Abstract
A region encompassing the rat aldolase B gene (aldB) promoter acts as a chromosomal origin of DNA replication (origin) in rat aldolase B-nonexpressing hepatoma cells. To examine replicator function of the aldB origin, we constructed recombinant mouse cell lines in which the rat aldB origin and the mutant derivatives were inserted into the same position at the mouse chromosome 8 by cre-mediated recombination. Nascent strand abundance assays revealed that the rat origin acts as a replicator at the ectopic mouse locus. Mutation of site C in the rat origin, which binds an Orc1-binding protein AlF-C in vitro, resulted in a significant reduction of the replicator activity in the mouse cells. Chromatin immunoprecipitation (ChIP) assays indicated that the reduction of replicator activity was paralleled with the reduced binding of AlF-C and Orc1, suggesting that sequence-specific binding of AlF-C to the ectopic rat origin leads to enhanced replicator activity in cooperation with Orc1. Involvement of AlF-C in replication in vivo was further examined for the aldB origin at its original rat locus and for a different rat origin identified in the present study, which contained an AlF-C-binding site. ChIP assays revealed that both replication origins bind AlF-C and Orc1. We think that the results presented here may represent one mode of origin recognition in mammalian cells.
Proteins involved in the initiation of DNA replication and the function of individual replication-related proteins appear to be conserved among eukaryotic species. However, mechanism of replication initiation seems to vary, even in the simple eukaryotic cells (1, 9, 14, 22). In budding yeast Saccharomyces cerevisiae, direct binding of the origin recognition complex (ORC) to specific short DNA sequences called ACS is a prerequisite for the initiation of replication (8, 38, 39, 51). In Schizosaccharomyces pombe, however, sequences required for replication initiation are larger than those in S. cerevisiae and lack common unique sequences such as ACS. In addition, the ORC in S. pombe shows lower sequence specificity in its DNA binding. It binds to multiple asymmetric AT-rich sites in the replication origins (12, 25, 47).
The heterohexameric ORC is an important initiator protein that provides a site (replication origin) on the chromosomes where additional factors, including Cdc6, Cdt1, and Mcm complex, can be recruited for the formation of prereplication complex (pre-RC) (7). Again, this functional feature of ORC appears to be common from yeast to human. Nevertheless, the mechanism of origin determination in higher eukaryotes differs from that in yeasts. For instance, mammalian ORC does not show sequence specificity in its DNA binding; even it binds to random sequences (24, 49). It is has also been shown that the DNA sequence is not a critical determinant for ORC-DNA binding in Drosophila (40). In addition, there are no unique sequences apparently conserved in the origin sequences thus far determined for mammalian cells (9, 22). Despite these facts, ORC does not bind randomly to chromosomes but does to discrete sites that serves as origins in vivo (6, 19, 23, 28). Therefore, it is still an open question how mammalian cells select chromosomal DNA sites as replication origins. To this point, You et al. (55) proposed that selective activation by T-rich single strand of Mcm helicase loaded onto chromatin may be a determinant for selection of initiation sites, since the T-rich stretches are often seen in replication origins. However, another explanation might not be excluded that ORC interacts with proteins that bind to particular DNA sequences in the origins and provides sites for pre-RC formation. Such a case has been shown for replication of the Epstein-Barr (EB) virus genome, where the virus protein EBNA1 binds to the viral origin (oriP) and recruits the host cell ORC (11, 15, 44). It was also shown that an artificial origin could be created at the GAL4 binding site in a plasmid DNA via binding of GAL4-ORC subunit fusion proteins (48). These results imply that indirect association of ORC with the origins via other DNA-binding proteins can provide sites for replication initiation. Such a mechanism, however, is yet to be generally accepted as a cellular event occurring at mammalian chromosomal origins. To address this issue, it is still important to clarify whether specific DNA sequences are needed for accurate, efficient, and controlled initiation of replication at each mammalian origin.
A region encompassing the promoter of the rat aldolase B gene (aldB) acts as a replication origin in vivo in the rat aldolase B-nonexpressing rat hepatoma cells (34, 56). The aldB origin, in a plasmid form, exhibited autonomously replicating sequence (ARS) activity in mammalian cells (33, 57). It has been shown that site C in the aldB origin, which behaved as an element of ARS, binds two closely related proteins termed AlF-C1 and AlF-C2 (53) (together we refer to them as AlF-C). AlF-C specifically bound in vitro to Orc1, the largest subunit of ORC (42). This interaction is mediated between a C-terminal domain of AlF-C and a domain in Orc1, which is located on the C terminal side of the BAH domain (10). The latter Orc1 domain was found in Orc1 from humans, rats, mice, and Chinese hamsters but was absent in those from Drosophila, Xenopus, and yeasts (42; Y. Saitoh et al., unpublished data). These observations implied distinguished roles of Orc1 in mammalian cells and led to speculation that the site C-binding protein AlF-C may be one candidate of sequence-specific DNA-binding proteins that bind to the origins and help associating ORCs nearby (42).
In the present study, we examined whether the aldB origin DNA directs initiation of replication when positioned at different chromosomal locations, that is, whether the initiation of replication from the origins depends basically on the DNA sequence. In these experiments, we focused on the function of site C, because it has been considered to be an important site for ORC association. The results showed that the rat aldB origin exhibited replicator activity at an ectopic mouse chromosomal location. Mutation of site C caused reduced binding of AlF-C, which paralleled a significant decrease in the replicator activity and in the binding of Orc1 to the origin. The possible involvement of AlF-C in replication initiation was further examined for the aldB origin at its original location in the rat chromosome and for a different origin identified in the present study, which contained the AlF-C-binding sequence. Chromatin immunoprecipitation (ChIP) assays revealed that both origins bind AlF-C and Orc1.
MATERIALS AND METHODS
Plasmid construction.
The lox targeting vector pBS226 (18) was kindly provided by Bristol-Myers Squibb Pharma Company. The aldB origin DNA, which corresponds to the 0.94-kb HindIII fragment extending from −675 to +263 of the aldB gene (33, 57), was inserted at the HindIII site of the pBS226 and designated pBS226/aldB (see also Fig. 1). Construction of the aldB origin DNA carrying mutation at site C was previously described (33). This mutant origin was inserted at the HindIII site of the pBS226 vector in the same orientation as pBS226/aldB (termed pBS226/mutC). The control pBS226 DNA without the aldB origin was designated pBS226/null.
FIG. 1.
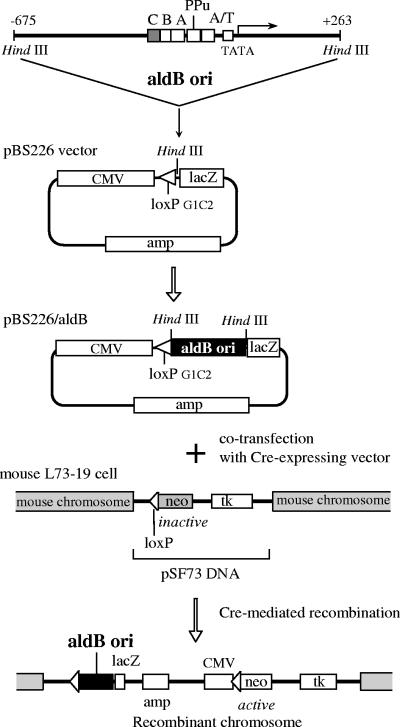
Cre recombinase-mediated site-specific insertion of the rat aldB origin DNA into a mouse chromosome. The structure of the 0.94-kb aldB origin DNA fragment and the targeting vector pBS226 are shown at the top. The transcription start site of the aldolase B gene is denoted as +1. The boxes represent cis-elements of transcription and/or ARS activity. Site C, which was examined for its involvement in replication initiation in the present study, is indicated by a gray box. The aldB origin DNA was inserted at the HindIII site juxtaposed to loxPG1C2 sequence in the pBS226 vector. The targeting vector pBS226 carrying the aldB origin or its derivatives was introduced into the mouse recipient L73-19 cells (18), which carry a single copy of pSF73 DNA containing loxP site in the chromosome, by cotransfection with a cre recombinase-expressing vector. The resulting mouse chromosomal DNA carrying the wild-type aldB origin (pBS226/aldB), the aldB origin with mutated site C (pBS226/mutC [see the text]), and the DNA lacking the 0.94-kb aldB origin (pBS226/null) are shown at the bottom. Cells carrying these recombinant chromosomes were termed cBS226/aldB, cBS226/mutC, and cBS226/null, respectively.
