Abstract
By using a genetic screen, we have isolated a mammalian cell line that is resistant to infection by retroviruses that are derived from the murine leukemia virus, human immunodeficiency virus type 1, and feline immunodeficiency virus. We demonstrate that the cell line is genetically recessive for the resistance, and hence it is lacking a factor enabling infection by retroviruses. The block to infection is early in the life cycle, at the poorly understood uncoating stage. We implicate the proteasome at uncoating by completely rescuing the resistant phenotype with the proteasomal inhibitor MG-132. We further report on the complementation cloning of a gene (MRI, modulator of retrovirus infection) that can also act to reverse the inhibition of infection in the mutant cell line. These data implicate a role for the proteasome during uncoating, and they suggest that MRI is a regulator of this activity. Finally, we reconcile our findings and other published data to suggest a model for the involvement of the proteasome in the early phase of the retroviral life cycle.
Keywords: host cell factors, proteasome, somatic cell genetics, suppressor
Retroviral infections cause pathology ranging from cancer to AIDS. The extent of infection is clearly mitigated by polymorphisms in the genomes of the host and virus. A classic example of this observation is the Fv1 gene in mice. The Fv1n allele allows for the replication of N-tropic murine leukemia virus (MLV), and the Fv1b allele permits replication of B-tropic MLV. Cloning and characterization of the Fv1 gene and the genomes of N- and B-tropic MLV reveal that polymorphisms within the Fv1 gene (1, 2) and a single amino acid in the viral capsid protein of MLV (3) are responsible for the observed permissive and restrictive phenotypes. Retroviral replication can also be restricted by the action of the host APOBEC3G/F proteins (for review, see ref. 4). Species differences for infection by human immunodeficiency virus type 1 (HIV-1) (5) have been exploited to clone a dominant restriction factor TRIM5α (6). Species differences in the TRIM5α sequence determine that it acts as a restriction factor for HIV-1 in simian cells, and it restricts N-tropic MLV in human cells (4). Another example is cyclin T1, which partners with HIV Tat (7), but a single amino acid difference in the murine protein renders it incompetent for Tat-mediated transactivation (8).
We set out to identify further host-cell proteins that may be involved in the early life cycle of retroviruses. Therefore, we mutagenized hamster V79-4 cells and selected clones that were resistant to infection by MLV and HIV-1 viral vectors. Here we report on the isolation and characterization of one clone that is refractory to infection by MLV, feline immunodeficiency virus (FIV), and HIV-1 viral vectors. The block is postentry and before reverse transcription at uncoating of the virus. The block can be reversed pharmacologically with the proteasome (protease) inhibitor MG-132. Furthermore, the mutant can be complemented by a cDNA coding for a protein of unknown function, which we have termed a modulator of retrovirus infection (MRI).
Results
Mutant 67-1 Cells Are Refractory to Infection by MLV, HIV-1, and FIV Viral Vectors.
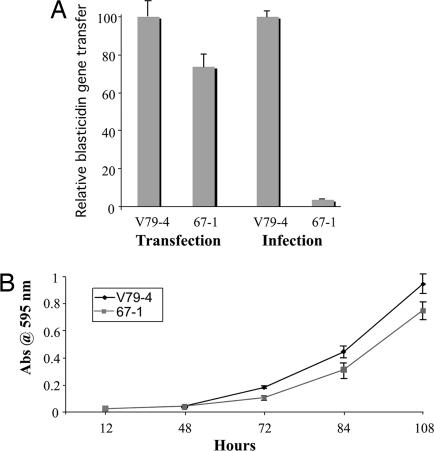
We initially mutagenized hamster V79-4 cells with the frameshift mutagen ICR-191 (an acridine half-mustard), and we multiply infected this population with an MLV-based retroviral vector that transduces the toxic gene barnase. Because these vectors recapitulate the early steps of the retroviral life cycle, we reasoned that cells that survive infection are either mutant in a cellular protein that is required for infection, or they simply escaped infection. We isolated and retested 96 surviving clones with an HIV-1-based vector that transduces EGFP to identify clones that are resistant to evolutionarily distant retroviruses. This approach reveals cellular pathways that are critical for retroviral infection of two distantly related retroviruses. The details of the mutagenesis and selection are reported elsewhere (9). One clone that was reconfirmed as resistant to infection was clone 67. We replated clone 67 at limiting dilutions, and we isolated 10 subclones to establish clonal cell lines. These analyses confirmed that the subclones are stable for the resistance phenotype. Fig. 1A illustrates an experiment that quantifies the level of resistance of clone 67-1 to infection by an HIV-1 viral vector transducing a gene for blasticidin resistance. The parental V79-4 and line 67-1 were either infected with the blasticidin HIV-1-based vector (CSII-Bsd), or the cells were transfected with vector DNA. After gene transfer, the cells were selected for resistance to blasticidin. Compared with wild-type (WT) cells, transfection of vector DNA in 67-1 cells resulted in 73% of WT levels of blasticidin-resistant colonies, whereas transduction of the marker resulted in only 3.8% of 67-1 cells becoming blasticidin-resistant. Because expression of the bsd gene conferring blasticidin resistance is dictated by the same regulatory elements regardless of the method of gene transfer, we conclude that the 67-1 cell line is not defective in proteins that are required for expression of the resistance gene. Experiments transfecting an EGFP expression plasmid or infection with an EGFP-transducing HIV-1 vector show similar results (data not shown). Fig. 1B illustrates the growth rate of V79-4 cells compared with 67-1 cells: 67-1 cells are slightly retarded for growth but not sufficiently to account for the resistance to infection by genetically marked HIV-1 virus.
