Abstract
Information must be shared and functions coordinated among the spatially distinct functional centers of the ribosome. To address these issues, a yeast-based genetic system enabling generation of stable strains expressing only mutant forms of rRNA was devised. The B1a bridge (helix 38) has been implicated in the subtle modulation of numerous ribosomal functions. Base-specific mutations were introduced into helix 38 at sites affecting the B1a bridge and where it contacts the aminoacyl-tRNA (aa-tRNA) D-loop. Both sets of mutants promoted increased affinities for aa-tRNA but had different effects in their responses to two A-site-specific drugs and on suppression nonsense codons. Structural analyses revealed an arc of nucleotides in 25S rRNA that link the B1a bridge, the peptidyltransferase center, the GTPase-associated center, and the sarcin/ricin loop. We propose that a series of regularly spaced “hinge bases” provide fulcrums around which rigid helices can reorient themselves depending on the occupancy status of the A-site.
The ribosome is a complex nanomachine that contains many discrete functional centers. The small subunit contains the mRNA decoding center, and the large subunit harbors the catalytic (peptidyltransferase) center, three tRNA binding pockets, and a factor binding region. Together, these subunits interact to coordinate a complex series of events resulting in the processive synthesis of polypeptides. The question of how spatially separated functional regions communicate their status to one another in a coordinated fashion remains one of the central unanswered questions in the field. Although the eukaryotic ribosome contains approximately 80 proteins, it is the rRNAs that truly determine the core functions of this machine. For example, its central catalytic activity, peptidyltransfer, is mediated entirely by rRNA. Likewise, the substructures involved in the selection of cognate aminoacyl-tRNAs (aa-tRNAs) at the decoding center, and the bulk movement of tRNAs and mRNAs through the ribosome are mostly formed by rRNA-based structures.
Although rRNAs comprise the heart of the ribosome, their molecular genetic manipulation has been much more challenging than for the ribosomal proteins. The major impediment to this line of research stems from the fact that rRNA genes have been amplified in all genomes. In Escherichia coli, strains exist in which all of the rRNA operons have been deleted, making such investigations possible in this model prokaryotic organism. It is also desirable to be able to perform similar studies with eukaryotic ribosomes. The simplest, most advanced eukaryotic model organism is the yeast Saccharomyces cerevisiae. In yeast, chromosome XII contains 100 to 200 tandemly arrayed copies of the RDN1 gene. RDN1 is an operon that contains RNA polymerase I transcribed 35S pre-mRNA, which is processed into 25S, 18S, and 5.8S rRNAs, plus RNA polymerase III transcribed gene encoding 5S rRNA. In yeast, two basic approaches have been used to address the problem of rRNA molecular genetics. One relies on coupling temperature-sensitive alleles of RNA polymerase I with a clone in which the 35S pre-rRNA is transcribed from a galactose-inducible RNA polymerase II-dependent promoter (20, 21). In theory, this approach should allow transcriptional shut off of wild-type rRNAs while inducing the synthesis of desired mutants. A parallel approach takes advantage of the observation that hygromycin can be used to select for cells that have spontaneously deleted the chromosomal RDN1 locus in favor of a plasmid-borne mutant encoding resistance to this translational inhibitor (38). The latest variations of these strains are called “rdn1ΔΔ” (22, 40).
In theory, these two platforms should simplify molecular genetic analyses of the rRNAs. As witnessed by the paucity of publications describing yeast rRNA mutants over the intervening years, it is clear that something has been missing. From experience gained in our laboratory and from informal sharing of information with other researchers, it became apparent that genes expressing mutant rRNA transcripts are heavily selected against. The result is that, during the time required for auxotrophic selection for the plasmids harboring the mutant ribosomal DNA (rDNA) genes, there is strong selection against mutant rRNAs expressed from them. This places extremely strong pressure for selection of recombinant plasmids expressing both the selectable auxotrophic marker and the wild-type rRNAs. As described here, the solution to this problem is to transcriptionally silence the mutant rDNA genes during the period required for establishment of the plasmid harboring them. Only after plasmid establishment is transcription of the mutant activated and, simultaneously, transcription of the wild type is turned off. When expression of the mutant is simultaneously coupled with selection for hygromycin resistance, it is possible to establish yeast cell lines expressing only mutant rRNAs.
Here, this approach was used to generate and characterize the effects of targeted mutations in helix 38 of yeast 25S rRNA. Two specific regions of this structure were chosen for mutagenesis based on (i) known interactions with the D-loop of aa-tRNA and (ii) involvement in forming the B1a intersubunit bridge with ribosomal protein S13 in E. coli (S15 in yeast) (43). A series of viable alleles expressing only mutant rRNAs were constructed and characterized both at the biochemical and functional genetic levels. Both classes of mutants promoted approximately twofold increases in ribosomal affinities for aa-tRNAs and subtle, nucleotide-specific alterations in 25S rRNA structure. At the functional level, however, the two mutants showed differences. Single point mutants in the aa-tRNA D-loop interacting region promoted distinct temperature-sensitive phenotypes, simultaneous hypersensitivity to anisomycin, resistance to paromomycin, and decreased rates of nonsense suppression. In contrast, only a triple-base mutant in the B1a bridge-forming region promoted discernible phenotypes. These characteristics were very different from those of the first class and included simultaneous resistance to both anisomycin and paromomycin, and increased rates of nonsense codon suppression. rRNA structural analyses examining in-line cleavage patterns showed that these functionally subtle mutants produced equally subtle changes in the flexibility of unpaired or “hinge” bases in 25S rRNA. Interestingly, although deprotection of hinge bases proximal to the sites of the mutations was similar for the two mutants, the mutations differentially affected 25S rRNA structure more distally. Specifically, the tRNA D-loop-interacting mutant appeared to increase rRNA flexibility in the direction of the GTPase-associated center, while the mutant in the B1a bridge-forming mutant altered rRNA structure along the A-site proximal side of the peptidyltransferase center. These studies implicate the tip of helix 38 as contributing to the ribosome's ability to distinguish between aa-tRNA and eRF1. They also highlight distinct functional differences between two spatially close regions of helix 38 and open the door to more detailed genetic and biochemical analyses aimed to help link rRNA sequence and structure with specific functions.
MATERIALS AND METHODS
Strains, media, reagents, and molecular methods.
E. coli strain DH5α was used to amplify plasmids. Yeast media and 4.7 MB plates were prepared as described previously (42); galactose media contained 2% galactose instead of glucose. Drug concentrations were as follows: doxycycline, 10 μg/ml; hygromycin B, 300 μg/ml; and anisomycin, 20 μg/ml. Yeast strain NOY1049 (MATa ade2-1 ura3-1 trp1-1 his3-11 leu2-3,112 can1-100 ΔrDNA::his3::hisG pNOY353 (GAL7-35S rDNA, 5S rDNA, TRP1 2μm amp), was kindly provided by M. Nomura (40). The yeast killer virus [L-A HN M1] was introduced into NOY1049 by cytoplasmic mixing (9) from JD759 (MATα kar1-1 arg1 thr1 [L-A HN M1]). Briefly, NOY1049 and JD759 cells were mixed in a ratio of approximately 1:20 on rich galactose medium and then incubated overnight at 30°C. Cells were subsequently streaked for single colonies on galactose-containing H-Arg medium and grown for 5 days at 30°C to eliminate the donor strain, i.e., JD759. Diploid cells were identified by their ability to grow on minimal medium. Colonies of NOY1049-derived cells containing the killer virus were identified by their ability to inhibit growth of a nonkiller diploid indicator strain on 4.7 MB plates containing 2% galactose as previously described (7). The resulting haploid, Killer+ strain derived from NOY1049, was designated JD1314. Its genotype is MATa ade2-1 ura3-1 trp1-1 his3-11 leu2-3,112 can1-100 ΔrDNA::his3::hisG [L-A HN M1] pNOY353.
