Abstract
Human artificial chromosomes (HACs) are promising reagents for the analysis of chromosome function. While HACs are maintained stably, the segregation mechanisms of HACs have not been investigated in detail. To analyze HACs in living cells, we integrated 256 copies of the Lac operator into a precursor yeast artificial chromosome (YAC) containing α-satellite DNA and generated green fluorescent protein (GFP)-tagged HACs in HT1080 cells expressing a GFP-Lac repressor fusion protein. Time-lapse analyses of GFP-HACs and host centromeres in living mitotic cells indicated that the HAC was properly aligned at the spindle midzone and that sister chromatids of the HAC separated with the same timing as host chromosomes and moved to the spindle poles with mobility similar to that of the host centromeres. These results indicate that a HAC composed of a multimer of input α-satellite YACs retains most of the functions of the centromeres on natural chromosomes. The only difference between the HAC and the host chromosome was that the HAC oscillated more frequently, at higher velocity, across the spindle midzone during metaphase. However, this provides important evidence that an individual HAC has the capacity to maintain tensional balance in the pole-to-pole direction, thereby stabilizing its position around the spindle midzone.
The centromere is an essential functional domain responsible for the correct inheritance of eukaryotic chromosomes during cell division. The centromere region has a number of specific functions (1, 7, 34, 38): (i) assembling centromere/kinetochore components CENP-A, CENP-B, CENP-C, CENP-H, hMis6 (CENP-I), hMis12, and CENP-F, as well as microtubule motor proteins (CENP-E and dynein-dynactin) and mitotic checkpoint proteins (Mad2 and BubR1); (ii) capturing spindle microtubules, which align chromosomes at the metaphase plate and maintain balanced tension; (iii) resolving sister chromatid cohesion at the point of metaphase/anaphase transition; and (iv) moving the resolved chromatids toward each spindle pole.
Even in the most simple and well-characterized centromere, that of the yeast Saccharomyces cerevisiae, more than 65 protein components are involved in centromere structure and function, and many of them are conserved between yeast and humans (31). Although it is not clear how these evolutionarily conserved centromere components are assembled at the specific site on the chromosomes, chromatin assembly involving CENP-A, a centromere-specific histone H3 variant, appears to be an essential step in incorporating the many other centromere components and, thus, in specifying the position of the centromere (15, 24, 47, 58, 61). Several lines of evidence also support the importance of epigenetic mechanisms. On stable dicentric chromosomes created by chromosome rearrangements, most active centromere markers do not assemble on the inactive centromere, despite the presence of human centromere-specific alphoid DNA (9, 57). A more striking phenomenon is the stable maintenance of a “neocentromere.” On rearranged chromosome fragments from patients, in rare cases, a functional centromere called a neocentromere forms in the complete absence of alphoid DNA (8).
Centromeric DNA organization is divergent among species, and these centromere structures are formed and maintained on species-specific centromere DNA in humans, mouse, rice, maize, and budding and fission yeasts (7, 23, 31, 40, 42, 57). The relevant centromere DNA sequence in the yeasts S. cerevisiae and Schizosaccharomyces pombe, if introduced as naked DNA into yeast cells, can induce the assembly of a functional centromere by a de novo mechanism (12, 19). All normal human centromeres are assembled and maintained at the megabase level of organization by repetitive loci composed of 171-bp repeat units of α-satellite (alphoid) DNA (18, 26, 63). The importance of alphoid DNA has also been demonstrated. It is required for the maintenance of truncated minichromosomes (22, 41) and in de novo assembly of human artificial chromosomes (HACs) (10, 11, 16, 21, 27, 39).
A yeast artificial chromosome (YAC) or bacterial artificial chromosome (BAC) clone containing a 60- to 70-kb α21-I alphoid array from chromosome 21 efficiently formed HACs, which bound CENP-A, CENP-B, CENP-C, and CENP-E in human cultured cells. These HACs were predominantly present at one copy per cell and segregated stably at 98.4 to 99.9% stability per cell division in the absence of selection. HACs were formed by a de novo mechanism, without acquisition of detectable host sequences, which in all cases involved multimerization of the input YAC/BAC DNA of about 30 to 50 copies (∼3 Mb), indicating that the repeated structure of alphoid DNA containing CENP-B binding sites (CENP-B box) and the YAC (BAC) arms including a selectable marker gene might be sufficient to establish mitotically stable chromosomal structures and centromere functions by a de novo mechanism (3, 27, 48). Thus, the first-generation HACs still have a crucial advantage for identifying the important structures required for a stable human chromosome and for analyzing mechanisms of centromere assembly. Moreover, in addition to functional kinetochore assembly, sister chromatid cohesion and its resolution mechanisms which involve passenger proteins (INCENP and Aurora B, e.g.) (34) and heterochromatin (HP1) (17, 60) might be required for the proper segregation of chromosomes. Thus, it is important to clarify whether de novo created HACs composed of the multimer alphoid DNA insert and YAC (BAC) vector arms have the capacity to carry out each step of chromosome segregation properly. However, HACs are very small, having negligible chromosome arms compared to host chromosomes, and thus are difficult to distinguish without fluorescent in situ hybridization (FISH) analysis of fixed and spread metaphase-arrested cells that have been treated with inhibitors of microtubule assembly (27, 36, 52). In addition, even natural chromosomes display increased numbers of abnormal kinetochores (merotelic attachment of kinetochore microtubules) at a 15-fold higher rate than that seen in normal PtK1 cells after treatment with an inhibitor of microtubule polymerization (6).
In this study, we used the green fluorescent protein (GFP)-Lac repressor fusion protein (GFP-LacR) and Lac operator system (51) to follow the segregation of individual HACs in living cells in detail. Time-lapse analyses of GFP-tagged HACs and host centromeres in living cells during mitotic cell cycles indicated that the HACs were properly aligned at the spindle midzone. Sister chromatids of the HAC separated with the same timing as the host sister centromeres and moved to each spindle pole with mobility similar to that of the host centromeres. Although the HACs oscillated more frequently, at higher velocity, around the spindle midzone than the host chromosome during metaphase, individual HACs had the capacity to maintain balanced tension in the pole-to-pole direction. Thus, a HAC composed of a multimer of input α-satellite YACs retains most of the functions expected of the centromeres on natural chromosomes.
MATERIALS AND METHODS
Cell lines.
HT1080 cells (ATCC CCL121) were cultured in Dulbecco's modified Eagle's medium (DMEM; Nissui) supplemented with 10% (vol/vol) fetal bovine serum, penicillin, streptomycin, and l-glutamine at 37°C in 5% CO2. To establish a cell line [HT1080GFP-LacI(11-3)] expressing the GFP-Lac repressor fusion protein (GFP-LacR), HT1080 cells were transfected with XmnI-linearized p3′ SS dimer Imp GFP plasmid (a gift from Andrew S. Belmont [51]) containing a GFP-Lac repressor fusion gene with the F9-1 promoter and the hygromycin gene with the thymidine kinase promoter by Lipofectamine (Invitrogen) according to the manufacturer's instructions. To construct cell lines expressing red fluorescent protein (RFP), CENP-C protein fused to RFP (DsRed1) (pRFPCENP-C [59]) was introduced by Lipofectamine transfection into the HT1080GFP-LacI(11-3) cells containing a HAC or a host centromere integration signal derived from α7C5opYAC DNA.
Replacement of the left arm of an alphoid YAC with the arm containing multiple Lac operators.
The left-arm replacement vector (pLmegaLacOpuro), a modified version of pMegaΔ (27), contains a SCAN linker (SalI and NotI sites), a telomere cassette (1.1 kb of mammalian TTAGGG telomere repeats) flanked by 0.3 kb of yeast TG(1-3) telomere repeats, and an SalI/ApaI fragment of 256 copies of the Lac operator derived from pSV2-DHFR-8.32 (where DHFR is dihydrofolate reductase; a gift from Andrew S. Belmont [51]) inserted at the SpeI, PvuII, and AatII sites, respectively, of YCp lac111 (14), which contains LEU2-ARS1-CEN4. Before the SalI/ApaI fragment containing 256 copies of the Lac operator was dissected, a 1.7-kb SalI fragment derived from pGKpuro (62) was inserted into the XhoI site of pSV2-DHFR-8.32. The SCAN linker consisted of 5′-CTAGTCGACCATCGATACCAATGCATTGGCGGCCG-3′ and 5′-CTAGCGGCCGCCAATGCATTGGTATCGATGGTCGA-3′ sequences.
In retrofitting the left YAC arm, 500 ng of pLmegaLacOpuro linearized with NotI and SalI and 500 ng of uncut YpSL1-Ura (Rad52 expression plasmid) were used to cotransfect 2 × 108 EPY305-5b cells (MATa rad52-D2000 leu2-D1 lys2 ade2-101 his3-D200 trp1::hisG ura3-52) (32), containing α7C5hTELYAC, by the lithium acetate method as previously described (27). The replacement of the left YAC arm of the retrofitted YAC (named α7C5opYAC) was confirmed by selection, the strain's growth requirements (Leu+ and Lys−), pulsed-field gel electrophoresis, and Southern hybridizations.
