Abstract
Chromatin remodeling at promoters of activated genes spans from mild histone modifications to outright displacement of nucleosomes in trans. Factors affecting these events are not always clear. Our results indicate that histone H3 acetylation associated with histone displacement differs drastically even between promoters of such closely related heat shock genes as HSP12, SSA4, and HSP82. The HSP12 promoter, with the highest level of histone displacement, showed the highest level of H3 acetylation, while the SSA4 promoter, with a lower histone displacement, showed only modest H3 acetylation. Moreover, for the HSP12 promoter, the level of acetylated H3 is temporarily increased prior to nucleosome departure. Individual promoters in strains expressing truncated versions of heat shock factor (HSF) showed that deletion of either one of two activating regions in HSF led to the diminished histone displacement and correspondingly lower H3 acetylation. The deletion of both regions simultaneously severely decreased histone displacement for all promoters tested, showing the dependence of these processes on HSF. The level of histone H3 acetylation at individual promoters in strains expressing truncated HSF also correlated with the extent of histone displacement. The beginning of chromatin remodeling coincides with the polymerase II loading on heat shock gene promoters and is regulated either by HSF binding or activation of preloaded HSF.
Chromatin changes at promoters of eukaryotic genes play an essential role in regulation of transcription. These changes can range from posttranslational modifications of single amino acid residues in histones to more widespread histone modifications and finally to nucleosome displacement from promoters. Although assembly of the transcription initiation complex is antagonized by the presence of nucleosomes at gene promoters, full nucleosome displacement from the promoter is not always observed. Nucleosome displacement events are usually accompanied by posttranslational modifications of histones. These posttranslational modifications, often occurring in a cascade manner influencing one another, lie at the foundation of the “histone code” hypothesis (25, 46). The most heavily characterized histone modifications are acetylation of lysines produced by the action of histone acetyltransferase (HAT)-containing complexes, such as SAGA, ADA, NuA3, NuA4, and others. The histone acetylation often leads to the loss of some histone-DNA bonds and to the formation of a distinct chromatin surface recognized by chromatin-remodeling coactivators bearing bromodomains (10, 22). These coactivators often belong to the class of ATP-dependent chromatin remodeling complexes, which include such multisubunit complexes as SWI/SNF, RSC, ISWI, and others. These chromatin remodeling complexes use the energy released from ATP hydrolysis to destabilize and finally push away promoter nucleosomes either in cis along the DNA (15, 31) or in trans, completely detaching histones from DNA (5, 28). It is not always clear if histone modifications are required for chromatin remodeling or vice versa, because different orders of recruitment of chromatin-remodeling and histone-modifying activities have been reported (1, 8, 11, 29, 41).
In cases where nucleosomes are removed from gene promoters, the comparison of experimental data for different gene promoters reveals a dissimilar extent of nucleosome displacement. In such well-documented cases as PHO5 and GAL promoters, completion of the nucleosome displacement process takes at least an hour and is in a range of 2- to 10-fold histone depletion relative to the initial level (13, 30, 41). The most remarkable example of extensive and fast chromatin remodeling so far described is nucleosome displacement at the HSP82 gene promoter. It starts to be detectable within 45 seconds of heat shock (52), and relative histone displacement is higher than for other genes (13, 30, 41). Similarly to the PHO5 promoter, a mild and transient increase in histone acetylation is detected at the HSP82 promoter prior to major nucleosome displacement (52). Yet, it is not clear if this histone acetylation is limited to the initial stages of chromatin remodeling or persists throughout the whole process and if the degree of histone acetylation changes.
Heat shock genes are primarily regulated by the broadly conserved heat shock factor (HSF), although regulators such as Msn2/4 and others were also shown to be involved for some HSP genes (6, 16). Yeast HSF, encoded by the essential HSF1 gene, contains two activation regions situated at the N and C termini of the HSF molecule. These activation domains differ in their relative activation potentials as well as in their functionalities during the time course of heat stress (38, 43). The activity of HSF is regulated via several distinct pathways, including monomer-trimer transition (37), phosphorylation, and other posttranslational modifications (21, 23, 45), as well as via repression by molecular chaperones interacting with HSF, thus blocking their own production (37, 47). It is assumed that the robust nucleosome displacement at heat shock gene promoters is regulated by HSF. Surprisingly, it was demonstrated that HSF bypasses in its function a number of critical coactivators and general transcription factors, such as TFIIA, TAF9 (a subunit of TFIID and SAGA), Kin28 (a vital subunit of TFIIH), Med 17, and Med22 (subunits of the Mediator complex) (3, 7, 33, 35), and even the C-terminal domain of polymerase II (Pol II) (34). Thus, the mechanisms of nucleosome displacement and initiation of transcription at heat shock gene promoters are not fully understood and may have unique characteristics.
In this study, by performing kinetics experiments we show that heat shock gene promoters differ from each other in the extent and timing of nucleosome displacement and histone H3 acetylation. While it has been previously shown that nucleosome displacement is preceded by transient histone acetylation at PHO5 (41) and HSP82 (52) promoters, we demonstrate that nucleosome displacement at HSP12, HSP82, and SSA4 promoters is accompanied by sustained and increasing acetylation of histone H3. The extent of H3 acetylation often correlates with the extent of chromatin remodeling. This correlation is observed not only between different genes but also for individual genes in strains bearing different truncations of the HSF activation domains. We also show that HSF is preloaded to HSP82 and SSA4 promoters in an inactive form, but not to the HSP12 promoter, and is activated upon heat shock mediating chromatin remodeling and Pol II recruitment.
MATERIALS AND METHODS
Yeast strains.
All Saccharomyces cerevisiae strains used in this study are derived from PS128 (MATa ade2-1 trp1-1 can1-100 leu2,3-112 his3-11 ura3 hsf1Δ2::LEU2 carrying HSF1 on a YCp50-based plasmid) (43). Truncated versions of HSF1 in a pNC160-based plasmid (43) were transformed into the PS128 strain, and the original plasmid containing full-size HSF1 and the URA3 marker was shuffled out by cultivation on 5-fluoroorotic acid-containing medium. The strain expressing myc-tagged H4 was obtained by transformation of PS128 with the plasmid pNOY436 (27).
Cultivation conditions.
Saccharomyces cerevisiae strains were cultivated at 30°C to early log phase in rich yeast extract-peptone-dextrose broth supplemented with 0.04 mg/ml adenine. Strains transformed with pNOY436 were grown similarly but in synthetic complete medium lacking tryptophan. For kinetics experiments, instantaneous upshift was achieved by rapidly mixing equal volumes of a 30°C culture with prewarmed 52°C medium and then incubating with shaking at 39°C for the times indicated.
ChIP analysis.
Chromatin immunoprecipitation (ChIP) was performed essentially as previously described (13) with the exception that protein A-magnetic beads instead of protein A-Sepharose beads were used to precipitate antigen-antibody complexes. Special attention was paid to the consistency of equal levels of sonication of cell lysates. Before immunoprecipitation, all samples were tested for the level of DNA fragmentation, and the mean size of DNA fragments was always 500 bp. Antibodies specific for the following epitopes were used: histone H3 total (ab1791; AbCam); myc (monoclonal antibody 9E10; Santa Cruz Biotechnology), diacetyl histone H3 (K9 and K14) (06-599; Upstate Biotechnology), acetyl histone H4(K16) and tetra-acetyl H4 (06-866; Upstate Biotechnology), Pol II-YSPT[pS]PS repeat of the Pol II C-terminal domain (the 4H8 monoclonal antibody recognizes both phospho- and nonphospho-Pol II; Upstate Biotechnology), HSF (rabbit antibody raised and characterized by us previously [12]). Immunoprecipitated DNA samples were used for real-time PCR with SYBR Green dye, and signals for individual gene promoters were normalized against the corresponding signal derived from the PHO5 promoter or from the chromosome V intergenic region (in the case of Pol II ChIPs) and to input and then against the non-heat-shock sample (0′), arbitrarily chosen as 1. Because we observed the loss of total histone signals upon heat shock, we also calculated the change in the acetylated histone/total histone ratio as acetylated histone abundance divided by total histone abundance, where relative abundance is an inverse value of relative displacement. For each DNA sample at least three consecutive dilutions of DNA were analyzed, making sure that the amplification rate was always optimal and the change in amplification signal was proportional to the change in the amount of DNA. In addition, no-DNA controls were always included, making sure that the primer-dimer formation was not detectable or never was comparable to the amplification from experimental samples. Experiments were typically repeated three times; error bars in the figures show standard deviations.
