Abstract
Although the regulation of several Bcl-2 family molecules, including Puma, Noxa, Bax, and Bid, by p53 has been studied intensively, the interplay between Bad (Bcl-2 antagonist of cell death) and p53 has not yet been reported thus far. Here, we report that p53 activates Bad transcription and expression through binding to a short conserved sequence located approximately 6.6 kb upstream of the translation start point. We also demonstrate that Bad physically interacts with cytoplasmic p53, thereby preventing p53 from entering the nucleus and resulting in reduced transcription of Bad. Moreover, Bad is able to direct p53 to the mitochondria and forms a p53/Bad complex at the mitochondria. Two lines of evidences support this hypothesis: first, when mitochondria purified from p53-deficient H1299 cells are incubated with p53 and either wild-type (wt) Bad or mutant Bad (this mutant binds p53 yet is unable to migrate to mitochondria), p53 can be detected only in mitochondria incubated with wt Bad and not in those incubated with mutant Bad; second, knockdown of Bad expression reduces mitochondrial localization of p53. The mitochondrial p53/Bad complex promotes apoptosis via activation and oligomerization of Bak. Elimination of Bad expression by RNA interference notably attenuates apoptosis induced by etoposide. Hence, our collective data provide the first evidence that Bad plays dual roles in both p53 transcription-dependent and -independent pathways.
Bad (Bcl-2 antagonist of cell death), a member of the Bcl-2 protein family, is a positive regulator of cell death and is localized in the cytoplasm under physiological conditions. Like other BH3-only proteins, Bad selectively displaces Bax to heterodimerize with Bcl-2 or Bcl-xL while promoting apoptosis (5, 25). The apoptotic activity of Bad is determined largely by its phosphorylation status at serine 112 (Ser112), Ser136, and Ser155 (5). In unstressed cells, Bad is hyperphosphorylated by several protein kinases, including protein kinase A and protein kinase B (AKT), and as a result is held onto by 14-3-3 (7, 11). However, in response to apoptotic stimuli, Bad is rapidly dephosphorylated and migrates to the mitochondria, where it induces cell death.
The p53 tumor suppressor is a pivotal mediator of cell cycle arrest and apoptosis in response to diverse stress stimuli. It transactivates various target genes, including those for p21waf1 (9), Bax (16), p53DINP1 (18), Bid (20), Puma (17), and many other proteins (21, 24). However, a steady input of evidence for the transcription-independent function of p53 in apoptosis has been accumulating recently (13, 15). Although a detailed mechanism has not yet been determined, recent data have revealed that upon an apoptotic stimulus, p53 translocates into mitochondria to interact directly with Bcl-2 or Bcl-xL to displace and activate proapoptotic Bax signaling (6, 15). Similarly, disruption of the association complex between Bak/Mcl-1 or Bak/Bcl-2 by p53 is also related to p53-dependent but transcription-independent apoptosis (14). In this study, we have identified a p53-responsive element located approximately 6.6 kb upstream of the ATG translational start codon of the Bad gene and have shown that p53 transcriptionally activates Bad expression through binding to this region. We provide evidence that Bad is part of a negative feedback loop control system serving to maintain the equilibrium level of Bad by reducing p53 nuclear entry. Excess Bad binds to p53 within the cytosol, preventing p53 from entering the nucleus for further transcription of bad. Moreover, we demonstrate that Bad is able to direct p53 to the mitochondria and facilitates the intrinsic mitochondrion-mediated apoptosis through activation and oligomerization of Bak.
MATERIALS AND METHODS
Reagents and antibodies.
The following antibodies were used in this study: monoclonal antibodies anti-Bad, anti-Bax, and anti-Bcl-xL (Santa Cruz Biotechnology, Santa Cruz, CA); anti-p53 (Oncogene, Manhasset, NY); anti-green fluorescent protein (GFP) (BD Biosciences, Palo Alto, CA); anti-Flag (Sigma, St Louis, MO); anti-cytochrome c (R&D Systems, Inc., Minneapolis, MN); anti-β-actin (Abcam, Cambridge, United Kingdom); anti-OxPhosComplex IV subunit IV antibody (COX IV; Molecular Probes, Eugene, Oregon); and anti-β-tubulin (Santa Cruz) and polyclonal antibody anti-Max (Santa Cruz). Hoechst 33342 and etoposide were purchased from Sigma, and 1,6-bismaleimidohexane (BMH) was purchased from Pierce Biotechnology (Rockford, IL).
RNA interference (RNAi) and apoptosis assays.
A549 (p53+/+) cells were cultured in 12-well plates to subconfluence (3 × 105 cells) and transiently transfected with small interfering RNA (siRNA) against Bad (Cell Signaling Technology, Danvers, Massachusetts) by using Oligofectamine (Invitrogen Cergy Pontoise, France). At 24 h posttransfection, the culture medium was replaced with fresh medium, followed by addition of etoposide at 50 μg/ml. After 24 h and 48 h of incubation, both floating and adherent cells were stained with Hoechst 33342 (2 μg/ml). Microscopic fields were selected randomly, and 10 fields were observed for each slide at a magnification of ×400. The percentages of apoptotic cells among the total number of cells were counted based on the numbers of apoptotic cells characteristic for DNA fragmentation and/or chromatin condensation under fluorescence microscopy. The data collected are the averages and standard deviations from three independent experiments.
Subcellular fractionation.
Cell fractionation was carried out using a protocol described previously (10). Briefly, cells were homogenized in 20 mM HEPES-KOH (pH 7.5), 10 mM KCl, 1.5 mM MgCl2, 1 mM sodium EDTA, 1 mM sodium EGTA, and 1 mM dithiothreitol in the presence of 250 mM sucrose and protease inhibitor cocktail (Roche Diagnostics, Meylan, France). Homogenates were centrifuged at 500 × g for 5 min at 4°C, and the supernatant was collected and centrifuged again at 10,000 × g for 20 min to obtain cytoplasmic and mitochondrial fractions. The mitochondrial pellet was washed and solubilized in TNC buffer (10 mM Tris acetate [pH 8.0], 0.5% Nonidet P-40, 5 mM CaCl2) containing protease inhibitors. The protein concentration was determined with a Micro-BCA kit (Pierce).
ChIP assay.
Chromatin immunoprecipitation (ChIP) assay was carried out essentially as described previously (12, 20). Briefly, A549 (p53+/+) (or HeLa) cells were first cross-linked with formaldehyde (final concentration, 1%) for 10 min at room temperature, and then the reaction was stopped with glycine (0.125 M). Nuclei were then isolated and resuspended in lysis buffer (50 mM Tris [pH 8.1], 10 mM EDTA, 1% sodium dodecyl sulfate, and protease inhibitors). The resulting chromatin was mixed with protein A/G-Sepharose beads conjugated either with anti-p53 antibody (Ab-2), or with anti-E2F1 antibody as a negative control. The bound DNA fragments were eluted as described by Hershko and Ginsberg (12) and amplified by PCR.