Fig. 1.
Comparison of gene transfer and growth. (A) V79-4 or mutant 67-1 cells were either infected or transfected with the HIV-1 vector transferring the blasticidin-resistance gene. The data are expressed relative to the gene transfer efficiency of WT cells. (B) Growth rate of V79-4 and 67-1 cells.
Resistance to Infection by MLV, HIV-1, and FIV in 67-1 Cells.
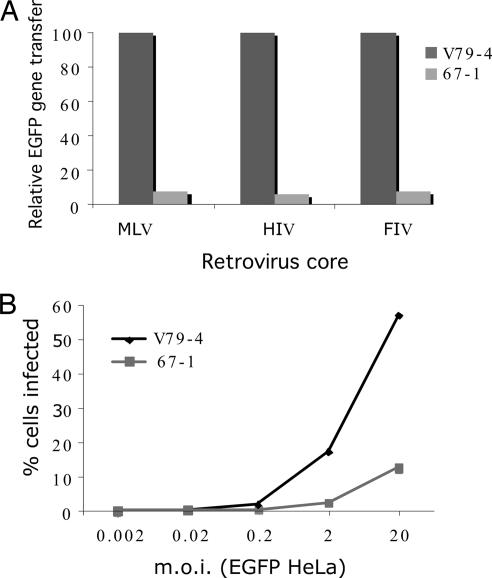
Other investigators have reported cellular mutants that are either resistant to both MLV and HIV-1 (10) cores or only to MLV cores (11). Because we selected with MLV vectors encoding barnase and subsequently screened with EGFP-marked HIV-1, we expected that 67-1 cells should be resistant to both cores. HIV-1 vectors used here are devoid of accessory proteins [except for Rev, Tat, and Vpu (ref. 12)]; hence, we additionally examined the properties of a virus that was produced in the presence of all of the HIV-1 accessory proteins. We also extended this analysis to FIV-based vectors. Fig. 2A illustrates that 67-1 cells are resistant to MLV, HIV-1 (produced with all accessory proteins), and FIV vectors with relative gene transfer efficiencies of 7.3%, 6.2%, and 7.4%, respectively. We conclude that the mutation in 67-1 cells is in a pathway that is common to these three retroviruses and that inclusion of HIV-1 accessory proteins is unable to rescue the resistance. All subsequent experiments were done by using HIV-1 vectors devoid of all accessory factors (except for Rev, Tat, and Vpu).
Fig. 2.
Resistance of mutant cells to infection. (A) V79-4 and 67-1 cells were infected with EGFP-marked MLV-, HIV-1-, and FIV-based vectors. The virus was produced in the presence of all accessory factors, and infection was measured by using flow cytometry to detect EGFP expression. These representative data are expressed relative to infection of the WT cells. (B) Infection of 67-1 cells is not readily saturable. V79-4 and 67-1 cells were infected with HIV-1 viral vectors transducing EGFP with increasing (by 1 log) multiplicities of infection (moi), and the extent of infection was measured by flow cytometry. Note that the titer of the EGFP virus was determined on HeLa cells.
Resistance to Infection Is Not Saturable in 67-1 Cells and Is Recessive.
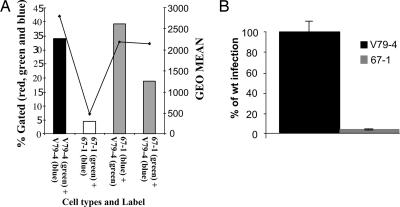
We next examined the properties of V79-4 and 67-1 cells by infecting with increasing moi of an HIV-1 EGFP viral vector. As shown in Fig. 2B, 67-1 cells are resistant to infection over a wide range (5 logs) examined. Although infection of V79-4 cells approaches saturation, 67-1 cells do not do so at the highest moi used, which suggests that the mutation may be recessive because dominant blocks to infection are overcome by challenge with an increasing virus load (see, e.g., ref. 13). Hence, we next performed cell-fusion experiments between WT V79-4 and mutant 67-1. Fig. 3A illustrates an example of such an analysis. V79-4 and 67-1 cells were labeled with the membrane dyes Oregon green or Vybrant DID, which mark cells green and blue, respectively. These differentially marked cells were then mixed (either as self–self or as V79-4 and 67-1 combinations) and fused by the addition of PEG. The fused cells were then infected with a Discosoma sp. red fluorescent gene (dsRED)-marked HIV-1 virus, and the homo- and hetrokaryons were analyzed by fluorescence-activated cytometry. Although 67-1 homokaryons exhibit a resistance to infection compared with V79-4 homokaryons (4.3% compared with 34%), this resistance is rescued in the V79-4 and 67-1 hetrokaryons (39.2% and 18.7%, respectively, in the reciprocal staining experiments). In repeat experiments, we routinely observed that in the reciprocal dyeing experiments the rescue is more pronounced when the 67-1 cells are labeled with Vybrant DID. We conclude from this experiment that the mutation causing the resistance in 67-1 cells is recessive.