Restriction endonucleases were purchased from MBI Fermentas (Vilnius, Lithuania), and the rapid DNA ligation kit was obtained from Roche (Nutley, NJ). PCR amplifications were made with PfuUltra Hotstart PCR master mix (Stratagene, La Jolla, CA). DNA oligonucleotides were from IDT DNA Technologies (Coralville, IA). Standard methods were used for molecular cloning (28). The QuikChange II XL site-directed mutagenesis kit (Stratagene) was used for oligonucleotide site-directed mutagenesis. Oligonucleotide design and reaction conditions were as recommended by the manufacturer. Total yeast RNA was isolated by acid phenol extraction and treated with RNase-free DNase (QIAGEN, Valencia, CA). The Titan One-Tube RT-PCR system (Roche) was used for reverse transcription-PCR (RT-PCR). [γ-32P]ATP (Perkin-Elmer, Wellesley, MA) T4 polynucleotide kinase, and avian myeloblastosis virus reverse transcriptase (Roche) were used for primer extension. Plasmid DNA and RT-PCR products were sequenced by Macrogen, Inc. (Seoul, South Korea).
Plasmid construction.
The yeast high-copy-number vector pRS426 (2) was used to provide the backbone for construction of the tetracycline-controllable rRNA expression plasmid pJD694. First, a second ClaI restriction site was introduced into pRS426 after the URA3 terminator at position 1321 by oligonucleotide site-directed mutagenesis. The resulting plasmid was digested with ClaI and then religated to remove the f1 ori-containing sequences. In order to reduce the likelihood of recombination of large plasmids during passage through E. coli during subsequent genetic engineering manipulations, the MB1 replicon was replaced with the low-copy-number replication origin from pBR322. To this end, a PCR fragment containing both the pMB1 and the rop gene (which reduces plasmid copy numbers in E. coli) was amplified from pBR322, digested with BsaB1/BtsI, and cloned into yeast vector at SmaI/BtsI restriction sites. Next, a TET cassette with two tetO repeats derived from pCM188 (10) was subcloned into the EcoRI/HindIII-digested vector backbone. For the next step, a 525-bp HindIII tailed PCR product containing the RDN5 gene amplified from pJD180 (31) was cloned into the HindIII site of the plasmid. The resulting plasmid (pJD690) served as an empty vector control. A 6,948-bp DNA fragment encoding the primary 35S rRNA transcript (from the transcription start at +1 to the enhancer region at +6922 [21]) and which contained flanking NotI and BamHI restriction sites was synthesized by PCR from pJD180. This product was cloned into NotI/BamHI-digested pJD690 downstream of the TET cassette and in the opposite transcriptional orientation relative to RDN5. The resulting plasmid was designated pJD692. A base substitution in RDN18 promoting hygromycin resistance (37) was introduced into pJD692 by site-directed mutagenesis to make the final construct, pJD694. This is also referred to as pTET. The RDN5 and 35S rDNA regions were confirmed by DNA sequencing. The complete sequence of pJD694 is available upon request. Subsequent mutations in rDNA encoding regions of pJD694 were introduced by oligonucleotide site-directed mutagenesis, and all mutations were confirmed by sequencing.
Yeast genetic manipulations.
Transformations of yeast strain JD1314 were performed according to an alkaline cation protocol (16). Yeast cells were grown at 30°C unless otherwise indicated. Transformants were selected after 7 days of growth on galactose-containing −Trp −Ura medium with doxycycline (gal −Trp −Ura +Dox). Colonies were then streaked on gal −Trp −Ura +Dox plates and incubated for 2 days. Under these conditions, only wild-type rRNA transcribed from the GAL7 promoter was expressed, allowing establishment of the pJD694-based plasmids while silencing transcription of mutant rRNAs. Cells were then replica plated on YPAD medium to turn off transcription of the wild-type 35S rRNA from the pGAL plasmid and to turn on transcription of mutant 35S rRNAs from the pJD694-based plasmids. Colonies were simultaneously replica plated onto control plates: YPAGal, YPAGal+Dox, and YPAD+Dox. On the following day, colonies were again replica plated on the same media and incubated for 2 days, thus eliminating residual growth of cells caused by wild-type ribosomes during the transition of rRNA transcription from GAL-regulated to TET-regulated promoters. Yeast cells growing on YPAD medium were then replica plated onto YPAD medium supplemented with hygromycin B (YPAD+Hyg) and grown for 2 days. This step was to select for a pure population of yeast expressing rRNA from the pJD694-based plasmids while simultaneously selecting against cells containing wild-type rRNA, which could either remain after switching rRNA expression templates and/or be caused by leaky expression from the GAL7 promoter. These growth conditions also select only for maintenance of the pJD694-based plasmids and encourage the spontaneous loss of pNOY353. Cells were subsequently passaged three more times on YPAD+Hyg and streaked for single colonies on the same medium to ensure elimination of pNOY353. After 7 days, colonies were replica plated onto YPAGal+Dox, Gal −Trp −Ura +Dox, or YPAD medium. After 2 days, clones that grew on YPAD plates but not on galactose-containing media were inoculated into 3 ml of liquid YPAD and cultured in a shaking incubator for 24 h. Subsequently, 2.5 μl of saturated yeast suspension was spotted onto either YPAGal, YPAGal+Dox, YPAD, or YPAD+Dox medium and grown for 2 days to assess the efficiency and/or quality of pGAL plasmid elimination. Pure cultures of cells that were viable in YPAD but not in both YPAGal+Dox and YPAD+Dox were used for subsequent genetic assays and isolation of total RNA. Aliquots of these cells were frozen at −80°C as a glycerol stock.
Genetic assays.
Genetic assays were performed with selected clones containing the pJD694-based plasmids as sole sources of rDNA. Overnight cultures of yeast were used for subsequent genetic assays. General growth defects were monitored by dilution spot assay. Five sets of 10-fold dilutions were made, and 2.5 μl of each suspension was spotted onto the YPAD medium to yield a range of spots from 105 to 101 CFU/spot. Plates were incubated at 20, 30, and 37°C. For drug disk assays, overnight yeast cultures were diluted to an optical density at 592 nm of 0.2, and 300-μl portions of the resulting suspensions were plated onto YPAD medium. Subsequently, 10-μl portions of solutions containing 2 mg of anisomycin/ml or 500 mg of paromomycin/ml were applied to 6-mm-diameter filter paper discs, and the discs were air dried and placed on a plated lawn. Plates were incubated at 30°C for 3 days, and the diameters of the growth inhibition zone were measured. A minimum of at least two independent assays were used. The ability of cells to suppress a UAA nonsense codon utilized the pYDL-UAA dual luciferase reporter constructs as previously described (13). At least three readings were taken for each assay, and all assays were repeated (n = 3 to 12) until the data were normally distributed to enable statistical analyses both within and between experiments (17).