Primers for real-time PCRs were selected from a significant number of primers based on the PCR efficiency. Only those primer pairs were used that gave an amplification rate of at least 1.9 per PCR cycle during the linear amplification and did not produce primer-dimers. The sequences of PCR primers used in this study were as follows (coordinates are relative to ATG): PHO5 (−214 to −192, −20 to −48), HSP12 (−304 to −279, −82 to −107), HSP82 (−193 to −167, −37 to −69), SSA4 (−307 to −279, −70 to −98), chromosome V intergenic region (GCAATCAACATCTGAAGAAAAGAAAGTAGT, CATAATCTGCTGAAAAATGGCGTAAAT).
RESULTS
Differential displacement of histones from heat shock gene promoters.
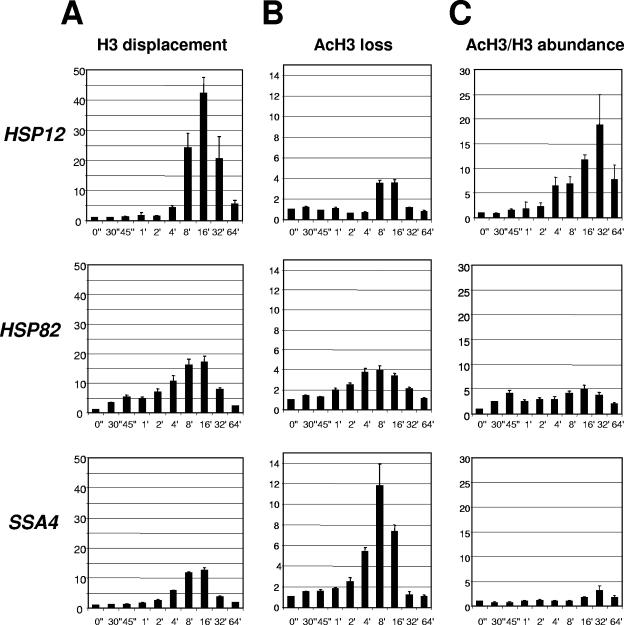
Upon induction of transcription, nucleosomes are displaced from heat shock gene promoters (9, 13, 52) as well as from PHO5 (5, 28) and Gal promoters (30). In most cases this was demonstrated using the ChIP technique and confirmed by other techniques. In these reports it was also demonstrated that the loss of histone signals during transcriptional induction is not likely the result of epitope masking affecting interpretation of ChIPs. In our experiments we performed time course experiments coupled with histone ChIP to test whether histone displacement occurred with similar kinetics and to a similar degree among highly related heat shock genes. The three genes analyzed, HSP12, HSP82, and SSA4, represent typical genes that are greatly induced upon heat shock. These genes express components of the molecular chaperone machinery of the cell. Yet, these genes exhibit interesting and important differences in their regulation. All of them are regulated by the principal transcriptional activator of heat shock genes, HSF. However, the promoters of these genes possess differently organized heat shock elements (HSEs), and additional regulators, such as Msn 2/4, are involved in regulation of HSP12 (6, 16). To monitor chromatin-remodeling events at indicated promoters, we used anti-H3 antibody raised against the C-terminal region of H3 (amino acids 125 to 135). This region is not known to be posttranslationally modified, thus allowing a measure of total histone H3 abundance. Utilization of this antibody for quantification of a change in total H3 was done previously by others (30, 32). The results of such experiments are traditionally presented as a drop (1 to 0, or 100% to 0%) in histone content during gene activation. This form of presentation restricts analysis of the data, since the closer the values are to 0 the more difficult it is to see fine differences. We decided to present results as an inverse value, which represents the degree of histone displacement and changes from 1 to ∞, thus better conveying fine differential chromatin remodeling. Figure 1A shows that, although all three gene promoters tested showed clear evidence of chromatin remodeling, the extent of histone displacement and kinetic profiles of this process are different. The HSP12 promoter showed the highest level of histone H3 loss, reaching a maximum of approximately 40-fold depletion relative to the non-heat-shock level, while the HSP82 and SSA4 promoters reached 18- and 13-fold depletion levels, respectively. Note that before heat shock all promoters showed occupancy with histone H3 equal to that of the PHO5 promoter, known to be fully occupied with nucleosomes under these conditions (Table 1). Another difference was in the timing of histone displacement. The HSP12 promoter was dormant for 2 minutes and started to show some changes only during the fourth minute after the temperature shift. Contrary to this, the HSP82 promoter showed threefold displacement of H3 after just 30 seconds of heat shock, which is consistent with the results obtained for this gene previously (52). The SSA4 promoter more closely resembled the HSP12 promoter, with chromatin changes lagging until the second minute after heat shock. Time course experiments coupled with ChIP thus indicate that the degree of chromatin remodeling and kinetic profiles of histone displacement are different for each heat shock gene promoter.
FIG. 1.
Kinetics of histone H3 displacement at heat shock gene promoters. (A) Fold displacement of histone H3 (y axis) over time (0 to 64 min) (x axis) relative to the non-heat-shock level (0"), which was arbitrarily set at 1. All real-time PCR values were normalized relative to the PHO5 promoter, which is known to contain positioned nucleosomes that do not change during heat shock (13). Values represent means ± standard deviations (n ≥ 3). (B) The same experiment as in panel A, except ChIPs were done with antibody raised against the acetylated form of H3. (C) Relative change of acetylated H3 (AcH3)/H3 abundance (y axis), calculated from the data in panels A and B. AcH3/H3 abundance = H3 displacement/AcH3 loss. (Note: the displacement [loss] value is an inverse value of abundance.)
TABLE 1.
Abundance of histones at indicated promoters before heat shock, relative to the PHO5 promoter
| Promoter | Strain expressing construct | Abundancea
|
|
|---|---|---|---|
| Total H3 | Acetylated H3 | ||
| HSP12 | WTb HSF | 1.1 ± 0.3 | 0.7 ± 0.1 |
| HSF(1-424) | 1.3 ± 0.2 | 0.6 ± 0.2 | |
| HSF(1-40Δ147-833) | 0.8 ± 0.2 | 0.9 ± 0.2 | |
| HSF(1-40Δ147-583) | 1.3 ± 0.2 | 0.9 ± 0.1 | |
| HSP82 | WT HSF | 0.8 ± 0.1 | 0.7 ± 0.1 |
| HSF(1-424) | 0.7 ± 0.2 | 0.6 ± 0.1 | |
| HSF(1-40Δ147-833) | 0.9 ± 0.1 | 1.1 ± 0.3 | |
| HSF(1-40Δ147-583) | 1.1 ± 0.1 | 0.8 ± 0.1 | |
| SSA4 | WT HSF | 0.8 ± 0.1 | 1.1 ± 0.1 |
| HSF(1-424) | 1.1 ± 0.2 | 1.1 ± 0.1 | |
| HSF(1-40Δ147-833) | 0.8 ± 0.2 | 1.1 ± 0.1 | |
| HSF(1-40Δ147-583) | 1.0 ± 0.1 | 1.2 ± 0.2 | |
Values (means ± standard deviations) were obtained by normalization of the real-time PCR signal of the specified promoter to the signal of the PHO5 promoter and to the input signal.
WT, wild type.
A higher level of chromatin remodeling often correlates with a higher level of histone H3 acetylation at HSP promoters.