Fig. 3.
Block to infection is recessive and not at the level of entry. (A) V79-4 and 67-1 cells were labeled with fluorescent membrane dyes [Oregon green or Vybrant DID (blue)], and V79-4 and 67-1 cells were either fused with PEG or self-fused. Cells were infected with an HIV-1 viral vector transducing dsRED, and the extent of infection was determined by flow cytometry. The extent of infection is expressed as the proportion of cells (percent, left; y axis) that are green, blue, and red compared with cells that are green and blue only. The extent of expression (also reflecting multiple infection events) of dsRED is quantified as the geometrical mean of the red fluorescence from the infected population (right; y axis). (B) V79-4 and 67-1 cells were infected with an HIV-1 vector transducing luciferase pseudotyped with the amphotropic MLV envelope. Data are expressed relative to the infection of WT cells.
The Block to Infection Is Not at the Level of the Entry Receptor.
The retroviral vectors used were pseudotyped with the vesicular stomatitis virus envelope (VSV-G) protein to mediate entry into hamster cells. To determine whether the 67-1 cells were deficient for VSV-G-mediated entry, we infected cells with an HIV-1-based luciferase vector pseudotyped with the MLV amphotropic 10A1 envelope. As illustrated in Fig. 3B, the average relative light units were significantly lower for 67-1 (4%) than V79-4 cells (100%). We conclude that the block to infection in 67-1 cells is not specific to VSV-G-mediated entry.
The Block to Infection Is at the Stage of Uncoating.
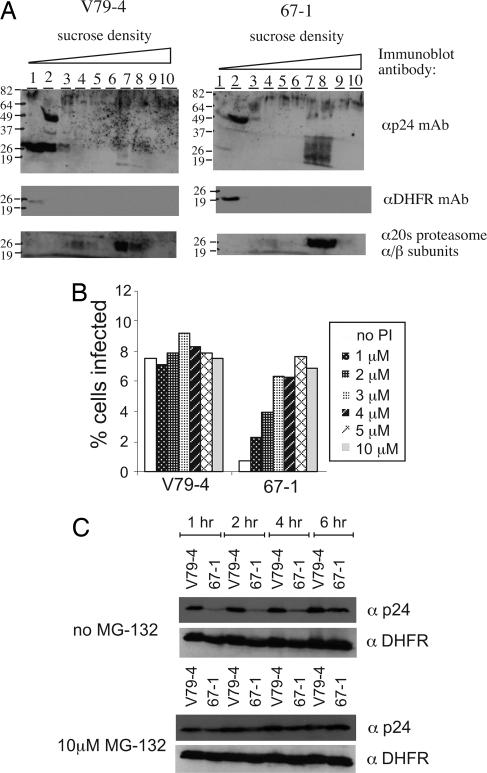
We next examined the fate of the incoming virus by following capsid (p24) protein on sucrose equilibrium density gradients 6 h postinfection in WT V79-4 and mutant 67-1 cells. This technique has been used previously to follow the dynamic nature of the reverse-transcription complex of MLV and HIV-1. Consistent with earlier reports (14), the majority of capsid protein is in the light fractions (1 and 2) in WT V79-4 cells after 6 h, presumably the result of dissolution of the core structure (uncoating) and movement of the majority of the capsid immunoreactivity into the low-density fractions. In contrast, the infection of 67-1 cells resulted in a significantly reduced amount of capsid in the low-density fractions at the 6-h time point (Fig. 4A). A loading control (dihydrofolate reductase protein, DHFR) verifies the integrity of the samples in the low-density fraction. In 67-1 cells we further observed p24 antigen reactivity in higher density fractions, and given the broad pattern of the signal, we hypothesized that the p24 may be targeted for degradation. Hence, we reprobed the immunoblot with an antibody (Ab) to the 20S proteasome α/β subunits. This analysis revealed that the capsid immunoreactivity in the higher density fractions and the proteasome were in the same fractions (7 and 8). Experiments tracking the reverse-transcription process with quantitative PCR (29) confirmed 10-fold fewer reverse-transcription products in 67-1 cells compared with V79-4 cells (see Fig. 8, which is published as supporting information on the PNAS web site). We conclude from this analysis that there is a defect in the uncoating or localization of uncoated virions in 67-1 cells.
Fig. 4.
