Abstract
The repair of chromosomal double-strand breaks (DSBs) is essential to normal cell growth, and homologous recombination is a universal process for DSB repair. We explored DSB repair mechanisms in the yeast Saccharomyces cerevisiae using single-strand oligonucleotides with homology to both sides of a DSB. Oligonucleotide-directed repair occurred exclusively via Rad52- and Rad59-mediated single-strand annealing (SSA). Even the SSA domain of human Rad52 provided partial complementation for a null rad52 mutation. The repair did not involve Rad51-driven strand invasion, and moreover the suppression of strand invasion increased repair with oligonucleotides. A DSB was shown to activate targeting by oligonucleotides homologous to only one side of the break at large distances (at least 20 kb) from the break in a strand-biased manner, suggesting extensive 5′ to 3′ resection, followed by the restoration of resected DNA to the double-strand state. We conclude that long resected chromosomal DSB ends are repaired by a single-strand DNA oligonucleotide through two rounds of annealing. The repair by single-strand DNA can be conservative and may allow for accurate restoration of chromosomal DNAs with closely spaced DSBs.
DNA double-strand breaks (DSBs) are common mediators of genetic change in all organisms and can lead to mutations, recombination, chromosome aberrations, and cell death. Repair in eukaryotes can occur by recombination between homologous DNAs and by end joining (29, 30). To directly address DSB repair mechanisms, many cellular studies have focused on the repair of enzymatically induced single DSBs or the repair of transfected broken plasmids (2, 13, 18, 30).
The repair of a DSB via nonhomologous end joining (NHEJ) involves little or no homology and is often mutagenic because of the misalignment of overhanging ends paired at microhomologies (14). NHEJ can also generate gross rearrangements by joining the wrong DSB ends (reference 11 and references therein).
There are two ways of establishing the interaction between two recombining DNA sequences in homologous recombination (HR): strand invasion and single-strand annealing (SSA) (2, 29, 44). Intrinsic to all HR mechanisms for DSB repair is 5′ to 3′ end resection that in the yeast Saccharomyces cerevisiae can extend many kilobases from the DSB ends. The retained 3′ ends of a DSB can prime DNA synthesis or serve as a template. If both ends “invade” an unbroken homologous sequence, the resulting repair enables chromosome integrity to be maintained. An invasion by only one of the 3′ ends and the subsequent replication (break-induced replication) could lead to chromosome aberrations, including chromosome rearrangements and/or loss of heterozygosity.
SSA enables DSB repair by a less complex mechanism of recombination since it does not require three-strand interaction such as that involved in strand invasion. Following resection, repeat DNAs that flank a DSB can be simply annealed. Repair is completed through the subsequent removal of nonhomologous tails, DNA synthesis to fill any single-strand gaps, and ligation. SSA is considered to be a nonconservative mechanism of HR since the intervening sequence between repeats is eliminated, generating a deletion, while the strand invasion mechanism is considered conservative (error free) (44). In cells with many long DNA repeats, such as human cells, where more than 50% of the genome is repeated sequence (17), the SSA mechanism could be an important source of genome change.
DSB-induced SSA between defined direct repeats has been characterized in yeast as well as in bacteria, plant cells, Xenopus laevis oocytes, and mammalian cells (3, 11, 20, 24, 33, 36, 45). As shown in yeast and in mammalian cells, SSA is a mechanism that can compete efficiently with the strand invasion mechanism for the repair of a DSB (19, 36, 47). Unlike the strand invasion mechanism, SSA does not require Rad51, while both mechanisms require the Rad52 protein. Rad52 can anneal single-strand DNAs (ssDNAs), and it mediates the loading of Rad51 onto ssDNAs for strand invasion (29).
The substrates for SSA are regions of ssDNA that are commonly found in cells as intermediates in many DNA repair processes. ssDNA is generated as a consequence of 5′ to 3′ resection of DSB ends (29). Replication fork collapse can also lead to long ssDNA regions (for details, see reference 23).
Given the many possibilities for DSB induction in cells as well as the frequent appearance of ssDNA and the potential for genomic change, it is important to understand the mechanisms by which ssDNA can initiate or participate in DNA recombination and DSB repair. Recently, we reported that ssDNA presented to a cell as oligonucleotides with homology to regions that are near or distant from the two sides of a chromosomal DSB can efficiently repair the break (37). Because there was a requirement for Rad52 protein and for homology between the DNAs, we concluded that the repair relied on homologous recombination. However, the molecular mechanism of this novel form of repair was unclear. We establish here that the repair can be accomplished through SSA of the oligonucleotide with resected ends and does not involve strand invasion, even when homologous regions are many kilobases away from a DSB. Furthermore, we found that the DSB activates recombination at distant sites and stimulates oligonucleotide targeting to just one side of the break in a strand-biased manner. Based on our findings, we conclude that ssDNA can patch a DSB through two strand-annealing events. This mechanism can explain the accurate repair of closely opposed DSBs (i.e., in clusters) in homologous DNAs where strand invasion repair cannot be utilized.
MATERIALS AND METHODS
Yeast strains and plasmids.
The strain FRO-1 (MATα ade5-1 his7-2 leu2-3,112 ura3-52) contains the CORE-I-SceI cassette (I-SceI gene under the GAL1 promoter, hygromycin resistance gene, the counterselectable KlURA3) and the I-SceI site (HOT site) in TRP5. FRO-1 also contains an Alu inverted repeat (Alu-IR) with 100% homology inserted into the LYS2 gene. FRO-101 is the same as FRO-1 except that the I-SceI site is truncated to 6 bp (TAGGGA) and is nonfunctional (COLD site). Strains containing the rad52-327 and rad52-408 alleles were derived from FRO-1, where the C-terminal part of RAD52 was replaced with the G418 resistance gene starting at codons 328 and 409, respectively. Single-mutant deletion strains contain the kanMX4 module in place of the chosen gene and are all derivatives of FRO-1. Strains with deletions of two genes were obtained by tetrad dissection of diploids derived from crossing FRO-1 (containing a chosen deletion) with E133a (MATa) (38), which is isogenic to CG379 (containing a replacement of RAD52 by kanMX4), and a CORE-I-SceI site in TRP5 containing the natMX4 gene in place of the hygromycin resistance gene, which provides resistance to nourseothricin (12). The diploid strains FRO-922 (wild type) and FRO-923 (homozygous for rad51 deletion) were derivatives of FRO-1 (wild type or Δrad51) crossed with E133a (wild type or with RAD51 replaced by kanMX4), where trp1-289 was made wild type and TRP5 was disrupted with LEU2. FRO-917 is a derivative of BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0), where the TRP5 gene was inactivated with a 31-bp insert by CORE replacement (39) by using the oligonucleotide e1.INS31 (Table 1). Strains FRO-870, -872, -874, and -876 were derived from FRO-917 containing a CORE-I-SceI cassette (with the hygromycin resistance gene) approximately 6, 10, and 20 kb upstream and 10 kb downstream, respectively, from the 31-bp insert in TRP5. The diploid strains FRO-888 and FRO-897 were generated by mating FRO-872 with FRO-879 and FRO-876 with FRO-879, respectively. FRO-879 is BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) (6), where TRP5 has been replaced by LEU2.
TABLE 1.