We next sought to determine if the acetylated form of histone H3 follows the same kinetic profile of loss as total H3 displacement. If for an individual gene the acetylated form of H3 showed an equivalent degree of loss with similar kinetics as total H3 displacement, it would indicate that there is no change in histone acetylation. It would also indicate that acetylated H3, which is a fraction of the total H3 pool, is lost in the same manner as total H3. However, as Fig. 1B shows, there is a clear difference in the profiles. The HSP12 promoter, while showing maximal displacement for total H3 (Fig. 1A), shows only modest loss of the acetylated form of H3 (Fig. 1B). At the same time, the SSA4 promoter shows a much higher loss of acetylated H3, which is equivalent to the extent of total H3 displacement level for this gene (note difference in scales between panels A and B). The HSP82 promoter, while similar to HSP12 in terms of the maximum level of acetylated H3 loss (Fig. 1B), is weaker than HSP12 for displacement of total H3 (Fig. 1A). It is important to note that signals for all three genes are generated from the same immunoprecipitated samples, thus rendering it unlikely that immunoprecipitation by anti-acetyl-H3 antibody was less efficient for HSP12 or HSP82 in comparison with SSA4. The observed difference between the degree of displacement for total and acetylated forms of H3 indicates that the H3 acetylation was significantly different for these genes. In fact, the lower level of displacement of acetylated H3 compared to total H3 at the HSP12 and HSP82 promoters indicates that the fraction of acetylated H3 relative to total H3 is increased for these genes during the time course of activation after heat shock.
Knowing the relative fold displacement for the total and acetylated forms of H3, which is an inverse value of relative abundance, we can calculate the change in the ratio of the acetylated form to total H3 (Fig. 1C). Since loss of the acetylated form of H3 is relatively minor in comparison to the displacement of total H3, as in the case of HSP12, the acetylated H3/total H3 ratio is higher than 1, which was arbitrarily chosen as the value of the non-heat-shock sample (0′). For the HSP12 promoter the fraction of acetylated H3 that remains on the promoter or in its vicinity steadily increases during the time course of heat shock. Enrichment of acetylated H3 reaches a maximum of 20-fold after 30 min of heat shock and recedes afterward. For the HSP82 promoter, enrichment of acetylated H3 is much less, not exceeding fivefold. For the SSA4 promoter acetylated H3 enrichment is minimal, reaching just threefold after 32 min of heat exposure. For all three promoters the maximum acetylated H3/total H3 ratio is shifted in comparison with the total H3 displacement profile, indicating that it took approximately 16 min longer to reach maximum relative acetylation in comparison with the maximum total H3 displacement. Comparing all three promoters we can conclude that the relative change in acetylation level of histone H3 was highest for the HSP12 promoter, which shows the highest level of chromatin remodeling, and was smallest for the SSA4 promoter with the lowest level of chromatin remodeling.
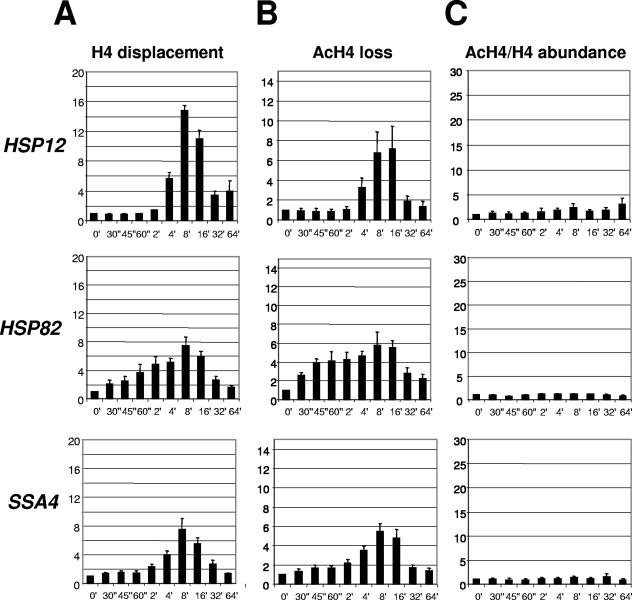
To test whether the degree of histone acetylation of histone H4 is similar to that of H3 in the nucleosome, we did analogous ChIPs utilizing anti-H4 antibodies. To test the kinetics of displacement of total H4, we used a strain expressing a myc-tagged version of H4 (27, 52) and used an antibody against the myc epitope. The reason for utilization of myc-tagged H4 is the unavailability of antibodies that recognize a region of histone H4 not modified in vivo. Figure 2A shows that the profiles of H4 displacement are similar to those of H3, with the HSP12 promoter showing the highest level and HSP82 and SSA4 promoters showing lower and approximately equal levels of H4 displacement. The absolute values for relative H4 displacement are lower than those of H3, possibly because of the differences in cross-linking abilities of H3 and H4 during the ChIP procedure. Contrary to what is observed for the acetylated form of H3, loss of the acetylated form of H4 almost parallels the kinetic profiles of total H4 for all three genes, indicating that the acetylation level of H4 did not change significantly. Accordingly, the ratio of acetylated H4 to total H4, showing the relative change in the acetylation level of histone H4, did not change significantly with heat shock (Fig. 2C). The HSP12 promoter revealed a minor two- to threefold increase in H4 acetylation, while HSP82 and SSA4 promoters did not display any change. Similar results, showing parallel changes in total and acetylated forms of H4 during the displacement process, were obtained with antibodies raised against acetylated K16 (Fig. 2) and the tetra-acetylated form of H4 (data not shown). Comparison of the results for H3 and H4 indicates that the observed effect of histone acetylation during displacement of nucleosomes is more specific to H3. A similar effect of higher acetylation of H3 in comparison to H4 during nucleosome displacement was reported for the Gal10 promoter (30).
FIG. 2.
Kinetics of histone H4 displacement at heat shock gene promoters. An experiment was performed as for Fig. 1, except ChIPs were done with anti-H4 antibodies. A strain expressing a myc-tagged version of H4 was utilized. (A) Displacement of total H4 (anti-myc antibody). (B) Loss of acetylated H4 (anti-tetra-acetyl H4 antibody). (C) Change in the acetylated H4 (AcH4)/total H4 ratio, calculated from data of panels A and B, as described in the Fig. 1 legend.
Deletion of activation domains in HSF drastically affects chromatin remodeling and histone H3 acetylation at heat shock gene promoters.