Uncoating assay and effect of infection with proteasome inhibitor. (A) Sucrose equilibrium density gradients analysis of HIV-1 capsid (p24) distribution 6 h after infection. Cellular cytoplamic extracts were fractionated on a 20–70% sucrose gradient, and fractions were collected from the top. Fractions were analyzed by immunoblotting, using a mouse anti-p24 Ab (αp24 mAb), mouse anti-dihydrofolate reductase Ab (αDHFR mAb), or rabbit anti-20S proteasome α/β subunits. (B) V79-4 and 67-1 cells were infected with an HIV-1 vector transducing EGFP in the absence or presence of increasing concentrations of the proteasome inhibitor (PI) MG-132. The extent of infection was quantified as the fractions of cells expressing EGFP by flow cytometry. (C) Capsids were stabilized by MG-132 in 67-1 cells. V79-4 and 67-1 cells were preincubated with 10 μM MG-132 for 4 h and then infected with an HIV-1 vector. At various times after infection (1, 2, 4, and 6 h) cell extracts were prepared, and p24 levels were assessed by immunoblot analysis (αp24). Equivalence in extract loading was checked by reprobing the blots with an Ab to dihydrofolate reductase (αDHFR).
Infection of 67-1 Cells Can Be Rescued by the Protease Inhibitor MG-132.
We next examined the ability of the protease/proteasome inhibitor MG-132 to rescue infection in 67-1 cells. V79-4 and 67-1 cells were preincubated with varying doses (1–10 μM) of the proteasome inhibitor MG-132 and infected with an HIV-1 EGFP viral vector. The result (Fig. 4B) shows that whereas MG-132 gives a modest increase in infection on WT V79-4 cells (1.2-fold increase at 3 μM), the effect of MG-132 is significant on 67-1 cells (a 10-fold increase at 5 μM), resulting in WT levels of infection. We next asked whether the rescue in titer correlated with the stability of p24 in 67-1 cells in the presence of MG-132. Cell lysates of WT V79-4 and mutant 67-1 cells were harvested at various times after infection with an HIV-1 vector in the presence and absence of MG-132. Fig. 4C illustrates that although the levels of p24 in WT cells are detectable at the early time point, they are reduced considerably in 67-1 cells. In the absence of MG-132, p24 in the mutant cells is decreased compared with WT V79-4 cells over the time course of the experiment. In contrast, in the presence of MG-132, p24 levels are rescued to WT levels in 67-1 cells. We conclude that the rescued titer in the presence of the proteasome inhibitor correlates with the stabilization of the capsid protein.
Cloning a Gene Complementing the Resistance Phenotype for 67-1.
MG-132 pharmacologically rescues the defect in 67-1 cells. We next embarked on a genetic complementation screen to identify cDNA clones that rescue the infection of HIV-1 vectors on 67-1 cells. To this end we used genome functionalization through arrayed cDNA transduction (GFAcT) (15). Approximately 11,000 full-length cDNA expression clones (in duplicate) were screened by transfection into the 67-1 cell mutant and the V79-4 WT cells. Twenty-four hours posttransfection the cells were infected with an HIV-1-based vector transducing luciferase at a titer that did not register above background for 67-1 cells but did give significant infection of V79-4 cells. After 48 h, the luciferase activity in each well was assayed by using the Bright-Glo reagent (Promega, Madison, WI) and a multiwell fluorescence reader. The data obtained from the 22,000 wells were collected and analyzed by using a number of criteria. cDNAs that conferred increased infection of both V79-4 cells and 67-1 cells were eliminated from further study because they are not specific for the rescue of the mutant. Approximately 94 clones, which possessed activity specific for the mutant cell line, were recovered from the master array for further validation. To determine whether the encoded proteins were specifically enhancing infection or modulating the surrogate luciferase readout, we cotransfected these cDNAs with the HIV luciferase vector. Additionally, we repeated the original infection assay to eliminate false positives from the high-throughput screen. One clone (A1) was reconfirmed as rescuing the infection defect in 67-1 cells, and it specifically increased luciferase activity when gene transfer was mediated by infection.
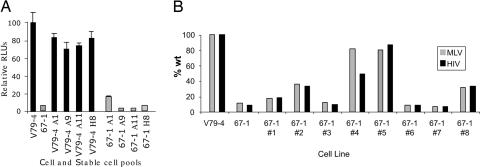
To verify this genetic complementation further, 67-1 cells were cotransfected with the expression plasmid for A1 and a puromycin-resistance marker, and the resistant pool was tested for infection with an HIV-1 vector transducing luciferase. Fig. 5A shows that 67-1 cells transfected with clone A1 are infected to a greater relative extent (17%) compared with 67-1 cells (6%) or 67-1 cells transfected with other clones (A9, 3%; A11, 3%; and H8, 6%). These data further illustrate the specificity of the rescue. Because the pool of cells likely exhibits variable expression of the A1 clone, individual puromycin-resistant clones were selected from the pool and tested for infection with EGFP-marked MLV and HIV-1. Fig. 5B shows a representative experiment from this analysis. Although 67-1 cells show the expected resistance to infection, puromycin-resistant clones exhibited varying levels of infection from no rescue (clone 7) to near complete rescue (clone 5 with 81% and 87% infection compared with WT for MLV and HIV-1 vectors, respectively). Random colony isolation from 67-1 cells did not yield any revertants (data not shown). Notably, V79-4 cells (or human 293 cells) transfected with clone A1 do not show enhanced infection by MLV or HIV-1 vectors (Fig. 5A), and clone A1 does not rescue infection in other resistant mutants we have generated (data not shown). Finally, although infection with an HIV-1 luciferase vector shows rescue in 67-1 A1 expression clones, transfection of the luciferase vector DNA did not result in enhanced expression in the clones compared with the parental 67-1 clone (data not shown). We conclude that A1 expression specifically rescues the infection defect in 67-1 cells.