Oligonucleotides used to insert CORE-I-SceI cassette and IROs used in this study
| Primer or IRO | Size | Sequence |
|---|---|---|
| Primersa | ||
| TRP5.IIr | 70-mer | 5′-GGCCAATATAAGAATACAAGGATTTGAAGTCTTCCCAGAATGTGGGATCGTTCGTACGCTGCAGGTCGAC |
| TRP5.SIr | 88-mer | 5′-CTTCATGCATGTCTAAGAGAGTTGGAAAAGGGTTTTGATGAAGCTGTCGC TAGGGATAACAGGGTAATTTGGATGGACGCAAAGAAGT |
| TRP5.1/2SIr | 76-mer | 5′-CTTCATGCATGTCTAAGAGAGTTGGAAAAGGGTTTTGATGAAGCTGTCGCTAGGGATTGGATGGACGCAAAGAAGT |
| 6KbUP.II | 70-mer | 5′-TATTTGTAGTTAGAAATTAGAACCGCAAATTTGTAAAACATCTATATCGT TTGGATGGACGCAAAGAAGT |
| 6KbUP.IS | 88-mer | 5′-CATATAAAAGATCAGAAAACAGTAATTAGATCTTTTCCATTACGACATTATAGGGATAACAGGGTAATTTCGTACGCTGCAGGTCGAC |
| 10KbUP.II | 70-mer | 5′-TCGTTCGTTATCCGAAGCTGGCCAATTGATACAATTAATTGACATCAGCA TTGGATGGACGCAAAGAAGT |
| 10KbUP.IS | 88-mer | 5′-AGATAATTTACCCTTGCTTTAAGCTGCGTATATCAAGTGCATTTGCTGTCTAGGGATAACAGGGTAATTTCGTACGCTGCAGGTCGAC |
| 20KbUP.II | 70-mer | 5′-TGTCGGTACTTCTGCCACTATCAGTATAAATAATTGCCGCCGTTCCATGA TTGGATGGACGCAAAGAAGT |
| 20KbUP.IS | 88-mer | 5′-ATTAATGAAGAATGGCCAAGATCAGAGTAAACCTTGCCATGCTCTCCATATAGGGATAACAGGGTAATTTCGTACGCTGCAGGTCGAC |
| 10KbDW.II | 70-mer | 5′-TTTGTCATTAGACACTTACCAGTTGATGTTTTCACTTTTTCTTTCCTTCC TTGGATGGACGCAAAGAAGT |
| 10KbDW.IS | 88-mer | 5′-CCGAGGTATTGTTGTATATACCACTACTCTGTGATTTTTTTTCACTCTTGTAGGGATAACAGGGTAATTTCGTACGCTGCAGGTCGAC |
| IROsb | ||
| e | 95-mer | 5′-CATGCATGTCTAAGAGAGTTGGAAAAGGGTTTTGATGAAGCTGTCGC∥G∥GATCCCACATTCTGGGAAGACTTCAAATCCTTGTATTCTTATATTGG |
| f | 95-mer | 5′-CCAATATAAGAATACAAGGATTTGAAGTCTTCCCAGAATGTGGGATC∥C∥GCGACAGCTTCATCAAAACCCTTTTCCAACTCTCTTAGACATGCATG |
| w | 95-mer | 5′-AATGGTGCGTTAGTTCACTGGGTTTATCCATATGCCAAATTGAGGGA∥C∥CCAAATGTTATTTCAACTATCAATGTTATGAGCTTAGCCGCCGTCGG |
| c | 95-mer | 5′-CCGACGGCGGCTAAGCTCATAACATTGATAGTTGAAATAACATTTGG∥G∥TCCCTCAATTTGGCATATGGATAAACCCAGTGAACTAACGCACCATT |
| e1.INS31 | 96-mer | 5′-GAGTTGGAAAAGGGTTTTGATGAAGCTGTCGCCCAAATCCTCAGCATAATGATTAGGTATGCACGATCCCACATTCTGGGAAGACTTCAAATCCTT |
| R1_6kbUP.e1 | 80-mer | 5′-TAGAAATTAGAACCGCAAATTTGTAAAACATCTATATCGT∥TAATGTCGTAATGGAAAAGATCTAATTACTGTTTTCTGAT |
| R2_6kbUP.f1 | 80-mer | 5′-ATCAGAAAACAGTAATTAGATCTTTTCCATTACGACATTA∥ACGATATAGATGTTTTACAAATTTGCGGTTCTAATTTCTA |
| R1_10kbUP.e1 | 80-mer | 5′-TCCGAAGCTGGCCAATTGATACAATTAATTGACATCAGCA∥GACAGCAAATGCACTTGATATACGCAGCTTAAAGCAAGGG |
| R2_10kbUP.f1 | 80-mer | 5′-CCCTTGCTTTAAGCTGCGTATATCAAGTGCATTTGCTGTC∥TGCTGATGTCAATTAATTGTATCAATTGGCCAGCTTCGGA |
| R1_20kbUP.e1 | 80-mer | 5′-TCTGCCACTATCAGTATAAATAATTGCCGCCGTTCCATGA∥TATGGAGAGCATGGCAAGGTTTACTCTGATCTTGGCCATT |
| R2_20kbUP.f1 | 80-mer | 5′-AATGGCCAAGATCAGAGTAAACCTTGCCATGCTCTCCATA∥TCATGGAACGGCGGCAATTATTTATACTGATAGTGGCAGA |
| R1_10kbDW.e1 | 80-mer | 5′-GTTGTATATACCACTACTCTGTGATTTTTTTTCACTCTTG∥GGAAGGAAAGAAAAAGTGAAAACATCAACTGGTAAGTGTC |
| R2_10kbDW.f1 | 80-mer | 5′-GACACTTACCAGTTGATGTTTTCACTTTTTCTTTCCTTCC∥CAAGAGTGAAAAAAAATCACAGAGTAGTGGTATATACAAC |
The first group of oligonucleotides comprises primers used to amplify the CORE-I-SceI cassette, containing 50 nucleotides used for the homologous targeting and 20 bases used for the amplification from the KlURA3 side (single underlines) or 18 bases containing the I-SceI site (bold) and 20 bases used for the amplification from the GAL1-I-SceI side (double underlines). Primers TRP5.IIr and TRP5.SIr were used to integrate the CORE-I-SceI cassette, together with a complete I-SceI site (HOT site) at the TRP5 locus; primer TRP5.1/2SIr was used in combination with TRP5.IIr to insert the CORE-I-SceI cassette, together with a truncated, nonfunctional I-SceI site (COLD site, bold) at the TRP5 locus. Primers 6KbUP.II, 6KbUP.IS, 10KbUP.II, and 10KbDW.IS were used to integrate the CORE-I-SceI cassette 6, 10, and 20 kb upstream and 10 kb downstream, respectively, from the TRP5 locus.
The second group of oligonucleotides comprises the IROs used in this study. Oligonucleotides e and f target the TRP5 gene by introducing a silent mutation that generates a BamHI site; w and c target the LYS2 gene by introducing a silent mutation that generates an AvaII site. Oligonucleotide e1.INS31 was used to target a 31-bp frameshift insertion (bold) in the TRP5 gene. Oligonucleotides of the series R1 and R2 delete the CORE-I-SceI cassette and repair a DSB generated at 6, 10, and 20 kb upstream or 10 kb downstream from the 31-bp insert in TRP5. The parallel vertical lines (∥) separate the two homologous tails of each IRO. When present, the new point mutation that is introduced is indicated in bold, and the resulting restriction site is underlined (BamHI or AvaII).
The CORE-I-SceI cassette used in this study was PCR amplified from plasmid pGSHU (37) using the pairs of primers described in Table 1. The integration of the CORE-I-SceI cassette in the yeast genome was carried out as previously described (37, 40).
Plasmids YEpNAT, YEpSc_rad52-327, and YEph_rad52-209 were constructed as follows. Plasmid pAG25 was used in PCRs to amplify natMX4, plasmid pRS316GALyRAD52 (E. Perkins, personal communication) was used to amplify the first 327 codons of yeast RAD52, and plasmid pZAThuRAD52 (E. Perkins, personal communication) was used to amplify the first 209 residues from the cDNA of human Rad52 isoform α (NM_002879). The natMX4 marker was placed upstream of yeast RAD52 and human Rad52 cDNA by fusion PCR. The gene natMX4 and the fused fragments were placed next to the GAL1 promoter on the YEp195SpGALFEN1-Ch vector (38) linearized by MscI and HindIII by in vivo gap repair recombination selecting for the circularized plasmid in the yeast E133 strain (38) (Ura+ and natMX4-resistant clones). Plasmids were rescued in Escherichia coli and confirmed by sequence analysis. For the selection of natMX4-resistant clones, cells were grown in yeast extract-peptone-dextrose-adenine (YPDA) or YPGal medium containing 100 μg/ml of nourseothricin (clonNAT; Werner BioAgents, Jena, Germany).
Genetic methods and standard media were described previously (reference 37 and references therein).
DSB induction and targeting with oligonucleotides.
DSB induction and targeting with oligonucleotides were performed as previously described (37, 40). Briefly, 1.5 ml of an overnight culture grown in rich YPDA medium was transferred into 50 ml of synthetic complete medium containing 2% galactose and incubated with vigorous shaking at 30°C for 4 h (7 h for experiments involving oligonucleotide targeting to the side of a DSB) (see Fig. 3) to express GAL1-I-SceI and induce a DSB. After incubation in galactose, cells were prepared for transformation. Transformation was done with 1 nmol of total oligonucleotides. Oligonucleotides (integrative recombinant oligonucleotides [IROs]) used to repair the I-SceI DSBs and inverted repeat DSB and to integrate the 31-bp insert in TRP5 are described in Table 1.
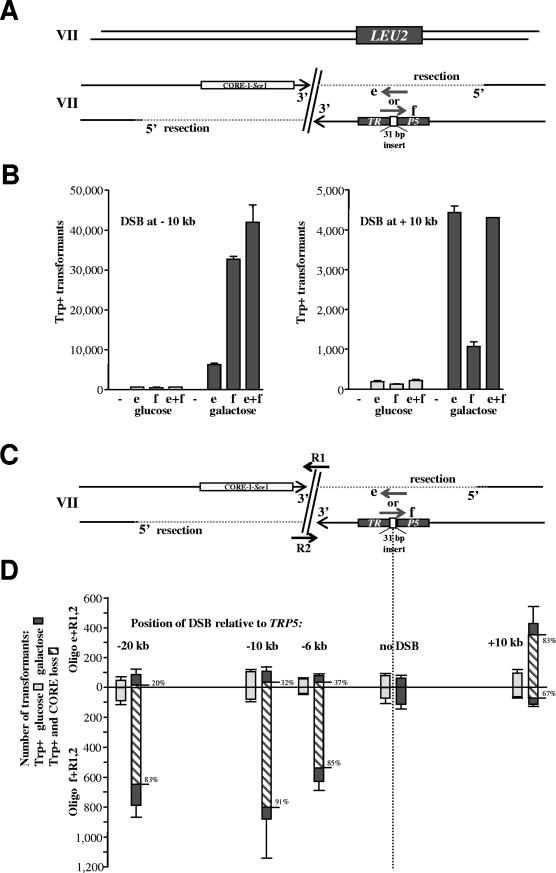
FIG.3.
