Abstract
Commitment to the melanocyte lineage is characterized by the onset of expression of the microphthalmia-associated transcription factor (Mitf). This transcription factor plays a fundamental role in melanocyte development and maintenance and seems to be crucial for the survival of malignant melanocytes. Furthermore, Mitf has been shown to be involved in cell cycle regulation and to play important functions in self-renewal and maintenance of melanocyte stem cells. Although little is known about how Mitf regulates these various processes, one possibility is that Mitf interacts with other regulators. Here we show that Mitf can interact directly with β-catenin, the key mediator of the canonical Wnt signaling pathway. The Wnt signaling pathway plays a critical role in melanocyte development and is intimately involved in triggering melanocyte stem cell proliferation. Significantly, constitutive activation of this pathway is a feature of a number of cancers including malignant melanoma. Here we show that Mitf can redirect β-catenin transcriptional activity away from canonical Wnt signaling-regulated genes toward Mitf-specific target promoters to activate transcription. Thus, by a feedback mechanism, Mitf can diversify the output of canonical Wnt signaling to enhance the repertoire of genes regulated by β-catenin. Our results reveal a novel mechanism by which Wnt signaling and β-catenin activate gene expression, with significant implications for our understanding of both melanocyte development and melanoma.
Melanocytes provide an excellent system to study complex regulatory networks. Their precursor cells, the melanoblasts, originate from the neural crest and then migrate along characteristic pathways to various destinations such as dermis and epidermis, the inner ear, and the choroid of the eye and hair follicles. The finding that the proliferation and migration of melanoblasts during development is highly similar to the proliferation and metastasis patterns of melanoma, a highly dangerous and increasingly common cancer, highlights the value of the melanocyte system as a model for addressing key issues of general significance in both development and cancer (45).
The microphthalmia-associated transcription factor (Mitf) protein is a key regulator of melanocyte development. Mitf is a basic helix-loop-helix leucine zipper (bHLH-Zip) transcription factor which plays an important role in the development of various cell types, ranging from neural crest derived melanocytes, mast cells, and osteoclasts to cells of the retinal pigment epithelia of the eye. The function of Mitf has been particularly well-studied in melanocytes (reviewed in reference 40), and recently it has been shown that Mitf is involved in the maintenance and self-renewal of melanocyte stem cells (28, 30, 32). In addition, MITF is expressed in nearly all cutaneous melanomas, and the gene has been shown to be amplified in 10% of melanomas, suggesting that activation or amplification of this important transcription factor contribute to tumorigenesis (14, 49).
Like other members of the bHLH-Zip family, Mitf can bind a subset of the canonical E-box sequence 5′-CANNTG-3′, either as a homodimer or as a heterodimer with one of its close relatives Tfe3, Tfeb, and Tfec (21). Mitf controls melanocyte development by regulating the expression of various different genes, ranging from genes involved in commitment and differentiation of the melanocyte lineage (summarized in reference 42) to genes important for regulation of cell cycle such as p21 (6), INK4a/ARF (30), and CDK2 (11). However, how Mitf controls and regulates these different events is not fully understood. One possibility is that Mitf recruits other key mediators to its target gene promoters. To determine the potential of Mitf to interact with other transcription factors and coregulators we performed a yeast two-hybrid screen. One of the genes found was β-catenin, the downstream mediator of the canonical Wnt signaling pathway.
Like Mitf, the Wnt/β-catenin signaling pathway has been implicated in both melanocyte development and in the formation of malignant melanoma. Signaling by this pathway is triggered by the binding of secreted Wnt growth factor proteins to Frizzled/low-density lipoprotein receptor-related protein receptor complexes. In the absence of Wnt signaling, β-catenin is degraded by a multiprotein complex containing Dishevelled, glycogen synthase kinase 3β (GSK-3β), axin or conductin, and the adenomatous polyposis coli tumor suppressor protein (3). In the presence of Wnt, the GSK-3β-dependent phosphorylation of β-catenin, which results in its degradation through the proteasome, is blocked. This results in the stabilization of β-catenin and subsequent nuclear translocation, where it can heterodimerize with one of the four members of the T-cell factor (Tcf/Lef1) family of HMG-box transcription factors (reviewed in references 31 and 5). Formation of β-catenin/Tcf complexes leads to the activation of specific target genes such as Ultrabithorax, siamois, or c-myc (summarized in reference 5).
A link between Wnt signaling and melanocyte formation was revealed by the finding that β-catenin and T-cell factors regulate Mitf expression positively at the transcriptional level (10). Ectopic Wnt3a upregulates Mitf expression and recruits β-catenin and Lef1 to the Lef1 binding sites of the Mitf promoter (43, 50). Another hint for the importance of Wnt signaling for melanocyte development and survival comes from the finding that β-catenin is expressed throughout melanocyte formation and in mature melanocytes (25). Deregulated activity of the Wnt/β-catenin pathway is observed in a significant fraction of primary human melanomas and may contribute to cancer formation by inducing Mitf expression (50).
Here, we report another level of cross talk between Wnt signaling components and Mitf. We show not only that β-catenin is involved in Lef1-dependent control of Mitf expression but also that it functionally interacts with the Mitf protein itself. Our findings indicate that Mitf can redirect β-catenin transcriptional activity away from β-catenin/Lef1-regulated genes toward Mitf-specific target promoters. The results unveil a new mechanism of regulating Mitf activity with major implications for our understanding of both melanocyte development and melanoma.
MATERIALS AND METHODS
Yeast two-hybrid screen.
For the yeast two-hybrid screen, a bait construct was generated using the bHLH-Zip part of mouse Mitf. Nucleotides 660 (amino acid 179) to 1125 (amino acid 332) from the mouse Mitf cDNA were PCR amplified using primers BamHI 1-1 (5′-GTCGTACATGTCAGGGATCCCTGGTGCTGTAC-3′) and EcoRI 1-18 (5′-CAGCAACTCCTGTCCAGAATTCCTTCCCAAC-3′) and cloned into the EcoRI/BamHI sites (underlined) of the pBTM116 vector to form a bait construct where the bHLH-Zip domain of Mitf was fused to the lexA DNA binding protein. The constructs were sequenced to confirm the absence of PCR-generated mutations. The prey library was generously donated by Stan Hollenberg. It was constructed from 9.5- to 10.5-day-old mouse CD1 embryos and consists of 350- to 700-bp-long fragments cloned into the NotI site of the pVP16 vector, a common library vector, containing a VP16 transactivation domain just N-terminal from the polylinker (22, 47). Once the bait was ready, it was transformed into the L40 yeast strain that includes the prey, and double selection was carried out by culturing in selective medium.
Plasmids: Mitf deletion constructs.
For mapping the interaction domain of Mitf with β-catenin, several mutant constructs were generated from a full-length Mitf-1m cDNA clone. The primers used for generating the MitfΔ28 and MitfΔ69 dilution constructs were the following (restriction sites are underlined): 5′miΔ69, 5′-TGAAAGCTTGCTATGCTGGAAATGCTA-3′; 3′miΔ69, 5′-GGAGAATTCACGCTGTGAGCTCCC 3′; 5′miΔ28, 5′-ACAAGCTTATGCTGGAAATGCTAGAGCTGGAGATGCAGGCTAGA-3′; and 3′miΔ28, 5′-GGGAATTCCACACGCATGCTCCGTTTCTTCTGC-3′. PCR products were digested with EcoRI/HindIII and ligated into full-length Mitf in pcDNA3.1. The Mitf Δhelix1 construct and the Mitf D222/D236N and Mitf D222N mutant constructs were generated using the megaprimer PCR amplification method, as described previously (36). The amplified products were digested with BamHI/PstI and ligated into full-length Mitf inserted in pUC19 vector. The full-length mutant Mitf constructs were digested with EcoRI/HindIII and subcloned into the pBK-CMV vector. The resulting pBK-CMV constructs were cloned into pcDNA3 by EcoRI/NotI digestion. All PCR-based constructs were verified by sequencing. To generate a mammalian expression vector for Mitf with six N-terminal copies of the Myc-epitope tag, the Mitf cDNA was inserted in frame into the XbaI/SnaBI sites of pCS2+MT (44).
Expression vectors for expressing the wild-type or constitutively active β-catenin, β-catenin with a single hemagglutinin (HA) epitope tag, or glutathione transferase (GST)-β-catenin fusions (amino acids 1 to 781, 1 to 284, 120 to 683, and 536 to 781, respectively, of β-catenin), as well as the TOPFLASH reporter have all been described previously (1, 2, 4, 18, 24, 46). To add two copies of the HA epitope tag to β-catenin, a ClaI/NotI restriction fragment from pCS2+LEFΔN-HA2 (46) was transferred to pCS2+β-catenin (24). Yeast shuttle vectors for VP16 fusions with amino acids 108 to 683, 108 to 553, and 536 to 683 of β-catenin were obtained by transferring SmaI/NotI restriction fragments from previously described β-catenin yeast expression constructs (18) to KpnI/NotI-cleaved pVP16 (22, 47). The resulting plasmids were used as starting material to construct VP16 fusions with β-catenin amino acid sequences 1 to 781, 1 to 683, and 108 to 781 by adding suitable restriction fragments from other plasmids containing β-catenin, as described above. Expression vectors for GST-β-catenin fusions (amino acids 1 to 683, 120 to 781, 120 to 553, and 536 to 683 of β-catenin) were made in an analogous manner by exchanging suitable restriction fragments between constructs previously described (1, 4, 18).