Luciferase reporter gene assay.
HeLa cells were transiently transfected with the indicated plasmids by using Lipofectamine 2000 (Invitrogen). A 422-bp DNA fragment containing the potential p53-binding sequence (ACCCAGGCCCTGGCTGGTCCT) was amplified by PCR with primers used in the ChIP assay and was cloned into pGL3-Basic vector (Promega). To construct a mutant vector, six bases (bases 8 to13) starting from the fourth nucleotide (C) within the putative 21-bp p53-binding region (p53-BR) were deleted by PCR-mediated site-directed mutagenesis as described by Song et al. (21a) The luciferase reporter assay was performed according to the manufacturer's instructions (Promega). The quantification of luciferase activities and calculation of relative ratios were carried out manually with a luminometer (Junior LB9509; Berthold Technologies). Transfection efficiency was normalized on the basis of the Renilla luciferase activity.
RESULTS
p53 transactivates Bad expression.
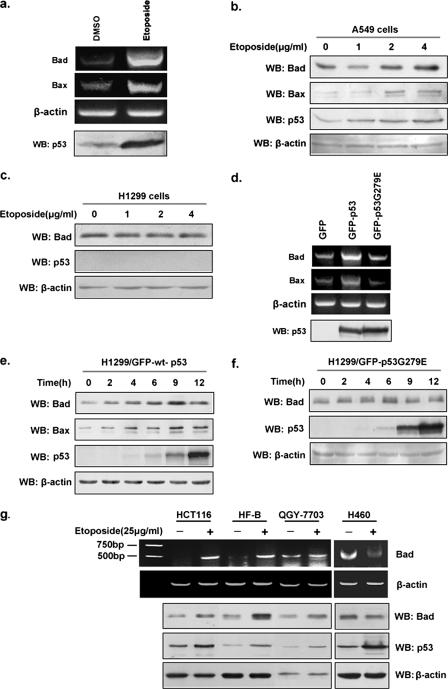
To determine whether p53 regulates the expression of the proapoptotic BH3-only protein Bad, reverse transcription-PCR (RT-PCR) and Western blot analyses were performed to examine the effects of endogenous p53 on Bad expression in A549 (p53+/+) cells treated with etoposide (50 μg/ml). When cells were treated with increasing amount of etoposide (0 to 4 μg/ml), a p53-induced increase in both Bad mRNA and protein levels were found in A549 cells (Fig. 1a and b) but not in p53-deficient H1299 (p53−/−) cells (Fig. 1c), indicating that the DNA-damage induced increases in Bad levels are p53 dependent. We next tested whether the increases in the levels of both Bad mRNA and protein were dependent on p53's transactivation activity. H1299 cells were transiently transfected with constructs expressing GFP, GFP-wild-type (wt) p53, or GFP-p53G279E (transcriptionally dead but DNA-binding-proficient p53) (23). As shown in Fig. 1d, the Bad mRNA levels in wild-type p53-expressing cells are higher than those in p53G279E-expressing cells. Similarly, levels of Bad protein showed a time-dependent increase in wild-type p53-expressing cells (Fig. 1e), whereas in p53G279E-expressing cells, Bad protein levels remain unchanged (Fig. 1f), indicating that wild-type p53 transactivation function is required for the up-regulation of Bad. We also noticed that Bad protein becomes detectable prior to p53 protein; this could be due to the quality of antibodies used. To confirm that transcriptional regulation of Bad by p53 was not restricted to A549 (p53+/+) cells, we examined the effect of p53 on Bad expression in four other cell lines. As shown in Fig. 1g, increased levels of both Bad mRNA and protein were observed in HCT116, QGY-7703, and HF-B (primary human fibroblast) cells treated with etoposide but not in those cells without treatment, indicating that the increased transcription of Bad is due to the up-regulation of p53. However, H460 cells stimulated with etoposide were devoid of Bad expression upon p53 induction. The p53-induced Bad expression occurs in the majority of but not all different cell lines tested (five out of six cell types). Thus, we hypothesize that this may represent a general phenotype. Nonetheless, different cell lines, such as H460 cells, could exhibit a cell-type-dependent variation in p53-induced Bad transcription.
FIG. 1.
p53 up-regulates Bad expression. a. The mRNA levels of Bad in A549 (p53+/+) cells treated with etoposide (50 μg/ml) or dimethyl sulfoxide (DMSO) for 24 h were compared by RT-PCR. Protein levels of p53 were examined by Western blotting (WB). b. A549 cells were treated as for panel a with various indicated concentrations of etoposide, followed by Western blot analysis. Bax was used as a positive control. c. H1299 (p53−/−) cells were treated with etoposide as described for panel b, and 24 h later, Western blot analysis of p53 and Bad was performed. d. H1299 cells were transiently transfected with pEGFP/C1, pEGFP/wt-p53, and pEGFP/mu-p53G279E individually; 24 h later, cells were harvested, total RNA was extracted, and RT-PCR was performed using the specific primers for Bad (5′-GGAATTCCATGTTCCAGATCCCAGAG-3′ and 5′-CTCGAGCTACTGGGAGGGGGCGGA-3′), Bax (5′-GCGAATTCCATGGACGGGTCCGGGGAG-3′ and 5′-CGCTCGAGGCCCATCTTCTTCCAGATG-3′), and actin (5′-GACCTGACTGACTACCTCATGAAGAT-3′ and 5′-GTCACACTTCATGATGGAGTTGAAGG-3′). Protein levels of wt p53 and mu-p53G279E were analyzed by Western blotting. e and f. H1299 cells were transfected with either pEGFP/wt-p53 or pEGFP/mu-p53G279E, and the expressions of p53, Bad, and Bax at the indicated time points were compared via Western blotting. g. HCT116, HF-B, QGY-7703, and H460 cells were treated with either etoposide (25 μg/ml) or DMSO, and 24 h later, treated cells were harvested and lysed for Western blot analysis of p53 and Bad proteins. Total RNA was prepared from the remaining cells, followed by RT-PCR using specific primers against Bad or β-actin.
p53 directly binds to the upstream region of Bad gene.
