Abstract
β-Catenin plays multiple roles in cell-cell adhesion and Wnt signal transduction. Through the Wnt signal, the cellular level of β-catenin is constitutively regulated by the multicomponent destruction complex containing glycogen synthase kinase 3β, axin, and adenomatous polyposis coli. Here, we present multiple lines of evidence to demonstrate that LZTS2 (lucine zipper tumor suppressor 2) interacts with β-catenin, represses the transactivation of β-catenin, and affects the subcellular localization of β-catenin. The LZTS2 gene is located at 10q24.3, which is frequently lost in a variety of human tumors. A functional nuclear export signal (NES) was identified in the C terminus of the protein (amino acids 631 to 641). Appending this motif to green fluorescent protein (GFP) induced nuclear exclusion of the GFP fusion protein. However, introducing point mutations in either one or two leucine residues of this NES sequence abolished the nuclear exclusion of the LZTS2 protein. The nuclear export of LZTS2 can be blocked by leptomycin B (LMB), an inhibitor of the CRM1/exportin-alpha pathway. Intriguingly, β-catenin colocalizes with LZTS2 in the cytoplasm of cells in the absence of LMB but in the nuclei of cells in the presence of LMB. Increasing the LZTS2 protein in cells reduces the level of nuclear β-catenin in SW480 cells. Taken together, these data demonstrate that LZTS2 is a β-catenin-interacting protein that can modulate β-catenin signaling and localization.
The Wnt/β-catenin signaling pathway plays critical roles in embryonic development and tumorigenesis (31). The Wnt ligands bind their receptors, the Frizzled receptors, activating different intracellular cascades through either the “canonical” or “noncanonical” pathways (31). The importance of β-catenin in tumorigenesis is supported by discoveries of mutations in both β-catenin and components of the destruction complex in a wide variety of human cancers (19, 33). The adenomatous polyposis coli (APC) tumor suppressor binds to β-catenin and the scaffold protein Axin to form the destruction complex that promotes glycogen synthase kinase 3β-mediated phosphorylation of β-catenin. It has been shown that more than 80% of colorectal cancers possess inactive mutants of the APC protein, which correlate with the increased levels of free β-catenin in the cancer cells (24, 34). Most of these mutations target the “mutation cluster region” in the center of the APC gene, resulting in truncated proteins incapable of binding Axin and other regulatory proteins or associating with microtubules, which abolishes the destruction complex and results in β-catenin accumulation (9, 33).
β-Catenin accumulates in the cytoplasm and nucleus in response to Wnt signaling (33). The nuclear shift of β-catenin reflects both an increase in total proteins and an enhanced nuclear targeting of β-catenin (2, 38). It has been shown that the central armadillo repeats of β-catenin directly bind the nuclear pore complex, mediating its own nuclear import (20). The CRM1/exportin nuclear export pathway has been proposed in regulation of the nuclear export of β-catenin. Amino acid sequences responsible for highly efficient nuclear export (NES) have been identified in an increasing number of proteins, such as the human immunodeficiency virus (HIV) type 1 Rev protein (8). The “classical” Rev-like NES is characterized by a leucine-rich sequence that binds CRM1/exportin to facilitate the nuclear export of the protein (10, 11). The tumor suppressor APC contains functional Rev-like NES motifs that are necessary for its leptomycin B (LMB)-sensitive nuclear exclusion (13, 27). It has been shown that ectopic and endogenous APC can associate with β-catenin in the nucleus and induce a translocation of β-catenin from the nucleus to the cytoplasm. While the precise role of APC in nuclear-cytoplasmic shuttling remains unclear, multiple lines of evidence suggest that it can affect the subcellular distribution and degradation of β-catenin (13, 27).
Although previous studies have suggested that the nuclear accumulation of β-catenin in colon cancer cells may correlate with a loss of nuclear export function of mutated APC (36), overwhelming evidence has suggested that it may also be regulated by an APC-independent mechanism (12, 18). Moreover, β-catenin nuclear export can occur even when the CRM1 export pathway is blocked by LMB (6). Additionally, the overexpression of CRM1 in an APC-mutated colon cancer cell line, SW480, stimulated the export of endogenous nuclear β-catenin (6). These findings suggest that other mechanisms may be involved in the regulation of the nuclear export of β-catenin, including a yet-undefined CRM1-dependent but APC-independent pathway.
The FEZ1/LZTS1 (leucine zipper, putative tumor suppressor 1) gene was identified by positional cloning from 8p21-22, a region that is frequently lost in human tumors (15). Although previous studies have shown that FEZ/LZTS1 may regulate cell mitosis, little is known about its biological roles in cells. Using a BLAST search, a sequence similar to that of FEZ1/LZTS1, named leucine zipper tumor suppressor 2 (LZTS2)/LAPSER1, was identified within human 10q24.3, which is frequently deleted in various human cancers (1). Like FEZ1/LZTS1, LZTS2 contains several domains and motifs, including leucine zippers. Overexpression of LZTS2 showed an inhibitory effect on cell growth (1).
Here, we present multiple lines of evidence in vitro and in vivo to demonstrate that LZTS2 is a novel β-catenin-interacting protein and represses β-catenin-mediated transcription on T-cell factor/lymphoid enhancing factor (TCF/LEF) and androgen receptor-mediated transcription. Particularly, we identified a Rev-like leucine-rich, CRM1/exportin-regulated NES sequence at the C terminus of LZTS2, through which LZTS2 can regulate the cellular localization of β-catenin. Our data show that LZTS2, acting as a β-catenin-interacting protein, regulates the cellular distribution and transcriptional activity of β-catenin through its nuclear export activity.
MATERIALS AND METHODS
Yeast two-hybrid system.
Yeast two-hybrid experiments were basically performed as described previously (43). The full-length armadillo repeat region of human β-catenin (amino acids [aa] 134 to 671) was fused in frame to the GAL4 DNA binding domain in the pGBT9 vector (Clontech, Palo Alto, CA). A cDNA library from human brain tissues was used in this screening (Clontech). The specificity of interaction with β-catenin was determined by a liquid β-galactosidase (β-Gal) assay. pGBT9 constructs with different β-catenin fragments and pGAD10 constructs with different LZTS2 fragments were created and used to confirm the interaction.
Plasmid construction.