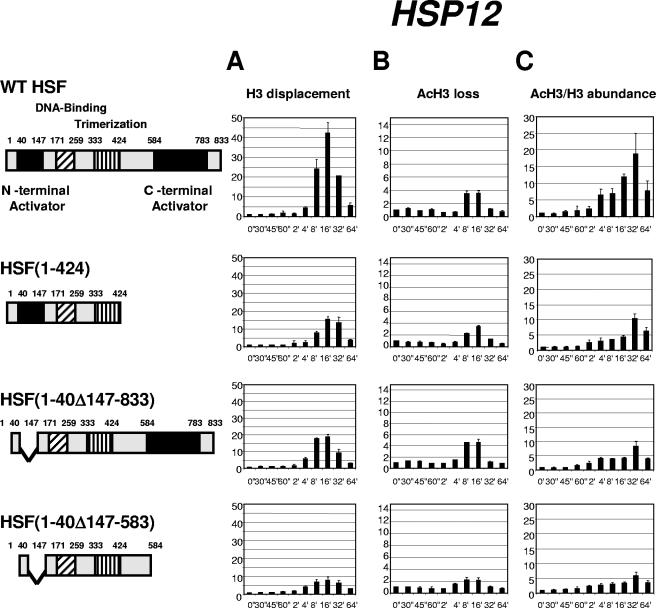
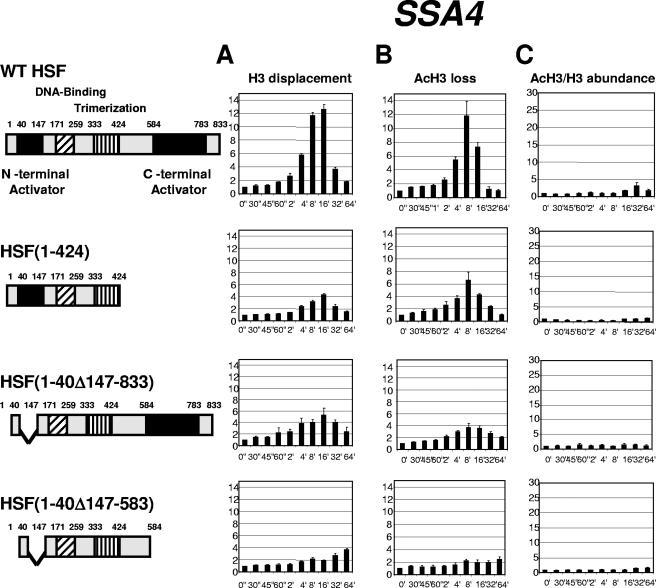
To test whether the correlation between the degree of histone H3 displacement and the acetylation level of H3 is dictated by the features of individual promoters recruiting different sets of transcription factors or is related to the function of HSF as a master regulator of heat shock genes, we utilized yeast strains expressing HSF containing deletions of its activation regions. This analysis is especially important for the HSP12 promoter, since the involvement of other activators, such as Msn2 and Msn4, was demonstrated for this gene (6, 16). Figure 3A demonstrates that the deletion of either one of the two activation regions in HSF [strains expressing HSF(1-424) or HSF(1-40Δ147-833)] has a modest effect on the total histone H3 displacement at the HSP12 promoter. Deletion of both HSF activation regions in the strain expressing HSF(1-40Δ147-583) has the strongest effect, decreasing the maximum H3 displacement from 40-fold to 8-fold. At the same time, profiles of loss for the acetylated form of H3 (Fig. 3B) were not affected as drastically. Only the strain expressing the maximally truncated HSF showed a twofold drop in comparison with the strain expressing wild-type HSF. Importantly, for all four strains tested the HSP12 promoter showed a decrease in the loss of the acetylated form of H3 below 1 at 2 minutes after heat shock. As the extent of loss is an inverse value of relative abundance, the drop in displacement below the initial non-heat-shock level means that the level of H3 acetylation was actually higher at this point in time than before heat shock. At this time point the displacement of total H3 had barely started. A burst of histone acetylation prior to nucleosome displacement was reported previously for the PHO5 and HSP82 genes (40, 52) and is considered an important step in nucleosome displacement. Our observation for the HSP12 promoter suggests that the onset of histone H3 acetylation precedes the beginning of nucleosome displacement by several minutes. Comparison of the ratios of acetylated H3 to total H3 for the four strains (Fig. 3C) shows that it was highest for the strain expressing wild-type HSF, dropped for the strains expressing either C- or N-terminally truncated HSF, and was significantly diminished for the strain expressing HSF depleted of both activation regions. Thus, the histone H3 acetylation level for the HSP12 promoter was highest for the strain with the highest level of histone displacement (wild type) and lowest for the strain expressing HSF(1-40Δ147-583), with only a modest level of total H3 displacement. For all four strains, the maximum H3 acetylation level was reached with a delay of approximately 16 min relative to maximum H3 displacement. The results described above also indicate that activation domains of HSF play a critical role in recruiting nucleosome remodeling activities, although the input of Msn2/4 is also possible since, when both regions in HSF were deleted, the level of chromatin remodeling at the HSP12 promoter still remained significant.
FIG. 3.
Kinetics of chromatin remodeling at the HSP12 promoter in strains expressing different versions of HSF. Data in panels A, B, and C are represented in the same format as in Fig. 1. Cartoons on the left represent maps of the HSF constructs expressed in the four different yeast strains. Note: expression of constructs represents the sole source of HSF.
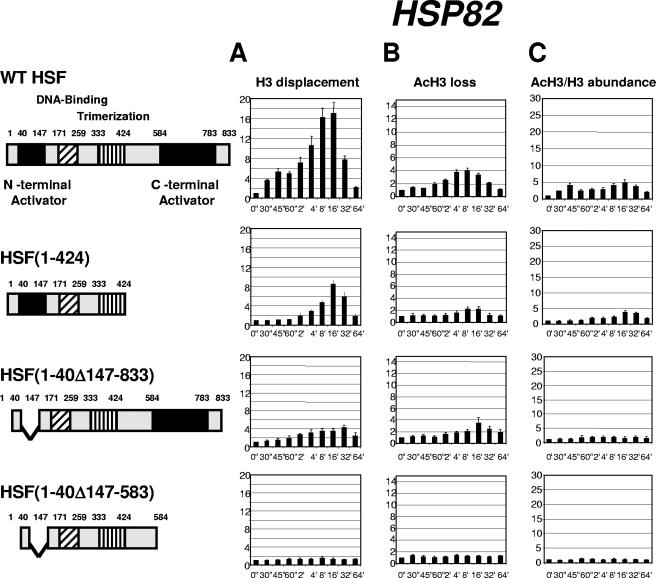
To extend this analysis, we examined the kinetics of chromatin rearrangements at the HSP82 promoter (Fig. 4), which is not known to be regulated by activators other than HSF (14). Although the effects of activation domain deletions were similar to that observed for the HSP12 gene, for some HSF deletions the histone displacement was more severely affected. The displacement profiles for total histone H3 showed that, despite being physically smaller, the N-terminal deletion in HSF(1-40Δ147-833) has a stronger effect on histone H3 displacement (Fig. 4A). For HSF(1-40Δ147-583), the histone displacement was completely abolished. This severe effect at the HSP82 promoter is consistent with the special role this gene plays for survivability at high temperatures for the yeast strain bearing the C-terminal activation domain deletion (36). Histone H3 acetylation for the HSP82 promoter was diminished for strains bearing deletions of individual activation regions and was completely abolished for HSF(1-40Δ147-583).
FIG. 4.
Kinetics of chromatin remodeling at the HSP82 promoter in strains expressing different versions of HSF. Data in panels A, B, and C are represented in the same format as in Fig. 1.
For the SSA4 promoter, which has the smallest degree of histone displacement relative to HSP12 and HSP82 genes, the kinetic profiles for total H3 and for its acetylated form parallel each other for all four strains tested (Fig. 5A and B), indicating that there is no significant change in the ratio of these forms of H3 during induction of SSA4. Correspondingly, the level of relative histone H3 acetylation is very modest, not exceeding fourfold even in strains expressing the full-size version of HSF. This low level of histone H3 acetylation is completely lost in the strain bearing the deletion of both activation regions in HSF (Fig. 5C).
FIG. 5.
Kinetics of chromatin remodeling at the SSA4 promoter in strains expressing different versions of HSF. Data in panels A, B, and C are represented in the same format as in Fig. 1.
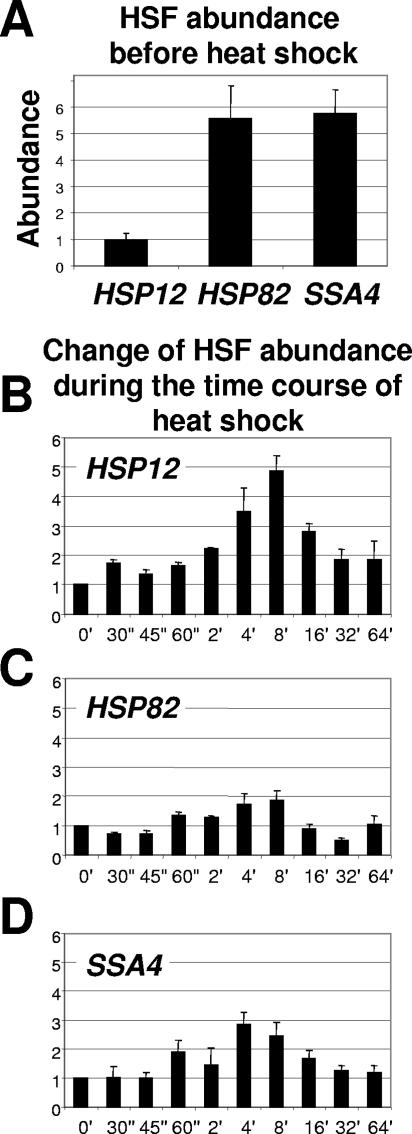
HSF abundance is different for the various HSP promoters before heat shock and is increased to different degrees upon heat shock.