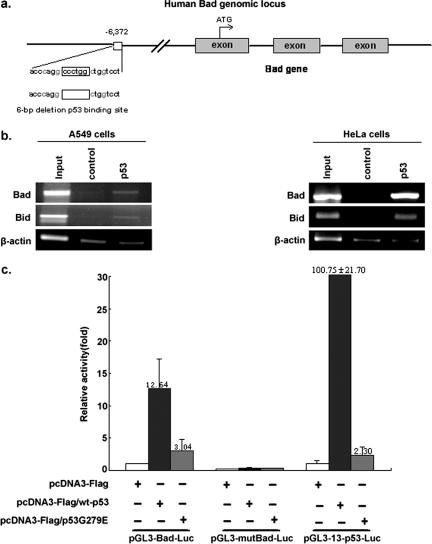
To understand the mechanism through which the Bad gene is up-regulated by p53, the Genomatix Promoter Inspector software was utilized. Inspection of the Bad promoter with the MatInspector (Genomatrix Inc, Munich, Germany) software revealed a potential 21-bp p53-binding sequence (ACCCAGGCCCTGGCTGGTCCT) located 6,613 bp upstream of the Bad translational start site (Fig. 2a).
FIG. 2.
p53 directly binds and activates the Bad upstream promoter region. a. Schematic representation of the bad genomic locus. The putative p53-binding region (p53-BR) is indicated. The deleted 6 bp within the putative p53-BR is shown by an empty box. Exons are represented as gray boxes. The number represents nucleotides upstream of the first nucleotide of exam I of the bad gene. b. HeLa cells (transfected with wt p53) and A549 (p53+/+) cells were treated with etoposide, followed by ChIP using an antibody against p53. Immunoprecipitation with an irrelevant anti-E2F1 antibody was carried out as a negative control. A known p53-targeted gene, the Bid gene, was used as a positive control. Primer pairs used for PCR in the ChIP assay were −6,657/−6,639 and −6,252/−6,235 for Bad and −6,752/−6,733 and −6,558/−6,540 for Bid (20). Primer pairs used for amplifying fragments of Bad, Bid, and actin were 5′-GGAAACCCGGTGGGGCCAC-3′ and 5′-ACCAGTAGCGGGTGGTC-3′, 5′-TTAAAGAATCCTTTGCGGC-3′ and 5′-GTGATTCTCCTGCTTCAG-3′, and 5′-GACCTGACTGACTACCTCATGAAGAT-3′ and 5′GTCACACTTCATGATGGAGTTGAAGG-3′, respectively. c. HeLa cells plated in 24-cell culture plates were individually cotransfected with 850 ng of reporter plasmid pGL3-Bad-Luc, pGL3-mutBad-Luc, or pGL3-13-p53-Luc in combination with 250 ng of pcDNA3-Flag, pcDNA3-Flag/wt-p53, or pcDNA3-Flag/mu-p53G279E expression vectors as indicated. Ten nanograms of Renilla vector pRL-CMV was used as a transfection internal control. The plasmid pGL3-13-p53-Luc, which contains an array of 13 p53-BRs upstream of the luciferase gene, was used as a positive control. The plasmid pGL3-mu-Bad-Luc contains an inactive p53-BR with a deletion in the binding site. Quantification of luciferase activities and calculations of relative ratios were performed manually with a luminometer. Error bars indicate standard deviations.
To test whether p53 can truly bind to this putative response element, a ChIP assay was performed. As shown in Fig. 2b, the p53-binding domain was amplified by PCR only in the presence of anti-p53 antibody and not in the presence of the irrelevant anti-E2F-1 antibody (negative control). A known p53-regulated gene, the Bid gene, was used as a positive control (Fig. 2b). As a negative control, DNA fragments corresponding to beta-actin were not enriched (bottom panel), yet a beta-actin band could be barley detected. This could be due to the nonspecific background binding between beads and chromosome DNA fragments. These results showed that endogenous p53 can specifically interact with this putative region. To further verify that this putative p53-binding region (p53-BR) was able to activate Bad gene expression, p53-dependent luciferase reporter plasmids pGL3-Bad-Luc (which contains p53-BR) and pGL3-mu-Bad-Luc (which contains a mutant [mu] p53-BR with a 6-bp deletion) were constructed. As shown in Fig. 2c, pGL3-Bad-Luc was activated by wt p53, producing an enhanced level of activation compared with either mock or p53G279E (Fig. 2c, left). As expected, pGL3-mu-Bad-Luc failed to be activated by wt p53, mu-p53G279E, or mock (Fig. 2c, middle). The plasmid pGL3-13-p53-Luc, which contains an array of 13 consecutive p53-binding sites upstream of the luciferase reporter gene, was used as a positive control (Fig. 2c, right). These data confirmed that the putative p53-BR upstream of the Bad gene is the functional p53-binding sequence.
Bad physically interacts with p53.
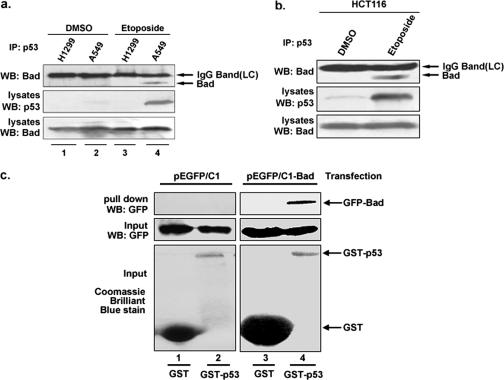
Whether p53 physically interacts with Bad has not yet been reported thus far. We therefore performed coimmunoprecipitation experiments, and the results are shown in Fig. 3a. The specific association of Bad and p53 was detected in the A549 (p53+/+) cells treated with etoposide, which up-regulates p53 expression, but not in p53- deficient H1299 (p53−/−) cells subjected to the same treatment (lanes 3 and 4). Neither A549 nor H1299 produced detectable amounts of endogenous Bad-p53 complex precipitates prior to drug stimulation (lanes 1 and 2). Similarly, the interaction between Bad and p53 was also detected in HCT116 cells treated with etoposide but not in untreated cells (Fig. 3b). To understand whether p53 directly binds to Bad, a glutathione S-transferase (GST) pull-down assay was performed (Fig. 3c). H1299 (p53−/−) cells were transfected with either GFP or GFP-Bad, and cell lysates from each of these transfected cells were mixed with GST and GST-p53 individually. GFP-Bad, but not GFP, was found to be pulled down by glutathione-Sepharose 4B beads associated with GST-p53 (Fig. 3c, lanes 2 and 4). Neither GFP-Bad nor GFP could be pulled down by glutathione-Sepharose 4B beads conjugated with GST alone (lane 1, 3). Collectively, these in vivo and in vitro data support our conclusion that Bad physically associates with p53.
