates mRNA decay by cleaving within the mRNA body (15, 38). Given the differences between these modes of mRNA decay, and considering the spatial segregation (be it in SGs or PBs) between translating and nontranslating mRNAs (4, 20), it is important to determine the relationship between PMR1 and these subcellular structures. The results presented here show that PMR1 and its substrate mRNA are recruited to SGs upon environmental stress (Fig. 1 and 2), but despite polysome disassembly, the PMR1-substrate mRNP complex remains intact (Fig. 3). Because of the toxicity of arsenite, arsenite-induced stress is generally limited to 30 to 45 min to avoid triggering apoptosis. Since we commonly assay PMR1-mediated decay of albumin mRNA at 24 to 48 h after transfection, it was not clear if treating cells with arsenite after this interval would produce detectable changes. Indeed, under the conditions used here, we saw neither an increase nor a decrease in the amount of albumin mRNA when cells expressing catalytically active PMR1 were treated with arsenite (Fig. 3D), indicating that PMR1-mediated mRNA decay is not accelerated by stress. At this point we cannot determine if it this process might be slowed by stress, as this will have to be addressed using less toxic agents.
The TIA C-terminal prion-like domain provides the self-aggregation that allows the TIA proteins to assemble stalled initiation complexes into SGs. Since such aggregation is the basis for SG formation, it was important to determine whether the recruitment of PMR1 represents a specific interaction of the mRNA endonuclease with one or more components of SGs or results from generalized nucleation of SG-associated mRNPs. Biochemical evidence of a specific recruitment of PMR1-containing mRNPs by TIA-1 is shown in Fig. 4, which shows that TIA-1 interacts with PMR60° only upon arsenite treatment. Importantly, the recovery of TIA-1 with PMR60° is unchanged upon digestion with an excess of RNase A. These data indicate that there is either a direct interaction between PMR60° and TIA-1 or an indirect interaction of these two proteins as part of a larger protein complex within SGs. SGs are forcibly disassembled by cycloheximide, which freezes translating ribosomes onto mRNA and thus shifts the overall dynamic equilibrium of mRNA between SGs and polysomes (21). Addition of cycloheximide to stressed cells disassembles SGs, returns the pattern of PMR60° staining to a diffuse distribution throughout the cytoplasm, and concurrently blocks the biochemical interaction of TIA-1 with PMR60o (Fig. 4), thus confirming the stress-specific nature of the interaction of PMR60° with TIA-1. The specificity of this interaction was verified (Fig. 4) in cells expressing the TIA-1-PRD, which comprises a glutamine-rich prion-like domain and functions as a dominant negative inhibitor of SG formation (14). Because TIA-PRD does not bind to PMR60° in arsenite-treated cells and prevents assembly of PMR60° into SGs (data not shown), it is likely that the PMR60o interaction with TIA-1 is mediated by sequences other than the TIA C terminus.
A major thrust of our research on PMR1 has been to map the domains responsible for its unique properties. The major polysome-targeting domain lies within the C-terminal ∼70 amino acids (38) and includes a phosphotyrosine at position 650, which together with downstream amino acids match a consensus Src homology 2 ligand (37). We now identify (Fig. 5) a new, stress-responsive domain in the first 50 amino acids that mediates binding toTIA-1 and recruitment of PMR60° into SGs. This domain likely defines a novel interacting site, as bioinformatics analyses predict only a putative phosphorylation site in this region. Because the interaction of PMR60° with TIA-1 is unaffected by RNase treatment, it seems likely that either PMR60° interacts directly with TIA-1 or the two proteins interact with a common partner. We were unsuccessful in attempts to recover in vitro-translated PMR60° by GST pulldown using bacterially expressed GST-TIA-1; this might be due to the lack of posttranslational modifications on either protein or to the involvement of another interacting protein(s). In this regard, it is interesting to note that phosphorylation of some SG-associated proteins regulates their recruitment to SGs. In one example, the arsenite-induced phosphorylation of TTP, an SG-associated RNA-destabilizing factor, promotes its rapid egress from SGs and concurrently inhibits its ability to promote ARE-mediated mRNA decay (30). Similarly, the phosphorylation of serine 149 in G3BP, another SG-associated protein, prevents its targeting to SGs (33). Mechanistically, however, these proteins differ from PMR1 in that the overexpression of either TTP or G3BP promotes SG assembly, whereas PMR1 overexpression does not. Moreover, other stimuli which promote SG assembly (energy starvation and heat shock) do not promote recruitment of PMR60° to SGs. As arsenite is known to activate a number of kinase cascades, the data suggest that phosphorylation of PMR60o within its N-terminal region may be required for its stress-specific interaction with TIA-1.