Cell culture and luciferase assay.
HEK293 (human embryonic kidney) cells and COS7 cells were maintained in Dulbecco minimal essential medium (Gibco/Brl), supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml and cultured in a humidified incubator at 37°C with 5% CO2. Human melanoma cell line 501mel cells were maintained in RPMI-1640 medium (Cambrex), supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. For luciferase assays, 2 × 105 HEK293 cells were seeded in 12-well plates and transfected with EXGEN 500 (Fermentas Lifescience) or calcium phosphate. For each luciferase assay, cells were transfected with 100 ng of reporter construct (either the tyrosinase promoter element that consists of DNA sequences containing 200 bp upstream of the transcription start site and 80 bp downstream [−200/+80 tyrosinase], four copies of the M-box[4×M-box], or the Tyrp-1 construct), 500 ng of pcDNA-3 Mitf, and increasing amounts of β-catenin construct, at a concentration ranging from 250 and 500 up to 750 ng of DNA. Total amounts of transfected DNA were kept constant at 2 μg by the addition of empty pcDNA3 vector. Transfection of 2 ng of pRenilla served as a transfection control and was used to normalize luciferase activity. At 24 h posttransfection, cells were lysed and assayed for firefly and Renilla luciferase activity as specified by the manufacturer (Promega). All transfection experiments were repeated at least twice and in triplicate each time. The normalized values were divided by the firefly luciferase activity of the empty vector alone, and the ratio was expressed as the relative induction (n-fold) over pcDNA3. For Mitf interference analyses, HEK293 cells were transfected as described previously (19) using 50 ng of activator plasmids and 1 ng, 5 ng, or 25 ng of Mitf expression construct. DNA amounts were kept constant by adding empty pCS2+ expression vector up to a final amount of transfected DNA of 360 ng. Firefly and Renilla luciferase activities were determined 40 h after transfection. Normalized values from at least three independent experiments were averaged. For each activator construct, they are expressed as percent activity relative to firefly luciferase expression in the absence of Mitf.
Colocalization studies.
For colocalization studies, COS7 cells were seeded on chamber slides and transfected with green fluorescent protein (GFP)-Mitf and c-Myc-tagged β-catenin using EXGEN 500 (Fermentas Lifescience). At 24 h posttransfection the cells were washed and fixed in 2% paraformaldehyde-phosphate-buffered saline (PBS) for 20 min at room temperature. After preincubation in PBS-0.1% Triton X-100, the cells were blocked in blocking buffer (PBS-3% milk powder) for 20 min. Anti-cMyc (9E10; Abcam) antibodies were used as primary antibodies at a concentration of 1:100 or 1:200 in blocking buffer for 60 min. After being washed three times in PBS, cells were incubated in anti-mouse Alexa 568 (Molecular Probes) antibodies at a dilution of 1:1,000 for 30 min, washed again three times with PBS, mounted in Fluormount-G (Southern Biotech), and sealed with nail polish. For nuclear staining, cells were incubated once with TOPRO-3 (Molecular Probes) for 15 min in between the washing steps. Fluorescence analysis was performed using a confocal Zeiss LSM 5 Pa microscope. The GFP-labeled Mitf was excited with an argon laser (488-nm excitation line with 515-nm long pass filter), and the ALEXA 568-conjugated anti-mouse antibody was simultaneously excited with an He-Ne laser (543-nm excitation line with 570-nm long pass barrier filter). Nuclear staining was visualized by excitation with an He-Ne laser at a 633-nm excitation line. For analysis of the Mitf Δhelix1 deletion mutant and the Mitf D222/236N and Mitf D222N (this mutation is known in mice as Mitfmi-vitilago or Mitfmi-vit) point mutations, COS7 cells were transfected with the different plasmids indicated and HA-tagged β-catenin and treated as described above. Localization of the Mitf mutants was analyzed by use of a mouse anti-Mitf antibody (C5; kind gift from D. Fisher) and the secondary anti-mouse Alexa 568 (Molecular Probes) antibody. Distribution of β-catenin was analyzed by use of a goat anti-β-catenin antibody (Santa Cruz) and a secondary anti-goat Alexa 483 (Molecular Probes) antibody as described above.
GST pull-down assays.
For GST pull-down experiments, radiolabeled wild-type or mutant Mitf cDNAs were transcribed and translated into proteins in vitro using a T7-based TNT system (Promega). For each binding reaction, a 2-μl aliquot of a 50-μl TNT reaction was used. GST pull-down assays, using 2 μg of GST or the various GST-β-catenin fusion proteins immobilized on glutathione-Sepharose beads, were performed as previously described (18). After preincubation in binding buffer containing 0.5% bovine serum albumin for 30 min at 4°C, [35S]methionine-labeled wild-type or mutant Mitf proteins were added, after which binding proceeded for 2 h at 4°C. Following extensive washing with binding buffer without bovine serum albumin, material retained on the glutathione-Sepharose matrix was eluted in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, separated by SDS-PAGE on 8% gels, and visualized by fluorography.
Western blotting and immunoprecipitation.
For in vitro coimmunoprecipitation of β-catenin and Mitf, 2 × 106 HEK293 cells were transfected by the calcium phosphate coprecipitation technique with 15 μg of expression vectors. Transfected cells were lysed 40 h after transfection in 1 ml of immunoprecipitation buffer (50 mM Tris-HCl, pH 7.6, 5 mM MgCl2, 0.1% Nonidet P-40) containing 150 mM NaCl. From each lysate 500 μl was supplemented with 1 μg of anti-HA antibody (3F10; Roche Applied Science), a 10-μl bed volume of protein G-Sepharose (Amersham Biosciences), and 2.5 μl of [35S]methionine-labeled Mitf protein synthesized in vitro as described above. Binding was allowed to occur for 4 h at 4°C before precipitates were recovered and washed once for 10 min with 1 ml of immunoprecipitation buffer with 120 mM NaCl and twice with immunoprecipitation buffer with 75 mM NaCl. Finally, precipitates were eluted from the protein G-Sepharose, and one half of each sample was analyzed by electrophoresis on 8% SDS-polyacrylamide gels and fluorography while the other half was analyzed by SDS-PAGE and Western blotting as previously described (20).
For coimmunoprecipitation of β-catenin and Mitf expressed in vivo, 2 × 106 HEK293 cells were transfected with 10 μg of each of the expression vectors for activated HA-tagged β-catenin and myc-tagged Mitf. Cells were stimulated with 40 mM LiCl for 2 h prior to harvest at 40 h posttransfection. Cells were lysed in 1 ml of immunoprecipitation buffer (50 mM Tris-HCl, pH 7.6, 5 mM MgCl2, 0.1% Nonidet P-40) containing 75 mM KCl, 1 mM dithiothreitol, 0.1 mM sodium orthovanadate, 10 mM NaF, and Complete protease inhibitor mix (Roche Applied Science). From each lysate 400 μl was supplemented with 1 μg of anti-myc antibody (9E10; Roche Applied Science), and a 10-μl bed volume of protein A-Sepharose (Amersham Biosciences). Binding was allowed to occur for 2.5 h at 4°C before precipitates were recovered and washed three times with 1 ml of immunoprecipitation buffer with 75 mM KCl for 10 min each. Finally, precipitates were eluted from the protein A-Sepharose and analyzed by electrophoresis on 8% SDS-polyacrylamide gels and by Western blotting as described previously (20), probing sequentially for Mitf-myc and β-catenin HA. For coimmunoprecipitation of endogenous proteins, 90% confluent 501mel cells were lysed in 1 ml of immunoprecipitation buffer (as above) and precleared with a 20-μl bed volume of protein A-Sepharose (Amersham Bioscience) for 1 h at 4°C. A total of 50 μl was taken as input sample; the remaining lysate was incubated with either 1 μg of anti-Mitf antibody (C5; Labvision) or 1 μg of anti-β-catenin (Santa Cruz) and a 20-μl bed volume of protein A-Sepharose (Amersham Bioscience) overnight at 4°C. Precipitates were recovered and washed three times with 1 ml of immunoprecipitation buffer for 10 min each. Material bound to the beads was eluted in SDS loading buffer, resolved by SDS-PAGE on 10% gels, transferred onto nitrocellulose, and analyzed by Western blotting, probing with monoclonal anti-Mitf (C5) and polyclonal anti-β-catenin (Santa Cruz).
Electrophoretic mobility shift assay.
For DNA binding studies, wild-type or mutant Mitf cDNAs were transcribed and translated into proteins in vitro using the T7-based TNT system (Promega). Equal volumes of the in vitro produced Mitf constructs were incubated with a 32P-labeled DNA probe (AAAGTCAGTCATGTGCTTTTCAGA) containing an E-box (underlined) in 20 μl of binding buffer (10 mM Tris, pH 7.5, 100 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 4% glycerol, 80 ng of salmon sperm/ml, 10% fetal calf serum, 2 mM MgCl2, and 2 mM spermidine). After a 20-min incubation on ice, the binding mix was loaded on a 6% nondenaturating PAGE gel and run for 2 h at 150 mA. The gel was dried and exposed to film overnight. Binding specificity was confirmed by supershift of Mitf proteins with an Mitf-specific antibody (C5; kind gift from D. Fisher).
Chromatin immunoprecipitation.