Recombinant cell lines.
The targeting vectors pBS226/aldB, pBS226/mutC, and pBS226/null were introduced into mouse L73-19 cells (a gift from S. Fukushige, Tohoku University). This cell line has been derived from mouse L cells lacking the thymidine kinase (tk) gene and has a single-copy insertion of pSF73 DNA carrying the loxP-neo target site in one of the chromosome (17, 18). Chromosomal site-specific recombination was carried out according to the procedures described by Fukushige and Ikeda (17), using the synthetic Cre recombinase (sCre) gene optimized for mammalian codon usage (26). The sCre gene fragment was inserted into EcoRI-XhoI site of pcDNA3.1Zeo(+) (Invitrogen). Then, 10 μg of each targeting vector and 20 μg of sCre recombinase expression vector were cotransfected into 107 cells by electroporation in 0.8 ml of the HeBS buffer containing 20 mM HEPES-KOH (pH 7.05), 137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, and 6 mM dextrose. Two days after transfection, stable transformants were selected in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, HAT (0.1 mM hypoxanthine, 0.4 μM aminopterin, and 16 μM thymidine), and 0.35 mg of Geneticin (G418; Invitrogen)/ml. The G418-resistant colonies grown at about 2 weeks after transfection were selected. Correct single-copy insertion of the targeting vectors into a mouse chromosome was confirmed by Southern blot analyses.
Isolation of RNA-primed short nascent DNA strands.
Total genomic DNA from each recombinant cell line was prepared as previously described (34). DNA was denatured by incubation in boiling water for 10 min, followed by rapid cooling in ice-cold water. Then, DNA was loaded onto a 5 to 25% linear sucrose gradient in Tris-EDTA (TE) containing 0.3 M NaCl and then centrifuged at 89,000 × g for 20 h at 20°C by using an SW28 rotor (Beckman). In a separate tube, heat-denatured DNA fragments ranging from 106 bp to 8,064 bp prepared from several kinds of plasmid DNA were simultaneously centrifuged for size markers. Fractions were collected from the top of the tube by using a fractionator (piston gradient fractionator; Biocomp Instrument, Inc.). The distribution of size markers was determined by agarose gel electrophoresis. DNA fractions with sizes ranging from 0.5 to 1 kb were pooled and precipitated in ethanol. The size-fractionated DNA was boiled for 2 min and phosphorylated by using T4 polynucleotide kinase and 50 μM ATP at 37°C for 30 min. As an internal control, linear plasmid DNA (pGL3; Promega), which was treated with calf intestinal alkaline phosphatase, was included. 5′-Phosphorylated DNA was extracted with phenol-chloroform, precipitated in ethanol, and incubated with 30 U of λ exonuclease (New England Biolabs) at 37°C for 3 h in 67 mM glycine-KOH buffer (pH 9.4) containing 2.5 mM MgCl2 and 50 μg of bovine serum albumin/ml. DNA was extracted with phenol-chloroform, precipitated in ethanol, and resuspended in TE. The exonuclease digestion was monitored by measuring the disappearance of the plasmid DNA, which was included in the reaction mixture as an internal control (34).
Quantitative real-time PCR.
Quantitative real-time PCR was performed with a LightCycler instrument (Roche Molecular Biochemicals) by using a ready-to-use hot start reaction mix (QuantiTect SYBR green PCR master mix; QIAGEN). PCRs were set up in 20 μl of the reaction mix including 0.5 μM concentrations of each primer and the fixed amounts of λ exonuclease-treated nascent strands (equivalent to 20 μg of host cell genomic DNA) as a template DNA. PCR were performed for 50 cycles under the standard condition recommended by Roche Molecular Biochemicals. The primers and their annealing temperatures are listed in Table 1. As a standard DNA for quantification, genomic DNA from the cBS226/aldB or dRLh84 cells was sequentially diluted to yield 50 ng, 5 ng, 0.5 ng, 50 pg, and 5 pg per reaction mix. After PCR, the cycle in which fluorescence from the real-time PCR crossed the set threshold was plotted against the logarithm of concentration to produce a standard curve according to the supplier's protocol (Roche Molecular Biochemicals). The relative abundance of the nascent strands was determined by extrapolation from the standard curve and was expressed as values relative to that derived from 5 pg of genomic DNA for each primer set.
TABLE 1.
Nucleotide sequences of PCR primers used in this study
| Primer set | Orientationa | Nucleotide sequence (5′ to 3′) | Product size (bp) | Annealing temp (°C) | Positionb |
|---|---|---|---|---|---|
| 1 | FD | ACATTCTCACCTGTCGTCCTGAGC | 163 | 55 | Mouse chromosome, 9.4 kb upstream of loxP site |
| RV | TCTTTGTTTGCCATGACCTTCACAG | ||||
| 2 | FD | AGCCCAGTAGAAGAGGCTGTTGG | 163 | 56 | Mouse chromosome, 1.5 kb upstream of loxP site |
| RV | CGGGTATCGGTGTCGAGTTTCTTG | ||||
| 3 | FD | GCGTTTCGGTGATGACGGTG | 160 | 63 | pSF73 vector, 0.5 kb upstream of loxP site |
| RV | ATCGCTACGTGACTGGGTCATGG | ||||
| 4 | FD | TCAGTCAACCCCCAACCCAGAG | 259 | 62 | aldB origin encompassing site C, rat specific |
| RV | CAATAACTCTGATTGGCGCGTG | ||||
| 5 | FD | CTGGCCGTCGTTTTACAACGTCGTGAC | 157 | 62 | lacZ juxtaposing to the aldB origin |
| RV | GCCATTCGCCATTCAGGCTGCGCAAC | ||||
| 6 | FD | TAACGCCAATAGGGACTTTCCATTGACGTC | 91 | 60 | 5′ portion of the CMV gene in pBS226 |
| RV | GGCATATGATACACTTGATGTACTGCCAAG | ||||
| 7 | FD | GGAACTGATGAATGGGAGCA | 158 | 55 | Between the neo and tk genes in pSF73 |
| RV | GGAAAGTCCTTGGGGTCTTC | ||||
| 8 | FD | GTGTTTGCCTGGGCCTTGGACGTCTTG | 177 | 63 | 3′ portion of the tk gene in pSF73 |
| RV | GCCAGGTCGCAGATCGTCGGTATGGAG | ||||
| 9 | FD | GACTCCCTTGTAGCTCTGAC | 112 | 50 | Rat aldB origin |
| RV | CCAGTAGGGAGGTGTTTATT | ||||
| 10 | FD | TCACAATCATCACTATGGCATCC | 113 | 55 | 2 kb upstream of site C in the rat aldB origin |
| RV | CCTCCTTTATCACCTGCCTCAG | ||||
| 11 | FD | TTTAGAGAACAGGCAGCAAGTGG | 119 | 53 | 0.4 kb upstream of site C in the rat aldB origin |
| RV | ATAAAGTCTACATACACAAGACACTGC | ||||
| 12 | FD | CAGATGAGTCTGTGGGTGAGTG | 115 | 60 | 4.5 kb downstream of site C in rat aldB origin (+4.5 kb) |
| RV | CTGTGTGGAAGGAGACTGATTG | ||||
| 13 | FD | CCAACGCTCTGGCTCGCTAC | 116 | 60 | 8 kb downstream of site C in rat aldB origin (+8 kb) |
| RV | ATGAAATTGAAAAGCGGGCTCTG | ||||
| 14 | FD | ACTGTGTGCCAGGCATGGTATTC | 129 | 60 | Site C in clone 52 DNA |
| RV | GTCTATATTCCTAAAGGGGAAGTTG | ||||
| 15 | FD | GCGCTAAGAAGGTGTTTCCTTG | 138 | 55 | 4 kb upstream of site C in clone 52 DNA (−4 kb) |
| RV | TGGAACCCAAGTCCCCTTTC | ||||
| 16 | FD | TTGCCATTCCTTTCTTATCCTTG | 113 | 55 | 2 kb upstream of site C in clone 52 DNA (−2 kb) |
| RV | TGCAGATTGGGTGGTCCTAAG | ||||
| 17 | FD | GACTTGCCAAAGGAATGGTGAC | 107 | 55 | 1 kb downstream of site C in clone 52 DNA (+1 kb) |
| RV | CCGAGCTGCTTGATTTGTTAGC | ||||
| 18 | FD | GCCTGTGCTCTGTCCCTCCT | 109 | 60 | 2 kb downstream of site C in clone 52 DNA (+2 kb) |
| RV | AGCCGCATGAACCTCTCTCT | ||||
| 19 | FD | TGGCTTTGTGGCTAGAATATGGA | 87 | 55 | 4 kb downstream of site C in clone 52 DNA (+4 kb) |
| RV | CACGTCACCCAACAGTAGCAAAG | ||||
| 20 | FD | ACAAGGACTCCCTGCCTCAA | 236 | 60 | Xic nonorigin region (amplicon 1 in reference 41) |
| RV | CCCGGTTTCCACACTGATTT | ||||
| 21 | FD | AGCCCTGTGTGCATTTAGCA | 242 | 61 | Xist origin region (amplicon 13 in reference 41) |
| RV | TGCAGAGGTTTTTGGCTGAA |
Preparation of anti-rat AlF-C and anti-rat Orc1 antibodies.