DSB-mediated stimulation of oligonucleotide targeting to the side of the break in a strand-dependent manner. (A) The diagram shows one copy of chromosome VII, where TRP5 has been replaced with the LEU2 gene (Fig. 2). The second copy of chromosome VII contains a TRP5 locus that has been inactivated by a 31-bp frameshift insertion and a CORE-I-SceI cassette that can generate a DSB either 10 kb upstream or 10 kb downstream of the TRP5::ins31 site. The oligonucleotides e and f can restore the Trp+ phenotype. The intact copy of chromosome VII can provide a template for the repair of the DSB as described for the experiments illustrated in Fig. 2. (B) Number of Trp+ transformants per 107 viable cells resulting from targeting 1 nmol of oligonucleotide e or f or both following no DSB induction (light gray bars) or induction of the DSB (7 h in galactose) (dark gray bars) that is either 10 kb upstream or 10 kb downstream from TRP5::ins31. The vertical light and dark gray bars correspond to the average values of three determinations; the ranges (error bars) identify the standard deviations. (C) Scheme to address possible resection and strand bias during targeting of the mutated TRP5 locus that is many kilobases (6, 10, and 20 kb downstream or 10 kb upstream) from an I-SceI-induced DSB in yeast haploid cells. The scheme and oligonucleotides are similar to those in panel A above, with the only difference being that repair oligonucleotides R1 and/or R2 were added to repair the DSB. (D) Number of Trp+ transformants per 107 viable cells resulting from targeting 0.33 nmol of oligonucleotide e or f to TRP5::ins31 with DSB induction (7 h in galactose) (dark gray bars) and without DSB induction (light gray bars) at a position 6, 10, or 20 kb upstream or 10 kb downstream from TRP5::ins31. Also included were 0.33 nmol of the repair oligonucleotide R1 and 0.33 nmol of R2 to repair the induced DSB. The frequency of CORE marker loss events due to the addition of R1 and R2 was about 1% of viable cells when a DSB was induced and ∼0.001% of viable cells in the absence of DSB induction (not shown). The vertical light and dark gray bars correspond to the average values from three to six determinations; the ranges (error bars) identify the standard deviations. The striped bars show the number of Trp+ clones that were without CORE markers, and the percentages of these clones are shown at the right of each bar. Trp+ clones that were without CORE were detected only for cells with a DSB transformed with R1 or R2. (E) Number of Trp+ clones per 107 viable cells resulting from transformation with oligonucleotide e or f (0.5 nmol) alone or together with the repairing oligonucleotide R1 or R2 (0.5 nmol) after cells were grown in glucose (no DSB; light gray bars) or in galactose for 7 h (DSB; dark gray bars). The DSB was generated 10 kb upstream from TRP5::ins31 (see scheme in panel C). The frequency of CORE marker loss due to the addition of R1 or R2 was about 0.3% of viable cells in which a DSB was induced and ∼0.0002% of viable cells in the absence of DSB induction (not shown). The vertical bars correspond to the average values from four determinations; the ranges (error bars) identify the standard deviations. The striped bars show the number of Trp+ clones that were without CORE markers, and the percentages of these clones are shown at the right of each bar. Trp+ clones that were without CORE were detected only for cells with a DSB transformed with R1 or R2.
Cells from each oligonucleotide transformation were diluted appropriately and spread directly to selective glucose Trp− or Lys− plates or to YPDA. To select for colonies that had lost the CORE markers, the YPDA plates were replica plated to 5-fluoroorotic acid medium after the first day of incubation. After 3 days, colonies from 5-fluoroorotic acid were replica plated to YPDA and YPDA containing hygromycin and on Trp− plates if applicable.
Cells from oligonucleotide transformations done in the presence of plasmid overexpressing yeast and human truncated Rad52 proteins were diluted appropriately and plated to YPGal medium containing nourseothricin in order to maintain expression of the rad52 allele and selection for the plasmid. After 3 days, the cells were replica plated to Trp− plates to select for the oligonucleotide repair events.
Analysis of I-SceI-induced chromosomal DSBs.
The chromosomal DNA from cells collected after I-SceI induction in galactose was embedded in agarose plugs using a CHEF genomic DNA plug kit from Bio-Rad (∼108 cells/ml of plug) according to the manufacturer's instructions. Chromosomal DNA was separated by transverse alternating field electrophoresis in a Gene Line II apparatus from Beckman. Intact chromosome VII, along with DSB fragments, was detected by Southern blot hybridization with a 32P-labeled, 300-bp ARO1-specific probe (for the left arm of chromosome VII) and a 300-bp RTS3-specific probe (for the right arm of chromosome VII). The amounts of unbroken chromosome VII and broken fragments were determined using a Molecular Dynamics PhosphorImager. The quantization of the broken chromosomal bands was carried out with the ImageQuant program.
RESULTS
DSB repair by a single-strand oligonucleotide occurs via SSA.
Previously, we demonstrated that a ssDNA oligonucleotide can repair a DSB induced in yeast by the site-specific I-SceI endonuclease or a spontaneous DSB generated at the site of an IR composed of human Alu sequences (37). In the wild-type yeast strain, the frequencies of oligonucleotide-targeting events after DSB induction exceeded the frequencies of transformation in cells without a DSB ∼5,000-fold for the I-SceI-induced break and 30-fold for the IR-induced break (37) (data not shown). Therefore, the vast majority of transformation events corresponded to the repair of a DSB by single-strand oligonucleotides. Here we investigated the genetic requirements in order to identify the processes involved in the repair of these two different types of DSBs by ssDNA.
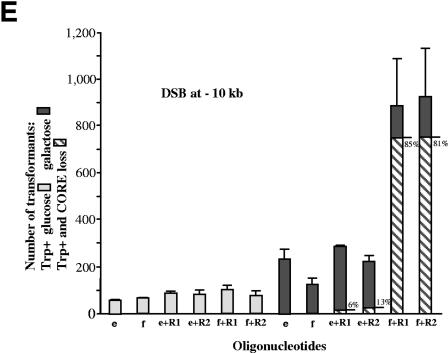
For this purpose, we constructed the haploid Saccharomyces cerevisiae strain FRO-1 in which both types of DSBs could be generated. This strain contains the self-generating DSB cassette “CORE-I-SceI” (37) composed of I-SceI endonuclease under the inducible GAL1 promoter, along with the I-SceI target scission site, integrated into the TRP5 locus (Fig. 1A). As shown in Fig. 1B, 26 to 38% of the DNA is cut within 4 h of induction in synthetic complete medium containing 2% galactose. The FRO-1 strain also contains the previously described Alu-IR (22, 37), which is located in the middle of the LYS2 gene. In strains of the same background, this IR leads to a spontaneous, hairpin-capped DSB in ∼2% of the cells (22) (Fig. 1C). A series of isogenic strains with deletions in DNA repair and recombination genes was also developed from FRO-1.
FIG. 1.
Systems for generating an inducible and a spontaneous chromosomal DSB. (A) Diagram of the I-SceI-inducible DSB system, presenting the scheme of the CORE-I-SceI cassette inserted into the middle of the TRP5 gene on chromosome VII. The position of the DSB is shown at the I-SceI site. 95-mers e and/or f with homology upstream and downstream from the CORE-I-SceI and the I-SceI site are used to target and repair the break and restore the TRP5 gene. The recombination product generating CORE loss Trp+ cells is shown. (B) Chromosome cleavage by I-SceI endonuclease in wild-type (WT) and various mutant strains as determined by pulsed-field electrophoresis. Cells grown in glucose do not induce I-SceI expression, while the expression of I-SceI in galactose results in a DSB. The following strains were tested for cleavage of chromosome VII: FRO-101, wild type, containing a COLD (truncated) I-SceI site; FRO-1, wild type containing a HOT (intact) I-SceI site; and mutant rad51, rad59, and rad52 strains derived from FRO-1. The DSB within TRP5 generates one band of 450 kb and one of 650 kb. Chromosome VII and fragments were detected by Southern hybridization. Each genotype is represented by two immediately adjacent lanes. Based on quantification by ImageQuant software, the broken chromosome VII represents 29% in the wild type, 32 to 33% in rad51, 26 to 27% in rad59, and 37 to 38% in rad52. A similar frequency of DSB formation was also obtained in the BY4742 background after I-SceI cutting within the TRP5 locus (not shown). (C) Diagram of the spontaneous DSB associated with an Alu-IR that generates hairpin-capped ends (for details, see reference 22). 95-mers w and/or c with homology upstream and downstream from the Alu-IR are used to target and repair the break and restore the LYS2 gene. The recombination product generating Lys+ cells without the Alu-IR is shown.