A yeast clone containing the truncated cDNA fragment of KIAA1813 (amino acids 118 to 669) was isolated in the screen. A BLAST search found that the sequence of KIAA1813 is identical with those of LZTS2, LAPSER, and several human expressed sequence tag cDNA clones. A full-length LZTS2 fragment was created by combining the cDNA fragment that encodes the amino-terminal region isolated from KIAA1813 cDNA and the yeast clone isolated from the yeast two-hybrid assay in the pcDNA3 vector with or without an amino-terminal FLAG epitope tag. Using this as a template, the C-terminal and internal deletions of LZTS2 clones were generated by PCR with specific primers containing the appropriate restriction enzyme sites. After cleavage, the fragments containing different portions of LZTS2 were cloned downstream of the GAL4 transactivation domain in the pACT2 vector (Clontech). Mutants of LZTS2 with single or double point mutations within the NES were generated by PCR-based mutagenesis. Plasmids encoding green fluorescent protein (GFP) fused to potential NES sequences from LZTS2 or from the human immunodeficiency virus type 1 Rev protein were generated by inserting double-stranded oligonucleotides into the pEGFP-C1 plasmid with appropriate restriction enzyme sites (Clontech). The oligonucleotide sequences included HIV-Rev (amino acids 75 to 83), 5′-TCGAGCTCTACCACCGCTTGAGAGACTTACTCTT-3′; LZTS2 (amino acids 134 to 143), 5′-TCGAGAGCTGGAGAAGAACATGGAGAAGATCCTGATC-3′; amino acids 389 to 397, 5′TCGAGAGCTGCTGCAGCTGCAGGTGTTCCAGCTG-3′; aa 471 to 478, 5′-AATTCAGCGCCACCCGCAGAGCCACCAGCTC-3; aa 571 to 580, 5′-TCGAGGCTTGCGGGCCCAGGTGGAGCGATTGCGGGTG-3′; and aa 631 to 642, 5′-TCGAGAGCTAGAGCAGGAGCTGCAGCAGCTCAGCCTGGAGCTG-3′. The pLentiviral LZTS2 expression constructs were generated in the pLenti-super vector (5).
Topflash (pGL3-OT) and Flopflash luciferease (pGL3-OF) reporters were obtained from Bert Vogelstein. pSV-β-galactosidase is a commercial reporter plasmid (Promega, Madison, WI).
Cell cultures and transfections.
The monkey kidney cell line, CV-1, human prostate cancer cell lines, PC3 and DU145, and human colon cancer cell line, SW480.7, were maintained in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum (HyClone, Denver, CO). An androgen receptor-positive prostate cancer cell line, LNCaP, was maintained in T-medium (Invitrogene, Carlsbad, CA) with 5% fetal calf serum.
Transient transfections were carried out using a Lipofectamine transfection kit or Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Transient transfection and reporter assays were performed as described previously (43). The relative light units from individual transfections were normalized by β-Gal activity in the same samples. Individual transfection experiments were done in triplicate, and the results are reported as mean RLU/β-Gal (± standard deviation).
In vitro binding assay.
Glutathione S-transferase (GST)-β-catenin fusion proteins were constructed in the pGEX-4T-1 vector (Amersham, Arlington Heights, IL). Expression and purification of GST fusion proteins were performed according to the manufacturer's instructions. Full-length human LZTS2 proteins were generated and 35S labeled by in vitro transcription and translation with a TNT-coupled reticulocyte lysate system (Promega). Equal amounts of GST fusion proteins coupled to glutathione-Sepharose beads were incubated with 35S-labeled proteins at 4°C for 2 h in the lysis buffer as described previously (43). GST fusion proteins were then eluted by incubating them with buffer containing 10 mM glutathione and 50 mM Tris-HCl (pH 8.0) for 10 min at room temperature. The bound proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by autoradiography.
Antibody production.
The polypeptide (amino acids 332-RDREAELQQLRDSLDEN-350) was chosen from the central region of LZTS2 and synthesized as a source of antigen for antibody production. A rabbit polyclonal, affinity-purified antibody against human LZTS2 was produced by Proteintech Group, Inc. (Chicago, IL). Antibody specificity for the LZTS2 protein was confirmed by Western blotting and enzyme-linked immunosorbent assay.
Preparation of whole-cell lysates, immunoprecipitation, and Western blotting.
Cells transfected with β-catenin and LZTS2 expression constructs were washed with phosphate-buffered saline (PBS) and resuspended in RIPA buffer (1% NP-40, 0.1% SDS, 50 mM NaF, 0.2 mM Na3VO4, 0.5 mM dithiothreitol, 150 mM NaCl, 2 mM EDTA, 10 mM sodium phosphate buffer, pH 7.2). Nuclear extracts were prepared according to the method of Dignam et al., with minor modifications (4).
Whole-cell lysates or nuclear extracts were first precleared with protein A-Sepharose beads for 1 h and then incubated with mouse or rabbit normal immunoglobulin G (IgG) or specific antibodies conjugated with preequilibrated protein A-Sepharose beads at 4°C for 2 h. The beads were collected by centrifugation and washed. Proteins were eluted by boiling in SDS sample buffer, resolved on 10% polyacrylamide gels, and transferred onto nitrocellulose membranes. Membranes were blocked with 5% milk in Tris-buffered saline with Tween for 1 h and then probed with anti-FLAG M2 (Sigma, St. Louis, MO) and anti-β-catenin (Signaling Transduction lab, Lexington, KY), followed by incubation with species-specific horseradish peroxidase-conjugated antibodies.
Immunofluorescence.
CV-1, PC3, or SW480.7 cells were plated onto chamber slides 16 h before transfection. pcDNA3-FLAG-LZTS2, pcDNA3-FLAG-LZTS2 (L638/640A), and pcDNA3-FLAG-APC were transfected with or without pCMG2-HA-β-catenin into cells with the Lipofectamine reagent (Invitrogen). Transfected cells were incubated for 24 h and fixed for 10 min (CV-1) or 30 min (PC3 and SW480) with 4% paraformaldehyde. Cells were then permeablized and blocked for nonspecific sites with 0.04% Triton X-100-5% goat serum-PBS buffer for 30 min and incubated with appropriate primary antibodies overnight at 4°C. Cells were washed three times, followed by incubation with appropriate secondary antibodies. Samples were also counterstained with 1 ng/ml 4′,6′-diamidino-2-phenylindole (DAPI). The pEGFP-C1-NES constructs were transfected into CV-1 or PC3 cells and were examined with an inverted epiflourescence microscope (Zeiss, Thornwood, NY) after 16 h.
RESULTS
LZTS2 is a novel β-catenin-interacting protein.
Using a bait construct containing the armadillo repeats (amino acids 134 to 671) of β-catenin, we employed a modified yeast two-hybrid system to identify proteins that interact with β-catenin, using a human brain library (16). Of 6.3 × 107 transformants, 133 grew under selective conditions and showed increased adenine and β-Gal production. Rescue of the plasmids and sequencing of the inserts revealed several different cDNAs, including several known β-catenin-interacting proteins, such as APC, E-cadherin, TCF/LEF, and ICAT. Importantly, three of these clones perfectly matched the partial sequence of the coding region of a novel protein, named LAPSER1 or LZTS2 (1). Sequence analysis of LZTS2 showed high sequence similarity to a putative tumor suppressor, FEZ1/LZTS1 (15).
Alignment of LZTS2 with FEZ1/LZTS1 and LZTS3, which was identified recently (39), showed significant sequence similarity among these proteins, particularly in the C-terminal potential leucine zipper regions (Fig. 1A). Interestingly, the nuclear export sequence identified in LZTS2 is not conserved in both the LZTS1 and the LZTS3 proteins but is highly conserved in the orthologs of LZTS2 across species (Fig. 1B). These observations suggest a potential role of LZTS2 in the regulation of nuclear and cytoplasmic trafficking, which may distinguish it from LZTS1 and LZTS3.