FIG. 3.
p53 physically interacts with Bad. a. Whole-cell extracts from A549 (p53+/+) and H1299 (p53−/−) cells, pretreated with etoposide (25 μg/ml) or without, were immunoprecipitated (IP) with mouse anti-p53 antibody, followed by immunoblotting (WB) with mouse anti-Bad antibody. The coprecipitated Bad is indicated (top panel). DMSO, dimethyl sulfoxide. b. HCT116 (p53+/+) cells, pretreated with etoposide (25 μg/ml) or without, were immunoprecipitated with mouse anti-p53 antibody, followed by immunoblotting with mouse anti-Bad antibody. The coprecipitated Bad is indicated (top panel). c. Direct binding between p53 and Bad in GST pull-down assay. GST or GST-p53 fusion proteins were isolated by affinity chromatography with glutathione-Sepharose beads and incubated with cell lysates from 293T cells transfected with either GFP-Bad or GFP-C1. Complexes were captured with glutathione-Sepharose, and the bound proteins were analyzed by immunoblotting with anti-GFP antibodies. Expression of GFP-Bad and GFP-C1 in the cell lysate is also shown. GST-p53 and free GST were detected by Coomassie brilliant blue staining (bottom panel).
Bad regulates a negative feedback loop for p53.
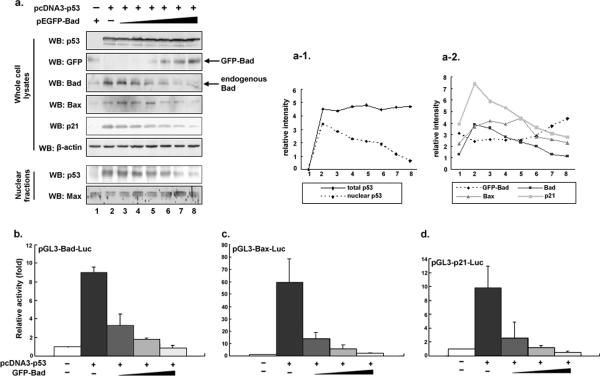
We next inquired whether a negative feedback loop existed for maintaining a proper balance between the levels of the transcription factor p53 and its transcriptional target Bad. If this feedback regulation was indeed present, we would expect that with increasing accumulation of Bad in the cytoplasm, more p53 should end up retained in the cytoplasm by binding to Bad, which would thus prevent p53 from entering the nucleus for further transcription of Bad. Nuclear fractions were prepared from H1299 (p53−/−) cells cotransfected with an equal amount of p53 and increasing amounts of GFP-Bad. The Western blotting results in Fig. 4a show that even if an equal amount of p53 was introduced, the levels of nuclear p53 declined with an accompanying increase of exogenous GFP-Bad. As a result, the levels of endogenous Bad, together with those of the two known p53 targets Bax and p21WAF1, decrease gradually (Fig. 4a).
FIG. 4.
Bad and p53 constitute a negative feedback loop which reduces p53-induced Bad expression. a. H1299 (p53−/−) cells were cotransfected with a constant amount of pcDNA3-p53 and an increasing amount of pEGFP-Bad (0, 0.01, 0.04, 0.10, 0.20, 0.30, and 0.40 μg/ml). At 24 h posttransfection, whole-cell lysates were prepared and subjected to immunoblot analysis with the indicated antibodies. β-Actin was used as a control for equal loading. The nuclear fractions were obtained from the same transfected cells as described previously (10). Max was used as a nuclear marker and loading control. The relative intensity of each protein band was quantified by PhosphorImager analysis provided by Eaglesight (Invitrogen) (a1 and a2). b to d. H1299 (p53−/−) cells were transiently cotransfected with pcDNA3-p53 together with one of the luciferase reporter constructs, pGL3-Bad-Luc (b), pGL3-Bax-Luc (c), or pGL3-p21-Luc (d), which carry the p53-reponsive element (p53-BR) derived from the Bad, Bax, and p21Waf1 promoter, respectively, in the presence or absence of increasing amounts of a pEGFP-Bad expression plasmid. The total amount of plasmid DNA per transfection was kept constant with pEGFP-C1. All transfections were performed in triplicate. Results are shown as fold induction of the firefly luciferase activity compared with control cells transfected with pEGFP-C1 alone. Error bars indicate standard deviations.
To confirm the inhibitory effects of Bad on p53 transactivation activity, H1299 (p53−/−) cells were transiently cotransfected with a constant amount of the expression plasmid pcDNA3-p53 together with p53-responsive Bad-luciferase (Fig. 4b), Bax-luciferase (Fig. 4c), or p21-luciferase (Fig. 4d) reporter constructs in the presence of increasing amounts of the expression plasmid pEGFP-Bad. GFP-Bad greatly reduces the ability of p53 to transactivate Bad, Bax, and p21 promoter activities in a dose-dependent manner. This can be best explained by the hypothesis that with increased Bad protein bound to cytoplasmic p53, less p53 enters the nucleus to transactivate Bad, Bax, and p21WAF1. These data indicate that cytoplasm/nuclear ratio of p53 is controlled by cytoplasmic Bad.
Bad directs p53 to mitochondria.
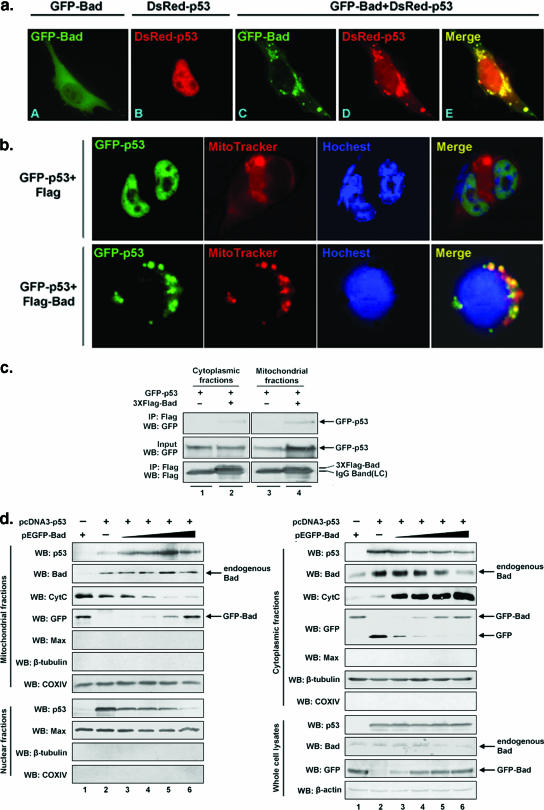
To examine whether p53 and Bad colocalize to the mitochondria in response to apoptotic stimuli, plasmids pDsRed-p53, pEGFP-Bad, and pDsRed-p53 plus pEGFP-Bad were transfected into H1299 (p53−/−) cells separately, and their fluorescence distribution patterns were compared. p53 is normally localized in the nucleus, whereas Bad is mainly found diffused across the cytoplasm. However, coexpression of p53 and Bad gave rise to a merged image characteristic of mitochondrial punctiform particles formed at an early apoptotic stage (Fig. 5a and b) (8). Particularly, in the absence of Bad, GFP-p53 is predominantly localized to the nucleus. However, upon overexpression of Bad, GFP-p53 was relocalized to the mitochondria in 30% of the GFP-positive cells examined (Fig. 5b), implying that the normally nucleus-localized p53 can be directed to mitochondria by Bad. To further investigate in detail the possibility that Bad interacts with p53 at the mitochondria, a coimmunoprecipitation procedure was performed. As shown in Fig. 5c, the association of Bad and p53 was indeed detected at the mitochondria as well as in the cytosol.