Chromatin immunoprecipitation assays were performed as described previously (13) with mouse monoclonal anti-Mitf antibody (C5), goat polyclonal anti-β-catenin antibody (C-18; Santa Cruz), rabbit polyclonal anti-LEF1 antibody (Santa Cruz), or 10 μl of nonspecific immunoglobulin G (Bio-Rad). After immunoprecipitation, samples were amplified by quantitative PCR for 30 cycles, taking care that the PCR was in the log phase of amplification, and analyzed by agarose gel electrophoresis. Primers used for amplification were the following: 5′ Brn-2, 5′-GAGGAGGGCTAGGAGGACTCC-3′; 3′ Brn-2, 5′-CGCGTAACTGTCAATGAAAAA-3′; 5′ Tyrp-1, 5′-TTCTCTTGCCTGGTTGCTCT-3′; 3′ Tyrp-1, 5′-TGTGGATTGCTGCCTGATAA-3′; 5′ tyrosinase, 5′-AATATCCTCTGTCCAATGC-3′; 3′ tyrosinase, 5′-ATAGTGAAGTTTTCATCTCC-3′; 5′ Hsp70 5′-CCTCCAGTGAATCCCAGAAGACTCT-3′; and 3′ Hsp70, 5′-TGGGACAACGGGAGTCACTCTC-3′.
siRNA treatment.
Small interfering RNA (siRNA)-mediated downregulation of Mitf in 501mel cells was essentially performed as described previously (6). The Mitf-specific sequences used were the following: 5′-r(AGCAGUACCUUUCUACCAC)d(TT)-3′ and 5′-r(GUGGUAGAAAGGUACUGCU)d(TT)-3′. The control siRNA sequences used were 5′-r(UUCUCCGAACGUGUCACGU)d(TT)-3′ and 5′-r(ACGUGACACGUUCGGAGAA)d(TT)-3′. After 3 days of siRNA treatment, chromatin immunoprecipitation assays were performed as described above.
RESULTS
Identification of β-catenin as an Mitf interacting partner.
To screen for novel Mitf interacting partners, we used the bHLH-Zip region of Mitf as bait in a yeast two-hybrid screen against a cDNA library prepared from 9.5- to 10.5-day-old mouse embryos in the library vector pVP16 (22, 47). A total of 5 × 107 colonies were screened, 895 of which turned out to grow in the absence of histidine. To further characterize the positive colonies, the nonspecific LexA-lamin control was used as a negative control against all His+ β-Gal+ colonies obtained in the original yeast two-hybrid screen. Of the 231 clones tested, 135 candidates did not turn blue in the β-Gal screen with the LexA-lamin control, suggesting specific interactions with the Mitf bait. Sequencing of these 135 clones revealed that they represent multiple copies of 27 different genes (data not shown). Among the clones identified were three copies each of the Tfe3, Ubc9, and Pias3 genes previously shown to interact with Mitf in vitro (21, 29, 51), confirming the robustness of our screen.
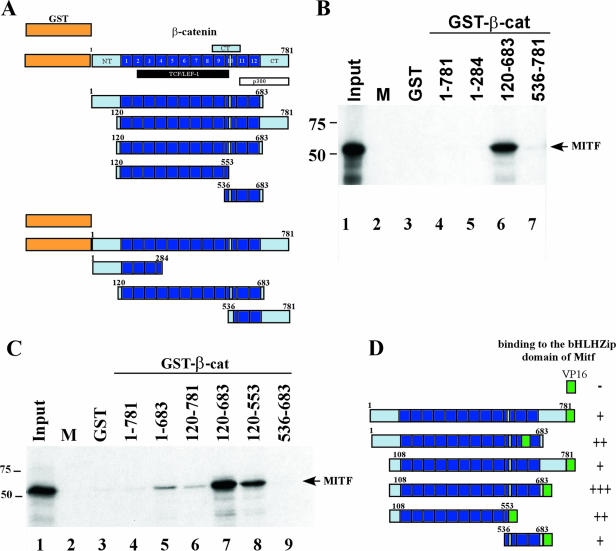
Among the 27 different cDNAs isolated in the screen, one clone harbored a fragment of mouse β-catenin, a well-known downstream target of the canonical Wnt signaling pathway. To confirm and to further characterize interactions between these proteins, we performed GST pull-down experiments. GST and GST-β-catenin fusions containing different portions of β-catenin (Fig. 1A) were used as baits to bind radiolabeled Mitf in vitro. Although GST fusions containing full-length β-catenin and β-catenin residues 1 to 284 or 536 to 781 did not interact with Mitf (Fig. 1B, compare lanes 4, 5, and 7 to lane 3), a β-catenin fragment consisting of the central Armadillo repeat region strongly interacted with Mitf (Fig. 1B, lane 6). These results suggest that Mitf interacts with the Armadillo repeats of β-catenin and that the N- and C-terminal extensions of β-catenin flanking the Armadillo repeats interfere with this interaction. These findings are in close agreement with previous reports showing that the C-terminal part of β-catenin regulates the binding affinity of β-catenin for diverse interacting partners in response to extracellular signals (16, 40). To address this issue in the context of the GST fusion proteins, we constructed an additional set of β-catenin deletion mutants lacking either the N-terminal or the C-terminal tail regions. In addition, the Armadillo repeat region was split into two parts to further define the interaction domain of Mitf. Indeed, removal of either N or C tail alone allowed a weak interaction between β-catenin and Mitf (Fig. 1C, compare lanes 5 and 6 to lane 4). Within the Armadillo repeats the interaction with Mitf appears to be largely mediated by amino acids 120 to 553. In this assay, amino acids 536 to 683 of β-catenin did not bind Mitf (Fig. 1C, lane 9).
FIG. 1.
Interactions between β-catenin and Mitf. (A) Schematic representation of the different β-catenin constructs used in the GST pull-down. Armadillo repeats are shown as numbered boxes. The binding domains of known β-catenin interacting partners, members of the Tcf/Lef1 transcription factor family, p300, and the C-terminal part of β-catenin are indicated below the full-length β-catenin protein. NT/CT: N-terminal and C-terminal tails of β-catenin, respectively. (B and C) Mapping the Mitf interaction domain in β-catenin. GST and GST-β-catenin fusion proteins were immobilized on glutathione-Sepharose beads, as indicated, and incubated with [35S]methionine-labeled Mitf. After extensive washing, material retained on the beads was analyzed by SDS-PAGE and fluorography, together with 10% of the input material. (D) Mapping the MITF interaction domain in β-catenin using the yeast two-hybrid assay. The different β-catenin constructs were tested for interactions with Mitf in a yeast two-hybrid assay. The different fragments used are outlined on the left, and their binding capacities are indicated on the right, as monitored by β-galactosidase activity: +++, high binding capacity; ++, good binding capacity; +, weak binding capacity; −, no binding capacity. β-cat, β-catenin; M, marker.
The results of these studies were confirmed by additional interaction analyses using the yeast two-hybrid system based on the original Gal4-Mitf bHLH-Zip bait and various β-catenin portions fused to the VP16 transactivation domain (Fig. 1D). Interactions were seen mainly with β-catenin mutants lacking the N-terminal and C-terminal extensions. As in the GST pull-down assay, the Armadillo repeat domain and its N-terminal part displayed the strongest interactions. In contrast to the GST binding experiments, however, the full-length β-catenin fusion and the C-terminal part of the Armadillo repeats were also able to interact with Mitf in the yeast system. Perhaps the two-hybrid system offers greater sensitivity compared to the GST pull-down to detect weak interactions. An alternative explanation is that bacterially expressed proteins such as the ones used in the GST assay are folded in a way unfavorable for Mitf binding. This is further supported by results of a mammalian two-hybrid assay, where full-length β-catenin is also able to interact with the bHLH-Zip domain of Mitf (data not shown). A third possibility is that modifications in vivo affect protein folding with resulting effects on interactions. Regardless, both series of experiments confirm the interaction of β-catenin and Mitf and localize the site of interaction to the Armadillo repeat domain of β-catenin. As Lef1 and other Tcf family members are not present in either assay, these experiments clearly show that the Mitf-β-catenin interaction does not take place through a scaffolding effect of Lef/Tcf.
Mitf and β-catenin coimmunoprecipitate from cell lysates.