Fig. 5.
Rescue of infection in 67-1 cells. (A) V79-4 and 67-1 cells cotransfected with a puromycin-resistance plasmid and candidate clone expression vectors were selected with puromycin, and the pool of surviving clones was tested for infection with a luciferase-marked HIV-1 virus. Data are expressed as relative light units (RLUs) compared with WT V79-4 cells. (B) Individual clones of puromycin-resistant clones were isolated and tested for infection with EGFP-marked MLV and HIV-1 viruses. Data are expressed as the percent of cells infected relative to V79-4 cells.
Clone A1 Codes for the MGC5242 Protein and Is an Overexpression Suppressor of the 67-1 Phenotype.
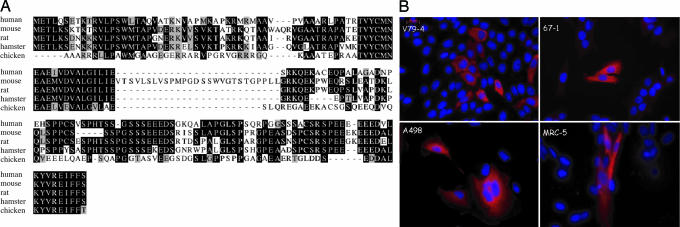
The identity of clone A1 was verified by sequencing as the cDNA for human protein MGC5242. This 157-aa protein has no known functions and no domains that would imply a function. The protein is conserved in mammalian species with an average 56% identity and 62% similarity to the human protein. An avian homolog has 34% identity and 51% similarity to the human protein (Fig. 6A). We have renamed clone A1 as MRI for modulator of retrovirus infection. Gene chip and Gene Expression Omnibus (GEO) database analyses [http://symatlas.gnf.org and www.ncbi.nlm.nih.gov/geo (use the search term BC000168)] reveal that the transcript for MRI is ubiquitously expressed. We next epitope-tagged the MRI protein and examined its intracellular localization. This analysis (Fig. 6B) reveals that MRI is a cytoplasmic protein and that it is unchanged in location between V79-4 and 67-1 cells. We confirmed the localization in human transformed (A498 kidney carcinoma) and primary (MRC-5 lung fibroblast) cells. The distribution is not uniform, and it appears very finely punctate in areas with possibly some associations with higher order structures. The localization does not change on infection with virus (data not shown). We next sequenced the mRNA coding MRI in V79-4 and 67-1 mutant cells, and we found no changes in the coding sequence between the two cell lines (the sequence is shown in Fig. 6A). Indeed, quantitative PCR and Northern blot analysis have been unable to detect a difference in the amount or size of the MRI transcripts between the cell lines (data not shown). Measurement of the steady-state MRI mRNA transcript levels by quantitative PCR in V79-4 and 67-1 cells compared with β-actin transcript revealed no significant differences (with changes in cycle number threshold values of 9.9 for V79-4 and 10.5 for 67-1). We conclude from this analysis that MRI is an overexpression suppressor of the mutation in 67-1 cells.
Fig. 6.
Sequence and localization of MRI. (A) Sequence comparisons of MRI protein from human, mouse, rat, hamster, and chicken. (B) Cellular localization of epitope-tagged MRI protein in hamster WT V79-4, mutant 67-1, human A498, and MRC-5 cells.
MRI Impacts HIV-1 Infection in Normal Human Cells.
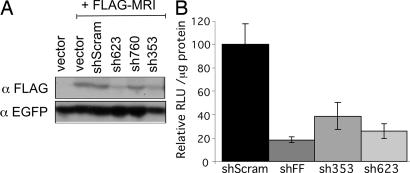
We next examined the effect of decreasing MRI expression on HIV-1 infection in normal human cells. We generated three short hairpin RNA (shRNA) targeting sequences (16) to human MRI, and we examined their potential to decrease expression of epitope-tagged MRI. Two of the three were effective in decreasing the expression of MRI in HEK 293 cells (sh353 and sh623; Fig. 7A). These two targeting sequences and controls (scrambled sequence shScram and a sequence targeting the luciferase gene shFF) were then transduced into human U87 cells, which are sensitive indicators of HIV-1 infection (17). The cells were infected with an HIV-1 luciferase vector, and the infection was quantified 48 h later. The data are illustrated in Fig. 7B. As expected, the luciferase-targeting sequence (shFF) effectively reduced luciferase expression compared with the scrambled sequence (18% of the scrambled sequence). The two sequences targeting MRI also had an effect on reducing infection to 39% (sh353) and 26% (sh623) relative to the scrambled sequence. We have observed similar results in another human cell line (HOS cells) and by using another reporter vector (HIV-1 dsRED; data not shown). We conclude that MRI depletion in normal human cells decreases HIV-1 viral vector infection.