FIG. 5.
Bad directs p53 to mitochondria, where they form protein-protein complexes. a. H1299 (p53−/−) cells were individually transfected with pDsRed-p53 (red), pEGFP-Bad (green), or pDsRed-p53 plus pEGFP-Bad, and 24 h later, cells were fixed and visualized by fluorescence microscopy (magnification, ×1,000). b. H1299 (p53−/−) cells were cotransfected with pEGFP-p53 and with either pcDNA3-Flag-Bad or pcDNA3, and 32 h later, cells were fixed and stained with MitoTracker Red and Hoechst 33342. Fluorescence images were captured with an Olympus DP71X camera system. c. Cytoplasmic and mitochondrial fractions from H1299 (p53−/−) cells transfected with pEGFP-p53 and with either pcDNA3-Flag-Bad or pcDNA3 were prepared as described in Materials and Methods, followed by immunoprecipitation (IP) with an anti-Flag antibody. Immunoblotting (WB) was performed to detect Flag-Bad-associated p53. d. A fixed amount of the p53 expression vector pcDNA3-p53 (0.1 μg/ml) and increasing amounts of the Bad-expressing vector pEGFP-Bad (0 to 0.4 μg/ml) were cotransfected into H1299 (p53−/−) cells. Empty pEGFP-C1 vector was added to keep the total plasmid DNA content constant. Twenty-four hours later, transfected cells were collected and subjected to cell fractionation. Each of the fractions from the different subcellular compartments were analyzed for the protein levels of p53, Bad, GFP-Bad, and cytochrome c (CytC) by immunoblotting. Max, tubulin, and COX IV were used as fractional markers and loading controls. Whole-cell lysates were also analyzed to ensure comparable levels of p53. e. H1299 cells were transfected with pEGFP, pEGFP-BadΔBH3, or pEGFP-wt-Bad. Cell lysates from each transfectant were incubated with GST-p53 fusion proteins. GST pull-down assay was performed as described for Fig. 3c. f. H1299 (p53−/−) cells were cotransfected with increasing amounts of pEGFP-BadΔBH3 and fixed amounts of pcDNA3-Flag-p53, and 24 h later, cells were harvested and subjected to cell fractionation. Each fraction was collected, and changes in p53 protein levels were detected by Western blotting (left panel). g. In vitro assay comparing the levels of mitochondrial p53 in the presence or absence of Bad. Equal amounts of isolated mitochondria from H1299 (p53−/−) cells were incubated with either p53 or Bad or with both proteins as indicated at 30°C for 30 min, followed by treatment with the cross-linker BMH (10 mM) at room temperature for another 30 min. The preincubated mixture containing p53 and Bad, which allows the formation of the p53/Bad complex, was also incubated with isolated mitochondria (lanes 5 and 10). The concentration of p53 used was 800 nM, and the concentration of Bad used was 1 μM. Mitochondrial supernatants and pellets were obtained as described previously (22) and immunoblotted with the indicated antibodies. Bovine serum albumin (BSA) was used as a negative control. COX IV was used as a loading control.  , nonspecific band; (pre-), mixture containing Bad and p53 preincubated overnight at 4°C. h. Equal amounts of isolated mitochondria from H1299 (p53−/−) cells were incubated with p53 and mutant BadΔBH3 as described above (see panel g). The mitochondrial soluble fractions (supernatant) and pellets were obtained and immunoblotted with the indicated antibodies.
, nonspecific band; (pre-), mixture containing Bad and p53 preincubated overnight at 4°C. h. Equal amounts of isolated mitochondria from H1299 (p53−/−) cells were incubated with p53 and mutant BadΔBH3 as described above (see panel g). The mitochondrial soluble fractions (supernatant) and pellets were obtained and immunoblotted with the indicated antibodies.
To validate whether Bad is truly able to direct p53 to mitochondria, H1299 (p53−/−) cells were cotransfected with a fixed amount of pcDNA3-p53 and an increasing amount of pEGFP-Bad. Nuclear, cytosolic, and mitochondrial fractions were prepared for Western blot analyses. As shown in Fig. 5d, increasing accumulation of both Bad and p53 in the mitochondrial fraction, with a corresponding decrease in their cytosolic levels, was detected. Concomitantly, levels of cytochrome c were gradually increased in the cytosol, accompanied by a corresponding decrease in cytochrome c in the mitochondria. However, we noticed that the level of GFP-Bad in cytosolic fractions does not exhibit a tendency towards a decline, which may be explained by the assumption that the total increasing amount of introduced GFP-Bad are given in excess and thus are more than enough to keep the remaining cytosolic GFP-Bad increasing.
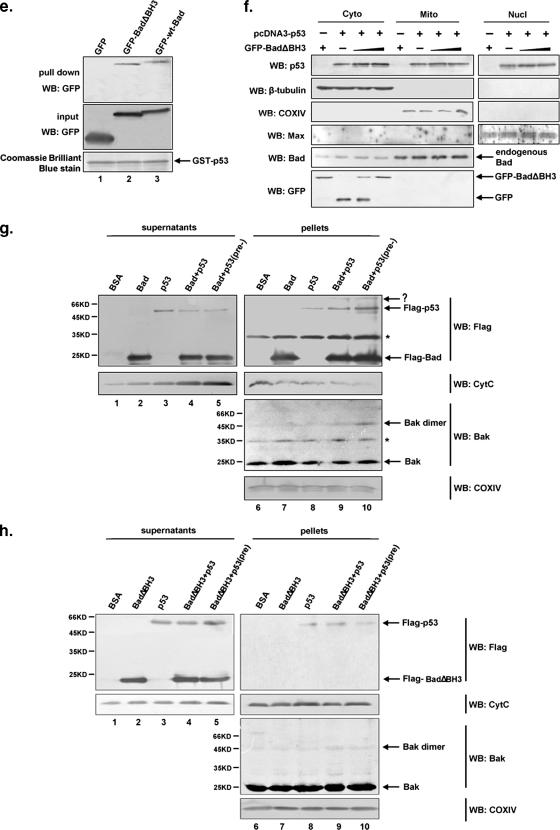
To further confirm that Bad-dependent translocation of p53 relies on the mitochondrial targeting of Bad, we constructed a BadΔBH3 mutant, which is known to lose the capability to localize to mitochondria (1, 25). Applying the GST pull-down method, we proved that BadΔBH3 is able to interact with p53 (Fig. 5e). H1299 (p53−/−) cells were cotransfected with a fixed amount of pcDNA3-p53 and an increasing amount of pEGFP-BadΔBH3. Nuclear, cytoplasmic, and mitochondrial fractions were prepared for Western blot analyses. As shown in Fig. 5f, increased expression of GFP-BadΔBH3 did not lead to a correspondingly increased level of either p53 or Bad in mitochondrial fractions, indicating that the translocation of Bad to the mitochondria is required for Bad-dependent mitochondrial targeting of p53.