Since we showed dependence of chromatin remodeling on HSF, we wanted to test if the abundance of HSF at these promoters is changing during the time course of heat shock. This is a controversial issue, because in higher eukaryotes binding of HSF to its cognate sites is heat inducible, while in yeast binding of HSF is believed to be constitutive (24, 44). Although recent data indicate that the majority of heat-inducible gene promoters in yeast are inducibly occupied by HSF, the occupation level can vary considerably (20). Indeed, Fig. 6 shows that occupation of the three promoters under investigation differs. Before heat shock the HSP12 promoter was not occupied by HSF, while those of HSP82 and SSA4 were (Fig. 6A). Correspondingly, the HSP12 promoter showed a significant increase in occupation of its promoter by HSF during the time course of heat shock (Fig. 6B), while occupation of the HSP82 and SSA4 promoters increased only slightly (Fig. 6C and D). Comparison of the kinetics of HSF loading (Fig. 6) for all three promoters with the kinetics of histone displacement (Fig. 1) indicates that the peak of HSF loading occurs earlier than the peak of histone displacement. This suggests that loading of HSF precedes histone displacement. These data are also consistent with our results of HSF deletion analysis and further strengthen the conclusion that histone displacement is HSF dependent. The comparison of the kinetics of HSF loading and the kinetics of histone displacement also suggests that HSF, although preloaded on the HSP82 and SSA4 promoters before heat shock, is probably inactive, because there is no detectable chromatin remodeling at any promoter before heat shock (Fig. 1 to 5) and, as we show below, Pol II is not recruited to any of the three promoters before heat shock.
FIG. 6.
Abundance of HSF is differentially increased at heat shock gene promoters upon temperature shift. (A) Abundance of HSF before heat shock at the indicated promoters. Real-time PCR signals for nonshocked samples were normalized to PHO5 and to input. (B) Change of HSF abundance at the HSP12 promoter during the time course of heat shock relative to the non-heat-shock level (0′), which was arbitrarily set at 1. (C and D) The same experiment as shown in panel B, except that signals were obtained for the HSP82 and SSA4 promoters, respectively.
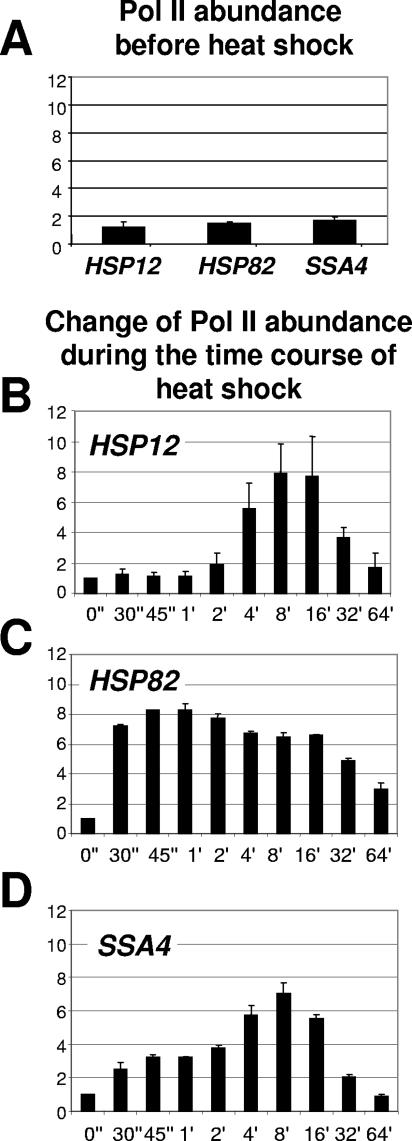
Pol II loading on the HSP promoters coincides with the beginning of chromatin remodeling.
The ultimate effect of heat shock on HSP genes is the initiation of transcription by Pol II. To establish if there is any connection between chromatin remodeling and change in abundance of Pol II at the analyzed promoters, we performed ChIPs with an anti-Pol II antibody that recognizes both phosphorylated and nonphosphorylated forms of Pol II. Figure 7A indicates that there is no Pol II detected at any promoter before heat shock. Absence of Pol II and presence of HSF (Fig. 6) at HSP82 and SSA4 promoters before heat shock further suggest that the prebound HSF exists in an inactive form. Upon heat shock all three promoters showed an increase in Pol II abundance, but with different kinetic profiles. The change in Pol II abundance at the HSP12 promoter occurred after the second minute of temperature shift and peaked between 8 and 16 minutes. In contrast, Pol II appeared at the HSP82 promoter in the first seconds after heat shock and stayed at a high level during the whole time course, decreasing somewhat after 30 min. For the SSA4 promoter, Pol II abundance gradually increased, peaking at 8 minutes after heat shock and decreasing afterwards. For all three promoters the increased level was approximately eightfold greater relative to the non-heat-shock level. Importantly, for all three promoters Pol II loading correlates with the onset of chromatin remodeling (Fig. 1).
FIG. 7.
Kinetics of Pol II loading on heat shock gene promoters. Data in panels A to D are represented in the same format as for Fig. 6, except that instead of normalizing to the PHO5 promoter normalization was done to the intergenic region of chromosome V (see Materials and Methods for primer sequences).
DISCUSSION
In this study we followed various processes taking place at three heat shock gene promoters during the time course of heat shock. The major findings were the following. First, comparing highly related inducible genes regulated by the same major heat shock gene activator, HSF, we found that these genes display different patterns of chromatin remodeling. While all studied promoters exhibited displacement of histones, the major difference is the degree of H3 acetylation occurring during the process of histone displacement. This difference is clearly seen if histone H3 acetylation is compared between the HSP12 and SSA4 promoters, with the former showing robust histone H3 specific acetylation and the latter showing very little effect. Second, the degree of histone H3 acetylation is often correlated with the degree of histone displacement. (i) Histone H3 acetylation increases during the time course of induction, with increasing displacement of histones for individual genes. (ii) The HSP12 promoter that displays the highest level of histone displacement also displays the highest level of histone H3 acetylation. (iii) Deletions of the activation domains in HSF decrease histone displacement and proportionally decrease histone H3 acetylation at individual promoters. Third, while significantly differing in amino acid sequences and functionalities, the two activation domains of HSF display an almost identical potential to mediate histone displacement and histone H3 acetylation at least at the HSP12 promoter. Fourth, HSF that mediates chromatin remodeling at analyzed promoters is activated via two distinct pathways, either by triggering activity of HSF already preloaded onto the promoter (HSP82 and SSA4) or by inducing HSF binding to the promoter (HSP12). These and other observations are discussed below.
Differential regulation of chromatin remodeling of heat shock gene promoters.
Upon heat induction, promoters of heat shock genes undergo drastic and very fast changes associated with the efficient eviction of nucleosomes (52). This chromatin remodeling surpasses in its speed and efficiency the well-characterized nucleosome displacement at the promoters of PHO5 (5, 28) and Gal10 (30) genes. Previous work on HSP82 (52) and PHO5 (41) showed that there is a mild transient histone acetylation prior to the major nucleosome displacement. At the HSP82 promoter, this acetylation has been shown as an approximately 1.5-fold increase in the abundance of acetylated forms of histones preceding major nucleosome displacement. Our results for the HSP82 promoter do not confirm such a transient and mild change of histone acetylation for the indicated promoter, probably because of a slight difference in the experimental approach. We used real-time PCR, while Zhao and coauthors (52) used conventional PCR. Although we used a very rigorous approach for selection of primers and control of optimal conditions for PCR (see Materials and Methods), we found that a change of 1.5-fold was often within the statistical error between repeated experiments. Although we cannot with absolute certainty detect any transient histone acetylation at the HSP82 promoter prior to histone displacement, we show that the histone H3-specific acetylation occurs during displacement. For the HSP82 promoter, this H3-specific acetylation peaks at a fivefold level of enrichment relative to the non-heat-shock level (Fig. 1). When we compare the H3 acetylation level for other promoters, it appears that it varies drastically, reaching a 20-fold increase of acetylated H3 fraction at the HSP12 promoter and only 3-fold for SSA4 (Fig. 1C). The dynamic character of H3 acetylation and the masking of this process by nucleosome loss could be the reason why histone acetylation during nucleosome displacement has not been previously reported for heat shock genes. Although we report an increase in acetylation of histone H3 during histone displacement, this increase in H3 acetylation is in reference to the fraction of histone H3 remaining at the promoter or in its vicinity during increasing histone displacement. The enrichment of the acetylated form of H3 could be a result of preferential displacement of the unacetylated form of H3, which would contradict the existing concept of gene activation, or a result of increased histone acetylation activity associated with nucleosome displacement as it was suggested previously (41, 52). Based on our experimental results we cannot determine if histone H3 acetylation is a by-product of chromatin remodeling or a required step as it was suggested previously. Yet, we see a significant difference in the level of H3 acetylation, especially if the analyzed promoters are compared to each other (Fig. 1). What could be behind such diverse variations in histone H3 acetylation and the extent of chromatin remodeling between heat shock gene promoters?