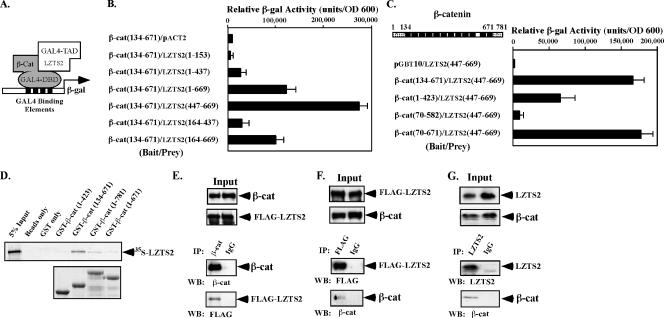
FIG. 1.
Alignment of LZTS proteins. (A) Alignment of the three LZTS protein sequences, including LZTS1, also called FEZ1 (AAD23840), LZTS2, also called LAPSER1/KIAA1813 (AAH58938), and LZTS3, also called ProSapip1 (AAH38860). Identical and similar residues are highlighted in black and gray, respectively. Asterisks denote leucine or similar hydrophobic residues with spacing consistent with leucine zipper domains. The NES sequence characterized in the text is underlined. (B) Alignment of similar NES sites in orthologs of hLZTS2 (AAH58938) from Xenopus tropicalis (AAH75457), Mus musculus (NP_663478), Canis familiaris (XP_543975), and Pan troglodytes (XP_521656).
The C-terminal LZTS2 and armadillo repeats of β-catenin are responsible for binding.
To confirm the interaction and map the binding regions of the proteins responsible for the interaction, we cotransformed various truncated constructs of LZTS2 and β-catenin into the yeast strain PJ69-4A (16) (Fig. 2A). A liquid β-Gal assay was performed to quantify the interaction. As shown in Fig. 2B, the LZTS2 mutants containing the C-terminal region, such as LZTS2(1-669), LZTS2(447-669), and LZTS2(164-669), showed an approximately 12-, 26-, and 10-fold induction compared to pVP16 alone, respectively. However, the LZTS2(1-437) and LZTS2(164-437) mutants showed virtually no interaction with the armadillo repeats of β-catenin, suggesting that the region between amino acids 447 and 669 is required for the interaction.
FIG. 2.
LZTS2 interacts with β-catenin in cells. (A) A schematic representation of the yeast two-hybrid assay for mapping the interaction between the β-catenin and LZTS2 proteins. (B) The cDNA fragments containing different portions of human LZTS2 were fused to a GAL4 transactivation domain, and the armadillo repeats of β-catenin were fused to the GAL4 DNA binding domain in pGBT9 vector. Numbers correspond to amino acid residues. Both of the plasmids were cotransformed into PJ69-4A cells as labeled in the figure and plated on SD-Ade-Leu-Trp plates or SD-Leu-Trp plates to monitor transformation efficiency. Three independent colonies were inoculated from each transformation for subsequent liquid β-Gal assays. The data for the liquid β-Gal assays are reported in relative units normalized by cell number (optical density at 600 nm [OD 600]). (C) The indicated fragments of β-catenin were examined by yeast two-hybrid assay for interaction with the C-terminal fragment of hLZTS2 (aa 447 to 669). (D) Equal amounts of GST-β-catenin fusion proteins were immobilized on a glutathione-Sepharose matrix. The binding of [35S]methionine-labeled LZTS2 to GST-β-catenin fusion proteins was analyzed by SDS-PAGE and visualized by autoradiography. (E and F) Whole-cell lysates of CV-1 cells transfected with full-length β-catenin and FLAG-tagged LZTS2 were immunoprecipitated with normal mouse IgG or a β-catenin (E) or FLAG (F) monoclonal antibody. The immunoprecipitates were analyzed by Western blotting with different antibodies as indicated. (G) Whole-cell lysates of SW480 cells were immunoprecipitated with the homemade LZTS2 antibody and normal rabbit IgG and analyzed by Western blotting.
Next, we examined whether the full armadillo repeat region of β-catenin is necessary for the interaction with LZTS2. The mutants, β-cat(1-423) and β-cat(70-582), produced no or much lower β-Gal activity than the other two mutants that contain the full-length armadillo repeat domain, β-cat(134-671) or β-cat(70-671) (Fig. 2C). In addition, full (134 to 671) or partial deletion (70 to 671) of the N-terminal transactivation domain of β-catenin did not significantly affect the interaction. This result suggests that the full-length armadillo domain of β-catenin is the primary binding region for LZTS2.
LZTS2 interacts with β-catenin in vitro and in vivo.
The physical interaction between LZTS2 and β-catenin was further assessed by GST pull-down experiments. A series of GST fusion proteins containing full-length β-catenin and truncated mutants was generated and immobilized onto a glutathione-Sepharose matrix. The binding of [35S]methionine-labeled LZTS2 to GST-β-catenin fusion proteins was analyzed by SDS-PAGE and visualized by autoradiography. As shown in Fig. 2D, the GST fusion protein containing full-length armadillo repeats, GST-β-cat(134-671), showed a more pronounced interaction with LZTS2 than full-length β-catenin or the other truncated mutants. The data are consistent with results of our yeast two-hybrid experiments and suggest a structural significance of the armadillo repeats for the interaction with LZTS2.
To confirm that full-length LZTS2 and β-catenin proteins are physically associated in intact cells, we carried out coimmunoprecipitation assays. β-Catenin and FLAG-tagged LZTS2 were coexpressed in CV-1 cells. Whole-cell lysates containing equal amounts of the proteins were immunoprecipitated with normal mouse IgG or a β-catenin or FLAG monoclonal antibody. As shown in Fig. 2E, the FLAG-LZTS2 protein was detected in the immunoprecipitates with the β-catenin antibody but not with IgG. Additionally, in the reverse coimmunoprecipitation assay, β-catenin was found specifically in the immunoprecipitates with the FLAG antibody (Fig. 2F). To detect the interaction between endogenous proteins, we produced a specific antibody against the LZTS2 protein and used it in the coimmunoprecipitation experiments. As shown in Fig. 2G, endogenous β-catenin was specifically detected in the immunoprecipitates pulled down by the LZTS2 antibody. Taken together, these data demonstrate a biologically relevant interaction between LZTS2 and β-catenin.
LZTS2 represses β-catenin-mediated transcription.
Transient transfection assays were performed to investigate the possible effect of LZTS2 on β-catenin-mediated transcription on TCF/LEF (25). First, we examined the repressive effect of LZTS2 on β-catenin-mediated enhancement of TCF-1 regulated transcription. Expression constructs of TCF-1 and β-catenin were transfected with pGL3-OT and pGL3-OF in three prostate cancer cell lines, LNCaP, PC3, and DU145. Coexpression of TCF-1 and β-catenin showed a transcription induction of the pGL3-OT promoter in all three cell lines (Fig. 3A, B, and C). Cotransfection of LZTS2 showed a dosage-dependent repression on OT promoters in all cell types examined. However, there was no significant change in the samples transfected with the OF promoter/reporter. These data provide the first line of evidence that demonstrates that LZTS2 represses the activity of β-catenin on TCF-mediated transcription.