Following galactose induction, the various strains were transformed with single-strand oligonucleotides (Table 1) designed to repair either the I-SceI break (e and/or f) or the spontaneous IR break (w and/or c), as described for Fig. 1A and C. Cells were selected either for loss of the CORE-I-SceI cassette (Trp+) or for deletion of the Alu-IRs (Lys+). The repair of both kinds of DSBs by single-strand oligonucleotides required the Rad52 protein (Table 2) (also see reference 37 for details of the repair of the I-SceI break). The deletion of RAD59, a RAD52 homolog, also greatly reduced repair. However, a small number of repair events (<0.1%) occurred independently from RAD52 (Table 2).
TABLE 2.
Impact of genes involved in single-strand annealing and strand invasion on DSB repair by single-strand oligonucleotides
| Genotype | Relative transformation frequency fora:
|
|||
|---|---|---|---|---|
| I-SceI break-targeting oligonucleotide
|
IR break-targeting oligonucleotide
|
|||
| e | f | w | c | |
| WT | 1 | 1 | 1 | 1 |
| rad52 | 0.00052b | 0.00088b | <0.0016b | <0.00078b |
| rad59 | 0.1b | 0.08b | 0.34 | 0.34b |
| rad52 rad59 | 0.000053b | 0.00011b | ND | ND |
| rad51 | 2.4b | 2.2b | 2.9b | 2.6b |
| rad52 rad51 | 0.000053b | 0.00053b | ND | ND |
| rad59 rad51 | 0.043b | 0.074b | ND | ND |
| rad54 | 1 | 1.6 | 1.9 | 1.3 |
| rad55 | 1.1 | 2.1b | 5.2b | 4.6b |
| rad57 | 2.7b | 2.7b | 4.6b | 3.2b |
| rdh54 | 2.2b | 1.6b | ND | ND |
| rad52-327c | 3.7b | 3.9b | 1.7b | 1.4b |
| rad52-408c | 3.4b | 3.2b | 1 | 1.2 |
Transformation frequency of single-strand oligonucleotides e and f used to target the I-SceI break associated with the CORE-I-SceI cassette or w and c used to target a spontaneous break associated with an Alu-IR relative to the wild type (WT). The relative transformation frequency was calculated by dividing the absolute frequency for each mutant by the absolute frequency for the WT obtained with the corresponding oligonucleotide. Absolute frequencies in the WT correspond to 93,606 for e, 129,847 for f, 636 for w, and 1,281 for c per 107 viable cells. All absolute values are provided in Tables S1 and S2 in the supplemental material. ND, not determined.
Statistically significant decrease or increase over the WT based on nonoverlapping 95% confidence intervals. Plasmid transformation frequencies are reduced about twofold in rad51, rad59, and rad51 rad59 and three- to fourfold in rad52, rad52 rad51, and rad52 rad59 mutants. When the frequencies of oligonucleotide targeting are normalized by plasmid transformation frequencies, their values remain different from that of the WT (statistically significant; data not shown).
Mutant alleles of RAD52 defective in Rad51 interaction that contain a C-terminal deletion from amino acids 328 (rad52-32) and 409 (rad52-408).
Rad52 is essential for both strand invasion and strand annealing (44) and is implicated in end joining between DSBs with complementary single-strand ends longer than eight bases (9). There was no detectable effect on DSB repair of mutations in the NHEJ genes YKU70 and LIG4 (Table 3), consistent with the view that nearly all single-strand oligonucleotide repair of a DSB is mediated through Rad52-dependent homologous recombination. Therefore, we investigated the role of genes specific for the genetic control of strand invasion. The Rad51 protein, like RecA of E. coli, is the recombinase that mediates strand invasion by ssDNA. The deletion of RAD51 as well as other genes involved in this mechanism (RAD55, RAD57, RAD54, and RDH54) did not reduce repair efficiency. In fact, targeting was increased as much as fivefold (Table 2), which is similar to what has been observed for direct repeat recombination (25). This increase appears to be due to sister chromatid competition for DSB repair (see below). The strong reduction in repair of the I-SceI break observed in rad52 and rad59 mutants and the increased frequency of DSB repair observed in the rad51 strain were not attributable to differences in DSB formation (Fig. 1B). Survival after transformation was ∼30 to 40% with or without oligonucleotides for the wild type and was not changed in any mutant tested.
TABLE 3.
Impact of genes involved in NHEJ and processing of recombination intermediates on DSB repair by single-strand oligonucleotides
| Genotype | Relative transformation frequency fora:
|
|||
|---|---|---|---|---|
| I-SceI break-targeting oligonucleotide
|
IR break-targeting oligonucleotide
|
|||
| e | f | w | c | |
| yku70 | 1.2 | 1.1 | 0.5b | 0.6b |
| lig4 | 1.1 | 1.5b | 0.8 | 0.6b |
| msh2 | 0.7 | 1.5 | 0.3b | 0.2b |
| msh3 | 0.5b | 0.8 | 0.3b | 0.2b |
| mlh1 | 1.2 | 1.8b | 1 | 1.2 |
| rad1 | 0.2b | 0.4b | 0.3b | 0.2b |
| rad10 | 0.2b | 0.7b | 0.2b | 0.2b |
See footnote a to Table 2.
Statistically significant decrease or increase over the wild type based on nonoverlapping 95% confidence intervals.
To further address the role of SSA versus strand invasion, rad52-327 and rad52-408 mutants with C-terminal truncations were examined. These mutants retain single-strand annealing function but are unable to recruit Rad51 to ssDNA (16, 26, 46). Both mutants were highly sensitive to gamma radiation in manners similar to that of a rad52-null strain, while their sensitivities to methyl methane sulfonate were intermediate between the wild-type and rad52-null strains (not shown) in manners similar to those of other C-terminal deletion mutants of RAD52 (1, 5, 46). As shown in Table 2, the efficiency of DSB repair by ssDNA in the rad52-327 and rad52-408 mutants was increased to levels comparable to those found in mutants of the Rad51 strand invasion mechanism. Thus, the primary pathway for single-strand oligonucleotide repair of a DSB occurs via the SSA function of Rad52.
Since large DNA regions must be lost in order to generate the Trp+ and Lys+ recombinants (Fig. 1A and C), we examined the roles of the RAD1, RAD10, MSH2, and MSH3 genes, which normally cleave the nonhomologous tails formed during HR (for details, see reference 29) (Table 3). While the impact was much smaller than that for the rad52 mutants, the Δrad1 and Δrad10 mutants reduced the repair of I-SceI and IR breaks by single-strand oligonucleotides up to fivefold. The Δmsh2 and Δmsh3 mutants affected only repair of the IR break. Partial dependence on Rad1/Rad10 and Msh2/Msh3 clippase is in agreement with the existence of alternative pathways for 3′ tail removal indicated for short (fewer than 30 nucleotides) 3′ tails or in situations with only one nonhomologous 3′ tail (7, 28). We note that in our experimental systems, only one long nonhomologous 3′ tail is formed per annealing of the oligonucleotide to the end of a DSB, and the nonhomologous tail on the other side can be very short (just 9 nucleotides of the I-SceI site).
Because of the large impact of the yeast RAD52 gene, we investigated whether the SSA function of the human Rad52 protein could also be characterized in our system. Amino acid homology between S. cerevisiae Rad52 and other eukaryote homologs in the N-terminal regions (27) suggests functional similarity. Since the human N-terminal domain of Rad52 corresponding to amino acids 1 to 209 can promote single-strand annealing in vitro (35), we asked if these same 209 residues of human Rad52 isoform α could enable oligonucleotide repair of a DSB in yeast. This sequence was placed under the GAL1 promoter on an episomal plasmid containing the nourseothricin selectable marker (12) in the Δrad52 mutant. Galactose induction results in both the expression of the human protein and the generation of a DSB at the I-SceI site within the TRP5 gene. After transformation with single-strand oligonucleotides, cells were plated to YPGal medium containing nourseothricin in order to maintain the expression of the rad52 allele and selection for the plasmid. Colonies formed on these plates were replica plated to Trp− plates to select for the oligonucleotide repair events.
As shown in Table 4, the overexpression of the human Rad52 N-terminal domain resulted in significant albeit limited levels of repair by the single-strand oligonucleotides. Moreover, we show that such repair mediated by the human Rad52 N-terminal domain is similar to repair mediated by the truncated yeast protein in that it is independent of Rad51 and partially dependent on Rad59. Our results with the human Rad52 isoform α containing the N-terminal 209 residues provide the first direct evidence for human Rad52 SSA activity in vivo. We conclude that the oligonucleotide DSB repair system we developed can serve as a sensitive genetic assay for detecting single-strand annealing capacity that can be extended to a variety of Rad52 homologs with potential SSA function.
