Abstract
Deltex1, Deltex2, and Deltex4 form a family of related proteins that are the mammalian homologues of Drosophila Deltex, a known regulator of Notch signals. Deltex1 is highly induced by Notch signaling in thymocytes, and overexpression of Deltex1 in T-cell progenitors can block Notch signals, suggesting that Deltex1 may play an important role in regulating Notch signals during T-cell development. A recent report found that T cells develop normally in mice carrying a targeted deletion in the Deltex1 gene (S. Storck, F. Delbos, N. Stadler, C. Thirion-Delalande, F. Bernex, C. Verthuy, P. Ferrier, J. C. Weill, and C. A. Reynaud, Mol. Cell. Biol. 25: 1437-1445, 2005), suggesting that other Deltex homologues may compensate in Deltex1-deficient T cells. We generated mice that lack expression of both Deltex1 and Deltex2 by gene targeting and further reduced expression of Deltex4 in Deltex1/Deltex2 double-deficient T-cell progenitors using RNA interference. Using a sensitive in vitro assay, we found that Notch signaling is more potent in cells expressing lower levels of Deltex proteins. Nevertheless, we were unable to detect any significant defects in thymocyte maturation in Deltex1/Deltex2 double-knockout mice. Together these data suggest that Deltex can act as a negative regulator of Notch signals in T cells but that endogenous levels of Deltex1 and Deltex2 are not important for regulating Notch signals during thymocyte development.
The cells of the mammalian immune system are continuously generated throughout life. Among these cells, T and B lymphocytes form the adaptive immune system (for general reviews, see references 1 and 68). T and B lymphocytes express antigen receptors that are generated by random rearrangement of the genes encoding the B-cell receptor (or immunoglobulin) or T-cell receptor (TCR), and lymphocyte precursors progress through a series of well-defined maturational stages that are dependent on this process. These two cell types are derived from a common progenitor, which originates in either the bone marrow or fetal liver, but further development into either the B- or T-cell lineage occurs in distinct anatomical locations. While B cells develop in the bone marrow, T-cell development occurs almost exclusively within the thymus (5, 47, 63).
T-cell development is a highly ordered process, and developing thymocytes progress through a series of distinct maturational stages that are linked to the stepwise acquisition of a functional TCR. Progression through these stages can be monitored by expression of cell surface markers, including the CD4 and CD8 coreceptor molecules. The most immature thymocytes express neither CD4 nor CD8 and are termed double negative (DN). DN thymocytes can be further separated into four distinct developmental stages (DN1 to DN4) defined by the expression of CD44 and CD25 surface molecules (21). DN1 thymocytes (CD44+ CD25−) comprise a mixed population of precursors that retains the capacity to differentiate into T, B, NK, and dendritic cells (3, 60, 73). DN2 thymocytes (CD44+ CD25+) lose the potential to differentiate into B cells but retain NK and dendritic-cell potential (30, 58), whereas DN3 thymocytes (CD44− CD25+) are committed to the T-cell lineage (68). Thymocytes begin to rearrange the genes encoding TCRβ, TCRγ, and TCRδ chains during the DN2/DN3 stage. During this stage, thymocytes that have successfully rearranged TCRβ express the pre-TCR, consisting of TCRβ complexed with pre-Tα. The pre-TCR mediates β-selection, following which thymocytes undergo a massive proliferative expansion as they progress to the DN4 (CD44− CD25−) stage. Only cells that have undergone β-selection are able to up-regulate CD4 and CD8 expression to enter the double-positive (DP) stage (24, 40, 45, 78), where they begin to rearrange the genes for TCRα and differentiate further into either the CD4 or CD8 single-positive (SP) lineage (22, 33, 38, 67).
While thymocyte maturation is controlled largely by signals through the TCR, Notch signals are also essential for promoting multiple stages of T-cell development (2, 61, 63). The Notch pathway is a conserved signaling mechanism that regulates cellular differentiation in a variety of tissue types throughout the life of multicellular organisms (7). Notch signals have been shown to influence cell proliferation (35, 80), apoptosis (14, 70), and developmental lineage choices (10, 49, 65). But the molecular mechanisms whereby Notch regulates these diverse functions have not been fully characterized. In mammals there are four Notch receptors (Notch1 to Notch4) and five Notch ligands which belong to either the Jagged or Delta class. Notch receptors signal through a common mechanism. Ligand binding induces a series of proteolytic cleavages within the intracellular domain of Notch. The terminal cleavage is mediated by a family of proteases, the presenilins, and releases the intracellular domain of Notch (Notch-IC), which enters the nucleus, where it activates the transcription of Notch-responsive genes by binding to CBF1/RBPJκ, a transcriptional activator/repressor that provides the primary mechanism for Notch signal transduction (55).
Notch signals have been proposed to influence multiple stages of T-cell development. Definitive studies utilizing both gain- and loss-of-function approaches revealed that Notch1 provides an essential nonredundant signal that promotes T-lineage commitment and prevents B-cell development within the thymus. Following T-lineage commitment, the dose of Notch signals delivered to T-cell precursors can influence their differentiation into the αβ versus γδ T-cell lineage, as reduced levels of Notch signaling within Notch1+/− precursors (79) or precursors harboring a conditional deletion of RBPJk/CBF1 (77) favor the production of γδ T cells within the thymus. Finally, there is evidence that Notch signals cooperate with signals through the pre-TCR to promote expansion and survival of DN3/DN4 thymocytes expressing a functional pre-TCR (11, 12, 29, 70). In addition to its role during early thymocyte development, Notch signaling has been proposed to affect the proliferation and differentiation of mature effector T cells into distinct Th1, Th2, or regulatory T-cell subsets, although the mechanisms whereby this occurs are less well understood (4, 17, 27, 28, 46, 53, 57, 69, 77).
Despite the preponderance of data demonstrating that Notch signals are essential for promoting T-lineage commitment and the expansion of T-cell precursors within the thymus, it remains uncertain how Notch signals promote these functions at the molecular level. Notch signaling induces expression of a number of target genes within T-cell progenitors (15), including Hes1, pre-Tα (66), Nrarp (39, 59), and Deltex1. These genes could promote T-cell maturation by performing T-cell-specific functions, as is the case for pre-Tα. Alternatively, these genes could have a role in altering the dose or quality of Notch signals within thymocytes as they progress through specific maturational stages. Consistent with the notion that Notch signals are highly regulated within thymocytes, there is evidence that the expression of Notch-responsive genes varies dramatically in DN, DP, and SP thymocyte populations (14, 15). Notch signals are subject to regulation on multiple levels (8). At the level of ligand binding, there is evidence that signals through the Jagged versus Delta class of Notch ligands transmit distinct signals to T-cell precursors (43). Within the Notch signal-receiving cell, a number of molecules regulate the transmission of Notch signals within the cytoplasm or nucleus (42), including Numb (19), Itch (52), Sel-10 (42), Nrarp (83), MINT (41), and Deltex.
Our lab has identified Deltex1 as a gene that is highly induced by Notch signaling in T cells, and expression of Deltex1 mRNA is highly regulated as thymocytes progress from the DN to DP to SP maturational stages. These data suggest that Deltex1 may have an important role in regulating Notch signals during T-cell development. Deltex encodes a putative E3-ubiquitin ligase that was first identified in Drosophila in a screen for mutations that could suppress the lethal phenotypes resulting from a Notch gain-of-function mutant (81). Deltex mutants share some of the phenotypic characteristics resulting from mutations in key components of the Notch pathway and exacerbate Notch loss-of-function phenotypes (23). Three mammalian homologues of the Drosophila Deltex gene have now been identified (37, 74) (the Deltex1, Deltex2, and Deltex4 genes), which code for a family of cytoplasmic proteins that contain three structural domains. Domain I contains two WWE repeats (6) that have been shown to physically interact with the Notch ankyrin domains (16, 50, 84). Domain II contains a proline-rich region that shares homology with SH3-binding domains, and domain III contains a RING finger domain commonly found in E3-ubiquitin ligases (34).
There are several lines of evidence that suggest that Deltex plays an important role in either regulating or transducing Notch signals. Initial genetic analysis in Drosophila suggested that Deltex acts as a positive regulator of Notch signals (23, 50, 51, 81). These observations are supported by evidence that either Notch-IC or Deltex can inhibit transcriptional activity of E-box-containing promoters (56) and that either Notch-IC or Deltex1 can inhibit the differentiation of a proneural cell line (82). However, Deltex can also inhibit Notch signals in some settings (20). Preliminary data suggested that in T cells, Deltex1 inhibits Notch signals as hematopoietic stem cell populations overexpressing Deltex1 differentiate preferentially into the B-cell lineage (31, 83). Although it is well established that Deltex interacts with the intracellular domain of Notch, the mechanism whereby Deltex regulates Notch signals either positively or negatively remains uncertain. Deltex has been proposed to inhibit the transcriptional activation of Notch-responsive genes (31) or to regulate transcription independently of Notch by binding to the transcriptional coactivator p300 (82) or by targeting MEKK1 for degradation (44). Other studies suggest that Deltex regulates the intracellular trafficking of Notch within endosomal compartments (25, 26). A more recent report revealed that in Drosophila cells Deltex can target Notch for ubiquitin-mediated proteasomal degradation via a trimeric complex including Deltex, Notch, and Kurtz (54). While it is not clear which of these pathways may be important within T-cell progenitors, the recent identification of a Deltex null mutant in Drosophila revealed that regulation of Notch through Deltex is highly cell type specific, as Notch signaling was altered in only a subset of tissues known to require Notch signaling for their development (20). Thus, regulation of Notch signals through Deltex is complex and highly influenced by the cellular context. Compounding this, analysis in mammalian cells is further complicated by the existence of at least three Deltex homologues.
In this study, we examined the role of Deltex during different stages of T-cell development by generating mice that are deficient in Deltex1 and Deltex2. We further reduced expression of Deltex4 in lymphoid progenitors from the Deltex1/Deltex2 double-deficient mice and examined the intensity of Notch signaling using a sensitive in vitro assay that allowed us to examine Notch signals during different stages of thymocyte maturation.
MATERIALS AND METHODS
Reverse transcriptase PCR (RT-PCR) TaqMan analysis.