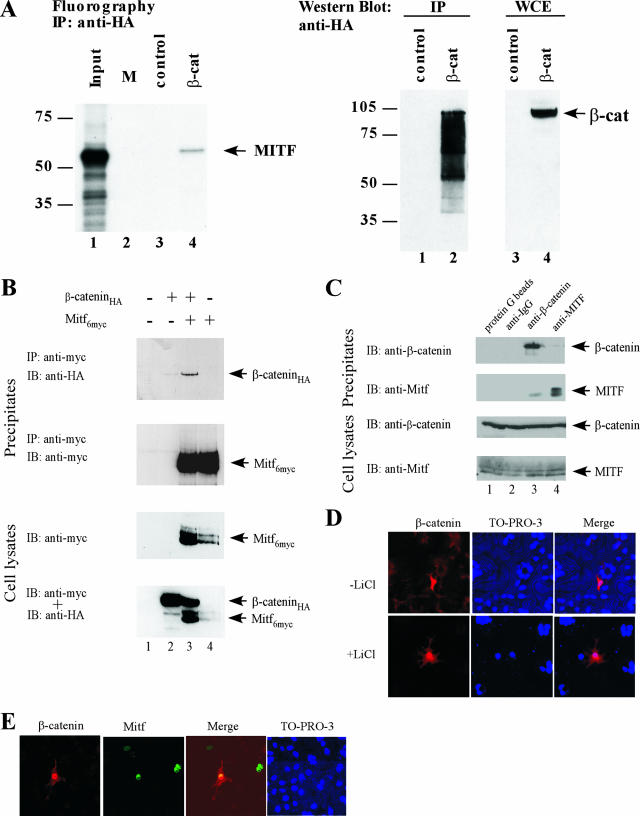
We next used coimmunoprecipitations to examine the physical interactions between Mitf and β-catenin. First, in vitro 35S-labeled recombinant Mitf protein was incubated with cell lysates of HEK293 cells transfected with β-catenin. A portion of the Mitf protein was pulled down upon precipitation of β-catenin, demonstrating that β-catenin and Mitf can interact in a complex protein mixture (Fig. 2A). To further analyze this interaction, HEK293 cells were transfected with expression vectors for HA-tagged β-catenin and myc-tagged Mitf. Prior to the preparation of cell lysates for immunoprecipitations, transfected cells were treated with LiCl to inhibit GSK-3β and thus alleviate its inhibitory influence on both Mitf and β-catenin (52). Immunoprecipitations were performed with an antibody directed against the myc epitope of Mitf. As shown, under these conditions β-catenin and Mitf coimmunoprecipitate with each other (Fig. 2B). To analyze this interaction with endogenously produced proteins, we repeated the assays in 501mel cells, which express both Mitf and β-catenin (50). When mouse monoclonal anti-Mitf antibodies were used, a fraction of β-catenin was precipitated (Fig. 2C). Similarly, when anti-β-catenin antibodies were used, a portion of the Mitf protein was precipitated.
FIG. 2.
Coimmunoprecipitation of Mitf and β-catenin. Coimmunoprecipitation assays were performed to test the interaction between Mitf and β-catenin. (A) Cell lysates from HEK293 cells, transfected with an HA-tagged β-catenin construct, were incubated with in vitro translated and 35S-labeled Mitf, precipitated with an HA-specific antibody, and analyzed for the presence of Mitf using SDS-PAGE and radiography. β-catenin expression and immunoprecipitation were confirmed by Western blot analysis using a β-catenin-specific antibody. (B) HEK293 cells were cotransfected with HA-tagged β-catenin and c-Myc-tagged Mitf constructs. Forty-eight hours after transfection, cells were incubated with 40 mM LiCl for 4 h and harvested. Cell lysates were precipitated with a c-myc-specific antibody and analyzed for the presence of Mitf and β-catenin using SDS-PAGE and Western blot analysis. (C) Cell lysates of 501mel cells were precipitated with either β-catenin- or Mitf-specific antibodies and analyzed for the presence of Mitf and β-catenin using SDS-PAGE and Western blot analysis. (D) Effects of LiCl stimulation on β-catenin localization. COS7 cells were transfected with an expression vector for c-Myc-tagged β-catenin. Twenty-four hours posttransfection, the cells were stimulated with either 40 mM LiCl or NaCl for 3 h, fixed in 2% paraformaldehyde, permeabilized with PBS-Triton X-100, and labeled for β-catenin with a mouse anti-cMyc antibody (Abcam) and a secondary anti-mouse Alexa 568 antibody (Molecular Probes). (E) To analyze the nuclear localization patterns of Mitf and β-catenin, COS7 cells were cotransfected with a c-Myc-tagged β-catenin construct and a GFP-tagged Mitf construct. Twenty-four hours posttransfection, cells were fixed in 2% paraformaldehyde, permeabilized with PBS-Triton X-100, and labeled for β-catenin with an anti-cMyc antibody (Abcam) and a secondary anti-mouse Alexa 568 antibody (Molecular Probes). IP, immunoprecipitation; IB, immunoblotting; β-cat, β-catenin; WCE, whole-cell extract; IgG, immunoglobulin G; M, marker.
To provide additional evidence for the interaction of β-catenin and Mitf in living cells, we studied the mutual influence of both proteins on their intracellular and nuclear localization patterns. First, COS7 cells were transfected with β-catenin or a c-Myc-tagged form of β-catenin, and the cellular distribution was analyzed by indirect immunofluorescence labeling. In unstimulated cells, β-catenin was located in the cytoplasm and in the cell membrane and was only sporadically located in the nucleus (Fig. 2D, upper panel). However, if transfected cells were stimulated with LiCl, which inhibits GSK-3β and thereby mimics Wnt signaling (26), β-catenin was withdrawn from the cell membrane and accumulated mainly in the nucleus (Fig. 2D, lower panel). Immunofluorescence analysis of COS7 cells transfected with an Mitf construct showed that Mitf localizes mainly in the nucleus, independent of LiCl stimulation (data not shown). Interestingly, immunofluorescence analysis of COS7 cells cotransfected with β-catenin and Mitf revealed that β-catenin accumulates in the nucleus where it colocalizes with Mitf, even without prior LiCl treatment (Fig. 2E). Apparently, Mitf can serve as a nuclear anchor for β-catenin, preventing the export of β-catenin and its further degradation. Combined with the binding studies described above, this result strongly suggests that Mitf and β-catenin can bind to each other in vitro and in vivo and identify β-catenin as a novel interaction partner of Mitf.
The helix domain of Mitf interacts with β-catenin.
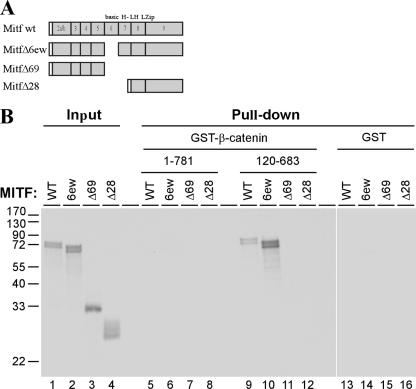
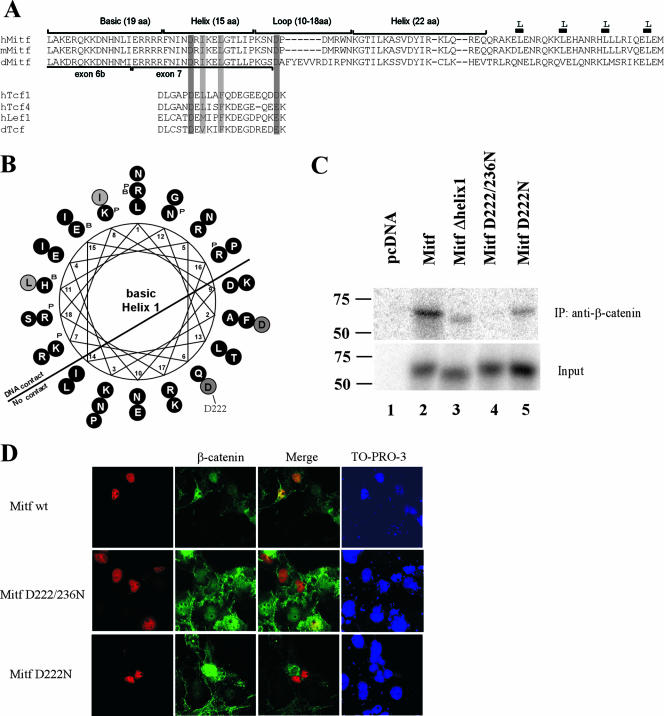
In order to map the region in Mitf which mediates its binding to β-catenin, several Mitf deletion constructs were generated (Fig. 3A) and used in GST pull-down experiments against GST fusions containing full-length β-catenin sequences or the Armadillo repeats only. As before, the Armadillo repeats were able to pull down the full-length Mitf protein (Fig. 3B, lane 9). Deletion of the basic domain of Mitf (Δ6ew) enabled full-length β-catenin to bind Mitf, suggesting the presence of an interfering domain located in the DNA binding domain of Mitf (Fig. 3B, lane 10). A construct containing only the N-terminal part of Mitf, up to the basic domain (Δ69), cannot be pulled down by either full-length β-catenin or the GST construct containing only the Armadillo repeats. Mitf Δ28, which lacks the basic domain and the helix 1 domain but still contains the helix 2 and the Zip domain cannot bind β-catenin either (Fig. 3B, compare lanes 11 and 12 with 9 and 10). In summary, these results indicate that the helix 1 domain is important for proper interactions between Mitf and β-catenin and that the basic domain somehow interferes with this association. To analyze the importance of the helix 1 domain in more detail, we did a sequence comparison of the Mitf protein from different species with known β-catenin interacting partners (Fig. 4A). The best-characterized β-catenin binding partners are among the members of the Tcf/Lef1 transcription factor family. Their binding domains have been analyzed by crystal structure and mutational analysis, and several amino acids important for the interaction have been identified (17, 48). Of special interest are two acidic residues, D16 and E29 (shown in Fig. 4A, numbered with respect to the hTCF4 sequence), which are thought to form salt bridges with the β-catenin protein (9). D16 is followed by several aliphatic residues, where the position of the first two residues is conserved between different members of the Tcf/Lef1 family (Fig. 4A, lower panel). A similar arrangement of amino acids is found in the β-catenin interaction domains of different groups of binding partners of β-catenin, including members of the cadherin and adenomatous polyposis coli tumor suppressor protein family (23). Sequence comparison of this domain with the presumptive interaction domain of the Mitf protein in different species revealed conservation of these important amino acids in the Mitf protein (Fig. 4A, upper panel). Amino acid residue D222 in helix 1 fulfills the criteria as a binding site for β-catenin, followed by several aliphatic amino residues in the correct spacing. Furthermore, another acidic residue, D236, is in close proximity and may build a salt bridge to β-catenin, as has been described for previously characterized binding partners of β-catenin (9, 23). When the Mitf sequence in question is projected as a helical wheel (Fig. 4B), the amino acid residues important for β-catenin contact are exposed on the same surface of the protein, further supporting the suggestion that they indeed participate in binding to β-catenin. In order to test the role of this domain in interactions with β-catenin, we created several mutations in this region including a complete deletion of helix 1 (Mitf Δhelix1) and single amino acid substitutions where D222 and D236 were changed to asparagine (Mitf D222/236N and Mitf D222N). In mice, the Mitf D222N mutation is known as Mitfmi-vit and results in gradual loss of pigmentation with age (reviewed in reference 40). We tested these mutant constructs in our in vitro coimmunoprecipitation system. Cell lysates of HEK293 cells transfected with β-catenin were incubated with the different radiolabeled Mitf constructs and precipitated with a β-catenin-specific antibody. Whereas the full-length Mitf protein precipitates efficiently with β-catenin (Fig. 4C), the Mitf Δhelix1 mutant shows significantly reduced binding to β-catenin. Interestingly, the Mitf D222/236N double mutant is unable to bind to β-catenin, whereas the Mitfmi-vit mutant binds β-catenin at similar levels as the Mitf Δhelix1 construct. Quantification of the precipitated proteins using the Typhoon image analyzer confirms reduced binding of Mitf Δhelix1 and Mitfmi-vit and no binding of Mitf D222/236N (data not shown). To further explore the importance of helix 1 and D222/236 for the interaction with β-catenin, we analyzed the effect of the different mutations on the nuclear localization pattern of β-catenin in COS7 cells (Fig. 4D). For these studies only the constructs Mitf D222/236N and Mitfmi-vit were used, because Mitf Δhelix1 failed to be expressed at sufficient levels and therefore could not be analyzed. In contrast, Mitf D222/236N and Mitfmi-vit were expressed, and both efficiently localized to the nucleus. Importantly, however, these Mitf mutants showed a much reduced capability to anchor β-catenin in the nucleus (Fig. 4D and data not shown). Taken together, these results confirm helix 1 as the main interaction domain of Mitf and emphasize the importance of amino acids D222/236 for mediating the binding to β-catenin.