Polysome profiles and aa-tRNA binding assays.
Lysates of cycloheximide-arrested yeast cells were sedimented through 7 to 47% sucrose gradients, and polysome profiles were determined by monitoring the A254 as previously described (27). Yeast phenylalanyl-tRNA was purchased from Sigma (St. Louis, MO), charged with [14C]Phe, and purified by high-pressure liquid chromatography as previously described (18). Ribosomes were isolated from isogenic wild-type and mutant cells, and aa-tRNA binding studies were carried out as previously described (18). Nonlinear regression analysis of binding data was performed with GraphPad Prism 4.0 software (GraphPad Software, San Diego, CA). Two models were tested: (i) binding with ligand depletion and (ii) binding with both ligand depletion and nonspecific binding. The terms used in the equations below are defined as follows: [tRNA], free tRNA concentration; [tRNA0], initial concentration of tRNA; [RS], free ribosome concentration; [RS0], initial ribosome concentration; [tRNA-RS], concentration of tRNA-ribosome complex; and NS, nonspecific binding.
In the first model, the concentration of free ligand is significantly different from the initial ligand concentration. Application of [tRNA] = [tRNA0] − [tRNA-RS] and [RS] = [RS0] − [tRNA-RS] substituted into the Kd definition equation (Kd = [RS] · [tRNA]/[tRNA-RS]; [tRNA-RS] = [RS] · [tRNA]/Kd) yields the following: [tRNA-RS]2 − [tRNA-RS]([RS0] + [tRNA0] + Kd) + [RS0][tRNA0] = 0.
Solution of the second-order equation results in [tRNA-RS] = (−b − b2−4c)/2, where b = −([RS0] + [tRNA0] + Kd) and c = [RS0][tRNA0]. The binding data was fitted into this equation constraining [RS0] and Kd to positive values.
In the second model (binding with both ligand depletion and nonspecific binding), nonspecific binding can be viewed as linear in the range of the [tRNA] concentrations used. Thus, [tRNA-RS] = ([RS] · [tRNA]/Kd) + NS[tRNA] (34). Application of [tRNA] = [tRNA0] − [tRNA-RS] and [RS] = [RS0] − [tRNA-RS] substituted into this equation produces the following: [tRNA-RS]2 − [tRNA-RS]([RS0] + [tRNA0] + Kd) + [RS0][tRNA0] = 0.
Solution of the second-order equation results in [tRNA-RS] = (−b − b2−4ac)/2a, where a = 1 + NS, b = −([RS0] + [tRNA0] + Kd + KdNS + 2[tRNA0]NS), and c = [tRNA0](KdNS + [tRNA0]NS + [RS0]). The binding data was fitted into this equation with [RS0], Kd, and NS constrained to positive values. Comparison of the results generated from both tests revealed that the second test provided the best fit to the equilibrium binding curves. Thus, the data generated using the second model (binding with ligand depletion and nonspecific binding) are reported in the Results.
Chemical protection and rRNA in-line cleavage analyses.
Chemical probing with dimethyl sulfate (DMS), kethoxal, and carbodiimide metho-p-toluenesulfonate (CMCT), followed by reverse transcriptase primer extension analyses of modified rRNAs, was performed as previously described (33), with the modification that reactions were performed at 30°C instead of 37°C. The following primers were used: H38 (5′-GCTCTTCATTCAAATGTCCACG-3′), 25-2 (5′-GACTTCCATGGCCACCG-3′), 25-4 (5′-TAAAGGATCGATAGGCC-3′), 25-6 (5′-AACCTGTCTCACGACGG-3′), 25-7 (5′-CCTGATCAGACAGCCGC-3′), and A108 (5′-AGATTGCAGCACC-3′). Primer extensions from H38 covered helix 38, primer 25-2 covered bases from helices 39 to 42, primer 25-4 allowed analyses spanning helices 83 to 87, primer 25-6 covered helices 89 to 93, primer 25-7 covered bases from helices 93 to 95 of 25S rRNA, and primer A108 from 5S rRNA. For rRNA in-line cleavage assays, 150 pmol of ribosomes was incubated at 30°C in 400 μl of 50 mM Tris (pH 7.4), 5 mM Mg(CH3COO)2, and 50 mM NH4Cl. At time points of 0, 3, 6, and 12 h, 100-μl aliquots of ribosome suspensions were removed, and rRNAs were phenol extracted and used for reverse transcriptase primer extension with the H38 DNA primer. Aliquots of the reactions were separated through 8% sequencing gels and imaged by using a Molecular Imager FX (Bio-Rad).
RESULTS
Development of a method to produce yeast strains with ribosomes containing only mutant rRNAs.
As discussed above, auxotrophic selection for a gene expressed from a plasmid concurrently transcribing mutant rRNAs confers strong pressure for selection of recombinant wild-type rDNA sequences derived from the original rRNA maintenance plasmid into the vector harboring the desired auxotrophic marker. The solution to this problem was to separate the processes of auxotrophic selection from rRNA transcription. To this end, two sets of plasmid constructs were used. In pNOY353 (pGAL), 35S rRNA transcription is under control of the GAL7 promoter, i.e., transcription can be induced in the presence of galactose, and repressed by glucose. In the pJD694-based plasmids (pTET), 35S rRNA transcription is under the control of a tetracycline-repressible promoter (Tetr). Thus, transcription of rRNAs can only occur in the absence of this drug. In addition, a mutation was introduced into a pJD694-based plasmid that confers recessive resistance to hygromycin (Hygr). This mutation is useful for further driving selection against the presence of the wild-type 18S rRNA if so desired.
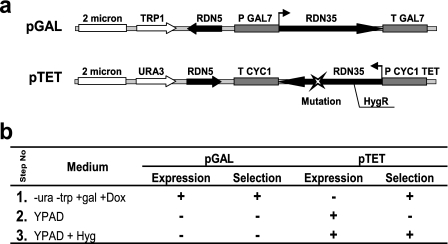
The genetic scheme is diagrammed in Fig. 1. Beginning with rdn1ΔΔ cells growing in galactose-containing medium expressing wild-type rRNAs from the galactose-inducible promoter on a high-copy TRP1 selectable plasmid (pNOY353/pGAL), cells are transformed with plasmid harboring mutant rDNA genes under control of the Tetr promoter on a high-copy URA3 selectable plasmid (pJD694/pTET), and transformants are selected for on medium lacking uracil and containing galactose and doxycycline (a stable tetracycline analog). This selects for the establishment and maintenance of the URA3-based pTET but prevents simultaneous selection against the 35S mutant allele that it contains. Once colonies have grown, these are transferred to rich medium (YPAD) containing glucose instead of galactose, conditions that allow transcription of the mutant 35S rRNA while shutting off transcription of the wild-type gene. In a third step, cells were replica plated onto rich glucose medium with hygromycin to further drive selection against remaining wild-type ribosomes and/or leaky expression of wild-type 35S transcripts. This protocol yielded colonies that were unable to grow in the presence of doxycycline, resistant to hygromycin, and viable with glucose as the sole carbon source. These results provided the initial proof of principle of the utility of the genetic manipulation strategy. pJD694 was used for subsequent oligonucleotide site-directed mutagenesis to create alleles containing mutations at specific bases of 25S rRNA.
FIG. 1.