RNA was isolated using STAT-60 (Tel-Test, Friendswood, TX) according to the manufacturer's instructions, and cDNA was generated using Moloney murine leukemia virus reverse transcriptase (Fermentas, Hanover, MD). cDNAs were normalized by TaqMan PCR (PE Applied Biosystems, Foster City, CA) for hypoxanthine phosphoribosyltransferase 1 (HPRT), as described previously (29). PCR was carried out on normalized cDNAs using Taq polymerase (Perkin Elmer, Boston, MA) and the following primers: for HPRT, 5′-TGGAAAGAATGTCTTGATTGTTGAA (forward), 5′-AGCTTGCAACCTTAACCATTTTG (reverse), and 5′-CAAACTTTGCTTTCCCTGGTTAAGCAGTACAGC (probe); for Hes1, 5′-TACCCCAGCCAGTGTCAACA (forward), 5′-TTCTTGCCCTTCGCCTCTT (reverse), and 5′-TGAGCACAGAAAGTCATCAAAGCCTATCATGG (probe); for pre-Tα, 5′-CTGCTTCTGGGCGTCAGGT (forward), 5′-TGCCTTCCATCTACCAGCA (reverse), and 5′-CCTTTCCGTCTCTGGCTCCACCCA (probe); for Deltex1, 5′-TGAGGATGTGGTTCGGAGGT (forward), 5′-CCCTCATAGCCAGATGCTGTG (reverse), and 5′-CGCCTGATGAGGACTGTACCATTTGCAT (probe); for Deltex2, 5′-CCCCTTACATCATCGACCTCC (forward), 5′-GCGCACAGACCTCATGGTG (reverse), and 5′-CAGCTGGACTCAGTTTCGCCAGAACACT (probe); and for Deltex4, 5′-GGGATTCTATAGTAAAGGCATGGC (forward), 5′-TCTATGTCCATTAGGGTCCAAGTTT (reverse), and 5′-TCTCACCTTTGCCAGCCCATCTCGTAA (probe).
Notch reporter assay.
The pGL2-8xCBF1 plasmid (a gift from S. D. Hayward, Johns Hopkins University, Baltimore, MD) was modified such that luciferase was replaced with green fluorescent protein (GFP) and introduced into Cos-7 cells. Plasmids containing the intracellular domains of Notch1, Notch2, and Notch3 fused to the Myc tag or the entire coding region of Deltex1, Deltex2, or Deltex4 fused to the Flag tag were introduced by transient transfection using FuGENE 6 (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions. GFP expression was monitored by fluorescence-activated cell sorting analysis (FACS) analysis after 24 h. The remaining cells used for FACS analysis were lysed in radioimmunoprecipitation assay buffer and analyzed by Western blotting using antibodies to Flag (M2; Stratagene, La Jolla, CA) or the Myc tag (9B11; Cell Signaling Technology, Danvers, MA). The expression vectors were constructed using pCMV-Tag vectors (Stratagene) by fusing the appropriate coding regions as follows: Notch1, accession no. NM008714 from bp 5329 to 8082; Notch2, accession no. D32210 from bp 5262 to 7579; Notch3, accession no. NM 008716 from bp 5064 to 7944). These inserts were fused in frame into CMV-Tag3. The entire coding regions of Deltex1 (NM_008052), Deltex2 (NM_023742), and Deltex4 (NM_172442) were fused into CMV Tag2. The epitope tag was cloned at the N terminus in the above vectors.
Targeted deletion of Deltex1 and Deltex2.
The genomic DNA for Deltex1 (NCBI GeneID, 14357) and Deltex2 (NCBI GeneID, 74198) was obtained by screening a BAC library (Research Genetics/Invitrogen Carlsbad, CA; clone RPCI-22 from the 129 mouse strain) using cDNA probes encompassing the first (Deltex1) or the first two coding exons (Deltex2). Genomic DNA was subcloned into pBluescript (Stratagene), and the Neo cassette flanked with Flp recombinase recognition sequences (FRT) was subcloned from PGK-neo FRT2-Lox2 DTA, a kind gift from Philippe Soriano (Fred Hutchinson Cancer Research Center, Seattle, WA). For Deltex1, the 1,098-bp region between the BglII and BamHI sites flanking the first coding exon was deleted. This region encompasses the first 87 amino acids (aa) (out of a total 627 aa), including the first WWE repeat of Deltex1. For Deltex2, the 3,846-bp region from KpnI to EcoRV encompassing the first two coding exons was replaced with the neo cassette. This region contains the ATG and encodes the first 301 aa of Deltex1 (out of a total 618 aa), including both WWE repeats. Targeting vectors were introduced into TC1 embryonic stem (ES) cells (derived from 129 mice), and neomycin-resistant clones were screened by Southern blotting. For Deltex1 knockout (Dx1-KO) mice, the targeted region, including the neomycin cassette, was deleted by transient transfection of a plasmid encoding Cre recombinase (PGK-Cre). Knockout mice were generated from ES cells using established methods (64). For Dx2-KO mice, the neo cassette was deleted by breeding mice to Flipper mice (18). Deltex1/Deltex2 double-knockout mice were generated by breeding the two strains together.
Typing Deltex1 and Deltex2 knockout mice.
PCR analysis was used for typing genomic DNA. Three PCR primers were included in a single reaction mix.
(i) Flipper PCR.
The flipper PCR detects wild-type (500 bp) or Flp (250 bp) DNA using the primers R1295, (5′-GCGAAGAGTTTGTCCTCAACC), R523 (5′-GGAGCGGGAGAAATGGATATG), and R26F2 (5′-AAAGTCGCTCTGAGTTGTTAT).
(ii) Deltex1 PCR.
The Deltex1 PCR detects wild-type (280 bp), 2Lox (314 bp), or knockout (494 bp) DNA using the primers F6 (5′-TGACAGCCTGGGGTATGATGC), 20R (5′-CGGCCTGAGTGGTGGTAGATC), and B3 (5′-GGTGGACGGGGAAGACTTTCTG).
(iii) Deltex2 PCR.
The Deltex2 PCR detects wild-type (631 bp) or knockout (300 bp) DNA using the primers F1 (5′-GATAGGGATCAAGAGTTGATC), R1 (5′-GGTCTTACCAGTGTTCTGGCG), and R2 (5′-CTCCACCTGGCTTTGCATGAG).
Southern blot on genomic DNA. (i) Deltex1.
Ten micrograms of genomic DNA from ES cells was digested with KpnI, and Southern blots were hybridized with probes recognizing the genomic DNA outside (probe 1) or inside (probe 2) the targeted region for Deltex1. Further analysis using restriction sites and probes inside and outside the targeted region confirmed that the endogenous gene is altered in the Dx1-KO ES cells (data not shown).
(ii) Deltex2.
Southern blots were performed as described above using 10 μg of genomic DNA from ES cells either digested with AccI or HindIII and hybridized with probes recognizing the genomic DNA outside (probe 1) or inside (probe 2) the targeted region for Deltex2.
Mixed bone marrow chimeras.
Bone marrow from 8-week-old donor mice, two Dx1-KO or two homozygous wild-type littermates (both Ly5.2), was pooled and mixed at a 1:1 ratio with wild-type competitor marrow from Pep3b/Boy (Ly5.1) mice (The Jackson Laboratory, Bar Harbor, ME). Bone marrow cells were depleted of mature T cells by complement lysis using antibodies to CD4, CD8, and Thy1.2. The donors were back-crossed twice to C57BL/6. The marrow was transferred into lethally irradiated recipients as follows. Recipients B6SJLF1/J (Ly5.1/5.2; The Jackson Laboratory) were irradiated with 1,000 rads and placed on antibiotic water 1 day prior to and for 3 weeks following bone marrow transfer. After 3 months, the hematopoietic cells derived from competitor (Ly5.1) or Dx1-KO (Ly5.2) littermates were detected using the antibodies CD45.1 (clone A20) and CD45.2 (clone 104) (BD Pharmingen, San Jose, CA). Proliferation of mature T cells was measured by dilution of carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes Inc. Eugene, OR). For CFSE labeling, cells were washed in phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin (BSA) and incubated at 37°C for 7 min in PBS containing 10 μM CFSE. The reaction was stopped by adding cold medium containing 1% BSA. The labeled cells were washed three times in complete medium and stimulated with plate-bound anti-CD3ɛ at 1 μg/ml or 0.2 μg/ml. Proliferation was measured as CFSE dilution by FACS analysis on day 2 and day 5.
Flow cytometric analysis for cell differentiation markers.
Expression of cell differentiation markers was analyzed by four-color flow cytometry using a FACSCalibur (BD Pharmingen). Cells were incubated with Fc block (24G2) and stained with the antibodies B220 (RA3-6B2), CD19 (ID3), CD44 (IM7), CD25 (7D4), CD4 (RM4-5), CD8 (53.67), NK1.1 (PK136), CD21 CD35 (7G6), and CD23 (B3B4).
Inhibition of Deltex4 by RNA interference (RNAi).
Two putative short interfering RNA (siRNA) sequences for Deltex4 beginning at position 562 or 1417 were cloned into the BamHI and SalI cloning sites of the LTRH retroviral vector (9) by generating the following oligonucleotides (underlining indicates sequences derived from Deltex4): 1447-19 For, 5′GATCCCCGAGGATTGTACCATCTGTATTCAAGAGATACAGATGGTACAATCCTCTTTTTGGAAC; 1447-19 Rev, 5′TCGAGTTCCAAAAAGAGGATTGTACCATCTGTATCTCTTGAATAC AGATGGTACAATCCTCGGG; 562-19 For, 5′GATCCCCGTCGGCATCACCATCCAGTTTCAAGAGAACTGGATGGTGATGCCGACTTTTTGGAAC; and 562-19 Rev, 5′TCGAGTTCCAAAAAGTCGGCATCACCATCCAGTTCTCTTGAAACTGGATGGTGATGCCGACGGG. The resulting vectors were sequenced, and DNA was transfected into the PhoenixE ecotropic retroviral packaging cell line to generate retroviral supernatants as described previously (83).
In vitro differentiation of fetal liver stem cells.