Finally, we consider the relationship between PMR1, SGs, and PBs. Previous work has shown that arsenite-induced SGs interact with PBs and that SG-PB interactions are also promoted by expression of activators of ARE-mediated mRNA decay such as TTP and BRF-1, which are themselves components of both SGs and PBs (21). Following arsenite treatment and recovery, PRM60o assembles into SGs and PBs but does not promote a consistent association between the two structures (Fig. 2G). This is very different from the fusion between SGs and PBs that results from overexpressing TTP or BRF-1 (21). Thus, it appears that during stress, PMR1 endonuclease-mediated decay is spatially segregated within SGs or PBs, while decapping-associated mRNA decay is sequestered in PBs. G3BP, another protein with endonucleolytic activity in vitro, is also routed to SGs but is excluded from PBs during stress. However, unlike PMR1, G3BP contains an RNA recognition motif and a self-aggregation domain (33), allowing it to nucleate SGs directly using a mechanism similar to that of the TIA proteins. Moreover, G3BP does not interact with TIA-1 and may nucleate some SGs that do not contain TIA-1 (33). PMR1 is the first known effector endonuclease containing a specific domain involved in targeting to SGs, supporting the notion that SGs are dynamic sites for mRNP sorting and raising the interesting possibility that they may determine the decay pathway for particular mRNPs.
Acknowledgments
We thank Jens Lykke-Andersen for antibody to Dcp1a, Jamal Tazi for GFP-G3BP, and members of the Schoenberg lab for their helpful comments.
This work was supported by NIH grants GM38277 to D.R.S. and AI33600 to N.K. Support for core facilities was provided by center grant P30 CA16058 to the OSU Comprehensive Cancer Center.
Footnotes
Published ahead of print on 18 September 2006.
REFERENCES
- 1.Anderson, P., and N. Kedersha. 2002. Stressful initiations. J. Cell Sci. 115:3227-3234. [DOI] [PubMed] [Google Scholar]
- 2.Andrei, M. A., D. Ingelfinger, R. Heintzman, T. Achesel, R. Rivera-Pomar, and R. Lührmann. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11:717-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashkirov, V. I., H. Scherthan, J. A. Solinger, J. M. Buerstedde, and W. D. Heyer. 1997. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 136:761-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brengues, M., D. Teixeira, and R. Parker. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310:486-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, C. Y., R. Gherzi, S. E. Ong, E. L. Chan, R. Raijmakers, G. J. Pruijn, G. Stoecklin, C. Moroni, M. Mann, and M. Karin. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107:451-464. [DOI] [PubMed] [Google Scholar]
- 6.Chernokalskaya, E., A. N. DuBell, K. S. Cunningham, M. N. Hanson, R. E. Dompenciel, and D. R. Schoenberg. 1998. A polysomal ribonuclease involved in the destabilization of albumin mRNA is a novel member of the peroxidase gene family. RNA 4:1537-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coller, J., and R. Parker. 2005. General translational repression by activators of mRNA decapping. Cell 122:875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cougot, N., S. Babajko, and B. Seraphin. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165:31-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eystathioy, T., E. K. L. Chan, S. A. Tenenbaum, J. D. Keene, K. Griffith, and M. J. Fritzler. 2002. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell 13:1338-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eystathioy, T., A. Jakymiw, E. K. L. Chan, B. Séraphin, N. Cougot, and M. J. Fritzler. 2003. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 9:1171-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferraiuolo, M. A., S. Basak, J. Dostie, E. L. Murray, D. R. Schoenberg, and N. Sonenberg. 2005. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 170:913-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallouzi, I., F. Parker, K. Chebli, F. Maurier, E. Labourier, I. Barlat, J. P. Capony, B. Tocque, and J. Tazi. 1998. A novel phosphorylation-dependent RNase activity of GAP-SH3 binding protein: a potential link between signal transduction and RNA stability. Mol. Cell. Biol. 18:3956-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gherzi, R., K. Y. Lee, P. Briata, D. Wegmuller, C. Moroni, M. Karin, and C. Y. Chen. 2004. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 14:571-583. [DOI] [PubMed] [Google Scholar]