Fig. 7.
Decreased MRI expression impacts infection. (A) Identification of effective shRNA targeting sequences to MRI. FLAG epitope-tagged MRI was cotransfected (+ FLAG-MRI) with an EGFP expression vector and various shRNA expression vectors (shScram random sequence; sh623, sh760, and sh353 targeting MRI; or empty vector) into human HEK 293 cells. MRI expression was assessed 48 h later by immunoblot analysis with an anti-FLAG Ab (αFLAG). Transfection efficiency was monitored by using an anti-EGFP Ab (αEGFP). (B) Human U87 cells expressing MRI shRNA are resistant to infection by HIV-1 vectors. ShRNA targeting a random sequence (shScram), the firefly luciferase gene (shFF), and targeting MRI (sh353 and sh623) were transduced into human U87 cells by using retroviral vectors. ShRNA-expressing cells were then infected with an HIV-1 vector transducing luciferase, and infection was monitored 48 h later. Data [relative light units (RLU)/μg of protein] are represented relative to the shScram sequence set at 100%. Standard deviations from the mean of triplicates are represented by the error bars.
Discussion
We present here the isolation and characterization of a hamster cell mutant that is resistant to infection by HIV-1, MLV, and FIV cores. We employed the hamster V79-4 cell line because it has been successfully used in a number of genetic screens, and it is haploid for one-third of its genome (18). Mutagenesis and a genetic selection generated a number of clones (9), one of which, 67-1, was characterized in more detail. The mutation affects gene delivery by infection and not by transfection. Other investigators have isolated cellular mutants that are resistant to MLV and HIV-1 cores (10) or MLV but not HIV-1 cores (11). The defect to infection in 67-1 cells is not readily saturable with increasing moi of virus in contrast to the resistance mediated by the TRIM5α proteins (13). Indeed, the defect in 67-1 cells is recessive, and this mutant cell is lacking a permissive factor rather than expressing a restriction factor that would exhibit a dominant phenotype. Using envelope proteins that enter by using different receptors and differing pH requirements (19), we further demonstrate that the block to infection is not at the level of entry. The block is at a stage before reverse transcription of the viral RNA, at the level of uncoating. Interestingly, none of the studies to date (9–11 and this work) have isolated mutant cells that are completely resistant (null) to infection, suggesting that these null mutants are lethal, that the screens have not reached saturation, or the that retroviruses use redundant pathways. The uncoating stage of the viral life cycle is poorly understood, and we examined the transition of the HIV-1 capsid protein from heavy to light sucrose fractions to follow this process. This technique has been reported previously to follow MLV and HIV-1 after infection (14, 20), and the analysis reveals that the majority of capsids in 67-1 cell are in higher densities of the gradient coincident with the proteasome. Indeed, we can rescue the infection defect of 67-1 cell with the proteasome/protease inhibitor MG-132. The proteasome has been implicated in the early and late stages of the retroviral life cycle (21–23). Treatment with proteasome inhibitors or genetic inactivation of the ubiquitin pathway modestly increases the retroviral titer, suggesting that the proteasome acts to restrict infection (21, 22, 24). Our observation of a genetic mutant that can be totally rescued by proteasome inhibitors also implicates the proteasome in the early phase of the retroviral life cycle. These observations suggest two models. In the first “nonspecific” model, the mutation in 67-1 cells retards the uncoating process and allows time for the proteasome to degrade the viral core. In wild-type cells, a fraction of viral cores are degraded, and proteasomal inhibitors rescue this fraction. This model explains the increase in titer of 67-1 cells and the modest increase in WT cells on incubation with inhibitors. A second “specific model” postulates that the incoming retroviral core specifically interacts with the proteasome possibly to use a nonproteolytic function (i.e., such as the protein-unwinding function of the 19S regulatory subunit) to uncoat. This model is not unprecedented with the involvement of the proteasome in dissolution of other macromolecular complexes using its nonproteolytic activities (25–27). An assumption of this model is that the viral core will recruit cellular protein(s) that will protect the core from degradation by the 20S proteasome subunit; nonetheless, a subset of cores will still be degraded. This fraction can be rescued on incubation with proteasome inhibitors. We speculate that in 67-1 cells the viral core is degraded by the proteasome because of a lack of a protecting cellular factor. This possibility is consistent with the rescue of the phenotype in 67-1 cells with MG-132. Notably, data from this and other studies are not sufficient to differentiate between these two models. We are currently testing predictions that differentiate between the models.