Mice that were heterozygous for both Deltex1 and Deltex2 were bred together, and embryos were removed from pregnant females on day 14 to obtain fetal liver. A single-cell suspension was generated from each liver by pipetting gently in 1 ml of PBS containing 0.5% BSA and 2 mM EDTA, and a 10-μl sample was removed to isolate genomic DNA. Each liver was genotyped by PCR analysis for both Deltex1 and Deltex2 (see above), and homozygous wild-type or Deltex1/Deltex2 double-knockout livers were pooled and depleted of mature lineage-positive cells by using Dynal magnetic bead separation (Dynal Biotech, Brown Deer, WI) and antibodies to Ly-76 (Ter119), Mac1 (M1/70), Gr1 (8C5), and B220 (RA3-6B2). The resulting lineage-negative cells were infected immediately with retroviral supernatants containing 5 μg/ml Polybrene, 100 ng/ml recombinant mouse stem cell factor (R&D Systems, Minneapolis, MN), and 5 ng/ml recombinant interleukin 7 (IL-7) (Peprotech, Rocky Hill, NJ). After 48 h, cells were sorted for expression of the human CD4 reporter (RPA-T4) and cKit (2B8). Sorted stem cells were plated on OP9 stromal cells and cultured in the presence of IL-7 and Flt3L as described above.
Cell culture for in vitro T-cell development.
Parental OP9 stromal cells and cells expressing Delta-1 were a kind gift from J. C. Zuniga-Pflucker, University of Toronto, Toronto, Ontario, Canada). In vitro T-cell development culture was carried out as described previously (43, 72). OP9 monolayers were prepared 1 day in advance by plating stromal cells at 2.5 × 104 cells/well in 24-well culture dishes, and stem cell populations were plated at 1,000 and 10,000 cells per well onto OP9 monolayers in RPMI medium (Gibco/Invitrogen, Carlsbad, CA) containing 10% fetal calf serum, supplemented with l-glutamine, β-mercaptoethanol, penicillin-streptomycin, and gentamicin. Growth medium was supplemented with 5 ng/ml recombinant IL-7 and Flt3L (Peprotech). The γ-secretase inhibitor X (Calbiochem, San Diego, CA) or 0.1% dimethyl sulfoxide carrier was added to selected wells on day 0 and replaced every 3 to 4 days.
Yeast two-hybrid assay.
Interaction between Deltex1 and Notch-IC was assessed using the Hybrizap 2 hybrid system (Stratagene, La Jolla, CA). The intracellular domain of Notch1 was fused to the Gal4 activation domain (pAD-GAL4-2.1) and the full-length coding region of Deltex1 (bp 1 to 1884) or deletion mutants lacking domain I (bp 74 to 523) or domain III (bp 1144 to 1884) were cloned into the pBD-GAL4-Cam expression vector. Both plasmids were transformed into Saccharomyces cerevisiae strains YRG-2 or PJ69-4A (32) and cultured on selective medium. Yeasts expressing both plasmids were selected for growth on medium lacking Leu and Trp, and yeasts harboring interacting proteins were identified by growth on medium lacking Leu, Trp, and His (for YRG-2) or Ade (for PJ694A).
RESULTS
Deltex1 is a Notch regulator that is induced by Notch signaling in T cells.
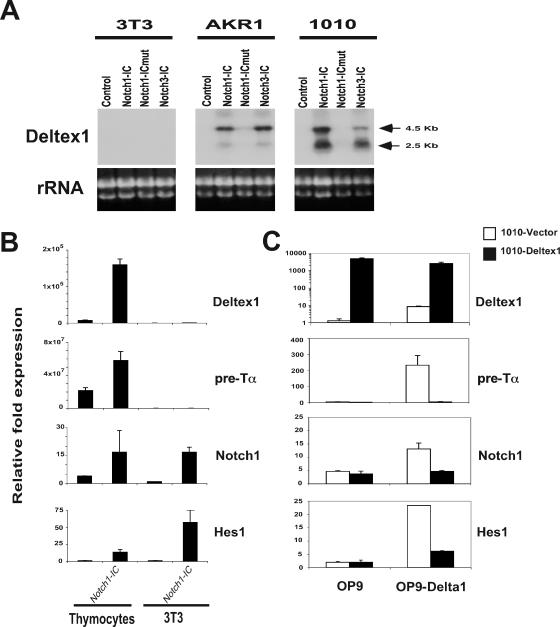
Deltex1, a known component of the Notch signaling pathway, is highly induced by Notch signaling, and expression of Deltex1 mRNA is regulated during the DN, DP, and SP stages of thymocyte maturation (14, 15). To test whether induction of Deltex1 mRNA is specific to T cells, and whether its induction results from CBF1/RBPJκ-dependent Notch signaling, we examined the expression of Deltex1 mRNA in 3T3 fibroblasts and in two thymoma cell lines that had been transduced with retroviruses expressing constitutively active forms of the Notch receptor (Fig. 1A). The active intracellular domains of Notch1 (Notch1-IC), Notch1 containing a three-amino-acid substitution (W1759FP to L1758AA) (75) within the RAM domain, which is required for binding to CBF1/RBPJκ (Notch1-ICmut), and Notch3 (Notch3-IC) were expressed in a retroviral vector that also encodes the human CD2 reporter. Transduced cells were identified by staining for the CD2 marker, and expression of Deltex1 mRNA was examined by Northern blotting. To confirm that Notch signaling was activated in 3T3 cells and to compare the level of gene induction in 3T3 cells to that seen in normal thymocytes, we examined expression of Deltex1, pre-Tα, Notch1, and Hes1 by TaqMan quantitative PCR analysis in 3T3 cells transfected with Notch1-IC versus thymocytes isolated from control or transgenic mice expressing Notch1-IC under the control of the Lck-proximal promoter (15). This analysis (Fig. 1B) revealed that although Notch1 and Hes1 were induced similarly in the two cell types, Deltex1 and pre-Tα were highly induced in the thymocytes but not in 3T3 cells. This analysis revealed that Deltex1 mRNA is highly induced by either Notch1-IC or Notch3-IC in T-lineage cells and that induction of Deltex1 mRNA is dependent on CBF1/RBPJκ.
FIG. 1.
Deltex1 is a Notch regulator that is induced by Notch signaling in T cells. (A) Deltex1 mRNA is induced by CBF1/RBPJκ-dependent Notch signaling in T-cell lines. The Northern blot shows expression of Deltex1 mRNA in two thymoma cell lines (AKR1 and 1010) and in 3T3 fibroblasts overexpressing the intracellular domains of Notch1 (Notch1-IC), Notch3 (Notch3-IC), and Notch1-mM2-2 (Notch1-ICmut), which contains a mutation in the RAM domain. Northern blots were hybridized with a probe detecting the 3′ untranslated region of Deltex1, and ethidium bromide staining of the ribosomal RNAs is shown as a loading control. (B) TaqMan RT-PCR analysis of Deltex1, pre-Tα, Notch1, and Hes1 on cDNA prepared from 3T3 cells transduced with control or Notch1-IC retrovirus (as described for panel A) or thymocytes derived from transgenic Lck-Notch-IC mice (15) or littermate controls. (C) Overexpression of Deltex1 inhibits ligand-dependent Notch signaling. RT-PCR analysis shows induction of Deltex-1, pre-Talpha, Notch1, and Hes1 mRNA in 1010 (open bars) or 1010 overexpressing Deltex1 (filled bars) cultured on stromal cells expressing empty vector (OP9) or Delta1 (OP9-Delta1).
Overexpression of Deltex1 inhibits Notch signals.
Deltex1 mRNA is highly expressed in subsets of thymocytes (13, 14), suggesting that Deltex1 may play an important role in regulating Notch signals during T-cell development. However, the function of Deltex1 remains uncertain, and Deltex has been reported to act as either a positive or negative regulator of Notch signals depending on the experimental system employed. To examine whether Deltex1 enhances or inhibits Notch signaling, we overexpressed Deltex1 in the 1010 thymoma cell line and compared the level of Notch signaling in control and Deltex1-expressing 1010 cells (Fig. 1C). 1010 cells expressing empty vector or Deltex1 were cultured on the stromal cell line OP9 or on OP9 expressing the Notch ligand Delta1 (OP9-Delta1) (43, 72), and the level of Notch signaling was measured by examining expression of pre-Tα, Notch1, and Hes1 mRNA by TaqMan RT-PCR analysis. This analysis revealed that Notch ligand-expressing cells can induce these genes and that overexpression of Deltex1 in 1010 effectively blocks the induction of all of the Notch-responsive genes we examined.
T-cell development is normal in Deltex1 knockout mice.
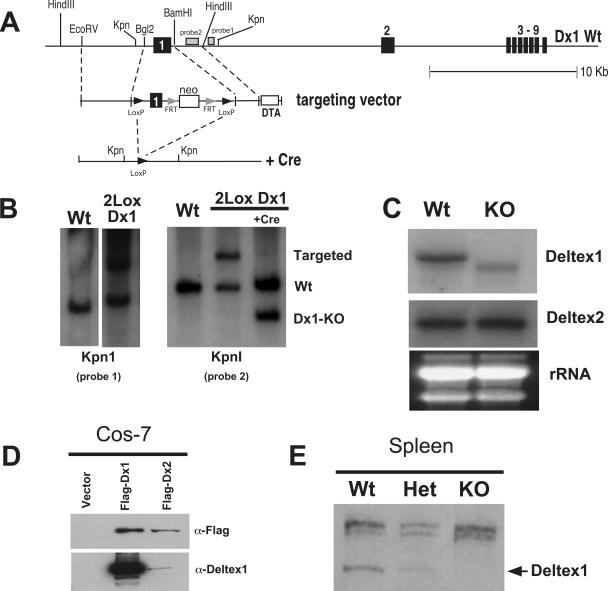
Although the above data reveal that overexpression of Deltex1 can inhibit Notch signals, it is not clear whether expression of endogenous Deltex1 in thymocytes is sufficient to prevent Notch signaling in vivo. To test this directly, we generated Dx1-KO mice. The first coding exon of Deltex1 was flanked by LoxP recombinase recognition sequences (36) using standard gene targeting technology in ES cells (64) (Fig. 2A). Results for one clone showing the targeted (2Lox Dx1) allele are shown in Fig. 2B. The targeted region was removed by transient transfection of the ES cells with Cre recombinase. Chimeric Dx1-KO founders were bred to C57BL/6 mice, and the progeny were interbred to generate homozygous Dx1-KO mice. Northern blot analysis of splenocytes from Dx1-KO mice revealed that the targeted allele produces a truncated mRNA (Fig. 2C). We examined expression of Deltex1 mRNA in splenocytes because Deltex1 mRNA is expressed only at low levels in DP thymocytes (which represent the majority of cells found in the thymus) (13, 14). Consistent with this, we were not able to detect Deltex1 protein by Western blot analysis of total thymocyte preparations. However, we were able to detect endogenous Deltex1 protein in splenocytes from wild-type but not Dx1-KO mice (Fig. 2E) using polyclonal antisera specific for Deltex1. The specificity of our antibodies for the C-terminal region of Deltex1 was confirmed by Western blot analysis of Cos7 cells transfected with recombinant Deltex1 or Deltex2 (Fig. 2D) or yeast cells expressing deletion mutants of Deltex1 (see Fig. S1A and B in the supplemental material). This analysis confirmed that although a truncated Deltex1 mRNA lacking the first coding exon of Deltex1 is transcribed in our Dx1-KO mice, expression of this RNA does not result in detectable Deltex1 protein. Even if very low levels of this truncated protein are expressed, this protein is unlikely to be functional, as it lacks the first WWE repeat within domain I, which is required for interaction with the ankyrin domains of Notch (16, 50, 84) (see Fig. S1C in the supplemental material).