TABLE 4.
The N-terminal domain of human Rad52 mediates DSB repair of single-strand oligonucleotides in yeast
| Genotype | Plasmid | Transformation frequency for oligonucleotidea
|
|
|---|---|---|---|
| e | f | ||
| rad52 | Vector | 7 (0-14) | 14 (0-28) |
| rad52 | Sc_rad52-327 | 35,300 (31,800-49,400) | 49,800 (42,400-76,900) |
| rad52 | Hs_rad52-209 | 233 (219-278) | 353 (254-409) |
| rad52 rad51 | Vector | <1 (0-6.8) | <1 (0-6.8) |
| rad52 rad51 | Sc_rad52-327 | 50,000 (50,000-50,700) | 45,300 (40,500-45,900) |
| rad52 rad51 | Hs_rad52-209 | 204 (171-337) | 250 (236-277) |
| rad52 rad59 | Vector | <1 (0-0) | <1 (0-5.8) |
| rad52 rad59 | Sc_rad52-327 | 4,800 (4,300-5,500) | 3,700 (3,600-6,800) |
| rad52 rad59 | Hs_rad52-209 | 107 (82-173) | 64 (53-134) |
Median and >95% confidence interval or range (in parentheses) from three to nine determinations of Trp+ transformant colonies per 107 viable cells. Cells were transformed with single-strand oligonucleotide e or f after the overexpression of I-SceI along with yeast or the human N-terminal domain of Rad52 in a yeast rad52-null, rad52 rad51, or rad52 rad59 FRO-1 background.
The Rad52 SSA oligonucleotide pathway for DSB repair is an efficient alternative to repair via strand invasion.
The enhanced DSB repair by SSA with oligonucleotides in the rad51 strand invasion mechanism mutants (Table 2) demonstrates that this mechanism involving simple annealing of short stretches of complementary DNAs can be highly efficient. Since only 30% of molecules have the DSB (Fig. 1A), in some cases repair could be accomplished through strand invasion of a sister chromatid that has vastly greater homology on both sides of the DSB. A lack of Rad51 would leave only the SSA oligonucleotide mechanism of repair. To establish that oligonucleotide-mediated repair can substitute for a strand invasion mechanism, RAD+ and rad51-null diploids that had the I-SceI DSB-generating cassette in the TRP5 gene on chromosome VII (37) were utilized. The TRP5 gene of the homologous chromosome was deleted by gene replacement with LEU2. Of the four possible mechanisms of DSB repair, strand invasion with a sister or homolog, NHEJ, or oligonucleotide-mediated SSA, only the last would result in Trp+ cells (Fig. 2A). The SSA repair would simultaneously result in CORE loss, as would recombination with the homolog, or complete loss of the broken chromosome. Repair by NHEJ or by strand invasion with the sister chromatid would not result in loss of the CORE.
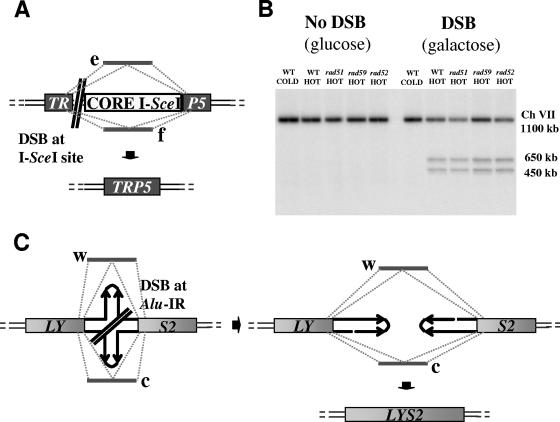
FIG. 2.
Oligonucleotide versus homologous chromosome-mediated DSB repair in diploid cells. (A) The diagram shows the position of the CORE-I-SceI cassette and the DSB in TRP5 in one copy of chromosome VII. TRP5 has been replaced with the LEU2 gene in the second copy of chromosome VII. 95-mers e and/or f with homology upstream and downstream from the CORE-I-SceI and the I-SceI break site are used to target and repair the break and restore the TRP5 gene. (B) CORE marker loss (light gray bars) and Trp+ colonies (dark gray bars) per 107 viable cells that arise after DSB induction in the TRP5 gene and transformation with e, f, or both (e+f) oligonucleotides or no oligonucleotides (−) in a wild-type (WT) (FRO-922) and in a rad51 homozygous (FRO-923) diploid. The vertical bars correspond to the average values from three to six determinations, and the range (error bars) identifies the standard deviation. The genotype of strains analyzed is indicated above the bars. In order to better compare the results, actual values are provided beneath the bars. Shown is the mean number of “CORE loss” colonies per 107 viable cells, followed by the mean number of Trp+ colonies and finally by the percentage of Trp+ colonies among all the CORE loss colonies. The possible consequences of a DSB in terms of phenotype are presented on the right side of the graph.
As previously reported (37), the efficiency of DSB repair (frequency of Trp+) by a mix of e and f oligonucleotides in wild-type diploid cells was 30-fold less than that for recombinational repair with the homologous chromosome (CORE loss only) (Fig. 2B). The frequency of Trp+ transformants with e, f, or both e and f oligonucleotides was 9- to 40-fold less than that in a haploid wild-type parent (compare Fig. 2B with Table S1A in the supplemental material for single oligonucleotides and with Table 1 in reference 37 for the mixed oligonucleotides). However, the deletion of the RAD51 gene in the diploid strain resulted in a 15- to 30-fold increase in Trp+ colonies compared to that in the RAD+ strain. Interestingly, the frequency of Trp+ transformant colonies in the rad51 diploid cells was comparable to that found in the haploid rad51 mutant. There was no detectable loss of viability in the rad51 mutant cells (16% in rad51 versus 15% in wild-type cells, with or without oligonucleotides). Therefore, the increase in frequency of Trp+ colonies reflected the increase in efficiency of oligonucleotide-mediated DSB repair. The higher repair efficiency with the combined e and f oligonucleotides than with the individual e and f oligonucleotides in the wild type and rad51 mutant (for details, see reference 37) is likely due to simultaneous annealing of complementary oligonucleotides to both sides of a DSB and/or to the formation of duplex oligonucleotide DNAs, which would be more resistant to degradation.
These results suggest that the presence of a homologous chromosome in the wild-type diploid cells provides an alternative pathway for the DSB repair, an option not available in the rad51 cells. Possibly, the opportunity to recombine with a homologous chromosome prevents additional rounds of cutting that might occur following repair between sister chromatids, thereby reducing the availability of a DSB for repair by the oligonucleotides. The increase in the frequency of oligonucleotide-targeted repair events in the absence of Rad51 could also be explained by Rad51 preventing oligonucleotide-directed annealing to ssDNA. We conclude that the Rad52 SSA oligonucleotide pathway for DSB repair is an efficient alternative to repair via strand invasion.
We note that SSA repair by an oligonucleotide(s) corresponded to at least 80% of the CORE loss events in the rad51 diploid (Fig. 2B, see the table under the graph). The CORE loss colonies that are also Trp− may be due to oligonucleotide-mediated repair with oligonucleotides that have mutations in the TRP5 sequence (37, 39) and/or to loss of the CORE markers independently from the oligonucleotides (e.g., deletion of the CORE region followed by telomere addition or chromosome fusion).
It is interesting that there is an overall reduction (as much as 80-fold) in the category of events in rad51 diploids that are due to only CORE loss. This is consistent with most CORE loss events in wild-type cells arising by strand invasion with the homologous chromosome. If in the absence of repair (strand invasion or SSA) the broken chromosome VII is lost, this either is a low-frequency event or leads to greatly reduced colony formation among cells with a DSB. (Note that a DSB occurs in ∼30% of cells.)
Oligonucleotide targeting to a DSB requires single-strand ends.
The role of ssDNA at the DSB ends during repair by an oligonucleotide could be addressed by comparing the genetic requirements for the repair of a hairpin-capped break associated with IRs (break with closed ends) versus the repair of an I-SceI-induced DSB (break with open ends). The IR-associated closed ends must be cut by the Mre11-Rad50-Xrs2 (MRX) complex in order to serve as a recipient for repair utilizing an unbroken chromosome (22). The I-SceI break results in four-base, 3′ OH overhangs (4, 31) and is available for 5′ resection. As shown in Table 5, repair of the IR-associated DSB is reduced over 100-fold in cells lacking the MRX complex. This contrasts with the modest (two- to fivefold) reduction in repair of the I-SceI break, consistent with our previous results (22). Thus, efficient DSB repair by a single-strand oligonucleotide requires DSB ends capable of forming single-strand regions. The single-strand ends could be created via resection or unwinding. The hairpin structures not only would prevent resection but also may impede the unwinding of the DNA ends to generate ssDNA regions.