Since all three promoters are regulated primarily by HSF, it is necessary to consider the HSE architecture in these promoters. A typical HSE contains three or more sequential inverted repeats of the sequence nGAAn. In natural promoters, this consensus sequence is rarely preserved. Currently, HSEs are separated into three groups: perfect, gapped, and stepped. A perfect HSE has all three inverted repeats in a contiguous array (nTTCnnGAAnnTTC) (2, 39, 48, 49). Gapped HSEs have two consecutive inverted sequences, with the third sequence separated by 5 bp (42). Stepped HSEs have 5-bp gaps separating all three modules (50). While the HSP82 and SSA4 promoters have a perfect match to the gapped HSE consensus, the HSP12 promoter has a stepped HSE with several mismatched nucleotides. Stepped HSEs, especially with deviation from consensus, are known to be bound by HSF in an inducible manner (14, 20), while perfect and stepped HSEs without mismatch are usually constitutively occupied by HSF in yeast (19, 24, 44). These notions are consistent with our results of HSF occupancy (Fig. 6), showing constitutive occupation for the SSA4 and HSP82 promoters and inducible occupation of the HSP12 promoter. The HSP82 and SSA4 promoters, although possessing similar types of HSEs, nevertheless have different types of regulation. While the HSP82 gene is known to change the expression level 20- to 40-fold upon heat shock (13, 14), the SSA4 gene is much more inducible, changing its expression 3,000-fold (reference 51 and our unpublished data from the same experiments reported in reference 13). The lower inducibility range of the HSP82 gene probably stems from the fact that this gene has a certain level of transcription before heat shock, which can be reduced 50-fold by deletion of its major HSE (14, 18). The SSA4 gene, in contrast, has no detectable level of basal transcription (51), which suggests that it is possibly repressed before the induction stimulus. This repression prior to heat shock has been shown for the similar highly inducible and related SSA3 gene, where it is likely determined by Spt2 (4). Another possibility for SSA4 repression could be the function of the URS1 sequence in the SSA4 promoter. Under non-heat-shock conditions, URS1 is usually bound by the Ume6-Sin3-Rpd3 repressor complex containing RPD3 histone deacetylase (26).
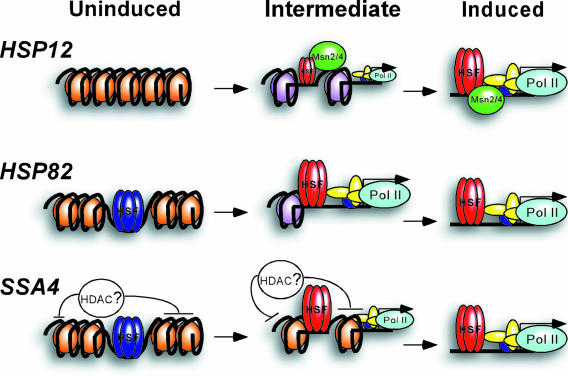
In light of the previously published experimental data described above, some aspects of differential chromatin remodeling at the three promoters we analyzed have now become clearer. Since the HSP82 promoter is probably poised for induction (HSF is preloaded and the promoter is not repressed, as probably is the case of SSA4), it shows the immediate beginning of chromatin remodeling, the onset of which coincides with the fast loading of Pol II. In contrast, the SSA4 promoter is possibly repressed by a factor causing a low level of histone H3 acetylation (Fig. 1 and 5) and delays recruitment of Pol II (Fig. 7D), even though HSF is preloaded. With the HSP12 promoter, the situation is different and is determined by the gradual loading of HSF, absent on this promoter before heat shock (Fig. 6A and B). To summarize (Fig. 8), our experiments as well as previously published data suggest that each gene is regulated in its unique way, which reflects the extent and kinetic profiles of chromatin remodeling taking place at each promoter. However, all three promoters display extensive histone displacement by a mechanism which is not yet determined and requires further analysis.
FIG. 8.
Dramatic histone displacement during activation of heat shock genes is not uniformly associated with robust histone H3 acetylation. The molecular model schematically represents differential processes at the three studied heat shock gene promoters during induction of transcription. Acetylated H3-enriched nucleosomes in the intermediate state at the HSP12 and HSP82 promoters are represented in purple. Nucleosomes with unacetylated H3 are shown in brown. Inactive HSF in the uninduced state is represented in blue, and activated HSF is shown in red. Intermediate levels of HSF and Pol II loading are represented by scaled-down cartoons for the corresponding proteins. Histone displacement is represented by a diminishing amount of nucleosomes in the intermediate state. Intermediate-state cartoons roughly represent the situation at promoters after 2 minutes of heat shock.
Correlation between histone H3 acetylation and histone displacement at heat shock gene promoters.
By analyzing differences in chromatin remodeling at three promoters, we observed that the level of histone H3 acetylation often correlated with the extent of histone displacement. First, this correlation is observed if we compare different time points of the kinetic experiments for the same promoter. The degree of histone displacement gradually increases over time for all three promoters tested. In each case, and especially obvious for the HSP12 and HSP82 promoters, the acetylation level of H3 also increases (Fig. 1C). Second, the three promoters analyzed in this study exhibited different extents of histone displacement. In this comparison we saw again that the HSP12 promoter with the highest level of histone displacement had the highest level of histone H3 acetylation, while the HSP82 and SSA4 promoters with lower levels of histone displacement displayed lower degrees of H3 acetylation. The correlations between histone displacement and histone H3 acetylation for the HSP82 and SSA4 promoters are not so clearly observed, because the relative level of histone H3 acetylation is relatively low and individual regulation of promoters by various factors (see above) is involved.
Correlation between the extent of chromatin remodeling and the acetylation of histone H3 is also noticeable when we compare events at an individual promoter in strains bearing differentially affected versions of HSF. It is most apparent for the HSP12 promoter, where the level of histone H3 acetylation is the highest (Fig. 3). Here, a deletion of either one of two HSF activation domains has a similar diminishing effect on histone displacement and histone H3 acetylation. Deletion of both activation regions in HSF also has strong effects on both histone H3 acetylation and displacement. For HSP82 and SSA4 promoters, similar trends are not so obvious, probably because the acetylation level of histone H3 is relatively low, even in the wild-type strain.
The H3-specific posttranslational modifications were shown to be important for subsequent changes in chromatin during transcriptional activation. In fact, similar H3-specific acetylation has been demonstrated for the Gal10 promoter (30). In this case a two- to threefold loss of histone H3 was paralleled by a mild increase in histone H3 acetylation. Mild histone acetylation preceding nucleosome loss at HSP82 and PHO5 promoters is considered to be an important step required for chromatin remodeling (40, 52). In our study we have demonstrated that histone H3 acetylation continues after histone displacement has been initiated and reaches a 20-fold increase at the HSP12 promoter. It is tempting to speculate that this robust histone H3-specific acetylation is an important step in chromatin remodeling at least for the HSP12 promoter. As suggested by the histone code hypothesis, increasing histone acetylation may additionally attract bromodomain-containing chromatin-remodeling complexes (22), promoting robust histone displacement at the HSP12 promoter. Another way to interpret the increase of the fraction of acetylated H3 associated with histone displacement is to assume that the ratio of HAT to its substrate is increasing during histone loss, thus increasing acetylation of the remaining histones. We do not favor this interpretation, because it suggests that histone acetylation plays a passive role in chromatin remodeling. Also, for the HSP12 and SSA4 promoters we see that histone H3 acetylation increases after histones start to return during attenuation. This is manifested as a shift in the maximum relative histone acetylation in comparison to the maximum histone H3 displacement, which is observed at the HSP12 promoter for all four strains expressing different variants of HSF (Fig. 3). For the SSA4 promoter, a similar effect is observed only for the wild-type strain, where histone H3 acetylation is still detectable (Fig. 5). This observation suggests that the balance between histone acetylation and deacetylation remains shifted toward the former, even during return of nucleosomes. This in turn may indicate that recruitment of HATs and recruitment of activities leading to the nucleosome displacement are separately regulated.