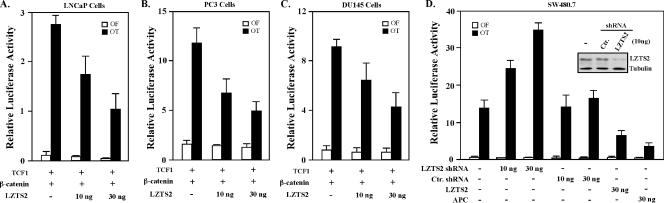
FIG. 3.
LZTS2 represses β-catenin-mediated transcription. (A) One hundred nanograms of pGL3-OT (OT) or pGL3-OF (OF), 25 ng of pcDNA3-β-gal, 5 ng of TCF1 expression vector, 20 ng of β-catenin, and various amounts of pcDNA3-FLAG-hLZTS2 as indicated were transfected into LNCaP cells. Cells were cultured for 24 h in the regular media and luciferase, and β-Gal activities were measured as indicated above. Similar experiments were repeated with PC3 cells (B) and DU145 cells (C). (D) Either 100 ng of pGL3-OT (OT) or pGL3-OF (OF), 25 ng of pcDNA3-β-gal, and other shRNA or expression constructs as indicated in the figure were transfected into the human colon cancer cell line SW480. Luciferase and β-Gal activities were measured in panel C.
Using the small-hairpin RNA (shRNA) interference approach, we examined the repressive effect of endogenous LZTS2. Transfection of the pGL3-OT reporter into the human colon cancer cell line SW480 showed high transcriptional activity, as reported previously (26). SW480 cells contain a truncated form of APC (amino acids 1 to 1,337) (29), which retains the sequences required to bind but not degrade β-catenin, resulting in a marked accumulation of β-catenin in cells. Knock-down of endogenous LZTS2 proteins attenuates the transcriptional repression mediated by the protein, showing a much greater luciferase activity in the reporter assays (Fig. 3D). The knockdown effect of the LZTS2 shRNA construct is specific, since a control shRNA vector appears to have no effect in the similar experimental setting. In addition, overexpression of the LZTS2 or APC protein resulted in a significant reduction of the pGL3-OT reporter (Fig. 3D), suggesting repressive roles for the proteins in the regulation of β-catenin transcriptional activity. Taken together, these data further demonstrate the role of LZTS2 in the regulation of β-catenin-mediated transcription.
LZTS2 localizes mainly in the cytoplasm, and its subcellular distribution is affected by leptomycin B.
Next, we examined the subcellular localization of the LZTS2 protein. Transfection with expression plasmids encoding FLAG-tagged LZTS2 in CV-1 cells showed that most of the exogenous LZTS2 proteins were exclusively located in the cytoplasm, surrounding the nuclear envelope (Fig. 4A). To confirm this observation, we generated a specific antibody to LZTS2 and used it to assess the cellular distribution of the endogenous protein. As shown in Fig. 4B, endogenous proteins showed a staining similar pattern to that of ectopically expressed LZTS2 proteins, confirming the validity of the overexpression models. To assess the mechanism of the exclusive cytoplasmic localization of LZTS2, we examined the effect of LMB, a specific inhibitor for CRM1/exportin-alpha-dependent nuclear export, on the subcellular distribution of LZTS2 (28). In the presence of 60 ng/ml LMB, either exogenous or endogenous LZTS2 show a clear nuclear retention in CV-1 and PC3 cells, respectively (Fig. 4C and D). These data suggest that the exclusive cytoplasmic localization of LZTS2 is the result of the specific export of LZTS2 proteins from the nuclei, which appears to be regulated by the CRM-1/exportin-alpha pathway.
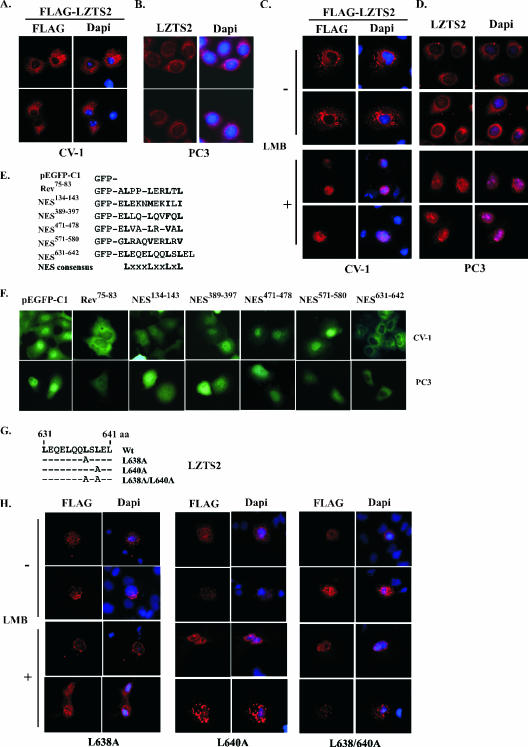
FIG. 4.
LZTS2 localizes in the cytoplasm and contains a leptomycin B-regulated NES motif. (A) The pcDNA3-FLAG-hLZTS2 expression vector was transfected into CV-1 cells. The ectopically expressed hLZTS2 was detected with FLAG monoclonal antibody and revealed by rhodamine-conjugated secondary antibody (red). The nuclei were counterstained with DAPI (blue). (B) Endogenous hLZTS2 was detected with a rabbit antibody in PC3 cells. (C) CV-1 cells were transfected with the pcDNA3-FLAG-hLZTS2 expression vector and then incubated in medium either with or without 60 ng/ml LMB. The ectopically expressed hLZTS2 was detected as in panel A. (D) PC3 cells were cultured in medium either with or without 60 ng/ml LMB for 24 h and then stained with the antibody against endogenous hLZTS2 (red). (E) The GFP fusion proteins containing the putative NES sequences of hLZTS2 and an HIV-Rev classical NES were generated in the pEGFP-C1 vector. Numbers correspond to amino acid residues. (F) The expression vectors containing the above GFP fusion proteins were transfected into CV-1 and PC3 cells. After 24 h of transfection, live cells were observed with an inverted fluorescence microscope. (G) A schematic representation of the NES sequence in hLZTS2. Either single or double mutations within the site were introduced. (H) The expression vectors of pcDNA3-FLAG-hLZTS2 containing either single or double point mutations were transfected into CV-1 cells. The cells were treated with LMB as indicated in panel C. The localization of the LZTS2 proteins was detected as in panel A.
A functional NES located in the C-terminal region of LZTS2.