The DNA fragment encoding the rat AlF-C1 N-terminal region (amino acids 1 to 155; database accession no. AB016536) was inserted into pGEX-6P-3 (named pGEX/AlF-C1-NTD) (GE Healthcare). The GST-tagged N-terminal portion of AlF-C1 in pGEX/AlF-C1-NTD was expressed in Escherichia coli BL21(DE3)/pLysS and purified through glutathione-Sepharose 4B (GE Healthcare) as previously described (42). The purified protein was digested with PreScission protease (GE Healthcare) for 16 h at 4°C. The glutathione S-transferase (GST) tag was removed by centrifugation for 15 min at 12,000 × g, followed by denaturation at 80°C for 10 min. The supernatant containing the AlF-C1 N-terminal fragment was recovered and used for raising antibodies in Japanese white rabbits. Anti-rat Orc1 antibodies were raised against synthetic rat Orc1 peptide from amino acid positions 376 to 396. Rat Orc1 cDNA has been cloned and sequenced in our laboratory (database accession no. BAC65338).
ChIP.
The ChIP assay was performed essentially as described by Ikeda et al. (21). Approximately 107 cells were fixed in 1% formaldehyde for 10 min at room temperature. After two washes with ice-cold phosphate-buffered saline (PBS), the cells were harvested and lysed in a lysis buffer (50 mM Tris-HCl [pH 8.0] containing 1% sodium dodecyl sulfate [SDS], 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 10 μg each of the proteinase inhibitors leupeptin, chymostatin, pepstatin A, and antipain/ml). The lysate was then sonicated to shear DNA to be an average of 1.5 kb. The chromatin fraction was obtained by centrifugation at 15,000 × g for 10 min. The supernatant was diluted 10-fold with immunoprecipitation buffer (16.7 mM Tris-HCl [pH 8.0] containing 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, and the protease inhibitors described above). For the ChIP assay, the sonicated chromatin preparation (usually 30 to 35 μg of DNA as measured by the absorbancy at 260 nm, after RNase treatment, proteinase treatment, phenol extraction, and ethanol precipitation) was first incubated with protein A-Sepharose to exclude proteins that nonspecifically bind to protein A. The pretreated chromatin was then incubated with rabbit anti-rat AlF-C (50 μl of serum), anti-rat Orc1 (15 μg of affinity-purified immunoglobulin G [IgG]), or control preimmune IgG (15 μg) at 4°C overnight using rotary shaker, followed by incubation with protein A-Sepharose for 2 h. After incubation, the protein A-Sepharose beads were washed sequentially with wash buffer (20 mM Tris-HCl [pH 8.0] containing 0.1% SDS, 1% Triton X-100, and 2 mM EDTA) containing 150 mM NaCl, with wash buffer containing 500 mM NaCl, with LiCl buffer (10 mM Tris-HCl [pH 8.0] containing 0.25 M LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, and 1 mM EDTA), and twice with TE. For the sequential ChIP assay, chromatins trapped by the first antibodies were detached from protein A-Sepharose, and the bound antibodies were inactivated by incubation for 15 min at room temperature in reimmunoprecipitation buffer (10 mM dithiothreitol, 1% SDS, and 0.1 M NaHCO3). The chromatin solution was then diluted with 10 times the volume of the immunoprecipitation buffer and incubated with the second antibodies. DNA in the immunoadsorbed chromatin was released from the beads by incubation at 65°C for 4 h with 10 mM Tris-HCl buffer (pH 8.0) containing 300 mM NaCl, 5 mM EDTA, and 0.5% SDS, followed by proteinase K digestion (200 μg/ml). DNA was extracted with phenol-chloroform, precipitated in ethanol, and resuspended in TE.
Cloning of genomic DNA fragments containing AlF-C-binding site.
DNA fragments that contain AlF-C-binding site were cloned essentially as described by Cuvier et al. (13). Rat genomic DNA was digested with HaeIII and RsaI. The digest was ligated with linker oligonucleotides (13) and then concentrated for site C-containing DNA fragments through GST pull-down, using GST-fused full-length AlF-C2. DNA fragments recovered were confirmed for their specific binding to AlF-C2 by using GST-fused AlF-C N-terminal domain (amino acids 1 to 147) as a negative control. The DNA fragments were then PCR amplified using the linker sequence as a primer. After seven cycles of the GST pulldown, DNA fragments was inserted into pUC19 and transfected into E. coli XL1-Blue. Each plasmid was selected for AlF-C-binding by electrophoretic mobility shift assay (EMSA) by using GST-fused AlF-C2.
RESULTS
Site-specific integration of the rat aldB origin fragments into a mouse chromosome.
Figure 1 shows the structure of the rat aldB origin, vectors used in the present study (pBS226 and pSF73 [14, 16]), and a strategy of site-specific recombination. The 0.94-kb aldB origin was inserted into the HindIII site of the loxP targeting vector pBS226 (referred to as pBS226/aldB). The vector pBS226/mutC carries the aldB origin that has a mutated site C (33). The mutation in site C was introduced by a three-base substitution at contact sites of AlF-C, which was previously determined by methylation interference assay (52). The inability of the mutant site C to bind AlF-C is shown in Fig. 2. In the competitive EMSAs, mutC DNA did not interfere with the binding of wild-type site C to either GST-fused AlF-C1 or AlF-C2. The pBS226 without the 0.94-kb aldB origin was termed pBS226/null. By coelectroporation with Cre recombinase expression vector, these DNA constructs were inserted into loxP site in the single copy of pSF73 DNA in a chromosome of the mouse acceptor cell L73-19 (17). The resulting recombinant cells (termed cBS226/aldB, cBS226/mutC, and cBS226/null) were directly selected by culturing in the presence of G418. Correct recombination should result in generation of a functional neomycin resistance gene, which would thus be G418 resistant due to insertion of the human cytomegalovirus (CMV) early promoter and initiator codon ATG just 5′ to the promoter- and ATG-less neomycin resistance gene at the acceptor site of the mouse chromosome (17, 18).
FIG. 2.