FIG. 3.
Mapping the β-catenin interaction domain in Mitf. (A) A schematic representation of the different Mitf deletion constructs used. Numbers indicate exons in the Mitf gene. basic HLH-LZip, region of the basic HLH leucine zipper domains. (B) Results of the GST pull-down assays. GST and GST-β-catenin fusion proteins were immobilized on glutathione-Sepharose beads as indicated and incubated with [35S]methionine-labeled Mitf and Mitf deletion mutants. After extensive washing, material retained on the beads was analyzed by SDS-PAGE and fluorography, together with 10% of the input material. WT, wild type.
FIG. 4.
Helix 1 of the Mitf bHLH domain provides critical contact points for the interaction with β-catenin. (A) Sequence comparison of the β-catenin binding domain of different Mitf homologues with known β-catenin binding domains of members of the Tcf/Lef1 transcription factor family. Protein alignments were carried out using ClustalW at the San Diego Supercomputer Center workbench (http://workbench.sdsc.edu/) and then further adjusted by eye. (B) To better visualize their potential functional importance, the positions of the conserved amino acids, helix 1, and the loop were also depicted in a helical wheel structure, using the sequence analysis program BIOEDIT. (C) To confirm the sequence analysis results, coimmunoprecipitation assays were performed. Cell lysates from HEK293 cells, transfected with an HA-tagged β-catenin construct, were incubated with the different in vitro translated and 35S-labeled Mitf deletion constructs, precipitated with an HA-specific antibody, and analyzed for the presence of the Mitf deletion construct using SDS-PAGE and radiography. β-Catenin expression and immunoprecipitation were confirmed by Western blot analysis using a β-catenin-specific antibody. (D) To analyze the importance of the D222 and D236 amino acids for the interaction with β-catenin, colocalization studies with the different Mitf mutant constructs were performed. COS7 cells were treated as before, and β-catenin was labeled with a goat anti-β-catenin antibody (Santa Cruz) and a secondary anti-goat Alexa 488 antibody (Molecular Probes). The nuclear localization of the different Mitf constructs was analyzed with a mouse anti-Mitf antibody and a secondary anti-mouse Alexa 568 antibody. IP, immunoprecipitation; wt, wild type.
Functional effects of Mitf and β-catenin interactions.
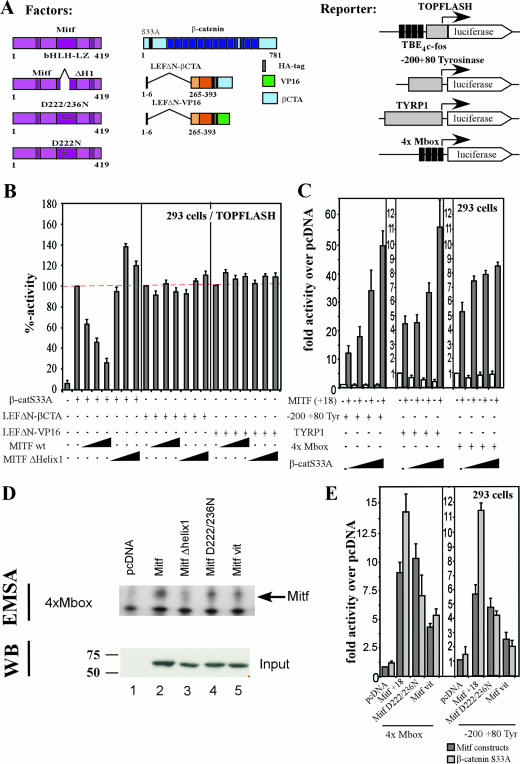
Members of the Lef1/Tcf family, in addition to other transcription factors such as Sox17, utilize β-catenin as a transcriptional coactivator which can lead to a cross-inhibition of different signaling pathways based on competitive interactions (12, 39, 41). Based on our findings that β-catenin and Mitf can interact, we asked whether a similar scenario applies to Tcf-mediated and Mitf-dependent transcriptional control. We first analyzed whether concomitant expression of Mitf affected the activation of the TOPFLASH reporter, a well-characterized reporter system for β-catenin and Tcf-induced transcription (Fig. 5A) (27). Expression of an activated form of β-catenin strongly activated the TOPFLASH reporter in 293 cells (Fig. 5B). Increasing amounts of Mitf reduced this activation to about 30% of its maximum. To determine whether this inhibition is due to an Mitf-mediated distraction of β-catenin from the TOPFLASH promoter, we activated the reporter with chimeric fusion proteins which consisted of the DNA-binding domain of Lef1 and two different transactivation domains derived from either the C terminus of β-catenin (Fig. 5A, LEFΔN-βCTA) or the VP16 protein (Fig. 5A, LEFΔN-VP16) (46). Because the LEFΔN portion lacks the β-catenin binding domain, both chimeras act independently of β-catenin. Moreover, the LEFΔN-βCTA fusion does not contain β-catenin sequences which interact with Mitf. We therefore expected that TOPFLASH activation by LEFΔN-βCTA and LEFΔN-VP16 would not be affected by Mitf coexpression, which turned out to be the case (Fig. 5B). So far, the results of our competition assays are consistent with the idea that Mitf affects β-catenin-dependent transactivation by distracting β-catenin from the TOPFLASH reporter through a direct interaction. If so, MITF mutants which are no longer capable of interacting with β-catenin should behave neutral in the competition test. The predicted behavior was indeed observed when we analyzed MITF Δhelix1 (Fig. 5B), further suggesting that MITF has the capacity to remove β-catenin from cognate Wnt regulated promoters.
FIG. 5.
Functional cooperation of Mitf and β-catenin. To test the functional relevance of the interactions, Mitf and β-catenin were tested in cotransfection assays in concert with β-catenin- or Mitf-specific reporter constructs. All the Mitf constructs contain the alternative 18-bp sequence just upstream of the basic domain (41a). (A) Schematic representation of the different constructs used. TBE4c-fos, four copies of the TCF binding element adjusted from the c-fos minimal promoter (gray box). (B) HEK293 cells were cotransfected with the TOPFLASH promoter construct and different activators, as indicated. The luciferase activity was measured, and the activity of the different activators was set as 100%. The influence of Mitf wild type (wt) and the Mitf Δhelix1 mutant construct on the activity of the different activators on the TOPFLASH promoter is shown as percent decrease. (C) HEK293 cells were cotransfected with either the −200/+80 tyrosinase promoter construct, the Tyrp-1 promoter construct, or a 4×M-box construct in concert with the different activators indicated. Mitf activity was measured as relative activity (n-fold) over pcDNA. (D) To determine whether the different Mitf mutant proteins are able to homodimerize and bind DNA, DNA band shift assays with in vitro translated proteins were performed. Binding specificity was confirmed by supershift with an Mitf-specific antibody. Input of equal amounts of the different proteins was confirmed by Western blotting. (E) To confirm the importance of the helix 1 region for the interaction with β-catenin, cotransfection studies with the different Mitf mutant constructs were performed. HEK293 cells were cotransfected with either the 4×M-box promoter construct or the −200/+80 tyrosinase construct and the different Mitf mutant constructs, with or without β-catenin.