Although HSF plays the major role in histone displacement and transient histone H3 acetylation, other stress-related transcription activators, such as Msn2 and Msn4 in the case of the HSP12 promoter, could be involved. In fact, while the deletion of both HSF activation regions practically eliminates H3 displacement at the HSP82 promoter, the H3 displacement level at the HSP12 promoter remains substantial. This may be attributable to the function of Msn2/4. The acetylation of histone H3 prior to the beginning of histone displacement observed only at the HSP12 promoter and at similar level for all four strains expressing variants of HSF (Fig. 3) might also be the function of Msn2/4. For the HSP82 promoter, where HSF is the major, if not the only, activator, double deletion of HSF activation regions virtually eliminates H3 displacement. This observation is also consistent with the previous report that it is the HSP82 gene that is critical for yeast survivability at elevated temperatures in the strain expressing C-terminally truncated HSF (36).
Alternative pathways of histone displacement.
An important point emerging from our data is that although the three coregulated genes we studied display robust histone displacement during induction, they reach their maximally histone-stripped state via two distinct chromatin-remodeling pathways. For the HSP12 and HSP82 promoters (especially for the former), chromatin changes are associated, and often correlated, with dramatic enrichment of promoter chromatin with acetylated histone H3, while for the SSA4 promoter they are not. Yet, all three promoters show dramatic depletion of histones. The possibility of distinct mechanisms differentially associated with histone H3 acetylation was demonstrated recently for the GAL10, BAT1, GDH1, and TGP1 genes (30). These genes are regulated by different activators and, for the last three genes, histone acetylation is observed without histone displacement. We show here different pathways of chromatin remodeling for very closely related and coregulated genes. Our data argue that the robust histone displacement from promoters is not necessarily associated with equally robust histone acetylation. The extremely high histone H3 acetylation in the case of the HSP12 promoter might be caused by cooperative action of HSF and Msn2/4 activators. The absence of significant histone H3 acetylation at the SSA4 promoter might be the action of a specific histone deacetylase. The intermediate behavior of the HSP82 promoter could be due to the absence of any other regulating factors except HSF. Whatever the cause of such differential behavior, we have demonstrated the existence of two distinct chromatin-remodeling pathways: one associated (for the HSP12 and HSP82 promoters) and the other one not associated with robust histone H3 acetylation (for the SSA4 promoter) during histone displacement (Fig. 8). This is an interesting observation, because it implies that a histone-acetylated platform recognized by bromodomain-containing chromatin-remodeling complexes is not uniformly required for the robust histone displacement at gene promoters during induction of transcription.
HSF activity is regulated via two distinct pathways.
Our data indicate that an important factor determining the differences in kinetic profiles of histone H3 displacement at the three promoters is the difference in abundance of HSF prior to heat shock. HSF is at low abundance at the HSP12 promoter before heat shock. That explains a delay in the beginning of chromatin remodeling for this gene. In contrast, HSF loading at the HSP82 and SSA4 promoters is already high before heat shock. Consistent with this is faster initiation of histone displacement at both promoters that is somewhat delayed at the SSA4 promoter, probably in connection with lower H3 acetylation. For the HSP12 promoter, where the kinetics of HSF loading is clearly defined, the peak of HSF loading precedes the peak of H3 displacement, suggesting that HSF is mediating this chromatin remodeling.
Differential loading of HSF raises an interesting issue of regulation of HSF activity. This activator is regulated at multiple levels: monomer-trimer transition with only the trimer being able to interact with the heat shock element sequence of promoter DNA (37); phosphorylation of HSF at multiple sites, some of which are activating and some repressing (21, 23, 45); and repressing interactions with molecular chaperones, many of which are products of heat shock genes, thus forming a feedback loop regulation (37, 47). One possibility for why HSF does not interact efficiently with the HSP12 promoter before heat shock is that this promoter contains a very degenerate HSE. The inducible binding of yeast HSF to such degenerate HSEs in a number of yeast promoters was demonstrated previously (14, 20). Of the several ways of regulating HSF activity mentioned above, monomer-trimer transition appears to be likely involved in DNA binding ability. This pathway of HSF regulation was demonstrated for higher eukaryotes but not for yeast. Our results showing heat-inducible binding of HSF to the HSP12 promoter and results from other groups (14, 17, 20) suggest that perhaps some form of regulated monomer-trimer transition functions in yeast as well as in higher eukaryotes. The activation of prebound HSF at the HSP82 and SSA4 promoters is likely explained by feedback loop regulation, as suggested previously (37, 47).
Another interesting aspect regarding the function of HSF is that for the HSP12 and SSA4 promoters the individual input of each of the two HSF activation domains in histone displacement seems to be almost equal (Fig. 3 and 5). This is even more surprising considering that these two domains do not have homology. It remains to be determined how these two different domains similarly organize specific recruitment of protein machineries leading to similar results in terms of chromatin remodeling.
Pol II loading on heat shock gene promoters precedes major chromatin remodeling.
Although HSF is sufficiently abundant at HSP82 and SSA4 promoters before heat shock, it appears to be inactive. This was apparent because chromatin remodeling was not initiated and none of the three promoters showed any presence of Pol II before a temperature shift. However, upon heat shock all three promoters showed an increase in Pol II loading. A striking example of this is observed at the HSP82 promoter. This promoter shows a sudden increase in Pol II abundance during the first 30 seconds of heat shock. Moreover, the level of Pol II at 30 seconds is not significantly different from the maximum at 1 min (Fig. 7C). At this time point chromatin remodeling is just in the beginning of its development (Fig. 4). A similar tendency is seen for the HSP12 and SSA4 genes. After 4 minutes the abundance of Pol II at the HSP12 promoter is close to maximum, while the displacement of H3 at this time point is very minor. The SSA4 promoter has a similar trend, although Pol II loading at this promoter is substantially delayed despite HSF being preloaded similarly to the HSP82 promoter. This delay could be connected to the anomalously low level of histone H3 acetylation at this promoter and/or this promoter being repressed before heat shock (see above). To summarize, it appears that Pol II loading on all three promoters coincides with or even slightly precedes the onset of chromatin remodeling. That could suggest that recruitment of a Pol II-containing protein complex (including the Mediator complex) brings major chromatin-remodeling activity to the promoters. This is supported by the fact that Pol II precedes major chromatin remodeling (compare Fig. 1A and 7B to D). To further define steps and the mechanism of chromatin remodeling at promoters of heat shock genes, it seems crucial to identify the enzymatic activities involved.
Acknowledgments
We thank David Gross, Keith Miskimins, Robin Miskimins, Barbara Goodman, Randall Morse, and Nikolai Barlev for critical reading of the manuscript and helpful suggestions and Peter Sorger and David Gross for plasmids and yeast strains.