(a) High-copy plasmids for episomal expression of rRNAs in rdn1ΔΔ cells. In the TRP1-selectable pGAL series, transcription of 35S pre-rRNA is driven from the GAL7 promoter, and transcription termination is effected by sequences in the GAL7 3′ untranslated region. In the URA3-selectable pTET series, transcription is controlled by a tetracycline-repressible promoter and transcription termination is mediated by sequences in the CYC1 3′ untranslated region. A mutation in 18S rRNA conferring recessive hygromycin resistance was added to enable selection for rRNAs produced from this plasmid. Mutations of interest are incorporated into rDNA sequences by oligonucleotide site-directed mutagenesis. (b) Three-step protocol for selection of cells expressing only mutant forms of rRNAs. Step 1 selects for the transformation of pGAL containing cells with pTET plasmid, while repressing expression of mutant rRNA species. During step 2, transcription of wild-type rRNA from pGAL is repressed, and transcription of mutant rRNA is derepressed. This step enables spontaneous loss of the pGAL. Step 3 is used to select against any wild-type rRNAs that may have resulted from recombination between the two plasmids or from leaky expression from pGAL or survived through multiple cell generations.
Generation of viable helix 38 mutants.
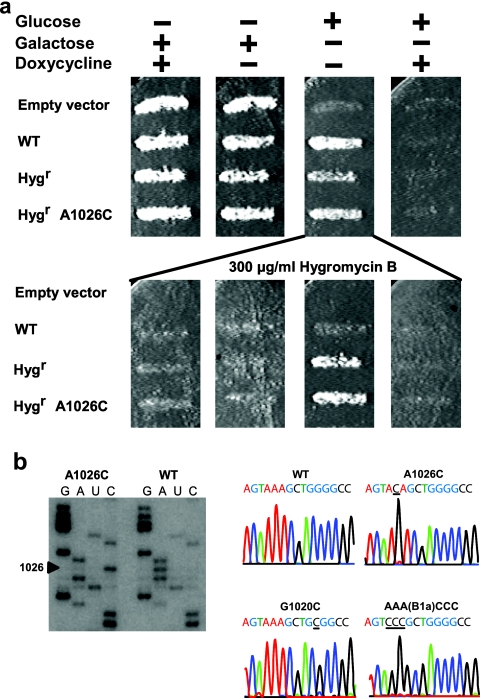
Selection of cognate aa-tRNA, is central to translational fidelity. Helix 38 is also known as the A-site finger because of its proximity to the ribosomal A-site, the pocket in the ribosome that is occupied by aa-tRNA during catalysis. The tip of the A-site finger also contacts a ribosomal protein in the small subunit (S13 in E. coli, S15 in yeast) to form the B1a intersubunit bridge (5). The C-terminal tail of S13 extends away from this bridge and inserts itself between the anticodon loops of the A- and P-site tRNAs at the decoding center (39). In light of this information, bases of helix 38 that make contacts with the D-loop of the aa-tRNA (Fig. 2, G1019 to G1021, highlighted in cyan), and those involved in the B1a intersubunit bridge (Fig. 2, A1025 through A1027, highlighted in green) were chosen for mutagenesis studies. All three possible substitutions were made for each of the six targeted bases. pJD694-derived plasmids were introduced into yeast, and cells were taken through the selection protocol described in Materials and Methods. An example of this using the A1026C allele is shown in Fig. 3a. Note that when cells capable of growth in glucose were replicated onto the hygromycin-containing set of media, only the hygromycin-resistant mutants were able to grow, and only when glucose was used as the carbon source and when transcription was derepressed in the absence of doxycycline. All of the mutants were viable, and direct sequence analysis of cellular rRNAs by reverse transcriptase extension of 32P-labeled DNA primers revealed that only the mutant rRNAs were expressed. An example of this is shown for the A1026C mutant in the left panel of Fig. 3b. Identical results were obtained from sequence analysis of PCR products generated from rRNAs extracted from cells (Fig. 3b, right panel).
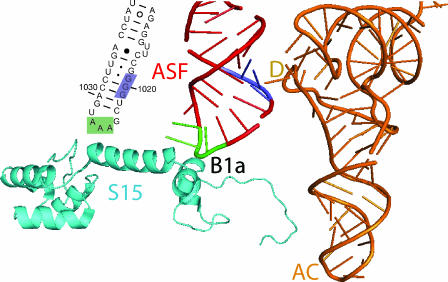
FIG. 2.
Locations of yeast 25S rRNA mutants characterized in the present study. Nucleotide sequence of the distal end of yeast helix 38 (the A-site finger [ASF]) is shown using S. cerevisiae 25S rRNA numbering. Mutagenized bases are blocked in color. The local atomic resolution structure of the E. coli ribosome (39) using PDB files 1PNS and 1PNU were modeled by using PyMOL (6). The aa-tRNA is depicted in orange, and the anticodon (AC) and D-loops (D) are indicated. Small ribosomal protein S15 is the yeast equivalent of E. coli S13. Mutagenized bases in helix 38 are colored.
FIG. 3.
Example of selection for cells expressing only mutant forms of rRNAs. (a) The top panel shows rdn1ΔΔ cells transformed with the indicated pTET plasmids grown on the indicated media. The bottom panel shows the growth phenotypes of cells in the presence of hygromycin B. Yeast expressing rRNA only from pTET plasmids (indicated by lines) were transferred on series of media containing hygromycin B. (b) Cells express only mutant 25S rRNA alleles. On the left is an autoradiogram of direct sequence analysis of rRNA for the A1026C allele. On the right are automated sequence tracings of cDNA clones made from rRNAs isolated from the indicated alleles.
The B1a and tRNA D-loop interacting mutants display distinctly different arrays of phenotypes.
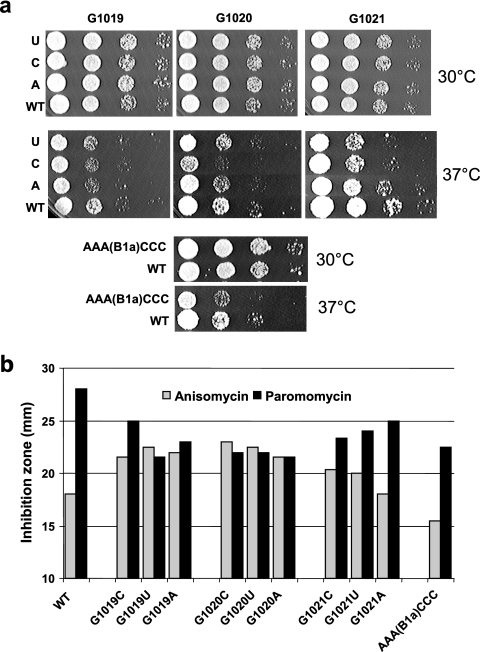
Preliminary characterization of the mutants revealed that all were remarkably healthy relative to isogenic wild-type cells. However, a mutant containing a duplication of approximately half of the A-site finger that was inadvertently generated during the mutagenesis process was inviable (data not shown), suggesting that large-scale insertions into this structure were able to significantly disrupt ribosomal function. Follow-up genetic analyses of viable alleles included assaying for heat and cold sensitivity; hypersensitivity to translational inhibitors known to interact with aa-tRNAs (anisomycin and paromomycin); suppression of near-cognate, noncognate, and nonsense mutations; and loss of the killer virus phenotype. Surprisingly, all single nucleotide substitutions involved in the B1a bridge (A1025 through A1027) were viable and showed no apparent phenotypes (data not shown). This observation prompted us to create a triple mutant, changing this stretch of three A's to CCC [mutant AAA(B1a)CCC]. Whereas the AAA(B1a)CCC allele did not affect cell growth at 30°C, it did result in reduced growth rates at 37°C relative to wild-type cells (Fig. 4a). This mutation also promoted simultaneous resistance to paromomycin and anisomycin, as scored by reductions in the sizes diameters of zones of growth inhibition surrounding filter discs containing these two drugs (Fig. 4b). A bicistronic dual luciferase reporter system was also used to monitor the effects of the AAA(B1a)CCC allele on recognition of a termination codon (nonsense suppression) (14). Rates of nonsense suppression in isogenic control wild-type cells were 2.43% ± 0.11%. The efficiency of nonsense suppression promoted by AAA(B1a)CCC allele was 4.01% ± 0.31%. Statistical analyses of the ratiometric data generated by these experiments (17) demonstrated that this (1.65 ± 0.05)-fold increase was highly significant (P < 0.01). None of the mutants in this region affected maintenance of the yeast killer virus (data not shown).