TABLE 5.
Oligonucleotide repair of a directly resectable I-SceI DSB and of a hairpin-capped DSB
| Genotype | I-SceI break-targeting oligonucleotide
|
IR break-targeting oligonucleotide
|
||||||
|---|---|---|---|---|---|---|---|---|
|
e
|
f
|
w
|
c
|
|||||
| No. of transformants per 107 cellsa | Relative frequencyb | No. of transformants per 107 cells | Relative frequency | No. of transformants per 107 cells | Relative frequency | No. of transformants per 107 cells | Relative frequency | |
| WT | 93,606 (85,560-107,459) | 1 | 129,847 (117,771-143,139) | 1 | 636 (511-795) | 1 | 1,281 (1,167-1,470) | 1 |
| mre11 | 28,709 (14,648-33,353) | 0.3c | 20,282 (13,776-21,752) | 0.2c | 0 (0-2) | 0.0016 | 2 (2-5) | 0.0016 |
| rad50 | 26,100 (21,138-34,770) | 0.3c | 40,608 (35,772-50,883) | 0.3c | 0 (0-0) | <0.0016 | 0 (0-3) | 0.00078 |
| xrs2 | 54,577 (43,337-71,083) | 0.6c | 44,484 (35,542-56,787) | 0.3c | 2 (0-2) | 0.003 | 4 (2-6) | 0.003 |
Median and >95% confidence interval, or range (in parentheses) when the number of repeated experiments was <6, of Trp+ transformant colonies per 107 viable cells, resulting from targeting single-strand oligonucleotide e or f to the I-SceI break at the CORE-I-SceI cassette or oligonucleotide w or c to a spontaneous break associated with an Alu-IR. Survival was ∼30 to 40% after transformation with or without oligonucleotides for the wild type and all mutant strains tested.
Transformation frequency relative to the wild type (WT). The relative transformation frequency was calculated by dividing the absolute frequency for each mutant by the absolute frequency for the WT obtained with the corresponding oligonucleotide. All values have statistically significant decreases from the WT based on nonoverlapping 95% confidence intervals.
Plasmid transformation frequency is reduced about threefold in mre11, rad50, and xrs2 mutants. When these frequencies are normalized by plasmid transformation frequency, there is no statistically significant difference from that of the WT.
Strand-biased oligonucleotide targeting to a site distant from a DSB.
As we previously demonstrated, a DSB can be repaired by oligonucleotides with homologies to sequences of up to 16 kb from the broken ends, suggesting that a DSB activates a large region for SSA recombination (37). It is also known that a DSB can stimulate triparental recombination, promoting efficient gene conversion of a locus distant from the break in yeast diploid cells (32). We hypothesize that in our system, such stimulation occurs via SSA. This is supported by the observation that in the absence of RAD51, there was a 3.7-fold increase in repair frequency by single-strand oligonucleotides capable of generating a 16-kb deletion around the DSB (data not shown). These results in combination with the requirements for repair of the open versus the hairpin-closed DSB ends led us to propose that recombination distant from the DSB might be due to the appearance of ssDNA. This, in turn, predicts that ssDNA formed on either side of a DSB increases the likelihood of targeting oligonucleotides complementary to only one side of a break.
To test the idea of DSB-stimulated targeting to one side of a break, we initially constructed two diploid strains (Fig. 3A), where one chromosome VII contained a mutated TRP5 gene inactivated by a 31-base frameshift insertion (TRP5::ins31) and the other was the LEU2 replacement chromosome analogous to that presented in Fig. 2A. The frequency of TRP5::ins31 reversions to Trp+ was <10−8. The CORE-I-SceI cassette was placed 10 kb upstream of the TRP5::ins31 target in one diploid strain (FRO-888) and 10 kb downstream of TRP5::ins31 in the other diploid strain (FRO-897). The induced DSB led to a 20- to 60-fold increase in targeting (Trp+) to the distant TRP5::ins31 site (Fig. 3B). We suggest that the homologous chromosome provided an opportunity for DSB repair in a manner similar to that for the experiments described in the legend for Fig. 2, and this enabled the events targeted to the side to be recovered. If targeting to the side of the break is stimulated by strand-specific resection (i.e., the 5′ to 3′ resection) there should be a strand bias since only the oligonucleotide that is complementary to the remaining strand would enable recombination. Indeed, the bias was observed in favor of the oligonucleotide complementary to the strand expected to remain intact after resection (Fig. 3B).
The findings for the diploid strains led us to examine the ability of a DSB to stimulate recombination at a distant site in yeast haploid cells under conditions where the DSB could be repaired directly. To further characterize resection and possible strand bias, we created a series of haploid strains (FRO-917) in which the CORE-I-SceI cassette, together with the I-SceI cutting site, was inserted ∼6 (FRO-870), 10 (FRO-872), and 20 kb upstream (FRO-874) or 10 kb downstream (FRO-876) from the TRP5::ins31 locus (Fig. 3C). Oligonucleotide e or f, together with a second pair of oligonucleotides referred to as repairing oligonucleotides R1 and R2 (Fig. 3C) that would repair the DSB and delete the CORE-I-SceI cassette, was transformed into cells grown in glucose (i.e., no-break induction) or incubated for 7 h in galactose. As shown in Fig. 3D, glucose-grown cells exhibited comparable low frequencies of Trp+ transformants for oligonucleotide e or f in all strains. In contrast, break induction led to a large increase in targeting with a strong strand bias in favor of the oligonucleotide that is complementary to the strand with a 3′ end if resection has occurred at the DSB. This was the f oligonucleotide when the break occurred upstream from the targeted TRP5::ins31 locus and was the e oligonucleotide for the downstream DSB. In the transformations where targeting to the side was stimulated by a DSB, the most Trp+ events were associated with CORE loss. The frequency of Trp+ events not associated with CORE loss was similar to the frequency of Trp+ events in the absence of a DSB (glucose). These findings are consistent with mechanisms that include the resection of a large region distant to the break during repair with oligonucleotides.
Interestingly, the presence of only one of the repairing oligonucleotides, R1 or R2, was sufficient to enhance strand-biased targeting. There was an approximate 7.5-fold increase in Trp+ transformants with the f oligonucleotide (Fig. 3C and E). However, there was no increased targeting by the e oligonucleotide.
The bias between oligonucleotides e and f in targeted recombination distant from the DSB suggests 5′ resection (at least 20 kb) from the DSB, which is consistent with resection detected by physical means (13, 21). Importantly, in every transformant that arose from a break that was repaired by an oligonucleotide(s) and in which there was oligonucleotide correction of sequence up to 20 kb away, the restoration of the ssDNA to the double-strand state must have occurred. The sufficiency of just a single R1 or R2 oligonucleotide to repair the resected DSB can be explained only if two separate SSA events lead to the recovery of Trp+ recombinants. These findings support the view that a DSB can be repaired by a ssDNA via a “template” mechanism, as discussed below (Fig. 4A).
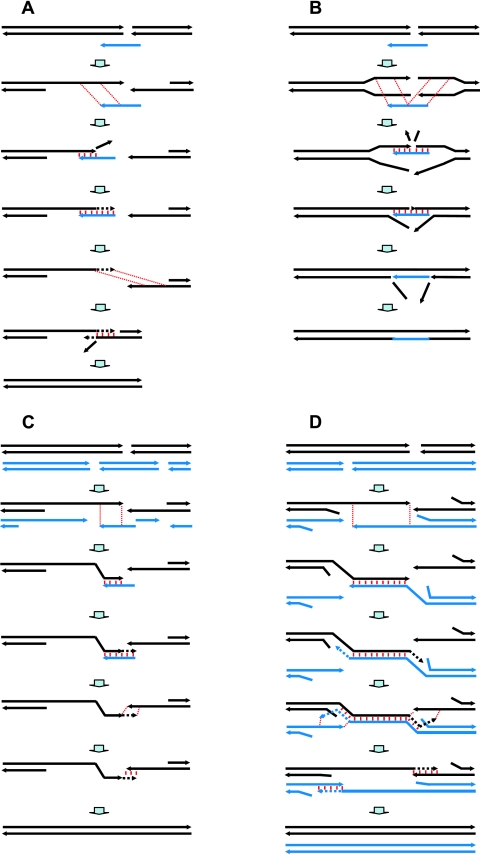
FIG. 4.