We have cloned an overexpression suppressor (MRI) that rescues the resistant phenotype of 67-1 cells. Given that 67-1 cells are not totally resistant to infection, but rather refractory, the application of the GFAcT technology is ideally suited for the complementation cloning of cDNA that rescues the infection. This single-well assay can accommodate the background infection (5–10%) that is apparent in 67-1 cells. The power of the technique is demonstrated by the isolation of a genetic suppressor of the mutant phenotype in 67-1 cells, MRI. Other approaches have also been applied to this problem with the cloning of suppressors to the resistant phenotype (28). Indeed, MRI is an overexpression suppressor, and it is very specific to the rescue of the 67-1 phenotype; it does not rescue other mutant cells lines that we have generated. Hence, it is gene-specific, and it is likely not an informational suppressor but acting in the same or a parallel pathway for infection. In support of this hypothesis, we demonstrate that MRI depletion in normal human cells results in a decrease in infection by HIV-1 vectors. Although there is no documented function or domains that may imply a function for MRI, it is tempting to speculate that its overexpression modulates the proteasome to prevent core degradation in 67-1 cells. We are currently testing this hypothesis.
Materials and Methods
Cell Lines.
V79-4 (CCL-93) cells were obtained from American Type Culture Collection (Manassas, VA). A male hamster lung fibroblast line and its derivatives (67-1; this work) were maintained in DMEM supplemented with 10% FBS and 50 units/ml each of penicillin and streptomycin.
Vectors and Plasmids.
Vectors and plasmids are described in the Supporting Materials and Methods, which is published as supporting information on the PNAS web site and are described elsewhere (9).
Viral Vector Production.
MLV, HIV, and FIV vectors were generated by transient transfection of multiple plasmids into 293T cells as described previously (29). Details are provided in Supporting Materials and Methods.
MG-132 Rescue Experiments.
MG-132 (BIOMOL, Plymouth Meeting, PA) was resuspended in DMSO and added at doses between 1 and 10 μM to 105 V79-4 or 67-1 cells for 2 h. After this time, cells were infected for 6 h with an EGFP-marked HIV-1 vector (CSII-EF-EGFP) at an moi of 0.5 (measured on HeLa cells) in the presence of the inhibitor. Cells were washed twice in culture medium and incubated for 72 h in medium without MG-132. After this time, the extent of infection was quantified by flow cytometry on a FACScalibur (BD Biosciences, San Jose, CA) with subsequent analysis using CellQuest Pro acquisition software (BD Biosciences).
Growth Curves, Colony Count Assay, and Luciferase Assay.
These procedures are described in Supporting Materials and Methods and ref. 9.
Equilibrium Density Gradients.
This preparation was done essentially as described in ref. 14.
Immunoblot Analysis.
Immunoblot analysis was performed as described in ref. 29. Mouse anti-p24 Ab (183-H12-5C) was obtained from Dr. Bruce Chesebro and Kathy Wehrly through the National Institutes of Health AIDS repository and used at a 1:3,000 dilution. Secondary Ab was HRP-conjugated goat anti-mouse IgG (Pierce, Rockford, IL), and detection was performed by using the Femto chemiluminescence reagents (Pierce) according to the manufacturer's instructions.
Cell-Fusion Assay.
Cells were plated 5 × 106 cells on 10-cm dishes, and after 18 h they were stained for 20 min with Oregon green (O34550; Invitrogen, Carlsbad, CA) or Vybrant DID (V22887; Invitrogen) probes according to the manufacturer's protocol. The cells were washed three times with PBS and allowed to recover for 4 h. The cells were removed from the plate with a non-trypsin-dissociation medium, and self–self or V79-4 and 67-1 cells were mixed in 15-ml conical tubes (Falcon, Brookings, SD) and concentrated by centrifugation for 5 min at 500 × g. To the pellet was added 1 ml of sterile 50% PEG 3000–3700 (Sigma, St. Louis, MO) and 2% glucose in PBS solution; after 1 min, 1 ml of PBS was added to the cells. After 45 s, 3 ml of PBS + 2% FBS was added to the cells, and cells were washed twice with pelleting (5 min at 500 × g) and resuspended in PBS + 2% FBS. Cells were plated (in DMEM without phenol red + 20% FBS) onto two 10-cm tissue culture dishes and allowed to recover for 6 h. At this time the cells were infected with a CSII-EF-dsRED lentiviral vector at an moi of 1 (infection units calculated on HeLa cells). The cells were analyzed 48 h later by flow cytometry using four-color differentiation on a FACSCalibur with subsequent analysis using CellQuest Pro acquisition software. Background leakage through the channels was compensated by subtraction of the background value from all samples.
High-Throughput Transfection and Reporter Assay.
For details, see Supporting Materials and Methods.
Northern Blot Analysis.
Northern blot analysis was performed as described in ref. 29.
Cellular Localization.
Immunocytochemisry was carried out essentially as described in ref. 29, and it is described in more detail in Supporting Materials and Methods.