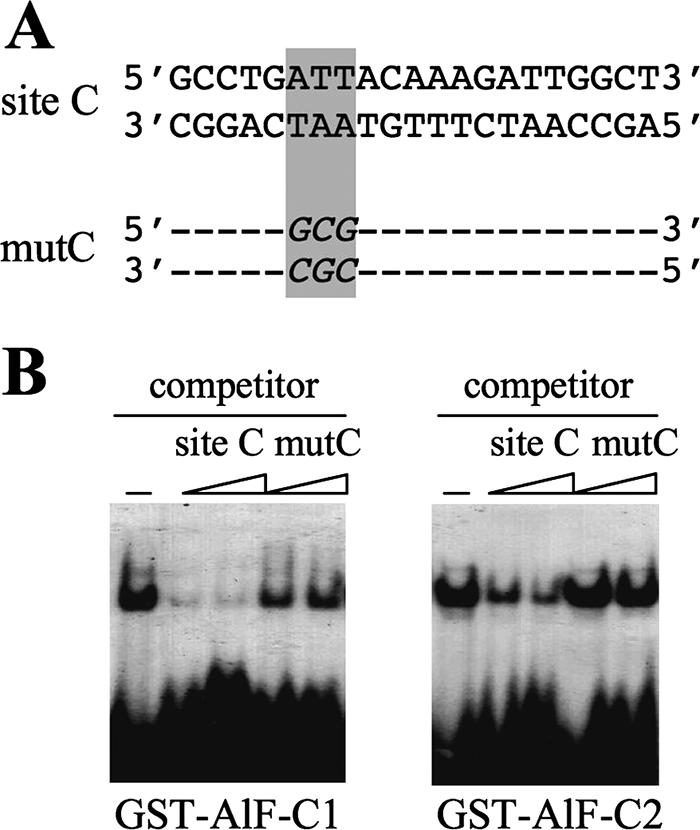
Mutation of site C in the aldB origin results in reduced binding of AlF-C proteins in vitro. (A) Nucleotide sequences of wild-type and the mutated site C DNAs (mutC). (B) Competitive gel EMSA. Bacterially expressed purified GST-fused AlF-C1 and AlF-C2 proteins (200 ng each) (42) were assayed for their binding to wild-type and mutC DNAs by competitive EMSA using 32P-labeled wild-type site C DNA (3 ng) as a probe. Site C and mutC DNAs used as competitors were in 100- and 200-fold molar excess amounts (from left to right).
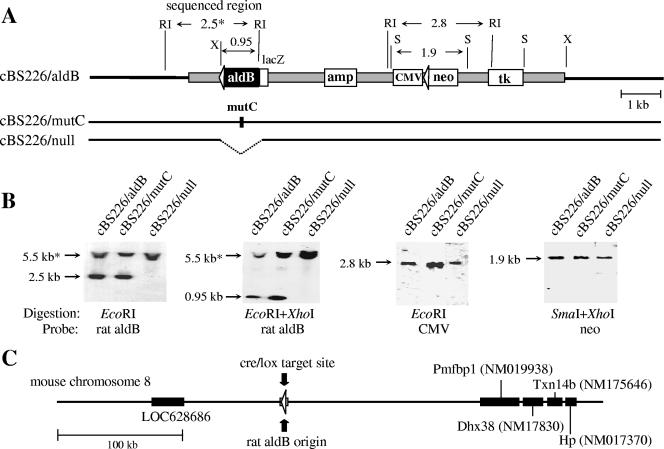
To see correct single-copy integration of the targeting vectors, chromosomal DNA from each recombinant cell line was analyzed by Southern blot hybridization. Figure 3A shows the restriction map anticipated after site-specific single-copy integration of pBS226/aldB, pBS226/mutC, and pBS226/null DNA into the recipient mouse cell chromosome. For each type of recombinant cells, a series of Southern blot analyses was performed. Representative results are shown in Fig. 3B. When genomic DNAs digested with EcoRI alone or EcoRI plus XhoI were hybridized with the probe specific for the aldB origin, 2.5- or 0.95-kb DNAs, respectively, that contain the aldB origin were detected in cBS226/aldB and cBS226/mutC recombinant cells but not in cBS226/null. The 5.5-kb EcoRI fragment (indicated by an asterisk) derived from the mouse aldB locus was also detected in each digest. When the EcoRI digests were hybridized with the probe specific for CMV DNA, the 2.8-kb DNA was detected. If correct single-copy integration of pBS226 vectors occurred at the chromosomal loxP site, the 1.7-kb EcoRI fragment in the recipient L73-19 cells should be replaced by a novel 2.8-kb EcoRI fragment. Similarly, a 1.1-kb SmaI-XhoI fragment in the recipient chromosome should be replaced by a novel 1.9-kb SmaI fragment after integration of the CMV DNA with the promoter-less neo gene. The results obtained from these experiments were all in agreement with the expected maps of recombination products.
FIG. 3.
Isolation of mouse recombinant cells carrying a single-copy insertion of the aldB origin or its derivatives. (A) Restriction map of the inserted DNA in the recombinant cells cBS226/aldB, cBS226/null, and cBS226/mutC. The gray box indicates pSF73 DNA. The nucleotide lengths of restriction fragments are shown in kilobases. RI, EcoRI; X. XhoI; S, SmaI. The 2.5-kb EcoRI fragment, marked by an asterisk, was cloned and sequenced (see the text). (B) Southern blots of DNA prepared from the recombinant cells. Genomic DNA from each recombinant cell was digested with the restriction enzymes indicated below the autoradiograms, blotted, and hybridized with the indicated probes. The 5.5-kb fragments marked by asterisks are those derived from the mouse aldB locus. (C) Insertion point of the targeting vector at chromosome 8, which was determined by sequencing the flanking region indicated in panel A (see the text). Black boxes with database accession numbers indicate transcription units (including hypothetical ones).
To further confirm this, the 2.5-kb EcoRI fragment (marked with an asterisk in Fig. 3A) was cloned from cBS226/aldB cells, and the entire nucleotide sequence was determined (data not shown). A database search (BLAST; the mouse genome) of the sequence revealed that the targeting vectors were inserted into a site in mouse chromosome 8 with neither the predicted transcription unit nor the reported replication origin sequences located close by (Fig. 3C).
The rat aldB origin directs replication initiation at an ectopic mouse locus.
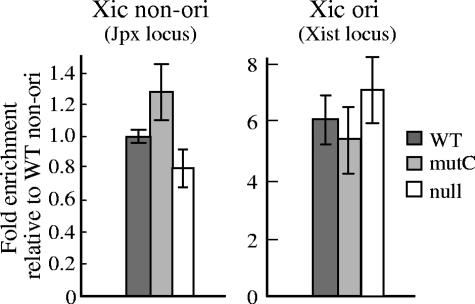
For the analysis of replication initiation activities, RNA-primed nascent DNA strands ranging from 0.5 to1.0 kb were prepared from each recombinant cell line. To validate the qualities of the nascent strand preparations, the presence of the nascent strands derived from previously reported known origins were tested. For this purpose, we analyzed mouse origin at Xist locus and the nonorigin region nearby in the X-inactivation center (Xic) using the same primer pairs as previously reported by Rowntree and Lee (41). The results showed that the known origin was detected at similar levels in all nascent strand preparations from cBS226/aldB, cBS226/mutC, and cBS226/null cells (Fig. 4). This indicates successful preparation of the nascent strands and that the qualities of the nascent strands do not significantly differ among the cell lines.
FIG. 4.
Validation of the nascent strand preparations from cBS226/aldB (WT), cBS226/mutC (mutC), and cBS226/null (null) cells used in the present study. Nascent strand abundances were assayed for known origin at the Xist locus, as well as the nearby nonorigin region at Jpx locus in the mouse X-inactivation center (Xic) region. Equivalent amounts of RNA-primed nascent strands with sizes ranging from 0.5 to 1.0 kb, which corresponded to the same numbers of cells, were subjected to quantitative real-time PCR. The primer sequences included primer pair 13 for Xist origin (named primer set 21 in the present study [Table 1]) and primer pair 1 for the nonorigin region (primer set 20 in the present study) that have been reported by Rowntree and Lee (see Table 1 in reference 41). The copy number was expressed relative to the nonorigin region in WT cells. The values are averages of two experiments. Error bars represent the range of measured values.