Previous studies have shown that the nuclear export of many large proteins by CRM1/exportin-alpha is mediated through a short leucine-rich motif, known as the NES sequence. Several Rev-like leucine-rich NES sequences appear in the LZTS2 protein (Fig. 1 and Fig. 4E). To test whether these NES sequences are involved in the regulation of the nuclear export of LZTS2, five double-stranded oligonucleotides encoding putative leucine-rich NES sequences were synthesized and inserted into a pEGFP-C1 vector to produce fusion proteins with GFP (Fig. 4E). Constructs containing either the HIV-Rev NES (75 to 83 aa) or the pEGFP-C1 vector were used as a positive or negative control, respectively. The GFP constructs were transfected into CV-1 and PC3 cells to test their nuclear export activities. As expected, the GFP construct containing the HIV-Rev NES sequence shows a clear nuclear exclusion pattern (Fig. 4F). Intriguingly, in the cells transfected with GFP-LZTS2(631-642), proteins appear exclusively in the cytoplasm, displaying a nuclear omission pattern similar to that of the GFP-Rev NES. In contrast, the other GFP fusion proteins were observed in both the cytoplasm and the nuclei. These data indicate that the sequence between amino acids 631 and 642 constitutes a functional NES sequence and is capable of targeting GFP proteins for export from the nuclei.
The consensus sequence of NES is defined as a set of critically spaced hydrophobic residues, usually leucines, such as L(XXX/XX)L(XX/XXX)LXL (10, 28). The sequence between amino acids 631 and 640 in LZTS2 shows a perfect match to the consensus NES sequence (Fig. 4E). To confirm that this sequence is responsible for the nuclear export of LZTS2, we replaced leucine with alanine at either aa 638 or 640 or both positions by a PCR-based, site-directed mutagenesis technique (Fig. 4G). These mutants and wild-type LZTS2 were ectopically expressed in CV-1 cells. Staining of transfected cells with anti-FLAG antibodies showed that FLAG-LZTS2 NES mutated proteins are mainly retained in the nuclei either in the presence or absence of LMB (Fig. 4H). In contrast, as we have observed above, wild-type LZTS2 was excluded from the nuclei (Fig. 4A and C). These data confirm that the sequence between 631 and 640 is a functional NES site and is required for the nuclear export of LZTS2 through the CRM1/exportin-alpha pathway.
LZTS2-dependent nuclear export of β-catenin requires an intact NES.
A physical interaction between LZTS2 and β-catenin has been demonstrated in this study. One possible mechanism for the regulation of β-catenin by LZTS2 is through the modulation of subcellular localization of β-catenin. To test this hypothesis, we cotransfected LZTS2 and β-catenin into CV-1 cells. As reported earlier (17), ectopically expressed β-catenin mainly localized in the nuclei (Fig. 5A). Interestingly, coexpression of LZTS2 with β-catenin showed that both proteins were excluded from nuclei and colocalized in the cytoplasm in the absence of LMB (Fig. 5B, top panels). However, most of the LZTS2 and β-catenin proteins colocalize and accumulate in nuclei in the presence of LMB, whereas some cells showed a nuclear and cytoplasmic distribution of the proteins (Fig. 5B, bottom panels). To further evaluate the effect of LZTS2 on the subcellular localization of β-catenin in a physiologically relevant cellular context, we examined the colocalization of endogenous LZTS2 and β-catenin in PC3 cells. The cells treated with 60 ng/ml LMB for 24 h showed an accumulation of both LZTS2 and β-catenin in the nuclei (Fig. 5D, bottom panels). In contrast, most β-catenin and LZTS2 colocalized in the cytoplasm of untreated cells (Fig. 5D, top panels).
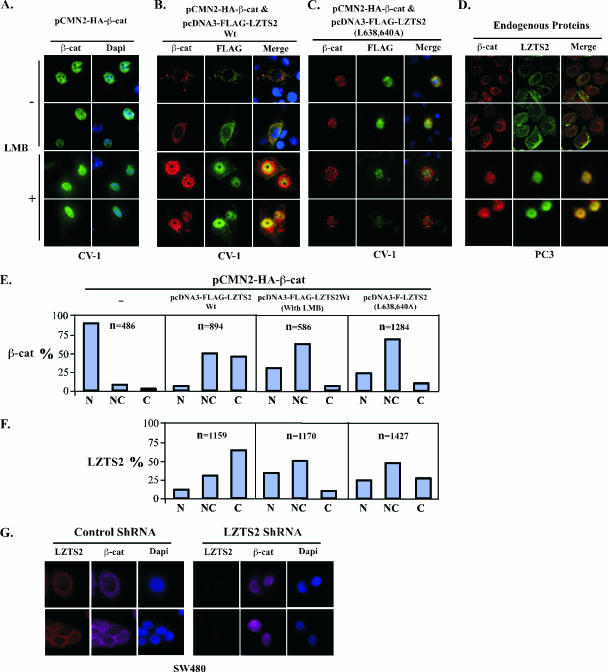
FIG. 5.
LZTS2 affects the cellular localization of β-catenin. (A) pcDNA3-HA-β-catenin was transfected into CV-1 cells and incubated with or without 60 ng/ml LMB. The subcellular localization of the β-catenin (β-cat) was monitored by hemagglutinin monoclonal antibody and fluorescein isothiocyanate-conjugated secondary antibody (green). (B) Both pcDNA3-HA-β-catenin and pcDNA3-FLAG-hLZTS2 were transfected into CV-1 cells. Cells were then cultured with or without 60 ng/ml LMB for 24 h. The ectopically expressed proteins were detected with FLAG or hemagglutinin antibody and revealed with rhodamine or fluorescein isothiocyanate-conjugated secondary antibody, respectively. (C) As described above, the mutant of LZTS2 was used in the experiment to examine the colocalization with β-catenin. (D) PC3 cells were cultured in medium either with or without 60 ng/ml LMB. Endogenous β-catenin and LZTS2 proteins were detected by the specific antibodies against each protein and revealed with appropriate secondary antibodies. (E and F) Percentages of cells in which the localization of the β-catenin and LZST2 proteins in the nuclei (N), cytoplasm (C), or both (N and C) were assessed in CV-1 cells as described in the above experiments. (G) SW480 cells were transfected with either the pBS/U6-LZTS shRNA or pBS/U6 vector as a negative control and then fixed and stained for endogenous LZTS2 and β-catenin after 48 h. The rabbit anti-LZTS2 or mouse anti-β-catenin antibody was developed with the Alexa-Fluor goat anti-rabbit 594 (Invitrogen) (red) or Alexa-Fluor donkey anti-mouse 647 (pink) antibody, respectively. DAPI was used for nuclear visualization.
In addition, we assessed the effect of the functional NES sequence of LZTS2 in shuttling of β-catenin. In the absence of LMB, β-catenin mainly colocalizes in the cytoplasm with wild-type LZTS2 (Fig. 5B, top panel). More than 80% of the β-catenin protein shifted from the nucleus to the cytoplasm relative to the samples transfected with β-catenin alone (Fig. 5A and E). However, this shift of the β-catenin protein from the nucleus to the cytoplasm was significantly reduced in cells treated with LMB or cotransfected with the LZTS2 NES mutant (L638/640A) in comparison with results for cells transfected with the wild-type protein (Fig. 5C and E). Analysis of the subcellular localization of the LZTS2 and β-catenin proteins showed a clear correlation between them (Fig. 5E and F). Taken together, these data suggest that LZTS2 enhances the nuclear export of β-catenin in cells through its C-terminal NES sequence, which is regulated by the CRM1/exportin-alpha pathway.