Supplementary Material
Acknowledgments
We thank Scott McIvor and Kathleen Conklin for helpful suggestions. N.S. dedicates this work to the memory of Prof. John Scaife. This work was supported by startup funds from the Institute of Human Genetics, University of Minnesota, the University of Minnesota Medical School, and National Institutes of Health Grant R21 AI60470 (to N.S.). S.A. received a student fellowship from the Institute of Human Genetics, University of Minnesota.
Abbreviations
- dsRED
Discosoma sp. red fluorescent gene
- FIV
feline immunodeficiency virus
- GFAcT
genome functionalization through arrayed cDNA transduction
- HIV-1
HIV type 1
- MLV
murine leukemia virus
- moi
multiplicity of infection
- MRI
modulator of retrovirus infection
- shRNA
short hairpin RNA
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Best S, Le Tissier P, Towers G, Stoye JP. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 2.Bishop KN, Bock M, Towers G, Stoye JP. J Virol. 2001;75:5182–5188. doi: 10.1128/JVI.75.11.5182-5188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozak CA, Chakraborti A. Virology. 1996;225:300–305. doi: 10.1006/viro.1996.0604. [DOI] [PubMed] [Google Scholar]
- 4.Goff SP. Mol Cell. 2004;16:849–859. doi: 10.1016/j.molcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J, Ferrigno P, Sodroski J. J Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 7.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 8.Garber ME, Wei P, KewalRamani VN, Mayall TP, Herrmann CH, Rice AP, Littman DR, Jones KA. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agarwal S, Nikolai B, Yamaguchi T, Lech P, Somia NV. Mol Ther. 2006;14:555–563. doi: 10.1016/j.ymthe.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 10.Gao G, Goff SP. Mol Biol Cell. 1999;10:1705–1717. doi: 10.1091/mbc.10.6.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruce JW, Bradley KA, Ahlquist P, Young JA. J Virol. 2005;79:12969–12978. doi: 10.1128/JVI.79.20.12969-12978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu K, Ma H, McCown TJ, Verma IM, Kafri T. Mol Ther. 2001;3:97–104. doi: 10.1006/mthe.2000.0238. [DOI] [PubMed] [Google Scholar]
- 13.Hatziioannou T, Cowan S, Goff SP, Bieniasz PD, Towers GJ. EMBO J. 2003;22:385–394. doi: 10.1093/emboj/cdg042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fassati A, Goff SP. J Virol. 2001;75:3626–3635. doi: 10.1128/JVI.75.8.3626-3635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chanda SK, White S, Orth AP, Reisdorph R, Miraglia L, Thomas RS, DeJesus P, Mason DE, Huang Q, Vega R, et al. Proc Natl Acad Sci USA. 2003;100:12153–12158. doi: 10.1073/pnas.1934839100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Ihrig MM, McManus MT, Gertler FB, et al. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 17.Bjorndal A, Deng H, Jansson M, Fiore JR, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman DR, Fenyo EM. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thacker J. Cytogenet Cell Genet. 1981;29:16–25. doi: 10.1159/000131547. [DOI] [PubMed] [Google Scholar]
- 19.McClure MO, Sommerfelt MA, Marsh M, Weiss RA. J Gen Virol. 1990;71:767–773. doi: 10.1099/0022-1317-71-4-767. [DOI] [PubMed] [Google Scholar]
- 20.Fassati A, Goff SP. J Virol. 1999;73:8919–8925. doi: 10.1128/jvi.73.11.8919-8925.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz O, Marechal V, Friguet B, Arenzana-Seisdedos F, Heard JM. J Virol. 1998;72:3845–3850. doi: 10.1128/jvi.72.5.3845-3850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei BL, Denton PW, O'Neill E, Luo T, Foster JL, Garcia JV. J Virol. 2005;79:5705–5712. doi: 10.1128/JVI.79.9.5705-5712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ott DE, Coren LV, Sowder RC, Jr, Adams J, Schubert U. J Virol. 2003;77:3384–3393. doi: 10.1128/JVI.77.6.3384-3393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Caballero D, Hatziioannou T, Zhang F, Cowan S, Bieniasz PD. J Virol. 2005;79:15567–15572. doi: 10.1128/JVI.79.24.15567-15572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferdous A, Kodadek T, Johnston SA. Biochemistry. 2002;41:12798–12805. doi: 10.1021/bi020425t. [DOI] [PubMed] [Google Scholar]
- 26.Nishiyama A, Tachibana K, Igarashi Y, Yasuda H, Tanahashi N, Tanaka K, Ohsumi K, Kishimoto T. Genes Dev. 2000;14:2344–2357. doi: 10.1101/gad.823200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sulahian R, Johnston SA, Kodadek T. Nucleic Acids Res. 2006;34:1351–1357. doi: 10.1093/nar/gkl012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao G, Goff SP. Retrovirology. 2004;1:30. doi: 10.1186/1742-4690-1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somia NV, Schmitt MJ, Vetter DE, Van Antwerp D, Heinemann SF, Verma IM. Proc Natl Acad Sci USA. 1999;96:12667–12672. doi: 10.1073/pnas.96.22.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.