The nascent strands were then subjected to quantification by real-time PCR using specific primer sets (Table 1) designed for eight map positions on the DNA insert and the flanking region in the mouse chromosome (Fig. 5, upper panel). Relative abundances of the nascent strands derived from each map position were determined by calculating from the standard curves of the real-time PCR amplification using known amounts of chromosomal DNA from cBS226/aldB cells, and the values were normalized to the nonorigin region at the Jpx locus in the mouse chromosome (Fig. 4). Normalization to the Xist origin did not significantly differ from that to the nonorigin region.
FIG. 5.
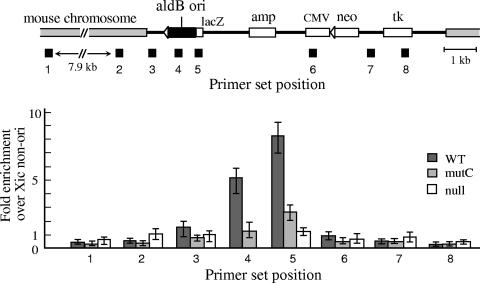
Relative abundance of newly replicated short nascent DNA in cBS226/aldB (WT), cBS226/mutC (mutC), and cBS226/null (null) cells. Equivalent amounts of the nascent strands from these cells, which corresponded to the same number of cells, were subjected to quantitative real-time PCR, using primer sets 1 to 8 designed for different map positions (indicated by black boxes below the map; the sequences and annealing temperatures are given in Table 1). The relative abundance of the nascent strands was determined from the standard curve calculated with known amounts of genomic DNA from cBS226/aldB cells as a template and normalized to the mouse nonorigin region in the Xic region (see Fig. 4). Values are averages of three independent experiments, and error bars represent the range of measured values.
As shown in Fig. 5, the relative abundance of the nascent strands derived from the aldB origin region (primer set positions 4 and 5) in the cBS226/aldB cells was significantly higher than those from other map positions. This is not due to the fact that the aldB origin was inserted adjacent to or within an origin zone of the mouse chromosome because the nascent strands derived from the adjacent regions in the mouse chromosome (positions 1 and 2) were significantly low. In addition, since the aldB promoters at the ectopic and the endogenous mouse loci are not active in the mouse L73-19 cells (data not shown), the effect of transcription may be negligible. In the cBS226/null cells (without the 0.94-kb aldB origin), the nascent strand abundance was low over the inserted DNA and the adjacent mouse chromosome. Thus, the rat aldB origin acts as a replicator at an ectopic mouse locus.
Mutation of site C in the ectopic aldB origin reduced replicator activity, which paralleled with reduced binding of AlF-C and Orc1.
We next examined whether site C in the rat aldB origin is required for efficient replicator activity. This site binds two closely related proteins AlF-C1 and AlF-C2 (AlF-C, together); the only difference lies in an insertion of 47 amino acids near C terminus in AlF-C1 (53). These two proteins have similar specificities in DNA binding and, importantly, have been shown to bind Orc1 (42). We therefore thought it very interesting to examine whether site C is required for replicator activity in vivo. For this purpose, site C was mutated so that AlF-C1 and AlF-C2 could not bind (Fig. 2). Then, the relative abundances of the nascent strands derived from the cBS226/mutC chromosome were determined. The results in Fig. 5 showed that the amounts of nascent strands derived from the mutant aldB origin significantly decreased compared to the wild-type origin.
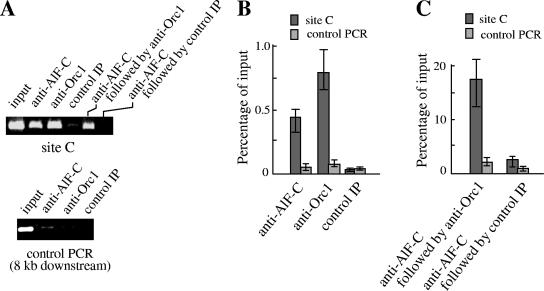
To examine whether binding of AlF-C is crucial for full replicator activity, ChIP assays were performed using anti-rat AlF-C and anti-rat Orc1 antibodies (Fig. 6). For this experiment, specific PCR primers were designed to selectively detect site C DNA in the ectopically located rat aldB origin in the mouse chromosome. Figure 6A shows that primer set 4 (see Table 1) amplifies site C in the rat origin but not endogenous site C (cBS226/null cells) at the mouse aldB locus, since the nucleotide sequence of the forward primer did not match that of the corresponding region in the mouse DNA. When the DNAs from the cBS226/aldB chromatins trapped by the anti-AlF-C or anti-Orc1 antibodies were subjected to PCR, primer set 4 amplified DNA fragments to a much higher extent versus control chromatins (control IP using preimmune IgG) (Fig. 6B). However, the antibody-bound cBS226/mutC chromatins contained site C DNA to a significantly lower extent. As expected, chromatin fragments from cBS226/null cells did not contain rat site C DNA.
FIG. 6.
ChIP assays for the rat aldB origin at an ectopic mouse locus. (A) PCR primers that selectively amplify site C region in the ectopic rat aldB origin but not the mouse cognate region. Primer set 4 (Table 1) amplifies DNA from cBS226/aldB, cBS226/mutC, and dRLh84 cells but not DNA from cBS226/null (no rat sequence). (B) Detection of the aldB origin fragment containing site C DNA in the immunoadsorbed chromatin. Equal amounts of chromatin fragments from cBS226/aldB, cBS226/mutC, and cBS226/null cells were first pretreated with protein A-Sepharose. The unbound chromatin was then incubated with anti-AlF-C or anti-Orc1 antibodies, and the resulting antibody-chromatin complexes were trapped by protein A-Sepharose. DNA in the complex was subjected to PCR for 35 cycles using primer set 4 under the following conditions: denaturing at 95°C for 10 s, annealing at 62°C for 10 s, and an extension reaction at 72°C for 20 s. For control experiments, equal amounts of chromatins were incubated with preimmune IgG (control IP). PCR products equivalent to the same amounts of the input chromatin were loaded onto 2.5% agarose gel. Note that the “input DNA” in the figure was one-tenth amount compared to other lanes. (C) Real-time PCR of DNA in the immunoadsorbed chromatins. DNA in the immunocomplex was prepared as described in panel B and subjected to real-time PCR using primer set 4 (left panel). Control PCR (right panel) for a region distal to site C was carried out using primer set 6 corresponding to CMV region. Values are averages of three independent experiments and are expressed as a percentage of input chromatin. Error bars represent the range of measured values. WT, cBS226/aldB; mutC, cBS226/mutC.
These results were confirmed by quantitative real-time PCR (Fig. 6C). The results showed that mutation of site C resulted in nearly threefold decrease in the chromatin-bound AlF-C and Orc1 proteins compared to those of wild type (cBS226/aldB). The control PCR (CMV region) showed that the immunoadsorbed chromatin does not contain DNA fragments located distal to the aldB origin. These results suggested that the mutation of site C caused less efficient binding of AlF-C, leading to less efficient binding of Orc1 to the rat origin. In this way, the mutation of site C might cause reduced replicator activity. Accordingly, the residual replicator activity of the mutant origin (Fig. 5) may be a reflection of the residual binding of AlF-C.
AlF-C and Orc1 bind to the aldB origin at its original location in the rat chromosome.
We next confirmed the binding of rat AlF-C and Orc1 to the aldB origin at its original rat locus. As shown in Fig. 7A, site C DNA was detected in either of the fragmented chromatins that bound to the anti-AlF-C antibodies or the anti-rat Orc1 antibodies, whereas it was not detected in the chromatins bound to preimmune serum (control IP). Control PCR experiments (Fig. 7A, lower panel) showed that the fragmented chromatin that bound to the antibodies does not contain DNA corresponding to about 8 kb downstream of the aldB gene (primer set 13 [see Table 1]). Thus, it is considered that both AlF-C and rat Orc1 bind to the aldB origin, at least within a 1.5-kb region, which is the average length of the fragmented chromatin. ChIP assays were then performed sequentially with the anti-AlF-C, followed by the anti-rat Orc1 antibodies, to confirm the cohabitation of the two proteins in the aldB origin complex. Chromatins bound to the anti-AlF-C antibodies were first trapped by protein A-Sepharose. After detachment of the chromatin-antibody complex from protein A-Sepharose and inactivation of the bound antibodies, the complexes were incubated again with the anti-rat Orc1 antibodies (see Materials and Methods). As shown in Fig. 7A, the chromatins trapped by the second antibody contained site C DNA. These observations were further examined by real-time PCR using the same primer sets (Fig. 7B and C). The results confirmed the cohabitation of the two proteins in the aldB origin complex.