Using the shRNA construct of LZTS2, we also assessed the effect of endogenous LZTS2 in regulation of cellular distribution of β-catenin. As shown in Fig. 5G, knockdown of endogenous LZTS2 proteins results in an accumulation of nuclear β-catenin proteins in SW480 cells. However, no specific effect was observed in cells transfected with a control shRNA vector. The above results are correlated with our previous observation from the transient transfection experiments (Fig. 3E), suggesting that the repressive effect of LZTS2 on β-catenin-regulated transcription is mainly mediated through regulation of β-catenin nuclear export.
LZTS2 affects endogenous β-catenin in SW480 cells.
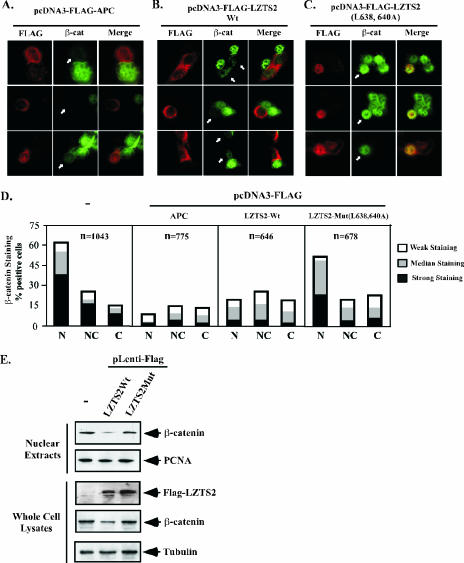
As described above, SW480 cells possess mutated APC (29), resulting in a marked accumulation of β-catenin in the nucleus of cells (26). Introducing wild-type APC into the cells increases the degradation of β-catenin in SW480 cells. In order to assess a direct role of LZTS2 in the regulation of β-catenin, we expressed both the wild type and the NES mutant of LZTS2 in SW480 cells. Full-length LZTS2, as well as APC, mainly localize in the cytoplasm of SW480 cells (Fig. 6A and B, left panels). As reported earlier (7), the nuclear level of β-catenin is significantly decreased in the cells transfected with wild-type APC (Fig. 6A). Intriguingly, overexpression of wild-type LZTS2 also reduces the level of nuclear β-catenin (Fig. 6B), but the LZTS2 NES mutant shows no significant effect on endogenous β-catenin (Fig. 6C). The intensity of β-catenin staining was measured and scored in the above cells (Fig. 6D). It appears that expression of the ectopic APC or LZTS2 wild-type protein reduces the nuclear level of β-catenin. In addition, the level of cytoplasmic β-catenin was also slightly reduced in the above cells.
FIG. 6.
LZTS2 regulates the cellular level of β-catenin. The FLAG-tagged expression vectors containing wild-type APC (A) or LZTS2 (B) or the LZTS2 NES mutant (C) were transfected into SW480 cells. Cells were stained with FLAG and β-catenin antibodies for the ectopically expressed proteins and endogenous β-catenin, as indicated by arrows. (D) Percentages of positively stained β-catenin cells were measured from the above experiments. The intensity and localization of β-catenin staining in the nuclei (N), cytoplasm (C), or both (N and C) were analyzed in cells transfected with different expression constructs as indicated. (E) SW480 cells were infected with the pLentiviral vectors for either wild-type or mutated LZTS2. The nuclear extracts and whole-cell lysates were prepared from the above cells and used for Western blotting.
Next, we performed the Western blot to analyze the level of nuclear and total β-catenin in cells infected with either wild-type LZTS2 or the NES mutant virus. As shown in Fig. 6E, an approximately 70% or 40% reduction of nuclear or total β-catenin, respectively, was observed in cells that express FLAG-tagged wild-type LZTS2 proteins. However, there was no notable change in the expression of PCNA and tubulin proteins used as controls. These results are consistent with the above immunostaining data and provide an additional line of evidence that demonstrates LZTS2's potential role in the regulation of the cellular level of β-catenin.
DISCUSSION
LZTS2, also named LAPSER, was originally identified as a homologue of FEZ1/LZTS1, a putative tumor suppressor (1). The LZTS2 gene is located on human chromosome 10 at 10q24.3, near 10q23.3, where PTEN, a tumor suppressor, was identified (21). Previous studies have suggested that more than one tumor suppressor may be harbored in this locus (35). Overexpression of LZTS2 showed an antiproliferative effect in several human tumor cell lines (1). However, there is little knowledge regarding the biological role and molecular mechanism of LZTS2 in the regulation of cell growth and tumorigenesis.
In this study, we provide several lines of evidence to explore a link between LZTS2 and the β-catenin-mediated cell signaling pathway. We first identified that LZTS2 interacts with β-catenin using a yeast two-hybrid approach. The interaction was further confirmed by both in vitro GST pulldown and coimmunoprecipitation experiments. The full-length armadillo repeats of β-catenin (amino acids 134 to 671) were mapped as being responsible for the interaction. Binding of LZTS2 to the central armadillo domain is consistent with previous findings, which have shown that the armadillo repeats provide a long positively charged groove for binding (14). In this study, we also showed that the C terminus of LZTS2 (aa 447 to 669) contains a unique leucine zipper region and is responsible for binding to β-catenin. Interestingly, a sequence similar to a β-catenin binding motif was found between amino acids 608 and 613 (EqVIrY) within a largely acidic region of LZTS2 (3). It should be noted that this region is very similar to the one of FEZ1/LZTS1, suggesting a potential link between FEZ1/LZTS1 and β-catenin.
In this study, we also showed a functional consequence of the interaction between LZTS2 and β-catenin. Expression of LZTS2 can repress β-catenin-mediated transcription on the TCF/LEF-regulated promoter. However, no significant repression was observed in transient-transfection assays when full-length and truncated LZTS2 were targeted through fusion to the GAL4 DNA binding domain (data not shown). These data indicate that LZTS2 may not contain an intrinsic transcriptional repressive domain, and the repression of β-catenin by LZTS2 may be mediated through other mechanisms. In the course of a search for the mechanisms by which LZTS2 regulates β-catenin-mediated transcription, we demonstrated that both exogenous and endogenous LZTS2 localize exclusively in the cytoplasm, with a perinuclear staining pattern (Fig. 4A to D). Using an immunofluorescence approach, we further observed that the nuclear exporting of LZTS2 is blocked by LMB, suggesting that the CRM-1-dependent pathway is involved in the regulation of the subcellular distribution of LZTS2. Several NES-like sequences appear in the LZTS2 protein. Expression of the GFP fusion proteins with these NES sequences in frame identified the C-terminal sequence (amino acids 631 to 642) as capable of regulating the nuclear export of the GFP fusion protein. Mutation of this NES site by substitution of either one or two leucines with alanine diminishes the nuclear export of LZTS2. Overall, these data suggest an important role of LZTS2 in nuclear and cytoplasmic trafficking.