Construction of an alphoid BAC containing multiple Lac operators.
To generate a BAC containing alphoid DNA (pBACα7C5), the 70-kb NotI fragment of α21-I alphoid DNA from α7C5hTELYAC was inserted into the NotI site of pBeloBAC11 (30). Next, the XhoI fragment containing 256 copies of Lac operators derived from pSV2-DHFR-8.32 (51) and the PvuII/EcoRI fragment of the SV2bsr gene derived from pSV2bsr (28) with a linker were inserted into the XhoI site of pBACα7C5. This BAC was named pBACαLacObsr.
YAC or BAC transfection.
YAC DNAs were purified by pulsed-field gel electrophoresis as previously described (27). A total of 25 ng of purified α7C5opYAC DNA was mixed with 100 ng of α7C5hTEL YAC DNA in 400 μl of 10 mM Tris (pH 7.4), 0.1 mM EDTA, and 100 mM NaCl and used to transfect 80% confluent HT1080LacI/GFP cells in 3.5-cm dishes, using Lipofectamine according to the manufacturer's instructions. BAC DNAs were purified using a QIAGEN large construction kit (QIAGEN). Using 4.5 μl of Lipofectamine, 0.4 μg of purified BAC DNAs (a 1:4 mixture of pBACαlacObsr and pBACα7C5) was used to transfect HT1080LacI/GFP cells. Blasticidin S-resistant cells were selected with 4 μg/ml blasticidin S (Kaken Seiyaku) for more than 3 weeks and then analyzed by FISH.
De novo HAC formation analyses by FISH.
Standard techniques for FISH were carried out for the alphoid YAC- and BAC-transformed cell lines as previously described (35). The probes used were p11-4 alphoid DNA (26) for the α21-I loci and pYAC4 (5) for YAC arm DNA. Pan-alphoid and intra- and inter-Alu sequences were described in our previous reports (27, 36). The BAC vector template was generated by PCR as described in our previous report (48). Plasmid DNAs and PCR products were labeled using a nick translation kit with digoxigenin 11-dUTP or biotin 16-dUTP (Roche Diagnostics). Images were captured using a cooled charge-coupled-device camera (PXL; Photometrics Ltd.) and analyzed by IPLab software (Signal Analytics).
Indirect immunofluorescence and simultaneous staining by FISH.
Indirect immunofluorescence and simultaneous staining by FISH were carried out as previously described (35). Cytospun mitotic cells were fixed in fixation solution (4% paraformaldehyde, 136.9 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.47 mM KH2PO4) for 15 min and washed two times in phosphate-buffered saline. The cells were then treated with 0.5% Triton X-100 and 0.1 M glycine for 5 min each. Antibodies used were anti-CENP-A (mAN1) (36), anti-CENP-B (2D8D8) (48), anti-CENP-C (CGp2) (59), and anti-GFP (Roche Diagnostics or Medical and Biological Laboratories) (44).
Live-cell observation.
Observation of live cells was carried out according to a modified version of a published method (20). Cells cultured in a 35-mm glass-bottom dished were stained with 1 μg/ml Hoechst 33342 (Invitrogen) for 10 min and washed twice with DMEM supplemented with 10% fetal bovine serum. The cells were cultured in phenol red-free DMEM supplemented with 10% fetal bovine serum, penicillin, and streptomycin at 37°C in a 5% CO2 atmosphere for 30 min. These cells were then placed in a microscopic stage chamber (Zeiss CZI-3, equipped with a temperature and CO2 control set and an objective heater). Observation was carried out, and images were taken using a Zeiss microscope (Axiovert 200; AttoArk Hg lamp controller adjusted to 50 W) equipped with a cooled charge-coupled-device camera (MicroMax; Princeton Instruments) coupled to MetaMorph (Universal Imaging) at 15 Z-steps in every 3- or 5-min capture period. The captured images were used to produce reconstructions of the image stacks with AutoDeblur (AutoQuant Imaging).
Data analyses of captured digital images from live cells containing GFP signals from different focal planes were stacked and produced the image of the HAC or the host chromosomal centromeres at each time point. Relative distance between the GFP signal and the spindle equator at the each time point was traced and plotted with Photoshop (Adobe). Velocities (μm/min) of HACs and host centromeres were calculated and plotted from the distances that GFP signals moved within each time interval (3 or 5 min).
Size analyses of minichromosomes and quantitation of alphoid YAC arms.
Metaphase cells containing minichromosomes were stained with 5 μg/ml propidium iodide after methanol/acetic acid (3:1) fixation. Fluorescence from 10 minichromosomes and chromosome 21 of each cell line was analyzed using IPLab. Minichromosome sizes were determined by quantitation of the fluorescence and comparison to the long arm of chromosome 21 (∼50 Mb). The copy numbers of the YAC on the minichromosomes were also determined to be 24 or 33 copies (including 3 or 10 copies of α7C5opYAC) by real-time PCR of the left and the right YAC arm DNA (43).
RESULTS
Generation of a HAC detectable with the GFP-LacR/LacO system.
To investigate the precise segregation of a HAC in living mitotic cells, we introduced a GFP-Lac repressor fusion protein (GFP-LacR) and Lac operator system into our HAC (Fig. 1). A 10-kb fragment containing 256 copies of the Lac operator sequence (LacO) was integrated into a HAC precursor YAC (α7C5hTEL) or BAC (pBACα7C5) containing 70 kb of centromere-competent 21-I alphoid DNA from the centromere region of chromosome 21. Derived YAC DNA (α7C5opYAC) or BAC DNA (pBACαLacObsr) was purified and introduced into human cultured HT1080 cells expressing GFP-LacR [HT1080GFP-LacI(11-3)]. As we obtained only low numbers of transformants with individual YAC or BAC DNA constructs containing the inserted LacO, we could not obtain transformants showing GFP signals in the repeated experiments (see Discussion). Next, purified α7C5opYAC DNA (or pBACαLacObsr DNA) containing the LacO fragment was mixed with original α7C5hTEL YAC DNA (or pBACα7C5, respectively) at a 1:4 molar ratio and introduced into HT1080GFP-LacI(11-3) cells. In the previous reports, despite the multimerized structure of input DNA molecules, HACs were formed efficiently from the mixture of input YAC or BAC DNA (16, 44). In the present result, among 17 analyzed stable transformants from the YAC DNA mixture, seven cell lines showed one to three GFP signal spots in the nuclei of almost all living cells (Fig. 2A). FISH analysis of metaphase spreads from these seven cell lines using a YAC vector probe and α21-I alphoid probe indicated that high-efficiency HAC formation as an extra minichromosome with GFP signals occurred in two cell lines (94% and 77% of spreads) (Table 1 and Fig. 2B and C). The other five cell lines showed the YAC integration signals in the region of either the telomere or the centromere of host chromosomes. The efficiency of HAC formation (about 30% of transformants) and the other fates of the introduced YAC are both very similar to our previous results using an original alphoid YAC without LacO sequence (27). We also obtained two HAC-containing cell lines (98% and 67% of spreads) from 24 analyzed cell lines derived from BAC DNA transfection. Our structural analyses of the two independent HACs derived from YAC mixture (α7C5YAC and α7C5opYAC) indicated that no signal showed an acquisition of the host chromosomal arm fragment on the both HACs with inter- and intra-Alu PCR probes by FISH analysis, while all host chromosome arms were detectable with these probes (Fig. 3A). No host centromeric alphoid DNA was detected on the HACs other than the alphoid DNA derived from chromosomes 13 and 21, which cannot be distinguished from the insert alphoid DNA of the YACs by FISH analysis with pan-alphoid probes (Fig. 3B). The copy number of the input alphoid YAC DNA on the two HAC cell lines analyzed by real-time PCR presents, in total, 24 or 33 alphoid YAC copies (corresponds to a total size of 2.4 or 3.4 Mb, respectively, including 3 or 10 copies of α7C5opYAC DNA). These data are consistent with the size estimated from the DNA content of the HACs: about 1/10 to 1/20 (about 3 to 5 Mb) of the length of the long arm of chromosome 21 (see Materials and Methods). Moreover, chromatin immunoprecipitation analysis showed that the essential marker for the functional centromere, CENP-A, directly assembled on the input alphoid YAC DNAs (see Fig. S1 in the supplemental material). In agreement with our previous structural and chromatin immunoprecipitation analyses of independent and reproducible HACs derived both from the same insert alphoid DNA and the synthetic α21-I alphoid DNA (27, 36, 43, 44, 48), all these results are consistent with nonacquisition of host chromosome sequences during HAC formation with amplified introduced DNAs. However, it would be difficult to exclude a possible presence of small host DNA sequence below the threshold of any detection methods.