FIG. 7.
ChIP assays for the aldB origin in the rat chromosome. (A) Chromatins were incubated with the antibodies and subjected to PCR using primer set 4, as in the legend to Fig. 6B. For sequential ChIP assays, chromatins were first incubated with the anti-AlF-C antibodies. The chromatin anti-AlF-C complexes were then incubated with the second antibody anti-rat Orc1 or control preimmune serum, after inactivation of the anti-AlF-C antibodies (see Materials and Methods). DNAs from the chromatins that bound to the anti-rat Orc1 antibodies were subjected to PCR using primer set 4 as in the legend to Fig. 6B. The input DNA in the figure was one-tenth the amount used in the other lanes. For control PCR experiments, primer set 13 (8 kb downstream of site C [see Table 1]) was used. (B) Quantitative real-time PCR of the ChIP experiments shown in panel A. Values are averages of three experiments, and expressed as percentage of input chromatin. Error bars represent the range of measured values. (C) Quantitative real-time PCR for DNA obtained from sequential ChIP experiments in panel A. Note that input DNA in this experiment corresponds to that of chromatins bound to the anti-AlF-C antibodies. Values and error bars are the same as in panel B.
AlF-C binds other replication origins in the rat chromosome in addition to the aldB origin.
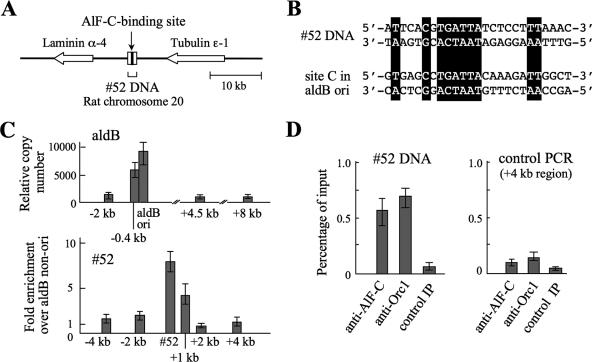
Next, we sought to isolate an additional rat origin that has a binding site for AlF-C to determine whether AlF-C and Orc1 bind to it. Consequently, a 302-bp DNA fragment termed clone 52 DNA (database accession no. AB272072) was isolated from the rat genomic DNA after enrichment through repetitive cycles of binding to GST-fused AlF-C and subsequent cloning in bacteria (data not shown). Nucleotide sequencing and a database search (Rat Genome Database [http://rgd.mcw.edu/]) revealed that the clone 52 DNA is located between the predicted laminin α-4 (database accession no. RGD1560062) and tubulin ɛ-1 (RGD1306048) genes at chromosome 20 (q12) (Fig. 8A). This DNA contained a nucleotide sequence similar to that of site C in the aldB origin (Fig. 8B), and its binding to AlF-C was confirmed by EMSA (data not shown).
FIG. 8.
Candidate replication origin that binds AlF-C and Orc1. (A) Chromosomal location of clone 52 DNA in the rat genome. Clone 52 DNA (302 bp) was cloned from DNA library of restriction digests of rat genomic DNA, which was prepared through repetitive cycles of binding to GST-fused AlF-C2. Horizontal arrows indicate transcription direction of predicted laminin α-4 (RGD15600629) and tubulin ɛ-1 (RGD1306048) genes. (B) Nucleotide sequences of AlF-C-binding sites in clone 52 DNA and the aldB origin (site C). Shaded regions indicate identical nucleotides between the two DNA sequences. (C) Relative abundance of nascent strands derived from the aldB origin (upper panel) and clone 52 DNA regions (lower panel). Experimental procedures are the same as in the legend to Fig. 5. For the detection of the aldB region, primer sets 9 (site C), 10 (2 kb upstream, −2 kb), 11 (0.4 kb upstream, −0.4 kb). 12 (4.5 kb downstream, +4.5), and 13 (8 kb downstream, +8 kb) were used (see Table 1). For the clone 52 DNA region, primer sets 14, 15, 16, 17, 18, and 19 were used, which correspond to site C in clone 52 DNA, 4 kb upstream (−4 kb), 2 kb upstream (−2 kb), 1 kb downstream (+1 kb), 2 kb downstream (+2 kb), and 4 kb downstream (+4kb) of site C in clone 52 DNA, respectively. The relative abundance was determined as described in Fig. 5, using genomic DNA from dRLh84 cells as standards. Values are averages of three independent experiments. The relative abundance of the nascent strands from the clone 52 DNA region was normalized to the aldB nonorigin region (+8 kb region). Error bars represent the range of measured values. (D) ChIP followed by real-time PCR analyses for the clone 52 DNA region using anti-AlF-C and anti-Orc1 antibodies. Equal amounts of sheared chromatin from dRLh84 cells were treated as described in the legend to Fig. 6. Values are averages of three independent experiments and are expressed as the percentage of input chromatin. Control PCR experiments (right panel) were performed with primer set 19 for the region about 4 kb apart from the clone 52 DNA. Values are averages of three independent experiments, and error bars represent the range of measured values.
Figure 8C shows nascent strand abundance assays for clone 52 DNA and the aldB origin; the latter was used as a reference for initiation activity of known origin. RNA-primed nascent strands ranging from 0.5 to 1.0 kb were prepared from the rat dRLh84 cells and subjected to real-time PCR using primer sets 9 to 13 for the aldB region and primer sets 14 to 19 for the clone 52 DNA region (see Table 1). As is apparent from the figure, the relative abundance of the nascent strands derived from clone 52 region was higher than those from the flanking regions, and was comparable to that from the aldB origin. Thus, a region encompassing or in the close vicinity of clone 52 DNA in the rat genome has replication initiation activity.
Binding of AlF-C and Orc1 to the clone 52 DNA region was then examined by ChIP, followed by real-time PCR using fragmented chromatins from the rat dRLh84 cells. As shown in Fig. 8D, both chromatin fragments that bound to anti-AlF-C or anti-Orc1 antibodies contained AlF-C-binding site derived from the clone 52 DNA region. The antibody-complexed chromatins did not contain DNA derived from several kilobases apart from the AlF-C-binding site (control PCR). Therefore, it can be said that the clone 52 origin region associated with both AlF-C and Orc1, similar to the aldB origin.
DISCUSSION
In the present study, we showed that the rat origin fragment functioned as a replicator in the ectopic mouse locus. Mutation of site C in the origin DNA resulted in a significantly decreased level of the replicator activity. ChIP assays suggested that the mutation caused insufficient binding of AlF-C and Orc1 to the mutant origin, leading to reduced replicator activity. In addition, two distinct rat replication origins, the aldB and clone 52 DNA origins that contain AlF-C-binding sequence, were associated with both AlF-C and Orc1 in vivo.
Replicator activity of mammalian origin sequence was first reported for the human β-globin origin (2). Similar results have been reported for a 2.4-kb human c-myc origin (30). Recently, three reports demonstrated the requirement of specific DNA sequences for mammalian replicator activity (3, 37, 50). Altman and Fanning (3) reported that defined sequence modules, including a target site of DNA-binding protein RIP60 and an AT-rich element, are required for initiation at the hamster dhfr origin at ectopic chromosomal location. Wang et al. (50) reported two independent replicators in the β-globin region, each of which contained short discrete sequences including an AT-rich stretch and an asymmetric purine/pyrimidine stretch that cooperatively promote replicator activity. Human lamin B2 replicator also has a modular structure, and binding of specific proteins to the sequence modules is required for replicator activity (37). A modular structure has also been expected for the aldB origin since, in addition to site C, an asymmetric polypurine/polypyrimidine stretch located about 60 bp apart from site C was required for efficient replication initiation in vivo (45).