An in vitro experiment was also performed to validate that Bad is able to direct p53 onto the mitochondria. Equal amounts of total isolated mitochondria from p53-deficient H1299 cells were incubated separately with purified Flag-p53 and either Flag-Bad or Flag-BadΔBH3 (which is competent in binding p53 yet is unable to migrate to mitochondria). After incubation, the mixtures were centrifuged, and the resulting soluble and pellet fractions were used for Western blot analysis. As shown in Fig. 5g and h, Flag-p53 accumulated much more significantly at the mitochondria when incubated with Flag-Bad than when incubated without Flag-Bad (Fig. 5g, lane 8 versus lane 9). In contrast, incubation with or without BadΔBH3 showed hardly any difference in mitochondrial accumulation of Flag-p53 (Fig. 5h, lane 8 versus lane 9). To examine whether the preformed Bad/p53 complex is more effective for translocation to the mitochondria than the Bad/p53 complex formed without preincubation, Bad and p53 were preincubated at 4°C overnight to allow for Bad/p53 complex formation before being added to isolated mitochondria. A more significant accumulation of mitochondrial p53 was indeed detected (Fig. 5g, lane 10), indicating that Bad directs p53 to the mitochondria via their direct physical interaction. Increases in cytochrome c release (Fig. 5g, lower panel, lanes 1 to 5) and Bak oligomerization (lanes 6 to 10, second panel from bottom) were also observed, which is in accordance with an increased mitochondrial accumulation of p53 and Bad.
Knockdown of Bad reduces mitochondrial localization of p53 and Bak oligomerization.
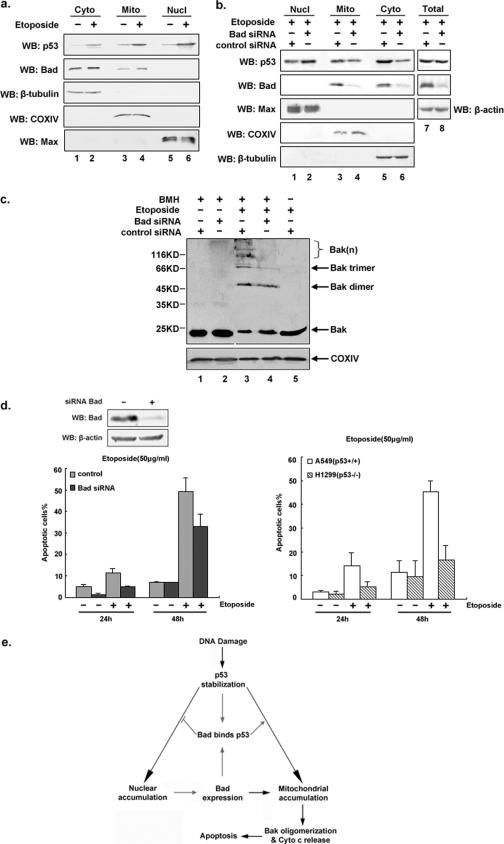
To verify that endogenous p53 and Bad accumulate on the mitochondria upon etoposide treatment under nonoverexpressing condition, A549 (p53+/+) cells were left untreated or treated with etoposide to allow for the up-regulation of p53, followed by preparation of cytoplasmic, nuclear, and mitochondrial fractions. As indicated in Fig. 6a, protein levels of both mitochondrial p53 and Bad were found to be higher in cells with etoposide treatment than in cells without treatment (top two panels, lanes 3 and 4). To directly demonstrate that p53 subcellular localization is affected by Bad, an RNAi strategy for the elimination of endogenous Bad was employed. A549 (p53+/+) cells were transfected with either Bad siRNA or a control siRNA, followed by etoposide treatment. Nuclear, cytosolic, and mitochondrial fractions and total cell lysates were prepared for Western blot analyses. Figure 6b shows that more than 90% of Bad expression had been suppressed compared to that in control cells. While the total amount of cellular p53 remained constant in Bad siRNA-treated and untreated A459 (p53+/+) cells, the subcellular distribution of p53 varied. Levels of both mitochondrial and cytosolic p53 were found to be reduced, whereas nuclear levels of p53 had increased, which might be explained by a reduction in cytosolic Bad (due to the application of siRNA) that would free more cytosolic p53 to enter the nucleus.
FIG. 6.
Suppression of Bad expression reduces mitochondrial localization of p53 and attenuates apoptosis induced by etoposide. a. Coaccumulation of p53 and Bad at the mitochondria under native nonoverexpressing conditions. A549 (p53+/+) cells were treated with or without etoposide (25 μg/ml) for 24 h. Harvested cells were fractionated into cytoplasmic, nuclear, and mitochondrial portions. Each fraction was analyzed by immunoblotting (WB) with antibodies against p53 and Bad. b. RNAi-induced ablation of Bad reduces mitochondrial localization of p53. A549 (p53+/+) cells were transfected with either Bad siRNA or control siRNA. At 24 h after transfection, cell lysates were treated with etoposide (50 μg/ml) for another 36 h, followed by cell fractionation and immunoblotting analysis. Total cell lysates were also analyzed to ensure comparable levels of p53 in both Bad siRNA- and control siRNA-treated cells. c. Bak oligomerization is reduced in Bad siRNA-treated cells. A549 (p53+/+) cells were transfected with either Bad siRNA or control siRNA for 24 h, followed by treatment with or without etoposide (25 μg/ml) for another 36 h. Isolated mitochondria from each transfectant were incubated with the cross-linker BMH (1 mM) or dimethyl sulfoxide in HE buffer (10 mM HEPES [pH 7.4], 1 mM EDTA) (6) at 37°C for 1 h before being subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting using anti-Bak antibody. COX IV was used as mitochondrial loading control. d. Cells with depleted Bad and p53 are more resistant to etoposide-induced apoptosis. A549 (p53+/+) cells were transfected with siRNA oligonucleotides targeting Bad. At 48 h posttransfection, cell lysates were prepared and subjected to immunoblot analysis for assessing the degree of silencing of Bad expression. A549 (p53+/+) cells were transfected with either mock or Bad-specific siRNA, and 24 h later, etoposide (50 μg/ml) was added and left for another 24 h and 48 h. Cells were stained with 2 μg/ml Hoechst 33342 for 10 min, and both floating and adherent apoptotic cells were scored for cell death (abnormal nuclei) based on abnormal apoptotic nuclear morphology under a fluorescence microscope (left). The effect of p53 on Bad-dependent apoptosis was also examined in the p53-deficient human lung carcinoma H1299 (p53−/−) cell line (right). Each set of experiments was repeated three times, and at least 1,000 cells were counted in each case. The error bars represent the standard deviations. e. Hypothetical model illustrating the dual role that Bad plays in p53 transcription-dependent and -independent pathways. Upon treatment with a DNA damage-inducing agent such as etoposide, p53 accumulates in the nucleus to directly bind to the upstream promoter region of Bad, thereby transactivating its gene expression. When sufficient Bad has accumulated in the cytoplasm, it associates with p53. On the one hand, this Bad/p53 complex prevents p53 from entering the nucleus and thus attenuates Bad expression; on the other hand, Bad directs cytosolic p53 to mitochondria and promotes apoptosis via activation of Bak oligomerization and cytochrome c release.