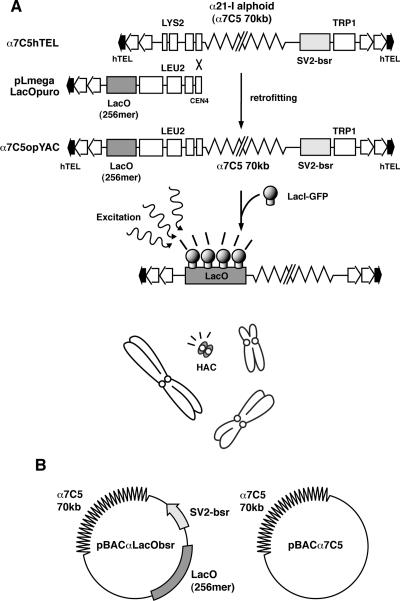
FIG. 1.
Alphoid YAC/BAC constructs with Lac operators. (A) α7C5hTEL YAC has 70 kb of α21-I alphoid DNA (α7C5). The left arm of the α7C5hTEL YAC was replaced with the vector (pLmegaLacOpuro) by homologous recombination in yeast. The resultant YAC vector (named α7C5opYAC) contains 256 copies of Lac operators on its left arm. The HAC containing α7C5opYAC DNA is made detectable by the binding of GFP fusion Lac repressor proteins (GFP-LacR) expressed in living cells. (B) Scheme of the BAC vectors containing α7C5 alphoid DNA. Both alphoid BACs contain 70-kb α21-I alphoid DNA derived from α7C5hTEL YAC. pBACαLacObsr contains 256 copies of Lac operators, as does the α7C5opYAC vector, and the SV2bsr gene as a selective marker.
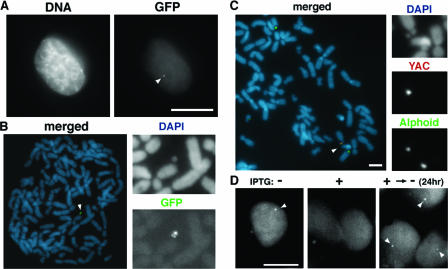
FIG. 2.
Artificial chromosomes containing α7C5opYAC DNA can be detected as green fluorescence signals of GFP-LacR. (A) Introduced α7C5opYAC was observed as a tiny green fluorescent spot of GFP-LacR (arrowhead in right frame) in a living transformed cell. DNA was counterstained with Hoechst 33342 (left). (B) GFP-LacR fusion protein specifically assembled on the minichromosome containing α7C5opYAC (arrowhead). Cytospun metaphase chromosomes were immunostained with anti-GFP antibody (green), and chromosomes were counterstained with DAPI (blue). (C) FISH analysis of a minichromosome containing α7C5opYAC (arrowhead). Cells were hybridized with a YAC vector probe (red) and α21-I alphoid DNA probe (green). Chromosomes were counterstained with DAPI (blue). (D) The association and dissociation of GFP-LacR with the minichromosome containing α7C5opYAC were reversibly controlled by IPTG treatment. The signal of GFP-LacR on the GFP-HAC (arrowhead in left frame) disappeared with 0.2 mM IPTG treatment for 2 h (middle frame). At 24 h after removal of IPTG, GFP-LacR signals were again observed (arrowheads in right frame). The data from cell line op29 were used. Scale bars, 10 μm.
TABLE 1.
Efficiency of HAC formation by transfection with alphoid YAC/BAC DNA containing LacOa
| Introduced DNA mix | No. of cell lines analyzed | Fate of transfected DNA by FISH analysis (no. of cell lines)
|
|||
|---|---|---|---|---|---|
| HAC formation | Host chromosomal integration regionb
|
||||
| Tel | Cen | Arm | |||
| α7C5opYAC-α7C5hTEL YAC | 7 | 2 | 4 | 1 | 0 |
| pBACαLacObsr-pBACα7C5 | 24 | 2 | 0 | 17 | 5 |
Cell lines were classified according to transfected DNA fate as measured by FISH and GFP analysis. In each category, more than 50% of FISH signals were on either minichromosomes (two 7C5hTEL cell lines) or host chromosomes.
Tel, telomeric; Cen, centromeric; Arm, chromosome interstitial integration sites.
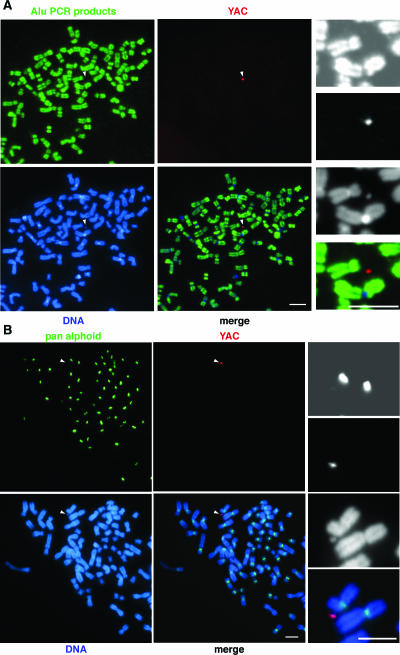
FIG. 3.
Structural analyses of the GFP-HAC by FISH. (A) Metaphase GFP-HACs were hybridized with a mixture of intra- and inter-Alu PCR products (green signals) and the YAC vector probe (red signals). DNA was counterstained with DAPI (blue signals). No positive signals were detected on the GFP-HACs (arrowhead) using Alu PCR probes, while the strong hybridization signals were detected on whole host chromosome arms (right upper frame). (B) Metaphase GFP-HACs were hybridized with the PCR-amplified pan-alphoid DNA probes (green signals) designed to detect all human centromeres and the YAC vector probe (red signals) and counterstained with DAPI (blue signals). The excess amount of nonlabeled α21-I alphoid DNA was added to the hybridization mixture as a competitor. The signals of pan-alphoid DNA were specifically competed out from the GFP-HAC (arrowhead) and chromosomes 13 and 21, confirming the absence of the other host centromeric alphoid DNA sequences on the GFP-HAC. Bars, 5 μm.
Distribution of centromere proteins relative to LacO/GFP-LacR on the HAC.
Next, we analyzed the correlation of the GFP-LacR-tagged sites with the sites of centromere protein assembly on the HACs and on the YAC integration site of the host centromere as a control, using indirect immunofluorescence staining with specific antibodies on fixed spreads of metaphase chromosomes. In the four HAC cell lines derived from the YAC and the BAC transformants, GFP-LacR signals overlapped with extrachromosomal DAPI (4′,6′-diamidino-2-phenylindole) signals and CENP-B (a specific binding protein of type-I alphoid DNA) staining regions and were always accompanied by staining for the inner kinetochore essential proteins, CENP-A and CENP-C, both of which assemble on only the active centromere/kinetochore (Fig. 4A). No other specific GFP signal was detected on the host chromosomes in these cell lines; there was a weak background of GFP-LacR over the entire nucleus and the chromosomes (Fig. 2A and B). In the cell line in which the YAC was integrated into the host centromere (op10), the GFP signal also overlapped with CENP-A, -B, and -C assembled at the canonical host centromere/kinetochore (Fig. 4B). Thus, centromere/kinetochore components (CENP-A, -B, and -C) assembled on HACs containing GFP-LacR bound to multimerized LacO.
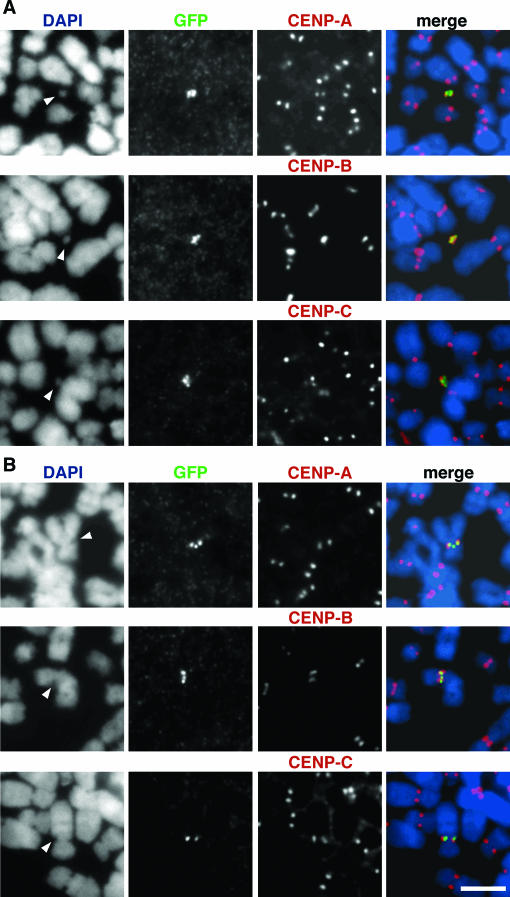
FIG. 4.
Assembly of centromere components on human artificial chromosomes containing α7C5opYAC (GFP-HAC). Metaphase GFP-HACs in the op29 cell line (A) and a GFP-host centromere in the op10 cell line (B) were analyzed by indirect immunofluorescence using anti-CENP-A, anti-CENP-B, or anti-CENP-C (all red, as indicated on the figure) and anti-GFP (green) antibodies. Chromosomes were counterstained with DAPI (blue). Arrowheads indicate HACs (A) or host centromeres integrated by α7C5opYAC (B). Scale bar, 10 μm.