Previous studies have suggested that the intracellular trafficking of β-catenin is regulated by the CRM/exportin nuclear export pathway (6, 13). Since LZTS2 interacts with β-catenin and its subcellular distribution is regulated by the CRM-dependent nuclear export signal through the intrinsic NES, we presented several lines of evidence to explore the possibility that LZTS2 regulates β-catenin nuclear export. First, the expression of wild-type LZTS2 increased the cytoplasmic level of endogenous or ectopically expressed β-catenin, and treating cells with leptomycin B blocked the effect of LZTS2, resulting in the nuclear retention of both the β-catenin and LZTS2 proteins. Moreover, the LZTS2 NES mutants showed no effect on β-catenin under similar experimental conditions. Furthermore, the expression of wild-type LZTS2 but not the NES mutant significantly reduced the level of endogenous nuclear β-catenin in SW480 cells. These data suggest that LZTS2 regulates the nuclear trafficking of β-catenin and affects the cellular level of β-catenin through an APC-independent degradation pathway in SW480 cells.
The tumor suppressor APC has been shown to affect the nuclear-cytoplasm shuttling of β-catenin (36). Several functional NES sequences have been identified within the APC protein. However, the significant exodus of β-catenin from the nucleus appears in APC-mutated tumor cells (6), which suggests that an additional CRM1-dependent export pathway may exist and be involved in the regulation of β-catenin. Our data that show the role of LZST2 in the regulation of β-catenin nuclear export agree with the previous observation and provide evidence demonstrating a novel regulatory pathway for β-catenin. Since cellular levels of β-catenin are tightly regulated in normal cells, mutations affecting the degradation of β-catenin can result in increasing the cellular level of β-catenin, which has been considered a key event in tumorigenesis (33). APC-dependent proteasomal degradation pathways for β-catenin are mediated through either the APC/Axin complex (32) or a p53-inducible pathway involving Siah-1 (22, 23). In addition, other APC-independent pathways have also been suggested to regulate the cellular level of β-catenin (37, 42). In this study, we observed that overexpression of LZTS2 reduces the level of β-catenin in SW480 cells. Given the fact that LZTS2 has been shown previously to inhibit the proliferation of human tumor cells (1), it would be interesting to further investigate whether the inhibitory role of LZTS2 in cell growth is mediated through the regulation of β-catenin.
The sequence analysis showed that LZTS2 is very similar to a putative tumor suppressor, FEZ1/LZTS1. The FEZ1/LZTS1 gene was mapped to chromosome 8p22, a region that is frequently deleted in human tumors (15). Previous studies have shown that the expression of FEZ1/LZTS1 was altered in multiple human malignancies (30, 40, 41). Currently, the precise role of FEZ1/LZTS1 as a tumor suppressor still remains unclear. Based on the sequence similarity between LZTS2 and FEZ1/LZTS1, defining the functions of these two proteins in tumorigenesis will be extremely important.
Acknowledgments
We are grateful for the various reagents received from Albert Brinkmann, Jan Trapman, and Bert Vogelstein, and we thank Xiaomeng Li, Jane Lee, and Meletios Verras for technical assistance.
This work was supported by National Institutes of Health grants CA70297, CA87767, and DK61002.
Footnotes
Published ahead of print on 25 September 2006.
REFERENCES
- 1.Cabeza-Arvelaiz, Y., T. C. Thompson, J. L. Sepulveda, and A. C. Chinault. 2001. LAPSER1: a novel candidate tumor suppressor gene from 10q24.3. Oncogene 20:6707-6717. [DOI] [PubMed] [Google Scholar]
- 2.Chan, S. K., and G. Struhl. 2002. Evidence that Armadillo transduces wingless by mediating nuclear export or cytosolic activation of Pangolin. Cell 111:265-280. [DOI] [PubMed] [Google Scholar]
- 3.Daniels, D. L., and W. I. Weis. 2002. ICAT inhibits beta-catenin binding to Tcf/Lef-family transcription factors and the general coactivator p300 using independent structural modules. Mol. Cell 10:573-584. [DOI] [PubMed] [Google Scholar]
- 4.Dignam, J. D., P. L. Martin, B. S. Shastry, and R. G. Roeder. 1983. Eukaryotic gene transcription with purified components. Methods Enzymol. 101:582-598. [DOI] [PubMed] [Google Scholar]
- 5.Dull, T., R. Zufferey, M. Kelly, R. J. Mandel, M. Nguyen, D. Trono, and L. Naldini. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eleftheriou, A., M. Yoshida, and B. R. Henderson. 2001. Nuclear export of human beta-catenin can occur independent of CRM1 and the adenomatous polyposis coli tumor suppressor. J. Biol. Chem. 276:25883-25888. [DOI] [PubMed] [Google Scholar]
- 7.Faux, M. C., J. L. Ross, C. Meeker, T. Johns, H. Ji, R. J. Simpson, M. J. Layton, and A. W. Burgess. 2004. Restoration of full-length adenomatous polyposis coli (APC) protein in a colon cancer cell line enhances cell adhesion. J. Cell Sci. 117:427-439. [DOI] [PubMed] [Google Scholar]
- 8.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475-483. [DOI] [PubMed] [Google Scholar]
- 9.Fodde, R., R. Smits, and H. Clevers. 2001. APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer 1:55-67. [DOI] [PubMed] [Google Scholar]
- 10.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390:308-311. [DOI] [PubMed] [Google Scholar]
- 12.Harada, N., Y. Tamai, T. Ishikawa, B. Sauer, K. Takaku, M. Oshima, and M. M. Taketo. 1999. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 18:5931-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson, B. R. 2000. Nuclear-cytoplasmic shuttling of APC regulates beta-catenin subcellular localization and turnover. Nat. Cell Biol. 2:653-660. [DOI] [PubMed] [Google Scholar]
- 14.Huber, A. H., W. J. Nelson, and W. I. Weis. 1997. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell 90:871-882. [DOI] [PubMed] [Google Scholar]