Based on our findings that Mitf and β-catenin can interact, we next asked whether Mitf was able to recruit β-catenin to Mitf-regulated promoters and employ β-catenin as transcriptional coactivator. Therefore, we measured activation of a luciferase reporter gene driven by the Mitf regulated −200/+80 tyrosinase promoter after coexpression of Mitf and β-catenin. As shown in Fig. 5C, while the −200/+80 tyrosinase promoter activity was stimulated by about 12-fold in the presence of Mitf only, coexpression of β-catenin enhanced reporter gene expression in a dose-dependent manner up to 45-fold. This effect of β-catenin was strictly dependent on the presence of Mitf. We also analyzed the activation patterns of Mitf and β-catenin on the Tyrp-1 promoter, another Mitf target promoter. Again, coexpression of Mitf and β-catenin resulted in an increased activation of the Tyrp-1 luciferase construct. This increase was less than what was observed with the short tyrosinase construct yet dose dependent in a similar manner. To exclude the possibility that the effects of β-catenin are due to interactions with other DNA binding factors, such as Lef1, we examined the effects of β-catenin on the ability of Mitf to activate an artificial promoter, where expression of luciferase is driven by four serially arranged M-box sequences. Mitf is able to bind and activate expression of this construct and, again, cotransfection of β-catenin results in a dose-dependent increase of the transcriptional activity of Mitf (Fig. 5C). Furthermore, cotransfection studies with a β-catenin construct unable to interact with Lef1 (β-catenin S33AΔ19) on the −200/+80 tyrosinase promoter and the artificial M-box promoter showed similar results (data not shown). This suggests that the interactions between Mitf and β-catenin are direct and do not need cofactors.
If Mitf indeed recruits β-catenin to enhance transcription from Mitf target promoters, mutations which interfere with the interaction of Mitf and β-catenin should abrogate the stimulatory effect of β-catenin coexpression. As dimerization is absolutely required to be able to transactivate, we first tested the ability of the different Mitf mutant proteins to homodimerize and bind DNA in DNA band shift assays with an E-box containing sequence as probe. Whereas Mitf wild-type, Mitf D222/236N, and the Mitfmi-vit mutant proteins bound the probe efficiently (Fig. 5D), the Mitf Δhelix1 mutant failed to do so. This is not surprising, as the lack of helix 1 results in truncation of the dimerization motif. We next analyzed the ability of β-catenin to cooperate with Mitf D222/236N and Mitfmi-vit in cotransfection assays. Both Mitf mutant constructs were able to activate luciferase expression from either the 4×M-box promoter (Fig. 5E, left panel) or the −200/+80 tyrosinase promoter construct (Fig. 5E, right panel). However, cotransfection of β-catenin failed to enhance reporter gene activity in the presence of the Mitf D222/236N and the Mitfmi-vit mutants. Taken together, the results of our binding assays and the functional analyses are fully consistent with the notion that MITF competitively redirects β-catenin transcriptional activity away from Wnt regulated genes to MITF target promoters.
ChIP analysis.
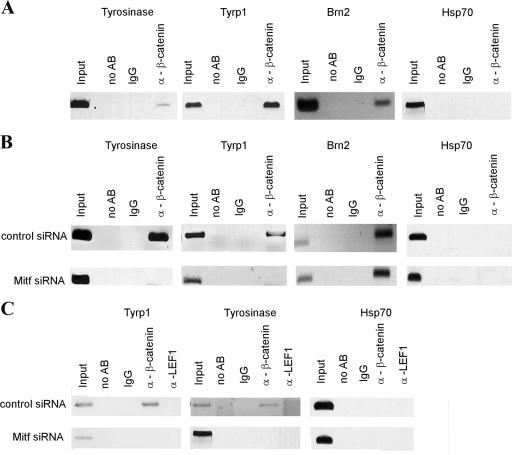
To further analyze the possibility that β-catenin activates Mitf target genes, we performed chromatin immunoprecipitation (ChIP) (8) analysis in 501mel cells with a β-catenin-specific antibody. Precipitates were analyzed for different Mitf downstream targets by PCR amplifying previously characterized Mitf binding sites including the E-box sequence CATGTG in the promoter region. As shown in Fig. 6A, β-catenin is able to precipitate the E-box sequences from the tyrosinase and Tyrp-1 promoters. To demonstrate that this promoter binding is Mitf dependent, we depleted Mitf from 501mel cells using siRNA and repeated the ChIP assay. Figure 6B shows that β-catenin clearly uses Mitf as a DNA binding partner; depleting Mitf from the cells results in the absence of Mitf target promoters when β-catenin is used in immunoprecipitations. The effect is specific for Mitf as binding of β-catenin to the known β-catenin target promoter Brn2 (15) through a Tcf binding site is not impaired by Mitf siRNA depletion. To further confirm that the observed effect is Mitf dependent and not mediated through interaction with Lef1, a control immunoprecipitation with a Lef1-specific antibody was performed. Figure 6C shows that Lef1 is not able to precipitate either the tyrosinase or the Tyrp-1 promoters. Amplifications with a primer pair specific for the Hsp70 promoter served as a negative control in all assays. These observations, combined with the results of our interference assays, lead us to conclude that Mitf can redirect β-catenin toward Mitf-specific target promoters, where it utilizes β-catenin as a coactivator and thereby competes with Lef1 for β-catenin.
FIG. 6.
Mitf recruits β-catenin to specific target genes. (A) Association of β-catenin with Mitf-regulated genes was analyzed by ChIP assays. 501mel melanoma cells were fixed in paraformaldehyde and harvested, and DNA was sheared by sonication. Cell lysates were precipitated with either a nonspecific antibody or a polyclonal β-catenin antibody. After precipitation, cross-links were reverted, and precipitated DNA was purified and analyzed via PCR for known Mitf binding sequences or for control sequences. (B) To confirm that the presence of β-catenin at the tyrosinase and Tyrp-1 promoters depends on Mitf-β-catenin interactions, Mitf was depleted using specific siRNAs prior to ChIP. (C) To confirm that the observed results were not dependent on a scaffold effect of Lef1, control ChIP assays with an anti-Lef1 antibody were performed on both control-depleted and Mitf-siRNA-depleted 501 cells. Cells were treated as above. IgG, immunoglobulin G; AB, antibody.
DISCUSSION
Mitf is a key regulator of melanocyte development and is also important for melanocyte stem cell maintenance (32). Recently, it has been shown that Mitf is involved in cell cycle regulation (6, 30, 11) as well as in regulating the expression of Tbx2, an important senescence repressor in melanoma (7, 45). How Mitf controls these different events is, however, poorly understood. It is likely that Mitf regulates different sets of target genes at different stages of development. Clearly, because of its pivotal role in melanocytes, both the levels and activity of Mitf itself need to be tightly controlled. We envision that this occurs through the activity of different signaling pathways and through dynamic interactions with other factors in a highly regulated and promoter-specific manner.
Several studies have shown that Mitf is a downstream target of Wnt signaling in melanocyte development. The Mitf promoter itself contains Tcf/Lef1 binding sites, and it has been shown that ectopic Wnt3a upregulates Mitf expression and recruits β-catenin and Lef1 to the Lef1 binding site of the Mitf promoter (43). Lef1 not only activates Mitf expression but can also interact with Mitf itself, in a positive feedback loop, to maintain Mitf expression levels at important stages of melanocyte development (35). Here, we show that β-catenin is not only involved in the transcriptional regulation of Mitf expression but can also interact directly with Mitf itself. As a functional consequence, overexpression of Mitf reduces β-catenin/Tcf-dependent gene expression. Conversely, overexpression of β-catenin enhances Mitf-dependent gene expression. These findings indicate that Mitf can redirect β-catenin transcriptional activity away from β-catenin/Lef1-regulated genes toward Mitf-specific target promoters. These results are further supported by ChIP analysis of different Mitf downstream promoters, showing that β-catenin is associated with E-box sequences which are the DNA-binding elements for Mitf.
Although other investigators (35, 51) have shown that Lef1, a well-characterized binding partner of β-catenin, can bind to Mitf through the bHLH-Zip domain, the interactions between Mitf and β-catenin described here are not Lef1 dependent. First, the interaction takes place in yeast and in the GST system where Lef1 is not present, and, second, β-catenin affects Mitf-mediated transcription from the artificial 4×M-box promoter which does not contain Lef1 binding sites. In addition, a β-catenin construct unable to bind to Lef1 is still able to interact with Mitf and increase its transcriptional activity. The location of the Mitf interaction domain in the N-terminal armadillo repeats of β-catenin additionally argues against an indirect interaction between β-catenin and Mitf through Lef1. Rather, these interactions suggest that Lef1 and Mitf may compete for binding to β-catenin (Fig. 4).
Indeed, Wnt signaling activates Mitf expression early in melanocyte development, which is a key event in melanocyte differentiation and maintenance. Mitf then activates targets further downstream according to the specific needs of the cell. These needs are in part specified through convergence of different signaling pathways on Mitf which fine-tune Mitf activity and protein level and thereby determine the fate of the cell. As one example, Lang et al. (28) have shown that PAX3 activates Mitf expression early in melanoblasts but at the same time prevents Mitf from activating downstream targets, thus preventing terminal differentiation. Mitf accumulates in the cell, and once the PAX3-mediated repression is relieved through external stimuli, Mitf drives the cell into further differentiation.