FIG. 2.
Targeted deletion of Deltex1. (A) Targeting strategy for Dx1-KO mice. The first coding exon of Deltex1 (between the indicated BglII and BamHI sites) was flanked by LoxP recombinase sites to generate 2Lox-Deltex1 knock-in ES cells. Deltex1 was deleted by transient transfection of ES cells with Cre recombinase. (B) Southern blot of genomic DNA isolated from wild-type (Wt) or 2Lox-Deltex1 knock-in ES cells (2Lox Dx1) before or after Cre-mediated deletion. Genomic DNA was digested with KpnI, and Southern blots were hybridized with the probes indicated in panel A. (C) Northern blot of 10 μg of total RNA isolated from the spleen of Deltex1 knockout (KO) or littermate control (Wt) mice, showing that Deltex1 knockout mice produce a truncated mRNA. The Deltex1 probe is localized within the 3′ end of the Deltex1 coding region. Blots were striped and rehybridized with a probe for Deltex2, and the ethidium bromide-stained gel showing the rRNA bands before transfer is shown as a loading control. (D) Rabbit polyclonal antiserum raised against the C-terminal region of Deltex1 detects recombinant Deltex1 but not Deltex2. A Western blot of Cos-7 cells transfected with Deltex1 or Deltex2 and probed with either anti-Flag (upper panel) or anti-Deltex1 (lower panel) is shown. (E) Western blot with rabbit polyclonal antisera against the C terminus of Deltex 1 of total splenocytes from Dx1-KO or littermate control mice.
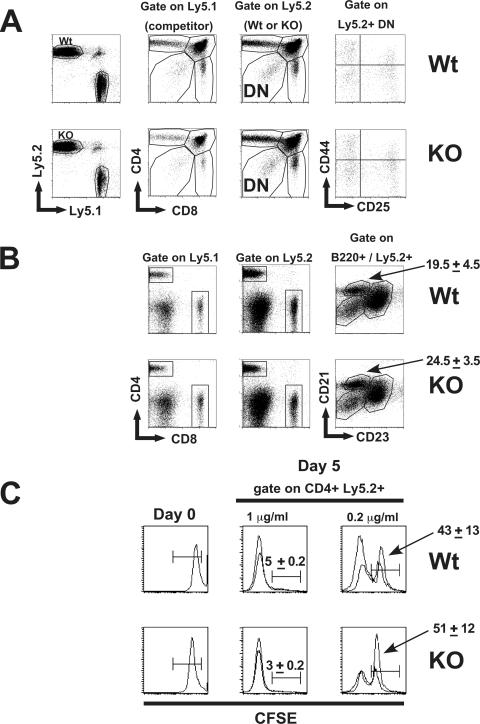
Preliminary analysis of Dx1-KO mice did not reveal any overt developmental abnormalities. In mice that had been back-crossed to C57BL/6 between two and six times, we could not detect any defects in the frequency of T cell, B cell, and myeloid cell types isolated from the bone marrow, thymus, and spleen among seven Dx1-KO and seven littermate mice in which Deltex1 was deleted either in the germ line or specifically in T-lineage cells through the expression of Cre recombinase under the control of the Lck-proximal promoter (data not shown). The absence of developmental abnormalities within the majority of B and myeloid cell populations found in the bone marrow is consistent with the notion that Notch signaling is not essential for development of these cell types. However, Notch signals are essential for T-cell development and for the development of marginal-zone B cells within the peripheral lymphoid organs (2, 47, 61-63). Therefore, we wanted to examine these cell populations using a more sensitive assay. To address the possibility that Dx1-KO cells may exhibit minor developmental abnormalities that may be overcome over the course of their differentiation, we generated mixed bone marrow chimeras in which Dx1-KO stem cells were forced to compete with normal bone marrow in vivo. Bone marrow from Dx1-KO or littermate controls that express the allelic marker Ly5.2 was mixed at a 1:1 ratio with bone marrow from wild-type mice that express Ly5.1 (competitor) and used to reconstitute lethally irradiated recipient mice. After 3 months, thymocyte and splenocyte populations were examined by FACS analysis for cell surface molecules. The results from this analysis, shown in Fig. 3A, reveal that stem cells from Dx1-KO mice differentiate normally into thymocytes, and normal ratios of CD4, CD8, and DN populations were present in the Ly5.2-versus-Ly5.1 fractions. Splenocytes from the chimeric mice described above did not reveal any defects in the differentiation of Dx1-KO stem cells into mature T or B cells based on their expression of CD4, CD8, or B220 (Fig. 3B). Notch signaling is essential for normal development of marginal-zone (MZ) B cells (71, 76), and Deltex1 has been reported to be highly expressed in these cells (71). Nevertheless, we did not observe any significant differences in the frequency of MZ B cells among the Dx1-KO cells (Fig. 3B). To test whether Dx1-KO T cells are able to proliferate in response to signals through the TCR, we labeled total splenocytes isolated from the above chimeric mice with CFSE and examined their proliferation in response to plate-bound anti-CD3 (Fig. 3C). This analysis did not reveal any major defects in the proliferative capacity of Dx1-KO T cells.
FIG. 3.
Normal T- and B-cell development in Dx1-KO stem cells. (A) Bone marrow pooled from pairs of Dx1-KO mice or littermate controls (both Ly5.2+) was mixed at a 1:1 ratio with bone marrow from normal (Ly5.1+) mice and used to reconstitute lethally irradiated recipients. After 3 months, thymus (A) was analyzed for expression of CD4+ and CD8+ cells among Ly5.1+ versus Ly5.2+ cells or for expression of CD44 and CD25 within the Ly5.2+ CD4− CD8− (DN) fraction. (B) Total splenocytes were analyzed for normal T- or B-cell development by expression of CD4, CD8, or B220. Immature, follicular, or marginal-zone B cells were identified within the B220+ fraction by expression of CD21 and CD23. Numbers are the average frequencies of marginal-zone cells found within the indicated region (two recipients per group). (C) In vitro proliferation of splenocytes from chimeric mice. Total splenocytes were labeled with CFSE and activated with plate-bound anti-CD3 at 1 μg/ml or 0.2 μg/ml. The level of CFSE fluorescence is shown for total splenocytes on day 0, or in the CD4+ Ly5.2+ fraction after 5 days of culture. Histograms show overlays for two mice analyzed per group, and numbers are the average values for the two mice analyzed.
Deltex1, Deltex2, and Deltex4 are expressed in thymocytes and can inhibit Notch signals.
The above data reveal that T-cell development is mostly normal in mice carrying a targeted deletion in the Deltex1 gene and could indicate that Deltex does not have an important role in regulating Notch signals during T-cell development. However, Deltex belongs to a family of at least four related molecules: Deltex1, Deltex2, Deltex3, and Deltex4. Deltex1 and Deltex2 were both shown to interact with the intracellular domain of Notch using a yeast two-hybrid assay, whereas Deltex3 is considerably smaller and does not interact with Notch-IC (37) (data not shown). In contrast, the most recently identified member, Deltex4, shares the highest degree of sequence similarity with Deltex1 (74) and is therefore likely to interact. Together, the above observations suggest that only Deltex2 and Deltex4 are likely to compensate for Deltex1 in our Dx1-KO mice. To address whether Deltex homologues are likely to compensate for loss of Deltex1, we first wanted to test if these molecules are expressed in thymocytes.
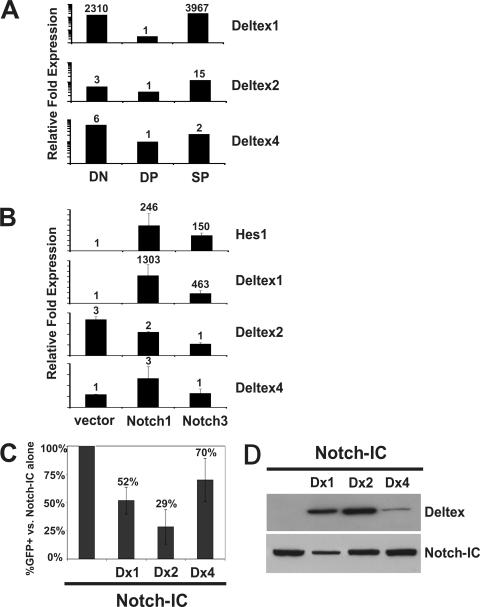
Our previous data revealed that Notch-responsive genes, including the Deltex1 gene, are highly expressed during the DN stages of thymocyte development, down regulated during the DP stage, and reinduced as thymocytes mature into the SP (CD4 or CD8 single-positive) stage (15). To test whether expression of other Deltex homologues is also regulated during thymocyte maturation, we sorted thymocytes from C57BL/6 mice based on their expression of CD4 and CD8 into DN, DP, and SP subsets to generate cDNA and examined expression of Deltex1, Deltex2, and Deltex4 mRNAs by TaqMan RT-PCR analysis (Fig. 4A). The mRNAs for all three Deltex homologues were readily detectable in thymocytes. Deltex1 exhibited the most dramatically regulated expression pattern, with a >2,000-fold difference in relative expression between the DN and DP stages. This expression pattern is highly characteristic of Notch-responsive gene expression, but to test more directly whether the Deltex mRNAs are induced by Notch signaling, we examined expression of Deltex mRNAs in the 1010 cell line transduced with retroviruses expressing empty vector, Notch1-IC, or Notch3-IC (Fig. 4B). While Hes1 and Deltex1 mRNAs were highly induced (100- to 1,000-fold) by Notch signals derived from either Notch1 or Notch3, expression of Deltex2 or Deltex4 was not changed by Notch signaling.