Stability of HACs containing GFP-LacR binding sites.
Stable HACs are formed from the multimerized structure of the input alphoid YAC/BAC DNA and have been shown to be composed of a regular multimerized structure of an input alphoid YAC DNA if linear DNA molecules terminating with telomere sequences are used (27, 36, 43). Consequently, we have focused our further analyses using the GFP-LacR/LacO system on YAC-based HACs. We obtained sublines of cells containing one copy (op21-510 and op21-516) and multiple copies (mainly two copies; op21-410 and op29-13) of HACs in the majority of cells by subcloning from the original cell lines (op21 and op29). The stability of the HACs was analyzed by observation of GFP signals in living cells (Fig. 2A); (this method can avoid errors due to spreading) and on cytospun metaphase spreads (Fig. 2B) or by FISH on metaphase spreads (Fig. 2C) during 10 weeks (70 days) of growth in nonselective medium. The cell population containing two to three HAC copies fluctuated in a small number, and then the cell populations with no HAC signal increased gradually during long culture. This result was consistent and independent of the detection method (Table 2) (the loss rate R = 0.0064 to 0.0090 for op21-510 and op21-516), suggesting that this small instability might be caused by irregular nondisjunction and consequent chromosome loss events during passage in culture. The cell sublines containing multicopy HACs (op21-410 and op29-13) also maintained a loss rate lower than an R value of 0.014 (data not shown). All the results showed that HACs were maintained substantially stably, with an efficiency of 98.6 to 99.4% per cell division. When isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the medium for 2 h, GFP-LacR signals disappeared from the HAC, but the signals on the HAC reappeared when IPTG was removed from the medium (Fig. 2D). Thus, GFP-LacR binding to the HAC is reversible. However, the stability of the HAC was not changed by preventing GFP-LacR binding to the HAC by adding IPTG for an additional 9 weeks (63 days) of culture (Table 2) (R = 0.0074). Thus, the stability of HACs containing the GFP-LacR/LacO system is similar to that of HACs based on an alphoid YAC without the GFP-LacR/LacO system (98.4 to 99.5% stability per cell division) (27) and can be followed by the GFP signal in living cells without using FISH analyses. The GFP signal from the YAC integrated into the host centromere (op10) was quite stable (R < 0.001; data not shown).
TABLE 2.
Comparison of HAC stability tested by living cell, cytospin, and FISH analysesa
| Cell line | No. of days | Analysis method | Total no. of metaphase cells | Fate of transfected DNA (%)b
|
HAC loss rate (R)c | ||||
|---|---|---|---|---|---|---|---|---|---|
| No. of HACs/cell
|
Integration signal | No signal | |||||||
| 1 | 2 | 3 | |||||||
| op21-510 | 0 | Living cell | 36 | 97 | 3 | 0 | ND | 0 | |
| 0 | Cytospin | 38 | 97 | 0 | 0 | 0 | 3 | ||
| 14 | Living cell | 52 | 88 | 4 | 0 | ND | 8 | ||
| 14 | FISH | 67 | 91 | 4.5 | 0 | 0 | 4.5 | ||
| 28 | Living cell | 57 | 86 | 3 | 0 | ND | 11 | ||
| 28 | FISH | 68 | 82 | 6 | 2 | 0 | 10 | ||
| 42 | Living cell | 53 | 85 | 0 | 0 | ND | 15 | ||
| 42 | FISH | 67 | 78 | 10 | 3 | 0 | 9 | ||
| 56 | Living cell | 64 | 61 | 6 | 5 | ND | 28 | ||
| 56 | FISH | 93 | 67 | 5 | 0 | 0 | 28 | ||
| 70 | Living cell | 53 | 57 | 5 | 0 | ND | 38 | 0.0064 | |
| 70 | FISH | 58 | 47 | 5 | 0 | 0 | 48 | 0.0090 | |
| op21-516 | 0 | Living cell | 23 | 100 | 0 | 0 | ND | 0 | |
| 0 | Cytospin | 34 | 94 | 3 | 0 | 0 | 3 | ||
| 70 | Cytospin | 63 | 57 | 3 | 0 | 0 | 40 | 0.0072 | |
| op21-410 | 0 | Living cell | 89 | 27 | 67 | 5 | ND | 1 | |
| 0 | Cytospin | 51 | 12 | 78 | 6 | 0 | 4 | ||
| op21-510 | 49d | FISH | 68 | 68 | 0 | 0 | 0 | 32 | 0.0074 |
| 112e | FISH | 66 | 41 | 2 | 0 | 0 | 57 | 0.0074 | |
Metaphase cells were prepared from cell lines after 0, 2, 4, 6, 8, or 10 weeks in the absence of blasticidin S selection. The fate of the HAC was scored by the observation of GFP signals in living cells (Living cell) or on metaphase spreads (Cytospin) or by FISH analysis on metaphase spreads (FISH).
Based on signals from minichromosomes or integrated DNA. In observations of living cells, GFP signals from minichromosomes and integrated DNA are not distinguishable (ND).
The HAC loss rate (R) was calculated by the following formula: N70 = N0 × (1 − R)70. N0 and N70 are the rates for HAC-containing cells in each cell line at the time points of day 0 and day 70 (10 weeks).
IPTG was absent during the culture.
IPTG was added at day 49.
Real time analyses of HACs in living mitotic cells.
Before starting our precise, real-time observation of HAC segregation, we measured the average periods for each mitotic phase in normal HT1080 cells under the same conditions used for GFP signal analysis. We used a fluorescent microscope with a stage chamber at 37°C and 5% CO2, in which HT1080 cells grow normally for at least 3 days. In a normal HT1080 cell, the pole-to-pole orientation of the spindle rotates from vertical to horizontal during prometaphase. We defined the period “prophase to prometaphase” expediently as from nuclear membrane breakdown to the end of the spindle's rotation to the horizontal position, “metaphase to anaphase onset” as from the end of spindle rotation to the start of sister chromatid separation, and “anaphase to telophase” as from anaphase onset to the end of cytokinesis. In normal HT1080 cells, on average, prophase to prometaphase was 40.6 min (n = 5), metaphase to anaphase onset was 42.2 min, and anaphase to telophase was 18.6 min; the total time of the mitotic phases was 101.4 min (Table 3). The most sensitive mitotic periods (from metaphase to anaphase onset in HT1080 cells) were not affected by continuous exposure to B excitation (Zeiss AttoArk at 50W) for 10 min or to GFP-specific excitation but were arrested by a 30-s continuous exposure to UV excitation (Table 3). The metaphase to telophase mitotic periods of HT1080-derived cells containing one or two copies of an HAC (op29 cell line) and the host centromere YAC integration signals (op10 cell line) were not significantly different from those of the parental HT1080 cells during our time-lapse analyses of GFP signals (Table 3).
TABLE 3.
Periods of mitotic phases of GFP-HAC-containing cells and normal HT1080 cells
| Cell line and treatment | Avg time (min) of mitotic period (range)a
|
|
|---|---|---|
| Metaphase to anaphase onset | Anaphase onset to telophase | |
| HT1080 | 42.2 (36-50) | 18.6 (13-23) |
| B excitation for 10 min | 38.2 (35-42) | 20.6 (16-27) |
| UV excitation for 5 s | 39.6 (33-45) | 22.0 (16-30) |
| UV excitation for 30 s | Arrest | |
| op29b | 38.4 (37-40) | 17.6 (12-21) |
| op29c | 40 | 18 |
| op10d | 37.2 (33-40) | 21.3 (15-30) |
Mean value from observations of five individual live cells. All values except that of HT1080 with UV excitation for 30 s showed no significant differences by Student's t test (P > 0.05).
Cell line with 1 HAC by GFP analysis.
Cell line with 2 HACs by GFP analysis. Data are from a single cell.
Cell line with host centromere YAC integration signal.
Timing of sister chromatid separation.
To analyze HAC segregation, we used two different approaches using living cells. One was coobservation of GFP-HAC signals and centromeres of host chromosomes, which were visualized by RFP fused to CENP-C (RFP-CENPC). The second method was real-time observation and comparison of GFP-HAC signals and GFP-host centromere signals during mitosis using time-lapse analyses.