Models of rejoining of DSB ends by ssDNA via two SSA events. Annealing interactions between cDNA regions are shown as dotted, red parallel lines; actual pairing is identified as red, parallel lines that are vertical; DNA synthesis is shown as a dotted thick black line with an arrow. (A) Template intermediate model. The first annealing interaction between the single-strand repair oligonucleotide (blue arrow) and the DSB ends is presented as dotted, red parallel lines. The repairing oligonucleotide pairs with the homologous region on the 3′ strand after resection of the 5′ strand. The nonhomologous sequence is clipped away, and DNA synthesis can occur to copy the rest of the oligonucleotide. The original oligonucleotide may be removed by a helicase function, and a second annealing interaction occurs with the other 3′ end of the DSB as shown by the thin, dotted red lines. Pairing of the 3′ strands, followed by clipping of nonhomologous tails and subsequent DNA synthesis and ligation, completes the repair. (B) Bridge intermediate model. The repairing oligonucleotide (blue arrow) anneals through two events with both the 3′ and 5′ unwound ends of a DSB. Cleavage of displaced ends, gap filling, and ligation complete the repair event. The oligonucleotide region is either displaced or ligated after the cleavage of nonhomologous tails and DNA synthesis. (C) Multiple chromosomal breaks or closely spaced DSBs in homologous DNA molecules can be repaired via multiple rounds of SSA in a manner similar to that presented in panel A when strand invasion might not be available or as an alternative to strand invasion. Individual small chromosomal fragments after resection provide opportunities for SSA repair. The multiple breaks might result from closely spaced primary events or even at replication forks during replication of DNA with closely opposed single-strand lesions. (D) Repair of closely opposed DSBs in homologous molecules. Following DSB formation, there is degradation of the 5′ strands allowing an annealing interaction of the complementary single strands. DNA synthesis extends the 3′ strands. Branch migration events, due to equilibrium between the extended 3′ ends and the unwound 5′ tails, trigger reannealing of the extended 3′ strands to their initial chromosomal ends. DNA synthesis and ligation complete the repair of the DSB. Both original homologous molecules are repaired without rearrangements.
DISCUSSION
Single-strand DNA is formed during DNA replication, recombination, and repair. The ability to efficiently repair unique spontaneous and enzyme-induced DSBs with exogenously added single-strand oligonucleotides has provided opportunities to address minimal requirements for ssDNA in homologous recombination repair, characterize mechanisms of repair by SSA, and propose how SSA may play a more general role in DSB repair.
Mechanisms of DSB repair by single-strand oligonucleotides.
Based on the following results, we concluded that SSA is the major mechanism of DSB repair by single-strand oligonucleotides: (i) only the SSA domain of Rad52 was required for repair; (ii) NHEJ functions were not required for repair; (iii) strand invasion functions mediated by RAD51, RAD55, RAD57, RAD54, and RDH54 were not required for repair and even inhibited repair by single-strand oligonucleotide; and (iv) repair with single-strand oligonucleotides required that the ends of the DSB be open for resection or unwinding since hairpin-capped ends of a DSB generated by a closely spaced IR were unable to serve as substrates for repair unless cleaved by the MRX complex.
The models previously proposed for oligonucleotide repair accommodated intermediate steps involving strand invasion and/or SSA (37). The present findings exclude strand invasion in the repair process. Consistent with a proposed requirement that nonhomologous DNA tails must be removed from cDNA structures, the presence of the four proteins Rad1, Rad10, Msh2, and Msh3, which can function collectively as a “clippase,” provides for more efficient repair with ssDNA (Table 3).
The molecular models for repairing a DSB by ssDNA via SSA are presented in Fig. 4A and B. The models are based on “bridge” and “template” intermediates proposed previously (37). Each model of DSB repair by ssDNA invokes two SSA events. In the bridge model, the same oligonucleotide must anneal to each side of the break, whereas in the template model, the second annealing event involves pairing between the DNA copied from the oligonucleotide and a homologous sequence on the other side of the DSB. Since repair can occur with single-strand oligonucleotides having homology several kilobases away from the break, long stretches of ssDNA must be available for SSA. These can be created by 5′ to 3′ resection from the ends of DSBs. If both ends of a DSB are resected, only the template model can provide for repair by a single-strand oligonucleotide.
We developed a novel system in which resection associated with DSB repair by single-strand oligonucleotides (R1 and/or R2) was assessed by the extent of strand bias in the targeting of a second single-strand oligonucleotide to a sequence distant from the DSB (Fig. 3C to E). The bias as well as the ability to target and modify chromosomal DNA to one side of a DSB was observed over a large distance from the DSB (6 to 20 kb). Moreover, we found that regardless of which side of the DSB was repaired by a single repairing oligonucleotide, R1 or R2, there was a strong bias for targeting to the 3′ strand, consistent with 5′ end resection (Fig. 3E). These results can be explained only by the template model (Fig. 4A). In fact, all DSB repair by single-strand oligonucleotides in our experiments could be attributed to the template mechanism. The biased stimulation of single-strand oligonucleotide targeting to the side of a DSB could be used to determine the extent and polarity of resection under different physiological conditions, in different genetic backgrounds, and also in organisms other than yeast. Furthermore, our results may also have implications for experiments of gene targeting. The possibility of stimulating gene targeting with oligonucleotides or vectors at sites many kilobases away from a break may significantly expand the opportunity for genome modification and may lead to applications in organisms beyond yeast, including mammalian cells.
However, the bridge mechanism (Fig. 4B) cannot be completely excluded. An analysis of CORE-I-SceI loss events, along with targeting to sites distant from the DSB, provides support for the bridge mechanism in at least some repair events. After DSB induction, 0.3% of all viable cells experienced CORE-I-SceI loss as a result of R1 or R2 targeting (Fig. 3E). Yet, 6 to 13% of Trp+ colonies lost the CORE markers when cells were transformed with e and R1 or with e and R2 (Fig. 3E) (note that e is complementary to the 5′ strand at the DSB). This frequency of CORE marker loss events among the Trp+ is at least 20-fold higher than that predicted for random CORE marker loss and Trp+ coincident events. As expected for cells grown in glucose (i.e., no DSB induction), there was no loss of CORE markers among the Trp+ clones. These results indicate that targeting to one side of a break can also occur on the 5′ strand. In the framework of the template model, this could occur if (i) the 3′ end of a break anneals with the complementary repairing oligonucleotide (R) and extends on it and (ii) there is unwinding in the targeted TRP5::ins31 region so that targeting to the 5′ strand can be stimulated. Alternatively, targeting to the 5′ strand can be explained by a bridge model for SSA across a DSB where the repairing oligonucleotide anneals to both sides of a break. The choice between the two mechanisms, bridge versus template, in repair by a single-strand oligonucleotide might be a function of the extent of 5′ end resection, where rapid and extensive resection would favor the template model.
Biological relevance of DSB repair via multiple rounds of strand annealing.
Using single-strand oligonucleotides, we showed that a single-strand DNA molecule can mediate the repair of broken chromosomal ends via two SSA events. Sequences at the broken chromosomal ends are patched precisely in that the sequences in the oligonucleotides determine the joining (breakpoint) in the repaired chromosome. SSA between direct repeats is generally considered a nonconservative mechanism of DSB repair because of the loss of intervening material (44). However, if the sequence carried by the single-strand oligonucleotide exactly matches the ends of the DSB, the SSA-mediated repair would be conservative (i.e., restore broken DNAs as they were before the damage), even in cases when the resected single-strand regions extended for several kilobases.
Our findings not only are relevant to exogenously added ssDNAs but also provide a model for the repair of DNAs that experience two or more closely spaced DSBs. A high density of DNA lesions caused by ionizing radiation can result in clustered DSBs (42). For example, clustered DSBs can be directly induced by high-linear energy transfer radiation generating short DNA fragments (as close as 0.1 to 2 kb) (reference 34 and references therein). In addition, closely opposed cytologically detected chromatid breaks (isochromatid breaks) are frequently observed after exposure to high-linear energy transfer radiation. In human fibroblasts, the rejoining of these isochromatid breaks occurs quickly and leads to the appearance of slower kinetics for chromatid-type break rejoining (for details, see reference 15). In situations of closely opposed DSBs on homologous molecules, recombinational repair via strand invasion may be excluded since resection would progressively convert DNA in the region of the breaks to single strands (Fig. 4C). In fact, the DSBs could even be rather distant. For example, DSBs separated by many kilobases could become refractory to the strand invasion mechanism in a short time given the reported resection rate in yeast of 4 kb/h (41).
As demonstrated by our experiments with single-strand oligonucleotides in diploid cells (Fig. 2), SSA can provide a good alternative to the strand invasion mechanism of recombinational repair. We suggest that closely spaced chromosomal DSBs in sister chromatids or even homologous chromosomes that cannot be repaired by strand invasion because of resection could be efficiently and accurately repaired through the multiple events described in the template model for SSA. The minimal region of homology for SSA repair has not been determined directly. However, we found that an oligonucleotide with a 10-base homology to one side of a break could participate in repair (37), suggesting that closely opposed DSBs could be repaired, even when the distance between them is very small. The model for ssDNA repair of a break through rounds of SSA could also be applied to the repair of just two closely opposed breaks in sister chromatids or homologous chromosomes (Fig. 4D) through resection, association of 3′ ends, replication, and reannealing of fragments.