Together, these results strongly suggest that specific DNA sequences, which might not be common and even vary among the origins distributed over chromosomes, are required for the determination of active origin sites in mammalian cells. Such origin-specific DNA sequences might be required, in part, to induce a local chromatin structure that activate origin function by facilitating access to initiator proteins such as the ORC. Because mammalian ORC does not show sequence specificity in DNA binding, it is not surprising that sequence-specific DNA-binding proteins or a set of such proteins may mediate the interaction of ORC and other initiator proteins with replication origins. The pre-RC formation by origin-specific DNA-binding proteins might be supported, in part, by the recent observation that a cellular protein Ku80 binds sequence specifically to lamin B2, β-globin, and c-myc origins prior to the assembly of the ORC complex (46). An observation that an artificial functional origin can be created at GAL4 binding sequence in a plasmid DNA via binding of GAL4-ORC subunit or GAL4-cdc6 fusion proteins may also imply a possibility of indirect association of ORC to origins. Both GAL4 fusion proteins recruited other proteins of the ORC-cdc6 complex on the artificial origin (48). Very recently, Atanasiu et al. (5) have reported a model in which telomere repeat factor 2 (TRF2), a protein that binds to DS region of EB virus oriP, stimulates replication from oriP by direct binding to Orc1. In this replication, TRF2 binds to Orc1 through a domain similar to that for AlF-C interaction. This observation is also consistent with the model presented here.
Replication initiation is controlled by a regulatory step involving the association of Orc1 subunit with other subunits of the ORC complex on chromatin (14). Mammalian Orc1 subunit appears to behave differently from those of yeasts. For example, the interaction of Orc1 with the ORC core complex (Orc2, -3, -4, and -5) in mammalian cells changes during cell cycle. The cell cycle-dependent interaction of Orc1 with other subunits may account for the cell cycle-dependent interaction of the functional ORC complex with chromatin; mammalian Orc1 binds to chromatin at late G1 to S phase, and then it is released from the chromatin (27, 29, 31, 35, 54). In addition, it has been shown that Orc1 is a target of E2F regulation during G1-to-S transition, whereas other ORC subunits are not (4). From these observations, it seems that Orc1 has a regulatory role in ORC function, and association of the Orc1 protein to the chromosomal origins appears to be crucial to the formation of functional pre-RC. In this regard, our observation that the binding of AlF-C to site C in the aldB origin is required for full replicator activity might be of particular interest.
Finally, with regard to the cellular control of replication initiation in mammalian cells, recent reports have suggested that origin determination and activation are under the control of development (36), transcriptional regulation (43), and regulatory elements of transcription (20, 32). This situation might be true of the aldB origin. Our previous observation that the aldB origin is activated in aldB-nonexpressing cells but not in expressing cells (34) suggests mutually exclusive control between transcription and replication initiation at the aldB locus and is in good agreement with the present observation that AlF-C binds to the aldB origin and enhances its initiation activity in the aldB-nonexpressing mouse L73-19 cells. Considering that AlF-C acts as a transcriptional repressor of the aldB gene (53), it seems that binding of AlF-C may switch the roles of the aldB promoter from transcription to replication initiation. Consistent with this speculation, AlF-C expression is high in aldB-nonexpressing rapidly dividing fetal liver cells and low in differentiated aldB-expressing liver cells in the rat. In addition, the expression increases during liver regeneration, which paralleled with transcriptional repression of the aldB gene (53).
Acknowledgments
We thank S. Fukushige (Tohoku University) for advice on Cre-lox recombination and for the generous gift of mouse recombinant L cell line 73-19 carrying a single copy of pSF73 DNA, S. Miyagawa (Osaka University) for the generous gift of an sCre recombinase gene optimized for expression in mammalian cells, and Bristol-Myers Squibb Pharma Company for providing a loxP targeting vector pBS226. We also thank M. Sakai and H. Ikeda (Hokkaido University) for valuable suggestions on the ChIP assay.
H.M. was a recipient of the student support program of COE to Iwate University from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 18 September 2006.
REFERENCES
- 1.Aladjem, M. I., and E. Fanning. 2004. The replication revisited: an old model learns new tricks in metazoan chromosomes. EMBO rep. 5:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aladjem, M. I., L. W. Rodewald, J. L. Kolman, and G. M. Wahl. 1998. Genetic dissection of a mammalian replicator in the human β-globin locus. Science 281:1005-1009. [DOI] [PubMed] [Google Scholar]
- 3.Altman, A. L., and E. Fanning. 2004. Defined sequence modules and an architectural element cooperate to promote initiation at an ectopic mammalian chromosomal replication origin. Mol. Cell. Biol. 24:4138-4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asano, M., and R. P. Wharton. 1999. E2F mediates developmental and cell cycle regulation of ORC1 in Drosophila. EMBO J. 18:2435-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atanasiu, C., Z. Deng, A. Wiedmer, J. Norseen, and P. M. Lieberman. 2006. ORC binding to TRF2 stimulates OriP replication. EMBO reports 7:716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Austin, R. J., T. L. Orr-Weaver, and S. P. Bell. 1999. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev. 13:2639-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 8.Biensky, A. K., and S. A. Gerbi. 1998. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279:95-98. [DOI] [PubMed] [Google Scholar]
- 9.Bogan, J. A., D. A. Natale, and M. L. DePamphilis. 2000. Initiation of eukaryotic DNA replication conservative or liberal? J. Cell Physiol. 184:139-150. [DOI] [PubMed] [Google Scholar]
- 10.Callebaut, I., J. C. Courvalin, and J. P. Mornon. 1999. The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication, and transcriptional regulation. FEBS Lett. 446:189-193. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associates with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 98:10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chuang, R. Y., L. Chretien, J. Dai, and T. J. Kelly. 2002. Purification and characterization of the Schizosaccharomyces pombe origin recognition complex: interaction with origin DNA and Cdc18 protein. J. Biol. Chem. 227:16920-16927. [DOI] [PubMed] [Google Scholar]
- 13.Cuvier, O., C. M. Hart, and U. K. Laemmli. 1998. Identification of a class of chromatin boundary elements. Mol. Cell. Biol. 18:7478-7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DePamphilis, M. L. 2003. The “ORC cycle”: a novel pathway for regulating eukaryotic DNA replication. Gene 310:1-15. [DOI] [PubMed] [Google Scholar]
- 15.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 16.Dhar, S. K., L. Delmolino, and A. Dutta. 2001. Architecture of the human origin recognition complex. J. Biol. Chem. 276:29067-29071. [DOI] [PubMed] [Google Scholar]
- 17.Fukushige, S., and J. E. Ikeda. 1996. Trapping of mammalian promoters by Cre-lox site-specific recombination. DNA Res. 3:73-80. [DOI] [PubMed] [Google Scholar]
- 18.Fukushige, S., and B. Sauer. 1992. Genomic targeting with a positive-selection lox integration vector allows highly reproducible gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 89:7905-7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert, D. 2001. Making sense of eukaryotic DNA replication origins. Science 294:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh, M., G. Liu, G. Randall, J. Bevington, and M. Leffak. 2004. Transcription factor binding and induced transcription alter chromosomal c-myc replicator activity. Mol. Cell. Biol. 24:10193-10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda, H., S. Nishi, and M. Sakai. 2004. Transcription factor Nrf2/MafK regulates rat placental glutathione S-transferase gene during hepatocarcinogenesis. Biochem. J. 380:515-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kearsey, S. E., and S. Cotterill. 2003. Enigmatic variations: divergent modes of regulating eukaryotic DNA replication. Mol. Cell 12:1067-1075. [DOI] [PubMed] [Google Scholar]