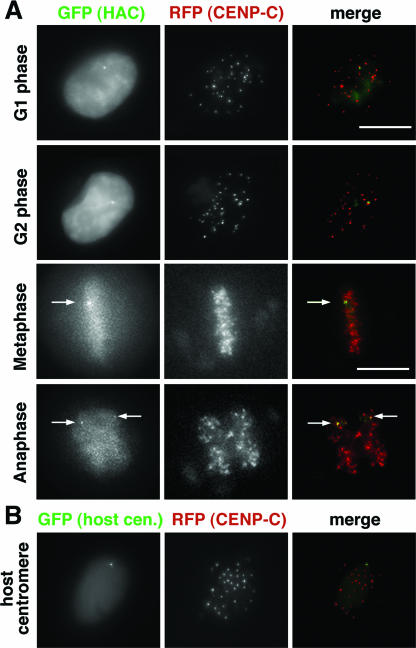
For the first approach, RFP-CENPC was expressed in the cell lines containing α7C5opYAC DNA. Through the cell cycle from G1 to G2 in a living cell, GFP signals on a HAC and on a centromeric integration site of a host chromosome were closely associated with one (own centromere) of the centromeres detected by RFP-CENP-C signals (Fig. 5A and B). During the periods from G2 to metaphase, GFP signals on the HAC were detected between a duplicated pair of RFP-CENP-C signals (Fig. 4A and 5A in metaphase). From the prometaphase to metaphase periods, GFP signals on the HAC aligned at the metaphase plate with the same timing as the host RFP-CENP-C centromeres. At anaphase onset, both GFP signals on the sister chromatids of the HAC separated into two signals at the same time as the sisters of the host centromeres and moved in the directions of the spindle poles. At least from the observations of a total of 200 GFP-HAC signals in live cells (op29) in metaphase and anaphase, we failed to detect a lagging HAC signal that was apart from host centromere movements or a nondisjunction signal (for example, 2:0 separation), consistent with the low loss rate. Thus, most of the centromeres of the artificial chromosomes function to align the chromosome accurately at the metaphase plate, maintain sister chromatid cohesion until the end of metaphase, and resolve the cohesion at the same time as host chromosome's sister centromere separation, in synchrony with the mitotic cell cycle progression to anaphase onset.
FIG. 5.
GFP-LacR signals on the HAC and the host centromere colocalized with CENP-C through the cell cycle. GFP-LacR (green) and CENP-C fused with RFP (red) on the GFP-HAC in op29 cell line (A) and the GFP-host centromere in op10 cell line (B) were observed in living cells. Arrows indicate GFP-HACs. Scale bars, 10 μm.
Dynamics and velocity of an HAC during metaphase and anaphase.
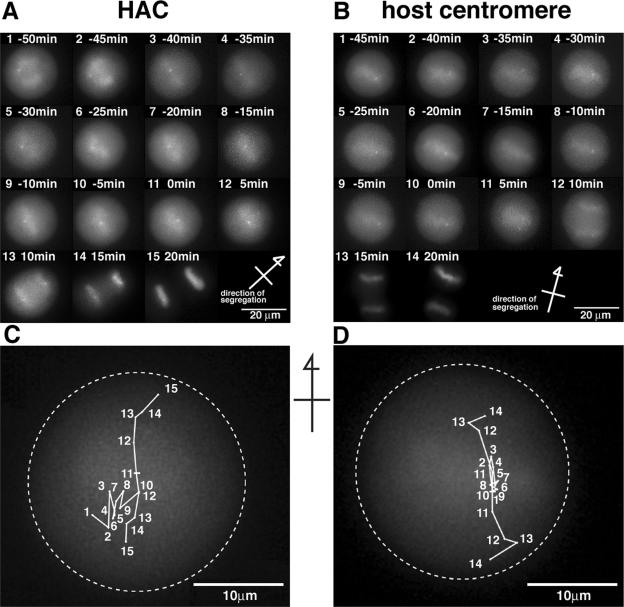
We asked if the centromere of the artificial chromosome controls the balance of tension between the sister kinetochores and spindle poles and if it has motor activities on spindle microtubules equivalent to those of the centromeres on host chromosomes (38, 45, 54). We observed the dynamics of the HACs by time-lapse analysis in living cells in mitosis. Time-lapse images of GFP-HAC and GFP-host centromeres in living cells during late prometaphase to telophase are shown in Fig. 6. GFP signals from a HAC and a host centromere were detected as single spots and closely colocated double dots, respectively, near the spindle equator, from the end of prometaphase to metaphase (Fig. 6A and B; see Fig. S2, S3, and S4 in the supplemental material). At anaphase onset (Fig. 6A and B, where time zero indicates the last time point before sister chromatid separation), the GFP signals from both were separated into two spots near the spindle equator (Fig. 6C and D; points 11 and 10, respectively, indicate HAC and host centromere signals) and moved toward each spindle pole. This timing and positioning are equivalent to those seen during host sister chromatid separation (Fig. 5).
FIG. 6.
Analysis of GFP-LacR signal trails during mitosis in live cells. Images from time-lapse analysis of live cells showing the GFP-LacR signals on GFP-HACs (A) and GFP-host centromeres (B). Scale bar, 20 μm. Time zero is the last time point at which the sister chromatids of the GFP-HAC or the GFP-host centromere were still associated, just before anaphase onset. Arrows indicate the direction of the chromosome movement during anaphase onset (pole-to-pole direction). The trails of GFP-HACs (C) and GFP-host centromeres (D) during mitosis are shown. Numbers in panels C and D correspond to frame numbers in panels A and B, respectively. Broken line indicates border of the mitotic cell surface. Scale bar, 10 μm. The data from op29 cell 1 (A and C) and op10 cell 1 (B and D) were used as examples.
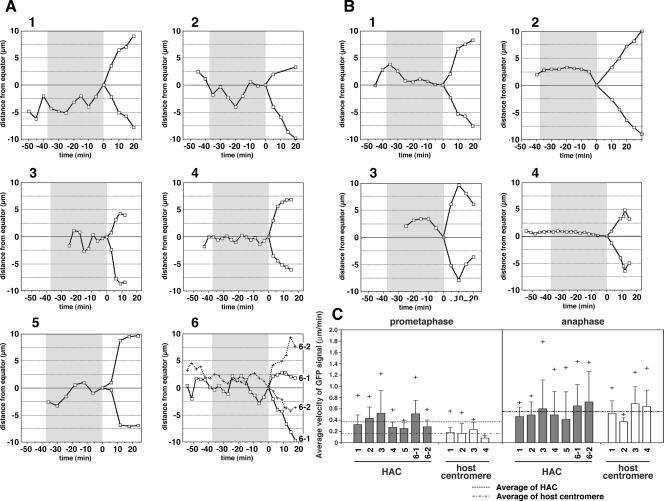
Displacements of individual GFP signals relative to the spindle equator in successive intervals (3 or 5 min) from the end of prometaphase to telophase (cytokinesis) are shown in Fig. 7 (y axis, distance from spindle equator; y = 0 at time zero). During this period, GFP-HAC moved faster and changed direction frequently (oscillation, 3 to 9 times; average, 6.0) across or around the spindle equator along the pole-to-pole axis (Fig. 7A), while the GFP-host centromere moved slowly and oscillated less (0 to 3 times; average, 1.3) (Fig. 7B). In the cell containing two HACs, two GFP-HAC signals also oscillated frequently and independently during metaphase periods, but the sister GFP signals from these two HACs separated with the same timing at anaphase onset (Fig. 7A, graph 6). Poleward-antipoleward oscillation around the spindle midzone is characteristic of metaphase centromere movements (33, 55, 56). Although more frequent oscillation across the spindle equator was observed with HACs during metaphase, the results indicated that the individual HACs stayed near the spindle midzone. During the late anaphase (telophase) period, some GFP signal tracks, both from sister HAC chromatids and host sister centromeres, showed turning from the poleward to the reverse direction (Fig. 7A, graph 3, and B, graphs 3 and 4), and these three-dimensional rotational movements of the gathered sister chromatids in the z-axial (depth of the stage) and the longitudinal (the equator) directions might be caused by cell-cell interactions and the directional change of cell division after anaphase. In these cases, the spindle pole-to-pole direction is not completely horizontal at time zero; sister centromere displacements at anaphase are shown to be nonsymmetrical to some degree (Fig. 7A, graphs 2, 3, 5, and 6; and B, graphs 3 and 4). In addition to these common movements of HACs and host sister centromeres, some HACs (Fig. 6C, numbers 1, 2, and 6-1; see Fig. S4A in the supplemental material) also gradually moved in the longitudinal direction during metaphase, compared to the simple oscillation at the same longitudinal position of the rest of the HACs and host centromeres (Fig. 6D; see Fig. S4B in the supplemental material).
FIG. 7.
The movement and the velocity of GFP-HACs and GFP-host centromeres during mitosis. The distances (one dimensional) from spindle equator (y = 0) to GFP-LacR signal of GFP-HACs (A) and GFP-host centromeres (B) at the each time point were plotted. The x axis represents time points. Time zero is the same as that described in the legend of Fig. 6. We regarded the stage from −37 min to 0 min as prometaphase-metaphase periods (gray area) and as a basis of the average length of prometaphase-metaphase under these culture conditions for these cell lines (Table 3). Velocities (μm/min) of HACs and host centromeres were calculated from the distances that GFP signals moved within each time interval of 3 or 5 min, including two-dimensional movement from one time point to the next time point; average values of each observation in metaphase or anaphase were plotted with bars in panel C. Dashed lines indicate the average velocity of GFP-HACs and GFP-host centromeres calculated from all the velocities measured along each observation. The bars on the columns indicate the standard deviations. +, a maximum value (velocity) during the observation. The data from cells A (graphs 1 to 6 correspond to op29 cells 1 to 6, respectively) and B (graphs 1 to 4 correspond to op10 cells 1 to 4, respectively) were used. The average velocity of HACs and the host centromeres during metaphase periods are significantly different (P < 0.01), while in anaphase, average velocities of the HACs and host centromeres were not significantly different (P > 0.9) by a Student's t test.