We also examined whether the Bad/p53-promoted apoptosis involves Bak oligomerization. A549 (p53+/+) cells were transfected with control siRNA or Bad-specific siRNA duplexes and further treated with or without etoposide (50 μg/ml). Isolated mitochondria were incubated with 1 mM BMH cross-linker and analyzed by Western blot analysis using an anti-Bak antibody. Cells transfected with Bad-specific siRNA displayed considerably less Bak-induced oligomerization than cells transfected with control siRNA (Fig. 6c, lane 3 versus lane 4).
Reduction of either p53 or Bad attenuates apoptosis induced by etoposide.
In order to evaluate the effects of Bad on p53-induced apoptosis, we eliminated Bad expression in A549 (p53+/+) cells by RNAi (Fig. 6d). At 24 h after siRNA transfection, cells were treated with or without etoposide (50 μg/ml) and incubation continued for a further 24 h and 48 h. For the apoptosis assay, cells were stained with Hoechst 33342 and scored under a fluorescence microscope for each time point. A549 (p53+/+) cells depleted of Bad expression were found to be more resistant to etoposide-induced cell death than control A549 (p53+/+) cells (33% versus 49% at 48 h). Similarly, p53-deficient H1299 cells (p53−/−, Bad+/+) showed reduced apoptosis compared to p53-competent A549 cells (p53+/+, Bad+/+). Furthermore, at 48 h after etoposide treatment, H1299 (p53−/−) cells underwent even less apoptosis than A549 cells (16% versus 45% at 48 h). Taken together, these results demonstrate that the presence of both p53 and Bad causes more etoposide-induced apoptosis than occurs in the absence of either p53 or Bad, implying that Bad may promote p53-induced and mitochondrion-mediated apoptosis during genotoxic stress.
DISCUSSION
The tumor suppressor p53 regulates the cell cycle and promotes apoptosis in response to cellular stresses through transcription-dependent and transcription-independent mechanisms. Transcriptional activation by p53 is critical for the induction of apoptosis (4). Numerous p53-inducible genes have been identified, such as the Bax (16), Puma (17), Bid (20), and p21waf1 (9) genes and many others. However, none of these candidate effectors alone can account for the complicated mechanisms underlying p53 transcription-dependent apoptotic signaling.
It has been demonstrated that p53 was able to translocate from the nucleus into the cytoplasm and even onto the mitochondria during DNA damage-induced and p53-dependent apoptosis in a variety of human and mouse cell systems. The p53 protein can associate with Bcl-xL to release Bid from the Bcl-xL/Bid complex and thereby indirectly activate Bax (1). Moreover, it has been reported that p53 can directly activate Bax to promote mitochondrion-mediated apoptosis (6).
Previous data suggest that Bad may contribute to p53-induced mitochondrion-mediated apoptosis, yet no known molecular mechanism has so far been proposed for those observations (2, 3). We have shown here that the proapoptotic gene bad is transcriptionally regulated by p53. Levels of Bad mRNA and protein are increased in response to the up-regulated expression of wt p53 but not with the mutant p53G279E, indicating that Bad induction requires a transactivation-competent p53. We have also noticed that in H1299 (p53−/−) cells, relatively lower levels of both Bad mRNA and protein can be detected. This may suggest the existence of yet unidentified transcriptional factors for regulating Bad expression. In addition, we identified a functional p53-binding element at the human bad genomic loci, residing roughly 6.6 kb upstream of the Bad translation start site ATG. The far-upstream position of the p53-binding element along the Bad promoter could be the reason for its unrecognized status as a potential p53-BR until now. Furthermore, we also found similar well-conserved p53-BRs approximately 7 kb upstream within the murine Bad gene locus (data not shown). Of note, it has been reported that in the human Bid gene, the p53-binding site also resides approximately 6 kb upstream of its ATG translational start site (20).
More importantly, we have found that Bad can directly associate with p53. Our unpublished data reveal that, unlike the case for 14-3-3, the p53/Bad interaction appears to be independent of phosphorylation of Bad on Ser112, Ser136, and Ser155 (P. Jiang and M. Wu, unpublished result). Overexpression of Bad in p53-transfected H1299 (p53−/−) cells not only reduces the level of nuclear p53 but also decreases the transcription of Bad mRNA. This negative feedback loop maintains the Bad protein at a physiological level to avoid unnecessary apoptosis in healthy cells.
We have also validated that p53 and Bad can cooperatively promote apoptosis induced by the DNA-damaging agent etoposide. Inhibition of Bad expression in A549 (p53+/+) cells by RNA interference results in reduced mitochondrial p53 and an increased resistance to etoposide. Ranger et al. had reported previously that upon treatment with etoposide, Bad+/− mouse embryo fibroblast cells showed much more pronounced apoptosis than Bad−/− mouse embryo fibroblast cells (19). However, they had not indicated that p53 is involved in this process. Thus, our data partially suggest, for the first time, the possible underlying mechanism.
In summary, we suggest a novel link between the nuclear transcriptional and cytoplasmic proapoptotic functions of p53, which are important for both p53- and Bad-mediated mitochondrial apoptotic signaling (Fig. 6e). In this model, p53 directly binds to the upstream region of the bad gene and regulates its transcription. The resultant increase in cytosolic Bad serves two distinctive functions. On the one hand, when Bad exceeds levels required for maintaining normal cell physiology, cytosolic p53 is neutralized by the excess Bad, thus preventing the former from entering the nucleus, followed by a resultant reduction in Bad transcription. On the other hand, Bad can direct p53 to the mitochondria, resulting in activation of Bak oligomerization and cytochrome c release, thereby leading to apoptosis. Our current data provide the first evidence that Bad plays dual roles in both p53 transcription-dependent and -independent pathways. Given the role of Bad in p53-mediated apoptosis shown here, targeting Bad in cancer gene therapy will be of particular interest.