- 14.Gilks, N., N. Kedersha, M. Ayodele, L. Shen, G. Stoecklin, L. M. Dember, and P. Anderson. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15:5383-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanson, M. N., and D. R. Schoenberg. 2001. Identification of in vivo mRNA decay intermediates corresponding to sites of in vitro cleavage by polysomal ribonuclease 1. J. Biol. Chem. 276:12331-12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollien, J., and J. S. Weissman. 2006. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313:104-107. [DOI] [PubMed] [Google Scholar]
- 17.Ingelfinger, D., D. J. Arndt-Jovin, R. Luhrmann, and T. Achsel. 2002. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8:1489-1501. [PMC free article] [PubMed] [Google Scholar]
- 18.Kedersha, N., and P. Anderson. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963-969. [DOI] [PubMed] [Google Scholar]
- 19.Kedersha, N., S. Chen, N. Gilks, W. Li, I. J. Miller, J. Stahl, and P. Anderson. 2002. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Cell. Biol. 13:195-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kedersha, N., M. R. Cho, W. Li, P. W. Yacono, S. Chen, N. Gilks, D. E. Golan, and P. Anderson. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151:1257-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kedersha, N., G. Stoecklin, M. Ayodele, P. Yacono, J. Lykke-Andersen, M. J. Fitzler, D. Scheuner, R. J. Kaufman, D. E. Golan, and P. Anderson. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kedersha, N. L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147:1431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laroia, G., R. Cuesta, G. Brewer, and R. J. Schneider. 1999. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284:499-502. [DOI] [PubMed] [Google Scholar]
- 24.Liu, J., M. A. Valencia-Sanchez, G. J. Hannon, and R. Parker. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7:719-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukherjee, D., M. Gao, J. P. O'Connor, R. Raijmakers, G. Pruijn, C. S. Lutz, and J. Wilusz. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21:165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pastori, R. L., J. E. Moskaitis, S. W. Buzek, and D. R. Schoenberg. 1991. Coordinate estrogen-regulated instability of serum protein-coding messenger RNAs in Xenopus laevis. Mol. Endocrinol. 5:461-468. [DOI] [PubMed] [Google Scholar]
- 27.Pastori, R. L., J. E. Moskaitis, and D. R. Schoenberg. 1991. Estrogen-induced ribonuclease activity in Xenopus liver. Biochemistry 30:10490-10498. [DOI] [PubMed] [Google Scholar]
- 28.Sen, G. L., and H. M. Blau. 2005. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 7:633-636. [DOI] [PubMed] [Google Scholar]
- 29.Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoecklin, G., T. Stubbs, N. Kedersha, S. Wax, W. F. Rigby, T. K. Blackwell, and P. Anderson. 2004. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23:1313-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taupin, J. L., Q. Tian, N. Kedersha, M. Robertson, and P. Anderson. 1995. The RNA-binding protein TIAR is translocated from the nucleus to the cytoplasm during Fas-mediated apoptotic cell death. Proc. Natl. Acad. Sci. USA 92:1629-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teixeira, D., U. Sheth, M. A. Valencia-Sanchez, M. Brengues, and R. Parker. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tourrière, H., K. Chebli, L. Zekri, B. Courselaud, J. M. Blanchard, E. Bertrand, and J. Tazi. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823-831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.van Dijk, E., N. Cougot, S. Meyer, S. Babajko, E. Wahle, and B. Seraphin. 2002. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21:6915-6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, Z., and M. Kiledjian. 2000. The poly(A)-binding protein and an mRNA stability protein jointly regulate an endoribonuclease activity. Mol. Cell. Biol. 20:6334-6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, Z., and M. Kiledjian. 2001. Functional link between the mammalian exosome and mRNA decapping. Cell 107:751-762. [DOI] [PubMed] [Google Scholar]
- 37.Yang, F., Y. Peng, and D. R. Schoenberg. 2004. Endonuclease-mediated mRNA decay requires tyrosine phosphorylation of polysomal ribonuclease 1 (PMR1) for the targeting and degradation of polyribosome-bound substrate mRNA. J. Biol. Chem. 279:48993-49002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, F., and D. R. Schoenberg. 2004. Endonuclease-mediated mRNA decay involves the selective targeting of PMR1 to polyribosome-bound substrate mRNA. Mol. Cell 14:435-445. [DOI] [PubMed] [Google Scholar]