FIG. 4.
Deltex1, Deltex2, and Deltex4 are expressed in T cells, but only Deltex1 is induced by Notch signaling in T cells. (A) Expression of Deltex family members in sorted DN (CD4− CD8−), DP (CD4+ CD8+), and SP (CD4+ and CD8+) thymocytes by TaqMan RT-PCR. Relative expression is shown for cDNAs normalized to the value for HPRT. (B) Expression of Hes1, Deltex1, Deltex2, and Deltex4 in 1010 expressing empty vector or the intracellular domain of Notch1 or Notch3. The relative expression of each gene was normalized to that for HPRT. (C) All three Deltex family members can inhibit Notch signals. Cos-7 cells expressing a CBF1/RBPJκ-responsive element (8xCBF1) linked to GFP were transiently transfected with Myc-tagged Notch-IC alone or cotransfected with Myc-Notch-IC and Flag-tagged Deltex1, Deltex2, or Deltex4, and the total percent GFP+ cells was determined by FACS analysis. Average inhibition of GFP expression in cells cotransfected with Deltex1, Deltex2, or Deltex4 compared to that in cells transfected with Notch-IC alone is shown. Error bars show average data from four independent transfections using Notch1-IC, Notch2-IC, or Notch3-IC. (D) Western blot of cells used for panel C to compare the levels of Notch-IC (anti-Myc) or Deltex (anti-Flag).
To test whether all three Deltex homologues could block Notch signals, we employed a reporter cell line carrying a stably integrated plasmid containing the CBF1/RBPJκ binding element (8xCBF1) linked to the gene for GFP. Overexpression of Notch-IC in these cells activates CBF1/RBPJκ, which can be detected by FACS analysis for GFP expression. We cotransfected Myc-tagged Notch-IC with Flag-tagged Deltex1, Deltex2, or Deltex4 and analyzed GFP expression after 24 h (Fig. 4C). The overall levels of Notch-IC and Deltex family members were monitored by Western blot analysis for the Myc or Flag tag (Fig. 4D). Activation of GFP expression in the presence of the three Deltex proteins was plotted as the percent GFP+ cells in cultures coexpressing Deltex and Notch-IC compared to cultures expressing Notch-IC alone. Although we did detect differences in the ability of Deltex homologues to repress Notch signaling, the degree of inhibition visible in Fig. 4C correlated well with the overall level of Deltex expression shown in Fig. 4D, suggesting that all three Deltex homologues are able to inhibit Notch signals similarly.
Targeted deletion of Deltex1 and Deltex2.
The above data demonstrating that Deltex1, Deltex2, and Deltex4 are expressed in thymocytes and that all three Deltex homologues are able to block Notch signals suggest that functional redundancy between the three Deltex homologues could mask any defects in Deltex1-deficient mice. Therefore, we attempted to reduce expression of all three Deltex homologues in T-cell progenitors using a combinatorial approach of conventional gene targeting and RNA interference.
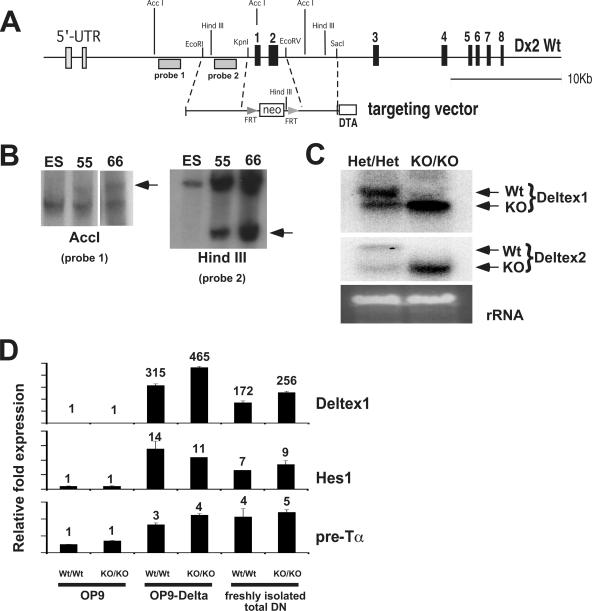
First, we generated Deltex2-deficient mice by gene targeting. The first two coding exons of Deltex2 were replaced with the gene for neomycin flanked by recognition sequences for the FLP recombinase (Fig. 5A). Two clones showing the targeted allele are shown in Fig. 5B. Cells from clone 66 were injected into blastocysts, and the resulting chimeric mice were bred to mice that express the FLP recombinase to remove the neomycin gene (18). We confirmed by Northern blot analysis that splenocytes from Dx2-KO mice produced a truncated mRNA (data not shown; see Fig. 5C for a similar analysis on Deltex1/Deltex2 double-knockout mice). Dx2-KO mice were viable and showed no overt developmental defects. We were also unable to detect any abnormalities in T- or B-cell development by FACS analysis for developmental markers (data not shown).
FIG. 5.
Generating Deltex1/Deltex2 double-knockout mice. (A) Targeting strategy for Dx2-KO mice. The first two coding exons of Deltex2 were replaced with neomycin, and the neomycin cassette was removed by breeding Dx2-KO mice to mice expressing the Flp recombinase. (B) Southern blot of genomic DNA isolated from ES cells used to generate Dx2-KO mice. Genomic DNA was digested with AccI or HindIII. Clone 66 was used to generate knockout mice. (C) Northern blot of splenocytes from Deltex1/Deltex2 doubly heterozygous (Het/Het) or double-knockout (KO/KO) mice. Deltex1/Deltex2 double-knockout mice produce truncated mRNAs for both Deltex1 and Deltex2. Northern blots were hybridized with probes that identify the 3′ coding region of either Deltex1 or Deltex2. (D) Notch-responsive genes are induced normally in thymocytes from Deltex1/Deltex2 double-knockout mice. Expression of Deltex1, Hes1, or pre-Tα in thymocytes from Deltex1/Deltex2 double knockout (KO/KO) or wild type littermate control (Wt/Wt) mice was analyzed by TaqMan RT-PCR. Deltex1 primers were designed within the 3′ untranslated region of Deltex1, which is outside the targeted region in Dx1-KO mice. cDNA was prepared from freshly isolated total DN or from sorted DN4 (Lin− CD44− CD25−) thymocytes cultured for 24 h on OP9 or OP9-Delta1. In cocultures, OP9 stromal cells were removed by magnetic cell sorting selection for CD45+ cells.
We bred the Dx2-KO mice to our Dx1-KO mice to generate double-knockout mice (Dx1-KO Dx2-KO). Since the genes for Deltex1 and Deltex2 are both found on chromosome 5, we bred homozygous Dx1-KO mice with homozygous Dx2-KO mice and bred the resulting double-heterozygous progeny to C57BL/6 mice to identify those that had undergone recombination such that both the Deltex1 and Deltex2 mutations were present on the same chromosome. This analysis revealed that recombination within the 15-Mb region between the Deltex1 and Deltex2 genes occurred with a frequency of approximately 1 out of 15. Northern blot analysis of splenocytes from mice that were heterozygous for both Deltex1 and Deltex2 or double-knockout mice revealed that the targeted Deltex2 allele produces a truncated mRNA (Fig. 5C). Although we were unable to detect the Deltex2 protein in normal splenocytes using a polyclonal antibody raised against the C-terminal region of Deltex2 (data not shown), the truncated Deltex2 mRNA produced in Deltex2-deficient mice is unlikely to code for a functional protein because the initiating ATG is deleted, and if a truncated protein is produced, it should not be functional because it lacks the first domain of Deltex2, which is essential for binding to Notch-IC (as shown for Deltex1 in Fig. S1 in the supplemental material). We were unable to detect any developmental abnormalities in the Dx1-KO Dx2-KO mice, and preliminary analysis did not reveal any defects in T- or B-cell development by FACS analysis (data not shown).
To test whether the intensity of Notch signaling is altered in thymocytes lacking both Deltex1 and Deltex2, we examined the expression of Notch responsive genes in thymocytes isolated from Dx1/Dx2 double-knockout mice or littermate controls. We compared expression of Hes1, Deltex1, and pre-Tα within the total DN subset of freshly isolated thymocytes that had been exposed to endogenous Notch ligands or in sorted DN4 thymocytes (Lin− CD44− CD25−) after 24 h of in vitro stimulation on OP9 stromal cells expressing empty vector (OP9) or the Notch ligand Delta1 (OP9-Delta1) (Fig. 5D). By this analysis, we were unable to detect any differences in the levels of Notch signaling in the Dx1/Dx2 double-knockout cells.
Notch signals are more potent in Deltex-deficient T-cell progenitors.
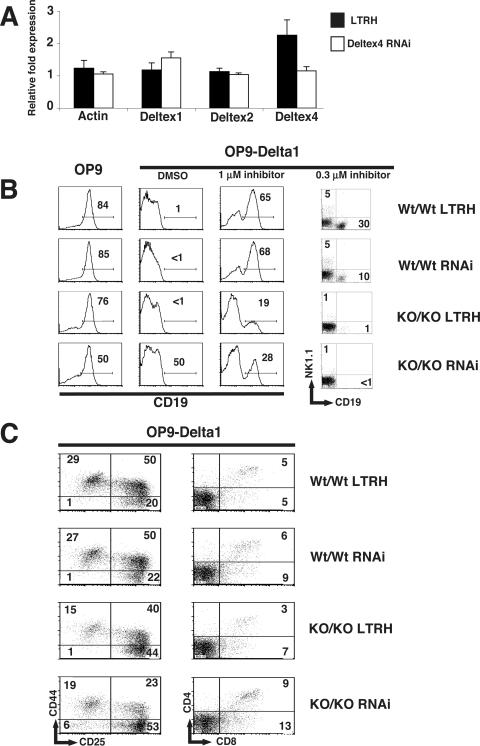
Although we were not able to detect any significant differences in expression of Notch-responsive genes in T-cell progenitors lacking both Deltex1 and Deltex2, it is possible that Deltex could regulate Notch signals independently of Notch-mediated transcriptional activation. Compounding this, our RT-PCR analysis shown in Fig. 4A revealed that Dx1-KO Dx2-KO cells likely express high levels of Deltex4. Therefore, we sought to reduce expression of Deltex4 in T-cell progenitors using RNAi. We generated two RNAi expression vectors (9) that each express an siRNA containing a 19-bp sequence derived from the coding region of Deltex4 and the extracellular domain of human CD4 as a marker for cells expressing the RNAi vector (see Materials and Methods). Coinfection of stem cell populations isolated from the fetal livers of C57BL/6 mice with retroviral supernatants containing the two Deltex4 siRNAs resulted in a 50% reduction of Deltex4 mRNA compared to cells transduced with the empty RNAi vector (LTRH) (Fig. 6A).