The velocities (μm/min) of HACs and host centromeres were calculated and plotted from the distances that GFP signals moved within each time interval of 3 or 5 min, including two-dimensional movement from one time point to the next time point; the average velocities calculated from all the velocities measured along each observation were plotted in Fig. 7C. The maximum velocity and the average velocity of HACs during metaphase periods were 0.40 to 1.22 μm/min and 0.38 μm/min, respectively, and were at least twice the host centromere velocities of 0.12 to 0.58 μm/min and 0.16 μm/min, respectively (P < 0.01). However, in anaphase, although both the maximum velocity and the average velocity of HACs and host centromeres increased, these HAC velocities (0.71 to 1.79 μm/min and 0.55 μm/min, respectively) approached and were not significantly different from those of the host centromeres (0.50 to 1.32 μm/min and 0.56 μm/min, respectively; P > 0.9) of the cell lines. Although frequent oscillation and higher velocity before anaphase onset were observed with HACs, the sister chromatids of the HACs and of the host chromosomes moved to the spindle poles with similar velocities in anaphase.
DISCUSSION
A HAC retains most of the functions expected of the centromere of a stable chromosome.
The following observations provide evidence that the HAC has a functional centromere that is indistinguishable from that of a natural chromosome. (i) Marker components for the centromere/kinetochore (CENP-A, -B, and -C), microtubule motor, kinetochore-dependent checkpoint (CENP-E and Mad2) (27, 48) (see Fig. S5 in the supplemental material), inner centromere, and heterochromatin (HP1α and Aurora B) (44) assembled on the HAC. (ii) The HAC can align accurately at the spindle midzone (Fig. 5, 6, and 7). (iii) The sister chromatids of the HAC are resolved at the same time as those of the natural chromosomes (Fig. 5, 6, and 7). (iv) The centromere of the artificial chromosome has motor activity (about 0.55 μm/min) on spindle microtubules equivalent to that of the centromeres on host chromosomes (about 0.56 μm/min) during anaphase (Fig. 7C). Consistent with the long-term stability of the artificial chromosomes in nonselective culture conditions (98.6 to 99.4% per cell division) (Table 2), all these results clearly indicate that de novo HACs retain most of the functions expected of the centromeres of natural chromosomes. Thus, it also implies that the multimer of the input alphoid YAC DNA can provide a common foundation for chromatin assembly and the structures required for a functional centromere and a stable chromosome.
HACs align at the metaphase plate accurately.
Laser microsurgery experiments demonstrated that a chromosome arm dissected from the proximal part of the chromosome containing a centromere moved rapidly in the direction of the spindle midzone (50). A chromosome arm receives the ejection force from the pole through the interaction between spindle microtubules and chromokinesin (Kid). Immunodepletion of a Kid homologue from Xenopus egg extract prevents normal chromosomal alignment (2, 13). In human cells, antibody-induced inhibition of Kid blocks chromosome oscillations, with the chromosome arm atypically extending towards the spindle poles during congression, while the chromosomes are aligned at the spindle midzone (33). These results indicate that human chromosomes can align and stabilize their position at the spindle midzone through kinetochore functions. The total size of a HAC (3 to 5 Mb) corresponds to less than only 1/10 of the long arm (∼50 Mb) of the small human chromosome 21. The smaller size and the negligible chromosome arm on the HAC imply that the HAC simply receives a smaller polar ejection force. Irrespective of this disadvantage of the HAC, our observation showed that HACs could align at the spindle midzone accurately, with mitotic periods similar to those of the original HT1080 cell line (Fig. 5, 6, and 7 and Table 3), indicating that the HAC can accomplish this mostly by kinetochore function. The main reason for the small instability of the HAC might be nondisjunction and consequent chromosome loss events during passage in the culture (Table 2). However, using the present system, we failed to determine how the nondisjunction and the loss occur or whether the lagging HAC is checked or unchecked by the mitotic checkpoint under normal, nonselective culture. CENP-E and Mad2, factors which are involved in the kinetochore-dependent checkpoint (7), can assemble on the artificial chromosomes derived from the alphoid YAC or BAC in metaphase-arrested cells (27, 48) (see Fig. S5 in the supplemental material), suggesting the possibility that an unattached HAC might also be monitored by the spindle checkpoint mechanism (see below).
The disadvantages of the smaller size of the HAC may be more serious if a HAC happens to be located outside of the spindle during prophase. It was reported that some HACs displayed increased instability (2 to 10%; mainly nondisjunction and lagging chromosomes) when cells were doubly treated with an inhibitor of microtubule polymerization (nocodazole) and then of cytokinesis (dihydrocytochalasin B) (52). The chromatin organization and balanced assembly of centromere/kinetochore components on de novo HACs may be more sensitive or dynamic, and thus affected, if the cellular and chromatin environment is drastically changed by such treatments. Indeed, even the natural chromosomes displayed merotelic kinetochores at a 15-fold higher rate (17%) than was seen in normal PtK1 cells after treatment with an inhibitor of microtubule polymerization (6), and mechanisms exist to correct improper kinetochore-microtubule attachments in the normal condition (54).
Frequent oscillations with larger velocities across around the spindle midzone with all HACs.
The control of tension generated by interactions between spindle microtubules and kinetochores at each side of the centromere is one of the major functions of the kinetochore and the inner centromere structure (7, 34, 38, 45, 54). Interestingly, more frequent oscillations with larger velocities across and around the spindle midzone were observed with all HACs, relative to those of natural chromosomes, during metaphase. It is conceivable that a smaller polar ejection force on chromokinesin because of a HAC's small size may decrease the stabilizing force of the HAC at the spindle midzone and, thus, may cause an increase in the frequency of the oscillations, with larger amplitudes, across the spindle equator. However, this interpretation is not consistent with the observation that antibody-induced inhibition of Kid blocks chromosome oscillations (33). The smaller size of an HAC may simply increase the HAC's degrees of freedom among the gathering chromosomes for movement in the spindle pole-to-pole direction and even for movement in the longitudinal direction in some cases. Otherwise, if there is a longer time lag between the tension and the counter-tension reactions at the centromere on the HAC than at the centromere on the host chromosome, the HAC may oscillate with larger amplitudes. However, actual HACs in living cells oscillate more frequently, with quicker motion, than the host chromosomes. The elastic nature of the inner centromere regions of human chromosomes has been demonstrated in living cells during oscillation (55). It is possible that the inner centromere region of the HAC derived from the multimerized structure of the 70 kb of alphoid DNA plus the 35 kb of YAC vector is less elastic than that of the natural centromere, based on the megabase-level order of the repetitive array, which is able to absorb the tension generated by polymerization or depolymerization of microtubule plus ends at the outer kinetochores (54). It could, therefore, generate the time margin just before applying the counter-tension at the outer kinetochore side, and as a result, a HAC with a less elastic structure between sister kinetochores might start to move quickly across the spindle equator if an unbalanced tension were generated and might frequently change direction if counter-tensions were applied. The resolution and sensitivity of our present observation system limit its use for shorter time-lapse observations of the distance between sister kinetochores, so further improvements of the system are required in order to determine which possibility is correct. However, in either case, the frequent oscillations around the spindle midzone indicate frequent generation of tension and counter-tension by frequent switching between polymerization and depolymerization of microtubule plus ends on the HAC. This is consistent with the HAC's having these major, precisely controlled functions of the kinetochore and the inner centromere structures.
The sister chromatids of the HAC are resolved coordinately with the natural chromosomes.
Recent analyses have indicated that, in addition to functional kinetochore assembly, sister chromatid cohesion and its resolution mechanisms involved with passenger proteins (INCENP and Aurora B, e.g.) are required for the proper segregation of the chromosomes (34). In fission yeast, cohesin is associated with a heterochromatin protein HP1 homolog (Swi6) (4, 46). In higher eukaryotes, the disruption of heterochromatin causes the loss of proper cohesion and missegregation of chromosomes (17, 60). The sister chromatids of the HAC are resolved with the same timing as those of natural chromosomes, synchronizing the progression of the mitotic cell cycle. The result indicates that the artificial chromosomes can maintain sister chromatid cohesion and can control its resolution mechanisms (Fig. 5, 6, and 7) and thus execute segregation processes properly, as required for a single chromosome in synchrony with the mitotic cell cycle. In addition to these results, the observation that all HAC sister chromatids (whether derived from the YAC-based linear or BAC-based circular constructs) that we have analyzed to date hold together in mitotic cells arrested by treatment with an inhibitor of microtubule assembly (Fig. 4) also strongly supports the conclusion that sister chromatid cohesion controlled by mitotic checkpoint mechanisms is coupled to the assembly of the functional centromere on the HAC.