This work was supported by grants awarded to A.M.E. from NSF (MCB-0352042) and from NIH (P20 RR016479) and the INBRE Program of the National Center for Research Resources.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Amin, J., J. Ananthan, and R. Voellmy. 1988. Key features of heat shock regulatory elements. Mol. Cell. Biol. 8:3761-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apone, L. M., C. A. Virbasius, F. C. Holstege, J. Wang, R. A. Young, and M. R. Green. 1998. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol. Cell 2:653-661. [DOI] [PubMed] [Google Scholar]
- 4.Baxter, B. K., and E. A. Craig. 1998. Suppression of an Hsp70 mutant phenotype in Saccharomyces cerevisiae through loss of function of the chromatin component Sin1p/Spt2p. J. Bacteriol. 180:6484-6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2004. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14:667-673. [DOI] [PubMed] [Google Scholar]
- 6.Boy-Marcotte, E., G. Lagniel, M. Perrot, F. Bussereau, A. Boudsocq, M. Jacquet, and J. Labarre. 1999. The heat shock response in yeast: differential regulations and contributions of the Msn2p/Msn4p and Hsf1p regulons. Mol. Microbiol. 33:274-283. [DOI] [PubMed] [Google Scholar]
- 7.Chou, S., S. Chatterjee, M. Lee, and K. Struhl. 1999. Transcriptional activation in yeast cells lacking transcription factor IIA. Genetics 153:1573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 9.Deckert, J., and K. Struhl. 2001. Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol. Cell. Biol. 21:2726-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 11.Dilworth, F. J., C. Fromental-Ramain, K. Yamamoto, and P. Chambon. 2000. ATP-driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR In vitro. Mol. Cell 6:1049-1058. [DOI] [PubMed] [Google Scholar]
- 12.Erkine, A. M., C. C. Adams, T. Diken, and D. S. Gross. 1996. Heat shock factor gains access to the yeast HSC82 promoter independently of other sequence-specific factors and antagonizes nucleosomal repression of basal and induced transcription. Mol. Cell. Biol. 16:7004-7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erkine, A. M., and D. S. Gross. 2003. Dynamic chromatin alterations triggered by natural and synthetic activation domains. J. Biol. Chem. 278:7755-7764. [DOI] [PubMed] [Google Scholar]
- 14.Erkine, A. M., S. F. Magrogan, E. A. Sekinger, and D. S. Gross. 1999. Cooperative binding of heat shock factor to the yeast HSP82 promoter in vivo and in vitro. Mol. Cell. Biol. 19:1627-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fazzio, T. G., and T. Tsukiyama. 2003. Chromatin remodeling in vivo: evidence for a nucleosome sliding mechanism. Mol. Cell 12:1333-1340. [DOI] [PubMed] [Google Scholar]
- 16.Ferguson, S. B., E. S. Anderson, R. B. Harshaw, T. Thate, N. L. Craig, and H. C. Nelson. 2005. Protein kinase A regulates constitutive expression of small heat-shock genes in an Msn2/4p-independent and Hsf1p-dependent manner in Saccharomyces cerevisiae. Genetics 169:1203-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giardina, C., and J. T. Lis. 1995. Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol. Cell. Biol. 15:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross, D. S., C. C. Adams, S. Lee, and B. Stentz. 1993. A critical role for heat shock transcription factor in establishing a nucleosome-free region over the TATA-initiation site of the yeast HSP82 heat shock gene. EMBO J. 12:3931-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross, D. S., K. E. English, K. W. Collins, and S. W. Lee. 1990. Genomic footprinting of the yeast HSP82 promoter reveals marked distortion of the DNA helix and constitutive occupancy of heat shock and TATA elements. J. Mol. Biol. 216:611-631. [DOI] [PubMed] [Google Scholar]
- 20.Hahn, J. S., Z. Hu, D. J. Thiele, and V. R. Iyer. 2004. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 24:5249-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashikawa, N., and H. Sakurai. 2004. Phosphorylation of the yeast heat shock transcription factor is implicated in gene-specific activation dependent on the architecture of the heat shock element. Mol. Cell. Biol. 24:3648-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 23.Hoj, A., and B. K. Jakobsen. 1994. A short element required for turning off heat shock transcription factor: evidence that phosphorylation enhances deactivation. EMBO J. 13:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jakobsen, B. K., and H. R. Pelham. 1988. Constitutive binding of yeast heat shock factor to DNA in vivo. Mol. Cell. Biol. 8:5040-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 26.Kadosh, D., and K. Struhl. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365-371. [DOI] [PubMed] [Google Scholar]
- 27.Keener, J., J. A. Dodd, D. Lalo, and M. Nomura. 1997. Histones H3 and H4 are components of upstream activation factor required for the high-level transcription of yeast rDNA by RNA polymerase I. Proc. Natl. Acad. Sci. USA 94:13458-13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korber, P., T. Luckenbach, D. Blaschke, and W. Horz. 2004. Evidence for histone eviction in trans upon induction of the yeast PHO5 promoter. Mol. Cell. Biol. 24:10965-10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krebs, J. E., M. H. Kuo, C. D. Allis, and C. L. Peterson. 1999. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 13:1412-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kristjuhan, A., and J. Q. Svejstrup. 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 23:4243-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langst, G., E. J. Bonte, D. F. Corona, and P. B. Becker. 1999. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97:843-852. [DOI] [PubMed] [Google Scholar]
- 32.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36:900-905. [DOI] [PubMed] [Google Scholar]
- 33.Lee, D., and J. T. Lis. 1998. Transcriptional activation independent of TFIIH kinase and the RNA polymerase II mediator in vivo. Nature 393:389-392. [DOI] [PubMed] [Google Scholar]
- 34.McNeil, J. B., H. Agah, and D. Bentley. 1998. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes Dev. 12:2510-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moqtaderi, Z., M. Keaveney, and K. Struhl. 1998. The histone H3-like TAF is broadly required for transcription in yeast. Mol. Cell 2:675-682. [DOI] [PubMed] [Google Scholar]
- 36.Morano, K. A., N. Santoro, K. A. Koch, and D. J. Thiele. 1999. A trans-activation domain in yeast heat shock transcription factor is essential for cell cycle progression during stress. Mol. Cell. Biol. 19:402-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morimoto, R. I. 1998. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 12:3788-3796. [DOI] [PubMed] [Google Scholar]
- 38.Nieto-Sotelo, J., G. Wiederrecht, A. Okuda, and C. S. Parker. 1990. The yeast heat shock transcription factor contains a transcriptional activation domain whose activity is repressed under nonshock conditions. Cell 62:807-817. [DOI] [PubMed] [Google Scholar]
- 39.Perisic, O., H. Xiao, and J. T. Lis. 1989. Stable binding of Drosophila heat shock factor to head-to-head and tail- to-tail repeats of a conserved 5 bp recognition unit. Cell 59:797-806. [DOI] [PubMed] [Google Scholar]
- 40.Reinke, H., P. D. Gregory, and W. Horz. 2001. A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol. Cell 7:529-538. [DOI] [PubMed] [Google Scholar]
- 41.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11:1599-1607. [DOI] [PubMed] [Google Scholar]
- 42.Santoro, N., N. Johansson, and D. J. Thiele. 1998. Heat shock element architecture is an important determinant in the temperature and transactivation domain requirements for heat shock transcription factor. Mol. Cell. Biol. 18:6340-6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorger, P. K. 1990. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell 62:793-805. [DOI] [PubMed] [Google Scholar]
- 44.Sorger, P. K., M. J. Lewis, and H. R. Pelham. 1987. Heat shock factor is regulated differently in yeast and HeLa cells. Nature 329:81-84. [DOI] [PubMed] [Google Scholar]
- 45.Sorger, P. K., and H. R. Pelham. 1988. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 54:855-864. [DOI] [PubMed] [Google Scholar]
- 46.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 47.Voellmy, R. 2004. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones 9:122-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao, H., and J. T. Lis. 1988. Germline transformation used to define key features of heat-shock response elements. Science 239:1139-1142. [DOI] [PubMed] [Google Scholar]
- 49.Xiao, H., O. Perisic, and J. T. Lis. 1991. Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell 64:585-593. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto, A., Y. Mizukami, and H. Sakurai. 2005. Identification of a novel class of target genes and a novel type of binding sequence of heat shock transcription factor in Saccharomyces cerevisiae. J. Biol. Chem. 280:11911-11919. [DOI] [PubMed] [Google Scholar]
- 51.Young, M. R., and E. A. Craig. 1993. Saccharomyces cerevisiae HSP70 heat shock elements are functionally distinct. Mol. Cell. Biol. 13:5637-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao, J., J. Herrera-Diaz, and D. S. Gross. 2005. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol. Cell. Biol. 25:8985-8999. [DOI] [PMC free article] [PubMed] [Google Scholar]