FIG. 4.
Phenotypic analyses of helix 38 mutants. (a) Tenfold dilution spot assays showing growth phenotypes of G1019N, G1020N, G1021N, and AAA(B1a)CCC alleles at 30 and 37°C. (b) Drug sensitivity phenotypes. Diameters of zones of cellular growth inhibition around 6-mm discs containing either 20 μg or anisomycin or 2.5 mg of paromomycin are indicated.
All of the single base substitutions in the tRNA D-loop interacting region (G1019 to G1021) were also viable. In contrast to the B1a region alleles, these mutants displayed a wider range of mutant phenotypes. Although none of these mutants displayed discernible growth-related phenotypes compared to isogenic wild-type control cells at 30°C, the G1020C, G1021C, and G1021U alleles grew more poorly at 37°C than wild-type cells (Fig. 4a). Interestingly, the mutants of the G1019 to G1021 series tended to display proportionally opposing phenotypes with regard to growth in the presence of anisomycin or paromomycin. In general, the degree of paromomycin resistance was matched by sensitivity to anisomycin (Fig. 4b). The most extreme example of this was observed with the G1020C allele, conferring the highest sensitivity to anisomycin and the greatest resistance to paromomycin. Since the G1020C allele produced the most pronounced phenotypes upon cells, this was chosen for more quantitative nonsense suppression measurements as described above. These studies determined that, in contrast to AAA(B1a)CCC, G1020C was hyperaccurate with regard to its ability to correctly recognize a nonsense codon, promoting 1.49% ± 0.11% nonsense suppression [(0.61 ± 0.02)-fold of the wild type, P < 0.01]. Similar to the AAA(B1a)CCC allele, none of the G1019 to G1021 mutants affected killer virus maintenance, suggesting that they did not affect maintenance of translational reading frame (data not shown).
Both the AAA(B1a)CCC and G1020C alleles result in increased ribosomal affinity for aa-tRNAs.
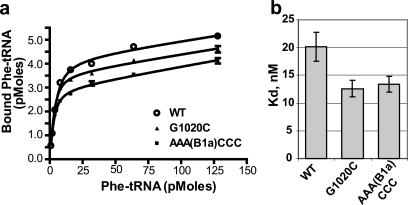
Given the physical interactions between the A-site and helix 38 and the effects of the two classes of mutants on translational fidelity, it was reasonable to hypothesize that ribosomes harboring these mutants may have altered affinities for aa-tRNAs. To test this, aa-tRNA binding to the ribosomal A-site was determined for purified wild-type ribosomes and for those containing the AAA(B1a)CCC and G1020C mutations. Care was taken to block nonspecific binding of [14C]Phe-tRNA to ribosomal E- and P-sites by preincubating ribosomes with uncharged tRNAs. The results of these experiments demonstrate that the Kd values for wild-type, G1020C, and AAA(B1a)CCC ribosomes were 20.2 ± 2.6, 12.6 ± 1.5, and 13.36 ± 1.4 nM, respectively (Fig. 5). Polysome profiles revealed no significant differences between the wild type and the two mutants (data not shown).
FIG. 5.
aa-tRNA binding studies. aa-tRNA binding to A-site of ribosome was carried out in 100-μl reaction mixtures containing 25 pmol of ribosomes primed with 25 μg of poly(U). Reaction mixtures were preincubated with 10 μg of uncharged tRNAs at 30°C for 30 min to ensure full occupation of P-sites by uncharged tRNA, after which increasing amounts of (1 to 128 pmol) of [14C]Phe-tRNA were added. Incubations continued for 10 min at 30°C to allow the formation of [14C]Phe-tRNA · 80S · poly(U) complexes. Reaction mixtures were applied onto nitrocellulose membranes, filters were washed with binding buffer, and radioactivity was measured by scintillation counting. Binding isotherms were fitted to the data (a) and Kd values (b) were calculated by using GraphPad Prism 4 as described in Materials and Methods.
rRNA structure analyses.
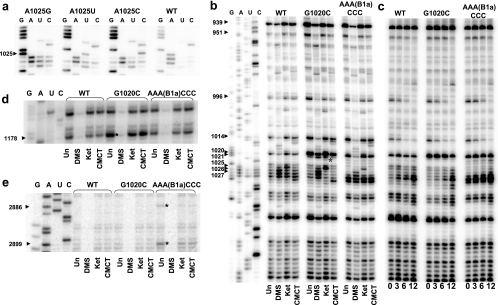
Initial inspection of reverse transcriptase sequencing reactions showed that mutations of any of the three A residues in the B1a region at the tip of helix 38 promoted strong barriers to progression of the reverse transcriptase through this sequence compared to the wild type. Examples of these are shown in Fig. 6a. Such patterns are typical of “in-line” cleavage, where disordering of normally structured RNA increases the frequency of spontaneous cleavage of an RNA phosphodiester linkage via an internal nucleophilic attack by the 2′ oxygen on the adjacent phosphorous center (32). These results suggest that the tip of helix 38 is normally highly structured inside of the ribosome, i.e., when participating in the B1a bridge, and that this is disrupted by the mutations. Similarly, a strong stop at G1020 was observed for the G1020C mutant independent of the presence or absence of chemical probes and, to a lesser extent, with AAA(B1a)CCC (Fig. 6b). This was expected for the G1020C mutant insofar as such a change should disrupt base pairing, increasing the probability of in-line cleavage. That this pattern was also observed for the AAA(B1a)CCC mutant suggests that the effect of the structural change at the tip of helix 38 is propagated up into the body of the helix itself.
FIG. 6.
rRNA structural analyses. Sequencing ladders and primer extensions were generated with avian myeloblastosis virus reverse transcriptase using rRNAs isolated from ribosomes. Un, untreated samples; DMS, ribosomes treated in vitro with DMS; Ket, ribosomes treated in vitro with kethoxal; CMCT, ribosomes treated in vitro with CMCT. (a) Direct sequencing of the distal tip of helix 38 from rRNAs extracted from cells expressing the wild-type, G1020C, or AAA(B1a)CCC alleles. (b) Structure probing of helix 38. Individual nucleotide positions of bases in mutant samples showing enhanced in line rRNA cleavage are indicated by arrows. The asterisk indicates a strong stop at nucleotide 1021 in the G1020C mutation. (c) Time course in-line cleavage experiments. Equal amounts of freshly prepared ribosomes purified from isogenic wild-type and mutant cells were incubated at 30°C in buffer without chemical modifying agents for 0, 3, 6, and 12 h, after which rRNAs were extracted, used for reverse transcriptase primer extension with the primer described above, and separated through an 8% acrylamide-urea gel. (d) Structure probing of helix 41. The asterisks indicate enhanced in-line cleavage at A1178 specific to the G1020C mutant. (e) Structure probing along helix 91. Asterisks indicate enhanced in line cleavage of bases in the AAA(B1a)CCC mutant.