- 15.Ishii, H., R. Baffa, S. I. Numata, Y. Murakumo, S. Rattan, H. Inoue, M. Mori, V. Fidanza, H. Alder, and C. M. Croce. 1999. The FEZ1 gene at chromosome 8p22 encodes a leucine-zipper protein, and its expression is altered in multiple human tumors. Proc. Natl. Acad. Sci. USA 96:3928-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, K., K. M. Pang, M. Evans, and E. D. Hay. 2000. Overexpression of beta-catenin induces apoptosis independent of its transactivation function with LEF-1 or the involvement of major G1 cell cycle regulators. Mol. Biol. Cell 11:3509-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kongkanuntn, R., V. J. Bubb, O. J. Sansom, A. H. Wyllie, D. J. Harrison, and A. R. Clarke. 1999. Dysregulated expression of beta-catenin marks early neoplastic change in Apc mutant mice, but not all lesions arising in Msh2 deficient mice. Oncogene 18:7219-7225. [DOI] [PubMed] [Google Scholar]
- 19.Korinek, V., N. Barker, P. J. Morin, D. van Wichen, R. de Weger, K. W. Kinzler, B. Vogelstein, and H. Clevers. 1997. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science 275:1784-1787. [DOI] [PubMed] [Google Scholar]
- 20.Kutay, U., F. R. Bischoff, S. Kostka, R. Kraft, and D. Gorlich. 1997. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell 90:1061-1071. [DOI] [PubMed] [Google Scholar]
- 21.Li, J., C. Yen, D. Liaw, K. Podsypanina, S. Bose, S. I. Wang, J. Puc, C. Miliaresis, L. Rodgers, R. McCombie, S. H. Bigner, B. C. Giovanella, M. Ittmann, B. Tycko, H. Hibshoosh, M. H. Wigler, and R. Parsons. 1997. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275:1943-1947. [DOI] [PubMed] [Google Scholar]
- 22.Liu, J., J. Stevens, C. A. Rote, H. J. Yost, Y. Hu, K. L. Neufeld, R. L. White, and N. Matsunami. 2001. Siah-1 mediates a novel beta-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol. Cell 7:927-936. [DOI] [PubMed] [Google Scholar]
- 23.Matsuzawa, S. I., and J. C. Reed. 2001. Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol. Cell 7:915-926. [DOI] [PubMed] [Google Scholar]
- 24.Miyoshi, Y., H. Nagase, H. Ando, A. Horii, S. Ichii, S. Nakatsuru, T. Aoki, Y. Miki, T. Mori, and Y. Nakamura. 1992. Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene. Hum. Mol. Genet. 1:229-233. [DOI] [PubMed] [Google Scholar]
- 25.Molenaar, M., M. van de Wetering, M. Oosterwegel, J. Peterson-Maduro, S. Godsave, V. Korinek, J. Roose, O. Destree, and H. Clevers. 1996. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell 86:391-399. [DOI] [PubMed] [Google Scholar]
- 26.Munemitsu, S., I. Albert, B. Souza, B. Rubinfeld, and P. Polakis. 1995. Regulation of intracellular beta-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc. Natl. Acad. Sci. USA 92:3046-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neufeld, K. L., F. Zhang, B. R. Cullen, and R. L. White. 2000. APC-mediated downregulation of beta-catenin activity involves nuclear sequestration and nuclear export. EMBO Rep. 1:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishi, K., M. Yoshida, D. Fujiwara, M. Nishikawa, S. Horinouchi, and T. Beppu. 1994. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J. Biol. Chem. 269:6320-6324. [PubMed] [Google Scholar]
- 29.Nishisho, I., Y. Nakamura, Y. Miyoshi, Y. Miki, H. Ando, A. Horii, K. Koyama, J. Utsunomiya, S. Baba, and P. Hedge. 1991. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science 253:665-669. [DOI] [PubMed] [Google Scholar]
- 30.Nonaka, D., A. Fabbri, L. Roz, L. Mariani, A. Vecchione, G. W. Moore, L. Tavecchio, C. M. Croce, and G. Sozzi. 2005. Reduced FEZ1/LZTS1 expression and outcome prediction in lung cancer. Cancer Res. 65:1207-1212. [DOI] [PubMed] [Google Scholar]
- 31.Nusse, R. 2003. Wnts and Hedgehogs: lipid-modified proteins and similarities in signaling mechanisms at the cell surface. Development 130:5297-5305. [DOI] [PubMed] [Google Scholar]
- 32.Polakis, P. 2002. Casein kinase 1: a Wnt'er of disconnect. Curr. Biol. 12:R499-R501. [DOI] [PubMed] [Google Scholar]
- 33.Polakis, P. 2000. Wnt signaling and cancer. Genes Dev. 14:1837-1851. [PubMed] [Google Scholar]
- 34.Powell, S. M., N. Zilz, Y. Beazer-Barclay, T. M. Bryan, S. R. Hamilton, S. N. Thibodeau, B. Vogelstein, and K. W. Kinzler. 1992. APC mutations occur early during colorectal tumorigenesis. Nature 359:235-237. [DOI] [PubMed] [Google Scholar]
- 35.Rasheed, B. K., R. E. McLendon, H. S. Friedman, A. H. Friedman, H. E. Fuchs, D. D. Bigner, and S. H. Bigner. 1995. Chromosome 10 deletion mapping in human gliomas: a common deletion region in 10q25. Oncogene 10:2243-2246. [PubMed] [Google Scholar]
- 36.Rosin-Arbesfeld, R., F. Townsley, and M. Bienz. 2000. The APC tumour suppressor has a nuclear export function. Nature 406:1009-1012. [DOI] [PubMed] [Google Scholar]
- 37.Sharma, C., A. Pradeep, L. Wong, A. Rana, and B. Rana. 2004. Peroxisome proliferator-activated receptor gamma activation can regulate beta-catenin levels via a proteasome-mediated and adenomatous polyposis coli-independent pathway. J. Biol. Chem. 279:35583-35594. [DOI] [PubMed] [Google Scholar]
- 38.Staal, F. J., M. Noort Mv, G. J. Strous, and H. C. Clevers. 2002. Wnt signals are transmitted through N-terminally dephosphorylated beta-catenin. EMBO Rep. 3:63-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teufel, A., A. Weinmann, P. R. Galle, and A. W. Lohse. 2005. In silico characterization of LZTS3, a potential tumor suppressor. Oncol. Rep. 14:547-551. [PubMed] [Google Scholar]
- 40.Toyooka, S., Y. Fukuyama, I. I. Wistuba, M. S. Tockman, J. D. Minna, and A. F. Gazdar. 2002. Differential expression of FEZ1/LZTS1 gene in lung cancers and their cell cultures. Clin. Cancer Res. 8:2292-2297. [PubMed] [Google Scholar]
- 41.Vecchione, A., H. Ishii, Y. H. Shiao, F. Trapasso, M. Rugge, J. F. Tamburrino, Y. Murakumo, H. Alder, C. M. Croce, and R. Baffa. 2001. Fez1/lzts1 alterations in gastric carcinoma. Clin. Cancer Res. 7:1546-1552. [PubMed] [Google Scholar]
- 42.Xiao, J. H., C. Ghosn, C. Hinchman, C. Forbes, J. Wang, N. Snider, A. Cordrey, Y. Zhao, and R. A. Chandraratna. 2003. Adenomatous polyposis coli (APC)-independent regulation of beta-catenin degradation via a retinoid X receptor-mediated pathway. J. Biol. Chem. 278:29954-29962. [DOI] [PubMed] [Google Scholar]
- 43.Yang, F., X. Li, M. Sharma, C. Y. Sasaki, D. L. Longo, B. Lim, and Z. Sun. 2002. Linking beta-catenin to androgen-signaling pathway. J. Biol. Chem. 277:11336-11344. [DOI] [PubMed] [Google Scholar]