- 23.Keller, C., E. M. Ladenburger, M. Kremer, and R. Knippers. 2002. The origin recognition complex marks a replication origin in the human TOP1 gene promoter. J. Biol. Chem. 277:31430-31440. [DOI] [PubMed] [Google Scholar]
- 24.Kong, D., T. R. Coleman, and M. L. DePamphilis. 2003. Xenopus origin recognition complex (ORC) initiates DNA replication preferentially at sequences targeted by Schizosaccharomyces pombe ORC. EMBO J. 22:3441-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong, D., and M. L. DePamphilis. 2002. Site-specific ORC binding, pre-replication complex assembly and DNA synthesis at Schizosaccharomyces pombe replication origins. EMBO J. 21:5567-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koresawa, Y., S. Miyagawa, M. Ikawa, K. Matsunami, M. Yamada, R. Shirakura, and M. Okabe. 2000. A synthesis of a new Cre recombinase optimal codon usage for mammalian systems. J. Biochem. 127:367-372. [DOI] [PubMed] [Google Scholar]
- 27.Kreitz, S., M. Ritzi, M. Baack, and R. Knippers. 2001. The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J. Biol. Chem. 276:6337-6342. [DOI] [PubMed] [Google Scholar]
- 28.Ladenburger, E. M., C. Keller, and R. Knippers. 2002. Identification of a binding region for human origin recognition complex protein 1 and 2 that coincides with an origin of DNA replication. Mol. Cell. Biol. 22:1036-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, C. J., and M. L. DePamphilis. 2002. Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle. Mol. Cell. Biol. 22:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malott, M., and M. Leffak. 1999. Activity of the c-myc replicator at an ectopic chromosomal location. Mol. Cell. Biol. 19:5685-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendez, J., X. H. Zou-Yang, S. Y. Kim, M. Hidaka, W. P. Tansey, and B. Stillman. 2002. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell 9:481-491. [DOI] [PubMed] [Google Scholar]
- 32.Mesner, L. D., and J. L. Hamlin. 2005. Specific signals at the 3′ end of the DHFR gene define one boundary of the downstream origin of replication. Genes Dev. 19:1053-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyagi, S., Y. Zhao, Y. Saitoh, and K. Tsutsumi. 2000. An overlapping set of DNA elements in the rat aldolase B gene origin/promoter regulates transcription and autonomous replication. Biochem. Biophys. Res. Commun. 278:760-765. [DOI] [PubMed] [Google Scholar]
- 34.Miyagi, S., Y. P. Zhao, Y. Saitoh, K. Tamai, and K. Tsutsumi. 2001. Replication of the rat aldolase B locus differs between aldolase B-expressing and non-expressing cells. FEBS Lett. 505:332-336. [DOI] [PubMed] [Google Scholar]
- 35.Natale, D. A., C. J. Li, W. H. Sun, and M. L. DePamphilis. 2000. Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M-G1 transition in mammals. EMBO J. 19:2728-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norio, P., S. Kosiyatrakul, Q. Yang, Z. Guan, N. M. Brown, S. Thomas, R. Riblet, and C. L. Schildkraut. 2005. Progressive activation of DNA replication in large domains of the immunoglobulin heavy chain locus during B-cell development. Mol. Cell 23:575-587. [DOI] [PubMed] [Google Scholar]
- 37.Paixao, S., I. N. Colaluca, M. Cubellus, F. A. Peverali, A. Destro, S. Giadrossi, M. Giacca, A. Falaschi, S. Riva, and G. Biamonti. 2004. Modular structure of the human lamin B2 replicator. Mol. Cell. Biol. 24:2958-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raghuraman, M. K., E. A. Winzeler, D. Collingwood, S. Hunt, L. Wodicka, A. Conway, D. J. Lockhart, R. W. Davis, B. J. Brewer, and W. L. Fangman. 2001. Replication dynamics of the yeast genome. Science 294:115-121. [DOI] [PubMed] [Google Scholar]
- 39.Rao, H., and B. Stillman. 1995. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc. Natl. Acad. Sci. USA 92:2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Remus, D., E. L. Beall, and M. R. Botchan. 2004. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 23:897-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowntree, R. K., and J. T. Lee. 2006. Mapping of DNA replication origins to noncoding genes of the X-inactivation center. Mol. Cell. Biol. 26:3707-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saitoh, Y., S. Miyagi, H. Ariga, and K. Tsutsumi. 2002. Functional domains involved in the interaction between Orc1 and transcriptional repressor AlF-C that bind to an origin/promoter of the rat aldolase B gene. Nucleic Acids Res. 30:5205-5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sasaki, T., S. Ramanathan, Y. Okuno, C. Kumagai, S. S. Shaikh, and D. M. Gilbert. 2006. The Chinese hamster dihydrofolate reductase replication origin decision point follows activation of transcription and suppresses initiation of replication within transcription units. Mol. Cell. Biol. 26:1051-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimotai, Y., H. Minami, Y. Saitoh, Y. Onodera, Y. Mishima, R. J. Kelm, Jr., and K. Tsutsumi. 2006. A binding site for Purα and Purβ is structurally unstable and is required for replication in vivo from the rat aldolase B origin. Biochem. Biophys. Res. Commun. 340:517-525. [DOI] [PubMed] [Google Scholar]
- 46.Sibani, S., G. B. Price, and M. Zannis-Hadjopoulos. 2005. Ku80 binds to human replication origins prior to the assembly of the ORC complex. Biochemistry 44:7885-7896. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi, T., E. Ohara, H. Nishitani, and H. Masukata. 2003. Multiple ORC-binding sites are required for efficient MCM loading and origin firing in fission yeast. EMBO J. 4:964-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeda, D. Y., Y. Shibata, J. D. Parvin, and A. Dutta. 2005. Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev. 19:2827-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vashee, S., C. Cvetic, W. Lu, P. Simancek, T. J. Kelly, and J. C. Walter. 2003. Sequence-independent DNA binding and replication initiation by human origin recognition complex. Genes Dev. 17:1894-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, L., C. M. Lin, S. Brooks, D. Cimbora, M. Groudine, and M. I. Aladjem. 2004. The human beta-globin replication initiation region consists of two modular independent replicators. Mol. Cell. Biol. 24:3373-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wyrick, J. J., J. G. Aparicio, T. Chen, J. D. Varnettm, E. G. Jennings, R. A. Yound, S. P. Bell, and O. M. Aparicio. 2001. Genome-wide distribution of ORC and MCM proteins in Saccharomyces cerevisiae: high-resolution mapping of replication origins. Science 294:2357-2360. [DOI] [PubMed] [Google Scholar]
- 52.Yabuki, T., S. Ejiri, and K. Tsutsumi. 1993. Ubiquitous factors that interact simultaneously with two distinct cis-element on the rat aldolase B gene promoter. Biochim. Biophys. Acta 1216:15-19. [DOI] [PubMed] [Google Scholar]
- 53.Yabuki, T., S. Miyagi, H. Ueda, Y. Saitoh, and K. Tsutsumi. 2001. A novel growth-related nuclear protein binds and inhibits rat aldolase B gene promoter. Gene 264:123-129. [DOI] [PubMed] [Google Scholar]
- 54.Yan, Z., J. DeGregori, R. Shohet, G. Leone, B. Stillman, J. R. Nevins, and R. S. Williams. 1998. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc. Natl. Acad. Sci. USA 95:3603-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.You, Z., Y. Ishimi, T. Mizuno, K. Sugasawa, F. Hanaoka, and H. Masai. 2003. Thymine-rich single-stranded DNA activates Mcm4/6/7 helicase on Y-fork and bubble-like substrates. EMBO J. 22:6148-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao, Y., R. Tsutsumi, M. Yamaki, Y. Nagatsuka, S. Ejiri, and K. Tsutsumi. 1994. Initiation zone of DNA replication at the aldolase B locus encompasses transcription promoter region. Nucleic Acids Res. 22:5385-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao, Y., S. Miyagi, T. Kikawada, and K. Tsutsumi. 1997. Sequence requirement for replication initiation at the rat aldolase B locus implicated in its functional correlation with transcriptional regulation. Biochem. Biophys. Res. Commun. 237:707-713. [DOI] [PubMed] [Google Scholar]