FIG. 6.
Notch signals are more potent in stem cells expressing low levels of Deltex1, Deltex2, and Deltex4. (A) Inhibition of Deltex4 mRNA in stem cells from fetal liver. Fetal liver cells from day 14 embryos were transduced with retroviral supernatants expressing empty vector (LTRH) or a pool of two Deltex4 siRNAs, and RNA was isolated after 4 days of culture. Expression of Deltex1, Deltex2, Deltex4, and β-actin was determined by TaqMan RT-PCR analysis. Relative cDNA expression for cDNAs was normalized to the value for HPRT. (B) Expression of CD19 on fetal liver stem cells derived from wild-type (Wt/Wt) or Deltex1/Deltex2 double-knockout (KO/KO) embryos infected with empty vector (LTRH) or the Deltex4 RNAis used for panel A (RNAi). Day 14 embryos were typed for Deltex1 and Deltex2 by PCR on genomic DNA, infected with the indicated retroviruses, and cultured in the presence of SCF and IL-7 for 2 days. Lineage-negative, human CD4+ stem cells were sorted and plated at 2,000 cells per well on OP9 or OP9-Delta1 in the presence of carrier (dimethyl sulfoxide) or increasing doses of a γ-secretase inhibitor. Expression of cell surface markers was assessed by FACS analysis on day 6 (histograms) or day 14 (dot plots). (C) Expression of T-lineage markers on fetal liver stem cells cultured on OP9-Delta1. DN subsets were analyzed by expression of CD44 and CD25 on day 6. CD4 and CD8 expression was examined on day 14. Numbers in quadrants indicate the frequency of cells in the quadrant shown. Data are representative of two independent experiments.
We used the above-described retroviruses to generate four stem cell populations expressing decreasing doses of Deltex. Mice that were heterozygous for both Deltex1 and Deltex2 were bred together to generate embryos that were homozygously wild type or homozygously knockout for both Deltex1 and Deltex2. The two groups of stem cells were infected with either the empty RNAi vector (LTRH) or a pool of the two Deltex4 RNAi vectors (RNAi). After 48 h, the transduced cells were sorted for expression of cKit and the retrovirally encoded human CD4 marker. We attempted to measure the intensity of Notch signaling within the four stem cell populations by examining their ability to differentiate into the T-cell lineage in vitro by culturing the cells on OP9 or OP9-Delta1 stromal cells in the presence of increasing doses of a γ-secretase inhibitor as described previously (43). Since all three Deltex homologues could inhibit Notch signaling in the in vitro assay (Fig. 4C), we hypothesized that Notch signals would be more potent within mutant T-cell progenitors that express reduced levels of Deltex1, Deltex2, and Deltex4.
First we examined whether Notch signals could inhibit B-lineage commitment equivalently in the stem cells that do and do not express Deltex proteins. Fetal liver stem cells differentiated efficiently into CD19+ B cells on OP9, whereas B-cell development was completely inhibited when these cells were cultured on OP9-Delta1 (Fig. 6B). When we added the γ-secretase inhibitor X to OP9-Delta1 cultures, Notch signals were partially blocked and fetal liver stem cells from wild-type mice expressing the empty vector or the Deltex4 siRNA were able to differentiate into either B cells or NK cells on OP9-Delta1. In contrast, we were not able to completely inhibit Notch signals in the double-knockout (KO/KO-LTRH) or triply deficient (KO/KO-RNAi) stem cells using the γ-secretase inhibitor. These data suggest that Notch signals are slightly more potent in the absence of Deltex.
Finally, we tested whether Notch signals could promote T-cell development equivalently in these stem cells. Notch signals promote the maturation of T-cell progenitors through the DN to DP stages of thymocyte development and are essential for promoting the proliferative expansion of thymocytes throughout these stages. T-cell maturation was largely unchanged in Dx1-KO Dx2-KO progenitors expressing the Deltex4 siRNA, although we did observe a slight increase in their maturation toward the DP stage after 14 days of culture on OP9-Delta1 (Fig. 6C). Since Notch signals are essential for promoting the proliferative expansion of DN3 thymocytes that have undergone β-selection (11, 12), we predicted that if Notch signaling is more potent in the absence of Deltex molecules, then the proliferation of T-cell progenitors would be enhanced in the Dx1-KO Dx2-KO progenitors expressing the Deltex4 siRNA. In contrast, we found that in cultures of Dx1-KO Dx2-KO stem cells expressing the Deltex4 siRNA, the total number of T cells recovered after 6, 14, or 20 days was similar to or slightly lower than that in wild-type controls (data not shown).
DISCUSSION
Deltex is a conserved member of the Notch signaling pathway that interacts with the intracellular domain of Notch receptors and has been shown to regulate Notch signals both positively and negatively in a variety of experimental systems. At the molecular level, Deltex has been proposed to regulate Notch signals by affecting the transcription of target genes (44, 82), altering the intracellular trafficking of Notch receptors (25, 26), or participating in a trimolecular complex that targets the Notch receptor for ubiquitin-mediated degradation (54). We identified the Deltex1 gene as a gene that is highly induced by Notch signals in T-cell precursors and demonstrated that expression of Deltex1 mRNA is dynamically modulated during different stages of thymocyte maturation. We and others have observed that Deltex1 mRNA is induced as thymocytes progress through the DN1 to DN3 stages (31), where T-lineage commitment is first established. Yet it has also been shown that overexpression of Deltex1 in T-cell progenitors inhibits Notch signals and prevents T-cell development. Together, these observations suggest that Deltex1 may have an important role in regulating Notch signals within developing T-cell precursors.
To test whether Deltex1 has an important role in regulating Notch signals during T-cell development, we generated knockout mice that were deficient in two of the three known mammalian Deltex homologues. In initial studies, we examined mice that were deficient in only Deltex1, and we were unable to detect any defects in thymocyte maturation in these mice. Our results are consistent with those published by Storck et al. (74) and extend their findings in several ways. First, we targeted the first coding exon of Deltex1, to be certain that Deltex1 expression is completely eliminated. (Storck et al. targeted the RING domain). Second, we tested the ability of Dx1-KO hematopoietic stem cells to differentiate in competitive mixed bone marrow chimeras. And finally, we tested whether the apparently normal development observed in Deltex1-deficient mice could result from functional redundancy with other Deltex homologues.
We examined the differentiation of T-cell precursors expressing decreasing doses of Deltex using a sensitive in vitro assay that allowed us to titrate the dose of Notch signals delivered to T-cell precursors by adding increasing doses of a γ-secretase inhibitor. This assay also permitted close monitoring of the temporal progression of thymocyte maturation through the DN to DP stages and examination of both cell autonomous and nonautonomous events. Considering the massive induction of Deltex1 mRNA in response to Notch signaling within thymocytes (Fig. 1B) and the dynamic expression of Deltex1 mRNA in DN, DP, and SP thymocyte subsets (Fig. 4A), we expected that among the three known Deltex homologues, loss of Deltex1 would have the greatest impact on thymocyte maturation. Therefore, we were very surprised to find that thymocyte development was completely normal in the absence of Deltex1 even in the context of Deltex2 deficiency. We were also not able to detect any significant differences in the intensity of Notch signals as determined by expression of Notch-responsive genes in thymocytes isolated from Deltex1/Deltex2 double-deficient mice (Fig. 5D). When we examined Notch-dependent inhibition of B-cell development, it did appear that Notch signaling is more potent in cells expressing lower levels of Deltex proteins. These data, shown in Fig. 6B, suggest that endogenous levels of Deltex proteins do block ligand-mediated Notch signals in a dose-dependent manner. However, since expression of Notch-responsive genes was not affected, Deltex may not regulate all aspects of Notch signaling equivalently.
Notch signals are essential for preventing B-cell development and for promoting thymocyte expansion (11, 12). Since the results shown in Fig. 6B suggested that Notch signals are more potent in Deltex-deficient progenitors, we expected Deltex-deficient progenitors to expand better than their wild-type controls. Surprisingly, we found a very minor defect in the expansion of Deltex-deficient progenitors (data not shown). These results suggest that Deltex may have opposing roles in Notch-induced lineage commitment and Notch-induced thymocyte expansion. The notion that Deltex may regulate a subset of Notch-mediated functions is consistent with data presented in at least three previously published reports. First, a recent report examining flies harboring a null mutation in the single Drosophila Deltex gene revealed that only a fraction of Notch-mediated functions were affected in the Deltex mutants and proposed that Deltex can enhance or inhibit Notch signaling in different settings (20). Maillard et al. revealed that within lymphoid cells, overexpression of Deltex1 can inhibit Notch signals required for T-lineage commitment (known to be dependent on Notch1), whereas Notch signals required for the development of marginal zone B cells (which are mediated through Notch2) are not inhibited by Deltex1 (48). Finally, Yun and Bevan proposed that overexpression of Deltex1 in bone marrow-derived stem cells inhibits Notch signals required for T-lineage commitment but does not affect thymocyte maturation through the early DN stages (83).
Our observation that Deltex inhibits some but not all Notch-mediated functions in our in vitro T-cell assay is interesting in light of the conflicting data suggesting a positive versus negative role for Deltex, and it will be of great interest to examine this issue further in other developmental settings. Nevertheless, the effect of Deltex appears to be extremely minor even in our stem cell populations expressing reduced levels of all three Deltex homologues. This result is consistent with observations made with Drosophila (20), where the phenotype observed in Deltex mutant flies was extremely mild. However, severe defects were observed when the Deltex null mutation was expressed in the context of additional mutations involving other Notch signaling components, and it will be interesting in future studies to examine the effect of Deltex deficiency in T cells lacking additional Notch signaling components.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant AI29802, the Howard Hughes Medical Institute, and predoctoral training grant GM07270.