Chemical probing experiments also revealed that the two mutants promoted numerous changes in rRNA structure at the tip of helix 38 (Fig. 6b). As expected, the G residue at position 1021 in the G1020C mutant showed increased kethoxal-specific deprotection (Fig. 6b, asterisk). Also as anticipated, the wild-type AAA sequence (bases 1025 to 1027) reacted strongly with DMS, G1024 reacted with kethoxal, and U1028 reacted with CMCT. The same pattern was observed for G1020C, suggesting that this mutation does not affect the structure of the tip of helix 38. In contrast, strong stops observed at positions 1028 and 1027 in untreated rRNA extracted from the AAA(B1a)CCC mutant support the hypothesis that an ordered structure normally present in this loop had been disrupted. More-distant base-specific changes in 25S rRNA consistent with in-line cleavage were also identified. The normally unpaired bases U939, A951, A966, and U1014 all showed increased in-line reactivity in both the G1020C and the AAA(B1a)CCC mutants, suggesting that the structural changes promoted by these mutations were propagated further along helix 38 and into nearby structures as well (Fig. 6b).
Over the course of a large number of chemical protection and RT primer experiments, we observed that the intensity of the bands at these sites increased in proportion with both of the ages of the ribosome preparations, i.e., weaker banding patterns were observed with freshly prepared ribosomes versus samples that had been stored at −80°C. These observations are consistent with the hypothesis that the mutants were affecting the in-line cleavage of specific unpaired bases in helix 38. To further investigate the in-line cleavage effect, a series of time course experiments were performed with untreated ribosomes. Equal amounts of freshly prepared ribosomes purified from isogenic wild-type and mutant cells were incubated at 30°C in buffer without chemical modifying agents for 0, 3, 6, and 12 h, after which rRNAs were extracted and used for reverse transcriptase primer extension using the H38 DNA primer. The results of this experiment are shown in Fig. 6c. Examination of the nonspecific bands in the lower portion of the figure shows that all samples contained equal amounts of rRNA. The remainder of the figure reveals base-specific increases or decreases in sensitivity to self-cleavage over time as at the same positions observed in the untreated, single-time-point samples shown in Fig. 6b. These results are consistent with the hypothesis that the G1020C and AAA(B1a)CCC mutants were exerting subtle, long-range effects on the structure of helix 38. Increased mutant specific in-line reactivities were also observed more distal to the sites of the mutations. The G1020C allele affected the stability of G1178, which is located in a bend of helix 41 (Fig. 6d). Increased in-line in AAA(B1a)CCC specific reactivity was also observed in helix 91 along the A-site proximal side of the peptidyltransferase center at bases A2886 and A2899 (Fig. 6e).
DISCUSSION
Robust system for rRNA structure-function studies in a model eukaryotic organism.
The atomic resolution pictures of ribosomes have driven home the point that rRNAs are the central players in this molecular machine. Unfortunately, the presence in genomes of multiple copies of rRNA encoding genes has complicated structure-function analyses of these molecules. rDNA gene amplification and the evolution of specialized RNA polymerases for rRNA transcription in eukaryotes is likely due to the fact that growing cells require large amounts of ribosomes (41). Although attempts to address this problem in a model eukaryotic system go back over 15 years, only a limited number of yeast mutants were originally identified, and no new ones have been generated for a decade. The problem of making yeast rRNA mutants has now been solved by separating selection for auxotrophic markers on plasmid vectors from selection for or against the rRNA transcripts encoded by genes into which they are cloned. The genetic system presented here is theoretically applicable to the any base in any of the four rRNAs, providing the tools to enable higher-resolution structure-function studies in this model eukaryote. As with all yeast mutants, however, a certain degree of recombination and/or reversion was observed to have occurred during the time when cells contained two plasmids, resulting in ∼10% of colonies containing only wild-type ribosomes. Thus, care must be taken to initially sequence a few samples of ribosomes extracted from cells derived from single colonies. However, once established, these alleles were as genetically stable as any other yeast mutants (i.e., spontaneous revertants and probable second-site suppressors were observed with normal frequencies). Second, at an observational level we noted that isogenic rdn1ΔΔ cells in which 35S rRNAs are transcribed from either pGAL or pTET tend to grow more slowly than cells in which 35S rRNAs are transcribed from the native promoter. This could be due to limiting quantities of RNA polymerase II, differences in relative promoter strengths, or differences in chromatin structure affecting rRNA transcription rates. Experiments are ongoing to investigate this phenomenon and to generate a more robust system for producing yeast cells that express only mutant forms of 25S rRNA.
Helix 38 structure-function analyses.
Studies in E. coli have examined both components of the B1a bridge. Deletion of the C-terminal tail of the small subunit partner S13 was viable, although cell doubling times were increased ∼2-fold, and in vitro studies showed that the P-site of this mutant had decreased affinity for tRNA (15). Reconstituted ribosomes completely lacking S13 were translationally competent but were refractive to thiol-modifying reagents, suggesting that S13 is involved in controlling EF-G-independent features of translocation (4). A follow-up study using S13-deficient E. coli revealed roles for this protein in subunit association, formation of translation initiation complexes, factor-dependent translocation, reading frame maintenance, and termination codon recognition (3). A complementary study focusing on the other side of the B1a bridge in E. coli showed that deletion of approximately one-third of the distal tip of helix 38 yielded ribosomes that were viable, and genetic analyses of this mutant revealed no significant functional differences in any of the basic steps of the elongation cycle (30). However, a small decrease in EF-G-stimulated GTPase activity was observed, and chemical probing experiments revealed changes in bases of 5S rRNA and in the large subunit rRNA located between the peptidyltransferase center and the elongation factor binding site. All of these findings support the hypothesis that this structure may be involved in the fine tuning of ribosomal activity (3) and that the B1a bridge may function as part of an allosteric communication pathway between the large and small subunits (30).