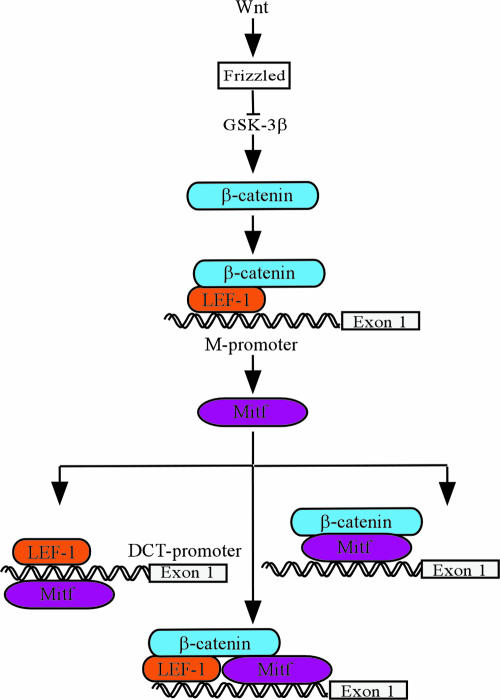
A considerable fraction of malignant melanomas possess a constitutively active Wnt/β-catenin signaling pathway, and β-catenin levels are elevated in a majority of melanomas (33, 34). Recent work has shown that Mitf is expressed in a majority of melanomas (38) and that the Mitf gene is amplified in a significant portion of melanomas (14). Thus, both Mitf and β-catenin play important roles in melanoma. Based on our new findings we present an extended model for how Wnt signaling, β-catenin, and Mitf cooperate during melanocyte differentiation and maintenance and possibly also in melanoma (Fig. 7). At early stages of melanocyte differentiation, Wnt signaling, e.g., through Wnt3a (43), results in accumulation of β-catenin, which then translocates into the nucleus. β-Catenin converts Lef1 from a repressor to an activator and activates Mitf expression from the Mitf-M promoter. Subsequently, Mitf upregulates its own expression through interaction with Lef1. Depending on the availability of Lef1 and active β-catenin, Mitf has at least three choices to activate transcription of distinct sets of downstream target genes and to diversify cellular behavior. First, Mitf can interact with Lef1 in the absence of β-catenin to activate melanocyte-specific genes, such as Mitf and DCT (37, 52). Second, Mitf can activate another set of downstream targets in concert with Lef1 and β-catenin. Or, third, Mitf can recruit β-catenin independently of Lef1 to Mitf-regulated genes important for melanocyte development and maintenance. Thus, Mitf may be able to integrate the output from the Wnt signal transduction pathway in multiple ways to determine the repertoire of genes regulated by Wnt and Mitf under different circumstances. Interestingly, the finding that the Mitfmi-vit mutant protein is able to bind to β-catenin but is unable to utilize this cofactor for transcription activation suggests that the interaction between the two proteins may be particularly important in melanocyte stem cells. Nishimura et al. (32) have recently shown that melanocyte stem cells are not properly maintained in Mitfmi-vit mutant mice, and β-catenin has also been shown to be important in melanocyte stem cells (28). Clearly, the role of these two proteins in melanocyte stem cell maintenance warrants further investigations. Olson et al. (32a) have shown that β-catenin is recruited by a tissue-specific homeodomain factor to promote cell lineage determination. In contrast to the common Wnt signaling pathway, they showed that in pituitary development β-catenin directly interacts with the specific homeodomain factor Prop1 to activate expression of the cell-lineage-determining transcription factor Pit1 and simultaneously directs repression of the lineage-inhibiting transcription factor Hesx1 through the TLE/Reptin/HDAC1 corepressor complex. In melanocytes, a similar situation is possible, where Mitf repression through PAX3 is withdrawn by β-catenin, which is then followed by the interaction of Mitf and β-catenin to activate genes important for melanocyte determination and development.
FIG. 7.
Model for the dynamic interactions between β-catenin and Mitf transcriptional activity. Mitf plays a key role at different stages of melanoblast and melanocyte development, probably by regulating different sets of target genes. To achieve this diversity, the activity of Mitf as a transcriptional activator is modulated either by regulating its protein level or by its engagement in changing and dynamic interactions with other factors in a highly regulated and promoter-specific manner. Moreover, signaling through the canonical Wnt signaling pathway provides several levels of regulation. Signaling through β-catenin activates Mitf expression. Depending on the protein levels of Mitf, Mitf can interact and cooperate either with Lef1 or β-catenin alone or in a complex with both to activate downstream targets.
Our results provide an important step to improve our understanding of these transcriptional regulatory networks and how they control cell identity. An important and interesting future task will be to define the parameters which govern the formation of the various possible complexes between Mitf, β-catenin, and Lef1 and to determine the spectrum of target genes regulated by each of the different combinations of these important transcription regulators.
Acknowledgments
We thank S. Shibahara for the −200/+80 tyrosinase and Tyrp-1 promoter constructs and H. Arnheiter for the 4×M-box promoter construct. We are particularly grateful to S. Carreira for assistance during the ChIP assays and D. Fisher for the C5 antibody.
This work was supported by grants from the Icelandic Research Council, the University of Iceland Research Fund, and the Bergthora Magnusdottir and Jakob J. Bjarnason Memorial Fund.
Footnotes
Published ahead of print on 25 September 2006.
REFERENCES
- 1.Aberle, H., S. Butz, J. Stappert, H. Weissig, R. Kemler, and H. Hoschuetzky. 1994. Assembly of the cadherin-catenin complex in vitro with recombinant proteins. J. Cell Sci. 107:3655-3663. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, A., O. Huber, and R. Kemler. 1998. Pontin52, an interaction partner of beta-catenin, binds to the TATA box binding protein. Proc. Natl. Acad. Sci. USA 95:14787-14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behrens, J., B. A. Jerchow, M. Wurtele, J. Grimm, C. Asbrand, R. Wirtz, M. Kuhl, D. Wedlich, and W. Birchmeier. 1998. Functional interaction of an axin homolog, conductin, with beta-catenin, APC, and GSK3β. Science 280:596-599. [DOI] [PubMed] [Google Scholar]
- 4.Bek, S., and R. Kemler. 2002. Protein kinase CKII regulates the interaction of beta-catenin with alpha-catenin and its protein stability. J. Cell Sci. 115:4743-4753. [DOI] [PubMed] [Google Scholar]
- 5.Bienz, M. 2005. β-Catenin: a pivot between cell adhesion and Wnt signalling. Curr. Biol. 15:R64-R67. [DOI] [PubMed] [Google Scholar]
- 6.Carreira, S., J. Goodall, I. Aksan, S. A. La Rocca, M. D. Galibert, L. Denat, L. Larue, and C. R. Goding. 2005. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature 433:764-769. [DOI] [PubMed] [Google Scholar]
- 7.Carreira, S., B. Liu, and C. R. Goding. 2000. The gene encoding the T-box factor Tbx2 is a target for the microphthalmia-associated transcription factor in melanocytes. J. Biol. Chem. 275:21920-21927. [DOI] [PubMed] [Google Scholar]
- 8.Chipysheva, T. A., V. I. Gel'shtein, V. D. Ermilova, V. Vishnevskaia Ia, and M. Vasil'ev Iu. 2003. Expression of cell adhesion molecules E-cadherin and beta-catenin in infiltrating breast carcinoma. Arkh. Patol. 65:3-7. (In Russian.) [PubMed] [Google Scholar]
- 9.Daniels, D. L., and W. I. Weis. 2002. ICAT inhibits beta-catenin binding to Tcf/Lef-family transcription factors and the general coactivator p300 using independent structural modules. Mol. Cell 10:573-584. [DOI] [PubMed] [Google Scholar]
- 10.Dorsky, R. I., D. W. Raible, and R. T. Moon. 2000. Direct regulation of nacre, a zebrafish MITF homolog required for pigment cell formation, by the Wnt pathway. Genes Dev. 14:158-162. [PMC free article] [PubMed] [Google Scholar]
- 11.Du, J., H. R. Widlund, M. A. Horstmann, S. Ramaswamy, K. Ross, W. E. Huber, E. K. Nishimura, T. R. Golub, and D. E. Fisher. 2004. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell 6:565-576. [DOI] [PubMed] [Google Scholar]
- 12.Easwaran, V., M. Pishvaian, Salimuddin, and S. Byers. 1999. Cross-regulation of beta-catenin-LEF/TCF and retinoid signaling pathways. Curr. Biol. 9:1415-1418. [DOI] [PubMed] [Google Scholar]
- 13.Galibert, M. D., S. Carreira, and C. R. Goding. 2001. The Usf-1 transcription factor is a novel target for the stress-responsive p38 kinase and mediates UV-induced tyrosinase expression. EMBO J. 20:5022-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garraway, L. A., H. R. Widlund, M. A. Rubin, G. Getz, A. J. Berger, S. Ramaswamy, R. Beroukhim, D. A. Milner, S. R. Granter, J. Du, C. Lee, S. N. Wagner, C. Li, T. R. Golub, D. L. Rimm, M. L. Meyerson, D. E. Fisher, and W. R. Sellers. 2005. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 436:117-122. [DOI] [PubMed] [Google Scholar]
- 15.Goodall, J., S. Martinozzi, T. J. Dexter, D. Champeval, S. Carreira, L. Larue, and C. R. Goding. 2004. Brn-2 expression controls melanoma proliferation and is directly regulated by β-catenin. Mol. Cell. Biol. 24:2915-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottardi, C. J., and B. M. Gumbiner. 2004. Distinct molecular forms of beta-catenin are targeted to adhesive or transcriptional complexes. J. Cell Biol. 167:339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham, T. A., C. Weaver, F. Mao, D. Kimelman, and W. Xu. 2000. Crystal structure of a beta-catenin/Tcf complex. Cell 103:885-896. [DOI] [PubMed] [Google Scholar]