From this point of view, the assembly of heterochromatin (trimethyl-H3K9 and HP1α) is indispensable to the control of the cohesion of the stable HACs. Indeed, our recent experiments showed that an additional insertion of a marker gene with a strong promoter/enhancer into the vector sequence significantly reduced the efficiency of HAC formation. Conversely, the additional transcriptional activities of these marker genes increased the assembly of the centromere proteins CENP-A, -B, and -C on the insert alphoid DNA at the ectopic integration site of the input alphoid BAC DNA (44). Therefore, the balanced assembly of heterochromatin as well as the assembly of the functional centromere/kinetochore structure is required for stable HAC formation (37, 43). In our present experiments, we obtained only a small number of transformants with α7C5opYAC DNA transfection alone. The existence of multiple LacO sequences or multiple LacR molecules that bind to the vector arm might incline the initial balance of centromere chromatin and heterochromatin on the introduced DNA to either side. The result also indicates that even this kind of YAC construct can form a stable HAC if the YAC DNA is mixed with an original alphoid YAC (α7C5hTel) DNA to compensate for the insufficient seed for chromatin assembly; thus, the number of the multiple LacO sequences is reduced to one-fifth (see similar results from the mixture of the different BAC constructs in reference 44). With the HAC formed from the YAC DNA mixture, sister chromatid cohesion controlled by a mitotic checkpoint mechanism functioned correctly and was resolved. Moreover, once a stable HAC was formed, the presence or absence of a large amount of LacR binding to the HAC did not affect its stability seriously, suggesting that having only one-fifth the number of multiple LacO sequences might not target a critical structure on the HAC. Otherwise, the chromatin assembly and self-maintenance mechanisms of the HAC permit considerable modulating capacity (plasticity) for ectopic protein binding. This kind of plasticity in HAC formation and centromere assembly are the most interesting properties in regard to extending the description of species-specific centromere DNA structure, the existence of active genes within centromere domains (42), and neocentromere formation (53), as well as for future applications of the HAC vector.
The only treatments we applied to the cells for HAC formation were transfection of a naked alphoid YAC/BAC DNA and selection. Our results indicate that HT1080 cells can generate quite elaborate mechanisms for stably replicating and segregating de novo chromosome systems. De novo centromere assembly in cultured human cells requires human centromere-specific alphoid DNA containing CENP-B binding sites (37, 48). However, CENP-B is not required for chromosome segregation itself in CENP-B knockout mice, human neocentromeres, and centromeres on Y chromosomes (9, 25, 29, 49). This is surprising, because the results strongly suggest that the elaborate chromosome systems for replication and segregation may be established semiautonomously if a nonessential but centromere-specific DNA-binding protein starts to assemble and thus changes the status of the naked input DNA. To date, a few cell lines combined with a particular satellite DNA are known to have this capacity. Despite these discrepancies, we have also generated de novo artificial chromosomes reproducibly in a mouse cell line; this assembly is dependent on the existence of the CENP-B/CENP-B box interaction on a satellite DNA (T. Okada and H. Masumoto, submitted for publication). Further investigation is required to determine how the cellular environment and chromatin assembly (of euchromatin, centromeric chromatin, and heterochromatin) and the nature of repetitive DNA are involved in the process.
Supplementary Material
Acknowledgments
We thank A. Belmont (University of Illinois) for giving us the multiple LacO/GFP-Lac repressor system, K. Hamazaki for GFP-HAC data analyses, K. Yoda for producing the anti-CENP-A antibody, N. Nozaki (Kanagawa Dental College) for producing anti-CENP-A and -B antibodies, and V. Larionov (NCI/NIH) for critical reading.
This work was supported by a grant-in-aid for Scientific Research on Priority Areas (B), Core Research for Evolutional Science and Technology (CREST), and Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Science, Sports, and Culture of Japan and by a grant-in-aid from the Cell Science Research Foundation. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Amor, D. J., and K. H. Choo. 2002. Neocentromeres: role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 71:695-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonio, C., I. Ferby, H. Wilhelm, M. Jones, E. Karsenti, A. R. Nebreda, and I. Vernos. 2000. Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell 102:425-435. [DOI] [PubMed] [Google Scholar]
- 3.Basu, J., G. Stromberg, G. Compitello, H. F. Willard, and G. Van Bokkelen. 2005. Rapid creation of BAC-based human artificial chromosome vectors by transposition with synthetic alpha-satellite arrays. Nucleic Acids Res. 33:587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard, P., J. F. Maure, J. F. Partridge, S. Genier, J. P. Javerzat, and R. C. Allshire. 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294:2539-2542. [DOI] [PubMed] [Google Scholar]
- 5.Burke, D. T., G. F. Carle, and M. V. Olson. 1987. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science 236:806-812. [DOI] [PubMed] [Google Scholar]
- 6.Cimini, D., B. Moree, J. C. Canman, and E. D. Salmon. 2003. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J. Cell Sci. 116:4213-4225. [DOI] [PubMed] [Google Scholar]
- 7.Cleveland, D. W., Y. Mao, and K. F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407-421. [DOI] [PubMed] [Google Scholar]
- 8.Craig, J. M., L. H. Wong, A. W. Lo, E. Earle, and K. H. Choo. 2003. Centromeric chromatin pliability and memory at a human neocentromere. EMBO J. 22:2495-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Earnshaw, W. C., H. Ratrie III, and G. Stetten. 1989. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma 98:1-12. [DOI] [PubMed] [Google Scholar]
- 10.Ebersole, T., Y. Okamoto, V. N. Noskov, N., Kouprina, J. H. Kim, S. H. Leem, J. C. Barrett, H. Masumoto, and V. Larionov. 2005. Rapid generation of long synthetic tandem repeats and its application for analysis in human artificial chromosome formation. Nucleic Acids Res. 33:e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebersole, T. A., A. Ross, E. Clark, N. McGill, D. Schindelhauer, H. Cooke, and B. Grimes. 2000. Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Hum. Mol. Genet. 9:1623-1631. [DOI] [PubMed] [Google Scholar]
- 12.Fitzgerald-Hayes, M., L. Clarke, and J. Carbon. 1982. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell 29:235-244. [DOI] [PubMed] [Google Scholar]
- 13.Funabiki, H., and A. W. Murray. 2000. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell 102:411-424. [DOI] [PubMed] [Google Scholar]
- 14.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 15.Goshima, G., T. Kiyomitsu, K. Yoda, and M. Yanagida. 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160:25-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grimes, B. R., D. Schindelhauer, N. I. McGill, A. Ross, T. A. Ebersole, and H. J. Cooke. 2001. Stable gene expression from a mammalian artificial chromosome. EMBO Rep. 2:910-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guenatri, M., D. Bailly, C. Maison, and G. Almouzni. 2004. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166:493-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haaf, T., and D. C. Ward. 1994. Structural analysis of alpha-satellite DNA and centromere proteins using extended chromatin and chromosomes. Hum. Mol. Genet. 3:697-709. [DOI] [PubMed] [Google Scholar]
- 19.Hahnenberger, K. M., M. P. Baum, C. M. Polizzi, J. Carbon, and L. Clarke. 1989. Construction of functional artificial minichromosomes in the fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 86:577-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haraguchi, T., D. Q. Ding, A. Yamamoto, T. Kaneda, T. Koujin, and Y. Hiraoka. 1999. Multiple-color fluorescence imaging of chromosomes and microtubules in living cells. Cell Struct. Funct. 24:291-298. [DOI] [PubMed] [Google Scholar]
- 21.Harrington, J. J., G. Van Bokkelen, R. W. Mays, K. Gustashaw, and H. F. Willard. 1997. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 15:345-355. [DOI] [PubMed] [Google Scholar]
- 22.Heller, R., K. E. Brown, C. Burgtorf, and W. R. Brown. 1996. Mini-chromosomes derived from the human Y chromosome by telomere directed chromosome breakage. Proc. Natl. Acad. Sci. USA 93:7125-7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henikoff, S., K. Ahmad, and H. S. Malik. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293:1098-1102. [DOI] [PubMed] [Google Scholar]