Although spatially close to one another, the two stretches of three nucleotides examined in the present study interact with different structures in the ribosome. This is reflected by both similarities and differences in their functional phenotypes. For example, neither class of mutants affected the ability of cells to maintain the yeast killer virus (a proxy monitor for changes in translational reading frame maintenance [see reference 7]). These observations are consistent with the E. coli helix 38 deletion studies, suggesting that this structure does not play a major role in elongation (30). However, the two classes of mutants examined in the present study did affect other aspects of translation, in some cases to similar extents, but dissimilarly in others. For example, heat sensitivity is suggestive of destabilization of intra- and/or intermolecular interactions, which is consistent with the observed frequencies of in-line cleavage of specific unpaired bases. The drug sensitivity and nonsense suppression assays were particularly interesting in that they yielded dissimilar and novel phenotypes. Anisomycin is known to promote decreased affinity for aa-tRNA by competing for their mutual binding site in the peptidyltransferase center (11, 12). Conversely, paromomycin can suppress nonsense mutations by increasing the affinity of noncognate aa-tRNAs termination codons in the decoding center (reviewed in reference 24). That both mutants were resistant to paromomycin but had opposite phenotypes in response to anisomycin suggests subtle, but differing functions of these two regions of the A-site finger in translational fidelity. That their effects on aa-tRNA affinity are similar may reflect cumulative effects on this function of the ribosome. However, aa-tRNAs interact with many different parts of the ribosome, suggesting that the changes in the quality of the interactions between the ribosome and aa-tRNA interactions may be more important than quantitative changes. The opposing effects of the mutants on the suppression of a nonsense codon are particularly informative. Like the S13 deletion studies (3), these findings implicate this region of helix 38 in factor-dependent ribosome function, i.e., eRF1/eRF3-mediated translation termination. Interestingly, although both the B1a bridge and G1020 may function as sensors of the A-site, they may differ in their specificity with regard to whether the A-site is occupied by aa-tRNA or eRF1. The in-line cleavage pattern of the AAA(B1a)CCC mutant showing increased flexibility of the hinge bases in helix 91 along the path of accommodation of the aa-tRNA (29) is particularly interesting in this aspect. We suggest that increased affinity of aa-tRNA in this mutant may be due to a more flexible rRNA environment, which may in turn lessen the ability of ribosomes to discriminate between a codon-anticodon base pair versus the eRF1-termination codon interaction. With regard to G1020C, we speculate that this mutant is more discriminatory with regard to the differences between the tRNA D-loop and the analogous topological region of eRF1. Thus, in this mutant, an increased ability to discriminate between the presence or absence of the D-loop may affect signal transmission to the GTPase-associated center, accounting for the altered reactivity of G1178 and the enhanced ability to discriminate between sense and nonsense codons. The opposing nonsense suppression and anisomycin sensitivity phenotypes and the allele-specific differences in in-line cleavage patterns (discussed below) presented by the two mutants in the present study provide supporting evidence for this model.
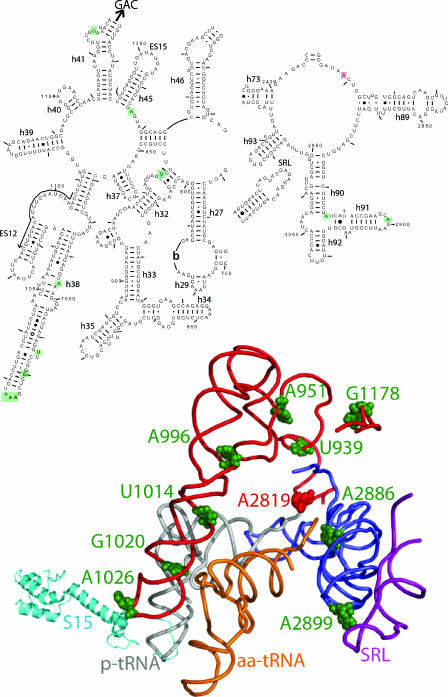
Significant rearrangements of rRNA elements have been demonstrated depending on the stage of the elongation cycle during which the ribosome samples were stalled (1, 8, 23, 35, 36, 44). More recently, a study examining elongation arrest by the SecM peptide in E. coli ribosomes has shown that these rRNA rearrangements proceed through a cascade of events (19). The question is, what is it about the architecture of rRNA that allows such cascades to occur, thus enabling the coordination of information sharing among spatially separate regions of the ribosome? It has been suggested that single-stranded regions of rRNAs, called RNA kink turns, can function to confer flexibility on more rigid helical rRNA protuberances. RNA kink-turn motifs have been likened to “molecular hinges” that allow large motions and/or smooth assembling of large portions of the ribosome (25, 26). When the bases in which in-line cleavage patterns are enhanced by the two mutants are mapped onto the atomic resolution ribosome structure, a striking pattern emerges: they appear to form a precisely spaced series of points forming an arc surrounding the tRNA binding pocket of the large subunit (Fig. 7). A simple “connect the dots” exercise suggests a large-scale communication bridge connecting the decoding center on the small subunit (the C-terminal tail of S15), with the large subunit through the B1a bridge, to the aa-tRNA D-loop, and up the body of the A-site finger. The allele-specific differences suggest a functional bifurcation. G1178 lies in a bend of helix 41, along the pathway to the GTPase-associated center. The increased reactivity of this base in the G1020C mutant suggests a functional link between the D-loop interacting region of helix 38 and the GTPase-associated center. A2886 and A2899 form a bend at the base helix 91 in the peptidyltransferase center and at its distal tip, respectively. This latter base interacts with the sarcin/ricin loop, which in turn interacts with elongation factors.
FIG. 7.
An arc of unpaired “hinge bases” facilitate information exchange among functional centers of the ribosome. The illustration at the top of the figure is a two-dimensional map showing the regions of yeast 25S rRNA of interest. Bases boxed in green show enhanced in-line cleavage in the 25S rRNA mutants. A2819 in the core of the peptidyltransferase center is blocked in red. Helices and expansion segments are indicated. The locations of every 50th base are labeled, and every 10th base is indicated by tick marks. The arrow indicates the direction to the GTPase-associated center (GAC). The lower diagram shows the positions of bases of interest modeled onto the atomic resolution structure of the E. coli ribosome (39). The labeling follows the conventions described in Fig. 2. The peptidyl-tRNA (p-tRNA) is gray, aa-tRNA is orange, the helices 32, 37, 38, and 41 are red, helix 91 is blue, and the sarcin/ricin loop (SRL) is purple. Hinge bases are shown as green spheres, and A2819 is shown in red.
We propose that the occupancy status of the A-site is sampled along these arcs. The C-terminal end of S15 (E. coli S13) samples the space between the A- and P-sites in the decoding center, and information pertaining to whether or not the A-site is occupied is transmitted to the large subunit through the B1a bridge. Ablation of this bridge by the AAA(B1a)CCC mutation impairs this communication pathway. We also suggest that G1020 in helix 38 functions as a sensor of the presence or absence of the aa-tRNA D-loop. We propose that the presence or absence of aa-tRNA (or of eRF1) influences the spatial orientation of these two sensors and that this is converted into physical information. This information is transduced through the arc of unpaired bases identified in the present study, which may serve as hinges around which rigid helices can reorient. The regular spacing of these hinges suggests that this functional arc is finely articulated, allowing a high degree of flexibility among the more rigid helical components. In fact, the term “A-site finger” is quite apt: like a finger it contains a series of joints that provide a large dynamic range of flexibility. Thus, information pertaining to the occupancy status of the A-site can be faithfully transmitted to other regions of the ribosome, i.e., the GTPase-associated center, which is activated consequent to proper occupation of the A-site, the peptidyltransferase center that interacts with the 3′ ends of the tRNAs, and the sarcin/ricin loop, which interacts with the elongation and release factors.
Acknowledgments
We thank the members of the Dinman laboratory, with special thanks to John Russ for his help in producing figures using PyMol, Alexey Petrov for help with biochemical assays, and Arturas Meskauskas for his keen insights.
This study was supported by a grant from the Public Health Service to J.D.D. (GM58859).
Footnotes
Published ahead of print on 25 September 2006.