Acknowledgments
We are grateful to Shamini Ayyadhury for editorial assistance. We thank Jörg Kobarg for supplying the GST-p53 plasmid and Ratna B. Ray for providing the pGL3-13-p53-LUC plasmid. We thank Wafik S. El-Deiry for his valuable suggestions on this study.
This research was supported by a 973 grant (2002CB713702) from the Ministry of Science and Technology of China, by grants from the National Natural Science Foundation of China (30530200, 30370308, 90208027, and 30121001), and by an ARC grant (ARC-3/05-M45080006) to K.H. from the Ministry of Education, Singapore.
Footnotes
Published ahead of print on 25 September 2006.
REFERENCES
- 1.Baptiste, N., and C. Prives. 2004. p53 in the cytoplasm: a question of overkill? Cell 116:487-489. [DOI] [PubMed] [Google Scholar]
- 2.Biswas, G., M. Guha, and N. G. Avadnoni. 2005. Mitochondria-to nucleus stress signaling in mammalian cells: nature of nuclear gene targets, transcription regulation, and induced resistance to apoptosis. Gene 354:132-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonini, P., S. Cicconi, A. Cardinale, C. Vitale, A. L. Serafino, M. T. Ciotti, and L. N. Marlier. 2004. Oxidative stress induces p53-mediated apoptosis in glia: p53 transcription-independent way to die. J. Neurosci. Res. 75:83-95. [DOI] [PubMed] [Google Scholar]
- 4.Chao, C., S. Saito, J. Kang, C. W. Anderson, E. Appella, and Y. Xu. 2000. p53 transcriptional activity is essential for p53-dependent apoptosis following DNA damage. EMBO J. 19:4967-4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chattopadhyay, A., C. W. Chiang, and E. Yang. 2001. BAD/BCL-xL heterodimerization leads to by pass of G0/G1 arrest. Oncogene 20:4507-4518. [DOI] [PubMed] [Google Scholar]
- 6.Chipuk, J. E., T. Kuwana, L. Bouchier-Hayes, N. M. Droin, D. D. Newmeyer, M. Schuler, and D. R. Green. 2004. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303:1010-1014. [DOI] [PubMed] [Google Scholar]
- 7.Datta, S. R., A. Katsove, L. Hu, A. Petros, S. W. Fesik, M. B. Yaffe, and M. E. Greenberg. 2000. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol. Cell 6:41-51. [PubMed] [Google Scholar]
- 8.Desagher, S., and J. C. Martinou. 2000. Mitochondria as the central control point of apoptosis. Trends Cell Biol. 10:369-377. [DOI] [PubMed] [Google Scholar]
- 9.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 10.Gottfried, Y., A. Rotm, R. Lotan, H. Steller, and S. Larisch. 2004. The mitochondrial ARTS protein promotes apoptosis through targeting XIAP. EMBO J. 23:1627-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harada, H., B. Becknell, M. Wilm, M. Mann, L. J. Huang, S. S. Taylor, J. D. Scott, and S. J. Korsmeyer. 1999. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol. Cell 3:413-422. [DOI] [PubMed] [Google Scholar]
- 12.Hershko, T., and D. Ginsberg. 2004. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J. Biol. Chem. 279:8627-8634. [DOI] [PubMed] [Google Scholar]
- 13.Kokontis, J. M., A. J. Wagner, M. O'Leary, S. Liao, and N. Hay. 2001. A transcriptional activation function of p53 is dispensable for and inhibitory of its apoptotic function. Oncogene 20:659-668. [DOI] [PubMed] [Google Scholar]
- 14.Leu, J. I., P. Dumont, M. Hafey, M. E. Murphy, and D. L. George. 2004. Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex. Nat. Cell Biol. 6:443-450. [DOI] [PubMed] [Google Scholar]
- 15.Mihara, M., S. Erster, A. Zaika, O. Petrenko, T. Chittenden, P. Pancoska, and U. M. Moll. 2003. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell 11:577-590. [DOI] [PubMed] [Google Scholar]
- 16.Miyashita, T., and J. C. Reed. 1995. The tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80:293-299. [DOI] [PubMed] [Google Scholar]
- 17.Nakano, K., and K. H. Vousden. 2001. PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 7:683-694. [DOI] [PubMed] [Google Scholar]
- 18.Okamura, S., H. Arakawa, T. Tanaka, H. Nakanishi, C. C. Ng, Y. Taya, M. Monden, and Y. Nakamura. 2001. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol. Cell 8:85-94. [DOI] [PubMed] [Google Scholar]
- 19.Ranger, A. M., J. Zha, H. Harada, S. R. Datta, N. N. Danial, A. P. Gilmore, J. L. Kutok, M. M. Le Beau, M. E. Greenberg, and S. J. Korsmeyer. 2003. Bad-deficient mice develop diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. USA 100:9324-9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sax, J. K., P. Fei, M. E. Murphy, E. Bernhard, S. J. Korsmeyer, and W. S. El-Deiry. 2002. BID regulation by p53 contributes to chemosensitivity. Nat. Cell Biol. 4:842-849. [DOI] [PubMed] [Google Scholar]
- 21.Schuler, M., and D. R. Green. 2005. Transcription, apoptosis and p53: catch-22. Trends Genet. 21:182-187. [DOI] [PubMed] [Google Scholar]
- 21a.Song, Z. Y., X. B. Yao, and M. Wu. 2003. Direct interaction between Survivin and Smac is essential for the anti-apoptotic activity of Survivin during Taxol-induced apoptosis. J. Biol. Chem. 278:23130-23140. [DOI] [PubMed] [Google Scholar]
- 22.Tan, K. O., N. Y. Fu, S. K. Sukumaran, S. Chan, J. H. Kang, K. L. Poon, B. S. Chen, and V. C. Yu. 2005. MAP-1 is a mitochondrial effector of Bax. Proc. Natl. Acad. Sci. USA 102:14623-14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomso, D. J., A. Inga, D. Menendez, G. S. Pittman, M. R. Campbell, F. Storici, D. A. Bell, and M. A. Resnick. 2005. Functionally distinct polymorphic sequences in the human genome that are targets for p53 transactivation. Proc. Natl. Acad. Sci. USA 102:6431-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vousden, K. H., and X. Lu. 2002. Live or let die: the cell's response to p53. Nat. Rev. Cancer 2:594-604. [DOI] [PubMed] [Google Scholar]
- 25.Yang, E., J. Zha, J. Jockel, L. H. Boise, C. B. Thompson, and S. J. Korsmeyer. 1995. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell 80:285-291. [DOI] [PubMed] [Google Scholar]