We thank Katherine Forbush for performing blastocyst injections and for her expert advice and assistance in generating Dx1-KO mice. We also thank Rong Xu for generating the Cos7 reporter cell line and Michael Deftos for his invaluable help throughout the project.
Footnotes
Published ahead of print on 21 August 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akashi, K., T. Reya, D. Dalma-Weiszhausz, and I. L. Weissman. 2000. Lymphoid precursors. Curr. Opin. Immunol. 12:144-150. [DOI] [PubMed] [Google Scholar]
- 2.Allman, D., J. C. Aster, and W. S. Pear. 2002. Notch signaling in hematopoiesis and early lymphocyte development. Immunol. Rev. 187:75-86. [DOI] [PubMed] [Google Scholar]
- 3.Allman, D., A. Sambandam, S. Kim, J. P. Miller, A. Pagan, D. Well, A. Meraz, and A. Bhandoola. 2003. Thymopoiesis independent of common lymphoid progenitors. Nat. Immunol. 4:168-174. [DOI] [PubMed] [Google Scholar]
- 4.Amsen, D., J. M. Blander, G. R. Lee, K. Tanigaki, T. Honjo, and R. A. Flavell. 2004. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell 117:515-526. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, G., and E. J. Jenkinson. 2001. Lymphostromal interactions in thymic development and function. Nat. Rev. Immunol. 1:31-40. [DOI] [PubMed] [Google Scholar]
- 6.Aravind, L. 2001. The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 26:273-275. [DOI] [PubMed] [Google Scholar]
- 7.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. [DOI] [PubMed] [Google Scholar]
- 8.Baron, M., H. Aslam, M. Flasza, M. Fostier, J. E. Higgs, S. L. Mazaleyrat, and M. B. Wilkin. 2002. Multiple levels of Notch signal regulation (review). Mol. Membr. Biol. 19:27-38. [DOI] [PubMed] [Google Scholar]
- 9.Barton, G. M., and R. Medzhitov. 2002. Retroviral delivery of small interfering RNA into primary cells. Proc. Natl. Acad. Sci. USA 99:14943-14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bray, S. 1998. Notch signalling in Drosophila: three ways to use a pathway. Semin. Cell Dev. Biol. 9:591-597. [DOI] [PubMed] [Google Scholar]
- 11.Ciofani, M., T. M. Schmitt, A. Ciofani, A. M. Michie, N. Cuburu, A. Aublin, J. L. Maryanski, and J. C. Zuniga-Pflucker. 2004. Obligatory role for cooperative signaling by pre-TCR and Notch during thymocyte differentiation. J. Immunol. 172:5230-5239. [DOI] [PubMed] [Google Scholar]
- 12.Ciofani, M., and J. C. Zuniga-Pflucker. 2005. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat. Immunol. 6:881-888. [DOI] [PubMed] [Google Scholar]
- 13.Deftos, M. L., and M. J. Bevan. 2000. Notch signaling in T cell development. Curr. Opin. Immunol. 12:166-172. [DOI] [PubMed] [Google Scholar]
- 14.Deftos, M. L., Y. W. He, E. W. Ojala, and M. J. Bevan. 1998. Correlating notch signaling with thymocyte maturation. Immunity 9:777-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deftos, M. L., E. Huang, E. W. Ojala, K. A. Forbush, and M. J. Bevan. 2000. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity 13:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diederich, R. J., K. Matsuno, H. Hing, and S. Artavanis-Tsakonas. 1994. Cytosolic interaction between deltex and Notch ankyrin repeats implicates deltex in the Notch signaling pathway. Development 120:473-481. [DOI] [PubMed] [Google Scholar]
- 17.Eagar, T. N., Q. Tang, M. Wolfe, Y. He, W. S. Pear, and J. A. Bluestone. 2004. Notch 1 signaling regulates peripheral T cell activation. Immunity 20:407-415. [DOI] [PubMed] [Google Scholar]
- 18.Farley, F. W., P. Soriano, L. S. Steffen, and S. M. Dymecki. 2000. Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28:106-110. [PubMed] [Google Scholar]
- 19.Frise, E., J. A. Knoblich, S. Younger-Shepherd, L. Y. Jan, and Y. N. Jan. 1996. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc. Natl. Acad. Sci. USA 93:11925-11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuwa, T. J., K. Hori, T. Sasamura, J. Higgs, M. Baron, and K. Matsuno. 2006. The first deltex null mutant indicates tissue-specific deltex-dependent Notch signaling in Drosophila. Mol. Genet. Genomics 275:251-263. [DOI] [PubMed] [Google Scholar]
- 21.Godfrey, D. I., J. Kennedy, T. Suda, and A. Zlotnik. 1993. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3−CD4−CD8− triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J. Immunol. 150:4244-4252. [PubMed] [Google Scholar]
- 22.Godfrey, D. I., and A. Zlotnik. 1993. Control points in early T-cell development. Immunol. Today 14:547-553. [DOI] [PubMed] [Google Scholar]
- 23.Gorman, M. J., and J. R. Girton. 1992. A genetic analysis of deltex and its interaction with the Notch locus in Drosophila melanogaster. Genetics 131:99-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayday, A. C., D. F. Barber, N. Douglas, and E. S. Hoffman. 1999. Signals involved in gamma/delta T cell versus alpha/beta T cell lineage commitment. Semin. Immunol. 11:239-249. [DOI] [PubMed] [Google Scholar]
- 25.Hori, K., M. Fostier, M. Ito, T. J. Fuwa, M. J. Go, H. Okano, M. Baron, and K. Matsuno. 2004. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development 131:5527-5537. [DOI] [PubMed] [Google Scholar]
- 26.Hori, K., T. J. Fuwa, T. Seki, and K. Matsuno. 2005. Genetic regions that interact with loss- and gain-of-function phenotypes of deltex implicate novel genes in Drosophila Notch signaling. Mol. Genet. Genomics 272:627-638. [DOI] [PubMed] [Google Scholar]
- 27.Hoyne, G. F., M. J. Dallman, and J. R. Lamb. 1999. Linked suppression in peripheral T cell tolerance to the house dust mite derived allergen Der p 1. Int. Arch. Allergy Immunol. 118:122-124. [DOI] [PubMed] [Google Scholar]
- 28.Hoyne, G. F., I. Le Roux, M. Corsin-Jimenez, K. Tan, J. Dunne, L. M. Forsyth, M. J. Dallman, M. J. Owen, D. Ish-Horowicz, and J. R. Lamb. 2000. Serrate1-induced Notch signalling regulates the decision between immunity and tolerance made by peripheral CD4+ T cells. Int. Immunol. 12:177-185. [DOI] [PubMed] [Google Scholar]
- 29.Huang, E. Y., A. M. Gallegos, S. M. Richards, S. M. Lehar, and M. J. Bevan. 2003. Surface expression of Notch1 on thymocytes: correlation with the double-negative to double-positive transition. J. Immunol. 171:2296-2304. [DOI] [PubMed] [Google Scholar]
- 30.Ikawa, T., H. Kawamoto, S. Fujimoto, and Y. Katsura. 1999. Commitment of common T/Natural killer (NK) progenitors to unipotent T and NK progenitors in the murine fetal thymus revealed by a single progenitor assay. J. Exp. Med. 190:1617-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izon, D. J., J. C. Aster, Y. He, A. Weng, F. G. Karnell, V. Patriub, L. Xu, S. Bakkour, C. Rodriguez, D. Allman, and W. S. Pear. 2002. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity 16:231-243. [DOI] [PubMed] [Google Scholar]
- 32.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jameson, S. C., K. A. Hogquist, and M. J. Bevan. 1995. Positive selection of thymocytes. Annu. Rev. Immunol. 13:93-126. [DOI] [PubMed] [Google Scholar]
- 34.Joazeiro, C. A., and A. M. Weissman. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549-552. [DOI] [PubMed] [Google Scholar]
- 35.Jundt, F., I. Anagnostopoulos, R. Forster, S. Mathas, H. Stein, and B. Dorken. 2002. Activated Notch1 signaling promotes tumor cell proliferation and survival in Hodgkin and anaplastic large cell lymphoma. Blood 99:3398-3403. [DOI] [PubMed] [Google Scholar]
- 36.Kilby, N. J., M. R. Snaith, and J. A. Murray. 1993. Site-specific recombinases: tools for genome engineering. Trends Genet. 9:413-421. [DOI] [PubMed] [Google Scholar]
- 37.Kishi, N., Z. Tang, Y. Maeda, A. Hirai, R. Mo, M. Ito, S. Suzuki, K. Nakao, T. Kinoshita, T. Kadesch, C. Hui, S. Artavanis-Tsakonas, H. Okano, and K. Matsuno. 2001. Murine homologs of deltex define a novel gene family involved in vertebrate Notch signaling and neurogenesis. Int. J. Dev. Neurosci. 19:21-35. [DOI] [PubMed] [Google Scholar]
- 38.Kisielow, P., and H. von Boehmer. 1995. Development and selection of T cells: facts and puzzles. Adv. Immunol. 58:87-209. [DOI] [PubMed] [Google Scholar]
- 39.Krebs, L. T., M. L. Deftos, M. J. Bevan, and T. Gridley. 2001. The Nrarp gene encodes an ankyrin-repeat protein that is transcriptionally regulated by the notch signaling pathway. Dev. Biol. 238:110-119. [DOI] [PubMed] [Google Scholar]
- 40.Kruisbeek, A. M., M. C. Haks, M. Carleton, A. M. Michie, J. C. Zuniga-Pflucker, and D. L. Wiest. 2000. Branching out to gain control: how the pre-TCR is linked to multiple functions. Immunol. Today 21:637-644. [DOI] [PubMed] [Google Scholar]
- 41.Kuroda, K., H. Han, S. Tani, K. Tanigaki, T. Tun, T. Furukawa, Y. Taniguchi, H. Kurooka, Y. Hamada, S. Toyokuni, and T. Honjo. 2003. Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity 18:301-312. [DOI] [PubMed] [Google Scholar]
- 42.Lai, E. C. 2002. Protein degradation: four E3s for the notch pathway. Curr. Biol. 12:R74-R78. [DOI] [PubMed] [Google Scholar]