Along this line, the bacterium Deinococcus radiodurans can tolerate high doses of ionizing radiation that result in hundreds of genomic DSBs (8). These bacteria are able to quickly rejoin many DSBs within the first 1.5 h following irradiation by a recA-independent (i.e., Rad51-strand invasion independent) recombination mechanism (10), and the repair can occur without induction of genomic deletions, insertions, or rearrangements (43). We suggest that the repair model via rounds of SSA for clustered chromosomal DSBs involving extensive resection (Fig. 4C and D) can explain the recA- and NHEJ-independent repair in D. radiodurans. In general, the conservative DSB repair by SSA described here operates with a minimum number of recombination-associated functions and it could provide genome integrity under conditions of extensive damage, including high levels of DSBs.
.
Supplementary Material
Acknowledgments
We thank Jim Westmoreland for help with TAFE and Tom Petes and Greg Stuart for critical reading of the manuscript and comments.
This work was supported by intramural research funds from the National Institute of Environmental Health Sciences (NIH).
Footnotes
Published ahead of print on 14 August 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Asleson, E. N., R. J. Okagaki, and D. M. Livingston. 1999. A core activity associated with the N terminus of the yeast RAD52 protein is revealed by RAD51 overexpression suppression of C-terminal rad52 truncation alleles. Genetics 153:681-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aylon, Y., and M. Kupiec. 2004. DSB repair: the yeast paradigm. DNA Repair 3:797-815. [DOI] [PubMed] [Google Scholar]
- 3.Baur, M., I. Potrykus, and J. Paszkowski. 1990. Intermolecular homologous recombination in plants. Mol. Cell. Biol. 10:492-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belfort, M., and R. J. Roberts. 1997. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 25:3379-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boundy-Mills, K. L., and D. M. Livingston. 1993. A Saccharomyces cerevisiae RAD52 allele expressing a C-terminal truncation protein: activities and intragenic complementation of missense mutations. Genetics 133:39-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 7.Colaiacovo, M. P., F. Paques, and J. E. Haber. 1999. Removal of one nonhomologous DNA end during gene conversion by a RAD1- and MSH2-independent pathway. Genetics 151:1409-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox, M. M., and J. R. Battista. 2005. Deinococcus radiodurans—the consummate survivor. Nat. Rev. Microbiol. 3:882-892. [DOI] [PubMed] [Google Scholar]
- 9.Daley, J. M., P. L. Palmbos, D. Wu, and T. E. Wilson. 2005. Nonhomologous end joining in yeast. Annu. Rev. Genet. 39:431-451. [DOI] [PubMed] [Google Scholar]
- 10.Daly, M. J., and K. W. Minton. 1996. An alternative pathway of recombination of chromosomal fragments precedes recA-dependent recombination in the radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 178:4461-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott, B., C. Richardson, and M. Jasin. 2005. Chromosomal translocation mechanisms at intronic Alu elements in mammalian cells. Mol. Cell 17:885-894. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 13.Haber, J. E. 2002. Uses and abuses of HO endonuclease. Methods Enzymol. 350:141-164. [DOI] [PubMed] [Google Scholar]
- 14.Hefferin, M. L., and A. E. Tomkinson. 2005. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair 4:639-648. [DOI] [PubMed] [Google Scholar]
- 15.Kawata, T., H. Ito, T. Uno, M. Saito, S. Yamamoto, Y. Furusawa, M. Durante, K. George, H. Wu, and F. A. Cucinotta. 2004. G2 chromatid damage and repair kinetics in normal human fibroblast cells exposed to low- or high-LET radiation. Cytogenet. Genome Res. 104:211-215. [DOI] [PubMed] [Google Scholar]
- 16.Krejci, L., B. Song, W. Bussen, R. Rothstein, U. H. Mortensen, and P. Sung. 2002. Interaction with Rad51 is indispensable for recombination mediator function of Rad52. J. Biol. Chem. 277:40132-40141. [DOI] [PubMed] [Google Scholar]
- 17.Lander, E. S., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 18.Liang, F., M. Han, P. J. Romanienko, and M. Jasin. 1998. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl. Acad. Sci. USA 95:5172-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liefshitz, B., A. Parket, R. Maya, and M. Kupiec. 1995. The role of DNA repair genes in recombination between repeated sequences in yeast. Genetics 140:1199-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, F. L., K. Sperle, and N. Sternberg. 1984. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol. Cell. Biol. 4:1020-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lobachev, K., E. Vitriol, J. Stemple, M. A. Resnick, and K. Bloom. 2004. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr. Biol. 14:2107-2112. [DOI] [PubMed] [Google Scholar]
- 22.Lobachev, K. S., D. A. Gordenin, and M. A. Resnick. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108:183-193. [DOI] [PubMed] [Google Scholar]
- 23.Lopes, M., M. Foiani, and J. M. Sogo. 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell 21:15-27. [DOI] [PubMed] [Google Scholar]
- 24.Maryon, E., and D. Carroll. 1991. Characterization of recombination intermediates from DNA injected into Xenopus laevis oocytes: evidence for a nonconservative mechanism of homologous recombination. Mol. Cell. Biol. 11:3278-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald, J. P., and R. Rothstein. 1994. Unrepaired heteroduplex DNA in Saccharomyces cerevisiae is decreased in RAD1 RAD52-independent recombination. Genetics 137:393-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milne, G. T., and D. T. Weaver. 1993. Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev. 7:1755-1765. [DOI] [PubMed] [Google Scholar]
- 27.Muris, D. F., O. Bezzubova, J. M. Buerstedde, K. Vreeken, A. S. Balajee, C. J. Osgood, C. Troelstra, J. H. Hoeijmakers, K. Ostermann, H. Schmidt, et al. 1994. Cloning of human and mouse genes homologous to RAD52, a yeast gene involved in DNA repair and recombination. Mutat. Res. 315:295-305. [DOI] [PubMed] [Google Scholar]
- 28.Paques, F., and J. E. Haber. 1997. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6765-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfeiffer, P., W. Goedecke, S. Kuhfittig-Kulle, and G. Obe. 2004. Pathways of DNA double-strand break repair and their impact on the prevention and formation of chromosomal aberrations. Cytogenet. Genome Res. 104:7-13. [DOI] [PubMed] [Google Scholar]
- 31.Plessis, A., A. Perrin, J. E. Haber, and B. Dujon. 1992. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics 130:451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray, A., N. Machin, and F. W. Stahl. 1989. A DNA double chain break stimulates triparental recombination in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 86:6225-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudin, N., and J. E. Haber. 1988. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol. Cell. Biol. 8:3918-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rydberg, B. 2001. Radiation-induced DNA damage and chromatin structure. Acta Oncol. 40:682-685. [DOI] [PubMed] [Google Scholar]
- 35.Singleton, M. R., L. M. Wentzell, Y. Liu, S. C. West, and D. B. Wigley. 2002. Structure of the single-strand annealing domain of human RAD52 protein. Proc. Natl. Acad. Sci. USA 99:13492-13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stark, J. M., A. J. Pierce, J. Oh, A. Pastink, and M. Jasin. 2004. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 24:9305-9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Storici, F., C. L. Durham, D. A. Gordenin, and M. A. Resnick. 2003. Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc. Natl. Acad. Sci. USA 100:14994-14999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Storici, F., G. Henneke, E. Ferrari, D. A. Gordenin, U. Hubscher, and M. A. Resnick. 2002. The flexible loop of human FEN1 endonuclease is required for flap cleavage during DNA replication and repair. EMBO J. 21:5930-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Storici, F., L. K. Lewis, and M. A. Resnick. 2001. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 19:773-776. [DOI] [PubMed] [Google Scholar]
- 40.Storici, F., and M. A. Resnick. 2006. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 409:329-345. [DOI] [PubMed] [Google Scholar]
- 41.Sugawara, N., and J. E. Haber. 1992. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 12:563-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sutherland, B. M., P. V. Bennett, O. Sidorkina, and J. Laval. 2000. Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc. Natl. Acad. Sci. USA 97:103-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sweet, D. M., and B. E. Moseley. 1976. The resistance of Micrococcus radiodurans to killing and mutation by agents which damage DNA. Mutat. Res. 34:175-186. [DOI] [PubMed] [Google Scholar]
- 44.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Symington, L. S., P. Morrison, and R. Kolodner. 1985. Intramolecular recombination of linear DNA catalyzed by the Escherichia coli RecE recombination system. J. Mol. Biol. 186:515-525. [DOI] [PubMed] [Google Scholar]
- 46.Tsukamoto, M., K. Yamashita, T. Miyazaki, M. Shinohara, and A. Shinohara. 2003. The N-terminal DNA-binding domain of Rad52 promotes RAD51-independent recombination in Saccharomyces cerevisiae. Genetics 165:1703-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, X., C. Wu, and J. E. Haber. 1997. Rules of donor preference in saccharomyces mating-type gene switching revealed by a competition assay involving two types of recombination. Genetics 147:399-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.