- 18.Hecht, A., C. M. Litterst, O. Huber, and R. Kemler. 1999. Functional characterization of multiple transactivating elements in beta-catenin, some of which interact with the TATA-binding protein in vitro. J. Biol. Chem. 274:18017-18025. [DOI] [PubMed] [Google Scholar]
- 19.Hecht, A., and M. P. Stemmler. 2003. Identification of a promoter-specific transcriptional activation domain at the C terminus of the Wnt effector protein T-cell factor 4. J. Biol. Chem. 278:3776-3785. [DOI] [PubMed] [Google Scholar]
- 20.Hecht, A., K. Vleminckx, M. P. Stemmler, F. van Roy, and R. Kemler. 2000. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 19:1839-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemesath, T. J., E. Steingrímsson, G. McGill, M. J. Hansen, J. Vaught, C. A. Hodgkinson, H. Arnheiter, N. G. Copeland, N. A. Jenkins, and D. E. Fisher. 1994. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 8:2770-2780. [DOI] [PubMed] [Google Scholar]
- 22.Hollenberg, S. M., R. Sternglanz, P. F. Cheng, and H. Weintraub. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 15:3813-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber, A. H., and W. I. Weis. 2001. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell 105:391-402. [DOI] [PubMed] [Google Scholar]
- 24.Huber, O., R. Korn, J. McLaughlin, M. Ohsugi, B. G. Herrmann, and R. Kemler. 1996. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech. Dev. 59:3-10. [DOI] [PubMed] [Google Scholar]
- 25.Jouneau, A., Y. Q. Yu, M. Pasdar, and L. Larue. 2000. Plasticity of cadherin-catenin expression in the melanocyte lineage. Pigment Cell Res. 13:260-272. [DOI] [PubMed] [Google Scholar]
- 26.Klein, P. S., and D. A. Melton. 1996. A molecular mechanism for the effect of lithium on development. Proc. Natl. Acad. Sci. USA 93:8455-8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korinek, V., N. Barker, P. J. Morin, D. van Wichen, R. de Weger, K. W. Kinzler, B. Vogelstein, and H. Clevers. 1997. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science 275:1784-1787. [DOI] [PubMed] [Google Scholar]
- 28.Lang, D., M. M. Lu, L. Huang, K. A. Engleka, M. Zhang, E. Y. Chu, S. Lipner, A. Skoultchi, S. E. Millar, and J. A. Epstein. 2005. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature 433:884-887. [DOI] [PubMed] [Google Scholar]
- 29.Levy, C., H. Nechushtan, and E. Razin. 2002. A new role for the STAT3 inhibitor, PIAS3: a repressor of microphthalmia transcription factor. J. Biol. Chem. 277:1962-1966. [DOI] [PubMed] [Google Scholar]
- 30.Loercher, A. E., E. M. Tank, R. B. Delston, and J. W. Harbour. 2005. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J. Cell Biol. 168:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson, W. J., and R. Nusse. 2004. Convergence of Wnt, beta-catenin, and cadherin pathways. Science 303:1483-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishimura, E. K., S. R. Granter, and D. E. Fisher. 2005. Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science 307:720-724. [DOI] [PubMed] [Google Scholar]
- 32a.Olson, L. E., J. Tolkuhn, C. Scafoglio, A. Krones, J. Zhang, K. A. Ohgi, W. Wu, M. M. Taketo, R. Kemler, R.Grosschedl, et al. 2006. Homeodomain-mediated beta-catenin-dependent switching events dictate cell-lineage determination. Cell 125:593-605. [DOI] [PubMed] [Google Scholar]
- 33.Provost, E., and D. L. Rimm. 1999. Controversies at the cytoplasmic face of the cadherin-based adhesion complex. Curr. Opin. Cell Biol. 11:567-572. [DOI] [PubMed] [Google Scholar]
- 34.Rubinfeld, B., P. Robbins, M. El-Gamil, I. Albert, E. Porfiri, and P. Polakis. 1997. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science 275:1790-1792. [DOI] [PubMed] [Google Scholar]
- 35.Saito, H., K. Yasumoto, K. Takeda, K. Takahashi, A. Fukuzaki, S. Orikasa, and S. Shibahara. 2002. Melanocyte-specific microphthalmia-associated transcription factor isoform activates its own gene promoter through physical interaction with lymphoid-enhancing factor 1. J. Biol. Chem. 277:28787-28794. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Schwahn, D. J., N. A. Timchenko, S. Shibahara, and E. E. Medrano. 2005. Dynamic regulation of the human dopachrome tautomerase promoter by MITF, ER-alpha and chromatin remodelers during proliferation and senescence of human melanocytes. Pigment Cell Res. 18:203-213. [DOI] [PubMed] [Google Scholar]
- 38.Selzer, E., V. Wacheck, T. Lucas, E. Heere-Ress, M. Wu, K. N. Weilbaecher, W. Schlegel, P. Valent, F. Wrba, H. Pehamberger, D. Fisher, and B. Jansen. 2002. The melanocyte-specific isoform of the microphthalmia transcription factor affects the phenotype of human melanoma. Cancer Res. 62:2098-2103. [PubMed] [Google Scholar]
- 39.Sinner, D., S. Rankin, M. Lee, and A. M. Zorn. 2004. Sox17 and beta-catenin cooperate to regulate the transcription of endodermal genes. Development 131:3069-3080. [DOI] [PubMed] [Google Scholar]
- 40.Solanas, G., S. Miravet, D. Casagolda, J. Castano, I. Raurell, A. Corrionero, A. G. de Herreros, and M. Dunach. 2004. β-Catenin and plakoglobin N- and C-tails determine ligand specificity. J. Biol. Chem. 279:49849-49856. [DOI] [PubMed] [Google Scholar]
- 41.Song, L. N., R. Herrell, S. Byers, S. Shah, E. M. Wilson, and E. P. Gelmann. 2003. β-Catenin binds to the activation function 2 region of the androgen receptor and modulates the effects of the N-terminal domain and TIF2 on ligand-dependent transcription. Mol. Cell. Biol. 23:1674-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Steingrimsson, E., K. J. Moore, M. L. Lamoreux, A. R. Ferre-D'Amare, S. K. Burley, D. C. Zimring, L. C. Skow, C. A. Hodgkinson, H. Arnheiter, N. G. Copeland, and N. A. Jenkins. 1994. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat. Genet. 8:256-263. [DOI] [PubMed] [Google Scholar]
- 42.Steingrimsson, E., N. G. Copeland, and N. A. Jenkins. 2004. Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 38:365-411. [DOI] [PubMed] [Google Scholar]
- 43.Takeda, K., K. Yasumoto, R. Takada, S. Takada, K. Watanabe, T. Udono, H. Saito, K. Takahashi, and S. Shibahara. 2000. Induction of melanocyte-specific microphthalmia-associated transcription factor by Wnt-3a. J. Biol. Chem. 275:14013-14016. [DOI] [PubMed] [Google Scholar]
- 44.Turner, G. 1994. Vectors for genetic manipulation. Prog. Ind. Microbiol. 29:641-665. [PubMed] [Google Scholar]
- 45.Vance, K. W., S. Carreira, G. Brosch, and C. R. Goding. 2005. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Res. 65:2260-2268. [DOI] [PubMed] [Google Scholar]
- 46.Vleminckx, K., R. Kemler, and A. Hecht. 1999. The C-terminal transactivation domain of beta-catenin is necessary and sufficient for signaling by the LEF-1/beta-catenin complex in Xenopus laevis. Mech. Dev. 81:65-74. [DOI] [PubMed] [Google Scholar]
- 47.Vojtek, A. B., S. M. Hollenberg, and J. A. Cooper. 1993. Mammalian Ras interacts directly with the serine/threonin kinas raf. Cell 74:205-214. [DOI] [PubMed] [Google Scholar]
- 48.von Kries, J. P., G. Winbeck, C. Asbrand, T. Schwarz-Romond, N. Sochnikova, A. Dell'Oro, J. Behrens, and W. Birchmeier. 2000. Hot spots in beta-catenin for interactions with LEF-1, conductin and APC. Nat. Struct. Biol 7:800-807. [DOI] [PubMed] [Google Scholar]
- 49.Widlund, H. R., and D. E. Fisher. 2003. Microphthalamia-associated transcription factor: a critical regulator of pigment cell development and survival. Oncogene 22:3035-3041. [DOI] [PubMed] [Google Scholar]
- 50.Widlund, H. R., M. A. Horstmann, E. R. Price, J. Cui, S. L. Lessnick, M. Wu, X. He, and D. E. Fisher. 2002. β-Catenin-induced melanoma growth requires the downstream target microphthalmia-associated transcription factor. J. Cell Biol. 158:1079-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, M., T. J. Hemesath, C. M. Takemoto, M. A. Horstmann, A. G. Wells, E. R. Price, D. Z. Fisher, and D. E. Fisher. 2000. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 14:301-312. [PMC free article] [PubMed] [Google Scholar]
- 52.Yasumoto, K., K. Takeda, H. Saito, K. Watanabe, K. Takahashi, and S. Shibahara. 2002. Microphthalmia-associated transcription factor interacts with LEF-1, a mediator of Wnt signaling. EMBO J. 21:2703-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]