- 24.Howman, E. V., K. J. Fowler, A. J. Newson, S. Redward, A. C. MacDonald, P. Kalitsis, and K. H. Choo. 2000. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA 97:1148-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hudson, D. F., K. J. Fowler, E. Earle, R. Saffery, P. Kalitsis, H. Trowell, J. Hill, N. G. Wreford, D. M. de Kretser, M. R. Cancilla, E. Howman, L. Hii, S. M. Cutts, D. V. Irvine, and K. H. Choo. 1998. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J. Cell Biol. 141:309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikeno, M., H. Masumoto, and T. Okazaki. 1994. Distribution of CENP-B boxes reflected in CREST centromere antigenic sites on long-range alpha-satellite DNA arrays of human chromosome 21. Hum. Mol. Genet. 3:1245-1257. [DOI] [PubMed] [Google Scholar]
- 27.Ikeno, M., B. Grimes, T. Okazaki, M. Nakano, K. Saitoh, H. Hoshino, N. I. McGill, H. Cooke, and H. Masumoto. 1998. Construction of YAC-based mammalian artificial chromosomes. Nat. Biotechnol. 16:431-439. [DOI] [PubMed] [Google Scholar]
- 28.Izumi, M., H. Miyazawa, T. Kamakura, I. Yamaguchi, T. Endo, and F. Hanaoka. 1991. Blasticidin S-resistance gene (bsr): a novel selectable marker for mammalian cells. Exp. Cell Res. 197:229-233. [DOI] [PubMed] [Google Scholar]
- 29.Kapoor, M., R. Montes de Oca Lunaqq, G. Liu, G. Lozano, C. Cummings, M. Mancini, I. Ouspenski, B. R. Brinkley, and G. S. May. 1998. The cenpB gene is not essential in mice. Chromosoma 107:570-576. [DOI] [PubMed] [Google Scholar]
- 30.Kim, U. J., B. W. Birren, T. Slepak, V. Mancino, C. Boysen, H. L. Kang, M. I. Simon, and H. Shizuya. 1996. Construction and characterization of a human bacterial artificial chromosome library. Genomics 34:213-218. [DOI] [PubMed] [Google Scholar]
- 31.Kitagawa, K., and P. Hieter. 2001. Evolutionary conservation between budding yeast and human kinetochores. Nat. Rev. Mol. Cell. Biol. 2:678-687. [DOI] [PubMed] [Google Scholar]
- 32.Kouprina, N., M. Eldarov, R. Moyzis, M. Resnick, and V. Larionov. 1994. A model system to assess the integrity of mammalian YACs during transformation and propagation in yeast. Genomics 21:7-17. [DOI] [PubMed] [Google Scholar]
- 33.Levesque, A. A., and D. A. Compton. 2001. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J. Cell Biol. 154:1135-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maiato, H., J. DeLuca, E. D. Salmon, and W. C. Earnshaw. 2004. The dynamic kinetochore-microtubule interface. J. Cell Sci. 117:5461-5477. [DOI] [PubMed] [Google Scholar]
- 35.Masumoto, H., K. Sugimoto, and T. Okazaki. 1989. Alphoid satellite DNA is tightly associated with centromere antigens in human chromosomes throughout the cell cycle. Exp. Cell Res. 181:181-196. [DOI] [PubMed] [Google Scholar]
- 36.Masumoto, H., M. Ikeno, M. Nakano, T. Okazaki, B. Grimes, H. Cooke, and N. Suzuki. 1998. Assay of centromere function using a human artificial chromosome. Chromosoma 107:406-416. [DOI] [PubMed] [Google Scholar]
- 37.Masumoto, H., M. Nakano, and J. Ohzeki. 2004. The role of CENP-B and alpha-satellite DNA: de novo assembly and epigenetic maintenance of human centromeres. Chromosome Res. 12:543-556. [DOI] [PubMed] [Google Scholar]
- 38.McIntosh, J. R., E. L. Grishchuk, and R. R. West. 2002. Chromosome-microtubule interactions during mitosis. Annu. Rev. Cell Dev. Biol. 18:193-219. [DOI] [PubMed] [Google Scholar]
- 39.Mejia, J. E., A. Willmott, E. Levy, W. C. Earnshaw, and Z. Larin. 2001. Functional complementation of a genetic deficiency with human artificial chromosomes. Am. J. Hum. Genet. 69:315-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mellone, B. G., and R. C. Allshire. 2003. Stretching it: putting the CEN(P-A) in centromere. Curr. Opin. Genet. Dev. 13:191-198. [DOI] [PubMed] [Google Scholar]
- 41.Mills, W., R. Critcher, C. Lee, and C. J. Farr. 1999. Generation of an approximately 2.4 Mb human X centromere-based minichromosome by targeted telomere-associated chromosome fragmentation in DT40. Hum. Mol. Genet. 8:751-761. [DOI] [PubMed] [Google Scholar]
- 42.Nagaki, K., Z. Cheng, S. Ouyang, P. B. Talbert, M. Kim, K. M. Jones, S. Henikoff, C. R. Buell, and J. Jiang. 2004. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36:138-145. [DOI] [PubMed] [Google Scholar]
- 43.Nakano, M., Y. Okamoto, J. Ohzeki, and H. Masumoto. 2003. Epigenetic assembly of centromeric chromatin at ectopic alpha-satellite sites on human chromosomes. J. Cell Sci. 116:4021-4034. [DOI] [PubMed] [Google Scholar]
- 44.Nakashima, H., M. Nakano, R. Ohnishi, Y. Hiraoka, Y. Kaneda, A. Sugino, and H. Masumoto. 2005. Assembly of additional heterochromatin distinct from centromere-kinetochore chromatin is required for de novo formation of human artificial chromosome. J. Cell Sci. 118:5885-5898. [DOI] [PubMed] [Google Scholar]
- 45.Nicklas, R. B. 1997. How cells get the right chromosomes. Science 275:632-637. [DOI] [PubMed] [Google Scholar]
- 46.Nonaka, N., T. Kitajima, S. Yokobayashi, G. Xiao, M. Yamamoto, S. I. Grewal, and Y. Watanabe. 2002. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 4:89-93. [DOI] [PubMed] [Google Scholar]
- 47.Oegema, K., A. Desai, S. Rybina, M. Kirkham, and A. A. Hyman. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohzeki, J., M. Nakano, T. Okada, and H. Masumoto. 2002. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 159:765-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez-Castro, A. V., F. L. Shamanski, J. J. Meneses, T. L. Lovato, K. G. Vogel, R. K. Moyzis, and R. Pedersen. 1998. Centromeric protein B null mice are viable with no apparent abnormalities. Dev. Biol. 201:135-143. [DOI] [PubMed] [Google Scholar]
- 50.Rieder, C. L., E. A. Davison, L. C. Jensen, L. Cassimeris, and E. D. Salmon. 1986. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J. Cell Biol. 103:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinett, C. C., A. Straight, G. Li, C. Willhelm, G. Sudlow, A. Murray, and A. S. Belmont. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135:1685-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rudd, M. K., R. W. Mays, S. Schwartz, and H. F. Willard. 2003. Human artificial chromosomes with alpha satellite-based de novo centromeres show increased frequency of nondisjunction and anaphase lag. Mol. Cell. Biol. 23:7689-7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saffery, R., H. Sumer, S. Hassan, L. H. Wong, J. M. Craig, K. Todokoro, M. Anderson, A. Stafford, and K. H. Choo. 2003. Transcription within a functional human centromere. Mol. Cell 12:509-516. [DOI] [PubMed] [Google Scholar]
- 54.Salmon, E. D., D. Cimini, L. A. Cameron, and J. G. DeLuca. 2005. Merotelic kinetochores in mammalian tissue cells. Philos. Trans. R. Soc. Lond. B 360:553-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shelby, R. D., K. M. Hahn, and K. F. Sullivan. 1996. Dynamic elastic behavior of alpha-satellite DNA domains visualized in situ in living human cells. J. Cell Biol. 135:545-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skibbens, R. V., V. P. Skeen, and E. D. Salmon. 1993. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J. Cell Biol. 122:859-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sullivan, B. A., M. D. Blower, and G. H. Karpen. 2001. Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2:584-596. [DOI] [PubMed] [Google Scholar]
- 58.Sullivan, K. F., M. Hechenberger, and K. Masri. 1994. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 127:581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuki, N., M. Nakano, N. Nozaki, S. Egashira, T. Okazaki, and H. Masumoto. 2004. CENP-B interacts with CENP-C domains containing Mif2 regions responsible for centromere localization. J. Biol. Chem. 279:5934-5946. [DOI] [PubMed] [Google Scholar]
- 60.Taddei, A., C. Maison, D. Roche, and G. Almouzni. 2001. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 3:114-120. [DOI] [PubMed] [Google Scholar]
- 61.Van Hooser, A. A., Ouspenski, I. I., H. C. Gregson, D. A. Starr, T. J. Yen, M. L. Goldberg, K. Yokomori, W. C. Earnshaw, K. F. Sullivan, and B. R. Brinkley. 2001. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114:3529-3542. [DOI] [PubMed] [Google Scholar]
- 62.Watanabe, S., N. Kai, M. Yasuda, N. Kohmura, M. Sanbo, M. Mishina, and T. Yagi. 1995. Stable production of mutant mice from double gene converted ES cells with puromycin and neomycin. Biochem. Biophys. Res. Commun. 213:130-137. [DOI] [PubMed] [Google Scholar]
- 63.Willard, H. F., and J. S. Waye. 1987. Hierarchical order in chromosome-specific human alpha satellite DNA. Trends Genet. 3:192-198. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.