- 43.Lehar, S. M., J. Dooley, A. G. Farr, and M. J. Bevan. 2005. Notch ligands Delta 1 and Jagged1 transmit distinct signals to T-cell precursors. Blood 105:1440-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, W. H., and M. Z. Lai. 2005. Deltex regulates T-cell activation by targeted degradation of active MEKK1. Mol. Cell. Biol. 25:1367-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacDonald, H. R., F. Radtke, and A. Wilson. 2001. T cell fate specification and αβ/γδ lineage commitment. Curr. Opin. Immunol. 13:219-224. [DOI] [PubMed] [Google Scholar]
- 46.Maekawa, Y., S. Tsukumo, S. Chiba, H. Hirai, Y. Hayashi, H. Okada, K. Kishihara, and K. Yasutomo. 2003. Delta1-Notch3 interactions bias the functional differentiation of activated CD4+ T cells. Immunity 19:549-559. [DOI] [PubMed] [Google Scholar]
- 47.Maillard, I., S. H. Adler, and W. S. Pear. 2003. Notch and the immune system. Immunity 19:781-791. [DOI] [PubMed] [Google Scholar]
- 48.Maillard, I., A. P. Weng, A. C. Carpenter, C. G. Rodriguez, H. Sai, L. Xu, D. Allman, J. C. Aster, and W. S. Pear. 2004. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood 104:1696-1702. [DOI] [PubMed] [Google Scholar]
- 49.Martinez Arias, A., V. Zecchini, and K. Brennan. 2002. CSL-independent Notch signalling: a checkpoint in cell fate decisions during development? Curr. Opin. Genet. Dev. 12:524-533. [DOI] [PubMed] [Google Scholar]
- 50.Matsuno, K., R. J. Diederich, M. J. Go, C. M. Blaumueller, and S. Artavanis-Tsakonas. 1995. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development 121:2633-2644. [DOI] [PubMed] [Google Scholar]
- 51.Matsuno, K., M. Ito, K. Hori, F. Miyashita, S. Suzuki, N. Kishi, S. Artavanis-Tsakonas, and H. Okano. 2002. Involvement of a proline-rich motif and RING-H2 finger of Deltex in the regulation of Notch signaling. Development 129:1049-1059. [DOI] [PubMed] [Google Scholar]
- 52.McGill, M. A., and C. J. McGlade. 2003. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 278:23196-23203. [DOI] [PubMed] [Google Scholar]
- 53.Minter, L. M., D. M. Turley, P. Das, H. M. Shin, I. Joshi, R. G. Lawlor, O. H. Cho, T. Palaga, S. Gottipati, J. C. Telfer, L. Kostura, A. H. Fauq, K. Simpson, K. A. Such, L. Miele, T. E. Golde, S. D. Miller, and B. A. Osborne. 2005. Inhibitors of gamma-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat. Immunol. 6:680-688. [PubMed] [Google Scholar]
- 54.Mukherjee, A., A. Veraksa, A. Bauer, C. Rosse, J. Camonis, and S. Artavanis-Tsakonas. 2005. Regulation of Notch signalling by non-visual beta-arrestin. Nat. Cell Biol. 7:1191-1201. [DOI] [PubMed] [Google Scholar]
- 55.Mumm, J. S., and R. Kopan. 2000. Notch signaling: from the outside in. Dev. Biol. 228:151-165. [DOI] [PubMed] [Google Scholar]
- 56.Ordentlich, P., A. Lin, C. P. Shen, C. Blaumueller, K. Matsuno, S. Artavanis-Tsakonas, and T. Kadesch. 1998. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol. Cell. Biol. 18:2230-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palaga, T., L. Miele, T. E. Golde, and B. A. Osborne. 2003. TCR-mediated Notch signaling regulates proliferation and IFN-gamma production in peripheral T cells. J. Immunol. 171:3019-3024. [DOI] [PubMed] [Google Scholar]
- 58.Pear, W. S., L. Tu, and P. L. Stein. 2004. Lineage choices in the developing thymus: choosing the T and NKT pathways. Curr. Opin. Immunol. 16:167-173. [DOI] [PubMed] [Google Scholar]
- 59.Pirot, P., L. A. van Grunsven, J. C. Marine, D. Huylebroeck, and E. J. Bellefroid. 2004. Direct regulation of the Nrarp gene promoter by the Notch signaling pathway. Biochem. Biophys. Res. Commun. 322:526-534. [DOI] [PubMed] [Google Scholar]
- 60.Porritt, H. E., L. L. Rumfelt, S. Tabrizifard, T. M. Schmitt, J. C. Zuniga-Pflucker, and H. T. Petrie. 2004. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity 20:735-745. [DOI] [PubMed] [Google Scholar]
- 61.Radtke, F., A. Wilson, B. Ernst, and H. R. MacDonald. 2002. The role of Notch signaling during hematopoietic lineage commitment. Immunol. Rev. 187:65-74. [DOI] [PubMed] [Google Scholar]
- 62.Radtke, F., A. Wilson, and H. R. MacDonald. 2004. Notch signaling in T- and B-cell development. Curr. Opin. Immunol. 16:174-179. [DOI] [PubMed] [Google Scholar]
- 63.Radtke, F., A. Wilson, S. J. Mancini, and H. R. MacDonald. 2004. Notch regulation of lymphocyte development and function. Nat. Immunol. 5:247-253. [DOI] [PubMed] [Google Scholar]
- 64.Ramirez-Solis, R., A. C. Davis, and A. Bradley. 1993. Gene targeting in embryonic stem cells. Methods Enzymol. 225:855-878. [DOI] [PubMed] [Google Scholar]
- 65.Rangarajan, A., C. Talora, R. Okuyama, M. Nicolas, C. Mammucari, H. Oh, J. C. Aster, S. Krishna, D. Metzger, P. Chambon, L. Miele, M. Aguet, F. Radtke, and G. P. Dotto. 2001. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 20:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reizis, B., and P. Leder. 2002. Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes Dev. 16:295-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robey, E., and B. J. Fowlkes. 1994. Selective events in T cell development. Annu. Rev. Immunol. 12:675-705. [DOI] [PubMed] [Google Scholar]
- 68.Rothenberg, E. V. 2000. Stepwise specification of lymphocyte developmental lineages. Curr. Opin. Genet. Dev. 10:370-379. [DOI] [PubMed] [Google Scholar]
- 69.Rutz, S., B. Mordmuller, S. Sakano, and A. Scheffold. 2005. Notch ligands Delta-like1, Delta-like4 and Jagged1 differentially regulate activation of peripheral T helper cells. Eur. J. Immunol. 35:2443-2451. [DOI] [PubMed] [Google Scholar]
- 70.Sade, H., S. Krishna, and A. Sarin. 2004. The anti-apoptotic effect of Notch-1 requires p56lck-dependent, Akt/PKB-mediated signaling in T cells. J. Biol. Chem. 279:2937-2944. [DOI] [PubMed] [Google Scholar]
- 71.Saito, T., S. Chiba, M. Ichikawa, A. Kunisato, T. Asai, K. Shimizu, T. Yamaguchi, G. Yamamoto, S. Seo, K. Kumano, E. Nakagami-Yamaguchi, Y. Hamada, S. Aizawa, and H. Hirai. 2003. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity 18:675-685. [DOI] [PubMed] [Google Scholar]
- 72.Schmitt, T. M., and J. C. Zuniga-Pflucker. 2002. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 17:749-756. [DOI] [PubMed] [Google Scholar]
- 73.Shortman, K., and L. Wu. 1996. Early T lymphocyte progenitors. Annu. Rev. Immunol. 14:29-47. [DOI] [PubMed] [Google Scholar]
- 74.Storck, S., F. Delbos, N. Stadler, C. Thirion-Delalande, F. Bernex, C. Verthuy, P. Ferrier, J. C. Weill, and C. A. Reynaud. 2005. Normal immune system development in mice lacking the Deltex-1 RING finger domain. Mol. Cell. Biol. 25:1437-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tamura, K., Y. Taniguchi, S. Minoguchi, T. Sakai, T. Tun, T. Furukawa, and T. Honjo. 1995. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su (H). Curr. Biol. 5:1416-1423. [DOI] [PubMed] [Google Scholar]
- 76.Tanigaki, K., H. Han, N. Yamamoto, K. Tashiro, M. Ikegawa, K. Kuroda, A. Suzuki, T. Nakano, and T. Honjo. 2002. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat. Immunol. 3:443-450. [DOI] [PubMed] [Google Scholar]
- 77.Tanigaki, K., M. Tsuji, N. Yamamoto, H. Han, J. Tsukada, H. Inoue, M. Kubo, and T. Honjo. 2004. Regulation of αβ/γδ T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity 20:611-622. [DOI] [PubMed] [Google Scholar]
- 78.von Boehmer, H., I. Aifantis, J. Feinberg, O. Lechner, C. Saint-Ruf, U. Walter, J. Buer, and O. Azogui. 1999. Pleiotropic changes controlled by the pre-T-cell receptor. Curr. Opin. Immunol. 11:135-142. [DOI] [PubMed] [Google Scholar]
- 79.Washburn, T., E. Schweighoffer, T. Gridley, D. Chang, B. J. Fowlkes, D. Cado, and E. Robey. 1997. Notch activity influences the αβ versus γδ T cell lineage decision. Cell 88:833-843. [DOI] [PubMed] [Google Scholar]
- 80.Weng, A. P., Y. Nam, M. S. Wolfe, W. S. Pear, J. D. Griffin, S. C. Blacklow, and J. C. Aster. 2003. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol. Cell. Biol. 23:655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu, T., and S. Artavanis-Tsakonas. 1990. deltex, a locus interacting with the neurogenic genes, Notch, Delta and mastermind in Drosophila melanogaster. Genetics 126:665-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yamamoto, N., S. Yamamoto, F. Inagaki, M. Kawaichi, A. Fukamizu, N. Kishi, K. Matsuno, K. Nakamura, G. Weinmaster, H. Okano, and M. Nakafuku. 2001. Role of Deltex-1 as a transcriptional regulator downstream of the Notch receptor. J. Biol. Chem. 276:45031-45040. [DOI] [PubMed] [Google Scholar]
- 83.Yun, T. J., and M. J. Bevan. 2003. Notch-regulated ankyrin-repeat protein inhibits Notch1 signaling: multiple Notch1 signaling pathways involved in T cell development. J. Immunol. 170:5834-5841. [DOI] [PubMed] [Google Scholar]
- 84.Zweifel, M. E., D. J. Leahy, and D. Barrick. 2005. Structure and Notch receptor binding of the tandem WWE domain of Deltex. Structure (Cambridge) 13:1599-1611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.