REFERENCES
- 1.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340-348. [DOI] [PubMed] [Google Scholar]
- 2.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Yeast 110:119-122. [DOI] [PubMed] [Google Scholar]
- 3.Cukras, A. R., and R. Green. 2005. Multiple effects of S13 in modulating the strength of intersubunit interactions in the ribosome during translation. J. Mol. Biol. 349:47-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cukras, A. R., D. R. Southworth, J. L. Brunelle, G. M. Culver, and R. Green. 2003. Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA:tRNA complex. Mol. Cell 12:321-328. [DOI] [PubMed] [Google Scholar]
- 5.Culver, G. M., J. H. Cate, G. Z. Yusupova, M. M. Yusupov, and H. F. Noller. 1999. Identification of an RNA-protein bridge spanning the ribosomal subunit interface. Science 285:2133-2135. [DOI] [PubMed] [Google Scholar]
- 6.DeLano, W. L. 2006. The PyMOL molecular graphics system. [Online.] http://www.pymol.org.
- 7.Dinman, J. D., and R. B. Wickner. 1992. Ribosomal frameshifting efficiency and Gag/Gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 66:3669-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank, J., and R. K. Agrawal. 2000. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406:318-322. [DOI] [PubMed] [Google Scholar]
- 9.Fried, H. M., and G. R. Fink. 1978. Electron microscopic heteroduplex analysis of “killer” double-stranded RNA species from yeast. Proc. Natl. Acad. Sci. USA 75:4224-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gari, E., L. Piedrafita, M. Aldea, and E. Herrero. 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13:837-848. [DOI] [PubMed] [Google Scholar]
- 11.Grollman, A. P. 1967. Inhibitors of protein biosynthesis. II. Mode of action of anisomycin. J. Biol. Chem. 242:3226-3233. [PubMed] [Google Scholar]
- 12.Hansen, J. L., P. B. Moore, and T. A. Steitz. 2003. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 330:1061-1075. [DOI] [PubMed] [Google Scholar]
- 13.Harger, J. W., and J. D. Dinman. 2004. Evidence against a direct role for the Upf proteins in frameshifting or nonsense codon readthrough. RNA 10:1721-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harger, J. W., and J. D. Dinman. 2003. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA 9:1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoang, L., K. Fredrick, and H. F. Noller. 2004. Creating ribosomes with an all-RNA 30S subunit P site. Proc. Natl. Acad. Sci. USA 101:12439-12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs, J. L., and J. D. Dinman. 2004. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res. 32:e160-e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meskauskas, A., A. N. Petrov, and J. D. Dinman. 2005. Identification of functionally important amino acids of ribosomal protein L3 by saturation mutagenesis. Mol. Cell. Biol. 25:10863-10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitra, K., C. Schaffitzel, F. Fabiola, M. S. Chapman, N. Ban, and J. Frank. 2006. Elongation arrest by SecM via a cascade of rRNA rearrangements. Mol. Cell 22:533-543. [DOI] [PubMed] [Google Scholar]
- 20.Nogi, Y., L. Vu, and M. Nomura. 1991. An approach for isolation of mutants defective in 35S rRNA synthesis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88:7026-7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nogi, Y., R. Yano, and M. Nomura. 1991. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc. Natl. Acad. Sci. USA 88:3962-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oakes, M., J. P. Aris, J. S. Brockenbrough, H. Wai, L. Vu, and M. Nomura. 1998. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J. Cell Biol. 143:23-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogle, J. M., F. V. Murphy, M. J. Tarry, and V. Ramakrishnan. 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111:721-732. [DOI] [PubMed] [Google Scholar]
- 24.Ogle, J. M., and V. Ramakrishnan. 2005. Structural insights into translational fidelity. Annu. Rev. Biochem. 74:129-177. [DOI] [PubMed] [Google Scholar]
- 25.Razga, F., N. Spackova, K. Reblova, J. Koca, N. B. Leontis, and J. Sponer. 2004. Ribosomal RNA kink-turn motif: a flexible molecular hinge. J. Biomol. Struct. Dyn. 22:183-194. [DOI] [PubMed] [Google Scholar]
- 26.Razga, F., M. Zacharias, K. Reblova, J. Koca, and J. Sponer. 2006. RNA kink-turns as molecular elbows: hydration, cation binding, and large-scale dynamics. Structure 14:825-835. [DOI] [PubMed] [Google Scholar]
- 27.Sachs, A. B., and R. W. Davis. 1989. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell 58:857-867. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Sanbonmatsu, K. Y., S. Joseph, and C. S. Tung. 2005. Simulating movement of tRNA into the ribosome during decoding. Proc. Natl. Acad. Sci. USA 102:15854-15859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sergiev, P. V., S. V. Kiparisov, D. E. Burakovsky, D. V. Lesnyak, A. A. Leonov, A. A. Bogdanov, and O. A. Dontsova. 2005. The conserved A-site finger of the 23S rRNA: just one of the intersubunit bridges or a part of the allosteric communication pathway? J. Mol. Biol. 353:116-123. [DOI] [PubMed] [Google Scholar]
- 31.Smith, M. W., A. Meskauskas, P. Wang, P. V. Sergiev, and J. D. Dinman. 2001. Saturation mutagenesis of 5S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:8264-8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soukup, G. A., and R. R. Breaker. 1999. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5:1308-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stern, S., D. Moazed, and H. F. Noller. 1988. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 164:481-489. [DOI] [PubMed] [Google Scholar]
- 34.Swillens, S. 1995. Interpretation of binding curves obtained with high receptor concentrations: practical aid for computer analysis. Mol. Pharmacol. 47:1197-1203. [PubMed] [Google Scholar]
- 35.Tama, F., M. Valle, J. Frank, and C. L. Brooks III. 2003. Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryo-electron microscopy. Proc. Natl. Acad. Sci. USA 100:9319-9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valle, M., A. Zavialov, J. Sengupta, U. Rawat, M. Ehrenberg, and J. Frank. 2003. Locking and unlocking of ribosomal motions. Cell 114:123-134. [DOI] [PubMed] [Google Scholar]
- 37.Velichutina, I. V., J. Y. Hong, A. D. Mesecar, Y. O. Chernoff, and S. W. Liebman. 2001. Genetic interaction between yeast Saccharomyces cerevisiae release factors and the decoding region of 18 S rRNA. J. Mol. Biol. 305:715-727. [DOI] [PubMed] [Google Scholar]
- 38.Venema, J., A. Dirks-Mulder, A. W. Faber, and H. A. Raue. 1995. Development and application of an in vivo system to study yeast rRNA biogenesis and function. Yeast 11:145-156. [DOI] [PubMed] [Google Scholar]
- 39.Vila-Sanjurjo, A., W. K. Ridgeway, V. Seymaner, W. Zhang, S. Santoso, K. Yu, and J. H. Cate. 2003. X-ray crystal structures of the WT and a hyper-accurate ribosome from Escherichia coli. Proc. Natl. Acad. Sci. USA 100:8682-8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wai, H. H., L. Vu, M. Oakes, and M. Nomura. 2000. Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res. 28:3524-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 42.Wickner, R. B., and M. J. Leibowitz. 1976. Two chromosomal genes required for killing expression in killer strains of Saccharomyces cerevisiae. Genetics 82:429-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292:883-896. [DOI] [PubMed] [Google Scholar]
- 44.Zavialov, A. V., and M. Ehrenberg. 2003. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell 114:113-122. [DOI] [PubMed] [Google Scholar]